Abstract
More than 100 genes have been associated with significantly increased risks of autism spectrum disorders (ASD) with an estimate of ∼1000 genes that may contribute. The new challenge is to investigate the molecular and cellular functions of these genes during neural and brain development, and then even more challenging, to link the altered molecular and cellular phenotypes to the ASD clinical manifestations. In this study, we used single-cell RNA-seq analysis to study one of the top risk genes, CHD8, in cerebral organoids, which models early neural development. We identified 21 cell clusters in the organoid samples, representing non-neuronal cells, neural progenitors, and early differentiating neurons at the start of neural cell fate commitment. Comparisons of the cells with one copy of a CHD8 knockout allele, generated by CRISPR/Cas9 editing, and their isogenic controls uncovered thousands of differentially expressed genes, which were enriched with functions related to neural and brain development, cilium organization, and extracellular matrix organization. The affected genes were also enriched with genes and pathways previously implicated in ASD, but surprisingly not for schizophrenia and intellectual disability risk genes. The comparisons also uncovered cell composition changes, indicating potentially altered neural differential trajectories upon CHD8 reduction. Moreover, we found that cell-cell communications were affected in the CHD8 knockout organoids, including the interactions between neural and glial cells. Taken together, our results provide new data and information for understanding CHD8 functions in the early stages of neural lineage development and interaction.
Keywords: Autism, ASD, CHD8, scRNA-seq, Cerebral organoid, Cell-cell communication, ciliary signaling
1. Introduction
CHD8 (chromodomain helicase DNA binding protein 8) is a ubiquitously expressed member of the CHD family of ATP-dependent chromatin-remodeling factors that play important roles in chromatin dynamics and transcription. It is one of the strongest risk genes whose loss-of-function mutations have been associated with autism spectrum disorder (ASD), a class of neurodevelopmental disorders characterized by persistent deficits in social communication/interaction and stereotypical behaviors/interests (DSM-5). It was initially discovered from studies of rare de novo mutations present in ASD probands but absent in their healthy siblings or parents, using whole exome or whole genome sequencing [[1], [2], [3], [4]]. The strong association has since been reproduced by many subsequent ASD genetics studies using large cohorts consisting of thousands of samples [[5], [6], [7], [8]]. Some studies further suggest that ASD individuals with CHD8 mutations may define a subgroup of ASD, with stereotypical ASD features, early brain outgrowth, and gastrointestinal syndromes [9,10].
Many studies have been carried out to understand CHD8 functions in neural and brain development, with additional work in non-neural contexts (e.g., cancers), using human cell, mouse, rat, or zebrafish models [9,[11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26]]. These studies found that CHD8 bound to thousands of genes, largely biased to gene promoters, and reduced CHD8 expression resulted in disruptions of gene networks involved in neurodevelopment, such as WNT signaling. A meta-analysis further defined the conserved roles of CHD8 across human, mouse, rat, in vivo and in vitro models. It showed that CHD8 reduction affected a large number of genes involved in fundamental biological processes (e.g., neuronal development and function, cell cycle, chromatin dynamics, and metabolism) [27]. The analysis also found that differentially expressed genes in the compared studies, however, were distinct and often not overlapping, indicating that genes targeted by CHD8 are likely context dependent, like other chromatin modifiers. Consistent with this, the reported molecular, cellular and behavior phenotypes in multiple Chd8 mouse models also varied, with mostly small changes [22]. One caveat in the transcriptomics analyses in those studies is the usage of bulk tissues, which may have masked cell type specific effects.
Our group and others have used cerebral organoids to study the role of CHD8 in multiple human cell types involved in early human neural and brain development [14,28,29]. Coupling the use of cerebral organoids with single cell RNA sequencing (scRNA-seq) [[28], [29], [30]], investigators have uncovered both common and cell type specific genes that were affected by CHD8 haploinsufficiency, generated using CRISPR editing to knock out one copy of CHD8 (CHD8+/−) or by introducing patient specific mutations. While the studies consistently point to a role of CHD8 in regulating neural development, the cell types and neural differentiation stages altered in CHD8-disrupted lines seem to vary. Moreover, differences among cell lines in the same study were also noticeable. Such variations, although not very satisfactory, are quite commonly observed in brain organoid studies, partially due to the sensitivity of the neural differentiation system to genetic background, differentiation protocols, and stochasticity.
Considering the variations in molecular and cellular phenotypes that were observed in previous CHD8 studies of both mouse models and human organoid models, we decided to perform an independent scRNA-seq analysis of the cerebral organoids that we generated from induced pluripotent stem cell lines (iPSCs) of a typically developing male individual and the corresponding isogenic lines with one copy of CHD8 knocked out using CRISPR-Cas9 gene-editing technology. The generation of the lines and bulk RNA-seq analysis of the cerebral organoids and 2D neural samples were described in our previous reports, which showed that genes highly expressed in inhibitory neurons were affected by CHD8 haploinsufficiency [13,14]. Here, we characterize the cell types in the organoids, neural differential trajectory and cell-cell communication, and their alterations upon CHD8 reduction. Distinct from studies conducted previously on cerebral organoids from longer culture periods [[28], [29], [30]], current work analyzes organoids after only 50 days of neural differentiation, thus providing complementary and new insights into the role of CHD8 in early brain development. Additionally, our study uncovered substantial alterations in intercellular communications, which have not been previously described.
2. Material and methods
2.1. Cerebral organoid preparation
Self-organizing cerebral organoids were derived from iPSCs using the STEMdiff™ Cerebral Organoid Kit developed by STEMCELL Technologies™ (catalogue # 08570), which is based on the protocol developed by Lancaster et al. [31]. The protocol uses defined, serum-free cell culture media. In the first step (days 0–5), embryoid bodies (EBs) were generated from iPSCs. iPSCs were grown in a 6-well plate to 70–80 % confluence. Areas of spontaneous differentiation were manually removed by scraping with a pipette tip. Medium was aspirated and the iPSC cells were washed with 1 mL of sterile phosphate-buffered saline (PBS). Following aspiration of PBS, 1 mL of Gentle Cell Dissociation Reagent was added and the cells were incubated at 37 °C for 8–10 min. Using a 1 mL pipettor, the cells were gently resuspended by pipetting up and down slowly 3–5 times. The cells were transferred to a sterile 50 mL conical tube. The well was rinsed with an additional 1 mL of EB Seeding Medium (EB Formation Medium + 10 μM Y-27632) and added to the tube containing cells. Following centrifugation of cells at 300×g for 5 min, supernatant was removed and discarded, and 1–2 mL of EB Seeding Medium was used to resuspend cells. The cells were counted in a hemocytometer using Trypan Blue staining. The volume of cells required to obtain 90,000 cells/mL was determined; this was added to an appropriate volume of EB Seeding Medium. Using a multi-channel pipettor, 100 μL of the cell suspension was added to each well of a 96-well round-bottom, ultra-low attachment plate (9000 cells/well). The cells were then incubated at 37 °C for 24 h, undisturbed, in a humidified incubator containing 5 % CO2. Under an inverted microscope, small EBs (∼100–200 μm) were visible. On days 2 and 4, 100 μL of EB Formation Medium was gently added to each well using a multi-channel pipettor. On day 5, the EBs reached a diameter of 400–600 μm. On day 5, induction medium at room temperature was prepared according to the manufacturer, and 0.5 mL was added to each well of a 24-well ultra-low attachment plate. 1–2 EBs were added to each well using a wide-bore 200 μL pipette tip, drawing up 50 μL from one well of the 96-well plate. Most of the medium was carefully removed by ejecting it back into the well, retaining EBs in the pipette tip. The EBs were dispensed into one well of the 24-well plate containing Induction Medium. The plates were then incubated at 37 °C for 48 h. On day 7, Matrigel® was thawed on ice for 1–2 h and used at 1.44 mL Matrigel® (15 μL/well x 96 wells).
Expansion Medium was prepared according to the manufacturer's instructions and warmed to room temperature. A UV-sterilized embedding surface (Parafilm®) was placed into an empty, sterile, 100 mm dish. EBs were drawn up using a wide-bore 200 μL pipette tip, along with 25–50 μL of medium, and transferred to the embedding surface. This was repeated until 12–16 EBs were collected on the embedding surface. Excess medium was removed from each EB by drawing up medium with a standard 200 μL pipette tip. 15 μL of Matrigel® drawn up in a cold 200 μL standard pipette tip was added drop by drop onto each EB. The EBs were repositioned to the center of the droplet using a new, cold 200 μL pipette tip, The plate was then placed in an incubator at 37 °C for 30 min to polymerize the Matrigel®. Sterile forceps were used to grasp the embedding surface containing Matrigel® droplets. The sheet was positioned directly above one well of a 6-well ultra-low adherent plate. Using a 1 mL pipettor, Expansion Medium was drawn up and the Matrigel® droplets were gently washed off the sheet and into the well. Using 3 mL of Expansion Medium/well, the process was repeated until all 12–16 Matrigel® droplets were in the well. The plates were then incubated at 37 °C for 3 days, after which embedded organoids were observed to develop expanded neuroepithelia (budding of the EB surface).
From days 10–40, the organoids were grown in Maturation Medium, which was prepared according to the manufacturer's instructions. Expansion Medium was slowly removed with a 5–10 mL serological pipette. This was replaced with Maturation medium (3mL/well). Maturation Medium was kept at room temperature prior to its addition to the organoids. The plate of organoids was placed on an orbital shaker in a 37 °C incubator. A full medium change was made every 72 h.
From day 40 to day 50, the organoids were cultured with STEMdiff™ Cerebral Organoid Maturation Kit (Catalog #08571) with a full medium change every 48 h. The organoids were harvested for single cell RNA sequencing on day 50.
2.2. Single cell RNA-seq data acquisition
Single cell RNA-seq libraries were prepared at the Einstein Genomics Core Facility using the Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1, according to the manufacturer's instructions (10x Genomics, Cat# PN-1000121), followed by paired-end sequencing on an Illumina NextSeq 500 platform. The four barcoded samples were pooled and sequenced together.
2.3. Single cell RNA-seq data analysis
Cell Ranger software (v3.0.2; 10X Genomics) [32] with default parameters was used to align the scRNA-seq reads to the human GRCh38 reference genome, identify cell barcodes, demultiplex samples, and generate gene expression count matrices. The count matrices were analyzed by the RISC (v1.0) [33] package for each of the four samples individually and then their integration. Cells with a minimum of 1000 and a maximum of 40,000 UMI (unique molecular identifier) counts, as well as a minimum of 200 genes were kept for further analysis. Data integration was performed by the RPCI (Reference Principal Component Integration) method [33], using the control 1 (“ctrl1”) sample as the reference and the top 20 principal components (PCs). Dimensionality reduction and cell clustering were performed on the integrated data by UMAP (Uniform Manifold Approximation and Projection) and Louvain methods, respectively, with the top 20 PCs, using RISC (default parameters). One small cell cluster very distant from other cells was identified as endothelial cells by marker genes (e.g., PECAM1) and removed from further analysis. Gene markers for each cluster were identified using AllMarker function and Quasi-Poisson method. Differential expression analysis between CHD8 KO and controls was run by comparing all the cells from the two KO samples with all cells from the two control samples, for each of the 21 clusters independently. Differentially expressed genes (DEGs) were determined using the scDEG function (using Negative Binomial option) at log2(fold change) > 1 or < −1 and adjusted p-value (padj) < 0.05. Differences in cell population abundance between CHD8 KO and controls were determined by miloR (v1.1.0) [34] using 30 k-nearest-neighbors (KNN) for graph building and 30 dimensions for KNN (K nearest neighbors) refinement.
2.4. Pseudotime trajectory analysis
Spliced/unspliced count matrices for each of the samples were generated by velocyto [35], using the GRCh38 as the reference with repeat DNA elements masked. The resulting loom files were further processed by scVelo (v0.2.4) [36] to compute and visualize the proportions of spliced/unspliced reads for each of the cell clusters within each sample. Variable genes were detected by minimum number of counts and dispersion, and data across cells were normalized by total library sizes and logarithm-transformed using default parameters through pp.filter_and_normalize function. Mean and variance of the data were calculated by nearest neighbors in PCA space using 30 PCs and 30 neighbors, and the graph was embedded into two dimensions UMAP. Clustering of the cells was based on the Louvain method. The estimation of RNA velocity was obtained by tl.velocity and tl.velocity_graph functions using default parameters, while projection of RNA velocity to the UMAP was obtained from a dynamic function (tl.umap) together with the RISC clustering. The tl.louvain function was also used to find clusters based on dynamic data to help cell type identification. Finally, the initial and terminal states were identified by CellRank (v1.5.1) [37] using tl.terminal_states and tl.initial_states.
2.5. Cell type annotation
Cell type identification was based on a combination of known markers, functional enrichment of markers, and pseudotime results. First, neuronal and non-neuronal cell types were separated by the expression of GAP43, STMN2, DCX (neuronal) and VIM, HES1, SOX2 (non-neuronal), as described by Tanaka et al. [38]. Neuronal clusters were then classified as excitatory cortical neurons through SLC17A7, TBR1 and NEUROD2 expression and inhibitory interneurons by SLC32A1, GAD1 and GAD2 markers. Neuronal progenitors were identified by high expression of corresponding marker genes and their positions in the pseudotime trajectory. Cells highly expressing TOP2A and MKI67 were annotated as neuroepithelial cells, while astrocytes were identified by high expression of GFAP and SLC1A3. Astrocytes progenitors were detected through lower expression of astrocytes markers using RNA velocity as support. In the same way, oligodendrocyte cells and their progenitors were identified by high and low OLIG1 expression, respectively. CRYM cluster was identified by unique CRYM expression and UPRC cells by DDIT3 expression, as well as enrichment of its markers in the GO:0006986 term (“response to unfolded protein”). Epithelial cells were identified by markers and the enrichment of “NS-moderated 16-33-Epithelial-Ciliated term” on ToppCell Atlas [39]. Lastly, mesenchymal cells were identified by PIFO and PCP4 expression and enrichment of markers in differentiating mesenchymal cilium from mesenchymal BMP cells.
2.6. Over representation analysis based on curated gene sets and Gene Ontology
Two over representation analyses were performed for DEGs between CHD8 KO and controls, for each of the cell types independently. In the first, EnrichGO function from clusterProfiler [40] was used to identify overrepresented GO terms in “biological processes,” with simplify function used to reduce redundancy. In the second, we tested the overrepresentation of our DEGs in gene sets associated with ASD, schizophrenia (SZ), intellectual Disability (ID), Attention Deficit Hyperactivity Disorder (ADHD), and neurodevelopmental disorders, for a total of seven gene sets curated in our previous work [41]. Fisher test was applied using GeneOverlap [42] package.
2.7. Pathway enrichment analysis
In addition to overrepresentation analysis, DEGs were also ranked by their expression fold changes and used for Gene Set Enrichment Analysis (GSEA, v4.2.2) [43,44]. Gene sets in the GO:“biological processes” were selected for enrichment analysis at FDR <25 % (default).
2.8. Cell-cell interaction
CellChat [45] software was used to infer cross-cell-type interactions between the 21 cell types in control and KO samples, including interactions involving ligand-receptor (L-R) pairs or other cell surface protein complexes. For that, normalized gene expression matrix from the RISC integration was loaded to CellChat, with cells from the two control samples merged to one control group while cells from the two KO samples to one KO group, and then computeCommunProb function was used to select genes expressed in >20 % of cells within one cell type for determining statistically significant (p < 0.05, permutation test) L-R interactions. From those L-R interactions, CellChat computed the interaction score for a signaling pathway by summing up the interaction strengths of all the L-R involved in that pathway across all cell-pairs. Based on these new signaling scores, dysregulated signaling were identified between control and KO groups using rankNet function (Wilcoxon test, p < 0.05), with a tolerance >0.05 for the differences in the relative contributions.
3. Results
3.1. Clustering and annotation of cell types
To model CHD8 function in early neural differentiation, we generated human cerebral organoids from two clones of a control iPSC line (“Ctrl1” and “Ctrl2”) and two derived lines (“KO1” and “KO2”) with a copy of the CHD8 gene edited by CRISPR-Cas9 (CHD8+/−; referred as heterozygous CHD8 knockout (KO) for simplicity). The original iPSC line was derived from a healthy male subject, and the two KO lines derived from it contained a 2-bp (KO1) and 10-bp (KO2) heterozygous deletion in the second exon encoding first few N-terminal amino acids of the CHD8 protein. The generation and characterization of the lines were described in our previous studies, which also confirmed a reduction of CHD8 protein in the KO lines [13,14]. The transcriptomic changes of CHD8 KO were also analyzed in neuron progenitors, differentiated neurons, and cerebral organoids by bulk RNA-seq analysis [13,14]. In this study, cerebral organoids after 50 days of neural induction were harvested for scRNA-seq analysis.
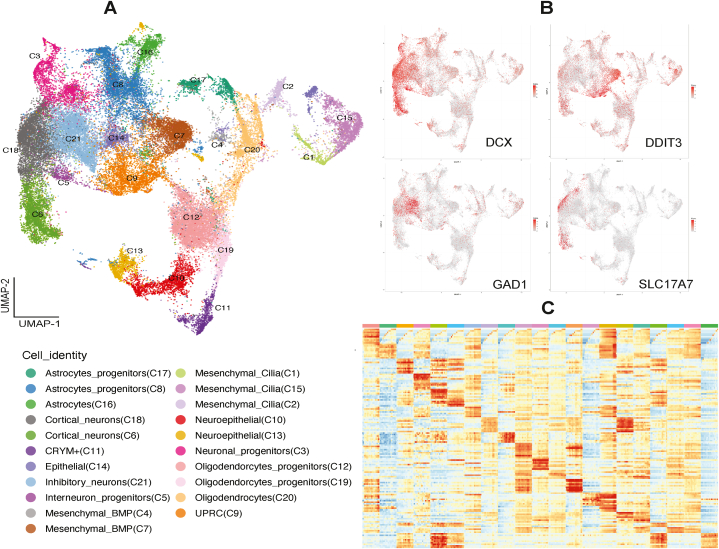
After low quality cells were removed, we obtained an average of 14,285 cells per sample, with an average of 25,398 reads and 2184 genes per cell (Supplementary Table S1). The cell level statistics was similar in all four samples. The data were then analyzed and integrated with RISC software, which was specifically designed for batch correction and integrating cells with expected treatment effects [33]. Additional filtering in RISC, to retain high quality cells, yielded an average number of 12,287 cells per sample, with maximal (13,723) and minimal (10,123) cells in KO2 and Ctrl2, respectively. Louvain clustering of the integrated data yielded 21 cell clusters, as visualized by dimension reduction and uniform manifold approximation and projection (UMAP) (Fig. 1A). Differential analysis between one cluster and the remaining cells identified marker genes expressed highly in each of the 21 cluster (Fig. 1B and C; Supplementary Table S2), which were used for cell type annotation, together with a set of markers reported in previous scRNA-seq studies of brain organoids [28,38,46]. While the transcriptomic distinctions of the 21 clusters were strongly supported by the data-derived markers (Fig. 1C), the expression of some known cell type markers was less distinct and appeared in a gradient (Supplementary Fig. S1).
Fig. 1.
Cell clustering and annotation of the integrated scRNA-seq data. A: UMAP of integrated data revealed 21 distinct cell clusters. B: UMAP plots for the expression of selected markers. C: Heatmap for the expression of top 10 marker genes (rows) computed for the 21 clusters (left to right columns, cluster 1 to 21). The names of the markers are not shown but can be found in Supplemental Table S2. Their expression patterns support transcriptomic differences of the 21 clusters. The cells in each cluster were binned (not equal size) and their averaged expression values were used for preparing the heatmap.
Five clusters showed high expression of neuronal GAP43, STMN2, DCX markers, while the remaining 16 clusters expressed high level of non-neuronal markers (Supplementary Fig. S1). Three of the five neuronal clusters were annotated as excitatory cortical neurons (Cortical_neurons (C6), Cortical_neurons (C18)) based on their expression of SLC17A7, TBR1 and NEUROD2 markers, with one of them (Neuronal_progenitors(C3)) determined to be neural progenitor due to lower SLC17A7 expression and their location in the early differentiation trajectories (see below; Fig. 2). Two clusters of interneurons were found, based on SLC32A1 expression, Interneuron_progenitors (C5) and Inhibitory_neurons (C21), with the latter expressing higher GAD1 and GAD2, canonical inhibitory neuron markers.
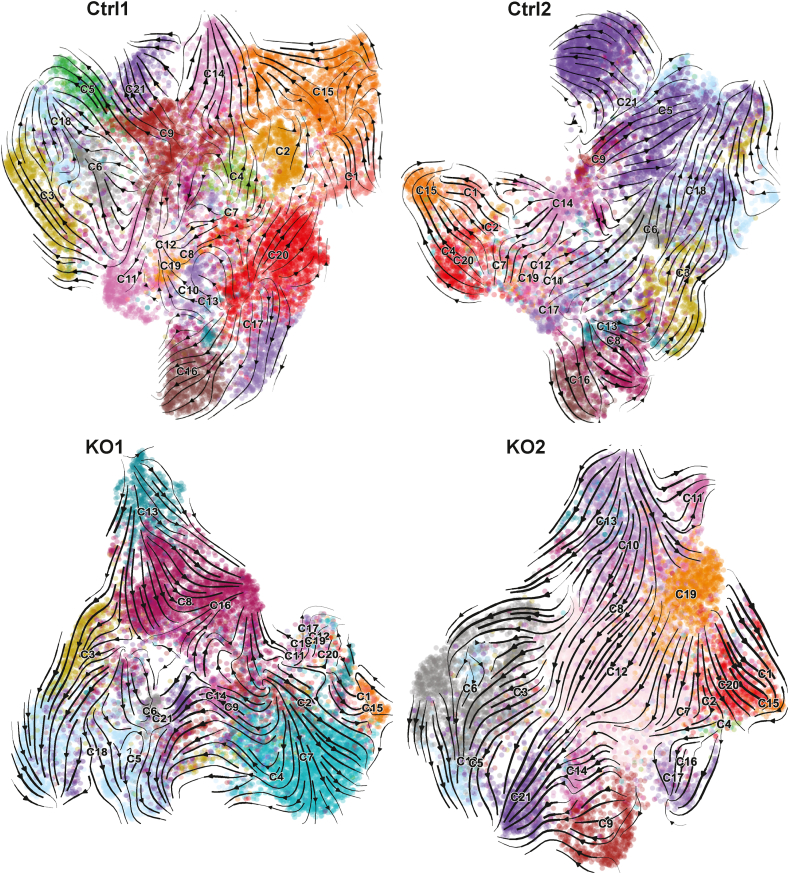
Fig. 2.
UMAP projection of cell differentiation trajectories predicted for each sample. RNA velocity trajectories were analyzed separately for each sample to avoid batch differences. The data were plotted as streamlines on the UMAPs computed by CellRank for each sample, to avoid potential artifacts introduced by co-embedding all samples together. The cells were colored by their cluster assignments in the integrated data (Fig. 1). Thus, some cells of the same color may become separated in these UMAPs, compared to the one in Fig. 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For non-neuronal clusters, TOP2A and MKI67 expression marked two neuroepithelial clusters (C10 and C13), while positive CRYM and DDIT3 expression were used for identifying CRYM+ (C11) and UPRC (C9) cells, respectively [38]. C9 markers were enriched for genes in the Gene Ontology (GO):0006986 (“response to unfolded protein”), supporting the cell identity. Three cluster of astrocytes, Astrocytes (C16), Astrocytes_progenitors (C8) and Astrocytes_progenitors (C17), with GFAP and SLC1A3 expression were identified, with the latter two expressing lower levels of these two markers than the former. RNA velocity, pseudotime trajectory analysis, and clustering based on scvelo dynamic data, indicated that C16 were more differentiated than C8 and C17 (Fig. 2), thus the latter two were designed as astrocyte progenitors. Similarly, three clusters of oligodendrocytes: Oligodendrocytes (C20), Oligodendroytes_progenitors (C12), Oligodendrocytes_progenitors (C19), were identified based on positive, but at different levels, OLIG1 and PDGFRA expression, with the annotation supported by trajectory data. Mesenchymal cells were separated into five clusters, using markers described in a previous study [38]. Marker genes for two of them showed significant enrichment in BMP pathway (Mesenchymal_BMP (C4), Mesenchymal_BMP (C7)), while the other three clusters expressed high levels of cilium genes (Mesenchymal_Cilia (C15), Mesenchymal_Cilia (C2), Mesenchymal_Cilia (C1)). Lastly, one cluster, Epithelial (C14), showed enrichment of epithelial markers (Fig. 1; Supplementary Table S2).
In short, scRNA-seq analysis indicate that the cerebral organoids from our differentiation protocols consist of a diverse array of neural progenitors, differentiating neurons, and non-neuronal cells (Supplementary Figs. S1 and S2). Given that the organoids were harvested after 50 days, most of the differentiating cell types were likely in immature states, just starting to express cell type specific genes and thus making it challenging to make definitive cell type assignment. For example, GAD2 was expressed in 11 % and 30 % of the cells in interneuron progenitors (C5) and inhibitory neurons (C21), respectively, while SLC17A7 was in 17 %, 22 % and 27 % of the neuronal progenitors (C3) and cortical neurons (C18, C6), respectively (Supplementary Fig. S1). As such, our data and results should be considered as models suitable for addressing CHD8 role in very early stages of neural differentiation, as described previously by Lancaster et al. [31].
3.2. Neural population composition affected by CHD8 knockout
With the cell type identification, we set out to infer the differentiating trajectory relationship of cells in each sample. We performed trajectory analysis based on RNA velocity and gene expression similarity between cell clusters, using CellRank [37]. RNA velocity predicts cell differentiation direction based on the ratio of splicing to unspliced RNAs [35]. Surprisingly, our scRNA-seq reads captured a high number of intronic reads, up to 45 % of unspliced reads in the Ctrl2 sample (Supplementary Fig. S3), making it easy to perform RNA velocity analysis.
We projected the inferred cell differentiation trajectory results to UMAP plots (Fig. 2), mainly to study global trends and sort through relationships between adjacent clusters in the same linages. The trajectories look largely similar for the four samples, suggesting that CHD8 reduction did not fully block neural differentiation potential, consistent with previous studies [28,29]. In three of the four samples (Ctrl2, KO1/KO2), Cortical_neurons (C18) and Inhibitory_neurons (C21) were identified as terminal states, as expected from known neural differentiation process. No terminal state, however, was confidently assigned for the Ctrl1 sample, despite similar RNA velocity support as for other samples (Fig. 2), suggesting that the computational prediction needs improvements. On the other hand, this trajectory analysis was very helpful for us to distinguish progenitor cells from their derivates, i.e., to establish local orders of a defined linage. Consistent with the expression analysis of marker genes (Fig. 1; Supplementary Fig. S1), progenitors of Cortical neurons, Astrocytes and Oligodendrocytes were located to the intermediate states of differentiation in the UMAPs, with supportive streamlines derived from RNA velocity (Fig. 2). The analysis also identified Neuroepithelial (C13) cells and UPRC (C9) cells as early states in all samples, while Oligodendrocytes_progenitors (C19) cells were considered as an additional initial state for KO2 sample. Overall, not perfect, but the trajectory analysis helps to correctly align cell types to known neural differentiation processes.
We next studied if the cell population differences between CHD8 KO and control samples could identify the points where differentiation is most regulated by CHD8, i.e., altered by CHD8 KO. We used miloR, which applies KNN graph based differential abundance analysis on data with replicates and is more robust than many other methods, such as simple proportion test of cell numbers in individual clusters [34]. Taking the integrated data, miloR constructed a KNN graph to represent the cell-cell expression similarity, partitioned the graph into partially overlapping neighborhoods, and then tested if the control or KO cells were statistically more abundant in each neighborhood using a negative binomial model. This avoids the caveat of separating cells into discrete clusters, which can be ambiguous for cells remaining at continuous differentiation. We found that most of the cell neighborhoods showed a decrease of abundance in the CHD8 KO samples (log fold change <0 in Fig. 3). Due to small sample size (two controls vs two KOs), the differences in many neighborhoods did not reach statistical significance, FDR <0.1 after adjusting for multiple testing, even though the fold changes appeared quite large (Fig. 3). The decreases were highly significant for the neighborhoods predominated by cells from Oligodendrocytes (C20), Neuronal_progenitors (C3), Mesenchymal_Cilia (C15), Astrocytes (C16) and Astrocytes_progenitors (C17) (red in Fig. 3). By contrast, Mesenchymal_BMP (C7) cell neighborhoods showed a significant increase in the KO samples (blue in Fig. 3). When the trends (regardless FDR) in all the detected cell types, especially the neural cells, were considered together, the data suggest that neuroepithelia (early progenitors C10 and C13) and cortical neurons (C6 and C18) were proportionally more abundant in the CHD8 KO samples than controls, while inhibitory neurons were less abundant, suggesting that CHD8 plays important roles in neuronal fate specification. If confirmed in future study, the data points to a scenario that early neurogenesis (e.g., lineage specification) may decelerate in CHD8 KO but the intermediate progenitors (e.g., C13), once produced, are accelerated to differentiate. This analysis also suggests that CHD8 could have important roles in glial cell differentiation, because astrocytes and mesenchymal cells were reduced in CHD8 KO.
Fig. 3.
Beeswarm plot showing the distribution of log-fold change in the abundance of cell types. Individual dots represent neighborhoods in a KNN graph representing cell-cell expression similarity constructed for all cells in the control and KO samples. For each of the dots/neighborhoods, the numbers of cells in the controls (n = 2) and KO samples (n = 2) were compared to determine the foldchange in frequencies and p values. The cluster identity of each dot/neighborhood was taken from the most frequent cluster names among cells within. The blue (only a few in C7) and red dots represent the cell neighborhoods with significantly higher and lower abundance in KO samples compared to control samples at FDR <0.1, with darker colors corresponding to lower p values. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Taken together, these analyses suggest that CHD8 plays important roles in both neural and non-neural cell fate specification, but the perturbation from its reduction may be subtle. The effects seem to be more robust in previous reports that analyzed organoids at later stages of differentiation [28,29], when the perturbed phenotypes become more stable.
3.3. Effects of CHD8 haploinsufficiency on cell type specific gene expression
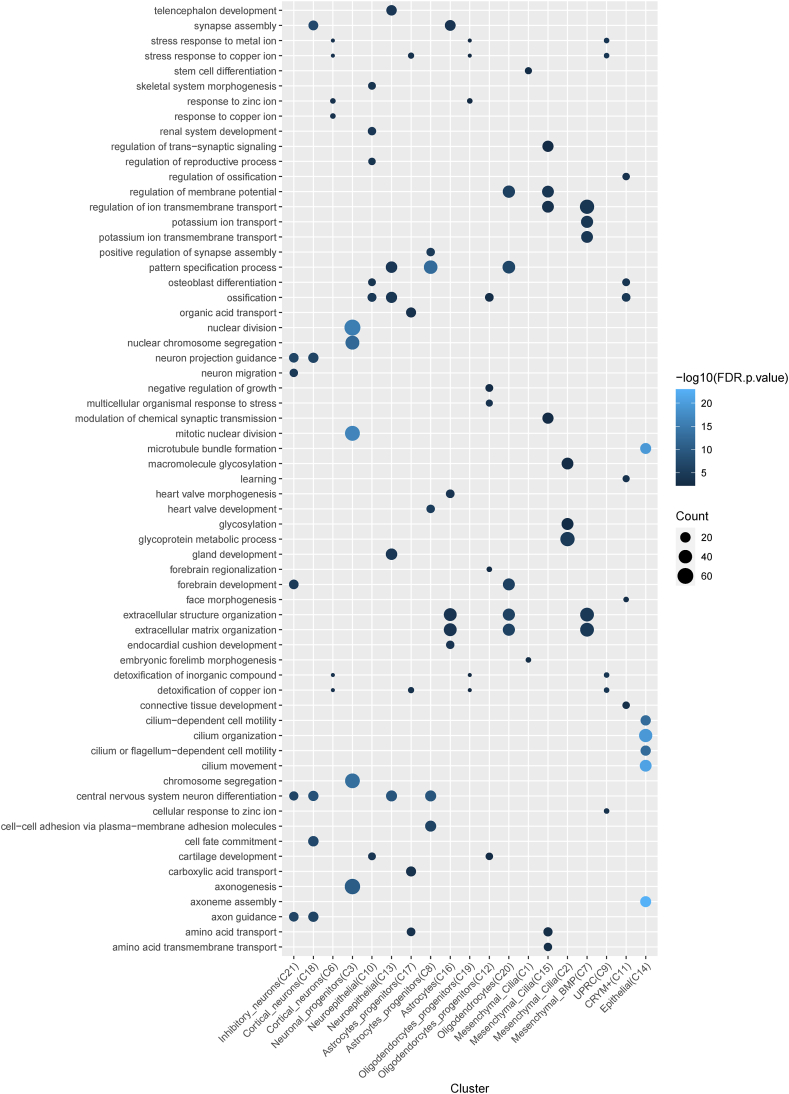
We next performed differential expression analysis between CHD8 KO and control samples, for each of the cell clusters independently. We found a total of 4913 unique genes affected by CHD8 haploinsufficiency in at least one of the cell types (padj <0.05 and fold change >2) (Supplementary Table S3). Mesenchymal_BMP (C7) had the most differentially expressed genes (DEGs) (n = 1005), while Cortical_neurons (C6) had the least (n = 35), indicating that CHD8 may have more important roles in some cell types than others. The other cortical neuron cluster (C18) had more DEGs (n = 248), 15 in common with C6, including NRNX3 and RBFOX1. We carried out GO enrichment analysis of the DEGs, and the top five most enriched terms for each of the cell types were shown in Fig. 4. The results demonstrated that GO terms such as synapse assembly, nuclear division, forebrain development, neuron projection guidance or axon guidance, were enriched in the DEGs detected in multiple neuronal cell types. For non-neuronal cell types, the enriched terms were related to axoneme assembly, central nervous system, neuron differentiation, cilium organization, extracellular matrix organization, modulation of chemical synaptic transmission or synapse assembly. The ECM difference was also observed in our previous bulk RNA-seq studies, while the axoneme and cilia formation abnormalities have been linked to the functions of ASD risk genes [[47], [48], [49], [50], [51]] (https://doi.org/10.53053/CMZZ2213).
Fig. 4.
Dot plot showing the top 5 enriched GO terms for the dysregulated genes in CHD8 KO for each of the cell clusters. The counts indicate the numbers of DEGs in a term.
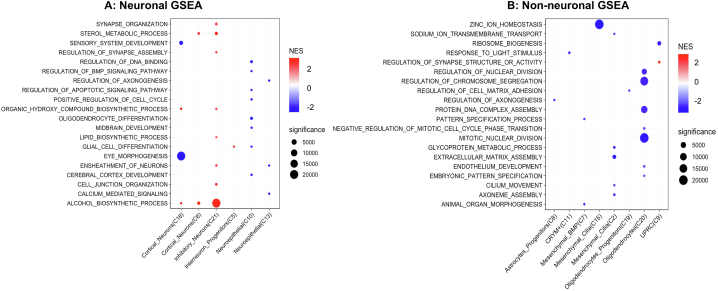
In addition to GO term enrichment analysis based on DEGs selected with thresholds, we applied Gene Set Enrichment Analysis (GSEA) to genes expressed in each cell type after they were ranked by expression changes between CHD8 KO and controls. From this analysis, inhibitory neurons showed upregulation of genes enriched in GO terms related to alcohol metabolomics, cell junction, lipid biosynthesis, regulation of synapse assembly or steroid biosynthesis (Fig. 5A; Supplementary Fig. S4). In contrast, most of the enriched gene sets in cortical neurons showed a negative enrichment (i.e., depletion) for sensory system development, nephron development or kidney and eye morphogenesis. Neuroepithelial cells showed downregulation for cell-cell adhesion, ensheathment of neurons, cerebral cortex development, glial cell differentiation and regulation of DNA binding and axonogenesis.
Fig. 5.
Dot plot showing top enriched pathways from GSEA for neuronal (A) and non-neuronal (B) cell clusters. Red and blue indicate higher and lower pathway activities, in the numeric form of normalized enrichment scores (NES), in the CHD8 KO samples compared to controls, respectively. Dot sizes indicate statistical significance defined as 1/FDR. A full version is shown in Supplementary Fig. 4. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Most of the enriched terms from GSEA for non-neuronal cells were found in the downregulated genes in CHD8 haploinsufficiency, with Oligodendrocytes having the most identified enriched pathways (Fig. 5B; Supplementary Fig. S4). Reduced expression genes in astrocytes showed an enrichment of eye/appendage development and regulation of axonogenesis. In mesenchymal cells, down-regulated genes were enriched in terms of axonome assembly, cilium movement, matrix assembly or cell substrate adhesion. CRYM+ and UPRC cells showed enrichment in response to radiation and light stimulus and response to abiotic stimulus and oxygens levels, respectively. Dysregulated genes in the oligodendrocytes were enrichment in cell cycle, chromosome organization, DNA repair, DNA replication, nuclear chromosome segregation or protein DNA complex assembly among others.
3.4. Cell-type effects of CHD8 haploinsufficiency on autism risk
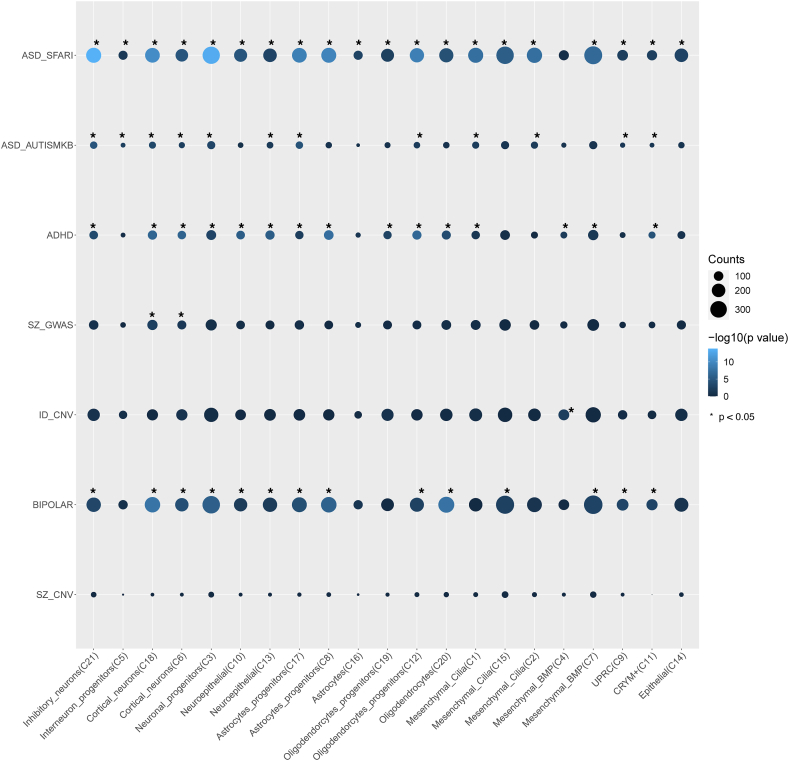
To explore how the dysregulated genes are related to autism and other neural disorder risks, we compared the cell type specific DEGs to gene sets associated with brain disorders (Fig. 6), including genes associated with ASD, SZ, bipolar disorder, ID, and attention deficit hyperactivity disorder (ADHD) [41]. Most of the cell types showed an enrichment of genes in the ASD_SFARI, ASD_AUTISMKB, ADHD and BIPOLAR databases, while only Cortical_neurons (C18 and C6) showed an enrichment in genes associated with SZ (SZ_GWAS). Interestingly, dysregulated genes in the Mesenchymal_BMP (C4) cluster were overrepresented with genes associated with ID (ID_CNV). It is interesting to see a strong enrichment with ASD genes but nearly no enrichment for SZ and ID risk genes. In total, 157 of the ASD SFARI genes showed differential expression in at least one of the 21 clusters, with the most in the neuron progenitor cluster (C3; n = 43) and astrocytes (C16; n = 34), including NRXN2/3, SHANK2 and SMARCA2. The altered expression of selected genes is shown in Supplementary Fig. S5.
Fig. 6.
Dot plot showing the over-representation between dysregulated genes and lists of genes implicated in several brain disorders. The p-values were from over-representation analysis by hypergeometric test. Asterisk (*) indicates nominal significance at p < 0.05.
3.5. Effects of CHD8 haploinsufficiency on cell-cell communication
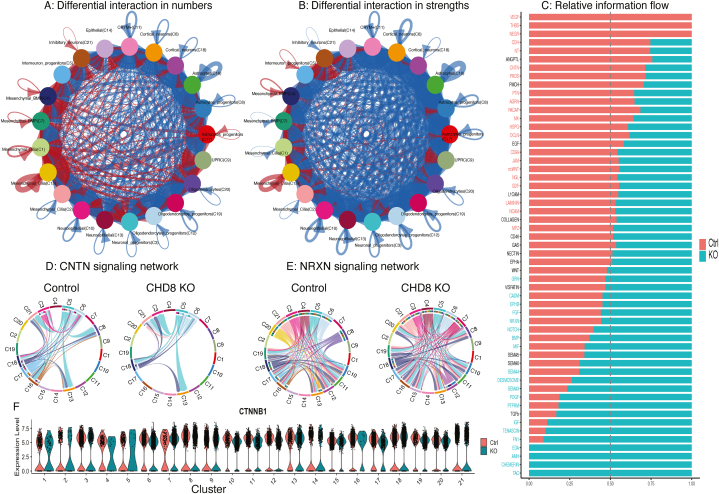
In addition to studying overall cell population changes and cell type specific effects, scRNA-seq data are also valuable for investigating cell-cell communications across different cell types [41]. We thus applied CellChat [45] and identified 12,763 ligand-receptor (L-R) interactions (and some non-L/R interactions) across all pairs of the 21 cell clusters in the control samples and slightly fewer interactions in the CHD8 KO samples (11,490) (Fig. 7). When these interactions in individual pairs of cell types were compared between control and KO cells, astrocytes (C16) showed the highest decrease in the number of interactions, specially, their interactions with cortical neurons and oligodendrocytes (Fig. 7A, Supplementary Fig. S6). On the other hand, mesenchymal cells showed the largest increase in the number of interactions. When interaction strengths were considered and compared, most pairs of cell types showed a reduction in interactions, with a global strength of 608.693 in the KO compared to 867.701 for the control samples. As shown in Fig. 7B and Supplementary Fig. S7, the majority of the cell types exhibited decreased interaction strengths, with only mesenchymal cells displaying an increase. Details of the L-R and other protein interactions and the corresponding cell-cell signaling differences are described in Supplementary Table S4. The noticeable changes involve some well-known surface proteins or ligands critical for neural development and function, including CNTN2, CNTNAP2, FGF, WNT, NRXN1, and RELN, which were also found in our differential expression analysis between controls and KOs (Supplementary Fig. S8).
Fig. 7.
Change in cell-cell communications between CHD8 KO and controls. A) Difference in the total number of L-R interactions in KO vs control samples, with red and blue indicating an increase or decrease, respectively, in the KO samples. B) Difference in the total strength of L-R interactions in KO vs control samples, with same color code as “B”. C) Relative information flow between KO and control cells for all 55 cell-cell signaling pathways detected by CellChat. The bars show the relative signaling strength in KO (red) vs controls (cyan). The signaling names in colors were statistically significant (paired Wilcoxon rank test; p < 0.05), with red and cyan for stronger in the control and KO, respectively. The absence of significantly altered cell-cell signaling is depicted in black. D,E) Chord diagrams plotting L-R interactions for CNTN (D) and NRXN (E) networks. F) Violin plot showing the expression of CTNNB1. The expression in C3 (neuronal progenitors) was significantly reduced in KO (log2-fold change = −0.3, padj <1.7e-29). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
It is difficult to go through all L-R interaction changes individually, but we can summarize the global difference involving all pairs of cell types in the entire organoids after cell-cell interactions in the same signaling pathways are merged and quantified. In total, the interactions detected by CellChat could be converged to 55 signaling pathways (Fig. 7C), with 3 (VEGF, THBS, NEGR) detected only in the control samples and 4 (EDA, AMH, CHEMERIN, TAC) only in the KO samples. Some of these altered signaling pathways may be important for the support that non-neural cells provide to the growth of neural cells. Using a paired Wilcoxon test (nominal p < 0.05), CellChat identified 21 signaling pathways with a global decreased activity (referred to as “information flow” in CellChat) in KO samples, while 20 displayed the opposite trend (Fig. 7C). Details of the involved genes (or proteins) and cell types can be found in the Supplementary Table S4. Interestingly, changes in the CNTN and NRXN signaling networks (Fig. 7D and E) were also observed in the brains from ASD subjects [41]. Non canonical WNT (ncWNT) signaling was significantly higher in controls, consistent with known interaction between CHD8 and Wnt/β-catenin signaling [13,14], but CTNNB1 was significantly differentially expressed only in the neuronal progenitors (C3) (Fig. 7F).
4. Discussion
After ASD risk genes are identified, deciphering their functional roles becomes a big challenge. CHD8 has been extensively studied since its discovery as a risk gene in 2012, with more than 200 publications. Here, we apply cerebral organoid and scRNA-seq technologies to investigate its function in early neural development and patterning at single cell resolution. Similar studies have been reported recently [[28], [29], [30]]. In one, two other ASD risk genes, SUV420H1 (also known as KMT5B) and ARID1B were studied together with CHD8, and all were found to play critical roles in neural development, as their haploinsufficiency conferred asynchronous development of GABAergic neurons and deep-layer excitatory neurons [29]. The effects, however, were dependent on iPSC lines and neural differentiation stages, with two of the four CHD8 lines showing an increase in GABAergic neurons and their progenitors 3.5 and 6 months post neural induction. This is consistent with our previous report of an increased expression of inhibitory neuronal markers (e.g., DLX1 and DLX6-AS1) in cerebral organoids generated by a different protocol [14]. Two other CHD8 lines, however, showed no significant difference in GABAergic interneurons [29]. Similarly, Villa et al. also found an increase in inhibitory neurons but a delay of excitatory neurons at day 60 and 120, leading to an imbalance of the two major neuron types [28]. Interestingly, they found that CHD8 haploinsufficiency caused an increased proliferation of the organoids, starting from day 10 and persisting to day 60, with an expansion of neural progenitors. While these two reports and our current study all reported a disruption in neural developmental trajectories and their dynamics, the effects on specific cell types seem different, suggesting the effects of CHD8 haploinsufficiency are likely modulated by genomic contexts, sex, differentiation protocols and stages of development. Considered that CHD8 is a ubiquitously expressed chromatin modifier and shows preferential binding to promoters of high expression genes [13,14], its reduction may have a large impact on genes that transiently change their expression at critical periods of cell fate specification or lineage bifurcation. What stages the impacts occur and on what genes, however, may depend on the expression of other regulators that functionally interact with CHD8. Consistent with this, one of the top functions enriched among ASD risk genes is chromatin modifiers.
In addition to the alterations in individual neuron cell types, our study identified substantial changes in non-neuronal cells and cell-cell communication in the cerebral organoids. The latter has not been investigated previously. Among the differential cell-cell interaction signaling, some are important for angiogenesis and vascular network patterning (e.g., VEGF, THBS, and FN1). Abnormal activations of such signaling may make the CHD8 deficient cells more prone to environmental changes, leading to asynchronous development disruptions. Along the same line, this may be a reason why variations of the KO effects were observed in different CHD8 studies; manifestations of CHD8 disruption may be sensitive to differentiation protocols and growth media (i.e., cell/environment interactions). As shown in Supplementary Fig. S8, we found that the expression of multiple genes encoding critical ligands and receptors were significantly altered by CHD8 reduction, including WNT4 and FGFR2. Although more studies are needed to follow up these findings, it is conceivable that non-neuronal cells and intercellular interaction networks can contribute to the increase of ASD risks in individuals with functional CHD8 mutations. It will be interesting to investigate if such signaling involves the cilia on the cells that we annotated as Mesenchymal_Cilia. Overall, our study suggests that CHD8 may orchestrate a complex network of molecular signaling events that are critical for the neural and brain development.
Two main limitations of our current study are small sample sizes and a single time point cross-section analysis. In addition, as the cerebral organoids were at a very early stage of differentiation, the separation of some cells into clusters may reflect more on the difference in transcriptomic states rather than cell types, suggesting some clusters may need to be merged. Nevertheless, our scRNA-seq findings and others together provide a broad survey of the cell type dependent and context-specific roles of CHD8, indicating that the impact of CHD8 mutations need more extensive analyses to map out the full functional spectrum.
Funding
This work was supported in part by grants to The Rose F. Kennedy Intellectual and Developmental Disabilities Research Center (RFK-IDDRC) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) at the NIH, USA (U54HD090260; P50HD105352).
Data availability statement
The integrated scRNA-seq data and clustering are publicly accessible in a R Shiny server (https://scviewer.shinyapps.io/CHD8_organoids/).
CRediT authorship contribution statement
Maider Astorkia: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis, Data curation. Yang Liu: Writing – review & editing, Software, Formal analysis, Data curation. Erika M. Pedrosa: Resources, Methodology. Herbert M. Lachman: Writing – review & editing, Writing – original draft, Funding acquisition, Conceptualization. Deyou Zheng: Writing – review & editing, Writing – original draft, Visualization, Supervision, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the supports from the Einstein Epigenomics Shared Facility, Einstein Computational Genomics Core, and Einstein High Performance Computing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34862.
Contributor Information
Herbert M. Lachman, Email: herb.lachman@einsteinmed.edu.
Deyou Zheng, Email: deyou.zheng@einsteinmed.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Talkowski M.E., Rosenfeld J.A., Blumenthal I., Pillalamarri V., Chiang C., Heilbut A., et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O ’ Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neale B.M., Kou Y., Liu L., Ma’ayan A., Samocha K.E., Sabo A., et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nat. 2012;485:242–245. doi: 10.1038/nature11011. 2012 4857397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X., Feliciano P., Shu C., Wang T., Astrovskaya I., Hall J.B., et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat. Genet. 2022;54:1305–1319. doi: 10.1038/s41588-022-01148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu J.M., Satterstrom F.K., Peng M., Brand H., Collins R.L., Dong S., et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54 doi: 10.1038/S41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beighley J.S., Hudac C.M., Arnett A.B., Peterson J.L., Gerdts J., Wallace A.S., et al. Clinical phenotypes of carriers of mutations in CHD8 or its conserved target genes. Biol. Psychiatr. 2020;87:123–131. doi: 10.1016/j.biopsych.2019.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015;6 doi: 10.1038/NCOMMS7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P., Lin M., Pedrosa E., Hrabovsky A., Zhang Z., Guo W., et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol. Autism. 2015;6 doi: 10.1186/S13229-015-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P., Mokhtari R., Pedrosa E., Kirschenbaum M., Bayrak C., Zheng D., et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism. 2017;8 doi: 10.1186/S13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnard R.A., Pomaville M.B., O'Roak B.J. Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front. Neurosci. 2015;9 doi: 10.3389/FNINS.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama Y., Nishiyama M., Shoji H., Ohkawa Y., Kawamura A., Sato T., et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- 17.Durak O., Gao F., Kaeser-Woo Y.J., Rueda R., Martorell A.J., Nott A., et al. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 2016;19:1477–1488. doi: 10.1038/nn.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platt R.J., Zhou Y., Slaymaker I.M., Shetty A.S., Weisbach N.R., Kim J.A., et al. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 2017;19:335–350. doi: 10.1016/j.celrep.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gompers A.L., Su-Feher L., Ellegood J., Copping N.A., Riyadh M.A., Stradleigh T.W., et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat. Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suetterlin P., Hurley S., Mohan C., Riegman K.L.H., Pagani M., Caruso A., et al. Altered neocortical gene expression, brain overgrowth and functional over-connectivity in Chd8 haploinsufficient mice. Cerebr. Cortex. 2018;28:2192–2206. doi: 10.1093/cercor/bhy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung H., Park H., Choi Y., Kang H., Lee E., Kweon H., et al. Sexually dimorphic behavior, neuronal activity, and gene expression in Chd8-mutant mice. Nat. Neurosci. 2018;21:1218–1228. doi: 10.1038/s41593-018-0208-z. [DOI] [PubMed] [Google Scholar]
- 22.Hui K., Katayama Y., Nakayama K.I., Nomura J., Sakurai T. Characterizing vulnerable brain areas and circuits in mouse models of autism: towards understanding pathogenesis and new therapeutic approaches. Neurosci. Biobehav. Rev. 2020;110:77–91. doi: 10.1016/j.neubiorev.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Hulbert S.W., Wang X., Gbadegesin S.O., Xu Q., Xu X., Jiang Y.H. A novel Chd8 mutant mouse displays altered ultrasonic vocalizations and enhanced motor coordination. Autism Res. 2020;13:1685–1697. doi: 10.1002/aur.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiménez J.A., Ptacek T.S., Tuttle A.H., Schmid R.S., Moy S.S., Simon J.M., et al. Chd8 haploinsufficiency impairs early brain development and protein homeostasis later in life. Mol. Autism. 2020;11 doi: 10.1186/S13229-020-00369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y., Zhang B., Ji P., Zuo Z., Huang Y., Wang N., et al. Changes to gut amino acid transporters and microbiome associated with increased E/I ratio in Chd8+/- mouse model of ASD-like behavior. Nat. Commun. 2022;13 doi: 10.1038/S41467-022-28746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombardo M.V., Moon H.M., Su J., Palmer T.D., Courchesne E., Pramparo T. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol. Psychiatr. 2018;23:1001–1013. doi: 10.1038/mp.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wade A.A., Lim K., Catta-Preta R., Nord A.S. Common CHD8 genomic targets contrast with model-specific transcriptional impacts of CHD8 haploinsufficiency. Front. Mol. Neurosci. 2019;11 doi: 10.3389/FNMOL.2018.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villa C.E., Cheroni C., Dotter C.P., López-Tóbon A., Oliveira B., Sacco R., et al. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 2022;39 doi: 10.1016/J.CELREP.2022.110615. [DOI] [PubMed] [Google Scholar]
- 29.Paulsen B., Velasco S., Kedaigle A.J., Pigoni M., Quadrato G., Deo A.J., et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602:268–273. doi: 10.1038/s41586-021-04358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin X., Simmons S.K., Guo A., Shetty A.S., Ko M., Nguyen L., et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science. 2020;370 doi: 10.1126/SCIENCE.AAZ6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;(8):1–12. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Wang T., Zhou B., Zheng D. Robust integration of multiple single-cell RNA sequencing datasets using a single reference space. Nat. Biotechnol. 2021;(39):877–884. doi: 10.1038/s41587-021-00859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dann E., Henderson N.C., Teichmann S.A., Morgan M.D., Marioni J.C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 2021;402(40):245–253. doi: 10.1038/s41587-021-01033-z. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., et al. RNA velocity of single cells. Nat. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. 5607719 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. 3812 2020. [DOI] [PubMed] [Google Scholar]
- 37.Lange M., Bergen V., Klein M., Setty M., Reuter B., Bakhti M., et al. Springer US; 2022. CellRank for Directed Single-Cell Fate Mapping. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka Y., Cakir B., Xiang Y., Sullivan G.J., Park I.H. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–1689.e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astorkia M., Lachman H.M., Zheng D. Characterization of cell-cell communication in autistic brains with single-cell transcriptomes. J. Neurodev. Disord. 2022;141(14):1–20. doi: 10.1186/s11689-022-09441-1. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen L IS of M at mount S. GeneOverlap: Test and visualize gene overlaps. R package version 1.26.0, http://shenlab-sinai.github.io/shenlab-sinai/.2020.https://bioconductor.org/packages/release/bioc/html/GeneOverlap.html (accessed 18 May2021).
- 43.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;343(34):267–273. doi: 10.1038/ng1180. 2003. [DOI] [PubMed] [Google Scholar]
- 45.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1–20. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhaduri A., Sandoval-Espinosa C., Otero-Garcia M., Oh I., Yin R., Eze U.C., et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nat. 2021;598:200–204. doi: 10.1038/s41586-021-03910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lasser M., Sun N., Xu Y., Wang S., Drake S., Law K., et al. Pleiotropy of autism-associated chromatin regulators. Development. 2023;150 doi: 10.1242/DEV.201515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willsey H.R., Xu Y., Everitt A., Dea J., Exner C.R.T., Willsey A.J., et al. The neurodevelopmental disorder risk gene DYRK1A is required for ciliogenesis and control of brain size in Xenopus embryos. Development. 2020;147 doi: 10.1242/DEV.189290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee B., Panda S., Lee H.Y. Primary ciliary deficits in the dentate gyrus of fragile X syndrome. Stem Cell Rep. 2020;15:454–466. doi: 10.1016/j.stemcr.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alhassen W., Chen S., Vawter M., Robbins B.K., Nguyen H., Myint T.N., et al. Patterns of cilia gene dysregulations in major psychiatric disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2021;109 doi: 10.1016/J.PNPBP.2021.110255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guglielmi G. Autism Research News. Spectr. News; 2023. Autism and the cell’s antennae | Spectrum. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The integrated scRNA-seq data and clustering are publicly accessible in a R Shiny server (https://scviewer.shinyapps.io/CHD8_organoids/).