Abstract
Background
The genus Hydrocotyle Tourn. ex L. is a key group for further study on the evolution of Apiales, comprising around 170 species globally. Previous studies mainly focused on separate sections and provided much information about this genus, but its infrageneric relationships are still confusing. In addition, the genetic basis of its adaptive evolution remains poorly understood. To investigate the phylogeny and evolution of the genus, we selected ten representative species covering two of three diversity distribution centers and exhibiting rich morphology diversity. Comparative plastome analysis was conducted to clarify the structural character of Hydrocotyle plastomes. Positive selection analyses were implemented to assess the evolution of the genus. Phylogenetic inferences with protein-coding sequences (CDS) of Hydrocotyle and 17 related species were also performed.
Results
Plastomes within Hydrocotyle were generally conservative in structure, gene order, and size. A total of 14 regions (rps16-trnK, trnQ-rps16, atpI-atpH, trnC-petN-psbM, ycf3-trnS, accD-psaI-ycf4, petA-psbJ, rps12-rpl20, rpl16 intron, rps3-rpl16 intron, rps9-rpl22, ndhF-rpl32, ndhA intron, and ycf1a) were recognized as hotspot regions within the genus, which suggested to be promising DNA barcodes for global phylogenetic analysis of Hydrocotyle. The ycf15 gene was suggested to be a protein-coding gene for Hydrocotyle species, and it could be used as a DNA barcode to identify Hydrocotyle. In phylogenetic analysis, three monophyletic clades (Clade I, II, III) were identified with evidence of rapid radiation speciation within Clade I. The selective pressure analysis detected that six CDS genes (ycf1b, matK, atpF, accD, rps14, and psbB) of Hydrocotyle species were under positive selection. Within the genus, the last four genes were conservative, suggesting a relation to the unique evolution of the genus in Apiales. Seven genes (atpE, matK, psbH, ycf1a, ycf1b, rpoA, and ycf2) were detected to be under some degree of positive selection in different taxa within the genus Hydrocotyle, indicating their role in the adaptive evolution of species.
Conclusions
Our study offers new insights into the phylogeny and adaptive evolution of Hydrocotyle. The plastome sequences could significantly enhance phylogenetic resolution and provide genomic resources and potential DNA markers useful for future studies of the genus.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05483-w.
Keywords: Hydrocotyle, Apiales, Positive selection, Adaptive evolution, Phylogeny
Background
The genus Hydrocotyle Tourn. ex L., belonging to Apiales with approximately 170 species, is widely distributed worldwide, with Australia, South America, and China as three diversity distribution centers [1]. This genus is a key group for further study on the evolution of Apiales. Hydrocotyle was once classified in Apiaceae in the traditional taxonomic studies due to the herbaceous habit, umbels, and two mericarps united fruits of species [2–5]. However, recent phylogenetic studies based on small molecular data, mainly from nuclear ribosomal DNA (nrDNA ITS and 26 S) and chloroplast DNA (cpDNA matK and rbcL), showed that Hydrocotyle was clustered with Araliaceae rather than Apiaceae [6–9]. Combining phylogenetic results with morphological evidence (the lignified endocarp, absence of tubing and carpel stalk in Hydrocotyle species), the genus was subsequently transferred to the family Araliaceae [10–12].
Hydrocotyle species are creeping, rooted at nodes, simple leaves with stipules, typically a single umbel inflorescence with petals valvate, fruit globose or ellipsoid, strongly flattened laterally [5, 12] (Fig. 1). This genus comprises both annual and perennial herbs that occur in variable habitat, such as mesic and aquatic environments [13] or seasonally dry and arid environments [14]. The genus was not given much attention in the early studies of Apiales. Species of Hydrocotyle are easily overlooked in the wild due to their short and prostrate growth, and the potential value of these plants is rarely exploited at present. With the transfer of Hydrocotyle to Araliaceae, the particularity and importance of Hydrocotyle in Apiales have been highlighted. Species of this genus were traditionally recognized based on leaf morphology, but leaf characters were widely variated and directly dependent on age and ecological factors [15]. Given this, species delimitation within Hydrocotyle is currently controversial. In recent years, most of the studies on this genus have focused on traditional taxonomic studies, and the discussions on the relationship between species were relatively few [16–22]. The reason was that few effective molecular markers have been obtained to reconstruct the phylogenetic relationship of this genus. The DNA barcode region (cpDNA trnH-psbA) was identified through extensive sampling and sequencing of a few Hydrocotyle species [23]. This region has been utilized in a few phylogenetic studies of the genus [14, 24]. Other DNA barcode regions, such as cpDNA trnL-trnF and nrDNA ITS/ETS, have also been employed in certain phylogenetic analyses [14, 24, 25]. Most phylogenetic studies were restricted to local groups with a small number of Hydrocotyle species, but they helped lay the groundwork for future integrative taxonomy [6, 11, 24, 25]. The study of Perkins (2019) on Hydrocotyle species in Australia combined molecular phylogenetic inference with morphological analyses, which set the boundaries among these species and provided new guidance for future systematic and taxonomic research within the genus [14]. Previous studies have shown that molecular phylogenetic studies were effective in the comprehensive classification of the genus Hydrocotyle. Therefore, it is necessary and urgent to construct a robust phylogenetic tree of this genus.
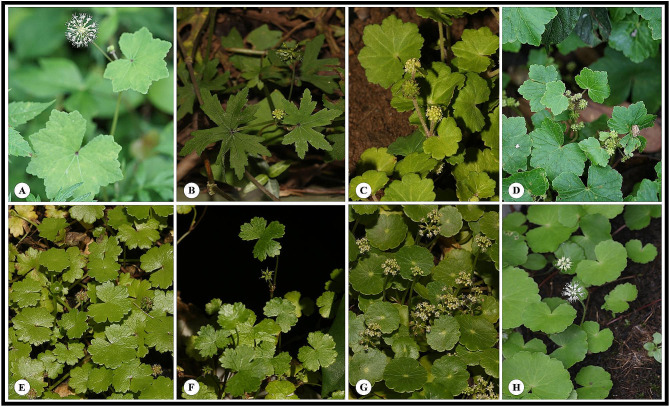
Fig. 1.
Morphological diversity of eight Hydrocotyle species. A, Hydrocotyle hookeri subsp. chinensis; B, H. dielsiana; C, H. pseudoconferta; D, H. nepalensis; E, H. sibthorpioides; F, H. sibthorpioides var. batrachium; G, H. verticillata; H, H. leucocephala
At present, plastome has been widely used in phylogenetic analysis of the order Apiales, especially for the family Apiaceae and Araliaceae [26–33]. Phylogenetic trees reconstructed by plastome data indicated improved supports than those inferred from small DNA markers. Plastome of nearly all Apiales has a highly conserved quadripartite structure composed of two copies of an inverted repeat region (IR) and two single copy regions, termed the large single copy (LSC) and small single copy (SSC). The phylogenetic location of Hydrocotyle has been identified as more closely related to Araliaceae by a few plastome studies, but the plastome of this genus has not been fully understood, because only a few have been reported until now [26, 34–36]. For the special phylogenetic position of Hydrocotyle in Apiales, it’s essential to study the plastome of this genus. In addition, understanding the plastome of different species of Hydrocotyle is helpful to screen out molecular markers suitable for the reconstruction of interspecific relationships within the genus.
Here, the newly sequenced plastomes of Hydrocotyle species, assembled from Illumina short reads, were presented. In combination with the previously released plastomes of this genus and the closely related species (belonging to Apiaceae and Araliaceae), we conducted comparative genomics and phylogenetic analyses on these data with the following aims: (1) to reveal the structural characteristics of Hydrocotyle plastome sequences; (2) to screen highly variable fragments suitable for phylogenetic reconstruction within the genus; (3) to investigate variations of the plastome structure among Hydrocotyle, Apiaceae, and Araliaceae; (4) to reconstruct robust phylogenetic relationships within Hydrocotyle and among Hydrocotyle, Apiaceae, and Araliaceae; (5) to investigate adaptive evolution patterns of protein-coding genes in Hydrocotyle. These results will provide insights into the evolutionary history of Hydrocotyle and the order Apiales as well as abundant information for future phylogenetic and population genetic studies.
Methods
Taxon sampling, DNA extraction, and genome sequencing
Six Hydrocotyle species were newly sequenced, which included two world-widespread species [Hydrocotyle sibthorpioides Lam. and Hydrocotyle sibthorpioides var. batrachium (Hance) Hand.-Mazz. ex R.H.Shan], two Pan-Himalaya endemic species [Hydrocotyle dielsiana H. Wolff and Hydrocotyle hookeri subsp. Chinensis (Dunn ex R.H.Shan & S.L.Liou) M.F.Watson & M.L.Sheh], and two South American species (Hydrocotyle verticillata Thunb. and Hydrocotyle leucocephalala Cham. & Schltdl.). These species contained a rich morphological diversity of the genus. The collections and voucher information for these species are provided in Table S1. Fresh and fully developed leaves were collected from disease-free plants, and timely dried in silica gel (Table S1). Total genomic DNA was extracted from leaf material using the Plant Genomic DNA Kit from Tiangen Biotech (Beijing) Co., Ltd., China. The quality and quantity of DNA were tested using 1% agarose gel electrophoresis, and the purity was detected by Nanodrop (OD 260/280 ratio). The high-quality DNAs were sequenced using the Illumina Novaseq 6000 platform at Novogene (Beijing, China), with paired-end reads 2 × 150 bp. DNA libraries were prepared using Rapid Plus DNA Lib Prep Kit for Illumina (RK20208).
Plastome assembly and annotation
Qualities of raw reads were checked by FastQC v0.11.9 [37]. The plastomes were assembled using NOVOPlasty v4.3.3 [38], a seed-extend-based de novo assembler and heteroplasmy/variance caller for short circular genomes. We used the Rubisco-bis-phosphate oxygenase (RUBP) sequences from the plastome of Hydrocotyle pseudoconferta Masam. (OK585058, which we reported early) as seed for plastome assembly, which has generated good results by the software developer [38]. The program Geneious v11.1.5 [39] was used to annotate the whole genomes, with gaps or degenerate bases that appeared in assembled genomes corrected by Sanger sequencing. Each species was annotated by comparing it against multiple reference genomes (Table S2) to obtain accurate annotations. The plastome maps were drawn using OGDRAW [40]. Raw data of the six newly obtained plastomes have been submitted to GenBank at NCBI (National Center for Biotechnology Information) BioProject PRJNA1035162. The plastome sequences have the accessions OR767307-OR767312 (Table 1).
Table 1.
Summary of major characteristics of the Hydrocotyle plastome sequences, including sequence length (bp), number of genes, GC content (%), and GenBank accession number
| Taxon | Whole genome | LSC | SSC | IRs | No. of CDS | No. of rRNA | No. of tRNA | GenBank accession No. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| length/bp | GC (%) | No. of Gene | length/bp | GC (%) | length/bp | GC (%) | length/bp | GC (%) | |||||
| Hydrocotyle dielsiana | 153,307 | 37.60 | 133 | 84,422 | 35.70 | 18,767 | 31.10 | 25,059 | 43.30 | 88 | 8 | 37 | OR767307 |
| Hydrocotyle hookerisubsp. Chinensis | 153,338 | 37.60 | 133 | 84,417 | 35.70 | 18,767 | 31.10 | 25,077 | 43.30 | 88 | 8 | 37 | OR767308 |
| Hydrocotyle leucocephala | 153,960 | 37.60 | 133 | 85,006 | 35.70 | 18,774 | 31.10 | 25,090 | 43.30 | 88 | 8 | 37 | OR767309 |
| Hydrocotyle nepalensis | 153,353 | 37.60 | 133 | 84,432 | 35.70 | 18,767 | 31.10 | 25,077 | 43.30 | 88 | 8 | 37 | MT561038 |
| Hydrocotyle pseudoconferta | 153,302 | 37.60 | 133 | 84,417 | 35.70 | 18,767 | 31.10 | 25,059 | 43.30 | 88 | 8 | 37 | OK585058 |
| Hydrocotyle sibthorpioides | 152,941 | 37.50 | 133 | 84,125 | 35.50 | 18,690 | 31.00 | 25,063 | 43.30 | 88 | 8 | 37 | OR767311 |
| Hydrocotyle sibthorpioides | 152,880 | 37.50 | 133 | 84,064 | 35.50 | 18,690 | 31.00 | 25,063 | 43.30 | 88 | 8 | 37 | NC_035502 |
| Hydrocotyle sibthorpioides var. batrachium | 152,663 | 37.60 | 133 | 83,962 | 35.70 | 18,585 | 31.10 | 25,058 | 43.30 | 88 | 8 | 37 | OR767310 |
| Hydrocotyle verticillata | 153,161 | 37.60 | 133 | 84,287 | 35.70 | 18,730 | 31.10 | 25,072 | 43.30 | 88 | 8 | 37 | OR767312 |
| Hydrocotyle verticillata | 153,207 | 37.60 | 133 | 84,352 | 35.70 | 18,739 | 31.10 | 25,058 | 43.30 | 88 | 8 | 37 | NC_015818 |
Note: Sequences newly obtained are indicated by bold font. LSC: Large single copy region; SSC: Small single copy region; IRs: Inverted repeat regions.
Comparative analysis of plastomes
Ten plastome sequences of Hydrocotyle were adopted for comparative analysis, the accession numbers of the previously released Hydrocotyle sequences are shown in Table S2. All sequences were derived from GenBank at NCBI up to July 25, 2023 (data released after were not used in our study because of unavailable specimens). The mVISTA program was used to conduct a sequence identity analysis of the ten plastome sequences of Hydrocotyle under LAGAN mode [41], using Hydrocotyle nepalensis Hook. as a reference. Mauve v2.4.0 with default settings within Geneious was used to identify large structural changes such as gene order rearrangements, inversions, and insertions in the plastome sequences [42]. The junction sites of LSC-IRA/B and SSC-IRA/B were compared with IRscope [43], and IR expansion or contraction in the Hydrocotyle genomes was also detected. To determine the nucleotide diversity (Pi) among the plastome of Hydrocotyle species, the sliding window analysis was conducted using DnaSP v.6.10 [44], with a step size of 200 bp and a window length of 600 bp. The Pi value meant the average number of nucleotide differences between two sequences randomly chosen, which estimated divergence among species [45].
Positive selection analysis
The CDS regions from ten Hydrocotyle species were extracted to calculate Pi and pairwise Ka/Ks with DnaSP v.6.10, and the average values were used to represent the Ka/Ks ratio of each gene. Identical CDS sequences (psbM, rps12, rps7, psaJ, and rpl36) were filtered out. The alignments of the remaining 76 CDS sequences were generated under MAFFT v7 [46] plug-in within Geneious. The ratio ω = Ka/Ks was used to measure the selective pressure, with ω > 1, ω = 1, and ω < 1 suggesting positive selection, neutral selection, and purifying selection, respectively [47]. Two datasets were used to conduct positive selection analyses. Within the genus Hydrocotyle, CDS sequences with pairwise Ka/Ks > 1 or only with Ka value will be focused on detecting positive selection driving protein evolution. The branch-site model [48] was utilized to detect positive selection in the foreground branch, which was designed according to genes.
Another dataset was used to identify positive selection in Hydrocotyle species compared to other family species under the branch-site model. A total of 80 CDS sequences from 29 species were contained in the analysis, with the genus Hydrocotyle specified as the foreground branch. The sources of all sequences, except for new sequencing, are displayed in Table S2. The Bayesian Empirical Bayes (BEB) [49] method was used to compute the posterior probabilities of amino acid sites under positive selection. The likelihood ratio tests (LRT) were implemented as a result. A gene with a P-value < 0.05 and positively selected sites was considered a positively selected gene. All these analyses were performed in the EasyCodeML v1.4 [50].
Sequence divergence of ycf15 gene
Sequences of the ycf15 gene in Hydrocotyle employed a GTG start codon similar to those in Araliaceae along with an intact open reading frame (ORF), which makes the gene more likely to be functional in this genus. However, multiple internal stop codons or GCG/GTA initial codons were detected in many other species within Apiaceae, suggesting that ycf15 may be disabled in these species. These findings thus raised our interest in further investigating the evolution of ycf15 in Apiales. Therefore, we selected the ycf15 coding sequences of Araliaceae and Hydrocotyle, and compared them to the sequences of the same regions in Apiaceae for analysis of nucleotide and protein-coding sequences.
Phylogenetic analysis
A total of 29 taxa were sampled for phylogenetic analysis, comprising eight taxa from Apiaceae, ten taxa from Hydrocotyle, nine taxa from Araliaceae, and two Torricellia DC. species from Torricelliaceae serving as outgroups (refer to Table S2). In total, 75 CDS sequences [except for ycf1a, which was very short in Schefflera delavayi (Franch.) Harms due to an internal stop codon] were extracted and used in phylogenetic analysis. Sequence matrices were aligned under MAFFT v7 [46] and manually adjusted in MEGA v7 [51]. The alignments were concatenated using MEGA v7 forming a super sequence matrix finally used for phylogenetic analysis. Two methods were adopted for phylogenetic analysis: Maximum likelihood analysis (ML) and Bayesian inference (BI). The best-fitting nucleotide substitution models were selected in jModelTest v2.1.4 [52] and finally determined by phylogenetic analysis software requirements. The ML analysis was performed with RAxML v8.2.4 [53] under the GTRGAMMA model and 1000 bootstrap replicates. BI analysis was executed under MrBayes v3.2 [54] to obtain the posterior support of phylogenetic relationships among taxa with the GTR + G + I substitution model. Two independent Markov chain Monte Carlo (MCMC) runs were performed, each with one cold chain and three heated chains for 10,000,000 generations. The first 25% were discarded as burn-in. MCMC convergence was reflected from the average standard deviation of split frequencies to approach zero. We used posterior probability (PP) and bootstrap support (BS) to measure the supports of the phylogenetic tree implemented under BI and ML methods, respectively. The final tree was viewed and edited in FigTree v1.4 [55].
Results
Structural characteristics of Hydrocotyle plastomes
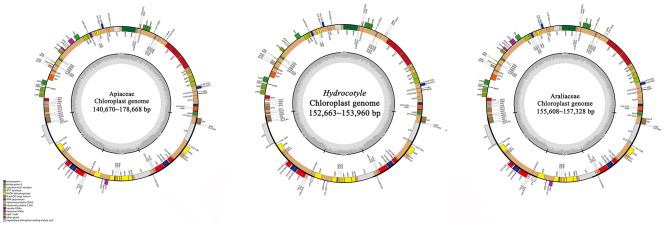
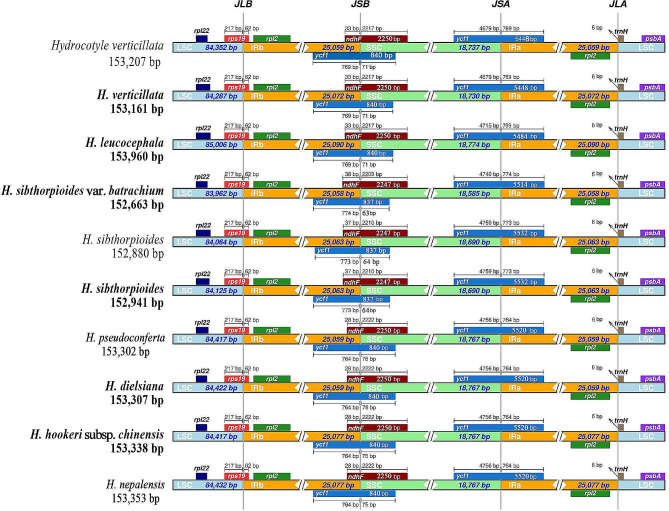
Plastome of Hydrocotyle species shared a typical quadripartite structure, with two inverted repeats regions (IRA and IRB), one large single copy region (LSC), and one small single copy region (SSC). The sizes of ten Hydrocotyle plastomes ranged from 152,663 bp to 153,960 bp, and their overall GC content ranged from 37.50 to 37.60% (Fig. 2; Table 1). Each plastome contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. A total of 18 genes have been detected owning two copies. These duplicated genes included seven protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and rps12), four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5), and seven tRNA genes (trnA-UGC, trnI-GAU, trnI-CAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) (Fig. 2). The junction of the LSC/IRB, IRB/SSC, SSC/IRA, and IRA/LSC were located on gene rps19, the overlap of ycf1b (the copy of ycf1 gene was largely located in IRB region) and ndhF, gene ycf1a (the copy of ycf1 gene was largely located in IRA region), and 6 bp before trnH, respectively (Fig. 3).
Fig. 2.
Circular gene maps for plastomes of Apiaceae, Hydrocotyle, and Araliaceae,representatives. Genes plotted outside the circle are transcribed counterclockwise, inside genes—clockwise. Genes are colored according to their function
Fig. 3.
Comparison of the LSC, SSC, and IR junction of plastomes among the ten Hydrocotylespecies. JLB: junction line between LSC and IRB; JSB: junction line between SSC and IRB; JSA: junction line between SSC and IRA; JLA indicates the junction line between LSC and IRA.
Comparative genomic analysis within Hydrocotyle
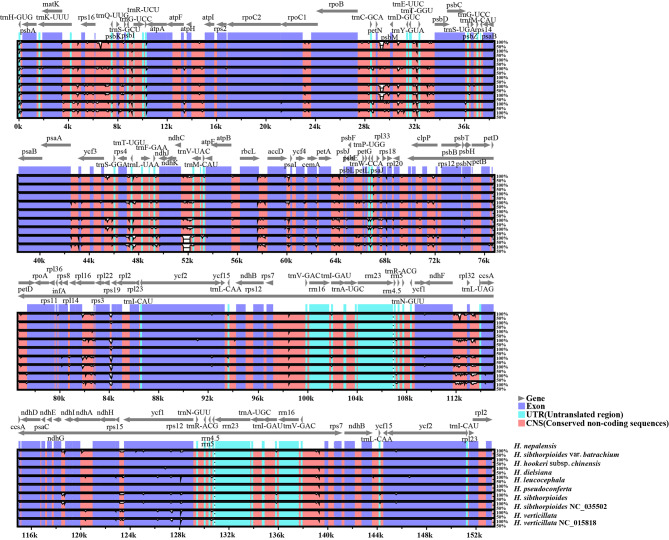
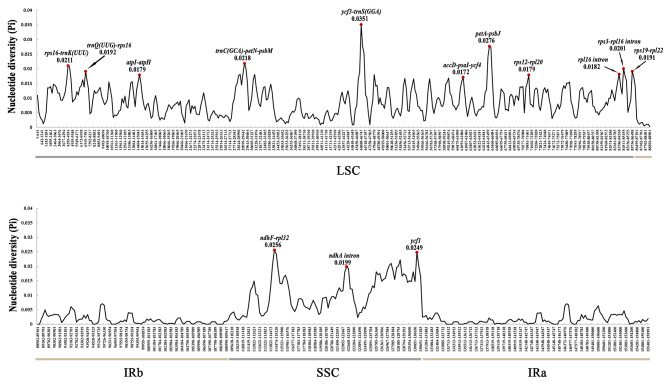
The aligned plastome sequences of ten Hydrocotyle species resulted in a matrix of 155,981 bp and showed high sequence similarity with 98.00% pairwise identity and 147,751 identical sites. Distances analysis suggested that identity among these species ranged from 96.49 to 99.97% (Table S3). Genes order was consistent without rearrangement as verified by Mauve analysis (Fig. S1). The mVISTA result revealed that coding regions (Exon) showed more sequence conservation than non-coding regions (CNS), and IR regions had less variation than the other two regions (Fig. 4). The nucleotide diversity (Pi) of ten Hydrocotyle plastome sequences has been calculated to assess the sequence divergence level. The Pi values ranged from 0 to 0.0351 across the ten plastomes, as indicated by the sliding window analysis (Fig. 5, Table S4). A total of 14 regions were recognized as hotspot regions with Pi > 0.0170. Most of these regions were located in the LSC region, and these included rps16-trnK, trnQ-rps16, atpI-atpH, trnC-petN-psbM, ycf3-trnS, accD-psaI-ycf4, petA-psbJ, rps12-rpl20, rpl16 intron, rps3-rpl16 intron, and rps9-rpl22. While the SSC region included ndhF-rpl32, ndhA intron, and ycf1a. The sequences within IR regions displayed very low Pi values. The nucleotide diversity of protein-coding sequences (CDSs) has been detected, except for five sequences that are identical across different species (psbM, psaJ, rpl36, rps7, and rps12). A total of 76 CDS regions were analyzed, Pi values ranged from 0.0006 to 0.0173, and CDS of ycf1a gene and atpE gene possessed Pi values more than 0.0150 (Table S5). The GC contents varied from 29.70 to 46.70% (Table S5).
Fig. 4.
mVISTA comparison of ten Hydrocotyle plastomes (H. nepalensis as reference). The percentage identity ranging from 50 to 100% is represented by the vertical scale.
Fig. 5.
The nucleotide diversity (Pi) of the ten Hydrocotyle plastomes. Regions with high Pi values (above 0.0170) were marked out.
Gene selective pressure analysis
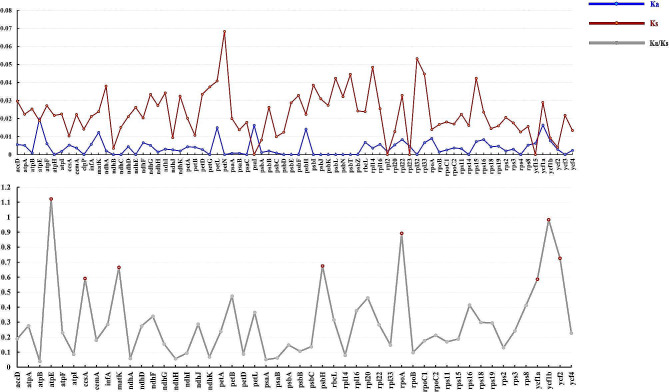
Ka, Ks, and their ratios Ka/Ks (ω) were also calculated to evaluate the selection pressure of the 76 protein-coding genes (Table S5, Fig. 6). The Ka values ranged from 0 to 0.0198, and all CDS genes possessed low Ka values with six genes over 0.0100 (atpE, matK, petL, psbH, psaI, and ycf1a). Ka = 0 without any non-synonymous mutations appeared in a large proportion of CDSs (21 CDSs: atpH, clpP, ndhB, ndhC, ndhE, petG, petN, psaC, psbD, psbE, psbF, psbI, psbJ, psbK, psbL, psbT, psbZ, rpl32, rps4, rps14, and ycf3). The Ks values ranged from 0 to 0.0682 and were generally distributed between 0.0100 and 0.0400, with eight genes over 0.0400 (rpl14, rpl32, rpl33, petN, petL, psbL, psbT, and rps15). Ks = 0 only with non-synonymous mutations but no synonymous mutations appeared in four genes (psaI, rpl2, rpl23, and ycf15; Table S6). The psaI gene showed non-synonymous mutations between South American species (H. verticillata and H. leucocephala) and the others. For gene rpl2, non-synonymous mutations were found between H. sibthorpioides (and the variety H. sibthorpioides var. batrachium) and the other species. The non-synonymous mutations of the rpl23 gene only occurred between H. leucocephala and other species, while those in the ycf15 gene only appeared between the newly sequenced H. verticillata and the other taxa. These four genes (psaI, rpl2, rpl23, and ycf15) should receive more attention in future expanded sampling studies.
Fig. 6.
The curves of the average Ka, Ks and Ka/Ks values of protein-coding regions (CDS) within Hydrocotyle plastomes in this study. CDSs with Ka/Ks > 0.5 were marked out with red circles in the Ka/Ks curve
Genes with either Ka = 0 or Ks = 0 were excluded when calculating the ratio ω = Ka/Ks. The ω values ranged from 0.0388 to 1.1224, with most genes below 0.5, indicating purifying selection. Eight genes were identified with ω > 0.5 (atpE, ccsA, matK, psbH, rpoA, ycf1a, ycf1b, and ycf2). The atpE gene had ω > 1 (1.1224) (Fig. 6, Table S5, S7). A total of seven genes (atpE, matK, rpoA, ycf1b, rpl16, rpl20, and rps16) exhibited Ka/Ks > 1 between certain taxa, atpE and rpoA showed some Ka/Ks values equal to 1 between taxa (Table S5). The details of the Ka and Ks for these genes were presented in Table S7 and Table S8. For the gene atpE, ω values between H. verticillata and the other species (except H. leucocephala) were greater than 1, indicating that the atpE gene might undergone positive selection in H. verticillata. ω > 1 were found in gene atpE, rpl20, and ycf1b between H. leucocephala and H. sibthorpioides var. batrachium. The ω values of atpE gene sequences between H. leucocephala and H. sibthorpioides were also detected greater than 1. For the matK gene, ω > 1 only existed between H. leucocephala and H. verticillata. H. nepalensis, H. dielsiana, H. pseudoconferta, and H. hookeri subsp. chinensis had a similar gene structure. Here, we defined them as Clade I. ω > 1 found in the rpl16 gene were between Clade I and H. sibthorpioides (Table S8). For the ycf1b gene, ω > 1 also existed between H. leucocephala and Clade I, as well as between H. leucocephala and the newly sequenced H. verticillata. For the rpoA gene, ω values between Clade I and H. verticillata were greater than 1, and ω = 1 existed between Clade I and H. sibthorpioides var. batrachium. ω > 1 for the gene rps16 existed between H. sibthorpioides and the newly sequenced H. verticillata population (Table S8).
A total of 15 CDSs (Ks = 0 or with pairwise Ka /Ks > 1) were selected for testing positive selection. When H. leucocephala was specified as the foreground branch, matK was certified as the positively selected gene, with both LRT P-value < 0.05 and positively selected sites present (Table S9). When the clade including H. leucocephala and two taxa of H. verticillata was designed as foreground branch, the LRT P-values of gene atpE and ycf2 were greater than 0.05. However, the BEB analysis detected several positively selected sites with a high posterior probability (> 95%) for both genes (Table S9).
To detect sites under positive selection in the CDS genes in the plastome of genus Hydrocotyle, a branch-site model analysis was conducted within Apiales. Six genes (ycf1b, matK, atpF, accD, rps14, and psbB) were tested LRT P-value < 0.05, among which four genes (ycf1b, matK, rps14, and psbB) exhibited some positively selected sites with a high posterior probability (> 95%). One positively selected site was detected in gene ndhJ with a high posterior probability (> 95%), but the LRT P-value of this gene was above 0.05 (Table S10).
The distribution of the ycf15 gene among Hydrocotyle apiaceae and araliaceae
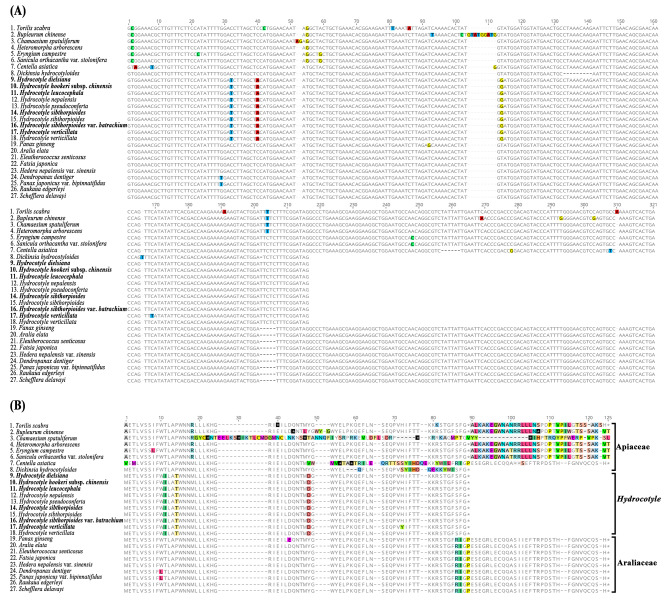
The results showed that the ycf15 gene of Hydrocotyle species employed a GTG start codon and possessed a complete intact ORF with a nucleotide sequence length of 204 bp (Fig. 7). We have conducted an investigation and tallied the specifics of the ycf15 gene in Apiaceae, Hydrocotyle, and Araliaceae. In most Araliaceae groups, the length of the nucleotide sequence of the ycf15 gene was 303 bp. However, a few were 330 bp (NC_028810) or 309 bp (NC_049886 and NC_049888). All sequences had intact ORF and were annotated locating between the ycf2 and trnL-CAA gene. Sequences of the same position (between ycf2 and trnL-CAA gene) in Apiaceae species were extracted and translated to protein sequences. Aligned protein sequences from 27 taxa were generated and displayed in Fig. 7B. This suggested that the ycf15 gene was highly conserved within Hydrocotyle and Araliaceae. The length of the ycf15 pseudogene varied in the subfamily of Apiaceae due to different insertion locations of stop codons (Fig. 7B). The ycf15 gene of subfamily Saniculoideae (Apiaceae) and the basal group of subfamily Apioideae had no internal stop codons inserted. Whether these genes are functional needs further study for the lack of start codons. Dickinsia hydrocotyloides Franch. was the only taxa with normal ycf15 genes in all the released data of Apiaceae. The ycf15 gene of D. hydrocotyloides employed a GTG start codon along with an intact ORF, which made it more likely to be functional. The length of this ORF was 195 bp, shorter than that of Hydrocotyle, and the insertion and deletion of a single base in the terminal sequence led to large changes in the protein-coding sequence (Fig. 7B). Whether these changes will cause functional changes remains to be studied.
Fig. 7.
Alignment of the ycf15 gene and translated sequences from the Apiaceae, Hydrocotyle, and Araliaceae species. (A) Alignment of the ycf15 gene sequences; (B) Alignment of the ycf15 translated sequences.
Phylogenetic analysis
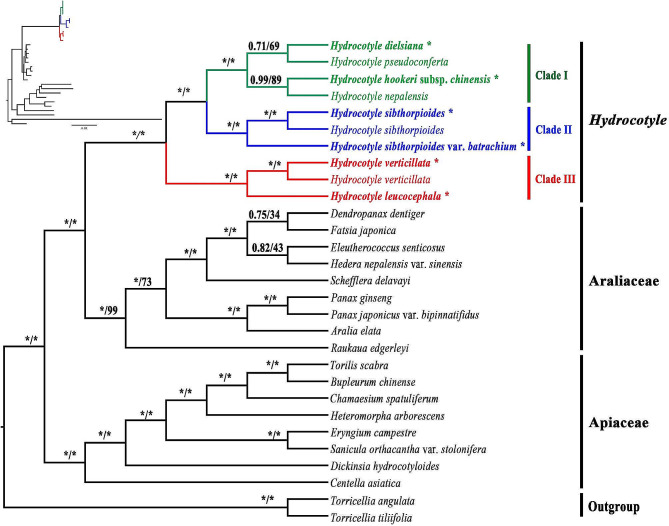
The phylogenetic tree constructed based on 75 protein-coding genes exhibited a clear evolutionary history of Hydrocotyle. Hydrocotyle was determined to be monophyletic in both ML and BI analyses with strong supports (BS = 100, PP = 1.00). This genus formed the closest sister group to Araliaceae (Fig. 8). Within Hydrocotyle, three monophyletic sister clades were recognized with high supports (BS = 100, PP = 1.00). Clade I comprised four species, including three Pan-himalaya endemic species (H. dielsiana, H. pseudoconferta, and H. hookeri subsp. chinensis) and one globally distributed species (H. nepalensis), which was consistent with the definition in the former section. Within this clade, two secondary clades were recognized: H. nepalensis and H. hookeri subsp. chinensis were gathered with high supports (BS = 89, PP = 0.99), while H. dielsiana was clustered with H. pseudoconferta with weak supports (BS = 69, PP = 0.71). The ML analysis showed clades within Clade I with very short branch lengths (Fig. 8). Clade II included H. sibthorpioides and the variant of this species (H. sibthorpioides var. batrachium). Two taxa of H. sibthorpioides were gathered with high supports (BS = 100, PP = 1.00), and then highly supported (BS = 100, PP = 1.00) to split with H. sibthorpioides var. batrachium. The two remaining South American species (H. verticillata and H. leucocephalala) constituted Clade III. These two species were strongly supported for separation (BS = 100, PP = 1.00). After separating with Hydrocotyle, the family Araliaceae rapidly differentiated into several genera with very short internal branches (Fig. 8). Phylogenetic relationships among Apiaceae taxa were highly supported (BS = 100, PP = 1.00).
Fig. 8.
Phylogenetic relationships inferred from 29 species based on 75 shared CDSs. Support values marked above the branches follow the order Bayesian inference (PP, posterior probability)/ maximum likelihood (BS, bootstrap support), * represent the best support (100%). Species with “*” in the right hand indicated these species were newly sequenced
Discussion
Comparison of Hydrocotyle apiaceae and araliaceae plastomes
In this study, a comprehensive comparative analysis of 10 plastomes from Hydrocotyle was implemented. The result indicated that plastomes of Hydrocotyle were highly identical in their structural organization, gene order, and gene content. Due to the special systematic position of this genus in Apiales, we have made a comparative analysis of the plastome of this genus and its close relatives, Apiaceae and Araliaceae. Plastome sizes of Hydrocotyle were smaller than those of Araliaceae, while sizes of the Apiaceae plastomes varied in a large scan (Fig. 2). There seemed to be no specific rule in plastome sizes among the three groups. We subsequently investigated the gene content of these taxa and found that the ycf15 gene greatly varied among Hydrocotyle, Apiaceae, and Araliaceae.
The ycf15 gene has long been questioned to encode protein [56–58]. Not all species contained the intact ycf15 gene, in some groups, the gene was disabled in case of separating by multiple internal stop codons [56, 59, 60] or even wholly lost [61, 62]. The intact copy of the ycf15 gene remains present in many species, e.g. Mognolia, Piper, and Camellia [61, 63–65]. The ycf15 gene was initially identified in the Nicotiana plastome, with GTG as the start codon [66, 67]. The expression information for the ycf15 gene in Nicotiana tabacum L. and Amborella trichopoda Baill. indicated that the GTG start codon in ycf15 was not edited into standard ATG [63]. In recent years, an ATG initial codon was detected in the Camellia ycf15 gene, which was suggested co-transcribed with ycf2 and antisense trnL-CAA [63]. Most studies have suggested that the ycf15 gene was located between ycf2 and trnL-CAA employing GTG or ATG as initial codons [33, 63, 68, 69].
We focused on the ycf15 gene because there were two annotation locations of this gene in Apiaceae species: one between the gene ycf2 and trnL-CAA [49], and the other between the gene rps7 and trnV-GAC [31, 70–72]. The nucleotide sequences of two kinds of ycf15 gene were extracted and compared. The comparison revealed significant variation between the sequences. Regardless of the annotation position, the gene was annotated as a pseudogene due to the absence of a start codon, except in the case of D. hydrocotyloides, which possessed a GTG start codon (Fig. 7A). The previous studies have shown that the ycf15 gene of Araliaceae species had intact ORF [61, 63]. Therefore, when annotating the gene of Hydrocotyle species, the annotation information of the ycf15 gene in these species was selected as reference. In our study, both Hydrocotyle and Araliaceae had intact ORF in the ycf15 gene and employed GTG as the start codon. However, the length of the former was significantly shorter than that of the latter. Due to the different gene lengths and non-synonymous mutations in multiple locations, the amino acid sequence encoded by the gene in the two groups varied (Fig. 7). Which indicated that ycf15 gene might undergone strong selection pressure during the evolution of Hydrocotyle and Araliaceae. The ycf15 genes in Apiaceae were special in that they had two annotation locations, one of which was the same as that in Hydrocotyle/Araliaceae, and the other was between rps7 and trnV-GAC. The latter was supported by some other findings [24, 31, 73], but this annotation has not been validated by transcription studies. Therefore, we focused on the former annotation results in this study. The ycf15 gene was re-annotated between ycf2 and trnL-CAA in all released sequences of Apiaceae and indicated high variability of this gene in Apiaceae. Unlike the ycf15 gene in Hydrocotyle/Araliaceae, this gene of Apiaceae employed GCG or GTA at the initial position instead of GTG/ATG (excepted D. hydrocotyloides), which suggested that they were pseudogenes and nonfunctional. Lengths of these ycf15 pseudogenes varied upon the position of internal stop codons, with the shortest being ~ 81 bp, indicating that the ycf15 pseudogene had undergone genetic degeneration in Apiaceae. Distribution of the ycf15 gene in the three groups further supports the evolutionary status of Hydrocotyle, which is more closely related to Araliaceae but independent of the transitional groups outside the two families. However, such a conjecture still needs further studies to clarify. In addition, the conservation of the ycf15 gene sequence within Hydrocotyle species and its difference between Hydrocotyle and Araliaceae indicated that this gene could serve as a DNA barcode for identifying Hydrocotyle and Araliaceae.
Promising DNA barcodes
Many efforts have been made to construct a robust phylogenetic framework of Hydrocotyle [14, 23–25]. The DNA fragments currently used in phylogeny reconstruction of Hydrocotyle included ITS, ETS, trnH–psbA, and trnL–trnF. The fragments trnK–rps16, rps16–trnQ, atpH–atpI, ycf3–trnS, ndhF-rpl32, petN-psbM, rpl16 intron, and ycf1 gene have previously been considered useful in resolving low-level relationships of Apiaceae and Araliaceae due to the high pi value [31, 33, 34, 68, 74]. In addition to these loci, we found that a total of six regions held relatively higher Pi values for Hydrocotyle: accD-psaI-ycf4, petA-psbJ, rps12-rpl20, rps3-rpl16 intron, rps9-rpl22, and ndhA intron. All the fragments had higher pi values than the two plastid genes [trnH–psbA (0.01556) and trnL–trnF (0.00791)] that were previously used to construct the phylogeny of Hydrocotyle. Therefore, we speculated that these fragments would play an important role in plant identification and reconstructing the global phylogenetic framework of Hydrocotyle.
Adaptive evolution
The Ka/Ks ratio (ω) is used to assess the selective pressure on protein-coding genes. A ω > 1 indicates that this gene has undergone strong positive selection. Such genes, which have been rapidly evolving recently, are of great significance to the evolution of species. The Ka/Ks calculating results in this study suggested that the gene atpE might be undergoing strong positive selection in the evolution of Hydrocotyle. The atpE gene encodes ATP synthase CF1 epsilon subunit [75], which produces ATP from ADP in the presence of a proton gradient across the membrane. The ω > 1 or Ka > 0 / Ks = 0 for this gene existed between clade III species and other species, several high posterior probability positively selected sites also have been detected in the clade. These indicated that the atpE gene has experienced positive selection in clade III species (H. verticillate and H. leucocephala). Combined with the function of this gene and the life habits of the two species, we hypothesized that the atpE gene under positive selection in H. verticillata and H. leucocephala might contribute to highly adaptive to temperature changes.
There was also a sort of relaxed selection with 0.5 < ω < 1 according to several studies [76–79]. Seven genes had 0.5 < ω < 1 and were considered to be under relaxed selection. The seven genes comprised one cytochrome synthesis gene (ccsA), one RNA polymerase subunits gene (rpoA), one photosynthesis maturase gene (matK), one gene (psbH) associated with photosystem II, and three conserved open reading frames (ycf1a, ycf1b, and ycf2). The ccsA gene encodes a protein required for heme attachment to c-type cytochromes [80]. The previous study suggested this gene was related to the geographical location of species [81]. However, the study of species allopatric distribution was not involved in our study, and there was no evidence to support such a conclusion. There was no significant positive selection in this gene among species according to the result of positive selection analysis. The rpoA gene encodes the alpha subunit of RNA polymerase in the plastome [66, 82]. The previous studies suggested that RNA polymerase could facilitate species to respond to changing environmental conditions by keeping the essential metabolic process to survive and regulating the process of gene transcription and expression [83, 84]. The rpoA gene with 0.5 < ω mainly existed between Clade I and other taxa, and two positively selected sites were detected in this clade although the LRT P-value was not significant. Considering the possible rapid differentiation within the clade, we suggested that the rpoA gene might be related to the internal species differentiation of Clade I. The matK gene is usually encoded in the trnK tRNA gene intron, which probably assisted in splicing its own and other chloroplast introns. This gene was proven to act on the photosynthesis pathway [26]. Although the ω value of this gene did not exceed 1, we found that it has an LRT P-value below 0.05 in the branch-site testing (H. leucocephala as foreground), and multiple positively selected sites were detected. These results suggested that matK should be experiencing strong positive selection in the H. leucocephala. Another gene involved in the photosynthesis pathway is the psbH gene, which obtained two positively selected sites with posterior probability exceeding 0.90 in H. sibthorpioides. The codings of ycf1 and ycf2 were enigmatic and their functions remained unclear for a long time [85]. Recent studies have proved that these two can encode proteins and are closely related to photosynthesis [79, 86, 87]. There was significant positive selection in ycf1a (LRT P-value < 0.05), even if the posterior probability of the site was not high. Although the LRT P-values of ycf1b and ycf2 were greater than 0.05, the positive selection sites of these two were with high posterior probability (> 0.90). Thus, these genes with a 0.5 < ω were necessary for photosynthesis. Species of Hydrocotyle were mainly distributed in grassy places or wet valleys, such an environment usually resulted in insufficient light for plants. The relaxed selective genes (rpoA, matK, psbH, ycf1a, ycf1b, and ycf2) may function in the growth of Hydrocotyle species in adaptation to a poor light environment.
The results of positive selection analyses indicated that six CDS genes (ycf1b, matK, atpF, accD, rps14, and psbB) of Hydrocotyle species were under significant positive selection. Within the genus, the two genes (ycf1b, and matK) experienced positive selection again in different taxa. Gene accD, rps14, atpF, and psbB were conservative within the genus, which indicated that these genes may be related to the unique evolution of this genus in Apiales. The ycf2 gene in Hydrocotyle species or within the genus species always experienced weak positive selection (LRT P-value > 0.05, with positive selection sites). These genes are likely to be extremely important in the evolution of Hydrocotyle, and we will pay more attention to them as we expand our sample in the future to get more statistically significant results.
Phylogenetic relationships among Hydrocotyle species
In recent years, several molecular studies on Hydrocotyle have been carried out [14, 24, 25]. Many studies have suggested that Hydrocotyle was a monophyletic group, but the relationships among species within the genus remain unsolved [6, 11, 88]. Low supports always existed in some clades no matter what kind of DNA fragment was used. In this study, plastome data was used for phylogenetic analysis of Hydrocotyle. A giant concatenated protein-coding sequence matrix was obtained, which contained richer information loci than the previous studies. A phylogenetic tree with significantly improved supports was constructed despite some short branches within the genus. Taxa of Hydrocotyle provided in this study were used in phylogenetic studies for the first time, except for H. sibthorpioides, H. nepalensis, and H. verticillata. The genus was strongly supported to divide into three clades. The short branch length and relatively low supports (PP/BS < 1/100) might indicate rapid radiation speciation of species within Clade I. In this clade, the leaf shapes of H. nepalensis, H. hookeri subsp. chinensis and H. pseudoconferta are highly similar (Fig. 1A, C, D). These species are challenging to differentiate in the absence of flowers and fruits. The leaf of H. dielsiana is palmately 5–7-divided and can be easily distinguished from the other three species. Morphological characteristics of the fruit and flower of this species are similar to those of H. hookeri subsp. chinensis (Fig. 1A, B). Both H. nepalensis and H. pseudoconferta have extremely shortened peduncles. However, the umbels of H. nepalensis are several fascicled in axils and stem tips, which of H. pseudoconferta are usually solitary at the nodes (Fig. 1C, D). Species of Clade I include rich morphological diversity, differentiation in this clade still needs to be further studied by sampling. Clade II consists of H. sibthorpioides and the variety. Leaves of species in this clade are glabrous or distally pubescent, the leaves and inflorescence are small, and each umbel is 5-8-flowered (Fig. 1E, F). Clade III contains two South American species and is geographically separated from the other two clades (Fig. 1G, H). Perkins (2019) has reconstructed the phylogenetic relationships among the annual species of Hydrocotyle and concluded that the life histories (annual and perennial) and presence/absence of floral bracts could be used to classify the genus [14]. These traits were not available in our study, because all of the taxa we used were perennially bracteate taxa. The systematic study of this genus still needs more taxa and more abundant evidence, including but not limited to morphological and molecular evidence.
Conclusions
Our work revealed that (1) the Hydrocotyle plastomes had similar structures. (2) The ycf15 genes of Hydrocotyle plastomes had intact open reading frames (ORF) and showed conservation within the genus. In Hydrocotyle plastomes, the length of the gene was 204 bp, which was shorter than those of ycf15 genes in Araliaceae plastomes (303 bp, 309 bp, and 330 bp). Within Apiaceae plastomes, ycf15 was suggested to be pseudogenes with variable lengths, with the exception of D. hydrocotyloides. The characteristics of the ycf15 gene indicated that it could be used as a DNA barcode to identify Hydrocotyle. (3) Six genes (ycf1b, matK, atpF, accD, rps14, and psbB) have been identified as evolving under positive selection in the genus Hydrocotyle. Among these, four genes (atpF, accD, rps14, and psbB) were conservative within the genus. This indicates that these four might be related to the unique evolution of the genus in Apiales. (4) Seven genes (atpE, matK, psbH, ycf1a, ycf1b, rpoA, and ycf2) were suggested to be under some degree of positive selection in different taxa within the genus Hydrocotyle, possibly contributing to species’ adaptability to the environment. (5) A total of 14 regions (rps16-trnK, trnQ-rps16, atpI-atpH, trnC-petN-psbM, ycf3-trnS, accD-psaI-ycf4, petA-psbJ, rps12-rpl20, rpl16 intron, rps3-rpl16 intron, rps9-rpl22, ndhF-rpl32, ndhA intron, and ycf1a) were recognized as promising DNA barcodes for phylogeny analyses of Hydrocotyle. (6) A phylogenetic tree with strong support has been reconstructed using plastome sequences. Many remain to be investigated on the phylogenetic relationships of Hydrocotyle species, notably improving the sampling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to You-Pai Zeng for collecting the species Hydrocotyle leucocephalala.
Author contributions
Conceptualization, J.W. and C-F S.; validation, B-C W., H-M L., W.Z.; data curation, B-C W. and J.W.; writing—original draft preparation, J.W.; writing—review and editing, C-F S.; funding acquisition, J.W. and C-F S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant no. 32200191), the Foundation of Jiangsu Key Laboratory for the Research and Utilization of Plant Resources (JSPKLB202214).
Data availability
The six plastomes generated in this study are available in NCBI (https://www.ncbi.nlm.nih.gov) with accession numbers OR767307-OR767312; see Table 1). Voucher specimens were identified by Jun Wen and deposited in NAS (Herbarium, Institute of Botany, Chinese Academy of Sciences, Jiangsu Province) with deposition numbers (NAS00638767, NAS00638751, NAS00638791, NAS00638796, NAS00638784, NAS00638788; Figure S2), and the collection information was listed in Table S1. Raw reads of these plastomes were uploaded to NCBI placing under project PRJNA1035162 with accession numbers SRR26661267-SRR26661272.
Declarations
Ethics approval and consent to participate
The plant materials used in the study were collected under permission. The collection of plant materials and use comply with relevant institutional, national, and international guidelines and legislation. This article does not contain any studies with human participants or animals and does not involve any endangered or protected species.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet; http://www.plantsoftheworldonline.org/ Retrieved 01 July 2022.
- 2.Koch WDJ. Generum tribuumque plantarum Umbelliferarum nova dispositio. Nova Acta Academiae Caesareae Leopoldino Carol Germanicae Naturae Curiosorum. 1824;12:55–156. [Google Scholar]
- 3.Drude CGO. Umbelliferae. In: Engler A, Prantl K, editors. Die Natürlichen P anzenfamilien 3. Leipzig: W. Engelmann; 1898. pp. 63–250. [Google Scholar]
- 4.Pimenov MG, Leonov MV. The genera of the Umbelliferae: a Nomenclator. Kew: Royal Botanical Gardens; 1993. p. 156. [Google Scholar]
- 5.Sheh ML, Watson MF, Cannon JFM. Hydrocotyle Linnaeus. In: Wu ZY, Raven PH, Eds. Flora of China. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis., 2005;14:14–7.
- 6.Chandler GT, Plunkett GM. Evolution in Apiales: nuclear and chloroplast markers together in (almost) perfect harmony. Bot J Linn Soc. 2004;144:123–47. 10.1111/j.1095-8339.2003.00247.x [DOI] [Google Scholar]
- 7.Plunkett GM, Soltis DE, Soltis PS. Clarification of the relationship between Apiaceae and Araliaceae based on matK and rbcL sequence data. Am J Bot. 1997;84:565–80. 10.2307/2446032 [DOI] [PubMed] [Google Scholar]
- 8.Plunkett GM. Relationships of the order Apiales to subclass Asteridae: a re-evaluation of morphological characters based on insights from molecular data. Edinb J Bot. 2001;58:183–200. 10.1017/S0960428601000567 [DOI] [Google Scholar]
- 9.Plunkett GM, Lowry IIPP. Relationships among ‘ancient araliads’ and their significance for the systematics of Apiales. Mol Phylogenet Evol. 2001;19:259–76. 10.1006/mpev.2000.0920 [DOI] [PubMed] [Google Scholar]
- 10.Plunkett GM, Chandler GT, Lowry IIPP, et al. Recent advances in understanding Apiales and a revised classification. S Afr J Bot. 2004;70:371–81. 10.1016/S0254-6299(15)30220-9 [DOI] [Google Scholar]
- 11.Nicolas AN, Plunkett GM. The demise of subfamily Hydrocotyloideae (Apiaceae) and the re-alignment of its genera across the entire order Apiales. Mol Phylogenet Evol. 2009;53:134–51. 10.1016/j.ympev.2009.06.010 [DOI] [PubMed] [Google Scholar]
- 12.Plunkett GM, Wen J, Lowry IIPP, Mitchell AD, Henwood MJ, Fiaschi P. Araliaceae. In: Kadereit JW, Bittrich V, editors. The families and genera of vascular plants XV. Flowering plants eudicots. Apiales, Gentianales (except Rubiaceae). Springer; 2018. pp. 413–46.
- 13.Mendoza JM, Fuentes AF. Hydrocotyle apolobambensis (Apiaceae), una especie nueva andina del noroeste de Bolivia. Novon. 2010;20:303–6. 10.3417/2007087 [DOI] [Google Scholar]
- 14.Perkins AJ. Molecular phylogenetics and species delimitation in annual species of Hydrocotyle (Araliaceae) from South Western Australia. Mol Phylogenet Evol. 2019;134:129–41. 10.1016/j.ympev.2019.02.011 [DOI] [PubMed] [Google Scholar]
- 15.Konstantinova AI, Yembaturova EY. Structural traits of some species of Hydrocotyle (Araliaceae) and their significance for constructing the generic system. Plant Divers. 2010;128:329–46. 10.1127/1869-6155/2010/0128-0015 [DOI] [Google Scholar]
- 16.Li R, Li H. A new species of Hydrocotyle (Umbelliferae) from western Yunnan, China. J Syst Evol. 2013;51:223–39. 10.1111/jse.12008_1 [DOI] [Google Scholar]
- 17.Henwood MJ. Hydrocotyle Rivularis: a new trifoliolate species from south-eastern Australia. Telopea. 2014;17:217–21. 10.7751/telopea20147840 [DOI] [Google Scholar]
- 18.Perkins AJ, Dilly ML. Hydrocotyle serendipita (Araliaceae), a new species of fire ephemeral from south-western Australia. Telopea. 2017;20:269–75. 10.7751/telopea11767 [DOI] [Google Scholar]
- 19.Perkins P AJ. Hydrocotyle spinulifera and H. dimorphocarpa (Araliaceae), two new western Australian species with dimorphic mericarps. Nuytsia. 2018;29:57–65. 10.58828/nuy00868 [DOI] [Google Scholar]
- 20.Perkins AJ, Hydrocotyle asterocarpa H. decorata and H. perforata (Araliaceae), three new Western Australian species with spicate inflorescences. Nuytsia. 2018;29:205–15.
- 21.Nery EK, Matchin-Viera ME, Camacho O, Caddah MK, Fiaschi P. Delimiting a constellation: integrative taxonomy of a star-shaped Hydrocotyle species complex (Araliaceae) from the Brazilian Atlantic forest. Plant Syst Evol. 2020;306:57. 10.1007/s00606-020-01682-8 [DOI] [Google Scholar]
- 22.Perkins AJ. Hydrocotyle simulans (Araliaceae), a new perennial species from south-eastern Australia. Phytotaxa. 2020;437:066–72. 10.11646/phytotaxa.437.2.3 [DOI] [Google Scholar]
- 23.Van De Wiel CCM, Van Der Schoot J, Van Valkenburg JLCH, et al. DNA barcoding discriminates the noxious invasive plant species, floating pennywort (Hydrocotyle ranunculoides lf), from non-invasive relatives. Mol Ecol Resour. 2009;9:1086–91. 10.1111/j.1755-0998.2009.02547.x [DOI] [PubMed] [Google Scholar]
- 24.Choi KS, Park SJ. Molecular phylogenetic studies of Korean Hydrocotyle L. Korean J Plant Resour. 2012;25:490–7. 10.7732/kjpr.2012.25.4.490 [DOI] [Google Scholar]
- 25.Karuppusamy S, Ali MA, Rajasekaran KM, et al. A new species of Hydrocotyle L. (Araliaceae) from India. Bangl J Plant Taxon. 2014;21:167–73. 10.3329/bjpt.v21i2.21356 [DOI] [Google Scholar]
- 26.Wen J, Xie DF, Price M, Ren T, Deng YQ, Gui LJ, et al. Backbone phylogeny and evolution of Apioideae (Apiaceae): new insights from phylogenomic analyses of plastome data. Mol Phylogenet Evol. 2021;161:107183. 10.1016/j.ympev.2021.107183 [DOI] [PubMed] [Google Scholar]
- 27.Xie DF, Xie C, Ren T, Song BN, Zhou SD, He XJ. Plastid phylogenomic insights into relationships, divergence, and evolution of Apiales. Planta. 2022;256:117. 10.1007/s00425-022-04031-w [DOI] [PubMed] [Google Scholar]
- 28.Valcárcel V, Wen J. Chloroplast phylogenomic data support eocene amphi-pacific early radiation for the Asian palmate core Araliaceae. J Syst Evol. 2019;57:547–60. 10.1111/jse.12522 [DOI] [Google Scholar]
- 29.Ji YH, Liu CK, Yang ZY, Yang LF, He ZS, Wang HC, et al. Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Mol Ecol Resour. 2019;19:1333–45. 10.1111/1755-0998.13050 [DOI] [PubMed] [Google Scholar]
- 30.Peng C, Guo XL, Zhou SD, He XJ. Backbone phylogeny and adaptive evolution of Pleurospermum s. l.: new insights from phylogenomic analyses of complete plastome data. Front Plant Sci. 2023;14:1148303. 10.3389/fpls.2023.1148303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin HH, Cai J, Liu CK, Zhou RX, Price M, Zhou SD, et al. The plastid genome of twenty-two species from Ferula, Talassia, and Soranthus: comparative analysis, phylogenetic implications, and adaptive evolution. BMC Plant Biol. 2023;23:9. 10.1186/s12870-022-04027-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen VB, Linh Giang VN, Waminal NE, Park HS, Kim NH, Jang W, et al. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J Ginseng Res. 2020;44:135–44. 10.1016/j.jgr.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong Z, Zhang R, Shi M, Song Y, Xin Y, Li F, et al. The complete plastid genome of the endangered shrub Brassaiopsis Angustifolia (Araliaceae): comparative genetic and phylogenetic analysis. PLoS ONE. 2022;17:e0269819. 10.1371/journal.pone.0269819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Downie SR, Jansen RK. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to Plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst Bot. 2015;40:336–51. 10.1600/036364415X686620 [DOI] [Google Scholar]
- 35.Ge L, Shen LQ, Chen QY, Li XM, Zhang L. The complete chloroplast genome sequence of Hydrocotyle sibthorpioides (Apiales: araliaceae). Mitochondrial DNA B. 2017;2:29–30. 10.1080/23802359.2016.1241676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen J, Zhou W, Wu BC, Li HM, Song CF. The complete chloroplast genome of Hydrocotyle pseudoconferta Masamune 1932 (Araliaceae). Mitochondrial DNA B. 2022;7:1199–200. 10.1080/23802359.2022.2090292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews S, Lindenbaum P, Howard B, Ewels P. FastQC: a quality control tool for high throughput sequence data. Cambridge (UK): The Babraham Institute; 2011. [Google Scholar]
- 38.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: denovo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:575–81. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–9. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darling ACE, Mau B, Blattner FR, Perna NT, Mauve. Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amiryouse A, Hyvonen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–1. 10.1093/bioinformatics/bty220 [DOI] [PubMed] [Google Scholar]
- 44.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 45.Rédei GP. Nucleotide diversity. Encyclopedia of Genetics, Genomics, Proteomics and Informatics. Dordrecht: Springer; 2008. p. 1378. [Google Scholar]
- 46.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang ZH, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–17. 10.1093/oxfordjournals.molbev.a004148 [DOI] [PubMed] [Google Scholar]
- 48.Yang Z, Dos RM. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28:1217–28. 10.1093/molbev/msq303 [DOI] [PubMed] [Google Scholar]
- 49.Yang ZH, Wong WSW, Nielson R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–18. 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- 50.Gao FL, Chen CJ, Arab DA, Du ZG, He YH, Ho SYW. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol. 2019;9:3891–8. 10.1002/ece3.5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Ohna SH, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Sys Biol. 2012;61:539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rambaut A. 2012. FigTree v1.4. University of Edinburgh, Edinburgh, UK http://tree.bio.ed.ac.uk/software/gtree/
- 56.Goremykin VV, Hirsch-Ernst KI, Wölfl S, Hellwig FH. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol. 2003;20:1499–505. 10.1093/molbev/msg159 [DOI] [PubMed] [Google Scholar]
- 57.Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–90. 10.1093/molbev/msl089 [DOI] [PubMed] [Google Scholar]
- 58.Steane DA. Complete nucleotide sequence of the chloroplast genome from the tasmanian blue gum, Eucalyptus globulus (Myrtaceae). DNA Res. 2005;12:215–20. 10.1093/dnares/dsi006 [DOI] [PubMed] [Google Scholar]
- 59.Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007;8:174. 10.1186/1471-2164-8-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu RS, Li P, Qiu YX. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front Plant Sci. 2017;7:2054. 10.3389/fpls.2016.02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao NN, Zhao ZL, Ni LH, Prospect. Identification of medicinal plant based on plastid gene ycf15. Chin Tradit Herb Drugs. 2017;48:3201–17. [Google Scholar]
- 62.Nguyen PAT, Kim JS, Kim JH. The complete chloroplast genome of colchicine plants (Colchicum autumnale L. and Gloriosa superba L.) and its application for identifying the genus. Planta. 2015;242:223–37. 10.1007/s00425-015-2303-7 [DOI] [PubMed] [Google Scholar]
- 63.Shi C, Liu Y, Huang H, Xia EH, Zhang HB, Gao LZ. Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in angiosperms. PLoS ONE. 2013;8:e59620. 10.1371/journal.pone.0059620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai ZQ, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, et al. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 2006;6:77. 10.1186/1471-2148-6-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011;54:663–73. 10.1139/g11-026 [DOI] [PubMed] [Google Scholar]
- 66.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–9. 10.1002/j.1460-2075.1986.tb04464.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, et al. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002;31:171–88. 10.1046/j.1365-313X.2002.01349.x [DOI] [PubMed] [Google Scholar]
- 68.Zhou J, Niu JM, Wang XY, Yue JR, Zhou SL, Liu ZW. Plastome evolution in the genus Sium (Apiaceae, Oenantheae) inferred from phylogenomic and comparative analyses. BMC Plant Biol. 2023;23:368. 10.1186/s12870-023-04376-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu RS, Hu K, Zhang FJ, Sun XQ, Chen M, Zhang YM. Pan-plastome of Greater Yam (Dioscorea alata) in China: Intraspecific Genetic Variation, Comparative Genomics, and phylogenetic analyses. Int J Mol Sci. 2023;24:3341. 10.3390/ijms24043341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai J, Qin HH, Lei JQ, Liu CK, He XJ, Zhou SD. The phylogeny of Seseli (Apiaceae, Apioideae): insights from molecular and morphological data. BMC Plant Biol. 2022;22:534. 10.1186/s12870-022-03919-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lei JQ, Liu CK, Cai J, Price M, Zhou SD, He XJ. Evidence from Phylogenomics and morphology provide insights into the phylogeny, Plastome Evolution, and taxonomy of Kitagawia. Plants. 2022;11:3275. 10.3390/plants11233275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tian R, Aou X, Song B, Li Z, He X, Zhou S. Plastid phylogenomic analyses reveal a cryptic species of Ligusticopsis (Apiaceae, Angiosperms). Int J Mol Sci. 2023;24:7419. 10.3390/ijms24087419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams AV, Boykin LM, Howell KA, Nevill PG, Small I. The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PLoS ONE. 2015;10:e0125768. 10.1371/journal.pone.0125768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Yu J, Chen YK, Wang ZC. Complete chloroplast genome sequence of the endemic and endangered plant Dendropanax Oligodontus: genome structure, comparative and Phylogenetic Analysis. Genes. 2022;13:2028. 10.3390/genes13112028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shinozaki K, Deno H, Kato A, Sugiura M. Overlap and co-transcription of the genes for the beta and epsilon subunits of tobacco chloroplast ATPase. Gene. 1983;24:147–55. 10.1016/0378-1119(83)90074-4 [DOI] [PubMed] [Google Scholar]
- 76.Xie DF, Yu HX, Price M, Xie C, Deng YQ, Chen JP et al. Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the chloroplast complete genome. Front Plant Sci. 2019;10. [DOI] [PMC free article] [PubMed]
- 77.Xie DF, Yu Y, Wen J, Huang J, He XJ. Phylogeny and highland adaptation of Chinese species in Allium section Daghestanica (Amaryllidaceae) revealed by transcriptome sequencing. Mol Phylogenet Evol. 2020;146:106737. 10.1016/j.ympev.2020.106737 [DOI] [PubMed] [Google Scholar]
- 78.Ren T, Li ZX, Xie DF, Gui LJ, Peng C, Wen J, et al. Plastomes of eight Ligusticum species: characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 2020;20:519. 10.1186/s12870-020-02696-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu X, Xie DF, Zhou YY, Cheng RY, Zhang XY, Zhou SD, et al. Phylogeny and adaptive evolution of subgenus Rhizirideum (Amaryllidaceae, Allium) based on plastid genomes. BMC Plant Biol. 2023;23:70. 10.1186/s12870-022-03993-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie ZY, Merchant S. The plastid-encoded ccsA gene is required for Heme attachment to Chloroplast c-type Cytochromes*. J Biol Chem. 1996;271:4632–9. 10.1074/jbc.271.9.4632 [DOI] [PubMed] [Google Scholar]
- 81.Huang X, Coulibaly D, Tan W, Ni Z, Shi T, Li H, et al. The analysis of genetic structure and characteristics of the chloroplast genome in different Japanese apricot germplasm populations. BMC Plant Biol. 2022;22:354. 10.1186/s12870-022-03731-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sijben-Muller G, Hallick R, Alt J, Westhoff P, Herrmann RG. Nucleic Acids Res. 1986;14:1029–44. 10.1093/nar/14.2.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. 10.1146/annurev.micro.54.1.499 [DOI] [PubMed] [Google Scholar]
- 84.Xie DF, Tan JB, Yu Y, Gui LJ, Su DM, Zhou SD, et al. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann Bot. 2020;125:1039–55. 10.1093/aob/mcaa024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Drescher A, Ruf S, Calsa TJ, Carrer H, Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000;22:97–104. 10.1046/j.1365-313x.2000.00722.x [DOI] [PubMed] [Google Scholar]
- 86.Nakai M. YCF1: a green TIC: response to the De Vries et al commentary. Plant Cell. 2015;27:1834–8. 10.1105/tpc.15.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kikuchi S, Asakura Y, Imai M, Nakahira Y, Kotani Y, Hashiguchi Y, et al. A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell. 2018;30:2677–703. 10.1105/tpc.18.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nicolas AN, Plunkett GM. Diversification times and biogeographic patterns in Apiales. Bot Rev. 2014;80:30–58. 10.1007/s12229-014-9132-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The six plastomes generated in this study are available in NCBI (https://www.ncbi.nlm.nih.gov) with accession numbers OR767307-OR767312; see Table 1). Voucher specimens were identified by Jun Wen and deposited in NAS (Herbarium, Institute of Botany, Chinese Academy of Sciences, Jiangsu Province) with deposition numbers (NAS00638767, NAS00638751, NAS00638791, NAS00638796, NAS00638784, NAS00638788; Figure S2), and the collection information was listed in Table S1. Raw reads of these plastomes were uploaded to NCBI placing under project PRJNA1035162 with accession numbers SRR26661267-SRR26661272.