Abstract
Background
Accumulating studies have highlighted the significant role of circulating metabolomics in the etiology of reproductive system disorders. However, the causal effects between genetically determined metabolites (GDMs) and reproductive diseases, including primary ovarian insufficiency (POI), polycystic ovary syndrome (PCOS), and abnormal spermatozoa (AS), still await thorough clarification.
Methods
With the currently most comprehensive genome-wide association studies (GWAS) data of metabolomics, systematic two-sample Mendelian randomization (MR) analyses were conducted to disclose causal associations between 1,091 blood metabolites and 309 metabolite ratios with reproductive disorders. The inverse-variance weighted (IVW) method served as the primary analysis approach, and multiple effective MR methods were employed as complementary analyses including MR-Egger, weighted median, constrained maximum likelihood (cML-MA), contamination mixture method, robust adjusted profile score (MR-RAPS), and debiased inverse-variance weighted method. Heterogeneity and pleiotropy were assessed via MR-Egger intercept and Cochran’s Q statistical analysis. Outliers were detected by Radial MR and MR-PRESSO methods. External replication and metabolic pathway analysis were also conducted.
Results
Potential causal associations of 63 GDMs with POI were unearthed, and five metabolites with strong causal links to POI were emphasized. Two metabolic pathways related to the pathogenesis of POI were pinpointed. Suggestive causal effects of 70 GDMs on PCOS were detected, among which 7 metabolites stood out for strong causality with elevated PCOS risk. Four metabolic pathways associated with PCOS mechanisms were recognized. For AS, 64 GDMs as potential predictive biomarkers were identified, particularly highlighting two metabolites for their strong causal connections with AS. Three pathways underneath the AS mechanism were identified. Multiple assessments were conducted to further confirm the reliability and robustness of our causal inferences.
Conclusion
By extensively assessing the causal implications of circulating GDMs on reproductive system disorders, our study underscores the intricate and pivotal role of metabolomics in reproductive ill-health, laying a theoretical foundation for clinical strategies from metabolic insights.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13048-024-01486-1.
Keywords: Genetically determined metabolites, Mendelian randomization, Primary ovarian insufficiency, Polycystic ovary syndrome, Abnormal spermatozoa
Introduction
The global incidence of infertility is 10% -15%, and reproductive disorders have risen to the forefront of health concerns worldwide [1, 2]. Currently, a declining trend in fertility rates is observed all over the world, with infertility not only profoundly affecting patients and their families but also imposing significant economic burdens on society [3]. Etiologically, infertility can arise from multiple factors, such as ovulatory disorders in women and sperm abnormalities in men. Primary ovarian insufficiency (POI) is a condition with medical, psychological, and reproductive impacts, occurring in at least 1% of women and leading to lifelong health issues and psychological stress [4]. As a critical cause of ovarian hormone deficiency and infertility, POI also links to an increased risk of cardiovascular diseases, osteoporosis, and a degree of cognitive decline [5]. Moreover, an association between POI and higher early mortality has been reported [6]. Disappointingly, due to the highly heterogeneous and multifactorial nature of POI, the biological mechanisms behind 90% of cases remain to be further elucidated [7]. Serving as the most common endocrine disorder affecting women of reproductive age (up to 6%-12%), polycystic ovary syndrome (PCOS) spans from adolescence through to post-menopause [8, 9]. Patients are not only challenged by the quintessential clinical features, including hyperandrogenism, ovulatory dysfunction, and the presence of polycystic ovaries, but also frequently suffer from various metabolic dysfunctions, such as insulin resistance and Type 2 diabetes, exerting substantial physical and psychological stress [10–12]. The etiology of PCOS is intricately complex and enigmatic, and its underlying biological mechanisms remain a focal point of research. Apart from that, male factors account for approximately 40% of infertility cases [1, 2]. Sperm morphology and motility, the ability to penetrate cervical mucus, as well as the capacity to enter the zona pellucida of the oocyte, are crucial for the successful fertilization of the sperm and oocyte. The impact of abnormal spermatozoa (AS) on delaying natural conception and elevating miscarriage risks underscores the urgent need for in-depth research in this area [13].
Metabolites, as the ultimate outputs of upstream genes and proteins, function as indicators of an individual's real-time physiological state and disease risk, thereby becoming a focal point for therapeutic strategies and management [14, 15]. Metabolomics leverages metabolites derived from human studies to map the connections between diseases and their metabolic pathways [16]. Its heightened sensitivity enables the detection of subtle biological changes, offering insights into the mechanisms underlying various physiological states, anomalies, and diseases [17]. Through genome-wide association studies (GWAS) extending to metabolic phenotypes, genetically determined metabolites (GDMs) forge a critical link between genetic variations and environmental triggers of diseases [18–21]. Metabolomics is increasingly harnessed to uncover the fundamental causes of complex diseases, such as cancer [22] and diabetes [23], significantly propelling the advancement of precision medicine. A plethora of studies have demonstrated the intricate link between reproductive function and metabolic health [24, 25]. Dysfunctions in the reproductive system are associated with deterioration in metabolic profiles, highlighting the intertwined nature of metabolism and reproduction [26]. Multiple metabolites including branched-chain amino acids [27], arachidonic [28], creatine metabolism [29], and vitamin D [30] metabolism, among others, have been proven to have an undeniable close association with the functionality of female ovaries and male sperm. Nonetheless, the limited scope and small sample sizes of these studies yield restricted insights into pathophysiological mechanisms. It is imperative to embark on novel research that encompasses extensive screenings of a wide spectrum of circulating metabolites, which are crucial for deciphering causality between GDMs and associated genetic variations underlying reproductive disorders.
Mendelian Randomization (MR) employs genetic variants, particularly single nucleotide polymorphisms (SNPs), as instrumental variables (IVs) to proxy exposure variables, facilitating the investigation of causal effects between exposures and distinct health outcomes [31]. Unlike traditional observational studies, which are susceptible to biases from small sample sizes, reverse causation, and the influence of potential environmental and societal confounders, MR offers a robust approach to unbiased detection of causal effects [32]. In scenarios where randomized controlled trials are impractical, causality is ambiguous, or confounding factors and reverse causality exists, MR presents a viable alternative for causal inference [33]. Over the past decade, the application of MR to publicly available GWAS data has seen a surge in popularity with significant results yielded [34, 35]. Herein, we leveraged the largest metabolite-associated GWAS data to date (encompassing 1,091 metabolites and 309 metabolite ratios) and conducted two-sample MR analyses to address research gaps in causal links between GDMs and reproductive disorders (comprising POI, PCOS, and AS) from a genetic variation perspective.
Methods
Study design
Exhaustive two-sample MR analyses were conducted to investigate the causal links between 1,400 GDMs and a spectrum of reproductive system diseases. Various robust MR analytical techniques were employed in our study to secure reliable causal inferences [36–39]. Sensitivity analyses were performed to ensure the robustness of our findings. To enhance the validation of the candidate GDMs and affirm the robustness of our findings, we leveraged GWAS data from another metabolomics study, encompassing 486 genetically influenced metabolites, for external replication in this research [19]. Moreover, enrichment analysis of metabolic pathways was performed to delve deeper into the mechanisms underlying reproductive system disorders. Figure 1A illustrates the three core principles essential for the effective execution of MR: (1) genetic variants strongly correlate with the exposure; (2) genetic variants are not associated with any potential confounders; (3) genetic instruments impact the outcome solely through effects on the exposure [40]. The workflow of our study design is depicted in Fig. 1B.
Fig. 1.
Study design overview. A Mendelian randomization (MR) analyses depend on three core assumptions. Assumption 1, genetic instruments are strongly associated with the exposures of interest; Assumption 2, genetic instruments are independent of confounding factors; Assumption 3, genetic instruments are not associated with outcome and affect outcome only via exposures. B Outline of the study design
GWAS data sources for GDMs and reproductive system disorders
Several GWAS studies on GDMs have been conducted to date, providing insights into the genetic influence on human metabolism [19, 41–44]. Previously, Shin et al. [19] identified 486 genetically influenced metabolites from a GWAS study on untargeted metabolomics with 7,824 participants, with related data retrievable from the IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/). Recently, the scope of metabolome-wide GWAS data has been significantly expanded to a more comprehensive range. The latest study by Chen et al. explored 1,091 blood metabolites and ratios of 309 metabolites, which is accessible through the GWAS Catalog database (https://www.ebi.ac.uk/gwas/) with catalog numbers listed as GCST90199621-GCS90201020 [44]. In our research, we utilized GWAS data from Chen et al. as the primary basis for exploratory analyses, while GWAS data from Shin et al. served as supplementary validation to enhance the generalizability and robustness of our conclusions.
Statistics on POI and PCOS were derived from the FinnGen consortium R9 release data (https://r9.finngen.fi/). For POI, the corresponding phenotype code utilized in this study was “E4_OVARFAIL”, with 469 cases and 200,581 controls. For PCOS, the designated phenotype identifier was “R9_E4_PCOS”, encompassing 1,424 cases and 200,581 controls. Additionally, summary statistics for AS were sourced from the FinnGen consortium R7 release data (https://r7.finngen.fi/). GWAS data for the “abnormal spermatozoa” phenotype were accessed, comprising 1,913 cases and 293,878 controls in total. Since the GWAS data on these reproductive disorders were publicly accessible as summary information, no further ethical approvals were necessary for their use.
Selection of instrumental variables
To ensure the precision and efficacy of MR analyses, a stringent selection process for IVs was applied to 1,400 metabolites, adhering to the three foundational assumptions of MR mentioned above. Genetic variants linked to metabolic traits were pinpointed using a genome-wide significance threshold of P < 1 × 10–5, ensuring IVs captured the predominant variance in metabolites. SNP pruning was executed using the clumping function in PLINK software (version v1.90). Independent SNPs (r2 < 0.1 within a 500 kb window) were chosen to generate IVs utilizing a clumping process with the European population from the 1,000 Genomes Project as the reference [45]. Furthermore, specific palindromic SNPs were excluded to avoid biased results due to IVs being selected improperly. To mitigate potential biases from weak instruments, the F-statistic (β2_exposure/SE2_exposure) was calculated to assess the strength of IVs, with an F-statistic above 10 indicating sufficient power for reliable MR analysis [46, 47]. These criteria were aligned with prior research recommendations and ensured the reliability and accuracy of the IVs used in this study [48].
Two-sample Mendelian randomization
In this study, the inverse-variance weighted (IVW) method served as the primary strategy for MR analysis. The IVW method is typically considered the gold standard for assessing causality, offering the most reliable test for the presence of causal effects [49]. Moreover, the flourishing development of MR methodologies in recent years has introduced a suite of innovative and effective analytical techniques into study strategy. To complement the IVW approach, various robust methods were employed in our analysis including MR-Egger, weighted median, constrained maximum likelihood (cML-MA), contamination mixture method, robust adjusted profile score (MR-RAPS), and debiased inverse-variance weighted method. Specifically, the MR-Egger method provides a more stable causal estimate even in the presence of invalid IVs by estimating the causal effect through the Egger regression's slope coefficient [50]. The weighted median approach offers protection against up to 50% of invalid IVs [51]. The cML-MA method, a constrained maximum likelihood and model averaging-based MR approach, is notably more powerful than MR-Egger in controlling both correlated and uncorrelated pleiotropic effects [52]. The contamination mixture method produces robust MR results even with invalid IVs and has shown the lowest mean squared error in various realistic scenarios [37]. MR-RAPS accounts for both systematic and idiosyncratic pleiotropy, providing robust inference for MR analysis with many weak instruments [38]. The Debiased Inverse-Variance Weighted Method eliminates bias from weak instruments, thus offering enhanced robustness [39].
In this study, the P-value from the IVW analysis was utilized as the primary metric for assessing the causal relationship, and six complementary methods were employed to further augment the assessment of MR findings. Consistent estimates (β values) across all methods indicated robust findings. Upon achieving uniform estimates across seven MR approaches, a metabolite was identified as a potential candidate with a possible causal link to reproductive system diseases when the IVW method indicated a significant difference (P < 0.05). When IVW and the six auxiliary MR tests all demonstrated a significant difference (P < 0.05), the candidate metabolite was deemed to exert a definitive strong causal relationship with reproductive disorders.
Sensitivity analysis
Diverse methodologies were utilized to assess the heterogeneity and pleiotropy of our findings. Cochran’s Q statistical analysis was applied to detect heterogeneity for IVW and MR-Egger method, with a Cochran Q test-derived P-value greater than 0.05 indicating no significant heterogeneity [50]. The presence of pleiotropy and potential biases due to ineffective IVs were evaluated by the MR-PRESSO global test and MR-Egger intercept, with a P-value greater than 0.05 suggesting the absence of pleiotropy [53]. Of note, Radial MR and the MR-PRESSO were performed to identify outliers, followed by a repetition of MR analysis after removing heterogeneous SNPs [54]. Furthermore, the MR-Steiger directionality test was employed to verify the accuracy of deduced causal direction, thereby preventing reverse causation.
External replication analysis
To verify whether consistent causal associations with diseases could be identified for the same metabolites based on GWAS data from different sources, we conducted replication analysis. For key metabolites identified in the primary analysis, we matched and performed external replication using publicly available data from Shin et al. which involved 486 serum metabolites [19]. For the causal association analysis of metabolites with the corresponding disease in the validation GWAS data, we also employed the IVW method and applied the same threshold criteria as used in the primary analysis.
Metabolic pathway enrichment analysis
To elucidate the biological mechanisms behind circulating GDMs influencing reproductive system diseases, we performed enrichment analysis of metabolic pathways associated with these candidate metabolites via MetaboAnalyst 6.0 (https://www.metaboanalyst.ca/). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was selected as the pathway library for this analysis to identify key metabolic pathways. The enrichment method utilized was the hypergeometric test, and the threshold for significance in metabolic pathway enrichment analysis was set at 0.05.
Statistical analysis
All MR analyses were conducted using R software (version 4.2.0). A variety of R packages including MendelianRandomization, MR-PRESSO, and Radial MR were employed in this study. Collectively, these methods established a systematic framework to comprehensively assess the causal relationships between circulating GDMs and reproductive system diseases. If the estimated causal effect P value for GDMs via the IVW method is less than 0.05, GDMs are defined as potential suggestive risk predictors for the disease. A strong causal relationship between the metabolite and the disease was confirmed when P values from the IVW method and six additional MR methods were all below 0.05. In all sensitivity analysis tests, a two-tailed P < 0.05 is considered statistically significant.
Results
Exploration of the causal effects of serum GDMs on POI
Preliminary IVW analysis identified 63 circulating metabolites causally linked to POI, concerning multiple metabolite categories including androgenic steroids, chemicals, creatine metabolism, fatty acid metabolism, amino acid metabolism, and nucleotide metabolism, etc. (Table S1). Of these, 34 serum metabolites were associated with an increased risk of POI, while 29 GDMs were deduced to potentially reduce the risk. To eliminate the influence of confounding factors, 63 causal effects were further evaluated using multiple complementary algorithms such as MR-Egger, weighted median, cML-MA, contamination mixture method, RAPS, and debiased inverse-variance weighted method (Table S2). All 63 metabolites demonstrated consistent causal directions across these supplementary algorithms. Heterogeneity was detected in only 4 of the 63 metabolites by Cochran's Q test with IVW random effect models applied accordingly, while the other GDMs displayed no heterogeneity among IVs (Table S3).
Based on horizontal pleiotropy analyses using MR-Egger intercept and MR-PRESSO global test, except for the 3beta-hydroxy-5-cholestenoate levels, P-values for the remaining 62 metabolites were all above 0.05, indicating a low risk of horizontal pleiotropy (Table S4). For 3beta-hydroxy-5-cholestenoate levels, five outlier variants were detected using Radial MR and MR-PRESSO methods (Fig. 2, Table S5). After removing these outliers, the protective causality of 3beta-hydroxy-5-cholestenoate levels on POI remained significant (OR: 0.578; 95% CI: 0.443–0.754; PIVW = 5.51E-5), and no pleiotropy or heterogeneity was present (Table S6). Moreover, MR Steiger directionality tests confirmed the accuracy of our causal direction inference for all 63 GDMs (Table S7).
Fig. 2.
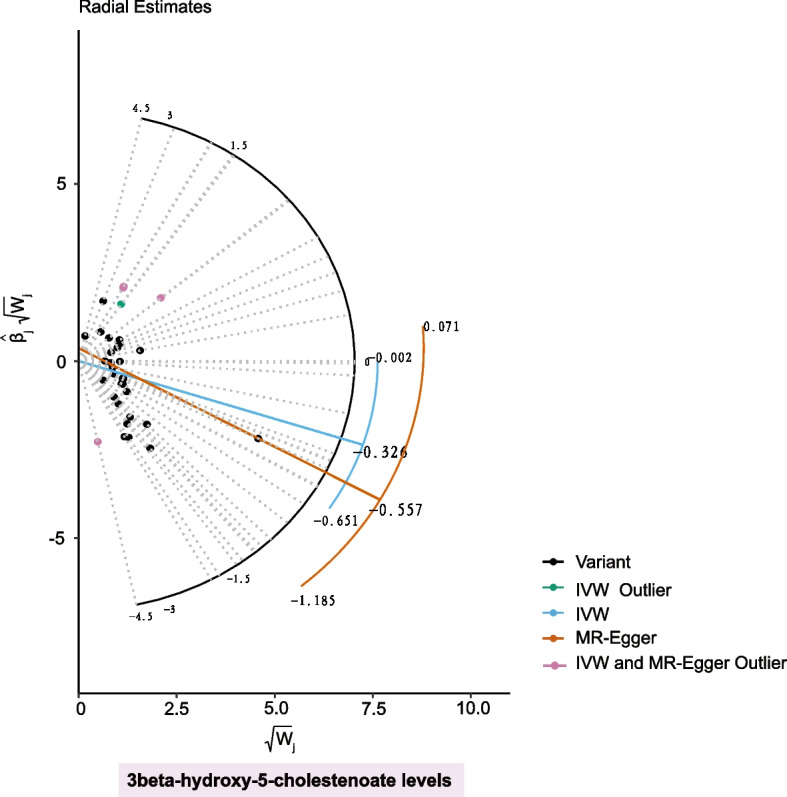
Radial plot demonstrating the identification of outlier variants for 3beta-hydroxy-5-cholestenoate levelsusing MR Radial method
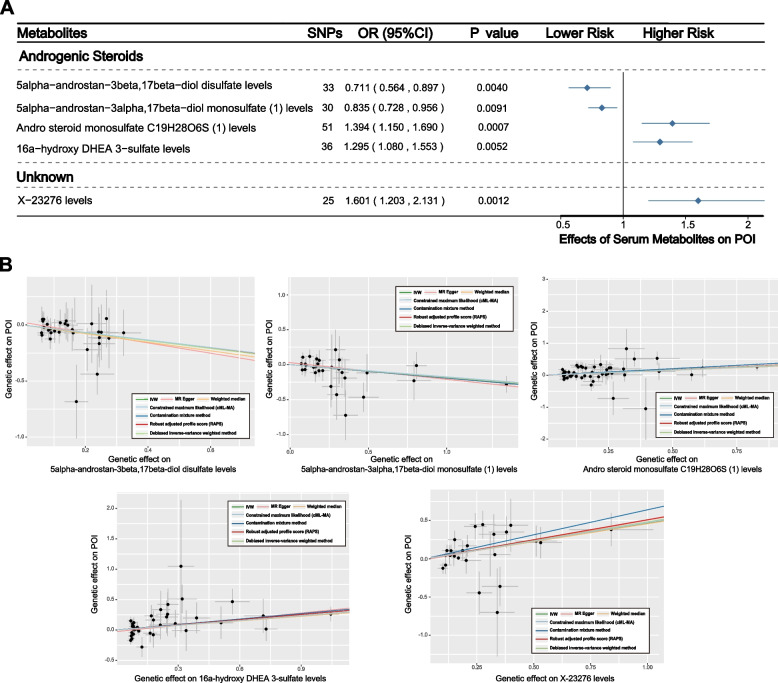
Notably, when considering significance across all seven MR methods, five serum metabolites stood out, including four androgenic steroids-related metabolites and one unknown metabolite (Fig. 3A). Of these, two metabolites were associated with increased risk of POI (Fig. 3B), specifically, andro steroid monosulfate C19H28O6S (1) levels (OR: 1.394; 95% CI: 1.150–1.690; PIVW = 0.001) and 16a-hydroxy DHEA 3-sulfate levels (OR: 1.295; 95% CI: 1.080–1.553; PIVW = 0.005), and X-23276 levels (OR: 1.601; 95% CI: 1.203–2.131; PIVW = 0.001). Additionally, two metabolites were revealed to be linked to a reduced risk of POI, namely 5alpha-androstan-3beta,17beta-diol disulfate levels (OR: 0.711; 95% CI: 0.564–0.897; PIVW = 0.004) and 5alpha-androstan-3alpha,17beta-diol monosulfate (1) levels (OR: 0.578; 95% CI: 0.443–0.754; PIVW = 0.009).
Fig. 3.
Causal associations between serum metabolites and POI: Results of MR analyses using multiple algorithms. A Forest plots showed the causal associations between serum metabolites and POI. The displayed P-values correspond to the IVW method. B Scatter plots of potential effects of single-nucleotide polymorphisms (SNPs) on five metabolites vs. POI, with the slope of each line corresponding to the estimated MR effect per method
Validation analyses of candidate metabolites were conducted to further corroborate the reliability of our findings. For several GDMs identified as potential risk predictors, we successfully matched a portion of the same metabolites in additional GWAS data, and the causal trends consistent with the primary analysis were successfully validated, as anticipated (Figure S1A-1D). Although the causal effects between some GDMs and diseases were not significant, these discrepancies might be attributed to significant variations in sample sizes across different GWAS data sources.
Moreover, based on known metabolites with suggestive causal links to POI, we identified two metabolic pathways potentially involved in POI pathogenesis (Table S7). Valine, leucine, and isoleucine biosynthesis (P = 0.025), and steroid hormone biosynthesis (P = 0.027) were detected as potential biological pathways involved in the development of POI. Further exploration of these pathways might provide clearer insights into the pathogenic mechanisms underlying POI.
Investigation of causal relationships between circulating metabolites and PCOS
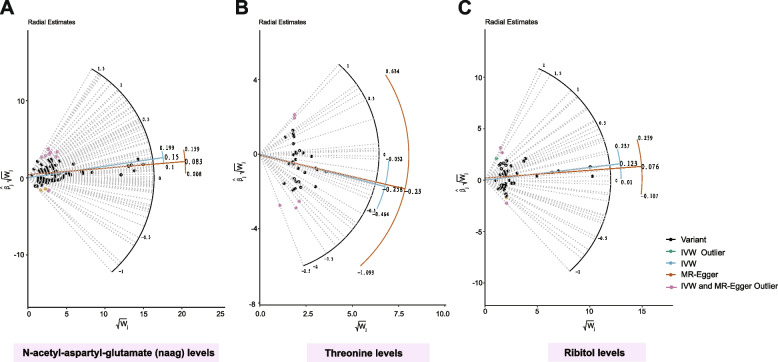
Initial IVW analysis unearthed 70 metabolites with potential causal links to PCOS, spanning categories including androgenic steroids, amino acid metabolism, partially characterized molecules, cardiovascular drugs, etc. (Table S9). Among these, 27 serum metabolites were identified as potential protective biomarkers for PCOS, while 43 GDMs were connected with an increased risk of PCOS development. Further assessment of these 70 causal associations employed a variety of robust methods, such as MR-Egger, weighted median, cML-MA, contamination mixture method, MR-RAPS, and debiased inverse-variance weighted method, each confirming causality estimates consistent with the initial IVW approach (Table S10). Significant heterogeneity was noted in analyses for N-acetyl-aspartyl-glutamate (NAAG) levels, threonine levels, and ribitol levels, prompting the application of the random-effect IVW model. In contrast, no heterogeneity was detected in analyses for the majority of GDMs (Table S11). Horizontal pleiotropy was also identified in analyses for N-acetyl-aspartyl-glutamate (NAAG) levels, threonine levels, and ribitol levels through MR-PRESSO global tests (Table S12). Radial MR and MR-PRESSO methods detected related outlier variants for N-acetyl-aspartyl-glutamate (NAAG) levels (Fig. 4A, Table S13), threonine levels (Fig. 4B, Table S15), and ribitol levels (Fig. 4C, Table S17). After the removal of these outliers, causal links between these three GDMs and PCOS were still significant, with no further detection of pleiotropy or heterogeneity (Tables S14, S16, S18). Additionally, MR Steiger directionality tests affirmed the precision of the causal effects' directionality for candidate metabolites (Table S19).
Fig. 4.
Radial plot demonstrating the identification of outlier variants for N-acetyl-aspartyl-glutamate (naag) levels, threonine levels, and ribitol levels using the MR Radial method
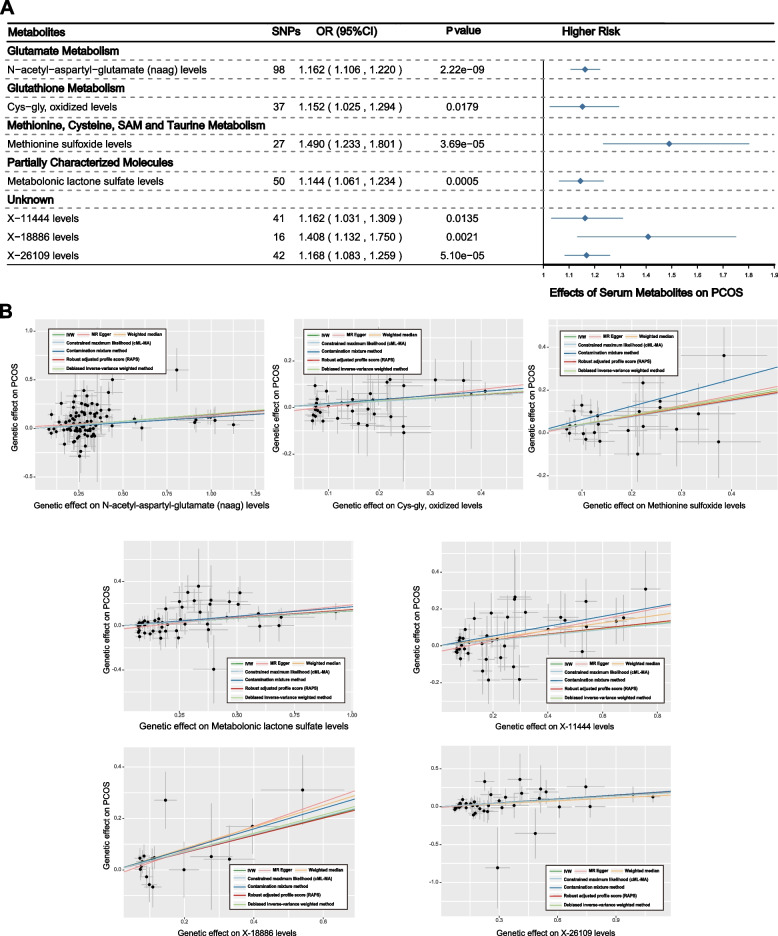
Upon considering the significance across all seven MR methods, seven serum metabolites were highlighted. Among them, three were unknown, and four known metabolites were involved in classes including glutamate metabolism, glutathione metabolism, methionine, cysteine, SAM, taurine metabolism, etc. (Fig. 5A). All these metabolites were associated with an increased risk of PCOS (Fig. 5B), namely, N-acetyl-aspartyl-glutamate (NAAG) levels (OR: 1.162; 95% CI: 1.106–1.220; PIVW = 2.22E − 09), cys-gly, oxidized levels (OR: 1.152; 95% CI: 1.025–1.294; PIVW = 0.0179), methionine sulfoxide levels (OR: 1.490; 95% CI: 1.233–1.801; PIVW = 3.69E − 05), metabolonic lactone sulfate levels (OR: 1.144; 95% CI: 1.061–1.234; PIVW = 0.0005), X-11444 levels (OR: 1.162; 95% CI: 1.031–1.309; PIVW = 0.0135), X-18886 levels (OR: 1.408; 95% CI: 1.132 -1.750; PIVW = 0.0021), and X-26109 levels (OR: 1.168; 95% CI: 1.083–1.259; PIVW = 5.10E − 05).
Fig. 5.
Causal associations between serum metabolites and PCOS: Results of MR analyses using multiple algorithms. A Forest plots showed the causal associations between serum metabolites and PCOS. The displayed P-values correspond to the IVW method. B Scatter plots of potential effects of single-nucleotide polymorphisms (SNPs) on five metabolites vs. PCOS, with the slope of each line corresponding to the estimated MR effect per method
Employing GWAS data from Shin et al. [19], replication analysis further substantiated the dependability of our results (Figures S1E-1G). Encouragingly, the protective causal associations of androsterone sulfate levels (Figure S1F, PIVW = 0.002) and 5alpha-androstan-3beta,17beta-diol disulfate levels (Figure S1G, PIVW = 0.005) with PCOS were robustly validated in the validation data.
Additionally, based upon known candidate metabolites, four latent biological pathways underneath the pathogenesis of PCOS were identified, including glutathione metabolism (P = 0.008), glycine, serine, and threonine metabolism (P = 0.011), pyrimidine metabolism (P = 0.0152), and valine, leucine and isoleucine biosynthesis (P = 0.040) (Table S20). Future in-depth research into these pathways is warranted to provide novel insights into the pathogenic mechanisms of PCOS.
Examination of causal relationships between serum metabolites and AS
Through IVW analysis, 64 serum metabolites were identified as exerting potential causal effects with AS, covering a variety of metabolic pathways including carotenoid, fatty acid, and bile acid metabolism (Table S21). Among these, 26 metabolites were noted for their potential in reducing the risk of AS, whereas 38 were implicated in increasing susceptibility to AS. Causal links between these candidate metabolites and AS were further evaluated using a series of sophisticated statistical models such as MR-Egger, weighted median, cML-MA, contamination mixture method, MR-RAPS, and debiased inverse-variance weighted method, which all affirmed the initial causality findings (Table S22). No heterogeneity was detected in most analyses, except in those concerning citrulline levels and 2'-o-methyluridine levels (Table S23), and there is no evidence of substantial pleiotropy affecting the results (Table S24). The accuracy of our causal direction inference was confirmed by MR Steiger directionality tests (Table S25).
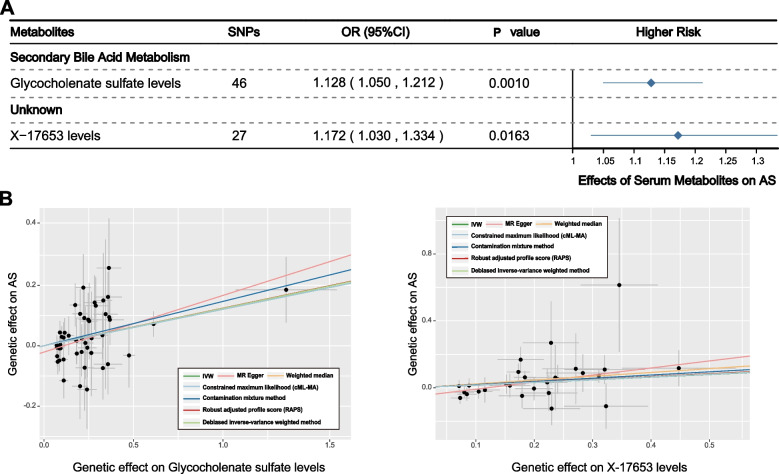
Founded on the IVW results, when the significance of six additional MR methods was considered, two metabolites emerged as particularly noteworthy (Fig. 6A). One GDM remained unknown, and the other was associated with secondary bile acid metabolism. Both glycocholenate sulfate levels (OR: 1.128; 95% CI: 1.050–1.212; PIVW = 0.001) and X-17653 levels (OR: 1.172; 95% CI: 1.030–1.334; PIVW = 0.0163) were identified as being linked to an increased risk of AS, indicating their potential as predictive biomarkers for the disease (Fig. 6B).
Fig. 6.
Causal associations between serum metabolites and AS: Results of MR analyses using multiple algorithms. A Forest plots showed the causal associations between serum metabolites and PCOS. The displayed P-values correspond to the IVW method. B Scatter plots of potential effects of single-nucleotide polymorphisms (SNPs) on five metabolites vs. PCOS, with the slope of each line corresponding to the estimated MR effect per method
To further strengthen our conclusions, replication analyses were conducted, confirming a similar causal association trend in alignment with the primary analysis between citrulline levels and AS (Figure S1H). In addition, insightful investigations based on known metabolites causally linked to AS identified three critical metabolic pathways potentially driving the onset of AS, involving sphingolipid metabolism (P = 0.0057), vitamin B6 metabolism (P = 0.0339), and caffeine metabolism (P = 0.0376) (Table S26). A deeper exploration of these pathways may enhance the clinical management and treatment strategies for AS.
Discussion
Based on the most extensive metabolomics GWAS data to date, our study employed systematic two-sample MR analyses to unveil the causal effects between 1,091 blood metabolites and 309 metabolite ratios with a range of reproductive disorders, including POI, PCOS, and AS. By deeply integrating genomics with metabolomics and utilizing a variety of robust MR analytical algorithms, we thoroughly explored the complex involvement of GDMs in the etiology of reproductive diseases. Latent causal relationships between 63 GDMs and POI were disclosed, and five metabolites with strong causal associations with POI were highlighted. Moreover, two key metabolic pathways implicated in the pathogenesis of POI were identified. Suggestive causal effects of 70 GDMs on PCOS were detected, with seven metabolites notably linked to an increased risk of PCOS, and four underlying pathogenic pathways were delineated. For AS, we identified 64 GDMs as potential biomarkers risk predictive, particularly pinpointing two metabolites with strong causal links to AS. Three critical pathways involved in AS's etiology were also uncovered. Rigorous sensitivity analyses were conducted to control for confounding factors and enhance the reliability of findings. In a systematic manner, our study broadened the scope of potential biomarkers and therapeutic targets for reproductive system diseases, enabling researchers and clinicians to comprehensively discern the metabolic landscape behind reproductive disorders.
In our study, the complex causal relationships between androgenic steroids and POI were particularly emphasized. Previous research has indicated that POI was associated with a significant reduction in testosterone and androgen precursor concentrations in women [55]. Some scholars suggested that androgens might exert a positive role in promoting preantral follicular growth and preventing follicular atresia, as well as in the activation of primordial follicles, although this viewpoint lacked substantial empirical support [56–58]. Our study pinpointed that certain androgenic steroid, such as 5alpha-androstan-3beta,17beta-diol disulfate and 5alpha-androstan-3alpha,17beta-diol monosulfate, exhibited latent protective effects against POI, supporting the beneficial role of androgens in restoring diminished ovarian reserve. However, it is noteworthy that our research also disclosed that steroids like andro steroid monosulfate C19H28O6S levels and 16a-hydroxy DHEA 3-sulfate levels might also increase the risk of POI, suggesting the potential adverse effects of androgen levels on POI. The bidirectional action of androgens uncovered in our study suggested their complex roles in ovarian health. Indeed, even though some clinical success has been observed in using dehydroepiandrosterone (DHEA) supplements as an androgen precursor to enhance fertility in women with low ovarian reserve in recent years [59], significant debate still persists regarding the overall effectiveness and mechanisms underlying androgen therapy [56]. A consensus has yet to be achieved, and robust scientific evidence and extensive, rigorous randomized trials supporting the beneficial impacts of androgens on ovarian physiology are still lacking [56]. Additionally, pathway enrichment analysis in our study also revealed the involvement of steroid hormone biosynthesis in the development of POI, further underscoring the importance of exploring the impact of steroids on this condition. Future studies are imperative to delve deeper into related mechanisms to better inform clinical strategies for POI, and thus provide more precise therapeutic options for patients.
Some GDMs with strong causal associations in promoting the progression of PCOS were focused on in our research. N − acetyl − aspartyl − glutamate (NAGG), a dipeptide comprised of glutamate and N-acetylaspartate linked via a peptide bond, functions as a neurotransmitter through the activation of the presynaptic metabotropic receptor 3 (GRM3), which is predominantly found in the brain [60]. Nevertheless, untargeted metabolomic analyses also detected the presence of NAAG in cumulus cells (CCs) and indicated its potential role in signaling [61]. As a supplement to previous research, our research further identified the potential promotive effects of circulating NAAG on the progression of PCOS. Furthermore, CCs in PCOS patients were documented to be subjected to oxidative stress. A reduction in glutathione within CCs was observed and was postulated to correlate with the quality of oocytes in women suffering from PCOS [62]. Glutathione is broken down into glutamate and Cys-Gly by the action of gamma-glutamyl transferase [63]. Our study's findings indicated that the buildup of oxidized Cys-Gly might contribute to the exacerbation of PCOS, thus corroborating similar results reported in existing literature. The pivotal role of glutathione metabolism was also unearthed in our analysis of biological pathways related to PCOS, potentially shedding light on therapeutic strategies for PCOS. Methionine sulfoxide has been established as a latent critical factor at the checkpoint of oogenesis progression [64], and our research further suggested its strong causal association with the advancement of PCOS. Metabolonic lactone sulfate was proved to be linked to several indicators of cardiometabolic health, such as obesity and insulin resistance [65], which are frequently implicated in PCOS [66]. Our study built on these observations, suggesting that higher levels of metabolonic lactone sulfate could contribute to an increased risk of PCOS, underscoring its significance in the underlying mechanisms of this condition.
A number of metabolites causally linked to AS were identified in this study, among which glycocholenate sulfate stood out due to its strong causality with AS. Prior research on glycocholenate sulfate primarily concentrated on its implications for cardiovascular conditions, notably atrial fibrillation [67, 68]. However, investigations within the context of reproductive disorders were limited. The causal relationship between elevated levels of glycocholenate sulfate and increased risk of AS is a novel finding that warrants further validation and exploration within a broader clinical dataset. Elevated serum creatinine levels in patients with chronic glomerulonephritis were associated with abnormal expression of IL-17 and IL-18, adversely affecting male semen quality and potentially contributing to infertility [69]. This appears to contradict our findings, where elevated creatinine levels are linked to a protective causal association with AS. Such discrepancies could be attributed to differences in sample size, disease status, or the varying impacts of different creatinine levels. This warrants further in-depth research to elucidate the underlying mechanisms. Additionally, pregnenolone sulfate, along with progesterone, has been identified as a primary steroid activating CatSper, the calcium channel crucial for sperm hyperactivation and fertility [70]. Our research further suggested that pregnenolone sulfate levels might potentially exacerbate AS. To enhance understanding of latent biological processes, future research could focus on exploring how pregnenolone sulfate impacts sperm function under different conditions and assess its interactions with sperm fertility.
Nonetheless, our research has some limitations. Firstly, the significant causal relationships identified require further validation through metabolomic experiments. Secondly, the lack of available detailed individual data impedes further stratified analyses within the population. Lastly, as this study relies on European data, our findings are not generalizable to other ethnic groups, thereby limiting the universality. Consequently, including individuals from diverse genetic backgrounds could enhance the generalizability of the findings. In addition, given that utilizing genetic variants associated with metabolites as instrumental variables only represents prolonged exposure conditions, short-term dietary interventions might produce different effects on the outcomes described.
Conclusion
In conclusion, our study utilized a systematical MR analysis framework to demonstrate for the first time the causal effects of 1,091 blood metabolites and 309 metabolite ratios on reproductive system disorders, paving new pathways for identifying potential metabolic mechanisms behind these disorders.
Supplementary Information
Supplementary Material 1. POI - Table S1. Primary results of MR analysis by IVW method (Serum metabolites on POI). Table S2. Results of complementary MR analysis via multiple algorithms (Serum metabolites on POI). Table S3. The heterogeneity of MR analysis (Serum metabolites on POI). Table S4. Horizontal pleiotropy of MR analysis (Serum metabolites on POI). Table S5. Outlier variants for 3beta-hydroxy-5-cholestenoate levels identified by Radial MR and MR-Presso methods. Table S6. Re-examining the causality after outliers excluded (3beta-hydroxy-5-cholestenoate levels on POI). Table S7. Estimation of the Steiger direction from serum metabolites to POI. Table S8. Metabolic pathways with significant enrichment of serum metabolites for POI. PCOS - Table S9. Primary results of MR analysis by IVW method (Serum metabolites on PCOS). Table S10. Results of complementary MR analysis via multiple algorithms (Serum metabolites on PCOS). Table S11. The heterogeneity of MR analysis (Serum metabolites on PCOS). Table S12. Horizontal pleiotropy of MR analysis (Serum metabolites on PCOS). Table S13. Outlier variants for N-acetyl-aspartyl-glutamate (naag) levels identified by Radial MR and MR-Presso methods. Table S14. Re-examining the causality after outliers excluded (N-acetyl-aspartyl-glutamate (naag) levels on PCOS). Table S15. Outlier variants for threonine levels identified by Radial MR and MR-Presso methods. Table S16. Re-examining the causality after outliers excluded (Threonine levels on PCOS). Table S17. Outlier variants for ribitol levels levels identified by Radial MR and MR-Presso methods. Table S18. Re-examining the causality after outliers excluded (Ribitol levels on PCOS). Table S19.Estimation of the Steiger direction from serum metabolites to PCOS. Table S20. Metabolic pathways with significant enrichment of serum metabolites for PCOS. AS - Table S21. Primary results of MR analysis by IVW method (Serum metabolites on AS). Table S22. Results of complementary MR analysis via multiple algorithms (Serum metabolites on AS). Table S23. The heterogeneity of MR analysis (Serum metabolites on AS). Table S24.Horizontal pleiotropy of MR analysis (Serum metabolites on AS). Table S25. Estimation of the Steiger direction from serum metabolites to AS. Table S26. Metabolic pathways with significant enrichment of serum metabolites for AS
Supplementary Material 2. Figure S1. External replication MR analysis between candidate metabolites and reproductive system diseases. (A-D) Forest plots showed causal associations of X-11470 levels (A), X-12007 levels (B),3-methyl-2-oxovalerate levels (C), and creatinine levels (D) with primary ovarian insufficiency (POI) using GWAS data from two different sources. (E-G) Forest plots showed the causal associations of 5-oxoproline levels(E), androsterone sulfate levels (F), and 5alpha-androstan-3beta,17beta-diol disulfate levels (G) with polycystic ovary syndrome (PCOS) in the validation set, confirming the results from the primary analysis. Similarly, (H) the forest plot showed the causal association between citrulline levels and abnormal spermatozoa (AS). The study denoted as "Yiheng Chen et al." pertains to the GWAS data of 1400 metabolites cited within our article. "Shin et al." refers to the study on 486 serum metabolites (PMID: 24816252), with GWAS data stored in the IEU Open GWAS project (met-a-561). The displayed p-values correspond to the IVW method. Higher risk corresponds to odds ratio > 1, and lower risk corresponds to odds ratio < 1. OR, odds ratio; SE, Standard Error.
Acknowledgements
We want to acknowledge all contributors for sharing the data involved in this study. We want to acknowledge all the participants and investigators of the FinnGen study, the researchers and participants of the original GWAS.
Authors’ contributions
SC, SHS and YHG contributed study design and data analysis. SC wrote and edited the manuscript. QLY, ZHZ, and FW edited the manuscript. CS, MSC, ZKZ and YM contributed project oversight and manuscript revisiting. JHL, WYS, KH, QLY, and YHG contributed manuscript revisiting. All authors read and approved the final manuscript.
Funding
2022 Annual’ Zhongyuan Talent Plan (Talent Cultivation Series)'.
Availability of data and materials
Publicly available datasets were analyzed in this study. Statistics on primary ovarian insufficiency and polycystic ovary syndrome were obtained from FinnGen consortium R9 release data (https://r9.finngen.fi/). Summary statistics of abnormal spermatozoa were obtained from FinnGen consortium R7 release data (https://r7.finngen.fi/). GWAS data for metabolites can be retrieved from and GWAS catalog database (https://www.ebi.ac.uk/gwas/home) and MRC Integrative Epidemiology Unit (https://gwas.mrcieu.ac.uk/).
Declarations
Consent for publication
All authors read and approved the final version of the manuscript for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuang Chen, Shihao Sun and Mingshu Cai contributed equally to this work and share senior authorship.
Contributor Information
Qingling Yang, Email: qingling531@163.com.
Yihong Guo, Email: 13613863710@163.com.
References
- 1.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, et al. The International Glossary on Infertility and Fertility Care, 2017. Fertil Steril. 2017;108:393–406. 10.1016/j.fertnstert.2017.06.005. 10.1016/j.fertnstert.2017.06.005 [DOI] [PubMed] [Google Scholar]
- 2.Dai M, Guo W, Zhu S, Gong G, Chen M, Zhong Z, Guo J, Zhang Y. Type 2 diabetes mellitus and the risk of abnormal spermatozoa: A Mendelian randomization study. Front Endocrinol (Lausanne). 2022;13:1035338. 10.3389/fendo.2022.1035338. 10.3389/fendo.2022.1035338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vollset SE, Goren E, Yuan CW, Cao J, Smith AE, Hsiao T, Bisignano C, Azhar GS, Castro E, Chalek J, et al. Fertility, mortality, migration, and population scenarios for 195 countries and territories from 2017 to 2100: a forecasting analysis for the Global Burden of Disease Study. Lancet. 2020;396:1285–306. 10.1016/s0140-6736(20)30677-2. 10.1016/s0140-6736(20)30677-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulam CB, Adamson SC, Annegers JF. Incidence of premature ovarian failure. Obstet Gynecol. 1986;67:604–6. [PubMed] [Google Scholar]
- 5.De Vos M, Devroey P, Fauser BC. Primary ovarian insufficiency. Lancet. 2010;376:911–21. 10.1016/s0140-6736(10)60355-8. 10.1016/s0140-6736(10)60355-8 [DOI] [PubMed] [Google Scholar]
- 6.Rocca WA, Grossardt BR, de Andrade M, Malkasian GD, Melton LJ 3rd. Survival patterns after oophorectomy in premenopausal women: a population-based cohort study. Lancet Oncol. 2006;7:821–8. 10.1016/s1470-2045(06)70869-5. 10.1016/s1470-2045(06)70869-5 [DOI] [PubMed] [Google Scholar]
- 7.Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med. 2009;360:606–14. 10.1056/NEJMcp0808697. 10.1056/NEJMcp0808697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman RJ, Dewailly D, Legro RS, Hickey TE. Polycystic ovary syndrome. Lancet. 2007;370:685–97. 10.1016/s0140-6736(07)61345-2. 10.1016/s0140-6736(07)61345-2 [DOI] [PubMed] [Google Scholar]
- 9.March WA, Moore VM, Willson KJ, Phillips DI, Norman RJ, Davies MJ. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum Reprod. 2010;25:544–51. 10.1093/humrep/dep399. 10.1093/humrep/dep399 [DOI] [PubMed] [Google Scholar]
- 10.Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7:219–31. 10.1038/nrendo.2010.217. 10.1038/nrendo.2010.217 [DOI] [PubMed] [Google Scholar]
- 11.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–9. 10.1038/ng.732. 10.1038/ng.732 [DOI] [PubMed] [Google Scholar]
- 12.Kandaraki E, Christakou C, Diamanti-Kandarakis E. Metabolic syndrome and polycystic ovary syndrome... and vice versa. Arq Bras Endocrinol Metabol. 2009;53:227–37. 10.1590/s0004-27302009000200014. 10.1590/s0004-27302009000200014 [DOI] [PubMed] [Google Scholar]
- 13.Gill, K., Jakubik, J., Rosiak-Gill, A., Kups, M., Lukaszuk, M., Kurpisz, M., Fraczek, M., and Piasecka, M. (2019). Utility and Predictive Value of Human Standard Semen Parameters and Sperm DNA Dispersion for Fertility Potential. Int J Environ Res Public Health 16. 10.3390/ijerph16112004. [DOI] [PMC free article] [PubMed]
- 14.Quinones MP, Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35:165–76. 10.1016/j.nbd.2009.02.019. 10.1016/j.nbd.2009.02.019 [DOI] [PubMed] [Google Scholar]
- 15.Assfalg M, Bertini I, Colangiuli D, Luchinat C, Schäfer H, Schütz B, Spraul M. Evidence of different metabolic phenotypes in humans. Proc Natl Acad Sci U S A. 2008;105:1420–4. 10.1073/pnas.0705685105. 10.1073/pnas.0705685105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. 10.1038/nm.3145. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridder L, van der Hooft JJ, Verhoeven S, de Vos RC, Bino RJ, Vervoort J. Automatic chemical structure annotation of an LC-MS(n) based metabolic profile from green tea. Anal Chem. 2013;85:6033–40. 10.1021/ac400861a. 10.1021/ac400861a [DOI] [PubMed] [Google Scholar]
- 18.Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4: e1000282. 10.1371/journal.pgen.1000282. 10.1371/journal.pgen.1000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50. 10.1038/ng.2982. 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikäinen LP, Kangas AJ, Soininen P, Würtz P, Silander K, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–76. 10.1038/ng.1073. 10.1038/ng.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. 10.1038/nature10354. 10.1038/nature10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. 10.1126/science.1193494. 10.1126/science.1193494 [DOI] [PubMed] [Google Scholar]
- 23.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. 10.1038/nm.2307. 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontana R, Della Torre S. The Deep Correlation between Energy Metabolism and Reproduction: A View on the Effects of Nutrition for Women Fertility. Nutrients. 2016;8:87. 10.3390/nu8020087. 10.3390/nu8020087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottsch ML, Clifton DK, Steiner RA. Galanin-like peptide as a link in the integration of metabolism and reproduction. Trends Endocrinol Metab. 2004;15:215–21. 10.1016/j.tem.2004.05.010. 10.1016/j.tem.2004.05.010 [DOI] [PubMed] [Google Scholar]
- 26.Roa J, Tena-Sempere M. Connecting metabolism and reproduction: roles of central energy sensors and key molecular mediators. Mol Cell Endocrinol. 2014;397:4–14. 10.1016/j.mce.2014.09.027. 10.1016/j.mce.2014.09.027 [DOI] [PubMed] [Google Scholar]
- 27.Chang AY, Lalia AZ, Jenkins GD, Dutta T, Carter RE, Singh RJ, Nair KS. Combining a nontargeted and targeted metabolomics approach to identify metabolic pathways significantly altered in polycystic ovary syndrome. Metabolism. 2017;71:52–63. 10.1016/j.metabol.2017.03.002. 10.1016/j.metabol.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczuko, M., Kikut, J., Komorniak, N., Bilicki, J., Celewicz, Z., and Ziętek, M. (2020). The Role of Arachidonic and Linoleic Acid Derivatives in Pathological Pregnancies and the Human Reproduction Process. Int J Mol Sci 21. 10.3390/ijms21249628. [DOI] [PMC free article] [PubMed]
- 29.Muccini, A.M., Tran, N.T., de Guingand, D.L., Philip, M., Della Gatta, P.A., Galinsky, R., Sherman, L.S., Kelleher, M.A., Palmer, K.R., Berry, M.J., et al. (2021). Creatine Metabolism in Female Reproduction, Pregnancy and Newborn Health. Nutrients 13. 10.3390/nu13020490. [DOI] [PMC free article] [PubMed]
- 30.Blomberg Jensen M. Vitamin D metabolism, sex hormones, and male reproductive function. Reproduction. 2012;144:135–52. 10.1530/rep-12-0064. 10.1530/rep-12-0064 [DOI] [PubMed] [Google Scholar]
- 31.Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization Jama. 2017;318:1925–6. 10.1001/jama.2017.17219. 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- 32.Minelli C, Thompson JR, Tobin MD, Abrams KR. An integrated approach to the meta-analysis of genetic association studies using Mendelian randomization. Am J Epidemiol. 2004;160:445–52. 10.1093/aje/kwh228. 10.1093/aje/kwh228 [DOI] [PubMed] [Google Scholar]
- 33.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89-98. 10.1093/hmg/ddu328. 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J, Wade KH, Timpson NJ, Evans DM, Willeit P, et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017;3:636–51. 10.1001/jamaoncol.2016.5945. 10.1001/jamaoncol.2016.5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White J, Swerdlow DI, Preiss D, Fairhurst-Hunter Z, Keating BJ, Asselbergs FW, Sattar N, Humphries SE, Hingorani AD, Holmes MV. Association of Lipid Fractions With Risks for Coronary Artery Disease and Diabetes. JAMA Cardiol. 2016;1:692–9. 10.1001/jamacardio.2016.1884. 10.1001/jamacardio.2016.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue, H., Shen, X., and Pan, W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am J Hum Genet 108, 1251–1269. 10.1016/j.ajhg.2021.05.014. [DOI] [PMC free article] [PubMed]
- 37.Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11:376. 10.1038/s41467-019-14156-4. 10.1038/s41467-019-14156-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, Q., Wang, J., Hemani, G., Bowden, J., and Small, D.S. Statistical inference in two-sample summary-data Mendelian randomizati on using robust adjusted profile score. arxiv:1801.09652 [stat.AP,math.ST,stat.TH,65J05, 46N60, 62F35].
- 39.Ye, T., Shao, J., and Kang, H. Debiased inverse-variance weighted estimator in two-sample summary-dat a Mendelian randomization. Ann. Statist. 49. 10.1214/20-aos2027.
- 40.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362: k601. 10.1136/bmj.k601. 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panyard DJ, Kim KM, Darst BF, Deming YK, Zhong X, Wu Y, Kang H, Carlsson CM, Johnson SC, Asthana S, et al. Cerebrospinal fluid metabolomics identifies 19 brain-related phenotype associations. Commun Biol. 2021;4:63. 10.1038/s42003-020-01583-z. 10.1038/s42003-020-01583-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kettunen J, Demirkan A, Würtz P, Draisma HH, Haller T, Rawal R, Vaarhorst A, Kangas AJ, Lyytikäinen LP, Pirinen M, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. 10.1038/ncomms11122. 10.1038/ncomms11122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borges CM, Fujihara CK, Malheiros D, de Ávila VF, Formigari GP, Lopes de Faria JB. Metformin arrests the progression of established kidney disease in the subtotal nephrectomy model of chronic kidney disease. Am J Physiol Renal Physiol. 2020;318:F1229-f1236. 10.1152/ajprenal.00539.2019. 10.1152/ajprenal.00539.2019 [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Lu T, Pettersson-Kymmer U, Stewart ID, Butler-Laporte G, Nakanishi T, Cerani A, Liang KYH, Yoshiji S, Willett JDS, et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat Genet. 2023;55:44–53. 10.1038/s41588-022-01270-1. 10.1038/s41588-022-01270-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. 10.1038/nature15393. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. 10.1002/gepi.21758. 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–52. 10.1093/ije/dyq151. 10.1093/ije/dyq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, Sole G, Lai S, Dei M, Mulas A, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52:1036–45. 10.1038/s41588-020-0684-4. 10.1038/s41588-020-0684-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess, S., Davey Smith, G., Davies, N.M., Dudbridge, F., Gill, D., Glymour, M.M., Hartwig, F.P., Kutalik, Z., Holmes, M.V., Minelli, C., et al. (2019). Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res 4, 186. 10.12688/wellcomeopenres.15555.3. [DOI] [PMC free article] [PubMed]
- 50.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. 10.1093/ije/dyv080. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–14. 10.1002/gepi.21965. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue H, Shen X, Pan W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am J Hum Genet. 2021;108:1251–69. 10.1016/j.ajhg.2021.05.014. 10.1016/j.ajhg.2021.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–89. 10.1007/s10654-017-0255-x. 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowden J, Spiller W, Del Greco MF, Sheehan N, Thompson J, Minelli C, Davey Smith G. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47:1264–78. 10.1093/ije/dyy101. 10.1093/ije/dyy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis SR. Androgens in premenopausal women and women with premature ovarian insufficiency. Climacteric. 2021;24:459–65. 10.1080/13697137.2020.1866530. 10.1080/13697137.2020.1866530 [DOI] [PubMed] [Google Scholar]
- 56.Prizant H, Gleicher N, Sen A. Androgen actions in the ovary: balance is key. J Endocrinol. 2014;222:R141-151. 10.1530/joe-14-0296. 10.1530/joe-14-0296 [DOI] [PubMed] [Google Scholar]
- 57.Magamage MPS, Zengyo M, Moniruzzaman M, Miyano T. Testosterone induces activation of porcine primordial follicles in vitro. Reprod Med Biol. 2011;10:21–30. 10.1007/s12522-010-0068-z. 10.1007/s12522-010-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith P, Steckler TL, Veiga-Lopez A, Padmanabhan V. Developmental programming: differential effects of prenatal testosterone and dihydrotestosterone on follicular recruitment, depletion of follicular reserve, and ovarian morphology in sheep. Biol Reprod. 2009;80:726–36. 10.1095/biolreprod.108.072801. 10.1095/biolreprod.108.072801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyman JH, Margalioth EJ, Rabinowitz R, Tsafrir A, Gal M, Alerhand S, Algur N, Eldar-Geva T. DHEA supplementation may improve IVF outcome in poor responders: a proposed mechanism. Eur J Obstet Gynecol Reprod Biol. 2013;168:49–53. 10.1016/j.ejogrb.2012.12.017. 10.1016/j.ejogrb.2012.12.017 [DOI] [PubMed] [Google Scholar]
- 60.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. 10.1074/mcp.M113.035600. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Moro Á, González-Brusi L, Querejeta-Fernández A, Padilla-Ruiz E, García-Blanco J, Bermejo-Álvarez P. Metabolomics analysis of human cumulus cells obtained from cumulus-oocyte complexes with different developmental potential. Hum Reprod. 2023;38:2187–95. 10.1093/humrep/dead181. 10.1093/humrep/dead181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turathum B, Gao EM, Yang F, Liu YB, Yang ZY, Liu CC, Xue YJ, Wu MH, Wang L, Grataitong K, Chian RC. Role of pyroglutamic acid in cumulus cells of women with polycystic ovary syndrome. J Assist Reprod Genet. 2022;39:2737–46. 10.1007/s10815-022-02647-1. 10.1007/s10815-022-02647-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vašková, J., Kočan, L., Vaško, L., and Perjési, P. (2023). Glutathione-Related Enzymes and Proteins: A Review. Molecules 28. 10.3390/molecules28031447. [DOI] [PMC free article] [PubMed]
- 64.Chou HY, Lin YH, Shiu GL, Tang HY, Cheng ML, Shiao MS, Pai LM. ADI1, a methionine salvage pathway enzyme, is required for Drosophila fecundity. J Biomed Sci. 2014;21:64. 10.1186/s12929-014-0064-4. 10.1186/s12929-014-0064-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das SK, Ainsworth HC, Dimitrov L, Okut H, Comeau ME, Sharma N, Ng MCY, Norris JM, Chen YI, Wagenknecht LE, et al. Metabolomic architecture of obesity implicates metabolonic lactone sulfate in cardiometabolic disease. Mol Metab. 2021;54: 101342. 10.1016/j.molmet.2021.101342. 10.1016/j.molmet.2021.101342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Purwar A, Nagpure S. Insulin Resistance in Polycystic Ovarian Syndrome. Cureus. 2022;14: e30351. 10.7759/cureus.30351. 10.7759/cureus.30351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alonso A, Yu B, Sun YV, Chen LY, Loehr LR, O’Neal WT, Soliman EZ, Boerwinkle E. Serum Metabolomics and Incidence of Atrial Fibrillation (from the Atherosclerosis Risk in Communities Study). Am J Cardiol. 2019;123:1955–61. 10.1016/j.amjcard.2019.03.017. 10.1016/j.amjcard.2019.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin, X., Li, J., Bose, D., Okamoto, J., Kwon, A., Jackson, A.U., Silva, L.F., Oravilahti, A., Stringham, H.M., Ripatti, S., et al. (2023). Metabolome-wide Mendelian randomization characterizes heterogeneous and shared causal effects of metabolites on human health. medRxiv. 10.1101/2023.06.26.23291721.
- 69.Zhang, H., Ying, Y., Chen, Y., Lu, X., and Huang, Y. (2017). Effect of chronic glomerulonephritis on the semen quality and cytokines in the semen of infertile males. Am J Reprod Immunol 77. 10.1111/aji.12598. [DOI] [PubMed]
- 70.Mannowetz N, Miller MR, Lishko PV. Regulation of the sperm calcium channel CatSper by endogenous steroids and plant triterpenoids. Proc Natl Acad Sci U S A. 2017;114:5743–8. 10.1073/pnas.1700367114. 10.1073/pnas.1700367114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1. POI - Table S1. Primary results of MR analysis by IVW method (Serum metabolites on POI). Table S2. Results of complementary MR analysis via multiple algorithms (Serum metabolites on POI). Table S3. The heterogeneity of MR analysis (Serum metabolites on POI). Table S4. Horizontal pleiotropy of MR analysis (Serum metabolites on POI). Table S5. Outlier variants for 3beta-hydroxy-5-cholestenoate levels identified by Radial MR and MR-Presso methods. Table S6. Re-examining the causality after outliers excluded (3beta-hydroxy-5-cholestenoate levels on POI). Table S7. Estimation of the Steiger direction from serum metabolites to POI. Table S8. Metabolic pathways with significant enrichment of serum metabolites for POI. PCOS - Table S9. Primary results of MR analysis by IVW method (Serum metabolites on PCOS). Table S10. Results of complementary MR analysis via multiple algorithms (Serum metabolites on PCOS). Table S11. The heterogeneity of MR analysis (Serum metabolites on PCOS). Table S12. Horizontal pleiotropy of MR analysis (Serum metabolites on PCOS). Table S13. Outlier variants for N-acetyl-aspartyl-glutamate (naag) levels identified by Radial MR and MR-Presso methods. Table S14. Re-examining the causality after outliers excluded (N-acetyl-aspartyl-glutamate (naag) levels on PCOS). Table S15. Outlier variants for threonine levels identified by Radial MR and MR-Presso methods. Table S16. Re-examining the causality after outliers excluded (Threonine levels on PCOS). Table S17. Outlier variants for ribitol levels levels identified by Radial MR and MR-Presso methods. Table S18. Re-examining the causality after outliers excluded (Ribitol levels on PCOS). Table S19.Estimation of the Steiger direction from serum metabolites to PCOS. Table S20. Metabolic pathways with significant enrichment of serum metabolites for PCOS. AS - Table S21. Primary results of MR analysis by IVW method (Serum metabolites on AS). Table S22. Results of complementary MR analysis via multiple algorithms (Serum metabolites on AS). Table S23. The heterogeneity of MR analysis (Serum metabolites on AS). Table S24.Horizontal pleiotropy of MR analysis (Serum metabolites on AS). Table S25. Estimation of the Steiger direction from serum metabolites to AS. Table S26. Metabolic pathways with significant enrichment of serum metabolites for AS
Supplementary Material 2. Figure S1. External replication MR analysis between candidate metabolites and reproductive system diseases. (A-D) Forest plots showed causal associations of X-11470 levels (A), X-12007 levels (B),3-methyl-2-oxovalerate levels (C), and creatinine levels (D) with primary ovarian insufficiency (POI) using GWAS data from two different sources. (E-G) Forest plots showed the causal associations of 5-oxoproline levels(E), androsterone sulfate levels (F), and 5alpha-androstan-3beta,17beta-diol disulfate levels (G) with polycystic ovary syndrome (PCOS) in the validation set, confirming the results from the primary analysis. Similarly, (H) the forest plot showed the causal association between citrulline levels and abnormal spermatozoa (AS). The study denoted as "Yiheng Chen et al." pertains to the GWAS data of 1400 metabolites cited within our article. "Shin et al." refers to the study on 486 serum metabolites (PMID: 24816252), with GWAS data stored in the IEU Open GWAS project (met-a-561). The displayed p-values correspond to the IVW method. Higher risk corresponds to odds ratio > 1, and lower risk corresponds to odds ratio < 1. OR, odds ratio; SE, Standard Error.
Data Availability Statement
Publicly available datasets were analyzed in this study. Statistics on primary ovarian insufficiency and polycystic ovary syndrome were obtained from FinnGen consortium R9 release data (https://r9.finngen.fi/). Summary statistics of abnormal spermatozoa were obtained from FinnGen consortium R7 release data (https://r7.finngen.fi/). GWAS data for metabolites can be retrieved from and GWAS catalog database (https://www.ebi.ac.uk/gwas/home) and MRC Integrative Epidemiology Unit (https://gwas.mrcieu.ac.uk/).