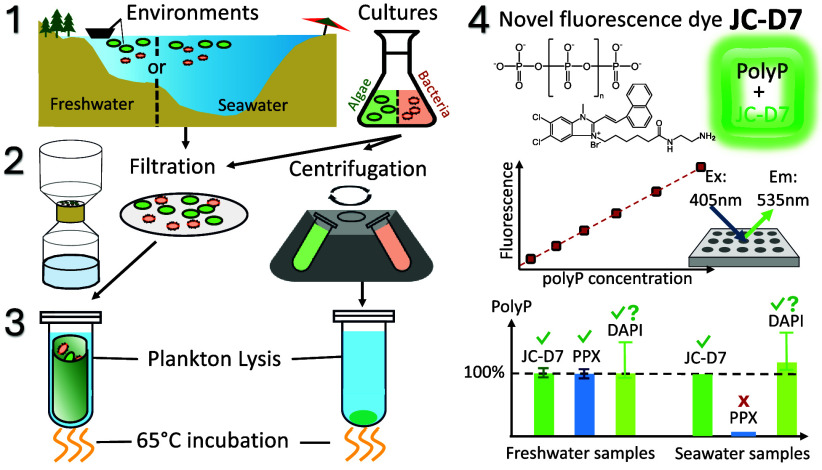
Abstract
Polyphosphate (polyP) is found in plankton of diverse aquatic ecosystems and is important for plankton ecology and biogeochemical cycling. However, our knowledge of polyP in aquatic environments is hindered by a lack of data due to the limitations of quantification methods. The estimate of polyP in model organisms using phenol-chloroform extraction followed by enzymatic hydrolysis is complicated and fails for environmental samples. The commonly used 4′,6-diamidino-2-phenylindole (DAPI) fluorescence method for environmental studies, on the contrary, severely overestimates polyP due to interference. In this paper, we develop a plankton lysis buffer to extract polyP and a quantification method using a novel polyP-specific fluorescence dye JC-D7. We test the methods using cultured algae and bacteria, as well as natural samples from marine and freshwater environments. We show that our plankton lysis extracts polyP with high recovery while requiring substantially less time and effort. Subsequent polyP quantification using JC-D7 fluorescence overcomes the interference encountered by the DAPI method and provides an accurate measurement of polyP down to <0.5 μmol L–1. This novel method enables more accurate quantification of polyP in aquatic environments and will profoundly enhance our knowledge of polyP, plankton ecology, and biogeochemistry.
Keywords: polyphosphate, fluorescence quantification, aquatic environments, planktonic samples, JC-D7 dye
Short abstract
Knowledge of polyphosphate in the environment is hindered by the lack of reliable quantification methods. This study presents a novel method enabling accurate quantification of polyphosphate in aquatic environmental samples.
Introduction
Polyphosphate (polyP) is a biologically important polymer present in diverse organisms and has many cellular functions.1 Accumulation of polyP in microorganisms has been widely used to remove phosphorus (P) in wastewater treatment facilities.2−6 PolyP transformation has also been suggested to be important for controlling water quality in agricultural landscapes.7,8 In natural aquatic environments, polyP is found to be strongly dynamic and has important ecological impacts: the intracellular metabolisms of polyP can help plankton adapt to ambient nutrient limitations;9−11 the accumulation and remineralization of polyP also regulate P availability.10,12−16 These processes are crucial in aquatic environments, as P is a common limiting nutrient for primary productivity and thus controls the trophic states and ecosystem functions.17
Our knowledge about the quantitative roles of polyP in P cycling and plankton ecology, however, is very limited, because data are extremely rare for natural environments.13 Most studies reporting environmental polyP only provided relative measurements due to the limitations of quantification methods.9,10,12,18−21 To extract polyP from environmental planktonic samples, a boiling-enzyme digestion extraction has been commonly used,22 followed by a fluorescence quantification using 4′,6-diamidino-2-phenylindole (DAPI).22−24 However, recent studies suggested that the boiling-enzyme method may not efficiently extract all polyP25 (also confirmed by our tests), despite being relatively convenient in terms of operation. More problematically, DAPI fluorescence often leads to a severe overestimation of polyP,12,13,18 because interferences from nucleic acids are difficult to remove.13 Some polysaccharides, such as sulfated polysaccharides and glycosaminoglycans, are also known to interfere with DAPI-polyP quantification.26 As a result, polyP concentrations estimated using this method are considered relative values: in many instances, the proportions of polyP in total particulate P were reported higher than 100%, and in some cases, even exceeded 300%,12,18 which is not realistic as polyP cannot constitute more than 100% of total particulate P.
A more robust method for polyP extraction from unicellular organisms involves a phenol-chloroform procedure, adapted from the DNA and RNA extraction technique, which removes lipids and proteins while retaining DNA, RNA, and polyP in the aqueous phase.27 PolyP is then quantified using exopolyphosphatase (PPX), an enzyme that exclusively hydrolyzes polyP to orthophosphate (PO43–), which is measured to calculate the amount of polyP in the original samples.27,28 The PPX method has been used for various unicellular model organisms including yeast S. cerevisiae, bacteria Escherichia coli, and algae Chlamydomonas reinhardtii.29−31 However, by far no published work has reported using the PPX method for environmental samples, and our numerous trials in various natural environments have failed to produce realistic results for polyP for reasons not known (see Results and Discussion).
Therefore, an improved method that enables efficient, sensitive, and accurate quantification of polyP in environmental planktonic samples is urgently needed to facilitate reliable polyP research in aquatic environments. In this paper, we develop and evaluate a new plankton lysing method designed to efficiently extract polyP from both cultural and environmental planktonic samples. We also develop a polyP quantification method using a new polyP-specific fluorescence dye. We compare these methods to the phenol-chloroform extraction and the PPX quantification techniques and show that our approach is equally efficient in polyP extraction and more sensitive in quantification. Most importantly, the method enables more accurate quantification of aquatic environmental polyP, the enigmatic compound that has yet to be fully studied in terms of its share in the P pool, contributions to P cycling, roles in plankton physiology and ecology, and more.
Materials and Methods
The methods we have developed include 1) the extraction of polyP in planktonic samples using a plankton lysis buffer solution, and 2) the fluorescence quantification of the extracted polyP using the polyP-specific dye JC-D7. This section first describes these methods and the samples used for testing (Table 1). The section also introduces the procedures of other methods we used to test against the performance of our new methods. These methods include phenol-chloroform polyP extraction,27 polyP quantification using PPX degradation (PPX method27), and polyP quantification using DAPI fluorescence (DAPI method22−24,32). Finally, the section presents the overall experimental design to assess the methods using cultured samples of algae and bacteria, as well as samples collected from natural freshwater and marine environments.
Table 1. Sample Descriptionsa.
| sample type | sample name | species (strain) | phylum | source and treatment | other information |
|---|---|---|---|---|---|
| freshwater algae | wild-type Chlamydomonas reinhardtii | C. reinhardtii (CC-1690) | Chlorophyte | from CRC, cultured in TAP medium | wild-type strain |
| freshwater algae | mutant C. reinhardtii | C. reinhardtii (CC-5322) | Chlorophyte | from CRC, cultured in TAP medium | vtc4 mutant, does not accumulate polyP |
| marine algae | Thalassiosira weissflogii | T. weissflogii | Diatom | isolated from Hong Kong coastal waters and cultured in Guildard’s (F/2) Medium | |
| marine algae | Ruttnera sp. | Ruttnera sp. (PLY510A) | Haptophyta | from RCC, cultured in Guildard’s (F/2) Medium | |
| freshwater bacteria | V. perlucida | Vogesella perlucida | Pseudomanodota | isolated from freshwater Kowloon Reservoir and cultured in R2A medium | |
| marine bacteria | Nautella sp. | Nautella sp. | Pseudomanodota | isolated from Hong Kong coastal waters and cultured in 2216E medium | |
| freshwater environment | Kowloon Reservoir | – | – | waters from freshwater Kowloon Reservoir were filtered through 0.2 μm PC membrane filters to collect particles | |
| marine environment | Hong Kong coast | – | – | coastal seawaters were filtered through 0.2 μm PC membrane filters to collect particles |
Abbreviations: CRC, Chlamydomonas Resource Center, University of Minnesota; ATCC, American Type Culture Collection; RCC, Roscoff Culture Collection.
Samples
To develop and test the methods, we used samples from both natural aquatic environments and pure cultures (Table 1). Natural water samples were collected from a freshwater reservoir (Kowloon Reservoir) and coastal seawaters in Hong Kong. To collect particulate matter, water samples were passed through a 75-μm mesh to remove large debris and then filtered through 0.2-μm pore-size polycarbonate membrane filters (47 mm, Sterlitech Corporation # PPF0247100; 0.2 μm PC filter hereafter).
Culture samples include various phytoplankton and bacteria that can be abundant in aquatic environments (Table 1). We also used a mutant strain of Chlamydomonas reinhardtii (CC-5321), which lacks the vtc-1 gene responsible for polyP synthesis and thus does not produce polyP,33 to compare with the wild-type Chlamydomonas reinhardtii strain (CC-1690) that can accumulate polyP. To collect cells from liquid cultures, samples were filtered through 0.2 μm PC filters and washed with NaCl solutions of 5 and 35 ppm for freshwater and marine cultures, respectively, to remove the residual solutes (e.g., phosphate) in the medium. While we primarily used filtration because the method is designed for working with environmental samples, we tested centrifugation as well (see Results and Discussion), as it can be more convenient for culture samples. For centrifugation, the cells were pelleted (12000 g, 2 min) to remove supernatants and then washed twice with NaCl solution (5 and 35 ppm for freshwater and marine cultures, respectively). All samples were preserved at −20 °C before polyP measurements. When collecting the C. reinhardtii cells by centrifugation, the samples were fixed with formaldehyde (0.5% v/v) before centrifugation to prevent cell mobility and resuspension. Our tests showed that this treatment did not affect subsequent polyP extraction and quantification.
Polyphosphate Standards
We use polyP standards of four different chain lengths, including n = 15, 45, 60, and 130. PolyP of n= 45 was from Sigma-Aldrich (sodium phosphate glass type 45, # S4379). The polyP standards in other chain lengths were generously provided by Dr. Toshikazu Shiba (RegeneTiss, Inc., Tokyo, Japan). The concentrations of polyP standards were verified by hydrolyzing the polyP into PO43– using persulfate digestion34 and subsequently measuring the PO43– concentration33 using either the molybdenum blue assay35 or the malachite green assay36 (see S1). Absorbance measurements were performed using transparent 96-well plates and a microplate reader (BMG ClarioStar Plus Microplate Reader). PO43– in the polyP standards without persulfate digestion was also measured to check for contamination of PO43–, which was found to be less than 0.4%. Therefore, the concentration of PO43– measured in the polyP standards after digestion, which breaks down polyP into PO43–, accurately reflects the original polyP concentration (in P units). To prepare working calibration standards for calibrating polyP concentrations in our samples (extracts), the standard solutions were adjusted to match the compositions of the sample solutions. All polyP standards, samples, and reagents hereafter were prepared using deionized water unless specified otherwise.
PolyP Extraction Using a Plankton Lysis Buffer (our method)
We developed a lysis assay to extract polyP from planktonic samples by modifying lysing solutions originally used for plant cells,37 bacteria,38 and diatoms.39 The lysing solution contains 100 μg mL–1 of proteinase K, 100 mmol L–1 of NaCl, 0.3% (v/v) of Triton X-100, 0.3% (v/v) of Tween 20, and 5 mmol L–1 of EDTA in a Tris-HCl buffer (30 mmol L–1, pH = 8.0). The components serve distinct purposes: EDTA acts as a chelating agent to remove metal cofactors of enzymes and helps disrupt the cell wall; Triton X-100 and Tween 20 permeabilize the cell membrane and remove peripheral proteins; NaCl increases the ionic strength and solubilizes proteins; proteinase K digests proteins that protect cell components during lysis. We refer to this lysing solution as “plankton lysis buffer” and the extraction method as “plankton lysing extraction” (or “plankton lysing method”) hereafter.
To extract polyP, cell samples collected on filters were fully immersed in the plankton lysis buffer, vortexed for 10s, incubated at 65 °C for 2 h, and ultrasonicated for 5 min. To extract polyP from cells collected using centrifugation, the cell pellets were resuspended in the plankton lysis buffer by vigorous mixing (e.g, using vortex), and the samples were incubated at 65 °C for 2 h and ultrasonicated for 5 min. The lysate was then collected using centrifugation (13000 g, 2 min) or filtration (0.2 μm PC filters) and stored at −20 °C before polyP quantification.
PolyP Extraction Using a Phenol-Chloroform Solution
The phenol extraction assay has been commonly applied to extract polyP in unicellular organisms.27 Thus, we compared this method with our methods. To extract polyP, the cells collected on the filters or pelleted in the microcentrifuge tubes were thoroughly mixed with 400 μL of AE buffer (50 mmol L–1 of sodium acetate buffer (pH 5.3) and 10 mmol L–1 of ethylenediaminetetraacetic acid (EDTA), stored at 4 °C), 300 μL of phenol (prepared by melting phenol solids at 60 °C), and 40 μL of 10% sodium dodecyl sulfate (SDS). The mixtures were then incubated at 65 °C for 5 min and immediately chilled on ice for 1 min. After incubation and cooling, 300 μL of chloroform was added to the sample, and the solution was vortexed for 10 s and inverted at least 5 times to ensure thorough mixing. We then allowed for 20 min for the reaction to proceed. These steps lyse the cells, denature proteins and lipids, and dissolve proteins, lipids, and other impurities into the organic (phenol-chloroform) phase. After lysis and reaction, we separated the aqueous and organic phases using centrifugation (13000 g, 2 min). The upper aqueous phase was transferred to a new microcentrifuge tube with 350 μL of chloroform for another round of cleaning (mixing, centrifugation, and separation as described above). The aqueous phases, after two rounds of cleaning, were treated with 4U of Deoxyribonuclease (DNase; TURBO, Thermofisher, AM2238) and 2 μL of Ribonuclease (RNase) Cocktail Enzyme Mix (Thermofisher, AM2286) that contains 1U of RNase A and 40 U of RNase T1, and incubated at 37 °C for 2 h to remove nucleic acids. TURBO DNase is more salt-tolerant than the DNase I that was originally used by Bru et al. (2016),27 and the addition of RNase T1 can remove fragments of RNA left from RNase A treatment. Finally, to precipitate polyP, the sample (now ∼400–420 μL of aqueous solution) was thoroughly mixed with 1 mL of 100% ethanol and 40 μL of sodium acetic acid (3 mmol L–1), both precooled at −20 °C. The mixture was left upright at −20 °C for 4 h to allow polyP precipitation. The polyP precipitates were then collected using centrifugation (13000 g, 20 min) and gently decanting the supernatant. The collected polyP precipitates were washed by mixing with 500 μL of precooled (4 °C) 70% ethanol, followed by removal of the ethanol using centrifugation (13000 g, 5 min). Any residual ethanol was removed using an additional centrifugation step and gently pipetting out the remaining supernatant. The solid phase (polyP precipitates) was dried in a desiccator and redissolved in deionized water before storage (at −20 °C) and subsequent analysis.
PolyP Fluorescence Quantification Using JC-D7 (our method)
To quantify polyP, we used the polyP-specific dye JC-D7 for staining. This novel dye was discovered to selectively stain polyP to generate fluorescence that can be used to visualize polyP in mammalian cells,40 but it has never been used for polyP quantification. JC-D7 (InvivoChem, USA, V22869, 87% purity) was dissolved in 100% dimethyl sulfoxide (DMSO) to prepare a JC-D7 stock solution of 1 mmol L–1. For the staining procedure, the JC-D7 stock solution was further diluted with 2-[4-(2-hydroxyethyl)piperazin-1-yl] ethanesulfonic acid (HEPES) buffer (25 mmol L–1, pH 8.0) to obtain a working staining reagent with JC-D7 concentration of 60 μmol L–1. This working JC-D7 solution was used to stain the polyP in samples (or standards) at a 1:1 ratio (v/v). Thus, the final staining mixture contained 30 μmol L–1 JC-D7 and 3% DMSO in 12.5 mmol L–1 of HEPES buffer. The samples were incubated for 5–10 min and fluorescence intensities were measured under excitation of 405 nm and emission of 535 nm, using 96-well black polypropylene plates (Greiner Bio-One, 650201) and a microplate reader (BMG ClarioStar Plus Microplate Reader).
PolyP Fluorescence Quantification Using DAPI
The DAPI quantification of polyP has been used for environmental samples due to its sensitivity and simple application,22,23 although it is known to overestimate polyP due to strong interferences.13,27 In this work, we also used DAPI staining to quantify polyP and compared the results with those obtained from our new method (JC-D7 staining). To quantify polyP using DAPI, samples (extracts) and polyP standards (with composition matching that of the samples) were stained with DAPI solution (100 μmol L–1, Thermo Scientific D1306) at a ratio of 10:1 (v/v; sample: DAPI) and incubated at room temperature for 10–15 min. The samples were measured for fluorescence intensities at excitation of 415 nm and emission of 550 nm in 96-well black polypropylene plates (Greiner Bio-One, 650201) using a microplate reader (BMG ClarioStar Plus Microplate Reader).
PolyP Quantification Using Exopolyphosphatase (PPX)
The quantification of polyP through the measurement of PO43– released from PPX hydrolysis of polyP has been proposed as a standard method for unicellular culture samples,27 and thus this technique is also compared to our new method. To prepare the yeast Saccharomyces cerevisiae PPX, we first transformed E. coli BL21(DE3) competent cells (ThermoFisher, # EC0114) using the plasmid with PPX gene (pKM263-ScPPX, Addgene plasmid #38327, Florian Freimoser) following the manufacturer’s instruction (see S2.1 for detailed procedures). The transformed E. coli competent cells were then cultured at 37 °C in LB medium (10% (w/v) Tryptone, 5% (w/v) Yeast Extract, and 10% (w/v) NaCl) containing 0.1 mg mL–1 of ampicillin to produce more E. coli cells containing the plasmid. To induce the expression of PPX in the culture, Isopropyl β- d-1-thiogalactopyranoside (IPTG; Sigma-Aldrich, #70527–3) was added to a final concentration of 1 mmol L–1, and the culture was incubated at 25 °C for 3 h.29
To collect the PPX produced, E. coli cells were pelleted (3000 g, 10 min) and the supernatant was removed. The cell pellets were frozen at −20 °C, resuspended in NEBExpress E. coli Lysis Reagent (New England Biolabs, # P8116S), and incubated on an orbital shaker at room temperature for 30 min to lyse the cells. The PPX in the lysate was then collected using centrifugation (16000g, 15 min) and purified using a HisPur Ni-NTA Spin Column (ThermoFisher, # 88226) following the manufacturer’s instruction (see S2.2 for detailed procedures). The concentration of the protein PPX in the final solution was determined to be ∼1.2 μg μL–1 by measuring the absorbance at 280 nm using a Nanodrop Spectrophotometer.41
To ensure accurate determination of polyP concentrations using the enzyme PPX, which relies on measuring the release of PO43– from the enzymatic degradation of polyP, we used dialysis to remove PO43– contamination in the PPX solution (see S2.2 for details). In brief, the solution was sealed into a dialysis bag, immersed in a TBS buffer (20 mmol L–1 Tris, 150 mmol L–1 NaCl, pH = 7.6), and dialyzed for 4 h at 4 °C. The dialysis was repeated using a fresh TBS buffer solution for another 4 h at 4 °C. After dialysis, the PPX solution was mixed with an equal volume of glycerol and stored at −20 °C for future usage (final PPX concentration ∼600 ng μL–1 for our PPX stock).
The quantification of polyP using PPX digestion was modified from Bru S. et al. (2016).27 In brief, 200 μL of polyP samples, extracted using phenol-chloroform (see above), were mixed with 300 μL of reaction buffer (20 mmol L–1 Tris-HCl, 100 mmol L–1 ammonium acetate, and 5 mmol L–1 magnesium acetate; pH = 7.5) and 120 ng of PPX. Although Bru et al. (2016)27 recommended using 50 ng of PPX, we applied a larger amount to promote the reaction. The mixture was incubated at 37 °C for 2 h, and the PO43– produced was determined using the molybdate-based methods (see S3 for details).35,36 To account for any PO43– originally present in the polyP samples or introduced during the treatment, each sample was treated with denatured PPX (heated at 85 °C for 5 min) as a control following the same procedure. The PO43– concentration measured in the control was subtracted from that measured in the samples treated with active PPX to calculate the polyP concentration in the sample.
Assessment of the Methods
To evaluate our methods, we first examined whether the polyP stained using JC-D7 can produce fluorescence signals that respond linearly to the concentrations of polyP in standard solutions. We further tested the effects of polyP chain length on the sensitivity and detection limits of the quantification, using polyP standards of different chain lengths (n= 14, 45, 60, and 130). The JC-D7 staining method was then compared against other methods for polyP quantification, including the PPX method27 and the DAPI fluorescence method.22−24 The former is widely used for culture studies of model organisms,29−31 while the latter has been used for natural planktonic samples.12,13,15,18−21,42 For consistency, we used the phenol-chloroform method to extract polyP from all samples being tested (see Results). To remove the interference from DNA and RNA, which compromises both DAPI and PPX methods,27 the extracts were treated with DNase and RNase for at least 2 h (see Methods).
We also evaluated polyP extraction using the plankton lysis buffer we designed, by comparing its extraction efficiency (recovery rate) to that of the commonly used phenol-chloroform method.27 Finally, the combined plankton lysis extraction and JC-D7 quantification method was tested, specifically whether the plankton lysis buffer composition interfered with the subsequent fluorescence quantification of polyP using JC-D7. In these assessments, polyP was measured in at least three replicate samples for each experiment. Analysis of Variance (ANOVA) was used to analyze the difference between the means of two or more methods (or experiments).
Results and Discussion
PolyP Quantification Using JC-D7 Staining
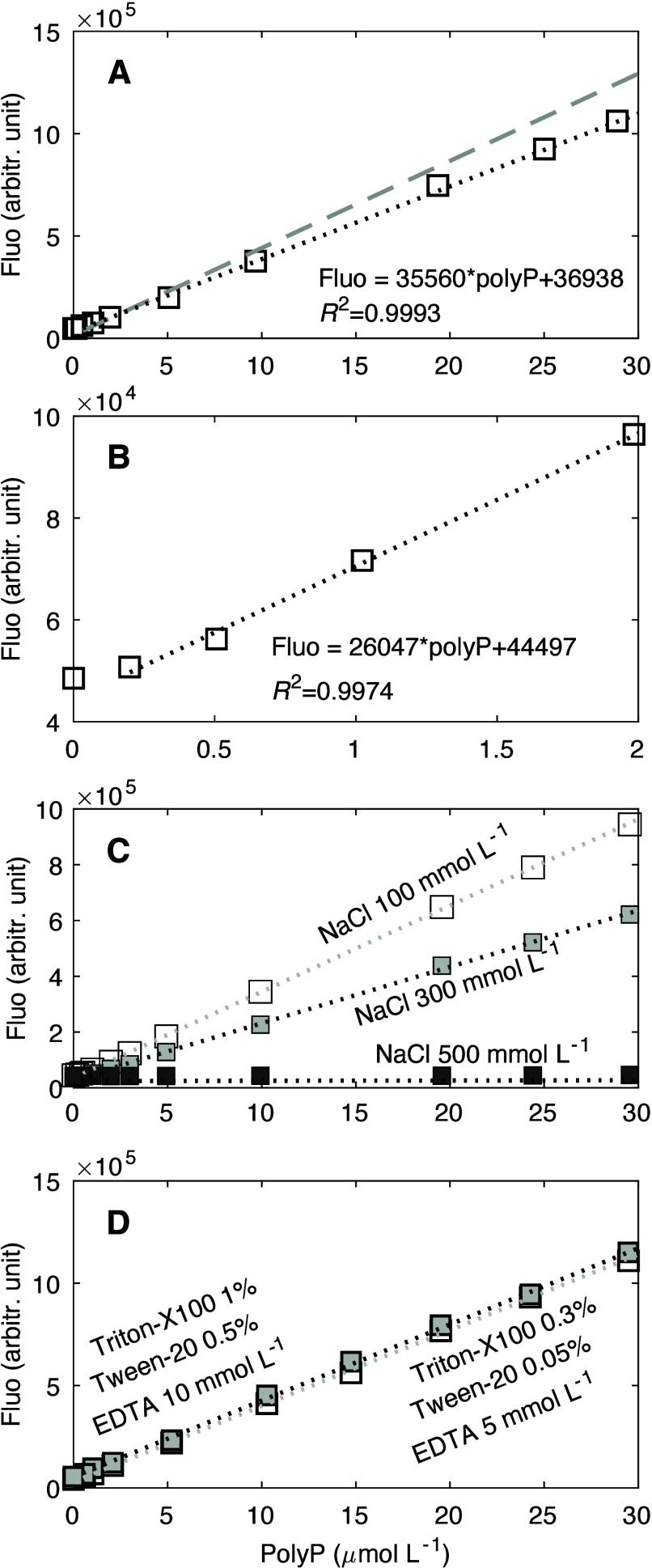
Our results indicate that the fluorescence intensity of polyP stained with JC-D7 responds linearly to the concentrations of polyP within the range of 0.5–36 μmol L–1 (R2= 0.9995; Figure 1A), under the conditions of 30 μmol L–1 JC-D7 and 3% DMSO in 12.5 mmol L–1 HEPES buffer (see Materials and Methods). The detection limit is below 0.5 μmol L–1. Linearity of the calibration decreases when the polyP concentrations exceed ∼50 μmol L–1, and fluorescence intensity reaches a maximum at polyP concentration of ∼100–120 μmol L–1 (Figure 1B). Variability in the concentrations of JC-D7 (e.g., 20–40 μmol L–1) does not change the sensitivity of the response (Figure 1C and Figure S1A–D); the fluorescence signals vary by ∼2.1 ± 0.6% (Table S1), and the slopes of the linear calibration curves vary by 3.8 ± 1.4% (Table S2). Our results also show that the chain length of polyP does not affect the linear response substantially (Figure 1D and Figure S1E–H): the fluorescence signals vary by an average of 5.0% (Table S3), and the slopes of linear calibration curves vary by <5.7 ± 1.0% (Table S2).
Figure 1.
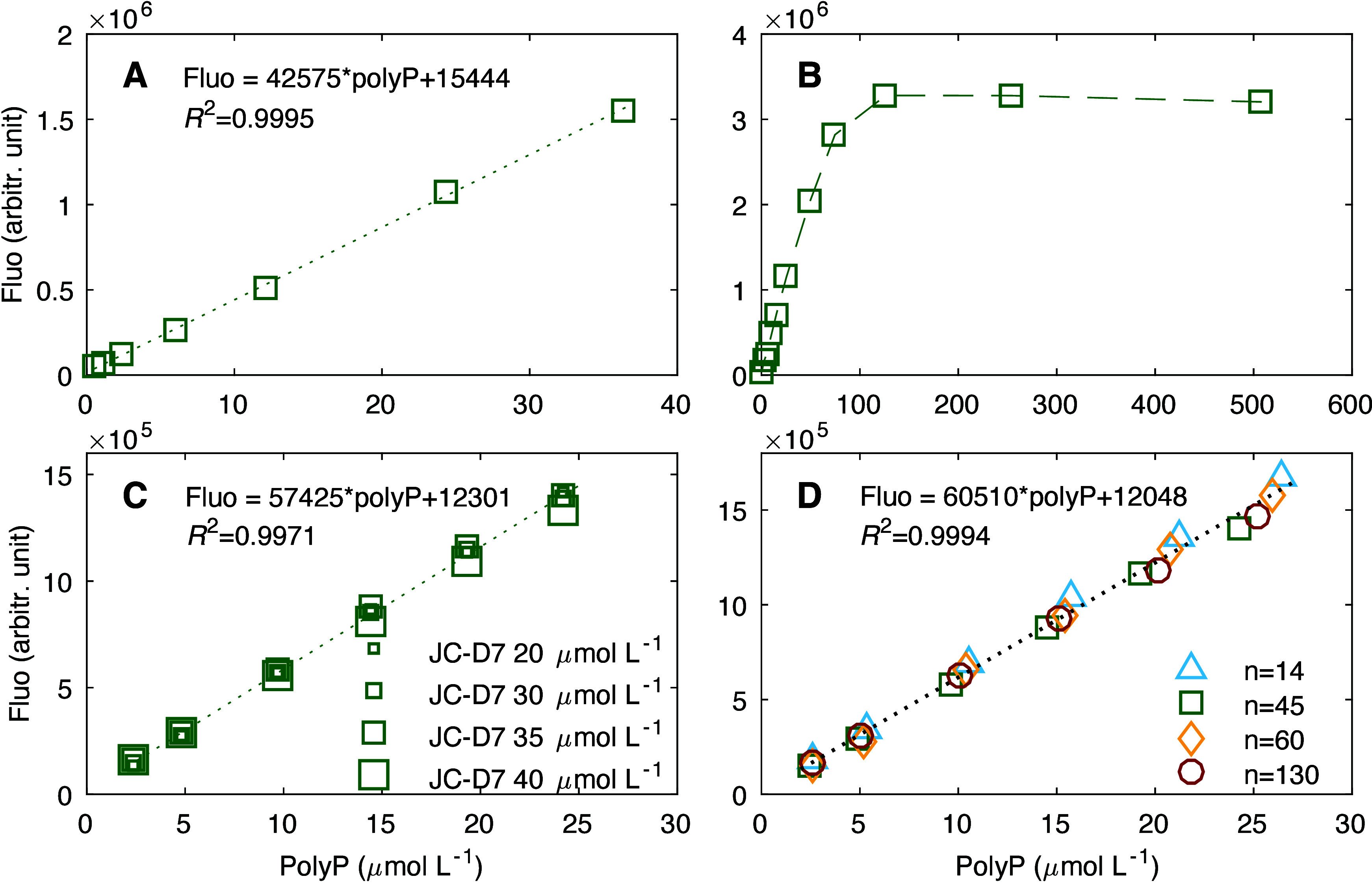
(A) Fluorescence intensity (Fluo, in arbitrary units) vs polyP concentration for polyP over a concentration range of 0.5–36 μmol L–1 and a chain length n of 45, stained by using 30 μmol L–1 JC-D7. Similar to panel A but for (B) polyP over a concentration range of 0.5–500 μmol L–1 , (C) polyP with a chain length n of 45 stained by using various concentration of JC-D7 (20–40 μmol L–1), and (D) polyP at various chain lengths (n = 14, 45, 60, and 130) stained by using 30 μmol L–1 JC-D7. PolyP was stained by using JC-D7 solutions dissolved in 3% DMSO and 12.5 mmol L–1 HEPES buffer. Fluorescence measurements were taken using 405 nm excitation and 535 nm emission.
Comparing the JC-D7 Staining Method with Other PolyP Quantification Methods
We tested our JC-D7-polyP quantification method against the PPX and the DAPI methods, using both algae and bacteria samples (Figure 2). Our results suggest that polyP estimates for the wild-type algae C. reinhardtii are consistent across the three methods (Figure 2A). For the mutant strain of the same algae species that cannot accumulate polyP, both PPX and JC-D7 consistently indicate undetectable polyP (Figure 2B; p = 0.69), while DAPI produces some polyP signals. The DAPI-polyP signal is likely an artifact due to the insufficient removal of DNA and RNA from the samples. Our tests show that when the time for DNase and RNase treatment is shortened, the DAPI signal becomes significantly higher for both the wide-type and mutant C. reinhardtii (Figure S2A, C). DAPI also overestimates polyP in the freshwater heterotrophic bacteria Vogesella perlucida, probably for similar reasons (Figure 2C), and this overestimation can be eliminated when the DNase and RNase treatment extend beyond 4 h (Figure S2F). The results for JC-D7 quantification, on the other hand, are consistently comparable to those of the PPX method (Figure 2C, p = 0.74). These results suggest that JC-D7 provides accurate quantification of polyP. The DAPI method can estimate polyP accurately if DNA and RNA are sufficiently removed, but this requires a longer treatment time which might be sample-specific (e.g., 2 h for our test of algae wild-type C. reinhardtii (Figure S2B) but >4 h for bacteria V. perlucida (Figure S2F), and in some cases longer than overnight (data not shown)). Nevertheless, the DAPI method can serve as a confirmation when producing results consistent with other measurements (i.e., PPX and JC-D7 methods).
Figure 2.
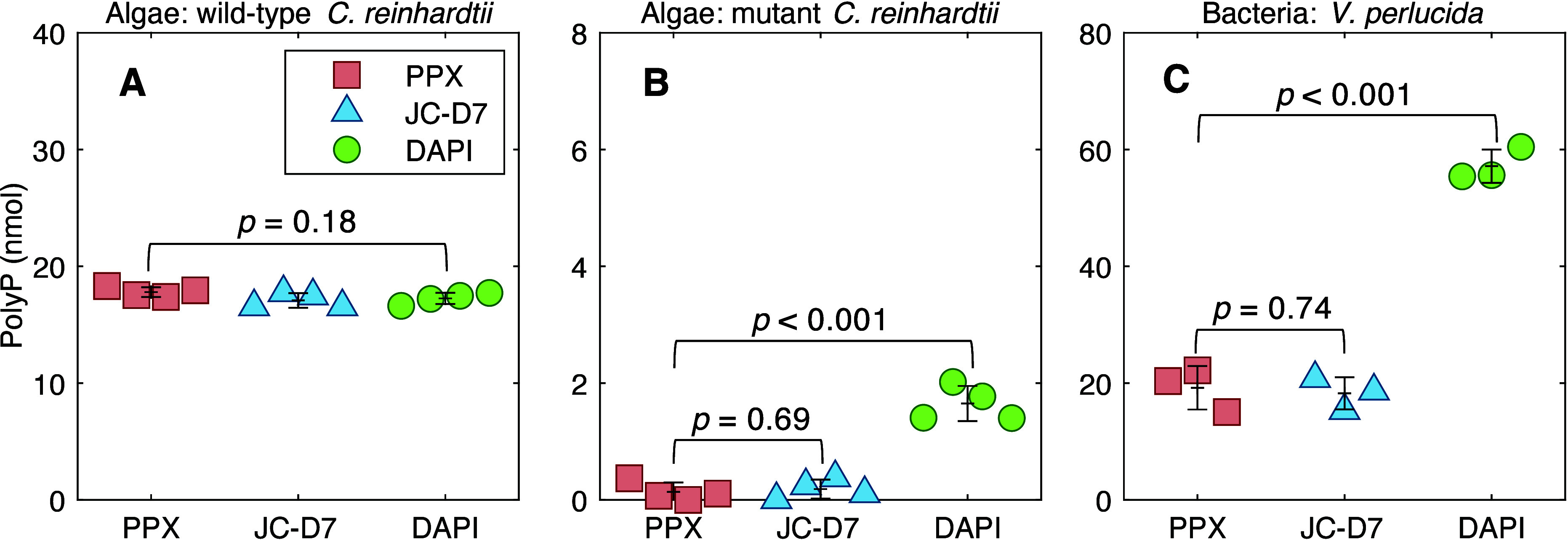
Comparison of polyP quantification using PPX, DAPI, and JC-D7. PolyP was extracted using the phenol-chloroform method from cultures of (A) a wild-type strain of Chlamydomonas reinhardtii (polyP accumulating) (∼5 × 105 cells), (B) a mutant strain of Chlamydomonas reinhardtii (lacking the ability to accumulate polyP33) (∼5 × 105 cells), and (C) heterotrophic bacterium Vogesella perlucida (∼5 × 106 cells). The results are the amount of polyP (nmol) in 1 mL of culture samples. Cross markers indicate average values, and error bars represent the standard deviations from the means. p values for ANOVA tests are shown in the figure.
While the PPX method has been used in other model organisms, including the freshwater algae C. reinhardtii (see our results Figure 2 and Figure S2), it has never been reported to be used for natural environments and seawater plankton cultures. We tested several environmental samples from various freshwater and marine ecosystems, as well as marine cultures, by using PPX to quantify polyP (extracted by the phenol-chloroform method). However, all the tests failed, producing negative or inconsistent results. We speculate that the enzyme PPX is sensitive to the complex composition of the lysates of environmental samples and saline cultures. On the other hand, the JC-D7 quantification method and DAPI method (after >6 h of DNA and RNA removal) can produce consistent measurements for our marine algae samples. It is very unlikely that both JC-D7 and DAPI have overestimated the same amount of polyP by staining some unknown compounds, in which case the produced fluorescence intensities by JC-D7 and DAPI staining these compounds would need to be exactly proportional to those produced by their binding with polyP, making such a scenario nearly impossible. This suggests that JC-D7 fluorescence can be used to reliably quantify polyP.
Effects of Plankton Lysis Buffer Composition on PolyP-JC-D7 Fluorescence
Our tests show that the designed plankton lysis buffer (see Methods) may decrease the sensitivity of the polyP-JC-D7 fluorescence response by ∼16% (Figure 3A), but the detection limit and the linear range are not affected (detection limit ∼0.5 μmol L–1·and linear range ∼0.5–30 μmol L–1). The detection limit can be lowered to about 0.2 μmol L–1, although sensitivity is reduced at this lower value (Figure 3B). Therefore, the polyP extraction using our designed plankton lysis buffer is suitable for subsequent quantification of polyP using JC-D7 fluorescence.
Figure 3.
Responses of JC-D7-polyP fluorescence intensity (Fluo) to the polyP concentrations under the influence of the plankton lysis solution composition. (A) Responses under the composition of the final designed plankton lysis buffer. The dotted line represents the linear curve compared to that without the lysis composition (dashed gray line). (B) Similar to panel A but at a lower range of polyP concentrations, which still shows a linear response, albeit a less sensitive one. (C) Responses under different levels of NaCl [100, 300, and 500 mmol L–1 (see full compositions in Table S5 for tests 6, 2, and 7, respectively)], suggesting that the NaCl concentration affects the linear response. (D) Effects of Triton X-100, Tween 20, and EDTA when the NaCl concentration remains the same (100 mmol L–1), suggesting that these three components of the lysis solution do not affect the JC-D7-polyP linear calibration.
We further tested how the calibration curve of polyP-JC-D7 fluorescence quantification responds to the variability of the lysis solution composition by using a Taguchi orthogonal array of experiment conditions (see the experimental design in Tables S4–5). This array included four parameters (four components of the lysis solution including Triton X-100, Tween 20, NaCl, and EDTA) and three levels of each parameter (component). Our results suggest that the effect of NaCl is substantial: higher concentrations of NaCl increase the detection limit, decrease the sensitivity, and narrow the linear range (Figure 3C, Figure S3, and Tables S5–8). Meanwhile, the effects of Triton X-100, Tween 20, and EDTA are small within the experimental range (Figure 3D, Figure S3, and Tables S5–8).
To test the possible interference from sulfated polysaccharides,26 we conducted several experiments comparing the fluorescence spectra of chondroitin sulfate and polyP stained by DAPI and JC-D7 (see Table S9). Results show that in Tris buffer, both DAPI and JC-D7 can stain chondroitin sulfate to produce fluorescence signals that are about 1–2 times those from polyP (Figure S4). Our plankton lysis, on the other hand, substantially suppresses the fluorescence of chondroitin sulfate stained by JC-D7 (Figure S4B), leading to much lower signals compared to those of polyP stained by JC-D7 (<3% at the emission of 550 nm; Figure S4C). This is likely an effect of the EDTA in the plankton lysis buffer:43 with the increasing levels of EDTA in the Tris buffer, the fluorescence signal of chondroitin sulfate decreases and eventually becomes negligible (Figure S5).
Plankton Lysing versus Phenol-Chloroform Method for PolyP Extraction
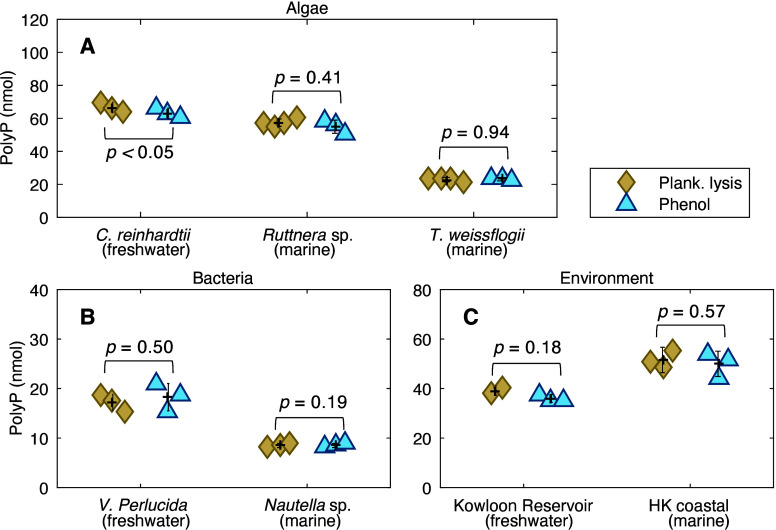
We tested the plankton lysing method against the phenol-chloroform method for polyP extraction, by using a variety of samples including algal cultures of C. reinhardtii (freshwater chlorophyte), Ruttnera sp. (marine haptophyte), and T. weissflogii (marine diatom), along with bacterial cultures of freshwater V. perlucida and marine Nautella sp., as well as freshwater and marine environmental samples. Our results show that the plankton lysis buffer can efficiently extract polyP from all types of planktonic samples (Figure 4). The amount of polyP extracted using the lysing solution is consistent with that obtained using the phenol-chloroform extraction not only for cultured planktonic samples (both algae and bacteria) but also for samples collected from both marine and freshwater aquatic environments (Figure 4).
Figure 4.
Comparison of polyP extracted using plankton lysis buffer and phenol-chloroform extraction in (A) algae, including C. reinhardtii (freshwater chlorophyte, ∼5 × 105 cells), Ruttnera sp. (marine haptophyte, ∼2 × 106 cells), and T. weissflogii (marine diatom, ∼2 × 107 cells), (B) bacteria, including V. perlucida (freshwater, ∼5 × 107 cells) and Nautella sp. (marine, ∼1.2 × 108 cells), and (C) freshwater environmental samples from Kowloon Reservoir (2000 mL of water sample) and marine environmental samples from Hong Kong coastal waters (2000 mL of water sample). The results are the amount of polyP (nanomoles) in 1 mL of culture samples or 2000 mL of environmental samples. For the sake of consistency, polyP quantifications are all conducted using polyP-JC-D7 fluorescence.
Efficiency of PolyP Extraction
Environmental samples typically have low plankton biomass density and need to be concentrated for polyP quantification, for example, by filtering a large volume (0.5–2L) of water through membrane filters.10,13 For laboratory cultures, cell density can become very high, especially when nutrients in the culture medium are abundant. To understand the efficiency of polyP extraction and to suggest a threshold of cell abundance at which the extraction can efficiently extract all polyP, we tested polyP extraction from culture samples across a range of cell abundances (Figure 5). For each test of a gradient of cell abundance, a series of samples were prepared by using dilutions from the same concentrated stock sample (e.g., 1 × 108 cells in 1 mL of samples). The polyP amount estimated in the most diluted samples (e.g., 2 × 105 cells in 1 mL of samples), if consistent among the results given by different methods (plankton lysis-JC-D7, phenol-JC-D7, and phenol-PPX), is considered accurate (100% extraction efficiency). This value can then be used as a reference to calculate the expected polyP amount in the series of more concentrated samples (e.g., 1 × 106, 1 × 107, and 1 × 108 cells in 1 mL of samples) using the appropriate dilution factors. PolyP amounts measured in the concentrated samples are then compared to the expected values to understand the efficiency (recovery) of the extraction (extracted polyP: total polyP). We tested samples prepared using both filtration and centrifugation: the former is commonly applied for environmental samples, and the latter is used more often for culture studies.
Figure 5.
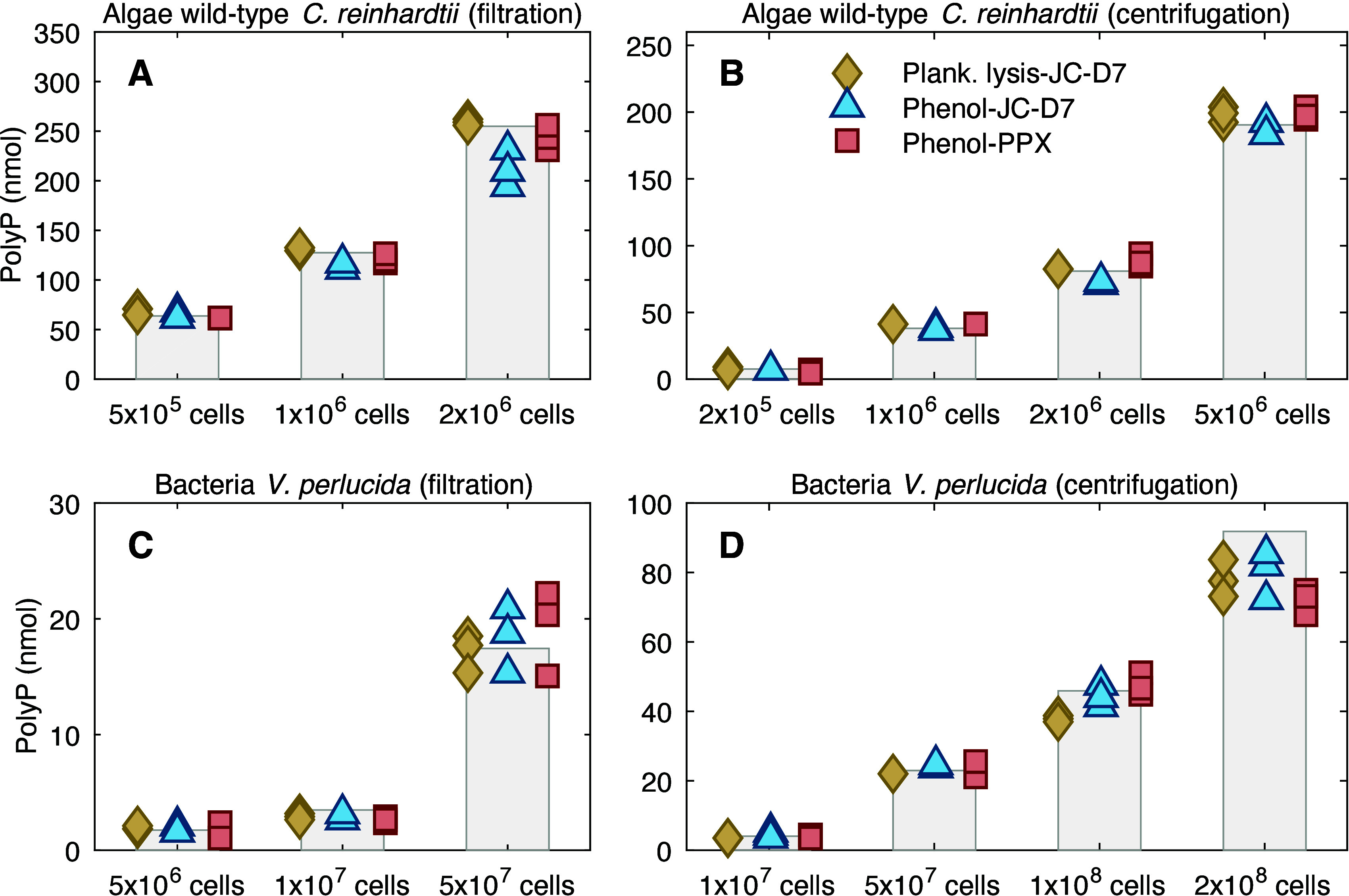
Efficiency of polyP extraction using plankton lysis buffer (plank. lysis) vs phenol-chloroform (phenol) from (A) wild-type C. reinhardtii collected using filtration, (B) wild-type C. reinhardtii collected using centrifugation, (C) V. perlucida collected using filtration, and (D) V. perlucida collected using centrifugation. The results are the amount of polyP (nanomoles) measured in 1 mL of culture samples of various cell quantities. The vertical bars represent the expected polyP amount in the samples (100% polyP), and the markers indicate results estimated using various extraction and quantification methods (e.g., plankton lysis-JC-D7 means polyP is extracted using the plankton lysis buffer and quantified using JC-D7 fluorescence).
Our result suggests that our plankton lysing method can extract polyP from Algae C. reinhardtii of at least 5 × 106 cells (cell quantity), with extraction efficiency comparable to the phenol-chloroform method (Figure 5A and 5B). For bacteria V. perlucida, the plankton lysis extraction can efficiently lyse polyP from up to 5 × 107 cells collected using both filtration (0.2 μm PC membrane) and centrifugation (Figure 5C and 5D). Cell abundance higher than 5 x107 cells leads to less efficient extraction using the plankton lysis buffer (82 ± 2% and 85 ± 6% for 1 × 108 and 2 × 108 cells, respectively; Figure 5D); phenol-chloroform extraction seems to perform better (97 ± 7% efficiency) at cell abundance of 1 × 108, but its efficiency also decreases when cell abundance reaches 1 × 108 (87 ± 7%, comparable to 85 ± 6% for our plankton lysing extraction).
Summary, Recommendations, and Future Work
Tables 2 and 3 summarize the comparisons among different polyP extraction and quantification methods for planktonic samples. The plankton lysing method we developed can efficiently extract polyP from cultured and natural samples across marine and freshwater environments. The efficiency of this extraction is comparable to the phenol-chloroform method but requires much less effort and time. The method also involves fewer toxic chemicals and enhances the reproducibility of results compared to phenol-chloroform extraction. The phenol-chloroform extraction involves the separation of organic and aqueous phases (both in liquid form) in the samples for two rounds, which potentially leads to sample loss, and thus the reproducibility strongly relies on the experience of the experimenter. Our plankton lysing method, on the other hand, involves only a final collection of the lysate, in which polyP is homogeneously dissolved, and thus sample loss would be minimal. Therefore, for environmental samples with limited sample amounts, we recommend our plankton lysing method due to its ease of operation and low risk of sampling loss. For culture samples with sufficient cell abundance, phenol-chloroform extraction can be used, but replicates are recommended to ensure reproducibility.
Table 2. Comparison of PolyP Extraction Methods for Planktonic Samples.
| method | efficiency | time (h) | summary of pros, cons, and recommendations |
|---|---|---|---|
| phenol-chloroform extraction27 | high for <5 × 106 algal cells and <1 × 108 bacterial cells | >8 | pros: high extraction efficiency (at least 5 × 106 cells for algae and 1 × 108 cells for bacteria) |
| cons: time-consuming and labor intensive, involves toxic chemicals, high risk of losing samples during the separation of aqueous and organic phases, reproducibility relies on the experience of the experimenter | |||
| recommendations: extractions of replicate samples to ensure reproducibility | |||
| plankton lysis buffer (this study) | high for <5 × 106 algal cells and <5 × 107 bacterial cells | 2.5 | pros: high extraction efficiency (at least 5 × 106 cells for algae and 5 × 107 cells for bacteria), easy operation and high reproductivity |
| cons: becomes less efficient when used for lysing bacterial pellets (collected via centrifugation) with >5 × 107 cells | |||
| recommendations: vigorous mixing during the extraction when working with cultured bacteria pellets |
Table 3. Comparison of PolyP Quantification Methods for Planktonic Samples.
| method | applicable samples | summary of pros, cons, and recommendations |
|---|---|---|
| DAPI fluorescence22 | all samples, cultured and environmental, freshwater and marine | pros: sensitive, easy operation, has been commonly used in environmental samples |
| cons: high risk of overestimation due to interferences | ||
| recommendations: should not be used as the sole method for polyP quantification | ||
| PPX digestion27 | only freshwater cultures | pros: has been commonly used for various model unicellular organisms. |
| cons: the activity of the enzyme in samples can be difficult to control and inhibitory molecules may cause interference, not working in marine plankton cultures and environmental samples | ||
| recommendations: not recommended for environmental samples and marine cultures, unless other methods are also used to validate the results | ||
| JC-D7 fluorescence (this study) | all samples, cultured and environmental, freshwater and marine | pros: sensitive, easy operation, stable, and can work in various extractants and buffers; polyP specific and has no nucleic acid interference |
| cons: has not been widely used (the dye is newly found). | ||
| recommendations: if lysing agents other than those recommended in this paper are used, the fluorescence response (linearity and detection limit) should be tested, especially for lysing reagents with high salt contents |
Our polyP quantification method using polyP-JC-D7 fluorescence is as convenient and sensitive as the commonly used DAPI method for environmental samples (Table 3), but it is better because the quantification is not affected by interferences as the DAPI method is. DAPI quantification can be used for comparison, but they are not recommended as the sole method due to the high risks of overestimation; overnight treatment with sufficient DNase and RNase is recommended but does not guarantee to remove all interference. The PPX method, combined with the phenol-chloroform extraction, can provide accurate measurements for freshwater cultures (both bacteria and algae). However, it fails for marine cultures and environmental samples due to the complicated lysate components that decrease the activity of the enzyme. Thus, the PPX method should only be used for freshwater cultured samples, and we recommend using at least one other method (e.g., JC-D7 fluorescence quantification) for verification. Therefore, for quantifying polyP in all planktonic samples (algae and bacteria, freshwater and marine), whether extracted using phenol-chloroform or the plankton lysis buffer, we recommend using the new JC-D7 fluorescence method, which is the most stable and reliable compared to others, especially for environmental applications.
Our present knowledge about polyP dynamics in aquatic environments is mostly obtained from polyP measurements using DAPI fluorescence. Future work should use this new method and compare it with the DAPI method, particularly for P-stressed environments where the proportions of polyP to total particulate P were measured to be >100% and thus were only regarded as relative values.12,18 This will help validate the new method and update our knowledge about P cycling in these P-stressed systems where polyP plays vital roles.12,13 How the method performs on more complex environmental samples, for example, sediments44 and solid polyP granules,9,45 should be tested. Possible interferences of JC-D7-polyP fluorescence from other polysaccharides besides chondroitin sulfate should also be investigated.
In summary, our method resolves issues of interference, avoids the high risk of polyP overestimation associated with previous methods, and provides an efficient, convenient, and accurate means of quantifying polyP in planktonic samples in both culture and natural systems. The application of this method across a wide range of aquatic environments will facilitate a deeper understanding of the metabolic functions of polyP and its roles in ecology, biogeochemical cycling, and environmental management.
Acknowledgments
The work is substantially supported by grants from the Research Grant Council (RGC) of the Hong Kong Special Administrative Region, China (Project No. 26305621 and 16303022 to JL and 26304723 to QZ). The work is also funded by the Center for Ocean Research in Hong Kong and Macau (CORE). CORE is a joint ocean research center between Laoshan Laboratory and the Hong Kong University of Science and Technology (HKUST). We thank Jing Sun and Xiaotian Zhou (HKUST) for their assistance in the collection and preparation of environmental samples. We thank Toshikazu Shiba (RegeneTiss, Inc) for providing polyP standards. Comments and suggestions from three anonymous reviewers have helped improve the paper.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.4c04545.
Experimental and method details for verification of polyP concentrations in standard solutions, preparation of exopolyphosphate (PPX), and quantification of PO43– after PPX degradation of polyP; polyP calibration curves of various chain lengths of polyP and under various levels of JC-D7; comparison of polyP quantification using PPX, DAPI, and JC-D7; and effects of lysis solution composition on polyP quantification and interference from chondroitin sulfate (PDF)
Author Contributions
X.Y. and J.L. designed the study with insights from R.G., Q.Z., and C.C.M.Y. X.Y. and R.G. conducted the sampling, culture experiments, and method testing. X.Y. and J.L. wrote the first draft of the paper, and the other co-authors helped to edit and revise the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Rao N. N.; Gómez-García M. R.; Kornberg A. Inorganic Polyphosphate: Essential for Growth and Survival. Annu. Rev. Biochem. 2009, 78 (1), 605–647. 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Powell N.; Shilton A. N.; Pratt S.; Chisti Y. Factors Influencing Luxury Uptake of Phosphorus by Microalgae in Waste Stabilization Ponds. Environ. Sci. Technol. 2008, 42 (16), 5958–5962. 10.1021/es703118s. [DOI] [PubMed] [Google Scholar]
- Gao H.; Liu M.; Griffin J. S.; Xu L.; Xiang D.; Scherson Y. D.; Liu W.-T.; Wells G. F. Complete Nutrient Removal Coupled to Nitrous Oxide Production as a Bioenergy Source by Denitrifying Polyphosphate-Accumulating Organisms. Environ. Sci. Technol. 2017, 51 (8), 4531–4540. 10.1021/acs.est.6b04896. [DOI] [PubMed] [Google Scholar]
- Li Y.; Rahman S. M.; Li G.; Fowle W.; Nielsen P. H.; Gu A. Z. The Composition and Implications of Polyphosphate-Metal in Enhanced Biological Phosphorus Removal Systems. Environ. Sci. Technol. 2019, 53 (3), 1536–1544. 10.1021/acs.est.8b06827. [DOI] [PubMed] [Google Scholar]
- Majed N.; Matthäus C.; Diem M.; Gu A. Z. Evaluation of Intracellular Polyphosphate Dynamics in Enhanced Biological Phosphorus Removal Process Using Raman Microscopy. Environ. Sci. Technol. 2009, 43 (14), 5436–5442. 10.1021/es900251n. [DOI] [PubMed] [Google Scholar]
- Huang R.; Wan B.; Hultz M.; Diaz J. M.; Tang Y. Phosphatase-Mediated Hydrolysis of Linear Polyphosphates. Environ. Sci. Technol. 2018, 52 (3), 1183–1190. 10.1021/acs.est.7b04553. [DOI] [PubMed] [Google Scholar]
- Saia S. M.; Carrick H. J.; Buda A. R.; Regan J. M.; Walter M. T. Critical Review of Polyphosphate and Polyphosphate Accumulating Organisms for Agricultural Water Quality Management. Environ. Sci. Technol. 2021, 55 (5), 2722–2742. 10.1021/acs.est.0c03566. [DOI] [PubMed] [Google Scholar]
- Ray K.; Mukherjee C.; Ghosh A. N. A Way to Curb Phosphorus Toxicity in the Environment: Use of Polyphosphate Reservoir of Cyanobacteria and Microalga as a Safe Alternative Phosphorus Biofertilizer for Indian Agriculture. Environ. Sci. Technol. 2013, 47 (20), 11378–11379. 10.1021/es403057c. [DOI] [PubMed] [Google Scholar]
- Li J.; Dittrich M. Dynamic Polyphosphate Metabolism in Cyanobacteria Responding to Phosphorus Availability. Environ. Microbiol 2019, 21 (2), 572–583. 10.1111/1462-2920.14488. [DOI] [PubMed] [Google Scholar]
- Li J.; Plouchart D.; Zastepa A.; Dittrich M. Picoplankton Accumulate and Recycle Polyphosphate to Support High Primary Productivity in Coastal Lake Ontario. Sci. Rep. 2019, 9 (1), 19563. 10.1038/s41598-019-56042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouviez M.; Oliveira da Rocha C. S.; Guieysse B. Intracellular Polyphosphate Is a P Reserve in Chlamydomonas Reinhardtii. Algal Res. 2022, 66, 102779 10.1016/j.algal.2022.102779. [DOI] [Google Scholar]
- Martin P.; Dyhrman S. T.; Lomas M. W.; Poulton N. J.; vAN Mooy B. A. S. Accumulation and Enhanced Cycling of Polyphosphate by Sargasso Sea Plankton in Response to Low Phosphorus. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (22), 8089–8094. 10.1073/pnas.1321719111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Gao R.; Huff A.; Katsev S.; Ozersky T.; Li J. Polyphosphate Phosphorus in the Great Lakes. Limnol. Oceanogr. Lett. 2024, 10.1002/lol2.10394. [DOI] [Google Scholar]
- Diaz J. M.; Ingall E. D.; Snow S. D.; Benitez-Nelson C. R.; Taillefert M.; Brandes J. A. Potential Role of Inorganic Polyphosphate in the Cycling of Phosphorus within the Hypoxic Water Column of Effingham Inlet, British Columbia. Global Biogeochem. Cycles 2012, 26 (2), n/a. 10.1029/2011GB004226. [DOI] [Google Scholar]
- Jentzsch L.; Grossart H.-P.; Plewe S.; Schulze-Makuch D.; Goldhammer T. Response of Cyanobacterial Mats to Ambient Phosphate Fluctuations: Phosphorus Cycling, Polyphosphate Accumulation and Stoichiometric Flexibility. ISME Commun. 2023, 3 (1), 6. 10.1038/s43705-023-00215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce N. J. T.; Parsons C. T.; Pomfret S. M.; Yates A. G. Periphyton Phosphorus Uptake in Response to Dynamic Concentrations in Streams: Assimilation and Changes to Intracellular Speciation. Environ. Sci. Technol. 2023, 57 (11), 4643–4655. 10.1021/acs.est.2c06285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecky R. E.; Kilham P. Nutrient Limitation of Phytoplankton in Freshwater and Marine Environments: A Review of Recent Evidence on the Effects of Enrichment. Limnol. Oceanogr. 1988, 33, 796–822. 10.4319/lo.1988.33.4part2.0796. [DOI] [Google Scholar]
- Hashihama F.; Saito H.; Shiozaki T.; Ehama M.; Suwa S.; Sugiyama T.; Kato H.; Kanda J.; Sato M.; Kodama T.; Yamaguchi T.; Horii S.; Tanita I.; Takino S.; Takahashi K.; Ogawa H.; Boyd P. W.; Furuya K. Biogeochemical Controls of Particulate Phosphorus Distribution Across the Oligotrophic Subtropical Pacific Ocean. Global Biogeochem Cy 2020, 34 (9), e2020GB006669. 10.1029/2020GB006669. [DOI] [Google Scholar]
- Martin P.; Lauro F. M.; Sarkar A.; Goodkin N.; Prakash S.; Vinayachandran P. N. Particulate Polyphosphate and Alkaline Phosphatase Activity across a Latitudinal Transect in the Tropical Indian Ocean. Limnol Oceanogr 2018, 63 (3), 1395–1406. 10.1002/lno.10780. [DOI] [Google Scholar]
- Diaz J. M.; Björkman K. M.; Haley S. T.; Ingall E. D.; Karl D. M.; Longo A. F.; Dyhrman S. T. Polyphosphate Dynamics at Station ALOHA, North Pacific Subtropical Gyre. Limnol Oceanogr 2016, 61 (1), 227–239. 10.1002/lno.10206. [DOI] [Google Scholar]
- Orchard E. D.; Benitez-Nelson C. R.; Pellechia P. J.; Lomas M. W.; Dyhrman S. T. Polyphosphate in Trichodesmium from the Low-phosphorus Sargasso Sea. Limnol Oceanogr 2010, 55 (5), 2161–2169. 10.4319/lo.2010.55.5.2161. [DOI] [Google Scholar]
- Martin P.; Van Mooy B. A. S. Fluorometric Quantification of Polyphosphate in Environmental Plankton Samples: Extraction Protocols, Matrix Effects, and Nucleic Acid Interference. Appl. Environ. Microbiol. 2013, 79 (1), 273–281. 10.1128/AEM.02592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J. M.; Ingall E. D. Fluorometric Quantification of Natural Inorganic Polyphosphate. Environ. Sci. Technol. 2010, 44 (12), 4665–4671. 10.1021/es100191h. [DOI] [PubMed] [Google Scholar]
- Aschar-Sobbi R.; Abramov A. Y.; Diao C.; Kargacin M. E.; Kargacin G. J.; French R. J.; Pavlov E. High Sensitivity, Quantitative Measurements of Polyphosphate Using a New DAPI-Based Approach. J. Fluoresc 2008, 18 (5), 859–866. 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- Lapointe A.; Spiteller D.; Kroth P. G. High Throughput Method for Extracting Polyphosphates from Diatoms. Endocytobiosis and Cell Research 2022, 31, 29–38. [Google Scholar]
- Lee W. D.; Gawri R.; Shiba T.; Ji A.-R.; Stanford W. L.; Kandel R. A. Simple Silica Column–Based Method to Quantify Inorganic Polyphosphates in Cartilage and Other Tissues. CARTILAGE 2018, 9 (4), 417–427. 10.1177/1947603517690856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru S.; Jiménez J.; Canadell D.; Ariño J.; Clotet J. Improvement of Biochemical Methods of PolyP Quantification. Microb Cell 2017, 4 (1), 6–15. 10.15698/mic2017.01.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ J. J.; Willbold S.; Blank L. M. Methods for the Analysis of Polyphosphate in the Life Sciences. Anal. Chem. 2020, 92 (6), 4167–4176. 10.1021/acs.analchem.9b05144. [DOI] [PubMed] [Google Scholar]
- Werner T. P.; Amrhein N.; Freimoser F. M. Novel Method for the Quantification of Inorganic Polyphosphate (IPoP) in Saccharomyces Cerevisiae Shows Dependence of IPoP Content on the Growth Phase. Arch. Microbiol. 2005, 184 (2), 129–136. 10.1007/s00203-005-0031-2. [DOI] [PubMed] [Google Scholar]
- Pokhrel A.; Lingo J. C.; Wolschendorf F.; Gray M. J. Assaying for Inorganic Polyphosphate in Bacteria. J. Vis. Exp. 2019, 143. 10.3791/58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudříková S.; Sadowsky A.; Metzger S.; Nedbal L.; Mettler-Altmann T.; Mojzeš P. Quantification of Polyphosphate in Microalgae by Raman Microscopy and by a Reference Enzymatic Assay. Anal. Chem. 2017, 89 (22), 12006–12013. 10.1021/acs.analchem.7b02393. [DOI] [PubMed] [Google Scholar]
- Kulakova A. N.; Hobbs D.; Smithen M.; Pavlov E.; Gilbert J. A.; Quinn J. P.; McGrath J. W. Direct Quantification of Inorganic Polyphosphate in Microbial Cells Using 4′-6-Diamidino-2-Phenylindole (DAPI). Environ. Sci. Technol. 2011, 45 (18), 7799–7803. 10.1021/es201123r. [DOI] [PubMed] [Google Scholar]
- Aksoy M.; Pootakham W.; Grossman A. R. Critical Function of a Chlamydomonas Reinhardtii Putative Polyphosphate Polymerase Subunit during Nutrient Deprivation. Plant Cell 2014, 26 (10), 4214–4229. 10.1105/tpc.114.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzumura M. Persulfate Chemical Wet Oxidation Method for the Determination of Particulate Phosphorus in Comparison with a High-Temperature Dry Combustion Method: Particulate Phosphorus Determination. Limnol. Oceanogr.: Methods 2008, 6 (11), 619–629. 10.4319/lom.2008.6.619. [DOI] [Google Scholar]
- Grasshoff K.; Kremling K.; Ehrhardt M. In Methods of Seawater Analysis; Grasshoff K., Kremling K., Ehrhardt M., Eds.; 1999. 10.1002/9783527613984 [DOI] [Google Scholar]
- Carter S. G.; Karl D. W. Inorganic Phosphate Assay with Malachite Green: An Improvement and Evaluation. J. Biochem. Biophys. Methods 1982, 7 (1), 7–13. 10.1016/0165-022X(82)90031-8. [DOI] [PubMed] [Google Scholar]
- Tsugama D.; Liu S.; Takano T. A Rapid Chemical Method for Lysing Arabidopsis Cells for Protein Analysis. Plant Methods 2011, 7 (1), 22–22. 10.1186/1746-4811-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehadul Islam M.; Aryasomayajula A.; Selvaganapathy P. R. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines 2017, 8 (3), 83. 10.3390/mi8030083. [DOI] [Google Scholar]
- Filloramo G. V.; Curtis B. A.; Blanche E.; Archibald J. M. Re-Examination of Two Diatom Reference Genomes Using Long-Read Sequencing. BMC Genom. 2021, 22 (1), 379. 10.1186/s12864-021-07666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova P. R.; Agrawalla B. K.; Elustondo P. A.; Gordon J.; Shiba T.; Abramov A. Y.; Chang Y.-T.; Pavlov E. V. In Situ Investigation of Mammalian Inorganic Polyphosphate Localization Using Novel Selective Fluorescent Probes JC-D7 and JC-D8. ACS Chem. Biol. 2014, 9 (9), 2101–2110. 10.1021/cb5000696. [DOI] [PubMed] [Google Scholar]
- Olson B. J. S. C.; Markwell J. Assays for Determination of Protein Concentration. Curr. Protoc. Protein Sci. 2007, 48 (1), 3.4.1–3.4.29. 10.1002/0471140864.ps0304s48. [DOI] [PubMed] [Google Scholar]
- Rier S. T.; Kinek K. C.; Hay S. E.; Francoeur S. N. Polyphosphate Plays a Vital Role in the Phosphorus Dynamics of Stream Periphyton. Freshw Sci. 2016, 35 (2), 490–502. 10.1086/685859. [DOI] [Google Scholar]
- Smith S. A.; Morrissey J. H. Sensitive Fluorescence Detection of Polyphosphate in Polyacrylamide Gels Using 4′,6-diamidino-2-phenylindol. ELECTROPHORESIS 2007, 28 (19), 3461–3465. 10.1002/elps.200700041. [DOI] [PubMed] [Google Scholar]
- Hupfer M.; Gloess S.; Grossart H. Polyphosphate-Accumulating Microorganisms in Aquatic Sediments. Aquat Microb Ecol 2007, 47, 299–311. 10.3354/ame047299. [DOI] [Google Scholar]
- Racki L. R.; Tocheva E. I.; Dieterle M. G.; Sullivan M. C.; Jensen G. J.; Newman D. K. Polyphosphate Granule Biogenesis Is Temporally and Functionally Tied to Cell Cycle Exit during Starvation in Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (12), E2440–E2449. 10.1073/pnas.1615575114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.