Abstract
Workflow managers play a critical role in the efficient planning and execution of complex workloads. A handful of these already exist within the world of computational materials discovery, but their dynamic capabilities are somewhat lacking. The PerQueue workflow manager is the answer to this need. By utilizing modular and dynamic building blocks to define a workflow explicitly before starting, PerQueue can give a better overview of the workflow while allowing full flexibility and high dynamism. To exemplify its usage, we present four use cases at different scales within computational materials discovery. These encapsulate high-throughput screening with Density Functional Theory, using active learning to train a Machine-Learning Interatomic Potential with Molecular Dynamics and reusing this potential for kinetic Monte Carlo simulations of extended systems. Lastly, it is used for an active-learning-accelerated image segmentation procedure with a human-in-the-loop.
Flexible and dynamic workflow manager with an emphasis on ease of use and powerful modular workflows.
1. Introduction
Attempting to do something for the first time is always a big undertaking. Whether this “something” is performed by following in the footsteps of others or by forging one's own path toward a working method, a systematic approach can be of great help. Trying a new method a number of times and with varying approaches, one should be able to reach a method encapsulating each step, which makes sense and is manageable to perform repeatedly.
In having discovered and implemented this method, one has found a workflow for the process. This workflow will contain both the order and nature of the actions to perform and the resources, both time and materials, required for each. By examining the overall workflow, one can split it into discrete tasks, each of which might show up multiple times throughout the workflow. These tasks can require different resources but be the same process, e.g., measuring an amount of a material, mixing chemicals in aqueous media, performing a computational single-point calculation, or training a machine learning (ML) model. Tasks can depend on the completion of other tasks, and they can run in parallel with each other. A collection of tasks can also be run over and over until some criterion is satisfied. Of course, it is important that each of these tasks is done correctly, but in the context of workflows, the input, output, required (man)power, and interconnections are far more important.
In summary, most methods can be represented by a workflow, which is a collection of connected (dependent) tasks with input, output and required resources. By completing all the tasks of the workflow, one arrives at the final result.
In our time, computers can be used to some extent in virtually all applications, and in this context, a lot of the book-keeping happening between tasks in a workflow can be performed using the computer itself. This allows one to focus on the tasks themselves, while the computer keeps track of results, inputs, and the order of tasks. Colloquially, a program that is able to keep track of the workflow is called a workflow manager.
Quite a selection of workflow managers exists,1,2 and in the space of computational materials science, AiiDA,3,4 Covalent,5 FireWorks,6 Jobflow,7 and MyQueue8 are already established workflow engines. They are all capable of running vast workflows with multiple workers and differing computational resources. Despite their usability in many situations, it is still hard to build truly dynamic workflows – where the structure changes while running – especially dynamic workflows that contain any sort of cyclicity of tasks. The need for dynamic workflows was determined to be one of the general workflow challenges at the workflow summit of 2022,9 urgently needed for ML/AI integrated workflow.
A particular case is active learning workflows, which have to run the training-selection-labeling until the underlying model is good enough.10 These workflows are hard to conceptualize in these established managers. Additionally, the procedures for setting up dynamic workflows in these managers are not very intuitive since the workflow structure is revealed as the workflow is running, which means that making errors in the construction is quite easy. Together, this results in a relatively high barrier to entry for more complex use cases, where the reliance on purely acyclic dependency graphs requires the user to devise creative ways to implement loops in their workflows.
With this in mind, we have created a different solution in which the user has to specify the structure of the workflow upfront using a small selection of very powerful modular building blocks. This workflow manager is called PerQueue – a contraction of Persistent Queue – and is supposed to be a lightweight self-contained program that runs directly on the resources where it is needed. With the dynamism of workflows within PerQueue, one can overcome the challenge of using acyclic dependency graphs with cyclicity and still uphold the FAIR principle. Supporting shareable dynamic workflows alongside generated databases with necessary metadata allows easier reproduction of the workflow outcome.
2. PerQueue architecture
PerQueue is written in the Python3 programming language, and it is designed to be a data-passing and persistence framework that utilizes the MyQueue8 package to submit jobs to the chosen queuing scheduler. The workflows, including the state and data for each task, are stored in an SQLite3 database that is stored locally to where PerQueue runs from. This means that PerQueue is meant to be installed on the (super)computer from where all its workflows will run, though a clever user will be able to start the jobs on remote systems.
Contrary to its competing workflow managers, dynamic and flexible workflows in PerQueue are defined as if they were not dynamic. Dynamic aspects are instead achieved by defining the kind of dynamism of each step of the workflow as part of its specification. An example is a step that has to run over and over until some condition is met, which is achieved by defining the step as being cyclical. This means that the workflow structure must be determined upfront, instead of being built only through execution. This allows the user to have a clear overview of the workflow already from the point of submission, instead of relying on their own code to correctly and dynamically build the workflow. While this requirement for defining the workflow structure up front might seem to limit flexibility, this is not the case for PerQueue. The flexible nature comes from being able to connect the workflow steps in a very fluid manner and from being able to call any code from inside Python, even other executables. The dynamic aspect comes from some of the workflow pieces defined within PerQueue, which are described later.
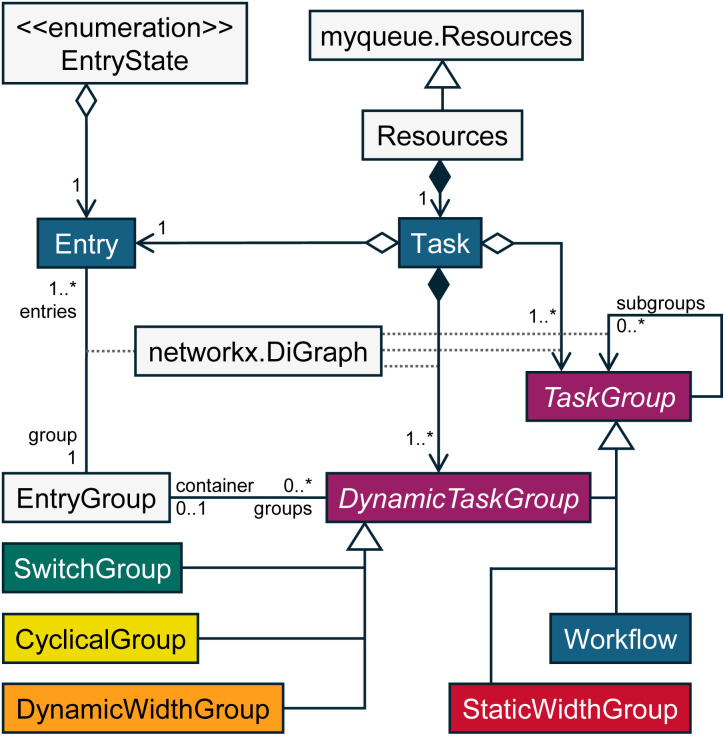
In the following sections, we explain the pieces that make up PerQueue. To guide the reader, the UML Class diagram in Fig. 1 gives a useful overview of the relations between these pieces.
Fig. 1. A UML Class diagram for the user-oriented classes of the PerQueue package. This describes the relations between different classes and visualizes how TaskGroups can contain both Tasks and other TaskGroups. Users only interact with the classes with colored boxes, where the rest are for the internals of PerQueue.
2.1. Architecture basics
In PerQueue, the workflow itself consists of single pieces of work, each denoted as a task and represented by a Task object. For creation, these require the path to the python script to run (enforced to exist), its (known) arguments and the allocated resources. Additionally, a custom name can be given to each Task.
A singular task can be submitted to PerQueue on its own, but the power of PerQueue only really shows when a collection of these are connected within a Workflow. Dependency linking is done by representing the workflow as a directed acyclic graph (DAG), where the roots denote the tasks that are started immediately, while the children only start when their parent tasks have succeeded.
One of the strengths of PerQueue is the level of abstraction that exists with workflow creation. The Workflow object can contain not only Task objects but also other Workflow objects, such that sub-workflows can be specified and reused without incurring memory overheads or code duplication.
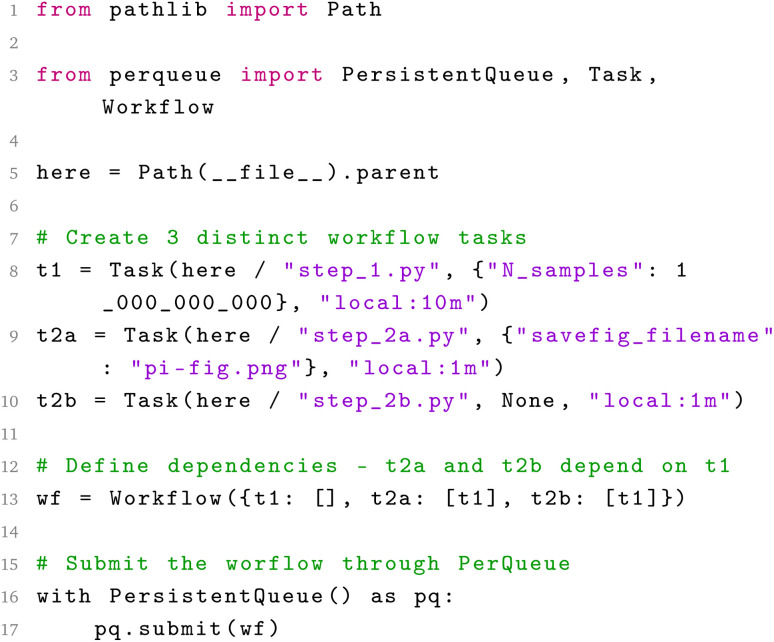
Submitting workflows to PerQueue is achieved by running a python script like the one shown in Chart 1. Once a Workflow or Task is submitted to the PerQueue manager, the manager runs a static compilation of the workflow, where sub-workflows are unpacked to result in as flat a structure as possible. Thereafter, all the unique Tasks are committed to the database together with all DynamicTaskGroups, described below.
Chart 1. Submitting a simple two-layer workflow with PerQueue, where two separate tasks depend on the same task, can be done by running this script with Python. This script checks that the 3 python scripts (“step_*.py”) exist and then runs them in the correct order as subprocesses. The arguments dictionaries are serialized with JSON and saved in the backing database. The code script for each “step_*.py” can be found in the ESI.†.
The manager then finds all the Tasks that have no dependencies or whose dependencies have been completed. It creates an Entry for each of these and sends the entry to its specified scheduler to run and get the result.
An Entry keeps track of a few things, most importantly which Task to execute, its current state, its PerQueue specific arguments, described later, and once completed, the data that the script returned. Unless inserted as arguments or returned as data, other inputs to and outputs of an Entry are not stored in the PerQueue database. A powerful feature of PerQueue is that the returned data of Entrys is both stored in the database and automatically forwarded to all subsequent tasks. This ensures a flexible and simple flow of data through any workflow. Moreover, it can ensure reproducibility, if the user stores all relevant parameters of the task in the output. The arguments and returned data for an Entry is de-/serialized as JSON, which is how the data is stored in the backing database. An extended JSON definition is used to support more datatypes such as NumPy arrays.
2.2. Statically resolved task groups
PerQueue ships with a handful of modular workflow pieces, which simplify the construction of workflows and are used for setting up the dynamic parts of those workflows.
The workflow pieces are collectively denoted by TaskGroup. These are then divided into two groups, that of StaticTaskGroups which each have functionality at workflow submission, and DynamicTaskGroups which each have dynamic behavior. The latter can only be resolved while the workflow is running, since they represent a choice to be made within the workflow.
At the time of workflow submission, PerQueue runs a static compilation, which entails running recursively through the built workflow. Here, it collects all Task objects and resolves all StaticTaskGroup objects by calling their resolve() method. This invocation recursively calls the resolve() method of contained TaskGroups and replaces them in the dependency graph with the new subgraph returned from the method call. What each TaskGroup returns when resolved is described in their subsection. While all StaticTaskGroups can be fully resolved at submission – such is their definition – any DynamicTaskGroup instead resolves its contents and then returns itself. In effect, DynamicTaskGroups are stored in the encapsulating Workflow alongside the Tasks, where they are ready to be processed before being run.
In this way, the static compilation flattens the user-specified workflow down to its simplest possible representation, consisting of only Tasks and DynamicTaskGroups, the latter of which contains only Tasks and possibly other DynamicTaskGroups.
This ensures that the workflow that PerQueue has to act on is as simple as possible, while the setup created from the user's perspective can be as flexible and complex as they want.
2.2.1. Workflow
The Workflow object is used for gathering tasks into a connected collection – the intended workflow. This means that the dependencies between tasks are defined within this object. A restriction here is that the tasks must form a DAG, which simply means that there can be no connections back to earlier tasks – any task must depend on only tasks that are not (eventual) dependencies of itself. This lack of cyclicity is necessary for static compilation, but it can easily be introduced with the use of a CyclicalGroup, which is described later.
The secondary job of the Workflow object is to create sub-workflows since these can be given to other TaskGroups, or simply be used for grouping tasks together. Due to the static compilation, this incurs no memory overhead, and since the sub-workflows are flattened by the compilation, it is very useful for workflow setup. When resolved, a Workflow returns its own dependency graph.
2.2.2. StaticWidthGroup
In cases where the workflow requires doing the same actions to a collection of data, where the only difference between the actions is their input, one can make use of a StaticWidthGroup (SWG). This takes either a single Task or a Workflow as its sub-workflow. The SWG will automatically create a number of parallel sub-workflows – the number given at creation as the width. Each sub-workflow, denoted as column, is given an index, such that the code can index back into a data array to get the right input.
When resolved, this group creates the columns described above and returns the disconnected graph generated by this operation, where each Entry contains a piece of data, pq_index, which is the column index of that/those task(s). Contrary to its dynamic counterpart, the DynamicWidthGroup, this group requires that the width is known at the time of workflow submission, which means it is useful for, e.g., triple determination of an experiment or a set number of endpoints for a Nudged Elastic Band (NEB)11 calculation.
2.3. Dynamic task groups
One of the main features of PerQueue is that of dynamic groups of tasks, which are TaskGroups that are resolved during the execution of the workflow itself. This allows one to include logic in the workflow that depends on the results obtained within the workflow itself. These are DynamicTaskGroups, which are the way dynamism is implemented within PerQueue.
Currently, PerQueue contains three types of DynamicTaskGroup. These are the CyclicalGroup, the DynamicWidthGroup and the SwitchGroup, each of which is explained below. What they have in common is that they take an input from their dependencies, which determines their behavior in a known way.
2.3.1. CyclicalGroup
The CyclicalGroup (CG) allows a group of tasks to run over and over again until a given condition is met, and the loop is terminated. This functionality is just like a while-loop with both a hard iteration limit and a break condition for each loop iteration. When created, the group takes a parameter, max_tries, that specifies the maximum number of times the group of tasks can run before PerQueue stops executing the loop. This hard limit is enforced to ensure that the workflow does stop at some point, raising an error, instead of trying forever on something that will possibly never exit. It is possible to effectively disable this limit by setting it to a very large number.
Working with a CG requires that at least one of the final tasks of the group returns the PerQueue constant CYCLICALGROUP_KEY as a boolean – True means stop the loop. If multiple tasks return this value, the loop continues if any of them returns False.
2.3.2. DynamicWidthGroup
The DynamicWidthGroup (DWG) is the dynamic version of the SWG, where the number of columns is determined from the task(s) immediately before the DWG in the workflow. This also means that no workflow can begin with a DWG, since there will be no task to give the required data. To give the width to the group, at least one of the dependency tasks must return the PerQueue constant DYNAMICWIDTHGROUP_KEY with the width as an integer, which must be the same if returned from multiple tasks.
Besides the dynamic nature of the DWG, it has exactly the same functionality as the SWG.
2.3.3. SwitchGroup
The SwitchGroup (SG) functions like a block of if-elseif-else clauses – also known as a switch or match statement – where the execution path is determined by the dependencies. At submission, one defines a dictionary of keys and Task or Workflow values, such that each key corresponds to a specific sub-workflow. A restriction here is that an empty path is not allowed due to how the workflows are stored in the database.
To control which path the workflow takes during runtime, at least one of the dependency tasks must return the PerQueue constant SWITCHGROUP_KEY with the key for the given path. To skip the SG – taking the empty path not allowed at creation – one can set the value of the SWITCHGROUP_KEY to the PerQueue constant SWITCHGROUP_SKIP. This acts as if the path was empty skipping the SG. In this way, the no-operation is still possible.
2.4. Filtering workflows
PerQueue has another capability, which is that it natively implements filtering workflow steps, where each Entry can mark itself as discarded upon finishing. This ensures that none of its dependents get to run since the task itself determined that it did not fulfill its filtering condition.
In this way, workflows in PerQueue can easily reduce resource usage, e.g., high-throughput workflows, where one is interested in performing only the relevant calculations.
3. Use cases
The following sections show the power and flexibility of PerQueue in real-world cases. These are deliberately chosen to be at different scales to illustrate the wide applicability of the workflow manager.
This starts with a workflow for high-throughput screening with filtering steps using Density Functional Theory (DFT). Secondly, a workflow that utilizes active learning to accelerate the training of machine-learning interatomic potentials. Thirdly, a workflow for cluster expansion and (kinetic) Monte Carlo simulations. Lastly, a workflow for active learning for segmenting electrode images with a human annotator is presented. The submission scripts for all four cases are available in the ESI.†
3.1. High-throughput screening for solid-state electrolytes
High-throughput computational screenings serve as robust frameworks for generating novel candidate materials tailored for specific applications.12,13 These screenings begin with the creation of a vast pool of starting systems, which are subsequently subjected to a series of steps that determine whether the material is still a good candidate. This is analogous to a dynamic funnel. Each step operates under defined conditions, allowing qualified systems to progress further along the screening pipeline. As the screening advances, there is an anticipated increase in the computational cost associated with calculations.
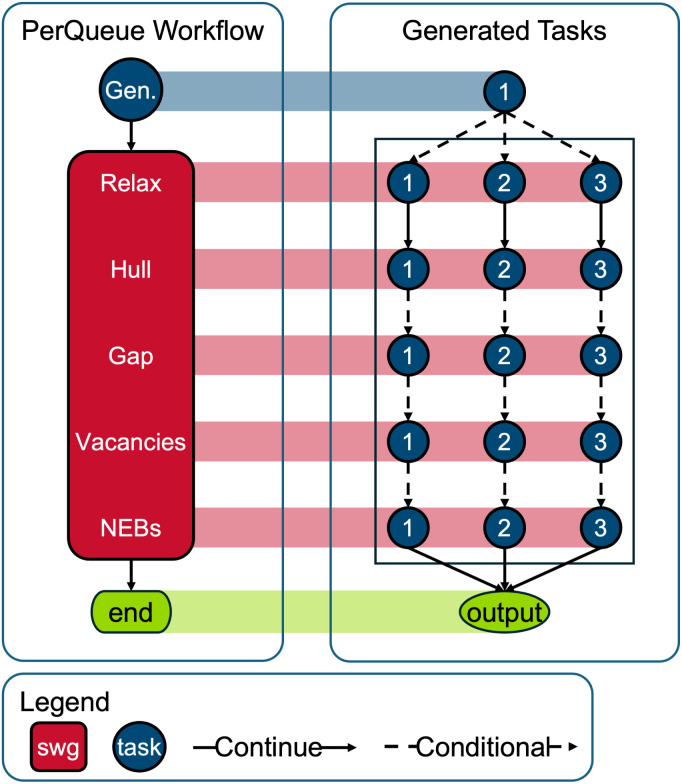
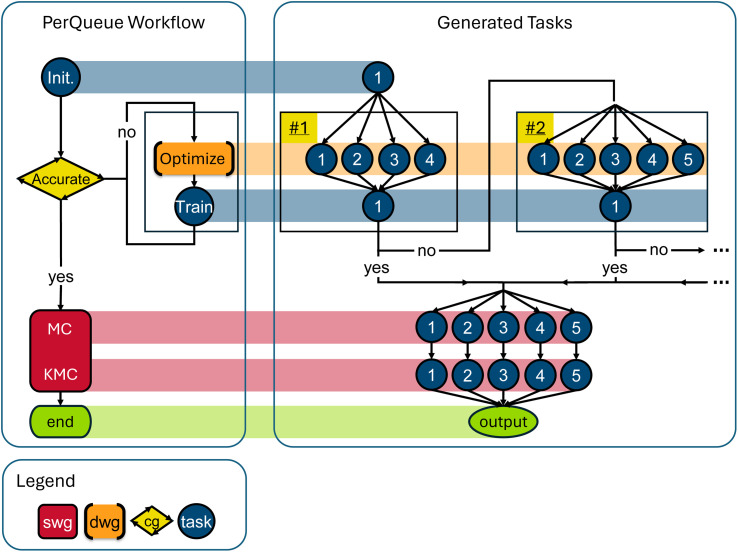
In this context, our objective is to explore a fraction of the vast chemical space in pursuit of adept solid-state electrolyte materials by means of DFT. A SWG is used to run the same workflow for three different vacancy locations in the material for each of the starting systems. At each “Conditional” connection in Fig. 2, that branch of the workflow can decide to stop, using the concept described in Section 2.4. The high-throughput aspect is implemented by creating one of these schematics for each of the thousands of decorations available by using different elements on the atomic positions of a prototype material. Consequently, by the final step, the number of candidate systems is significantly reduced, facilitating the utilization of resource-intensive techniques such as NEBs.
Fig. 2. Schematic of a PerQueue workflow tailored for high-throughput computational screenings. A dashed arrow indicates that the workflow can stop at that step, since a criterion is not upheld. The StaticWidthGroup is used to run the same sub-workflow for three different vacancy locations. The high-throughput aspect is implemented by instantiating this schematic for each of the thousands of decorations.
To streamline this process, a comprehensive workflow manager proves indispensable. Such a manager should not only facilitate the tracking of past and forthcoming steps but also empower users by enabling automated decision-making based on the generated results. PerQueue stands out in this capacity by abstracting complex workflows into simple parallel graphs, whose data passing allows for enhanced traceability through robust provenance tracking. PerQueue efficiently handles multiple parallel instances, essential for conducting high-throughput computational screenings. Moreover, the inclusion of descriptors within the manager's framework aids in identifying deviations from desired criteria. This enables users to sift through non-conforming results and reduce unnecessary workload. Armed with this detailed insight, users can make informed decisions despite the large amount of generated data points, thus optimizing the screening process.
In essence, the synergy between high-throughput computational screenings and advanced workflow management tools like PerQueue marks a significant development in material discovery methodologies. By automating repetitive tasks and providing enhanced decision-making capabilities, these tools not only expedite the screening process but also empower researchers to explore novel material landscapes with greater efficiency and precision.
3.2. Active learning for machine-learning interatomic potentials using molecular dynamics
The use of machine learning in computational chemistry has increased significantly due to its ability to combine the accuracy of ab initio methods with the efficiency of classical force fields.14 The main requirement for these force fields is a high-quality data set. This means additional tasks must be performed before the ML model can be used to calculate properties of the system with traditional computational methods. These extra steps contain data generation, training of the ML model itself, and often relies on active learning which is a machine learning algorithm that iteratively adds more data to the training set which maximally benefits the model, greatly cutting down the total amount of data needed.15 This can lead to complex workflows, e.g., the methodology used in CURATOR16 for efficient data generation, which can be effectively managed by PerQueue.
CURATOR is an autonomous batch active learning workflow for constructing Machine-Learned Interatomic Potentials (MLIPs). The workflow efficiently explores the potential energy surface (PES) to generate training data by employing four steps. First, a MLIP is trained on an initial data set, which is then used to simulate the PES through methods such as Molecular Dynamics (MD). This is done for a given number of configurations, e.g., structures with different vacancy concentrations. Using batch active learning, informative batches from the exploration are selected to calculate the true DFT energy for these images. The images are added to the training set, such that the model can be retrained. This is continued for several iterations, until the trained model reaches the required accuracy.
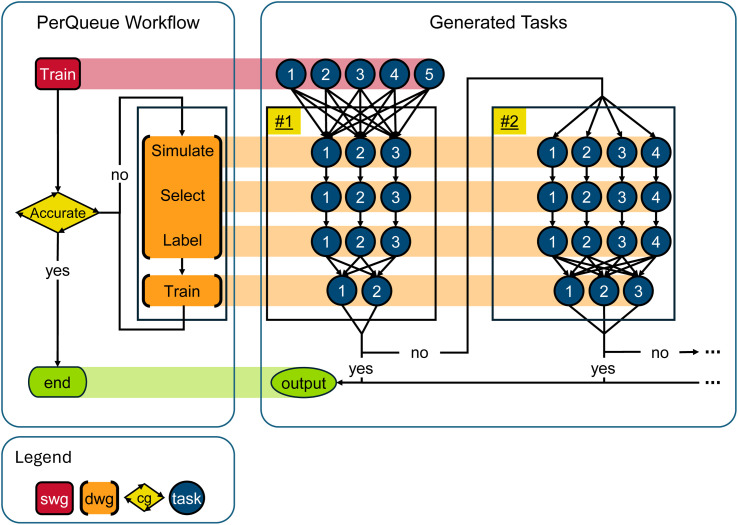
To provide a seamless user experience, these four steps, train-simulate-select-label, can be encapsulated in a workflow using the PerQueue engine (Fig. 3). A priori, it is unknown how many iterations are needed to reach the required accuracy, which the CyclicalGroup solves by starting a new iteration if the accuracy is not reached after the model is trained on the new data. Additionally, it might be beneficial to dynamically change which configurations to explore. For example, if the model is extremely accurate for a given vacancy concentration, little new information is gained by exploring that concentration, so it should be stopped. By using a DynamicWidthGroup for the simulate-select-label steps the exploration is stopped for well-learnt configurations. Finally, it is common to use an ensemble of models with different hyperparameters for the MLIP to provide a measure of uncertainty. By also using a DynamicWidthGroup for the train step, the ensemble size can be updated during the workflow, in case the user wishes so.
Fig. 3. Schematic of a PerQueue workflow for efficient data generation to construct machine-learned interatomic potentials (MLIP). The method involves four steps, train-simulate-select-label, to explore the potential energy surface. These are run continuously, adding new data to the data set at each cycle, which results in a progressively more accurate MLIP. First, a number of models are trained using a StaticWidthGroup giving an ensemble for an initial MLIP. Then a CyclicalGroup is entered, which ends when the MLIP reaches the accuracy criterion. Within the CyclicalGroup, the four steps are implemented as DynamicWidthGroups. Within an iteration the width of simulate-select-label is fixed to the number of configurations, but using DynamicWidthGroups allows dynamically updating the number of configurations and trained models between iterations.
Thus, PerQueue both reduces the work required by the user through autonomous handling of task submission, while also giving complete control and flexibility to the user if the workflow has to be modified while in progress.
3.3. Cluster expansion for Monte Carlo simulations
Following the ML/MD workflow presented above, one can expand this to even larger system sizes. Considering a system of micrometer scale, we would like to find the most stable configuration and model its dynamics. Doing so is possible through either MD or Monte Carlo (MC) simulations.
The most stable configurations of the system are required for MD simulation to model the dynamics, while MC methods can both find the most stable configuration and model the dynamics.17 Using purely ML would be too computationally demanding, due to its nonlinear scaling, which is why this use case employs another method called Cluster Expansion (CE),18 which shows linear scaling. With a trained CE model, each MC will be performed in microseconds, making CE the ideal tool to perform MC simulations of larger systems.
The workflow illustrated in Fig. 4 is used to efficiently obtain a trained CE model. The workflow autonomously trains a CE model for a particular system, only based on the structure file for that system and the chosen CE model parameters. Importantly one can choose what method should be used to perform geometry optimizations. That can either be pure DFT,19 which should be used for smaller sizes, or machine learning interatomic potentials. These MLIPs can either be one like the one described in Section 3.2 (CURATOR16) or a foundation model.20
Fig. 4. Schematic for PerQueue workflow for Cluster Expansion (CE) model training and Monte Carlo (MC) simulation workflow. The workflow is initiated with the system's structure file, CE parameters, MC parameters and employed optimization method – either Density Functional Theory (DFT) or a trained machine learning potential – within a single Task. The next part of the workflow is done with a CyclicalGroup, with geometry optimizations within a DynamicWidthGroup, such that the number of optimizations in each iteration can be varied. Each geometry optimization is used to re-train the CE model. The CyclicalGroup stops when the CE model has reached convergence, defined by the cross-validation score from the training. Following this, a StaticWidthGroup contains MC simulations for low energy structures and kinetic Monte Carlo (kMC) to simulate the dynamics of interest. The group width corresponds to either several permuted systems or a single system.
Using the dynamic nature of PerQueue for this workflow allows for on-the-fly limiting the search space of the CE models training. Implementation-wise, this means that the number of randomly generated structures to optimize during each iteration of a CG, as well as the convergence criteria, can be changed dynamically during model training by using a DWG. The dynamic framework also autonomously changes the method,18 used to generate structures for the training, based on the convergence value. The CE model training method can also be changed for each iteration. This makes the training dynamic, which is needed since a purely autonomous CE model training can be truly difficult. Obtaining an accurately trained CE model initiates the MC simulation, where simulations for one or several systems are run in parallel by employing a SWG. This enables the annealing process to be performed efficiently by minimizing the number of MC steps needed for each temperature. A kinetic Monte Carlo (kMC) simulation, using the low energy structure for a specific temperature from the MC simulation, is then run to simulate the dynamics of the structure based on the diffusion barriers of each movement.21
PerQueue manages all this within a single workflow, orchestrating the workflow autonomously while allowing for on-the-fly human interaction when needed. Thus, PerQueue is able to autonomously and efficiently train a CE model and perform meso-scale simulations.
3.4. Active learning for efficient image segmentation
With 3D imaging techniques such as X-ray nano-holo-tomography, high-resolution images of electrode material microstructures can be obtained.22–24 Being able to quantify these microstructures in terms of the proportions of active and inactive material, their geometric organization, and the development of cracks as the battery is cycled, is important to gain insights into the electrochemical properties of the battery.
To do so, it is necessary to attribute each pixel in a slice to the different phases, referred to as segmentation, with high accuracy. Unfortunately, this process is still reliant on expert annotation. This is time-intensive and slows down research. Luckily, deep learning methods for segmentation have reached high accuracy and present a viable alternative. However, they also require large amounts of data. For this task we use a U-Net architecture and show that a U-Net trained on an extensive dataset can reach human-level accuracy.25
Naturally, the workflow includes alternating steps of model training, selection of new samples to be added to the training set and annotation of those samples by an expert.
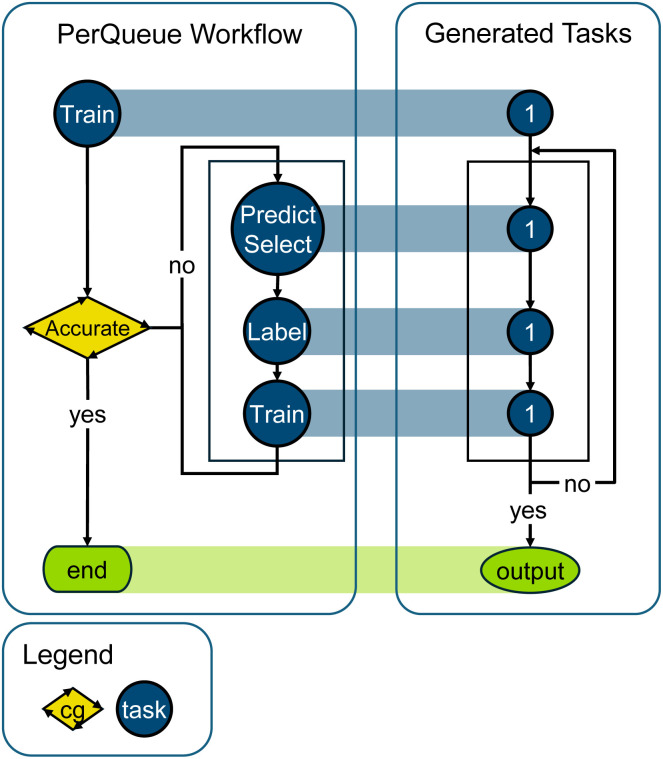
In this process, the deep learning model is initially trained with roughly annotated data. Once the model has converged to a good accuracy on this data, new data points where the model is highly uncertain are chosen to be annotated and added to the training set. The expert annotator then has to annotate the samples, after which training is re-initiated. The training continues until the model has converged and the new accuracy is calculated. The entire workflow is shown in Fig. 5.
Fig. 5. Schematic for the PerQueue workflow for segmentation with active learning. A single Task is responsible for training the segmentation model, and inside a CyclicalGroup, the main process of active learning using the expert annotator for labeling is easily handled by just three single Tasks.
By utilizing a CG, PerQueue allows repeating the process until the model is as accurate as required for the subsequent task without needing to set a fixed number of repetitions in advance. The process alternately requires GPU resources and an action by the expert annotator. By being able to switch between processes, GPU resources can be reserved only when they are needed for training and not in the time intensive step of labeling. If multiple networks are to be trained for an ensemble of neural networks, this can easily be done without needing to restructure the process by simply introducing a SWG and a DWG in the workflow.
Since the annotation is reliant on a human expert, the task called Label would currently involve a waiting loop that checks whether the annotations have been added to the respective folder. If the task were to involve both a human and computational resources, e.g., to utilize a pre-trained model for segmentation with anchor points to accelerate the high-accuracy segmentation, it would be advantageous to have the human task incorporated explicitly in the workflow.26 Similarly, it would be necessary if the human task has an expected waiting time exceeding the maximally allowed process time.
4. Outlook
Beyond the examples shown above, which comprise materials modeling and artificial intelligence, the design of PerQueue is able to leverage the power of Materials Acceleration Platforms (MAPs). MAPs disruptively accelerate the materials discovery by AI-orchestrating theoretical and experimental capabilities from many different sources and sites.27,28 In MAPs, the next experiment or simulation depends on the result of the previous step. This means that, to be efficient, MAPs require a dynamic workflow able to interact with different techniques and to handle data from various sources.29 PerQueue is one of the first examples of this. Among many examples of MAPs, FINALES is a battery problem-agnostic framework for preparation and characterization of electrolytes, cell assembly and testing, early lifetime prediction, and ontologized data storage.30 PerQueue could be used to increase the autonomy of the whole system by having complex workflows run dynamically with minimally required user interaction.
As can be seen from the above sections, PerQueue is already a quite capable piece of software for managing and running flexible and dynamic workflows, but it could be improved further.
One of the things that could still be implemented into the program is the concept of a ManualTask, in which the workflow pauses and waits for user input. This could be very powerful in connection with workflows that contain both computational resources and laboratory or other manual labor, where a human is required to be in the loop. Currently, this can be handled by a waiting loop within the task that only exits when the user gives it some input – the result of the manual task.
Additionally, PerQueue is a young piece of software, and where its competitors can run asynchronously with many service providers to get results from many different places, PerQueue is stuck on its local system, which makes remote calls impractical. Allowing PerQueue to live as a service that can communicate with many providers could be very interesting. At the moment, PerQueue is also very much tailored for single user workflows, so it could use better capabilities for multi-user workflows. In conjunction with this, the scaling capabilities of PerQueue have not been tested yet, so it could be of interest to stress-test the software for its ability to support large-scale concurrency in the future.
5. Conclusions
Here, we have introduced the software PerQueue, which is a new workflow manager written in Python3. PerQueue is a highly flexible workflow manager that focuses deeply on dynamic workflows, whose structure is defined up front to reduce the number of mistakes generally encountered in dynamic workflows. The concept of PerQueue, and the method by how it achieves its goals, is by treating the workflow setup as a building blocks session, where predefined modular blocks connect to make a full workflow.
These building blocks allow PerQueue workflows to exhibit cyclicity, dynamic width of parallel sub-workflow and powerful conditional control, with filtering workflows natively supported.
All this is backed by a local database that stores the state and returned data of all the tasks of the workflow and keeps track of the dependency graphs that prescribe the order of operations.
Having introduced this new piece of software for workflow management, its usage in four different computational use cases has been showcased. These deliberately cover multiple scales encountered within materials discovery. They go from a high-throughput study with DFT over MLIPs trained with active learning for MD and kMC applications within extended systems, and ending with image segmentation of microstructures of synthesized materials accelerated by active learning ML.
In summary, the authors hope that PerQueue will aid multiple researchers in accelerating their work processes in a simple but powerful way, leveraging easily accessible dynamic functionality and data passing between each task.
Data availability
The source code of PerQueue is available as a repository on GitLab (https://gitlab.com/asm-dtu/perqueue). PerQueue is licensed through the GPL-v3 license due to its use of MyQueue.8
Author contributions
Benjamin H. Sjølin: conceptualization, methodology, software, writing – original draft, visualization. William S. Hansen: validation, investigation, writing – original draft, visualization. Armando A. Morin-Martinez: validation, investigation, writing – original draft, visualization. Martin H. Petersen: validation, investigation, writing – original draft, visualization. Laura H. Rieger: investigation, writing – original draft, visualization. Tejs Vegge: resources, supervision, funding acquisition, writing – review & editing. Juan M. G. Lastra: supervision, funding acquisition. Ivano E. Castelli: conceptualization, visualization, supervision, project administration, funding acquisition, writing – review & editing.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
BHS, AAMM, IEC, MHP, and JMGL acknowledge extensive support from the Independent Research Fund Denmark (BHS, IEC: Research Project 1, project “Rational Design of High-Entropy Oxides for Protonic Ceramic Fuel Cells”, HERCULES, grant no. 1032-00269B. AAMM, IEC: Research Project 2, project “Nano-Engineered Solid State Ionic Metal Oxides for Near-Room Temperature Oxygen Conductivity”, NEMO, grant no. 1032-00261B. MHP, JMGL: Research Project 2, project “Data-driven quest for TWh scalable Na-ion battery”, TeraBatt, grant no. 2035-00232B). WSH and JMGL acknowledges that this project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 101054572). LHR acknowledges support from the Energy Technology Development and Demonstration Programme (“ViPES2X: Fully AI-driven Virtual Power Plant for Energy Storage and Power to X”). TV acknowledges funding from the Pioneer Center for Accelerating Materials Discovery (CAPeX), DNRF Grant P3. This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no. 957189 (BIG-MAP). It is also part of BATTERY 2030+ initiative under grant agreement no. 957213.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00134f
References
- Schaarschmidt J. Yuan J. Strunk T. Kondov I. Huber S. P. Pizzi G. Kahle L. Bölle F. T. Castelli I. E. Vegge T. Hanke F. Hickel T. Neugebauer J. Rêgo C. R. C. Wenzel W. Adv. Energy Mater. 2021;12:2102638. doi: 10.1002/aenm.202102638. [DOI] [Google Scholar]
- Amstutz P., Mikheev M., Crusoe M. R., Tijanić N. and Lampa S., Existing workflow systems, Common Workflow Language Wiki, 2024, https://s.apache.org/existing-workflow-systems, accessed July 2024
- Huber S. P. Zoupanos S. Uhrin M. Talirz L. Kahle L. Häuselmann R. Gresch D. Müller T. Yakutovich A. V. Andersen C. W. Ramirez F. F. Adorf C. S. Gargiulo F. Kumbhar S. Passaro E. Johnston C. Merkys A. Cepellotti A. Mounet N. Marzari N. Kozinsky B. Pizzi G. Sci. Data. 2020;7:300. doi: 10.1038/s41597-020-00638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrin M. Huber S. P. Yu J. Marzari N. Pizzi G. Comput. Mater. Sci. 2021;187:110086. doi: 10.1016/j.commatsci.2020.110086. [DOI] [Google Scholar]
- Cunningham W., Esquivel A., Jao C., Sanand S., Hasan F., Bala V., Venkatesh P., Rosen A. S., Tandon M., Ochia O. E., Welsch D., Kanem J., Prabaharan A., Ghukasyan A., Horowitz H., Li R., Neagle S. W., Kostadinov V., Dutta S., Rao P. U., Boltuzic F., Kulkarni U., Hughes A., Gurram R., Mukesh A. and Kashyap A. R., AgnostiqHQ/covalent: v0.232.0, 2024, 10.5281/zenodo.5903364 [DOI]
- Jain A., Ong S. P., Chen W., Medasani B., Qu X., Kocher M., Brafman M., Petretto G., Rignanese G., Hautier G., Gunter D. and Persson K. A., Concurrency and Computation: Practice and Experience, 2015, vol. 27, pp. 5037–5059 [Google Scholar]
- Rosen A. S. Gallant M. George J. Riebesell J. Sahasrabuddhe H. Shen J.-X. Wen M. Evans M. L. Petretto G. Waroquiers D. Rignanese G.-M. Persson K. A. Jain A. Ganose A. M. J. Open Source Softw. 2024;9:5995. doi: 10.21105/joss.05995. [DOI] [Google Scholar]
- Mortensen J. J. Gjerding M. Thygesen K. S. J. Open Source Softw. 2020;5:1844. doi: 10.21105/joss.01844. [DOI] [Google Scholar]
- da Silva R. F., Badia R. M., Bala V., Bard D., Bremer P.-T., Buckley I., Caino-Lores S., Chard K., Goble C., Jha S., Katz D. S., Laney D., Parashar M., Suter F., Tyler N., Uram T., Altintas I., Andersson S., Arndt W., Aznar J., Bader J., Balis B., Blanton C., Braghetto K. R., Brodutch A., Brunk P., Casanova H., Lierta A. C., Chigu J., Coleman T., Collier N., Colonnelli I., Coppens F., Crusoe M., Cunningham W., Kinoshita B. d. P., Di Tommaso P., Doutriaux C., Downton M., Elwasif W., Enders B., Erdmann C., Fahringer T., Figueiredo L., Filgueira R., Foltin M., Fouilloux A., Gadelha L., Gallo A., Saez A. G., Garijo D., Gerlach R., Grant R., Grayson S., Grubel P., Gustafsson J., Hayot-Sasson V., Hernandez O., Hilbrich M., Justine A., Laflotte I., Lehmann F., Luckow A., Luettgau J., Maheshwari K., Matsuda M., Medic D., Mendygral P., Michalewicz M., Nonaka J., Pawlik M., Pottier L., Pouchard L., Putz M., Radha S. K., Ramakrishnan L., Ristov S., Romano P., Rosendo D., Ruefenacht M., Rycerz K., Saurabh N., Savchenko V., Schulz M., Simpson C., Sirvent R., Skluzacek T., Soiland-Reyes S., Souza R., Sukumar S. R., Sun Z., Sussman A., Thain D., Titov M., Tovar B., Tripathy A., Turilli M., Tuznik B., van Dam H., Vivas A., Ward L., Widener P., Wilkinson S., Zawalska J. and Zulfiqar M., Workflows community summit 2022: a roadmap revolution, arXiv, 2023, preprint, arXiv:2304.00019, 10.48550/ARXIV.2304.00019 [DOI]
- Settles B., Active Learning Literature Survey, University of Wisconsin–Madison Computer Sciences Technical Report, 2009, vol. 1648 [Google Scholar]
- Henkelman G. Jónsson H. J. Chem. Phys. 2000;113:9978–9985. doi: 10.1063/1.1323224. [DOI] [Google Scholar]
- Bölle F. T. Mathiesen N. R. Nielsen A. J. Vegge T. Garcia-Lastra J. M. Castelli I. E. Batteries Supercaps. 2020;3:488–498. doi: 10.1002/batt.201900152. [DOI] [Google Scholar]
- Sjølin B. H. Jørgensen P. B. Fedrigucci A. Vegge T. Bhowmik A. Castelli I. E. Batteries Supercaps. 2023;6:e202300041. doi: 10.1002/batt.202300041. [DOI] [Google Scholar]
- Unke O. T. Chmiela S. Sauceda H. E. Gastegger M. Poltavsky I. Schütt K. T. Tkatchenko A. Müller K.-R. Chem. Rev. 2021;121:10142–10186. doi: 10.1021/acs.chemrev.0c01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. Li C. Yao Z. Appl. Sci. 2022;12:8103. doi: 10.3390/app12168103. [DOI] [Google Scholar]
- Yang X., Petersen M. H., Sechi R., Hansen W. S., Norwood S. W., Krishnan Y., Vincent S., Busk J., Cornet F. R. J., Winther O., Garcia Lastra J. M., Vegge T., Hansen H. A. and Bhowmik A., CURATOR: building robust machine learning potentials for atomistic simulations autonomously with batch active learning, ChemRxiv, 2024, preprint, 10.26434/chemrxiv-2024-p5t3l [DOI]
- Zhang X. Sluiter M. H. F. J. Phase Equilib. Diffus. 2015;37:44–52. doi: 10.1007/s11669-015-0427-x. [DOI] [Google Scholar]
- Chang J. H. Kleiven D. Melander M. Akola J. Garcia-Lastra J. M. Vegge T. J. Phys.: Condens. Matter. 2019;31:325901. doi: 10.1088/1361-648X/ab1bbc. [DOI] [PubMed] [Google Scholar]
- Hafner J. J. Comput. Chem. 2008;29:2044–2078. doi: 10.1002/jcc.21057. [DOI] [PubMed] [Google Scholar]
- Batatia I., Benner P., Chiang Y., Elena A. M., Kovács D. P., Riebesell J., Advincula X. R., Asta M., Avaylon M., Baldwin W. J., Berger F., Bernstein N., Bhowmik A., Blau S. M., Cărare V., Darby J. P., De S., Della Pia F., Deringer V. L., Elijošius R., El-Machachi Z., Falcioni F., Fako E., Ferrari A. C., Genreith-Schriever A., George J., Goodall R. E. A., Grey C. P., Grigorev P., Han S., Handley W., Heenen H. H., Hermansson K., Holm C., Jaafar J., Hofmann S., Jakob K. S., Jung H., Kapil V., Kaplan A. D., Karimitari N., Kermode J. R., Kroupa N., Kullgren J., Kuner M. C., Kuryla D., Liepuoniute G., Margraf J. T., Magdău I.-B., Michaelides A., Moore J. H., Naik A. A., Niblett S. P., Norwood S. W., O'Neill N., Ortner C., Persson K. A., Reuter K., Rosen A. S., Schaaf L. L., Schran C., Shi B. X., Sivonxay E., Stenczel T. K., Svahn V., Sutton C., Swinburne T. D., Tilly J., van der Oord C., Varga-Umbrich E., Vegge T., Vondrák M., Wang Y., Witt W. C., Zills F. and Csányi G., A foundation model for atomistic materials chemistry, arXiv, 2024, preprint, arXiv:2401.00096, 10.48550/ARXIV.2401.00096 [DOI]
- Canepa P. ACS Mater. Au. 2022;3:75–82. doi: 10.1021/acsmaterialsau.2c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. Villanova J. Su Z. Tucoulou R. Fleutot B. Delobel B. Delacourt C. Demortière A. Adv. Energy Mater. 2021;11:2003529. doi: 10.1002/aenm.202003529. [DOI] [Google Scholar]
- De Angelis S. Villanova J. Jørgensen P. S. Esposito V. Bowen J. R. Acta Mater. 2024;273:119965. doi: 10.1016/j.actamat.2024.119965. [DOI] [Google Scholar]
- Villanova J. Laurencin J. Cloetens P. Bleuet P. Delette G. Suhonen H. Usseglio-Viretta F. J. Power Sources. 2013;243:841–849. doi: 10.1016/j.jpowsour.2013.06.069. [DOI] [Google Scholar]
- Ronneberger O., Fischer P. and Brox T., Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Springer International Publishing, Cham, 2015, pp. 234–241 [Google Scholar]
- Docherty R., Squires I., Vamvakeros A. and Cooper S. J., SAMBA: a trainable segmentation web-app with smart labelling, arXiv, 2023, preprint, arXiv:2312.04197, 10.48550/ARXIV.2312.04197 [DOI]
- Flores-Leonar M. M. Mejía-Mendoza L. M. Aguilar-Granda A. Sanchez-Lengeling B. Tribukait H. Amador-Bedolla C. Aspuru-Guzik A. Curr. Opin. Green Sustainable Chem. 2020;25:100370. doi: 10.1016/j.cogsc.2020.100370. [DOI] [Google Scholar]
- Stier S., Kreisbeck C., Ihssen H., Popp M. A., Hauch J., Malek K., Reynaud M., Carlsson J., Gold L., Goumans F., Todorov I., Räder A., Bandesha S. T., Wenzel W., Jacques P., Arcelus O., Garcia-Moreno F., Friederich P., Maglione M., Clark S., Laukkanen A., Cabanas M. C., Carrasco J., Castelli I. E., Stein H. S., Vegge T., Nakamae S., Fabrizio M. and Kozdras M., The Significance of Accelerated Discovery of Advanced Materials to Address Societal Challenges, 2023, https://zenodo.org/record/8012140
- Castelli I. E. Arismendi-Arrieta D. J. Bhowmik A. Cekic-Laskovic I. Clark S. Dominko R. Flores E. Flowers J. Ulvskov Frederiksen K. Friis J. Grimaud A. Hansen K. V. Hardwick L. J. Hermansson K. Königer L. Lauritzen H. Le Cras F. Li H. Lyonnard S. Lorrmann H. Marzari N. Niedzicki L. Pizzi G. Rahmanian F. Stein H. Uhrin M. Wenzel W. Winter M. Wölke C. Vegge T. Batteries Supercaps. 2021;4:1803–1812. doi: 10.1002/batt.202100117. [DOI] [Google Scholar]
- Vogler M. Busk J. Hajiyani H. Jørgensen P. B. Safaei N. Castelli I. E. Ramirez F. F. Carlsson J. Pizzi G. Clark S. Hanke F. Bhowmik A. Stein H. S. Matter. 2023;6:2647–2665. doi: 10.1016/j.matt.2023.07.016. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code of PerQueue is available as a repository on GitLab (https://gitlab.com/asm-dtu/perqueue). PerQueue is licensed through the GPL-v3 license due to its use of MyQueue.8