Abstract
Retrons are a retroelement class found in diverse prokaryotes that can be adapted to augment CRISPR-Cas9 genome engineering technology to efficiently rewrite short stretches of genetic information in bacteria and yeast; however, efficiency in human cells has been limited by unknown factors.
We identified non-coding RNA (ncRNA) instability and impaired Cas9 activity as major contributors to poor retron editor efficiency. We re-engineered the Eco1 ncRNA to incorporate an exoribonuclease-resistant RNA pseudoknot from the Zika virus 3’ UTR and devised an RNA processing strategy using Csy4 ribonuclease to liberate the sgRNA and ncRNA. These modifications yielded a ncRNA with 5’- and 3’-end protection and an sgRNA with minimal 5’ extension. This strategy increased steady-state ncRNA levels and rescued Cas9 activity leading to enhanced efficiency of the Eco1 retron editor in human cells. The enhanced Eco1 retron editor enabled the insertion of missense mutations in human cells from a single integrated lentivirus, thereby ensuring genotype-phenotype linkage over multiple cell divisions.
This work reveals a previously unappreciated role for ncRNA stability in retron editor efficiency in human cells. Here we present an enhanced Eco1 retron editor that enables efficient introduction of missense mutations in human cells from a single heritable genome copy.
INTRODUCTION
Germline and somatic genetic variants play an important role in human health and disease1–4. CRISPR-Cas9 technology has revolutionized the manipulation of genetic information by enabling RNA-programmable delivery of enzymatic activities to targeted locations in the genome, facilitating new genome editing technologies that can interrogate the relationship between genotype and phenotype in living systems5. When delivered via lentivirus at a low multiplicity of infection (MOI) to ensure a single integration per cell, linked genotype-phenotype relationships can be maintained over multiple cell divisions and the fate of individually edited cells and their progeny can be tracked. This significantly simplifies the scalable parallelization of forward genetic screens6,7, retrieval of rare clones from a population of cells8–10, and somatic editing in genetically engineered models of cancer11–14.
Of the approaches developed to introduce single nucleotide variants (SNVs), only base editing and prime editing systems have been adapted to lentiviral delivery. Base editors can instill SNVs with high efficiency but require distinct and exquisitely engineered editing enzymes to generate different types of mutations, limiting their application in large-scale screens15. In contrast, prime editors offer a greater potential to efficiently introduce a wide spectrum of mutation types, including small insertions; however, each prime editing guide RNA requires extensive design effort to yield efficient editing, complicating the construction of large pegRNA libraries16–19. There remains a need for additional methods to complement current genome editing tools that have the properties of broad editing potential, predictable activity, and compatibility with lentiviral delivery.
Retrons are prokaryotic retroelements that have been adapted as genome editing tools by harnessing their ability to generate homology-directed repair templates in situ20,21. A retron minimally consists of a non-coding RNA (ncRNA) that is reverse transcribed into multicopy single-stranded DNA (msDNA) by its cognate primer-independent reverse transcriptase22,23. Arbitrary sequences of interest can be inserted into the ncRNA stem loop, where they are reversed transcribed into RT-DNA. Fusion of protein and RNA components of retrons to their respective CRISPR-Cas9 counterparts results in targeted, templated DNA repair24. However, the application of retrons as genome editing tools in human cells has been limited due to low efficiency25–27.
Previous efforts to improve retron editor efficiency have focused on the cellular abundance of RT-DNA26,28. Here we determined that retron ncRNA and sgRNA fusion transcripts are unstable in human cells. These unstable ncRNAs exhibit limited templated editing efficiency and Cas9 endonuclease activity, ultimately resulting in a lack of activity in the context of lentiviral delivery. Through iterative optimization and rational design, we developed a ncRNA architecture derived from the Eco1 (Ec86) retron with enhanced genome editing efficiency, broadening the utility of retrons as genome editing tools in human cells.
MATERIALS AND METHODS
Cell culture.
Parental HEK293T cell lines and their derivatives were cultured in high-glucose DMEM with L-glutamine and sodium pyruvate (Gibco) and supplemented with 10% Foundation™ fetal bovine serum (Gemini Bio-Products). K562 cell lines were grown in RPMI 1640 Medium supplemented with L-glutamine (Gibco) and 10% Foundation™ fetal bovine serum (Gemini Bio-Products). Both HEK293T and K562 parental lines were obtained from ATCC. All cell lines were grown at 37°C and 5% CO2. Huh-7.5 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Fisher Scientific, catalog no. 11995065) supplemented with 0.1 mM nonessential amino acids (NEAAs; Fisher Scientific, catalog no. 11140076) and 10% fetal bovine serum (FBS; HyClone Laboratories, lot. #AUJ35777).
Plasmid cloning.
All plasmids, oligos, and gBlocks used in this study are listed in Supplementary Table 1. Plasmids were transformed and propagated in NEB Stable Competent E. coli (New England Biolabs). All ncRNA expressing plasmids were cloned from lentiGuide-Puro (lentiGuide-Puro was a gift from Feng Zhang, Addgene #52963) using the PacI restriction enzyme cloning sites followed by NEBuilder HiFi DNA Assembly (New England Biolabs) of synthesized gBlock™ gene fragments (Integrated DNA Technologies). Cas9-RTwt expression plasmid was cloned from pBZ210 (pBZ210 was a gift from Hunter Fraser, Addgene #170185) by XbaI and PciI double digestion (New England Biolabs) to excise the chimeric sgRNA-retron ncRNA, followed by blunting with DNA Polymerase I Large Klenow fragment (New England Biolabs). The resulting dsDNA blunt ends were then ligated using a T4 ligase (New England Biolabs). Cas9-RTmut was generated by site-directed mutagenesis. TR-Csy4 and TR-Csy4-H29A were gifts from Aravind Asokan (Addgene #80601, Addgene #80602).
In vitro transcription.
T7 expression plasmids were linearized by digestion with EcoRI (New England Biolabs) and then purified by phenol:chloroform extraction followed by ethanol precipitation. A 5μL in vitro transcription reaction was set up with 250ng of linearized plasmid template, 0.375μL of T7 RNA Polymerase (New England Biolabs) and 100mM each of ATP, GTP, CTP, and UTP and then incubated overnight at 37°C. In vitro transcribed RNA was then purified by phenol:chloroform extraction and ethanol precipitation.
Plasmid transfection and electroporation.
Lipofectamine 3000 was used for all transient transfections of HEK293T-BFP cells. Briefly, 2e5 HEK293T-BFP cells were reverse transfected with 750ng of either Cas9-RTwt or Cas9-RTmut, and 650ng of plasmids expressing the retron editor ncRNA. For all experiments incorporating Csy4 cleavage, 750ng of either Csy4 wild-type or Csy4-H29A was added to the transfection mix, and constructs without a Csy4 recognition site were co-transfected with 750ng pUC19 (New England Biolabs) to balance total transfected mass. Cells were replated in a 12-well plate approximately 16–24 hours after transfection. The Neon NxT Electroporation System was used to electroporate K562-BFP ncRNA-expressing cell lines with Cas9-RT plasmids and one of Csy4, Csy4-H29A, or pUC19 to balance total transfected mass. 5e5 cells were pelleted at 300 RCF for 5 minutes and resuspended in Neon NxT R buffer and then mixed with 2500ng of the Cas9-RT plasmid and 2500ng of TR-Csy4/TR-Csy4-H29A/pUC19 plasmid. Cells were then electroporated using the following parameters in a 10μL Neon NxT tip: 1050V, 20ms pulse width, 2 pulses. Cells were added to 500μL of pre-warmed RPMI media in a 48 well plate after pulsing.
Flow cytometry.
All flow cytometry experiments were done on a BD FACSymphony™ A5 Cell Analyzer. To prepare samples, cells were centrifuged at 350 RCF for 5 minutes and the cell pellets were resuspended in DPBS (Gibco) supplemented with 1% Foundation™ fetal bovine serum (Gemini Bio-Products). Cell populations were analyzed to determine the proportion of BFP and GFP positive cells and each condition was run with three biological replicates. Transfected cells were selected using the mCherry fluorescent reporter on the Cas9-RT expression plasmids. All gates were drawn using untransfected control samples.
Virus production.
T25 flasks were seeded with 2e6 HEK293T cells 24 hours prior to transfection. On the following day, a transfection solution was made up of 200μL OptiMEM (Gibco), 3μg of the lentiviral vector, 2μg of psPAX2, 1μg of pMD2.G, and 12μg of PEI 25K (Polysciences). psPAX2 and pMD2.G were gifts from Didier Trono (Addgene #12260, Addgene #12259). After mixing, the transfection solution was vortexed for 15 seconds, incubated at room temperature for 15 minutes, and added to cells drop wise. The media was replaced 24 hours after transfection. At 72 hours post-transfection the viral supernatant was filtered through a 0.45μm syringe filter and immediately stored at −80°C.
Lentivirus transduction.
The HEK293T-BFP cell line was made by limiting dilution lentivirus transduction of BFP dest clone lentivirus and selected with 6μg/mL blasticidin (Gibco) for one week. BFP dest clone was a gift from Jacob Corn (Addgene #71825). Cells were then single-cell sorted into a 96-well plate for the top 2.5% fluorescent cells. Single-cell clones were expanded and two clonal lines were randomly selected. Low-copy integrated ncRNA HEK293T cell lines were generated by limiting dilution lentivirus transduction. 5e5 cells were plated in a 6-well plate and limiting volumes of viral supernatant were added to each well, ranging from 50μL to 1μL. 24 hours after the addition of virus, the cells were selected using 2μg/mL puromycin (Gibco) and grown out on puromycin for one week. For every HEK293T cell line generated, 1μL of virus was sufficient to yield sparse colonies for subsequent outgrowth. Low-copy integrated ncRNA K562 cell lines were generated by limiting dilution spinfection. Briefly, 1e6 K562 cells were plated in a 6-well plate with 2mL of RPMI 1640 media. After plating, 20μL of lentivirus and 2μL of polybrene (Sigma-Aldrich) were added directly to each well and cells were centrifuged at 930 RCF for 2 hours at room temperature. After centrifugation, 2mL of RPMI 1640 media was added to each well and cells were placed back in the incubator. Selection for infected cells started 24 hours after spinfection using 2μg/mL puromycin. The K562-BFP cell line used in this study was a gift from Dr. Chris Richardson.
Northern blotting.
1e6 HEK293T cells were first reverse transfected in a 6-well plate with 1μg of Cas9-RT (either Cas9-RTwt or Cas9-RTmut) and 1μg of ncRNA expressing plasmid using Lipofectamine 3000 according to the manufacturer’s protocol. Cells were incubated for 48h and then whole-cell RNA was extracted using TRIzol reagent (Invitrogen) followed by two chloroform washes. The aqueous layer was loaded onto a Monarch Total RNA Miniprep Kit (New England Biolabs) column and purified according to the manufacturer’s protocol. To radiolabel ssDNA probes, 0.15mCi ATP [𝛾-32P] (Perkin Elmer) was mixed with 200ng of the probe and T4 kinase (New England Biolabs) and then incubated at 37°C for 60 minutes. Labeled probes were then centrifuged for 30 seconds at 16,000 RCF in a MicroSpin G-50 column (Cytiva) and eluted in 80μL of TNES buffer (50mM Tris, 400mM NaCl, 20mM EDTA, 0.5% SDS). Probe sequences are listed in Supplementary Table 1. For electrophoretic size separation, 25ng of in vitro transcribed RNA and 7.5μg of total whole-cell RNA was loaded per well onto a Novex 10% TBE-Urea 10-well gel (Invitrogen) and run for 5 hours at constant 120V. RNA was then transferred to a Nytran SuPerCharge (Cytiva) nylon membrane using a Bio-Rad Trans-Blot Turbo (Bio-Rad) at 200mA for 60 minutes. Membranes were blocked at 65°C for 1 hour in 30mL of 6X SSC 7% SDS blocking buffer. After blocking, membranes were incubated overnight at 42°C with 30μL of the radiolabeled probe. Membranes were washed and exposed for 48h before imagining. For U6 loading controls, membranes were stripped using 0.1% SDS 2mM EDTA stripping buffer followed by three washes and then reprobed using a U6 loading control probe. Loading control membranes were exposed for 24 hours before imaging.
Statistics and reproducibility.
All experiments were performed in triplicate. Outcomes here are presented as the mean ± standard deviation. For all editing outcome assays, statistical significance was determined by unpaired t tests using the Holm-Šídák method for multiple testing correction. All p-values referred to here in the study are presented as two-sided.
RESULTS
Eco1 retron editor efficiency is inferior to synthetic templates in human cell lines
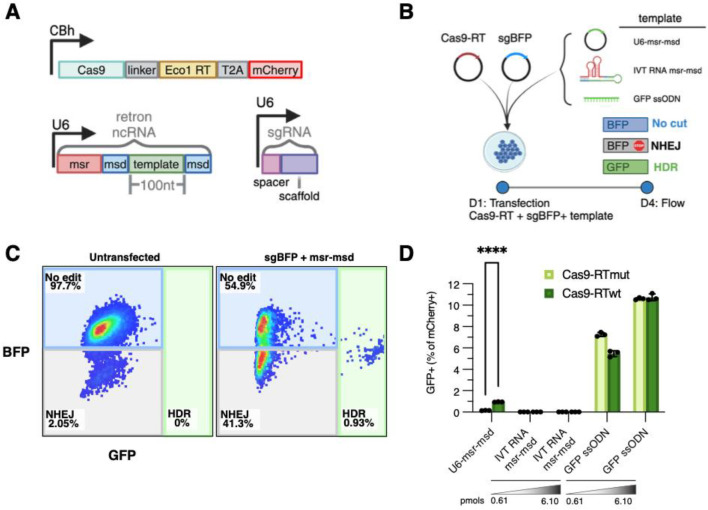
To investigate the potential causes of low retron editor efficiency in human cells, we employed the well-studied Eco1 retron as a model retron for iterative enhancement of genome editing efficiency through rational design. Non-coding RNA plasmids were designed to express the Eco1 ncRNA fused to an sgRNA and included a 100nt repair template internal to the msd sequences as previously described29,30. Retron editor ncRNAs were expressed from the U6 RNAPIII promoter and flanked by a polyT transcription termination signal (Fig. 1A)31. For direct RNA transfection, T7 polymerase in vitro transcribed (IVT) RNA was generated from a plasmid flanked by an HDV ribozyme sequence intended to generate precise 3’ ends. The apparent molecular weight of these IVT RNA products as measured by agarose gel size separation was consistent with full-length transcript (Supplementary Fig. 1). The protein-coding plasmid expresses Cas9 fused to a codon optimized Eco1 reverse transcriptase by a 33 amino acid linker and is followed by a T2A ribosome skipping sequence and mCherry. This construct allows for simultaneous tracking of transfected cells by red-orange fluorescence (Fig. 1A). To control for background levels of plasmid templated repair, we constructed a catalytically dead Eco1 mutant (DD196NN, RTmut)32 and confirmed that the mutant is inactive by qPCR of retron RT-DNA after RNA transfection (Supplementary Fig. 2).
Figure 1. Comparison of Eco1 retron to ssODN templated repair efficiency in a model genome editing assay.
(A) Schematic of Cas9-RT, msr-msd, and sgBFP expression vectors. (B) Experimental setup for transfection-based retron editing. HEK293T cells were transfected with Cas9-RT and ncRNA expressing plasmids and assessed for editing outcomes by flow cytometry 72h post-transfection. (C) Representative flow cytometry plots showing gates used to capture editing outcomes. Gates were drawn using an untransfected control. (D) Percentage of GFP positive cells by retron template source. In vitro transcribed RNA and GFP ssODN were transfected at two different concentrations, either 0.610pmols or 6.10pmols. Error bars denote standard error of the mean.
To compare retron editor designs, we measured precision genome editing rates using an established assay based on the conversion of a cDNA encoding blue fluorescent protein (BFP) to green fluorescent protein (GFP) by missense mutation of the chromophore29,33. ncRNA vectors were designed to encode an sgRNA targeting the BFP chromophore (sgBFP) and a repair template encoding GFP and disrupting the protospacer adjacent motif (PAM) of sgBFP (Fig. 1A). This system is disadvantageous for absolute editing efficiency due to the large number of different transfected components; however, it simplifies direct comparison of different retron editor designs. We benchmarked retron editors delivered as plasmid DNA or as IVT ncRNA against a synthetic 100 nucleotide single-stranded oligo (ssODN) repair template encoding the same template sequence as the plasmid-expressed ncRNA. We co-transfected the Cas9-RT expression plasmid with either sgBFP alone, two independent plasmids expressing U6-sgBFP and U6-msr-msd with a 100 nt GFP repair template, U6-sgBFP with IVT msr-msd RNA containing GFP template, or U6-sgBFP with a GFP ssODN repair template (Fig. 1B). After 72h we quantified editing outcomes (Fig. 1C). While the plasmid-expressed ncRNA edited the BFP locus to 0.95±0.01%, consistent with prior literature25–27, transfected RNA did not generate any successful editing events. In contrast, the ssODN repair template successfully edited 10.73±0.15% of transfected HEK293T cells (Fig 1D). These results are consistent with previous reports demonstrating poor retron editor efficiency in human cells25–27 and indicate that the Eco1 retron produces insufficient suitable homology template compared to ssODN when expressed either from a plasmid or by direct ncRNA transfection.
Efficient genome editing favors ncRNA architectures encoding sgRNAs with native 5’ ends
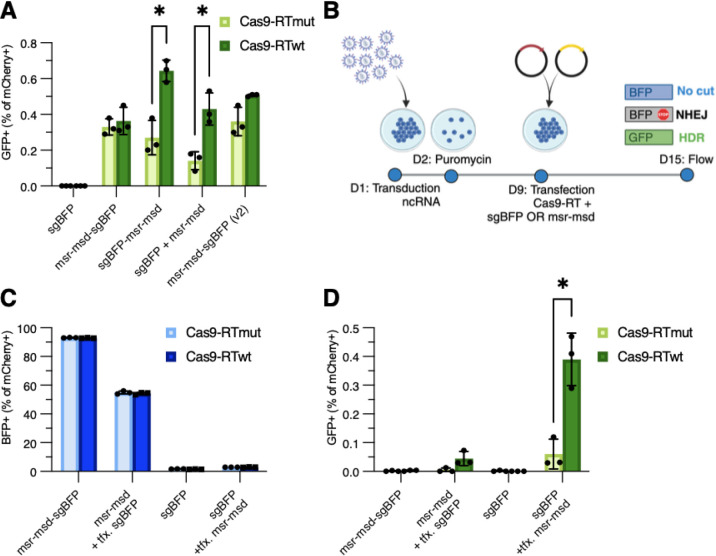
To determine which elements of ncRNA architecture most strongly affected editing efficiency, we systematically tested different ncRNA designs by transient transfection of retron editor plasmids. Initially we compared editing efficiencies of cis versus trans expression of the sgRNA and retron template. In this case, cis refers to a fusion transcript of sgRNA and retron msr-msd components while trans refers to expression of sgRNA and retron msr-msd as two separate transcripts. For editing in the context of a fusion RNA, we were interested in how orientation of the sgRNA relative to msr-msd might impact retron editing efficiency (ie., msr-msd-sgBFP vs. sgBFP-msr-msd). We were also motivated to investigate a number of additional sequence features that have been proposed to improve retron-mediated genome editing efficiency. These additional sequence features include a 14 nt extension of the msr-msd a1/a2 inverted repeats, 6 nt extension of the msd stem sequence, a 12 nt CAA-repeat linker between the retron msr-msd and sgRNA, and an alternative template design encoding the non-template strand of GFP26 (msr-msd-sgBFP (v2)). We observed that with Cas9-RTwt the msr-msd-sgBFP orientation showed no significant increase in GFP+ cells as compared to Cas9-RTmut (0.36±0.07% versus 0.37±0.04%) regardless of previously published ncRNA enhancements, indicating that the GFP+ cells likely result from plasmid-templated repair in this context (Fig. 2A). However, trans expression of the sgRNA and msr-msd from two different plasmids (sgBFP + msr-msd) and an sgBFP-msr-msd orientation had a marginal but statistically significant increase in editing efficiency (0.43±0.08% and 0.64±0.06% respectively). These results demonstrate that the overall architecture of the retron ncRNA favors an sgRNA with a native 5’ end. This aspect of retron editor design has a greater impact on editing efficiency in human cells than previously published improvements of the Eco1 msr-msd primary sequence26.
Figure 2. Genome editing efficiency of ncRNA architectures and delivery strategies.
(A) HDR editing outcomes from transfection-based editing assay as indicated by percentage of GFP+ cells. msr-msd-sgBFP (v2) represents an editor ncRNA with proposed improved structures. (B) Experimental setup for integrated ncRNA retron editing. Cells were transduced with ncRNA expressing lentivirus vectors and selected with puromycin 24h later. After one week of outgrowth, cells were transfected with plasmids expressing Cas9-RT and the corresponding ncRNA component in the case of integrated msr-msd or sgBFP. (C, D) NHEJ and HDR editing outcomes from an integrated ncRNA-based editing assay as indicated by percentage of BFP+ or GFP+ cells. Components that were expressed from a transfected plasmid are indicated (tfx), while all other ncRNA components were integrated into the genome by lentivirus. Error bars denote standard error of the mean.
Transfected retron ncRNA efficiently complements integrated sgBFP in trans
The importance of the relative abundance of the sgRNA and retron RNA template is an aspect of retron-mediated genome editing that has not previously been explored. We exploited our finding that retron ncRNA components are functional in trans to investigate the relative contribution of each component at steady-state abundance of the ncRNA-sgRNA fusion (Fig. 2A). We reasoned that this system would allow us to interrogate the relationship between ncRNA abundance and retron editing efficiency under conditions where one component is limiting. We tested this by integrating at low MOI one of the retron editor ncRNA components, either the sgRNA or msr-msd template, and transfecting the other component to induce overexpression relative to the low copy integrated component (Fig. 2B). We were surprised to observe that when integrated and expressed with Cas9-RT, sgBFP disrupted BFP at a mean frequency of 98.28±0.14%, while conversely, the msr-msd-sgBFP fusion was unable to disrupt BFP at a frequency greater than 6.93±0.15% (Fig. 2C). Efficient BFP disruption was restored when sgBFP was expressed with msr-msd template in trans either by transfection or integration. Overall, templated editing efficiency from integrated constructs was less than 0.1% except when sgBFP was integrated in the genome and the retron msr-msd template was overexpressed by plasmid transfection (Fig. 2D). Under these conditions templated repair overwhelmingly originates from RT-DNA, evidenced by a significant increase in GFP+ cells with Cas9-RTwt as compared to Cas9-RTmut which establishes the level of plasmid templated repair. These results suggest that the msr-msd template levels could be limiting in human cells and that retron-mediated editing efficiency might be improved by increasing the abundance of the ncRNA. These results also indicate that fusion of the sgRNA to the retron msr-msd inhibits Cas9 nuclease activity, potentially by interfering with efficient RNP assembly due to misfolding or steric hindrance35.
Steady-state levels of ncRNA are reduced in human cells
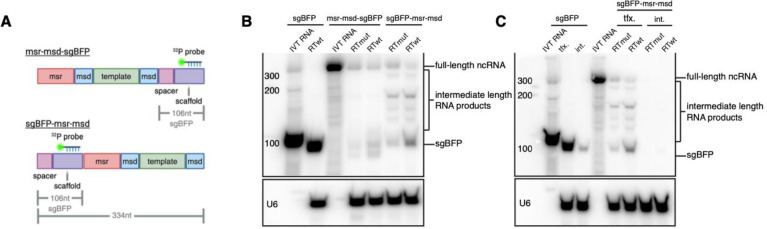
To detect steady-state levels of transfected retron ncRNAs, we designed 32P radiolabeled ssDNA probes to detect the sgBFP scaffold sequence of either a 106 nt sgBFP transcript or the 334 nt full-length sgBFP and msr-msd fusion transcript by northern blot (Fig. 3A). Pilot northern blot experiments revealed that co-delivery of Cas9 was required to stabilize sgRNAs (Supplementary Fig. 3). Based on this result, Cas9-RT plasmid was co-transfected in all subsequent experiments. We transfected plasmids expressing either sgBFP, msr-msd-sgBFP, or sgBFP-msr-msd retron ncRNAs and extracted whole-cell RNA for northern blot analysis. The abundance of sgBFP fused in either orientation to msr-msd ncRNA was significantly lower than sgBFP alone (Fig. 3B). We also observed that while the U6 snRNA loading control showed no evidence of RNA degradation, there was a consistent pattern of intermediate length ncRNA products that were not observed in in vitro transcribed RNA. We hypothesized that intermediate-length products originated through degradation by endogenous nucleases rather than early transcription termination, as they were present even when probing the 3’ end of the ncRNA.
Figure 3. Analysis of ncRNA integrity and quantity when expressed from plasmid and lentiviral vectors.
(A) Schematic of both 3’ and 5’ sgBFP ncRNA orientations showing location of the radiolabeled probe in the scaffold sequence. (B) Northern blot of retron editor ncRNA levels expressed from transfected plasmids for sgBFP, msr-msd-sgBFP, and sgBFP-msr-msd Radiolabeled probe targeted the scaffold sequence of the sgRNA. (C) Northern blot showing RNA levels of sgBFP or sgBFP-msr-msd when expressed from either transient transfection of a plasmid or low copy integration of a lentivirus vector.
Given that editing efficiency from a low-copy lentivirus integration demonstrated inferior editing efficiency compared to transfection, it seemed likely that steady-state ncRNA levels might be further reduced in this context. We probed for ncRNA expression in a low-copy lentivirus integration cell line with a sgBFP-msr-msd ncRNA construct and compared expressed RNA to a cell line with low-copy integrations of sgBFP alone (Fig. 3C). We detected a band corresponding to full-length sgBFP when expressed from a transfected plasmid and from an integrated locus; however, we were unable to detect the full-length sgBFP-msr-msd ncRNA when integrated despite being expressed from the same type of plasmid as sgBFP (Fig. 3C). Taken together, these results were indicative of ncRNA degradation by endogenous nucleases.
xrRNA knot rescues Eco1 retron editor ncRNA steady-state levels
Free 5’ and 3’ RNA ends are substrates for exoribonucleases and RNA sensing pathways36. We reasoned that protecting free ncRNA ends might stabilize and increase ncRNA abundance. We tested three distinct strategies of RNA protection found in nature: RNA circularization37, 3’ polyadenylation38, and RNA pseudoknots39.
To generate a circular RNA (circRNA) construct, we took advantage of flanking tRNA ligation sequences as endogenous tRNA ligase activity has previously been used to extend sgRNA half-life in cells40. Polyadenylation of RNAPIII-expressed SINE transcripts has been achieved by the addition of a canonical AATAAA polyA signal to the 3’ end of the transcript, and we reasoned that we could similarly take advantage of the stability provided by a polyA tail38. Finally, we tested three additional approaches incorporating stability-enhancing RNA pseudoknots on the 5’ end that all have been previously used to stabilize the 3’ end of pegRNA transcripts and improve prime editing efficiency18,41.
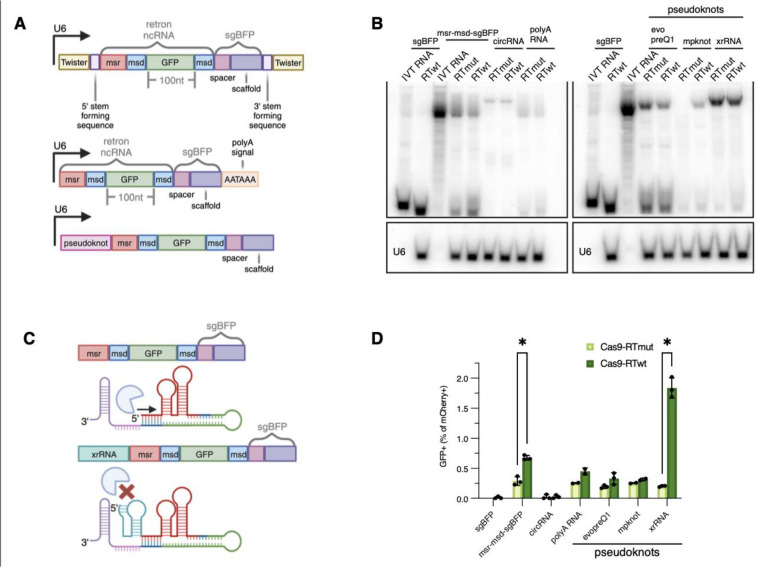
We tested 5 distinct ncRNA plasmids with the intention of circularizing, polyadenylating, or pseudoknot-protecting the retron editor ncRNA (Fig. 4A) and tested the degree to which each construct stabilized Eco1 retron editor transcripts within the cellular environment. Whole-cell RNA was extracted from HEK293T cells transfected with the different secondary structure-containing ncRNAs alongside either Cas9-RT or Cas9-RTmut and detected the products by northern blot (Fig. 4B). Of the approaches tested, the xrRNA pseudoknot at the 5’ end of the retron editor ncRNA strikingly increased RNA abundance and reduced the total amount of intermediate-length products that were detected. Other approaches tested provided little to no improvement in full-length ncRNA abundance. Given that the xrRNA pseudoknot evolved in nature to block nuclease-mediated degradation of an RNA genome42,43 a similar mechanism could explain the increase in RNA abundance in this context (Fig. 4C).
Figure 4. Evaluation of strategies to rescue ncRNA expression.
(A) Diagrams of retron editor ncRNA designs incorporating stabilizing elements. Top: circRNA design using Twister ribozyme sequences and tRNA stem forming ligation sequences. Middle: polyA RNA design incorporating a polyA AATAAA sequence before the RNAP III termination sequence. Bottom: ncRNA designs with a protective pseudoknot on the 5’ end before the retron template. (B) Northern blots of retron editor ncRNAs incorporating stabilizing elements. Radiolabeled probe targeted the scaffold sequence of the sgRNA. (C) Proposed mechanism of xrRNA-mediated RNA stability. Unprotected editor ncRNAs are susceptible to nuclease-driven decay while an xrRNA pseudoknot on the 5’ end of the ncRNA blocks degradation by cellular nucleases. (D) GFP editing outcomes for ncRNA constructs incorporating stability-enhancing elements. Error bars denote standard error of the mean.
Next, we compared editing efficiencies of these different stability enhancing ncRNAs to msr-msd-sgBFP. When transfected with Cas9-RTwt, the xrRNA-msr-msd-sgBFP reached GFP editing rates as high as 1.83±0.16%, representing a nearly 3-fold improvement over the native unprotected ncRNA (Fig. 4D). No other ncRNAs tested improved templated editing rates, while the circular RNA design exhibited reduced performance due to significantly impaired Cas9 activity, resulting in <0.1% editing efficiency.
Encouraged by these results, we tested whether improvements provided by the xrRNA pseudoknot in the transient transfection context could rescue templated genome editing when expressed from a lentivirus transduced at low MOI. Despite the improvements in RNA abundance and editing efficiency provided by grafting xrRNA in the context of transient plasmid transfection, this design showed low BFP disruption rates of 13.06±0.28% and an absence of templated GFP editing as a result (Fig. 5A). The lack of Cas9 activity in these cells despite ncRNA stabilization indicates further limitations on editing ncRNA efficiency.
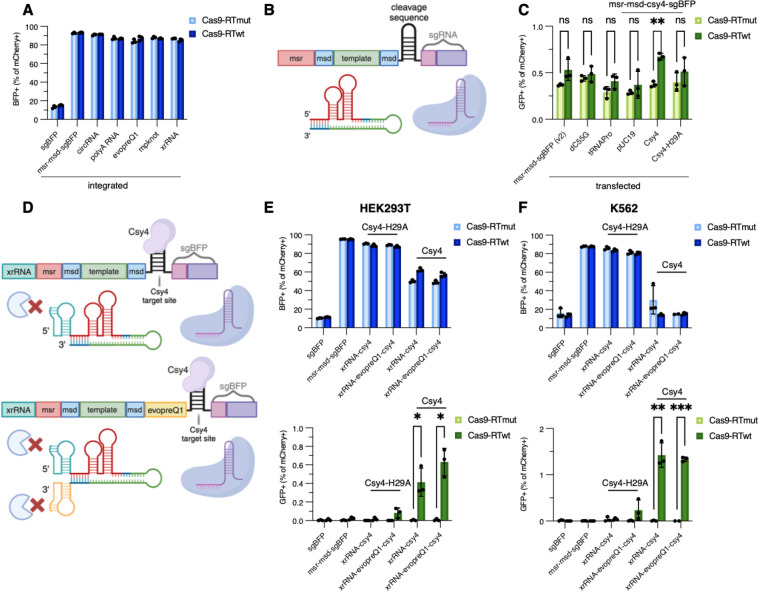
Figure 5. Optimization of post-transcriptional processing of ncRNAs to restore a native sgRNA 5’ end.
(A) BFP cutting efficiency of low-copy integrated structural ncRNAs. (B) Diagram of processed retron editor ncRNA. Either tRNA or Csy4 cleavage sequence allows separation of the retron template from sgRNA after transcription. (C) GFP editing efficiency of transfected processed retron editor ncRNAs. dC55G: mutated proline tRNA for improved processing, tRNAPro: full proline tRNA sequence, Csy4-H29A: nuclease deficient Csy4 mutant. (D) Diagram of pseudoknot protected and Csy4-protected retron editor ncRNAs. xrRNA-csy4 contains a single xrRNA pseudoknot on the 5’ end of the retron template, while xrRNA-evopreQ1-csy4 contains two pseudoknots that flank the retron template for both 5’ and 3’ protection after Csy4 processing. (E) Editing of BFP (top) and GFP (bottom) before and after ncRNA optimization in low-copy integrated HEK293T cells. (F) Same as Figure 5E done in low-copy integrated K562 cells. Error bars denote standard error of the mean.
Csy4 cleavage and pseudoknot protection enable Eco1 retron editing from an integrated lentivirus
Based on prior literature reporting that sgRNA activity is reduced by 5’ extensions35 and given our results demonstrating reduced BFP disruption in the context of 5’ sgBFP fusions, we reasoned that Cas9 activity might be improved by enzymatically cleaving the msr-msd retron template from the sgRNA after expression, thereby liberating the sgRNA and minimizing 5’ sgRNA extension (Fig. 5B). We explored multiple strategies to cleave the msr-msd-sgBFP including tRNA sequences that have natural processing of their 5’ and 3’ ends by cellular RNases44, as well as the Csy4 endoriboendonuclease that cleaves at a specific 20 nt stem loop sequence45. We tested three processable retron editor ncRNAs: two tRNA sequences and one Csy4-cleavable transcript. tRNAPro represents a full-length prolyl tRNA sequence between the template and sgRNA, while dC55G incorporates an engineered prolyl tRNA with reduced internal promoter activity and improved RNA processing46.
When co-transfected with Cas9-RT and a plasmid expressing wild-type Csy4, the Csy4-cleavable retron editor ncRNA (msr-msd-csy4-sgBFP) showed marginal improvements in RT-dependent templated repair as compared to an unprocessed ncRNA (Fig. 5C). This improvement is Csy4 cleavage dependent, as increased editing was not observed when co-transfected with a plasmid expressing a nuclease deficient Csy4 mutant (Csy4-H29A). Processing the sgRNA to minimize 5’ extension serves to further improve editing efficiency of the Eco1 retron editor ncRNA (Fig. 5C).
To overcome the problems of low steady-state RNA and reduced Cas9 nuclease activity of fusion ncRNA retron editor architectures, we combined both strategies of pseudoknot protection and Csy4 cleavage into a single ncRNA construct: xrRNA-msr-msd-csy4-sgBFP (Fig. 5D), a ncRNA expressing lentiviral vector that has an xrRNA pseudoknot grafted to the 5’ end of the msr-msd template and a Csy4 recognition site interposed between the template and the sgRNA. We also tested xrRNA-msr-msd-evopreQ-Csy4-sgBFP, which incorporates an additional pseudoknot on the 3’ end of the template that is exposed after Csy4 cleavage for additional nuclease protection of the mature msr-msd (Fig. 5D). When tested in low-copy HEK293T cell lines, the enhanced Eco1 xrRNA-msr-msd-evopreQ-csy4-sgBFP construct led to a 9-fold increase in Cas9 cutting along (Fig. 5E, top) with a 27-fold increase in editing rates when compared to the native Eco1 retron editor ncRNA (Fig. 5E, bottom).
We further validated these improvements in the K562 chronic myelogenous leukemia cell line. When the enhanced Eco1 retron xrRNA-msr-msd-evopreQ1-csy4-sgBFP ncRNA vector was delivered by lentivirus at a low MOI along with transiently transfected Cas9-RT and wild-type Csy4, we observed BFP disruptions rates of 84.83±1.16% (Fig. 5F, top) and missense mutations rates of 1.33±0.03% versus <0.01% using the unoptimized ncRNA construct (Fig. 5F, bottom).
DISCUSSION
Retron-mediated genome editing is an emerging biotechnology that complements existing tools for precision rewriting of genomic DNA. Retrons generate short ssDNA in situ for templated repair, leading to predictable editing outcomes and enabling applications that require large and complex DNA edits in diverse sequence contexts. Previously, retron editors have been used to introduce missense mutations in human cells but have been limited by poor efficiency when compared to orthologous systems expressed in bacteria and yeast24,47.
In this study, we identified unstable Eco1 ncRNA as a factor influencing low efficiency of templated genome editing in human cells. To overcome this limitation, we engineered a series of retron ncRNA variants to incorporate stability-enhancing secondary structures including a nuclease-resistant RNA pseudoknot from the Zika virus 3’ UTR that robustly rescued steady-state RNA levels in the context of transfected and stably integrated constructs. We further identified impaired Cas9 endonuclease activity in the context of a chimeric msr-msd-sgRNA fusion transcript and evaluated strategies to restore the native ends of both RNAs. The optimal approach used the Csy4 riboendonuclease to enzymatically process the sgRNA and retron ncRNA components, which rescued Cas9 activity. Combining these approaches enabled precision genome rewriting from a lentiviral vector integrated at low copy where templated repair was not previously observed.
The utility of genome editing technologies based on CRISPR-Cas systems has been expanded by fusing various enzymes to Cas9 to recruit specialized chemistry to target DNA sequences. In the case of prime editing, this also requires extending the sgRNA from the 3’ end to encode information for reverse transcriptase priming and the intended template16. When attached to Cas9, these 3’ components are exposed to degradation by endogenous nucleases41. Previous studies have identified approaches to protect the 3’ ends of pegRNAs including extension by RNA secondary structures and overexpression of the small RNA-binding exonuclease protection factor La48. In this study, stabilization of the sgRNA was not required as it is protected when co-expressed with Cas9; instead, we employed a strategy to protect both the 5’ and 3’ ends of the Eco1 ncRNA, which in this context significantly outperformed complementary a1/a2 extension. Recent studies have shown that 3’ truncated pegRNAs are preferentially loaded into Cas9 and that pegRNAs with strong 5’ and 3’ complementarity could circularize into a structure similar to retron ncRNA:sgRNA fusions49,50. These findings are consistent with our data that ncRNA:sgRNA fusions and circRNAs are relatively inefficient Cas9 guide RNAs.
Genome editing tools that rely on double-strand breaks to introduce templated missense mutations are limited by a general preference for NHEJ as a repair mechanism, which biases editing outcomes away from the intended edit51,52. It is likely that significant improvements to the overall efficiency of retron-mediated genome editing could be made by inhibiting alternative repair pathways by transient pharmacologic inhibition or by using a single or double strand nicking strategy53. We expect that it will be possible to expand the potential genome editing space of retron editors by: using PAM-expanded Cas variants, taking advantage of the intrinsic RNase activity of Cas12a to streamline RNA processing54, and implementing alternative promoter strategies to overcome the processivity limitations of the U6 promoter31. Further improvements in efficiency may also be realized through rational design, directed evolution, or metagenomic studies aimed at increasing the efficiency and processivity of reverse transcription in human cells or by increasing the efficiency of the msDNA structure as a substrate for templated repair. Since the msDNA arises from the ncRNA, we expect RT improvements to complement ncRNA stabilization.
This study investigated the limitations of Eco1 retron-mediated genome editing in human cells. Through systematic modifications and comparative assessments, we have identified key factors influencing the efficiency of a model retron editor, leading to ncRNA architecture and RNA stability enhancements. Incorporating RNA-stabilizing pseudoknots and Csy4 cleavage significantly improved the efficiency of this retron editing platform. This work underscores the importance of continued innovation in designing genome editing tools to overcome current limitations and expand their utility in biomedical research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues Drs. Jef Boeke, Ben tenOever, and Timothee Lionnet as well as the members of the Rice and Poirier labs for their helpful discussions. We thank Sanjana Venkittu for technical assistance with qPCR. Some instrumentation was provided by NYU Langone’s Small Instrument Fleet.
FUNDING
This work was funded by a pilot award from the NYU Laura & Isaac Perlmutter Cancer Center Support Grant’s Developmental Project Program P30CA016087 (JTP). The Cytometry and Cell Sorting Laboratory (RRID: SCR_019179) is partially supported by the Laura and Isaac Perlmutter Cancer Center support grant P30CA016087.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
NYU and Rockefeller University have filed a patent application directed to the subject matter described in this paper and the application is currently pending.
REFERENCES
- 1.Claussnitzer M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020). 10.1038/s41586-019-1879-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sud A., Kinnersley B. & Houlston R. S. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer 17, 692–704 (2017). 10.1038/nrc.2017.82 [DOI] [PubMed] [Google Scholar]
- 3.Consortium I. T. P.-C. A. o. W. G. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). 10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dagogo-Jack I. & Shaw A. T. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15, 81–94 (2018). 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 5.Anzalone A. V., Koblan L. W. & Liu D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38, 824–844 (2020). 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- 6.Wang T., Wei J. J., Sabatini D. M. & Lander E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalem O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umkehrer C. et al. Isolating live cell clones from barcoded populations using CRISPRa-inducible reporters. Nat Biotechnol 39, 174–178 (2021). 10.1038/s41587-020-0614-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z. Y. et al. Lineage-coupled clonal capture identifies clonal evolution mechanisms and vulnerabilities of BRAF(V600E) inhibition resistance in melanoma. Cell Discov 8, 102 (2022). 10.1038/s41421-022-00462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman D. et al. CloneSifter: enrichment of rare clones from heterogeneous cell populations. BMC Biol 18, 177 (2020). 10.1186/s12915-020-00911-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Rivera F. J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). 10.1038/nature13906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annunziato S. et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev 30, 1470–1480 (2016). 10.1101/gad.279190.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciampricotti M. et al. Rlf-Mycl Gene Fusion Drives Tumorigenesis and Metastasis in a Mouse Model of Small Cell Lung Cancer. Cancer Discov 11, 3214–3229 (2021). 10.1158/2159-8290.CD-21-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platt R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees H. A. & Liu D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770–788 (2018). 10.1038/s41576-018-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anzalone A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652 e5629 (2021). 10.1016/j.cell.2021.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson J. W. et al. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 40, 402–410 (2022). 10.1038/s41587-021-01039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould S. I. et al. High-throughput evaluation of genetic variants with prime editing sensor libraries. Nat Biotechnol (2024). 10.1038/s41587-024-02172-9 [DOI] [PubMed] [Google Scholar]
- 20.Simon A. J., Ellington A. D. & Finkelstein I. J. Retrons and their applications in genome engineering. Nucleic Acids Res 47, 11007–11019 (2019). 10.1093/nar/gkz865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang S. & Sternberg S. H. Genome editing with retroelements. Science 382, 370–371 (2023). 10.1126/science.adi3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yee T., Furuichi T., Inouye S. & Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell 38, 203–209 (1984). 10.1016/0092-8674(84)90541-5 [DOI] [PubMed] [Google Scholar]
- 23.Mestre M. R., Gonzalez-Delgado A., Gutierrez-Rus L. I., Martinez-Abarca F. & Toro N. Systematic prediction of genes functionally associated with bacterial retrons and classification of the encoded tripartite systems. Nucleic Acids Res 48, 12632–12647 (2020). 10.1093/nar/gkaa1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon E. et al. Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing. Cell 175, 544–557 e516 (2018). 10.1016/j.cell.2018.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B., Chen S. A., Lee J. & Fraser H. B. Bacterial Retrons Enable Precise Gene Editing in Human Cells. CRISPR J 5, 31–39 (2022). 10.1089/crispr.2021.0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez S. C., Crawford K. D., Lear S. K., Bhattarai-Kline S. & Shipman S. L. Precise genome editing across kingdoms of life using retron-derived DNA. Nat Chem Biol 18, 199–206 (2022). 10.1038/s41589-021-00927-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong X. et al. Precise genome editing without exogenous donor DNA via retron editing system in human cells. Protein Cell 12, 899–902 (2021). 10.1007/s13238-021-00862-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattarai-Kline S. et al. Recording gene expression order in DNA by CRISPR addition of retron barcodes. Nature 608, 217–225 (2022). 10.1038/s41586-022-04994-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson C. D., Ray G. J., DeWitt M. A., Curie G. L. & Corn J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34, 339–344 (2016). 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- 30.E., S. et al. Functional genetic variants revealed by massively parallel precise genome editing. Cell 175, 544–557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Z., Herrera-Carrillo E. & Berkhout B. Delineation of the Exact Transcription Termination Signal for Type 3 Polymerase III. Mol Ther Nucleic Acids 10, 36–44 (2018). 10.1016/j.omtn.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma P. L., Nurpeisov V. & Schinazi R. F. Retrovirus reverse transcriptases containing a modified YXDD motif. Antivir Chem Chemother 16, 169–182 (2005). 10.1177/095632020501600303 [DOI] [PubMed] [Google Scholar]
- 33.DeWitt M. A., Corn J. E. & Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods 121–122, 9–15 (2017). 10.1016/j.ymeth.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jinek M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamata M., Suzuki H. I., Kimura R. & Suzuki A. Optimization of Cas9 activity through the addition of cytosine extensions to single-guide RNAs. Nat Biomed Eng 7, 672–691 (2023). 10.1038/s41551-023-01011-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilusz C. J., Wormington M. & Peltz S. W. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2, 237–246 (2001). 10.1038/35067025 [DOI] [PubMed] [Google Scholar]
- 37.Litke J. L. & Jaffrey S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat Biotechnol 37, 667–675 (2019). 10.1038/s41587-019-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ustyantsev I. G. et al. [Polyadenylation of Sine Transcripts Generated by RNA Polymerase III Dramatically Prolongs Their Lifetime in Cells]. Mol Biol (Mosk) 54, 78–86 (2020). 10.31857/S0026898419040165 [DOI] [PubMed] [Google Scholar]
- 39.Staple D. W. & Butcher S. E. Pseudoknots: RNA structures with diverse functions. PLoS Biol 3, e213 (2005). 10.1371/journal.pbio.0030213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M., Xu J., Meng J. & Huang X. Synthetic Circular gRNA Mediated Biological Function of CRISPR-(d)Cas9 System. Front Cell Dev Biol 10, 863431 (2022). 10.3389/fcell.2022.863431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang G. et al. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat Commun 13, 1856 (2022). 10.1038/s41467-022-29507-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pijlman G. P. et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4, 579–591 (2008). 10.1016/j.chom.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 43.MacFadden A. et al. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat Commun 9, 119 (2018). 10.1038/s41467-017-02604-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor J. P. & Peebles C. L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol 11, 425–439 (1991). 10.1128/mcb.11.1.425-439.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haurwitz R. E., Jinek M., Wiedenheft B., Zhou K. & Doudna J. A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 (2010). 10.1126/science.1192272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knapp D. et al. Decoupling tRNA promoter and processing activities enables specific Pol-II Cas9 guide RNA expression. Nat Commun 10, 1490 (2019). 10.1038/s41467-019-09148-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farzadfard F. & Lu T. K. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014). 10.1126/science.1256272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan J. et al. Improving prime editing with an endogenous small RNA-binding protein. Nature 628, 639–647 (2024). 10.1038/s41586-024-07259-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ponnienselvan K. et al. Reducing the inherent auto-inhibitory interaction within the pegRNA enhances prime editing efficiency. Nucleic Acids Res 51, 6966–6980 (2023). 10.1093/nar/gkad456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W. et al. Enhancing CRISPR prime editing by reducing misfolded pegRNA interactions. Elife 12 (2024). 10.7554/eLife.90948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieber M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79, 181–211 (2010). 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue C. & Greene E. C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet 37, 639–656 (2021). 10.1016/j.tig.2021.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riesenberg S. et al. Efficient high-precision homology-directed repair-dependent genome editing by HDRobust. Nat Methods 20, 1388–1399 (2023). 10.1038/s41592-023-01949-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fonfara I., Richter H., Bratovic M., Le Rhun A. & Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). 10.1038/nature17945 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.