Abstract
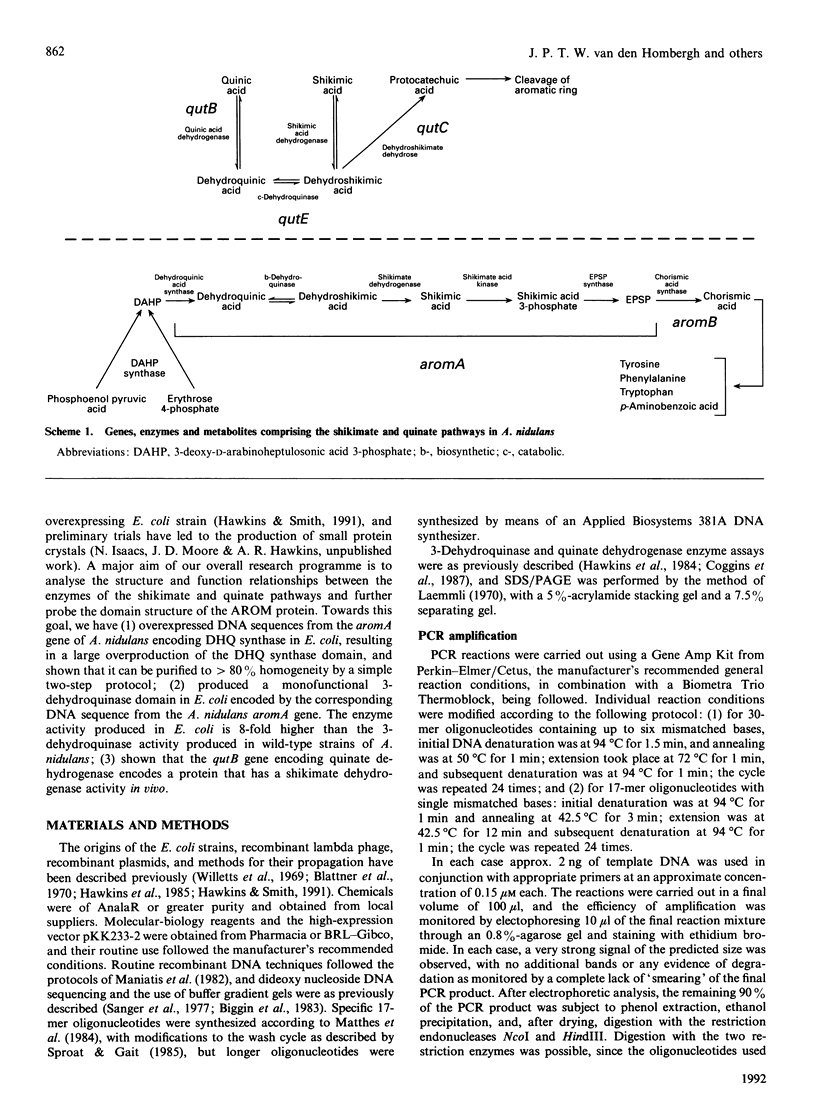
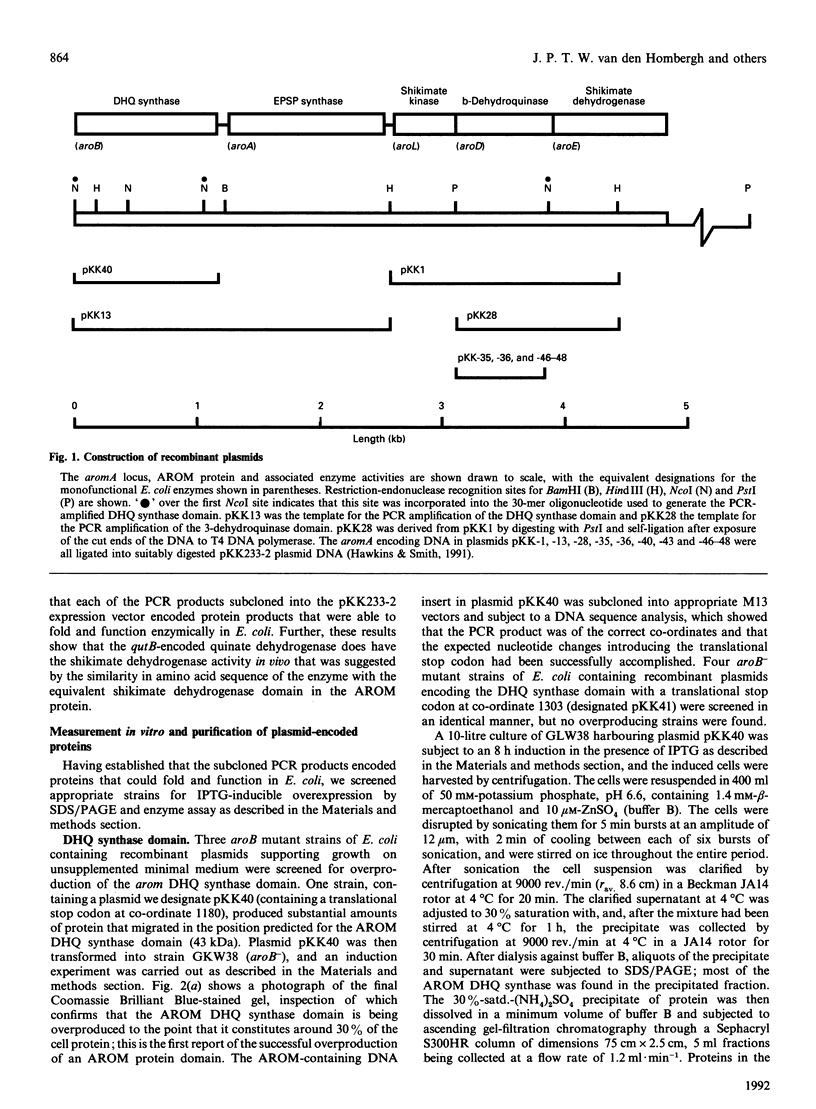
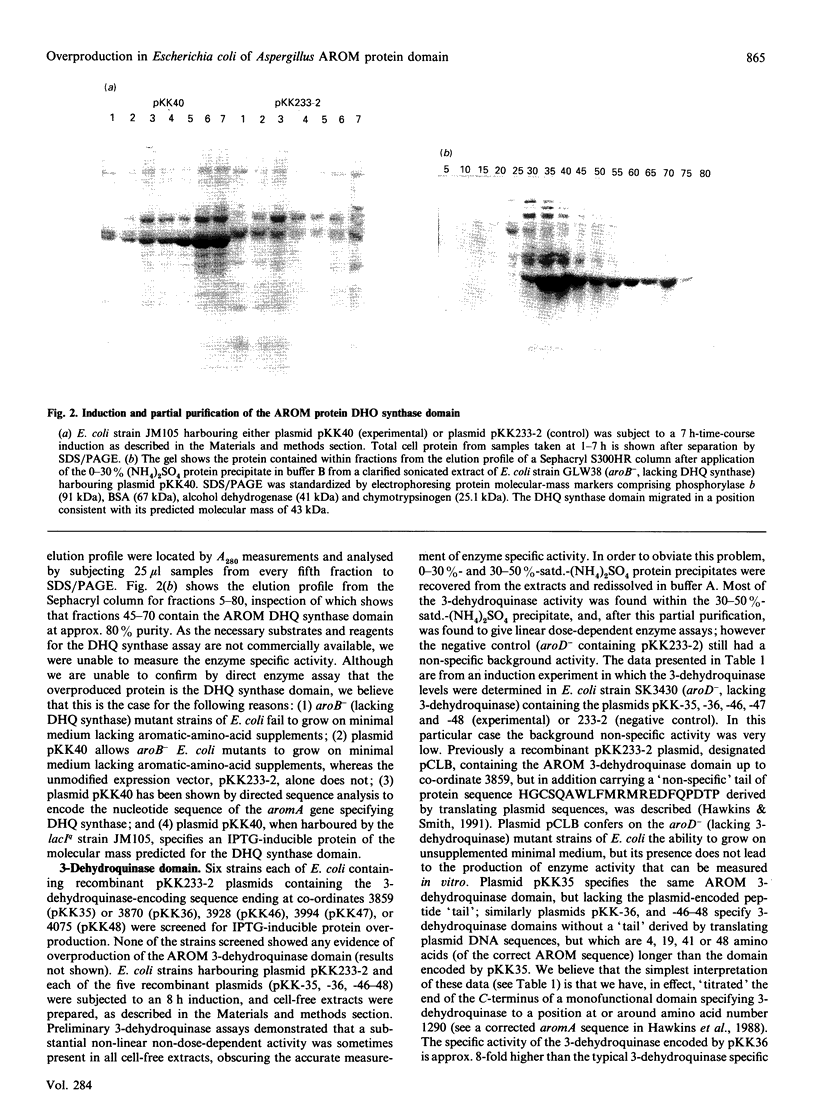
The pentafunctional AROM protein of Aspergillus nidulans is encoded by the complex aromA locus and catalyses steps 2-6 in the synthesis of chorismate, the common precursor for the aromatic amino acids and p-aminobenzoic acid. DNA sequences encoding the 3-dehydroquinate synthase (DHQ synthase) and 3-dehydroquinase domains of the AROM protein have been amplified with the inclusion of a translational stop codon at the C-terminus by PCR technology. These amplified fragments of DNA have been subcloned into the prokaryotic expression vector pKK233-2 and expressed in Escherichia coli. As a result, the DHQ synthase domain is overproduced in E. coli, forming 30% of total cell protein, and can be purified to greater than 80% homogeneity by a simple two-step protocol. The 3-dehydroquinase domain is produced at a specific activity 8-fold greater than the corresponding activity encoded by the aromA gene in A. nidulans. The qutB gene of A. nidulans encoding quinate dehydrogenase has similarly been subjected to PCR amplification and expression in E. coli. The quinate dehydrogenase is not overproduced, but is active in E. coli as a shikimate dehydrogenase, as the presence of the qutB gene allows the growth of an E. coli mutant strain lacking shikimate dehydrogenase on minimal medium lacking aromatic-amino-acid supplementation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anton I. A., Duncan K., Coggins J. R. A eukaryotic repressor protein, the qa-1S gene product of Neurospora crassa, is homologous to part of the arom multifunctional enzyme. J Mol Biol. 1987 Sep 20;197(2):367–371. doi: 10.1016/0022-2836(87)90130-6. [DOI] [PubMed] [Google Scholar]
- Beri R. K., Whittington H., Roberts C. F., Hawkins A. R. Isolation and characterization of the positively acting regulatory gene QUTA from Aspergillus nidulans. Nucleic Acids Res. 1987 Oct 12;15(19):7991–8001. doi: 10.1093/nar/15.19.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Hawkins A. R. Nucleotide sequence encoding the biosynthetic dehydroquinase function of the penta-functional arom locus of Aspergillus nidulans. Nucleic Acids Res. 1985 Nov 25;13(22):8119–8128. doi: 10.1093/nar/13.22.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Smith M., Hawkins A. R. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res. 1986 Mar 11;14(5):2201–2213. doi: 10.1093/nar/14.5.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Duncan K., Graham L. D., Coggins J. R. Identification of the active-site lysine residues of two biosynthetic 3-dehydroquinases. Biochem J. 1991 Apr 1;275(Pt 1):1–6. doi: 10.1042/bj2750001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Chaudhuri S., Lambert J. M., Lumsden J., Nimmo G. A., Smith D. D. The arom multifunctional enzyme from Neurospora crassa. Methods Enzymol. 1987;142:325–341. doi: 10.1016/s0076-6879(87)42044-2. [DOI] [PubMed] [Google Scholar]
- Da Silva A. J., Whittington H., Clements J., Roberts C., Hawkins A. R. Sequence analysis and transformation by the catabolic 3-dehydroquinase (QUTE) gene from Aspergillus nidulans. Biochem J. 1986 Dec 1;240(2):481–488. doi: 10.1042/bj2400481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Chaudhuri S., Campbell M. S., Coggins J. R. The overexpression and complete amino acid sequence of Escherichia coli 3-dehydroquinase. Biochem J. 1986 Sep 1;238(2):475–483. doi: 10.1042/bj2380475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe T., Servos S., Hawkins A., Dimitriadis G., Young D., Dougan G., Charles I. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Mol Gen Genet. 1991 Sep;228(3):385–392. doi: 10.1007/BF00260631. [DOI] [PubMed] [Google Scholar]
- Grant S., Roberts C. F., Lamb H., Stout M., Hawkins A. R. Genetic regulation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. J Gen Microbiol. 1988 Feb;134(2):347–358. doi: 10.1099/00221287-134-2-347. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Francisco Da Silva A. J., Roberts C. F. Cloning and characterization of the three enzyme structural genes QUTB, QUTC and QUTE from the quinic acid utilization gene cluster in Aspergillus nidulans. Curr Genet. 1985;9(4):305–311. doi: 10.1007/BF00419960. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Francisco da Silva A. J., Roberts C. F. Evidence for two control genes regulating expression of the quinic acid utilization (qut) gene cluster in Aspergillus nidulans. J Gen Microbiol. 1984 Mar;130(3):567–574. doi: 10.1099/00221287-130-3-567. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Roberts C. F. Structure of the Aspergillus nidulans qut repressor-encoding gene: implications for the regulation of transcription initiation. Gene. 1992 Jan 2;110(1):109–114. doi: 10.1016/0378-1119(92)90452-u. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Smith M., Keyte J. W., Roberts C. F. Molecular organisation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. Mol Gen Genet. 1988 Oct;214(2):224–231. doi: 10.1007/BF00337715. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Smith M. Domain structure and interaction within the pentafunctional arom polypeptide. Eur J Biochem. 1991 Mar 28;196(3):717–724. doi: 10.1111/j.1432-1033.1991.tb15870.x. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R. The complex Arom locus of Aspergillus nidulans. Evidence for multiple gene fusions and convergent evolution. Curr Genet. 1987;11(6-7):491–498. doi: 10.1007/BF00384611. [DOI] [PubMed] [Google Scholar]
- Kinghorn J. R., Hawkins A. R. Cloning and expression in Escherichia coli K-12 of the biosynthetic dehydroquinase function of the arom cluster gene from the eucaryote, Aspergillus nidulans. Mol Gen Genet. 1982;186(1):145–152. doi: 10.1007/BF00422927. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb H. K., Bagshaw C. R., Hawkins A. R. In vivo overproduction of the pentafunctional arom polypeptide in Aspergillus nidulans affects metabolic flux in the quinate pathway. Mol Gen Genet. 1991 Jun;227(2):187–196. doi: 10.1007/BF00259670. [DOI] [PubMed] [Google Scholar]
- Lamb H. K., Hawkins A. R., Smith M., Harvey I. J., Brown J., Turner G., Roberts C. F. Spatial and biological characterisation of the complete quinic acid utilisation gene cluster in Aspergillus nidulans. Mol Gen Genet. 1990 Aug;223(1):17–23. doi: 10.1007/BF00315792. [DOI] [PubMed] [Google Scholar]
- Lamb H. K., van den Hombergh J. P., Newton G. H., Moore J. D., Roberts C. F., Hawkins A. R. Differential flux through the quinate and shikimate pathways. Implications for the channelling hypothesis. Biochem J. 1992 May 15;284(Pt 1):181–187. doi: 10.1042/bj2840181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes H. W., Zenke W. M., Grundström T., Staub A., Wintzerith M., Chambon P. Simultaneous rapid chemical synthesis of over one hundred oligonucleotides on a microscale. EMBO J. 1984 Apr;3(4):801–805. doi: 10.1002/j.1460-2075.1984.tb01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproat B. S., Gait M. J. Chemical synthesis of a gene for somatomedin C. Nucleic Acids Res. 1985 Apr 25;13(8):2959–2977. doi: 10.1093/nar/13.8.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington H. A., Grant S., Roberts C. F., Lamb H., Hawkins A. R. Identification and isolation of a putative permease gene in the quinic acid utilization (QUT) gene cluster of Aspergillus nidulans. Curr Genet. 1987;12(2):135–139. doi: 10.1007/BF00434668. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]



