Abstract
Mfn2 is a mitochondrial outer membrane fusion protein with the additional role of tethering mitochondria to the ER. Here, we describe a novel connection between Mfn2 and calcium release from mitochondria. We show that Mfn2 controls the mitochondrial inner membrane sodium-calcium exchange protein NCLX, which is a major source for calcium release from mitochondria. This discovery was made with the fungal toxin Phomoxanthone (PXA), which induces calcium release from mitochondria. PXA-induced calcium release is blocked by a chemical inhibitor of NCLX, while NCLX and Mfn2 deletions both also prevent PXA-induced calcium release. CETSA experiments show that PXA directly targets Mfn2, which likely controls NCLX through physical interactions since co-immunoprecipitation and proximity ligation assays show increased association between Mfn2 and NCLX upon treatment with PXA. Interactions between Mfn2 and NCLX also increase when cells are treated with mitochondrial ROS-inducing conditions, such as oligomycin treatment of respiring cells, while the interactions do not increase in Oma1 −/− cells. It seems likely that opening of cristae by Oma1-mediated cleavage of Opa1 promotes translocation of NCLX from cristae to the rim where it can come into contact with Mfn2 thus promoting PXA-induced calcium release from mitochondria. These results therefore delineate a pathway that connects ROS produced inside mitochondria with calcium release and signaling in the cytosol.
Introduction
The Mitofusins Mfn1 and Mfn2 mediate mitochondrial outer membrane fusion in mammals (Labbe et al., 2014). Mfn1 and Mfn2 are members of the dynamin family of proteins with similar domain structures. The Mitofusins are anchored in the mitochondrial outer membrane through transmembrane segments, unlike other members of the dynamin family which transiently associate with membranes(Santel and Fuller, 2001). Mutations in both Mitofusins cause mitochondrial fusion defects (Chen et al., 2003). Mfn1 and Mfn2 can act together in heteromeric complexes, but they also have distinct phenotypes suggestive of functional differences (Chen et al., 2003). Mfn1 supports chemical stress-induced mitochondrial hyperfusion (SIMH) (Tondera et al., 2009), while Mfn2 has been observed in spots where fission occurs at mitochondria associated membranes (MAM) (Karbowski et al., 2002) and it acts as a tether to promote association between mitochondria and ER (de Brito and Scorrano, 2008). As a consequence of the tethering function, Mfn2 also affects other processes, including transfer of coenzyme Q (Mourier et al., 2015) and phosphatidyl serine (Hernandez-Alvarez et al., 2019), as well as control of metabolic functions connected with the MAM (Sebastian et al., 2012). Mutations in Mfn2 are the leading cause of axonal forms of hereditary peripheral neuropathies known as Charcot-Marie-Tooth disease type 2. It is unclear which of the different Mfn2 functions are most relevant for the CMT2 disease.
While investigating effects of the phomoxanthone PXA, a fungal metabolite that induces mitochondrial fission, apoptosis and release of mitochondrial calcium, we found that fission and apoptosis, but not calcium release, could be stopped by mutating Drp1 (Bohler et al., 2018). The remaining effects of PXA in Drp1 KO cells were extensive loss of mitochondrial calcium and contraction of the mitochondrial matrix to form small condensed fragments that were connected by constricted mitochondrial outer membrane tubules like beads on a string. In the present study, we first pursued the possibility that some of the other dynamin family members on mitochondria were responsible for the constrictions in Drp1 KO cells by making double knockouts with Drp1 and treating those with PXA. To our surprise, the mitochondria of Drp1-Mfn2 DKO cells were fully resistant to PXA-induced calcium release and condensation of the mitochondrial matrix. In light of earlier results showing Mfn2-dependent reversal of NCLX in isolated mitochondria (Samanta et al., 2018), these new results raise the possibility that PXA-induced calcium release is mediated by NCLX under control of Mfn2.
Results and discussion:
Mfn1 and Mfn2 mutations have opposite effects on PXA-induced mitochondrial matrix contractions.
We previously observed mitochondrial fragmentation and apoptosis as a result of rapid calcium release from mitochondria induced by PXA (Bohler et al., 2018). Apoptosis and fission of the mitochondrial outer membrane can be prevented by mutations in Drp1, but PXA still induces localized constrictions due to contraction of the mitochondrial matrix through excessive calcium release. This “beads on a string” phenotype with numerous localized constrictions even without Drp1 prompted us to investigate possible contributions of other mitochondrial dynamics proteins to this process.
To observe the effects of Mfn1 and Mfn2 on PXA induced constrictions without complications from apoptosis, we conducted our morphological analyses in double knockout (DKO) MEFs in which Mfn1 or Mfn2 mutations were combined with a Drp1 mutation and in Drp1-Mfn2 HeLa cells. The validity of the knockouts was confirmed with Western blots (Fig. S1a–b). To our surprise, the effects of PXA on Drp1-Mfn1 and Drp1-Mfn2 DKO cells were very different. Mfn2-Drp1 DKO cells were fully resistant to PXA-induced contraction, while Drp1-Mfn1 DKO cells showed even more matrix contraction than Drp1 KO cells, to the point that mitochondrial fission was induced even without Drp1 (Fig. 1a). Similar suppression of the contractive effects of PXA was observed in Drp1-Mfn2 DKO HeLa cells (Fig. 1b). The PXA-induced matrix retractions result in separation of mitochondrial outer membrane and matrix fluorescent signals, as is apparent with scatterplots (Fig. S1b). This level of separation enabled quantification of the morphological changes with Manders’ coefficients for overlap of a mitochondrial outer membrane marker (Tom20) and a mitochondrial matrix marker (Hsp60). These Manders’ coefficients show significantly reduced overlap between mitochondrial matrix and outer membranes upon treatment of Drp1 KO cells, but not in Drp1-Mfn2 DKO cells, while the effects in Drp1-Mfn1 DKO cells were enhanced (Fig. 1c–d). The results suggest that Mfn1 inhibits PXA-induced contraction, while Mfn2 promotes this effect. It was previously shown with siRNA that Mfn1 and Mfn2 also have opposing effects on calcium uptake and release in cardiomyocytes (Inagaki et al., 2023), in line with the differences in mitochondrial morphology observed here with PXA induced mitochondrial calcium release. The opposing effects may reflect the different functions of Mfn1 and Mfn2 as well as their ability to heterodimerize and thus antagonize each other’s functions. We conclude that Mfn2 is required for PXA-induced mitochondrial matrix contraction and fission, while Mfn1 antagonizes that effect.
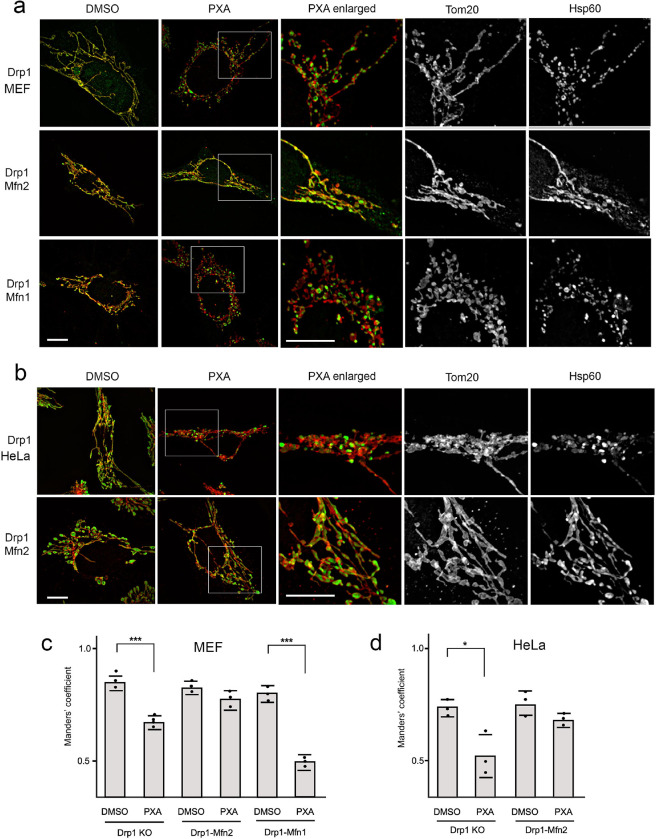
Fig. 1. PXA induced morphological effects on mitochondria are controlled by Mitofusins.
(a) MEFs with the indicated genotypes were treated with DMSO or 10uM PXA for 30 min at 37 °C and labeled with Tom20 (red) and Hsp60 (green) antibodies for immunofluorescence. PXA induces contraction of the mitochondrial matrix in Drp1 KO cells, but these contractions are suppressed in Drp1-Mfn2 DKO cells. Drp1-Mfn1 DKO cells show the opposite effect. There contractions are enhanced to the point that outer membrane fission also occurred. The boxed areas in PXA images cells were enlarged to show these changes more clearly. Scale bar is 10 μm. (b) Similar effects were observed with Drp1 KO and Drp1-Mfn2 DKO HeLa cells. (c, d) Manders’ coefficients of Tom20 and Hsp60 colocalization were used to quantify the matrix contractions. The averages (with SD) for three independent experiments in which 50 images were analyzed for each condition show significantly reduced Manders’ coefficients in PXA-treated Drp1 KO and Drp1-Mfn1 DKO cells, but not in Drp1-Mfn2 DKO cells.
PXA induces calcium release by NCLX.
We focused on calcium release through NCLX, because of a previously discovered connection between Mfn2 and NCLX(Samanta et al., 2018). Effects of PXA on calcium release from mitochondria were monitored with a fluorescent protein targeted to mitochondria (R-Cepia3mt ) and the vital dye Rhod-2AM. Tracings of mitochondrial calcium in individual cells were obtained with the fluorescent protein using a spinning disc confocal microscope (Fig. 2a–b) and for larger fields of cells with the vital dye using a plate reader (Fig. S2a–b). A HeLa cell NCLX KO cell line, generated with CRISPR/Cas9, was confirmed with a Western blot in which the band was identified through a comparison with NCLX siRNA cells (Fig. S2a). In line with previous results obtained with WT cells (Bohler et al., 2018), PXA induces rapid calcium release from mitochondria in Drp1 KO cells (Fig. 2a–b, Fig. S2b–c). However, this release is blocked in Mfn2-Drp1 DKO cells, in NCLX KO cells, as well as in cells transfected with NCLX shRNA, and in cells treated with the NCLX inhibitor CGP37157. We conclude that PXA induced calcium release from mitochondria requires Mfn2 and NCLX functions.
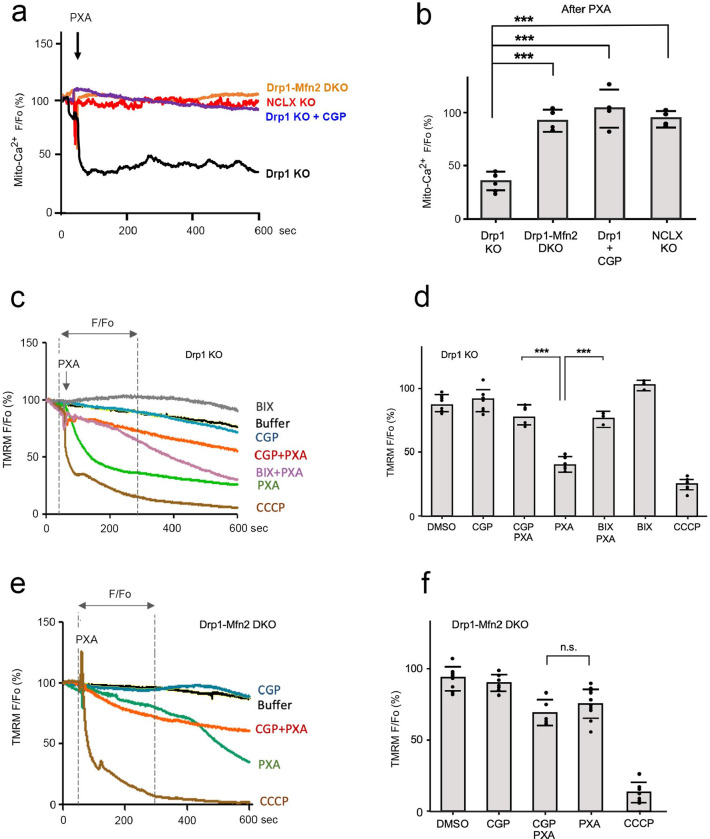
Fig. 2. Mfn2 regulates mitochondrial calcium efflux and membrane potential through NCLX.
(a) Tracings of mitochondrial matrix Ca2+ in Hela cells obtained with the Ca2+ sensitive fluorescent probe matrix3mt-R-CEPIA. Drp1 KO, Drp1-Mfn2 DKO, Drp1-NCLX DKO cells were recorded, as well as Drp1 KO cells that were pre-incubated for 30 min at 37°C with 10uM CGP37157. Baselines (Fo) were established with 60 sec tracings in HBSS media and then Ca2+ release was induced by perfusion with 10uM PXA in HBSS followed by 600 sec of further recording. (b) Changes in mitochondrial Ca2+ levels reflected by the relative fluorescence (F/Fo) at 600 sec. Averages are shown with SD for the data points in the histogram and significance was determined with a Student’s T test. (c) Effects of PXA on mitochondrial membrane potential (ΔΨ) determined with Drp1 KO HeLa cells loaded with 25 nM TMRM. Where indicated, cells were preincubated for 30 min with 100 nm BIX (NHE1 inhibitor) or 10uM CGP37157 (NCLX inhibitor). Baselines and tracings after perfusion with or without 10uM PXA were obtained as in panel a. Perfusion with 10uM CCCP was used as control for dissipation of ΔΨ. (d) Averages of F/Fo determined at 300sec after perfusion with SD for the data points in the histogram and significance was determined with a Student’s T test. (e, f) Tracings and histograms of TMRM fluorescence obtained as for panel c and d, but with Drp1-Mfn2 DKO cells instead of Drp1 KO cells.
We then monitored mitochondrial membrane potential, as this was also shown to be reduced by treating cells with PXA, possibly as an indirect consequence of calcium release. NCLX exchanges calcium for sodium and then excess sodium in mitochondria is exchanged for protons from the intermembrane space by the sodium/proton exchanger NHE1 as illustrated in Fig. S2d. As a result, the proton gradient decreases with calcium release and that lowers the membrane potential. Our results show that PXA-induced loss of membrane potential is blocked by mutations in Mfn2 and by the NCLX inhibitor CGP37157 (Fig. 2c–f). Loss of membrane potential is also slowed by the NHE1 inhibitor BIX, consistent with the connection between NCLX and NHE1 functions and membrane potential. We conclude that loss of mitochondrial membrane potential is an indirect consequence of NCLX activation by PXA.
PXA directly targets Mfn2, which then forms a complex with NCLX.
We used a cellular thermal shift assay (CETSA) to determine whether Mfn2 or NCLX are targeted by PXA. In this assay, dodecyl maltoside solubilized MEF extracts were incubated with PXA and then subjected to denaturation with a gradient of temperatures. Denatured proteins were removed by centrifugation and proteins that remained in solution were analyzed by Western blots. We tested the effects on denaturation temperatures using MEF cells that overexpress Mfn2-FLAG and cells that lack Mfn1 or Mfn2. Mfn2-FLAG was shown to be functional by rescuing the mitochondrial morphology defect of Mfn2 KO cells (Fig. S3a). We also verified identification of NCLX bands on Western blots of the MEFs using KO cells, because the available NCLX antibodies detect several cross-reacting bands (Fig. S3b). Results obtained with CETSA show that PXA causes a shift in the denaturation temperatures of Mfn1 and Mfn2 but not of NCLX or SLC25A46 (a protein that associates with the Mitofusins and Opa1) (Steffen et al., 2017) (Fig. 3a–b). The denaturation temperatures of Vinculin, which was used as a control, is also unaffected by PXA. The effects were more pronounced in a cell line that overexpresses Mfn2, in line with direct effects of PXA on Mfn2. The interaction of PXA with Mfn1 suggests that this compound may bind to more proteins, but this does not obviate the requirement for Mfn2 in PXA induced calcium release.
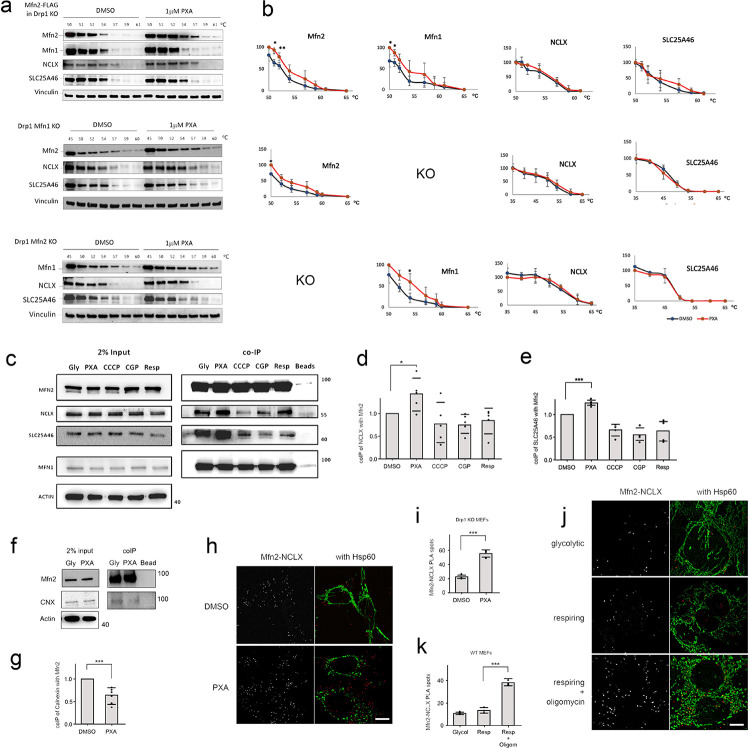
Fig. 3. PXA targets Mitofusins and that targeting induces interactions between Mfn2 and NCLX.
(a) CETSA of dodecyl maltoside extracts from Drp1-Mfn2 DKO + Mfn2-FLAG, Drp1-Mfn1 DKO and Drp1-Mfn2 DKO MEFs. These extracts were incubated with DMSO or 1uM PXA and subjected to heat denaturation for 3 min at the indicated temperatures, followed by removal of denatured proteins by centrifugation and blot analysis of the supernatants with antibodies against Mfn2, Mfn1, NCLX and SLC25A46, as well as Vinculin as control. The upper panel was made with Drp1-Mfn2 DKO cells that stably express exogenous Mfn2-FLAG, while the lower two sets of blots were made with endogenous proteins. (b) Band intensities determined with densitometric scans of blots as shown in panel a. The intensities were normalized to 45 or 50°C. The points are averages with SD from 3 independent experiments. (c) Co-immunoprecipitation (coIP) of endogenous NCLX and SLC25A46 with Mfn2-FLAG using FLAG antibody attached to beads. Cells were grown under glycolytic conditions (Gly) and treated for 30 min with DSMO, 10 μM PXA, 10 μM CCCP or 10 μM CGP37157, or grown under respiring conditions (galactose). These cells were incubated with DSP cross-linker and subjected to coIP, followed by Western blot analysis. Actin was used as a loading control. (d) Densitometry of the coIPs, normalized to the levels of Mfn2 for each condition. Averages of 5 independent experiments are shown with SD and results of a Student’s T test. (e) Densitometry of SLC25A46 coIPs as described for NCLX in panel d. (f) CoIP of Calnexin with Mfn2-FLAG after treatment with PXA under glycolytic conditions as described in panel c. (g) Densitometry of Calnexin coIP with Mfn2-FLAG as in panels d and e. (h) PLA of Mfn2-myc and NCLX-HA (red spots) in glycolytic Drp1 KO MEFs treated with DMSO or PXA. Mitochondria were detected by immunofluorescence with a chicken antibody against Hsp60 (green). The size marker is 10 μm. (i) Average numbers of PLA spots per cell from 3 independent experiments with SD and results of a Student’s T test. PLA spots in 35 cells were counted for each experiment. (j) PLA of Mfn2-myc and NCLX-HA (red spots) in glycolytic or respiring Drp1 KO MEFs treated with DMSO or oligomycin as indicated. (k) Average numbers of PLA spots for these conditions determined as in panel i.
To determine whether Mfn2 controls NCLX by physical interactions, we conducted co-immuno-precipitation experiments with epitope tagged versions (Mfn2-FLAG and NCLX-HA) and chemical crosslinking. Mfn2-myc is functional as shown by rescue of the fusion defect in Mfn2 KO cells (Fig. S3a). Western blots show an increase of co-immunoprecipitation of NCLX with Mfn2 after treatment with PXA, as well as that of SLC25A46 (Fig. 3c–e). In contrast, co-immunoprecipitation of Calnexin was reduced upon treatment with PXA, suggesting that Mfn2 dissociates from the MAM with these treatments (Fig. 3f–g). The association of Mfn2 and NCLX was further investigated with a Proximity Ligation Assay (PLA). Cells were transfected with Mfn2-myc and NCLX-HA constructs and grown under glycolytic or respiring conditions after which they were processed for PLA with myc and HA antibodies along with immunostaining for mitochondria with chicken anti-Hsp60 antibody. The results show increased numbers of PLA spots in glycolytic cells treated with PXA (Fig. 3h–i) and in respiring cells treated with oligomycin (Fig. 3j–k). It should be noted that the PLA spots do not always coincide with Hsp60 fluorescence because the matrix appear more contracted than the membranes on which Mfn2 and NCLX reside. We conclude that the numbers of Mfn2 and NCLX proteins that are in close proximity increase with PXA and with oligomycin.
Oma1 and Opa1 control Mfn2-NCLX association and calcium release.
The results described thus far show increased association of Mfn2 and NCLX under stress inducing conditions. These stressors may induce proteolytic cleavage of Opa1 by the Oma1 protease on the mitochondrial inner membrane, which is induced by oligomycin in respiring cells most likely through ROS (MacVicar and Lane, 2014). Cleavage of Opa1 is known to open up mitochondrial cristae, which would allow for movement of NCLX from cristae where it has been shown to reside by immunoelectron microscopy (Palty et al., 2010) to the rim where it can come into contact with Mfn2. To test whether this sequence of events indeed controls the association of NCLX with Mfn2, we monitored the effects of Oma1 KO cells on ROS induced changes in mitochondrial morphology, interactions between Mfn2 and NCLX and calcium release. Knockout of Oma1 in HeLa cells was confirmed by Western blot (Fig. S4a). Oma1 KO suppresses the morphological effects induced by oligomycin in respiring cells, as observed with immunofluorescence microscopy and quantified using algorithms for aspect ratios (Fig. 4a–b). The effects of Oma1 KO on the association between Mfn2 and NCLX was determined with PLA. The marked increase in PLA spots that is induced by oligomycin in respiring cells is fully suppressed in Oma1 KO cells (Fig. 4c–d). Similar results were obtained with glycolytic cells treated with oligomycin and antimycin A (Fig. S4c–d). To verify that Oma1-mediated cleavage of Opa1 is responsible for the association of Mfn2 and NCLX in oligomycin treated cells, we also tested the effects of oligomycin in Drp1-Opa1 DKO cells. Our results show a higher number of spots than in WT cells, but no further increase upon treatment with oligomycin (Fig. 4e–f). These data support a pathway in which L-Opa1 impedes Mfn2-NCLX association, but that impediment is removed by Oma1-mediated cleavage of Opa1 upon treatment with oligomycin. To confirm that Oma1 affects calcium release, we monitored mitochondrial calcium under ROS inducing conditions in wildtype and Oma1 KO HeLa cells. We observed calcium release upon treatment of respiring cells with oligomycin and this calcium release was blocked by treatment with the NCLX inhibitor CGP37157 as well as in Oma1 KO cells (Fig. 4g–h).
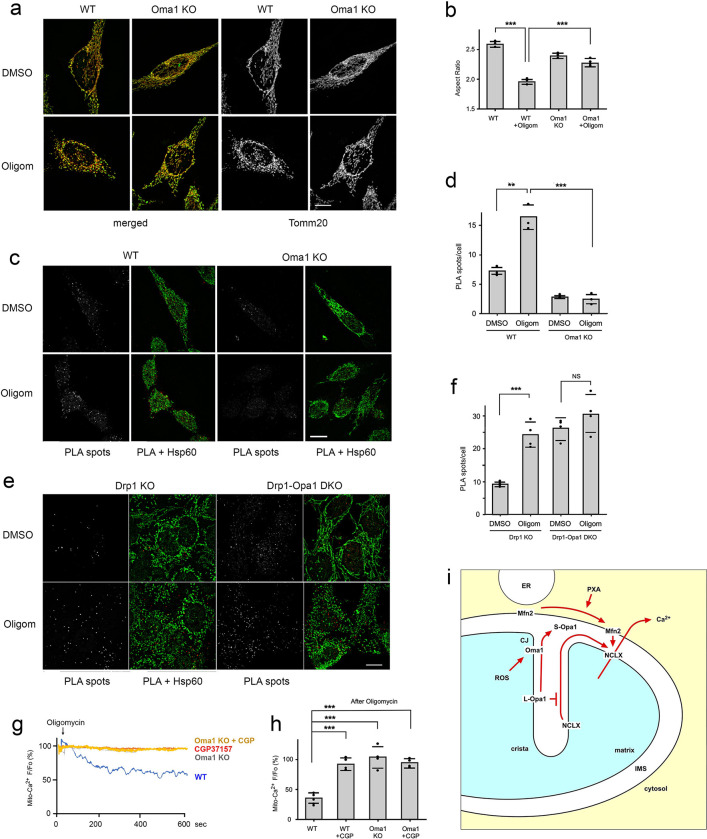
Fig. 4. Oma1 controls Mfn2-NCLX association and calcium release.
(a) Immunofluorescence of WT and Oma1 KO HeLa cells treated for 30 min with DMSO or 10 μM Oligomycin in respiring conditions. Merged images show Tomm20 (red) and Hsp60 (green), while the black and white images show Tomm20 labeling separately. Scale bar is 10 μm. (b) Aspect ratios of mitochondrial outer membranes (Tom20) under the conditions for panel b. Averages are shown for 3 independent experiments with SD and the results of Student’s T tests. Aspect ratios were determined for each condition with mitochondria in 25 cells per experiment. (c) PLA of Mfn2-myc and NCLX-HA (red spots) in WT and Oma1 KO HeLa cells grown under respiring conditions and treated for 30 min with DMSO or 10 μM Oligomycin. Mitochondria were detected by immunofluorescence with a chicken antibody against Hsp60 (green). Scale bar is 10 μm. (d) Average numbers of PLA spots per cells treated with DMSO or Oligomycin from 3 independent experiments with SD and results of a Student’s T test. (e) PLA of Mfn2-myc and NCLX-HA (red spots) in Drp1 KO and Drp1/Opa1DKO MEFs grown as in panel d under respiring conditions and treated for 30 min with DMSO or 10 μM Oligomycin. Mitochondria were detected by immunofluorescence with a chicken antibody against Hsp60 (green). Scale bar is 10 μm. (f) Average numbers of PLA spots per cells treated with DMSO or Oligomycin from 3 independent experiments with SD and results of a Student’s T test. (g) Tracings of mitochondrial matrix Ca2+ in WT and Oma1 KO Hela cells grown under respiring conditions and treated with 10 μM Oligomycin. Where indicated with CGP, cells were pre-incubated for 30 min at 37°C with 10uM CGP37157. (h) Changes in mitochondrial Ca2+ levels reflected by the relative fluorescence (F/Fo) at 600 sec. Averages are shown with SD for the data points in the histogram and significance was determined with a Student’s T test. (i) outline of pathway for PXA and ROS induced association of Mfn2 and NCLX, which connects stress in the mitochondrial matrix to cytoplasmic signaling through mitochondrial Ca2+ release (CJ, Cristae Junction).
Conclusions
Together these results describe a sequence of events in which ROS activates Oma1, which then opens up cristae by cleaving Opa1 and that allows for transfer of NCLX from cristae to the rim. NCLX can then associate with Mfn2 to promote calcium release from mitochondria (Fig. 4i). This pathway fits well with observations from other labs describing calcium release induced by mitochondrial stress. Those observations include acute damage induced actin assembly (ADA) in an NCLX and Oma1-dependent manner (Chakrabarti et al., 2022), frequent Oma1-dependent localized constrictions in the mitochondria of neuronal processes (Cho et al., 2017) and the block in CCCP-induced fission in Oma1 knockout cells (Quiros et al., 2012). The pathway depicted in Fig. 4i also provides a framework for the effects of heterozygous Opa1 loss of function mutations, which cause dominant optic atrophy through degenerative effects on retinal ganglion cells. A mouse model with a heterozygous Opa1 mutation showed sustained mitochondrial calcium release, which induces excessive mitophagy thereby leading to cell death (Zaninello et al., 2022). In this case, sustained calcium release may reflect increased association of NCLX with Mfn2 due to an inability to retain NCLX in cristae as a result of partial loss of Opa1 function. The pathway, which starts with triggers from within mitochondria, contrasts with the effects of external triggers, such as calcium released by the ER or extracellular calcium influx upon treatment with the calcium ionophore ionomycin. These external triggers promote INF2-dependent actin assembly at the MAM (Korobova et al., 2014; Korobova et al., 2013) and calcium uptake by mitochondria, which then leads to fission, but they do not depend on Oma1, as shown previously (Chakrabarti et al., 2018), in contrast with the Mfn2 and NCLX dependent process that we describe here, which does depend on Oma1 and thereby relies on internal triggers.
The pathway controlling calcium release from mitochondria through Mfn2 may also be relevant for diseases caused by mutations in Mfn2. The main Mfn2 disease is an axonal peripheral neuropathy known as Charcot Marie Tooth disease type 2A (CMT2A), but there are also effects of Mfn2 mutations in other tissues, such as cardiomyocytes (Franco et al., 2023). The cellular processes affected by mutations in Mfn2 include mitochondrial fusion and transport defects (Stuppia et al., 2015; Zhou et al., 2019), tethering to ER in the MAM (Bernard-Marissal et al., 2019) and mitophagy (Franco et al., 2023). It is not yet known whether disruptions of NCLX-mediated calcium release under control of Mfn2 also contribute to CMT disease, but it has been shown that deficiencies in Pink1-dependent phosphorylation of Mfn2 cause mitochondrial calcium overload, which could lead to necrosis (Gandhi et al., 2009) and conversely that NCLX activation by PKA can compensate for the calcium overload caused by loss of Pink1 (Kostic et al., 2015). More research is needed to untangle these different pathways. Our results, nevertheless, do show that Oma1 activation promotes the association between Mfn2 and NCLX and calcium release thus establishing a signaling pathway that connects ROS produced inside mitochondria with calcium release to the cytosol followed by mitochondrial fission.
Materials and methods
Antibodies and chemicals:
Rabbit polyclonal anti-Mfn1, NCLX, SLC25A46, and Tomm20 antibodies were from ProteinTech. Rabbit polyclonal anti-Mfn2 and Oma1 antibodies were from Cell Signaling. Rabbit polyclonal anti-Vinculin and Myc antibodies were from Sigma. Mouse monoclonal anti-Tubulin and Actin antibodies were from Sigma. Mouse monoclonal anti-CNX, Opa1 and Drp1 antibodies were from BD Pharmingen. Mouse monoclonal anti-Hsp60 antibodies were from Abcam. Mouse monoclonal anti-FLAG antibodies were from Proteintech. Mouse monoclonal anti-HA antibodies were from Millipore. Chicken polyclonal ant-Hsp60 antibodies were from EnCor. Oligomycin, Antimycin, CGP37157, CCCP, TMRM, DSP, SPDP and Dodecyl-maltoside were from Sigma. PXA was from Adipogene. Rhod-2AM was from Biotium. FLAG Magnetic Beads were from Pierce. BIX was from Tocris.
DNA constructs
The Mfn2–16xmyc, pCMV R-Cepia3mt and px459 plasmids were from Addgene (#23213, #140464 and #62988, respectively). The pPB EF1A>hNCLX-3xHA and pPB EF1A>hMfn2–3xFlag and pBase plasmids were from Vectorbuilder. The pCDNA3 hMfn2-FLAG plasmid was a kind gift from Gerald Dorn (Washington University). Target sites for gene deletions were identified using the Boutros Lab Website (http://www.e-crisp.org/E-CRISP/designcrispr.html) or as recommended by the Vectorbuilder website. The gRNAs were cloned in the px459 plasmid or in a custom vector from Vectorbuilder. The gRNAs for human Mfn2, Drp1 and Oma1 were: AGAGGCGGTTCGACTCATCA, TGGTTCATGAGGAAATGCAA and GGCTGGCATGGTTCATTTGT, respectively. The gRNAs for mouse Drp1 and Opa1 were CAGTGGGAAGAGCTCAGTGC and GTCAGACTGCTTTTGGAAAA. A pool of 3 gRNAs was used for human NCLX (ATCCTTGTCCGGGTCCACGA, TAACCAGCACGCCAGCGCCT and CCTGGATCTACCAACGGCAA) and for mouse NCLX (CCGACAAGGACGATCGGAAT, AATTCCGATCGTCCTTGTCG and ACCGGTTGTGGACCCCGACA), respectively. NCLX expression was knocked down in MEFs with shRNA as described (Palty et al., 2010) and in HeLa cells with mixture of 2 predesigned Dicer-Substrate siRNAs from IDT (cat nr. hs.Ri.SLC8B1.13.2 and hs.Ri.SLC8B1.13.3.
Cell culture, transfections and gene knockouts.
HeLa cells were from James Wohlschlegel (Dept. of Biological Chemistry, UCLA) and MEFs were from David Chan (Dept of Biology, CalTech). Cells were grown in glycolytic medium (DMEM with 10% FBS with 1% Pen/Strep and 4.5 g/l glucose) or respiring medium (DMEM without glucose and supplemented with 4.5g/L galactose, 10% FBS, 1% PS, 5mM sodium pyruvate and 2mM L-glutamine) as indicated. All cell lines were periodically checked for Mycoplasm. Transient transfections were done with jetPRIME following manufacturer’s instructions (Polyplus). For siRNA, cells were grown in 6cm dishes, transfected with 50nM oligonucleotides using RNAimax (Invitrogen) and analyzed 72h later. For gene knockouts, 1.2μg/well was transfected into 6 well plates cells, followed by selection at increasing concentrations or puromycin from 1μg to 5 μg/ml for 1–2 days. Surviving colonies were isolated and analyzed with Western blots. A stable cell line that expresses human Mfn2 with 3XFLAG under the EF1alpha promoter was generated with a PiggyBac construct from Vectorbuilder. This construct was transfected into Drp1/Mfn2 DKO MEFs along with the PBase vector from Vectorbuilder at equal concentrations (2μg/per well in a 6 well plate). At 24h post transfection, cells were subjected to puromycin selection at increasing concentrations from 1μg to 5 μg/ml for 1–2 days. After selection, surviving colonies were isolated with cloning rings, expanded and analyzed for Mfn2 expression levels by Western blotting.
Immunoblotting and immunofluorescence:
Total cell lysates for Western blots were made with RIPA buffer. Samples were subjected to SDS-PAGE, transferred to PVDF membranes, blocked with 5% non-fat milk and incubated overnight at 4°C with primary antibodies. Membranes were then washed with TBST and incubated with secondary antibodies. Chemiluminescent bands were detected with a BioRad scanner. For immunofluorescence images, cells were grown on 12mm coverslips, fixed for 15min with 4% paraformaldehyde in PBS, and permeabilized for 15min with 0.25% Triton X-100 in PBS, blocked for 1h with BSA in PBS-T and incubated with primary antibodies. Secondary antibodies were Alexa Fluor 488-, 594- or 647-conjugated goat anti-mouse or rabbit IgG (Invitrogen).
Proximity ligation assays were conducted with Duolink as recommended by the manufacturer (Sigma-Aldrich). Cells were seeded on coverslips and at 60% confluency they were transfected with Mfn2-myc and NCLX-HA overnight. The cells were then treated for 5–10 min with 10μM PXA or for 20–30min 10μM oligomycin or DMSO as indicated, after which they were processed for the PLA as described(Alam, 2018), but with the following modification: After the fixation and washing, cells were permeabilized with 0.3% Triton-X100 for 10 minutes at room temperature and then washed twice for 5 min with PBS. Along with the primary antibodies for PLA, we also added chicken anti-HSP60 antibody (EnCor Biotechnology Inc) and anti-chicken Alexa Fluor 488 as secondary antibody, along with the primary and secondary antibodies for PLA.
Microscopy was with a Marianas spinning disc confocal from Intelligent Imaging, which uses an Axiovert microscope (Carl Zeiss Microscopy) with 40x/1.4 and 100x/1.4 oil objectives, a CSU22 spinning disk (Yokogawa), an Evolve 512 EMCCD camera (Photometrics) and a temperature unit (Okolab). For live cell imaging, cells were grown in glass bottom dishes (MatTek) and imaged in situ at 37°C. Fiji software was used to determine Manders’ coefficients and to generate scatterplots. Aspect ratios of mitochondria were determined with Tomm20 fluorescence images. First, cells within the images were demarcated with FIJI ImageJ software (https://fiji.sc/). Mitochondria were then demarcated with CellProfiler cell image analysis software (https://cellprofiler.org/), while background noise was suppressed with a median filter. Mitochondria were segmented with a global, three-class, Otsu-thresholding method, minimizing the weighted-variance to shape. The aspect ratio of each object was then determined as the quotient of Major Axis Length over Minor Axis Length. Data represent the average aspect ratio for 25 cells in 3 independent experiments using WT and Oma1 KO HeLa cells.
Mitochondrial Ca2+ levels in HeLa cells were determined with cells that were transiently transfected with pCMV R-Cepia3mt (Addgene #140464). These transfected cells were first perfused for 30 min with HBSS to record a baseline signal. Where indicated 10 μM CGP37157 or 10 μM BIX was included with the perfusion. Mitochondrial Ca2+ release was then triggered by adding 10 μM PXA or 10 μM Oligomycin. Using 3I software, fluorescence intensities of individual cells was determined once per second over a period of 10 min after the addition of PXA or Oligomycin. Mitochondrial calcium levels in MEFs were determined with a Tecan Spark 10M multimode plate reader equipped with an injector as previously described (Martinez et al., 2017). In brief, MEF DRP1 KO or double MFN2/DRPI KO cells were plated on Corning 96 Flat black, clear bottom wells and loaded with 1 μM Rhod2-AM for 30 min at 37 °C using a modified Krebs–Ringer’s solution containing (126 mM NaCl, 5.4 mM KCl, 0.8 mM MgCl2, 20 mM HEPES, 1.8 mM CaCl2, 15 mM glucose, with pH adjusted to 7.4 with NaOH and supplemented with 0.1% BSA) After dye loading, cells were washed three times with fresh dye-free Krebs–Ringer’s solution, followed by additional incubation of 30 min to allow for the de-esterification of the residual dye. Kinetic live-cell fluorescent imaging was performed to monitor Ca2+ transients. Rhod2-AM was excited at 552 nm wavelength light and imaged with a 570 nm. After establishing a baseline, cells were triggered with PXA at a final concentration of 10 μM. Kinetic measurements were taken at ~ 5 s interval. Traces of Ca2+ responses were analyzed and plotted using KaleidaGraph. The rate of ion transport was calculated from each graph (summarizing an individual experiment) by a linear fit of the change in the fluorescence over time (ΔF/dt), as previously described (Assali et al., 2020; Taha et al., 2024).
Mitochondrial membrane potential was detected with TMRM (Thermo-Fisher Scientific). Cells plated in 35 mm glass bottom dishes (MatTek) were rinsed with 10 mM HEPES buffered HBSS, pH 7.3 with 15 mM glucose and subsequently incubated with 25 nM TMRM for 30 min at 37°C, followed by 3 washes with PBS and destaining for 30min at 37°C in 1.8mM CaCl2, 120mM NaCl, 5.4mM KCl, 0.8mM MgCl2, 20mM HEPES, 15mM glucose, adjusted to pH 7.3 with NaOH. Fluorescence images were acquired for a field of cells with 10x objective. Where indicated, cells were treated for 30 min at 37°C with 10 μM CGP37157 or 100 nM BIX before adding 10 μM PXA. As reference for uncoupling, cells were treated with 10 μM CCCP.
Cellular Thermal Shift Assay (CETSA)
MEFs cultured to 1.0 × 107 cells in a 15 cm dish were washed with PBS and harvested in 2 ml lysis buffer (1.8mM CaCl2, 120mM NaCl, 5.4mM KCl, 0.8mM MgCl2, 20mM HEPES, 15mM glucose, 0.5% Dodecyl-Maltoside, adjusted to pH 7.3 with NaOH). Equal volumes of cell lysates were transferred to 1.5 ml Eppendorf tubes for incubation with 1 μM PXA or DMSO for 4 min at RT. After this incubation, 100 μl aliquots of cell lysates were transferred to PCR tubes and heated on a gradient PCR machine for 3 min at different temperatures as indicated and then cooled to 25°C for 2 min, followed by centrifugation for 30 min at 20.000g in 4°C. Supernatants were then transferred to new tubes with sample buffer. For each temperature, 10 μl (approximately 30 μg protein) was analyzed by SDS-PAGE and Western blotting. Band intensities were quantified with densitometry and ImageJ.
Co-immunoprecipitation
MEFs stably expressing Mfn2-FLAG were treated for 5 minutes at 37°C with DMSO, 10μM PXA, 10μM CCCP or 10μM CGP37157 as indicated. These cells were then washed with ice cold PBS with 0.9 mM CaCl2 and 0.5 mM MgCl2 and crosslinked for 2h on ice with 1 mM DSP and 1 mM SPDP. The crosslinkers were then quenched by incubating for 15 minutes on ice with 20 mM Tris-Cl (pH7.4) followed by a wash with ice cold PBS with 0.9 mM CaCl2 and 0.5 mM MgCl2 and resuspending in RIPA buffer. Co-immunoprecipitations were conducted with 1 mg protein (determined with BCA assay). 50 μl of magnetic FLAG beads (Pierce/Sigma) per sample were washed twice with 1X TBST and then incubated with protein samples for 45min at 4°C on a rotor. Beads were then collected using a magnetic stand and the supernatant was discarded. The beads were washed twice with 1X TBST and then once with ddH2O. Finally, the beads were resuspended in 50μl of 2X sample buffer, heated for 5 min at 95°C and analyzed with Western blots.
Supplementary Material
Acknowledgements
CMK was supported by NIH grants R01GM61721, R01GM073981 and R01DK101780. AMvdB was supported by NIH grants U01GM109764 and R01NS120690.
References
- Alam M.S. 2018. Proximity Ligation Assay (PLA). Curr Protoc Immunol. 123:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assali E.A., Jones A.E., Veliova M., Acin-Perez R., Taha M., Miller N., Shum M., Oliveira M.F., Las G., Liesa M., Sekler I., and Shirihai O.S.. 2020. NCLX prevents cell death during adrenergic activation of the brown adipose tissue. Nat Commun. 11:3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard-Marissal N., van Hameren G., Juneja M., Pellegrino C., Louhivuori L., Bartesaghi L., Rochat C., El Mansour O., Medard J.J., Croisier M., Maclachlan C., Poirot O., Uhlen P., Timmerman V., Tricaud N., Schneider B.L., and Chrast R.. 2019. Altered interplay between endoplasmic reticulum and mitochondria in Charcot-Marie-Tooth type 2A neuropathy. Proc Natl Acad Sci U S A. 116:2328–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohler P., Stuhldreier F., Anand R., Kondadi A.K., Schlutermann D., Berleth N., Deitersen J., Wallot-Hieke N., Wu W., Frank M., Niemann H., Wesbuer E., Barbian A., Luyten T., Parys J.B., Weidtkamp-Peters S., Borchardt A., Reichert A.S., Pena-Blanco A., Garcia-Saez A.J., Itskanov S., van der Bliek A.M., Proksch P., Wesselborg S., and Stork B.. 2018. The mycotoxin phomoxanthone A disturbs the form and function of the inner mitochondrial membrane. Cell Death Dis. 9:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., Fung T.S., Kang T., Elonkirjo P.W., Suomalainen A., Usherwood E.J., and Higgs H.N.. 2022. Mitochondrial dysfunction triggers actin polymerization necessary for rapid glycolytic activation. J Cell Biol. 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., Ji W.K., Stan R.V., de Juan Sanz J., Ryan T.A., and Higgs H.N.. 2018. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol. 217:251–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 160:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B., Cho H.M., Jo Y., Kim H.D., Song M., Moon C., Kim H., Kim K., Sesaki H., Rhyu I.J., Kim H., and Sun W.. 2017. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat Commun. 8:15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito O.M., and Scorrano L.. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456:605–610. [DOI] [PubMed] [Google Scholar]
- Franco A., Li J., Kelly D.P., Hershberger R.E., Marian A.J., Lewis R.M., Song M., Dang X., Schmidt A.D., Mathyer M.E., Edwards J.R., Strong C.G., and Dorn G.W.. 2023. A human mitofusin 2 mutation can cause mitophagic cardiomyopathy. Elife. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Wood-Kaczmar A., Yao Z., Plun-Favreau H., Deas E., Klupsch K., Downward J., Latchman D.S., Tabrizi S.J., Wood N.W., Duchen M.R., and Abramov A.Y.. 2009. PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell. 33:627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Alvarez M.I., Sebastian D., Vives S., Ivanova S., Bartoccioni P., Kakimoto P., Plana N., Veiga S.R., Hernandez V., Vasconcelos N., Peddinti G., Adrover A., Jove M., Pamplona R., Gordaliza-Alaguero I., Calvo E., Cabre N., Castro R., Kuzmanic A., Boutant M., Sala D., Hyotylainen T., Oresic M., Fort J., Errasti-Murugarren E., Rodrigues C.M.P., Orozco M., Joven J., Canto C., Palacin M., Fernandez-Veledo S., Vendrell J., and Zorzano A.. 2019. Deficient Endoplasmic Reticulum-Mitochondrial Phosphatidylserine Transfer Causes Liver Disease. Cell. 177:881–895 e817. [DOI] [PubMed] [Google Scholar]
- Inagaki S., Suzuki Y., Kawasaki K., Kondo R., Imaizumi Y., and Yamamura H.. 2023. Mitofusin 1 and 2 differentially regulate mitochondrial function underlying Ca(2+) signaling and proliferation in rat aortic smooth muscle cells. Biochem Biophys Res Commun. 645:137–146. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Lee Y.J., Gaume B., Jeong S.Y., Frank S., Nechushtan A., Santel A., Fuller M., Smith C.L., and Youle R.J.. 2002. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 159:931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Gauvin T.J., and Higgs H.N.. 2014. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 24:409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V., and Higgs H.N.. 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339:464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic M., Ludtmann M.H., Bading H., Hershfinkel M., Steer E., Chu C.T., Abramov A.Y., and Sekler I.. 2015. PKA Phosphorylation of NCLX Reverses Mitochondrial Calcium Overload and Depolarization, Promoting Survival of PINK1-Deficient Dopaminergic Neurons. Cell Rep. 13:376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol. 30:357–391. [DOI] [PubMed] [Google Scholar]
- MacVicar T.D., and Lane J.D.. 2014. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J Cell Sci. 127:2313–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Martinez N.A., and Silva W.I.. 2017. Measurement of the Intracellular Calcium Concentration with Fura-2 AM Using a Fluorescence Plate Reader. Bio Protoc. 7:e2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier A., Motori E., Brandt T., Lagouge M., Atanassov I., Galinier A., Rappl G., Brodesser S., Hultenby K., Dieterich C., and Larsson N.G.. 2015. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J Cell Biol. 208:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R., Silverman W.F., Hershfinkel M., Caporale T., Sensi S.L., Parnis J., Nolte C., Fishman D., Shoshan-Barmatz V., Herrmann S., Khananshvili D., and Sekler I.. 2010. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci U S A. 107:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros P.M., Ramsay A.J., Sala D., Fernandez-Vizarra E., Rodriguez F., Peinado J.R., Fernandez-Garcia M.S., Vega J.A., Enriquez J.A., Zorzano A., and Lopez-Otin C.. 2012. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. The EMBO journal. 31:2117–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta K., Mirams G.R., and Parekh A.B.. 2018. Sequential forward and reverse transport of the Na(+) Ca(2+) exchanger generates Ca(2+) oscillations within mitochondria. Nat Commun. 9:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A., and Fuller M.T.. 2001. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 114:867–874. [DOI] [PubMed] [Google Scholar]
- Sebastian D., Hernandez-Alvarez M.I., Segales J., Sorianello E., Munoz J.P., Sala D., Waget A., Liesa M., Paz J.C., Gopalacharyulu P., Oresic M., Pich S., Burcelin R., Palacin M., and Zorzano A.. 2012. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc Natl Acad Sci U S A. 109:5523–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J., Vashisht A.A., Wan J., Jen J.C., Claypool S.M., Wohlschlegel J.A., and Koehler C.M.. 2017. Rapid degradation of mutant SLC25A46 by the ubiquitin-proteasome system results in MFN1/2-mediated hyperfusion of mitochondria. Mol Biol Cell. 28:600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuppia G., Rizzo F., Riboldi G., Del Bo R., Nizzardo M., Simone C., Comi G.P., Bresolin N., and Corti S.. 2015. MFN2-related neuropathies: Clinical features, molecular pathogenesis and therapeutic perspectives. J Neurol Sci. 356:7–18. [DOI] [PubMed] [Google Scholar]
- Taha M., Assali E.A., Ben-Kasus Nissim T., Stutzmann G.E., Shirihai O.S., Hershfinkel M., and Sekler I.. 2024. NCLX controls hepatic mitochondrial Ca(2+) extrusion and couples hormone-mediated mitochondrial Ca(2+) oscillations with gluconeogenesis. Mol Metab. 87:101982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., Ehses S., Krause F., Chan D.C., Alexander C., Bauer C., Youle R., Langer T., and Martinou J.C.. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28:1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaninello M., Palikaras K., Sotiriou A., Tavernarakis N., and Scorrano L.. 2022. Sustained intracellular calcium rise mediates neuronal mitophagy in models of autosomal dominant optic atrophy. Cell Death Differ. 29:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Carmona S., Muhammad A., Bell S., Landeros J., Vazquez M., Ho R., Franco A., Lu B., Dorn G.W. 2nd, Wang S., Lutz C.M., and Baloh R.H.. 2019. Restoring mitofusin balance prevents axonal degeneration in a Charcot-Marie-Tooth type 2A model. J Clin Invest. 129:1756–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.