Abstract
Dissecting biological pathways highlighted by Mendelian gene discovery has provided critical insights into the pathogenesis of Parkinson’s disease (PD) and neurodegeneration. This approach ultimately catalyzes the identification of potential biomarkers and therapeutic targets. Here, we identify PSMF1 as a new gene implicated in PD and childhood neurodegeneration. We find that biallelic PSMF1 missense and loss-of-function variants co-segregate with phenotypes from early-onset PD and parkinsonism to perinatal lethality with neurological manifestations across 15 unrelated pedigrees with 22 affected subjects, showing clear genotype-phenotype correlation. PSMF1 encodes the proteasome regulator PSMF1/PI31, a highly conserved, ubiquitously expressed partner of the 20S proteasome and neurodegeneration-associated F-box-O 7 and valosin-containing proteins. We demonstrate that PSMF1 variants impair mitochondrial membrane potential, dynamics and mitophagy in patient-derived fibroblasts. Additionally, we develop models of psmf1 knockdown Drosophila and Psmf1 conditional knockout mouse exhibiting age-dependent motor impairment, with diffuse gliosis in mice. These findings unequivocally link defective PSMF1 to early-onset PD and neurodegeneration and suggest mitochondrial dysfunction as a mechanistic contributor.
INTRODUCTION
Parkinson’s disease (PD), the second-most prevalent neurodegenerative disorder worldwide, is projected to grow exponentially with no cure currently available.1,2 Notably, up to 15% of PD cases are linked to mono- and biallelic pathogenic or risk variants in single genes.3 Monogenic forms of PD and parkinsonism have unveiled crucial pathomechanisms that also underpin sporadic forms, highlighting a complex neurobiological network.1,4 This involves α-synuclein synthesis and dynamic equilibrium (SNCA), homeostasis of cytoplasmic components and organelles via the autophagy-endolysosomal system (GBA1, LRRK2, VPS35), proteostasis through the ubiquitin-proteasome system (PRKN, FBXO7), mitochondrial functions (PRKN, PINK1, FBXO7), oxidative stress response (DJ-1) and neuroinflammation regulation (LRRK2).1,4,5 Disruption in these pathways culminates in the loss of dopaminergic neurons in the substantia nigra pars compacta, manifesting clinically as parkinsonism.1,5 Exploring biopathways highlighted by Mendelian gene discovery fosters the identification of biomarkers and development of disease-modifying strategies for PD, as exemplified with LRRK2 inhibitors.6–8
Perturbations of the ubiquitin-proteasome system and mitochondrial processes are solidly recognized as pivotal, interconnected drivers of neurodegeneration in PD, ultimately contributing to dopaminergic neuron vulnerability through proteotoxicity and bioenergetic failure.9–13 Within this framework, PSMF1, which encodes the proteasome regulator PSMF1/PI31,14,15 emerges as a significant gene.
PSMF1 is a highly conserved 31-kDa protein which is ubiquitously expressed across human tissues.15,16 In vitro, PSMF1 inhibits peptide hydrolysis by the 20S proteasome through its C-terminal proline-rich domain, via either direct binding or competitive binding to the 20S activating particles PA700 and PA28.15,17 However, PSMF1 can also stimulates 26S proteasome-mediated proteolysis in vitro.18 In vivo studies demonstrate a biological function of PSMF1 in promoting protein breakdown. PSMF1 can stimulate 26S assembly,19 and Psmf1 inactivation in Drosophila and mice causes accumulation of poly-ubiquitinated aggregates, which are exclusive substrates of the 26S proteasome.18–21 Furthermore, PSMF1 promotes proteolysis in Drosophila, mouse, yeast and plants.18,22–24 Finally, it mediates fast transport of proteasomes between neurosomes and synapses and is required for synapse maintenance and neuronal survival in Drosophila and mice.20,22 Notably, PSMF1 interacts with key proteins implicated in human neurodegeneration. For instance, PSMF1 is a high-affinity binding partner of F-box only protein 7 (FBXO7), whose genetic defects cause juvenile-onset PD/parkinsonism,25 through their respective N-terminal FBXO7/PSMF1 (FP) domain.26 In human fibroblasts, the ablation of this heterodimer by an FBXO7 missense variant leads to reduced expression and stability of PSMF1 and FBXO7 as well as proteasomal and mitochondrial dysfunction.27 Intriguingly, FBXO7 prevents PSMF1 from proteolytic cleavage in Drosophila and mouse, suggesting that FBXO7 loss may cause proteasomal impairment by PSMF1 inactivation.18,28 Additionally, PSMF1 directly binds valosin-containing protein (VCP),29 whose defective functions are linked to amyotrophic lateral sclerosis, frontotemporal dementia and Huntington disease,30,31 with VCP and PSMF1 acting as in vitro up- and down-regulator of proteasomal activity, respectively.29 Finally, PSMF1 variants have also been associated with Alzheimer’s disease.32,33
This study identifies PSMF1 as a new gene implicated in PD and early human neurodegeneration. By extensive data mining and functional validation, we establish that biallelic PSMF1 missense and loss-of-function variants cause a phenotypic spectrum ranging from early-onset PD to perinatal lethality with neurological manifestations, with clear genotype-phenotype correlation. We define the organellar and cellular consequences of PSMF1 deficiency in vitro through mitochondria live imaging of patient-derived cultured fibroblasts. Finally, we explore its pathological consequences in vivo in Drosophila and mouse models.
RESULTS
Identification of biallelic PSMF1 variants in affected subjects from 15 unrelated, ethnically diverse families
In a nonconsanguineous European family (Pedigree A; Fig.1a) with the male proband (A-II-2) affected by early-onset parkinsonism, patient-parent trio exome sequencing (ES) revealed he was compound heterozygote for one missense variant (c.724C>G, p.Arg242Gly) and one splice variant (c.282+2T>A) in the candidate gene PSMF1 (NM_006814.5). Through initial data sharing, we identified a consanguineous West Asian family (Pedigree B; Fig.1a) with two siblings (B-II-3; B-II-4) manifesting with early-onset PD and harboring the same PSMF1 missense variant detected in Pedigree A in the homozygous state.34 To replicate the association of biallelic PSMF1 variants with PD/parkinsonism and further delineate PSMF1-related disorder, we applied a genotype-first approach by interrogating large next-generation sequencing datasets from collaborative networks and platforms, including Queen Square Genomics (Pedigrees G-J-K), 100,000 Genomes Project, UK Biobank (UKB), The Accelerating Medicines Partnership program for Parkinson's disease (AMP-PD), Solve-RD, CENTOGENE (Pedigrees B-C-L), GeneDx, Baylor Genetics, Genesis (Pedigree E), ClinVar, VarSome, GeneMatcher (Pedigrees H-I-N-O),35 and from several diagnostic and research genetic laboratories worldwide (Pedigrees C-D-E-F-K-M). Pedigrees identified from multiple sources were deduplicated.
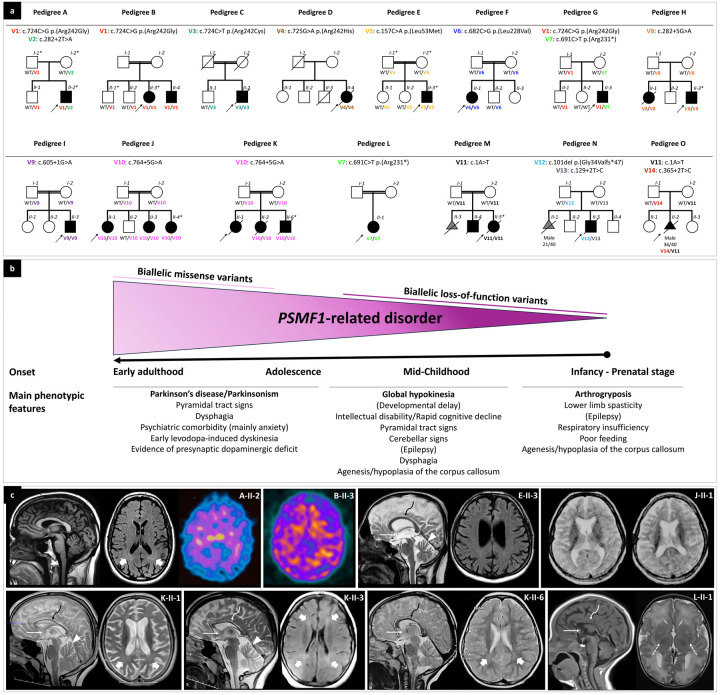
Figure 1. Partial pedigrees and phenotypic features of individuals with biallelic PSMF1 variants.
(a) Partial pedigrees of PSMF1 families. Simplified pedigrees of 15 unrelated PSMF1 families are presented, with the results of segregation analysis of PSMF1 variants. In the diagrams, males are represented by squares, females by circles and fetuses by triangles. Fetal gender and gestational age at death (n weeks/40 weeks) are reported below their symbol, if available. Consanguineous marriages are denoted by double lines between symbols, while probands are marked with an arrow. Individuals exhibiting a neurological disease phenotype are represented by black filled symbols. Fetuses aborted for undetermined cause (M-II-3, N-II-1) are represented by a gray filled symbol. Roman numerals indicate generations, whereas Arabic numbers denote individuals within each generation. Variants are referenced to PSMF1 transcript NM_006814.5 and displayed with different font colors (see Fig. 2a and Supplementary File 1). “WT” designates the wild-type PSMF1 allele. An asterisk identifies dermal fibroblast donors. Fully identifying pedigrees are available upon request to the corresponding author. (b) Genotype-phenotype correlation in PSMF1-related disorder. Schematic representation of the core phenotypic features associated with biallelic variants in PSMF1, with indication of age at onset and main clinical manifestations in subgroups of subjects (see main text). Increasingly severe phenotypes correspond to progressively more deleterious mutational effects. (c) Neuroimaging features of PSMF1-related disorder. Brain MRI with sagittal T1- or T2-weighted images and axial FLAIR or T2-weighted images of individuals A-II-2 (25–30 years), E-II-3 (25–30 years), J-II-1 (10–15 years), K-II-1 (10–15 years), K-II-3 (10–15 years), K-II-6 (0–5 years) and L-II-1 (infancy). Mild-to-moderate enlargement of the cerebral subarachnoid spaces is noted in subjects A-II-2, E-II-3, J-II-1, K-II-1, and K-II-3. Enlargement of the subarachnoid spaces of the superior cerebellar vermis is present in individuals E-II-3, K-II-1, and K-II-3. Additional faint T2/FLAIR-signal alterations are visible in the periventricular white matter in individuals A-II-2, K-II-1, K-II-3, and K-II-6 (thick arrows). There is hypoplasia of corpus callosum (curved arrows) and small anterior commissure (thin arrows) in subjects E-II-3, J-II-1, K-II-1, K-II-3, K-II-6, and L-II-1. There is lack of myelination of the posterior limbs of the internal capsules in patient L-II-1 (dashed arrows), associated with foci of T2 hyperintensity at the level of the putamina. DaTscan of subjects A-II-2 and B-II-3 reveals severely reduced tracer uptake in the striatum bilaterally, with increased background activity.
Biallelic PSMF1 variants cause a phenotypic spectrum from early-onset PD or parkinsonism to perinatal lethality with neurological manifestations
A total of 22 affected subjects from 15 unrelated families of European, Asian and African ancestry were identified (Fig.1a). Genotype was determined for all but two individuals (K-II-1, M-II-4; DNA not available), who were included due to striking clinical resemblance to their affected siblings carrying homozygous PSMF1 variants in the context of parental consanguinity. One aborted fetus (O-II-2) was included based on PSMF1 variants identified on reanalysis of the ES data from prenatal genetic testing and phenotypic overlap with the proband of Pedigrees M-N, who died during infancy. Pregnancy losses in Pedigrees M-N were excluded due to insufficient pheno-genotypic information. Clinical and neuroimaging features are presented in Fig.1 and Table 1.
Table 1.
Simplified phenotypic features and genotype of 22 subjects with PSMF1-related disorder belonging to 15 families
| Pedigree | Pedigree A | Pedigree B | Pedigree C | Pedigree D | Pedigree E | Pedigree F | Pedigree G | Pedigree H | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | A-II-2 | B-II-3 | B-II-4 | C-II-2 | D-II-4 | E-II-3 | F-II-1 | G-II-3 | H-II-1 | H-II-3 | |
| Sex/Current age | M/25–30 yrs | F/35–40 yrs | M/35–40 yrs | M/Deceased (40–45 yrs) | F/Deceased (65–70 yrs) | M/30–35 yrs | M/30–35 yrs | M/15–20 yrs | F/Deceased (15–20 yrs) | M/15–20 yrs | |
| Parental consanguinity | No | Yes | Yes | Yes | No | Yes | Yes | No | No | No | |
| Family history | Unremarkable | One sibling affected | One sibling affected | PD; deaths in childhood | Unremarkable | Unremarkable | Unremarkable | Unremarkable | One sibling affected | One sibling affected | |
| Core phenotype | Parkinsonism | Parkinsonism | Parkinsonism | Parkinsonism | Parkinsonism | Parkinsonism | Parkinsonism | Parkinsonism | Parkinsonian-pyramidal syndrome | Spastic-ataxia syndrome | |
| Age at onset | 20–25 yrs | 15–20 yrs | 15–20 yrs | 25–30 yrs | 45–50 yrs | 15–20 yrs | 0–5 yrs | 0–5 yrs | 0–5 yrs | 0–5 yrs | |
| Symptom at onset | Bradykinesia, dysphagia, anxiety | Right hand tremor | Right hand tremor | Micrographia | Left upper limb tremor | Hand tremor, left foot dragging, anxiety | Dystonic head tremor |
Clumsiness, frequent falls without fall reflex |
Limb spasticity | Limb spasticity | |
| Neurological manifestations | |||||||||||
| Motor features | Parkinsonism. Eye movement abnormalities. Dysarthria. Gait festination. Balance difficulty. Pyramidal signs. Mild cerebellar signs. |
Parkinsonism. Dysarthria. Freezing of gait. Balance difficulty. |
Parkinsonism. Blepharospasm. Dysarthria. Postural and kinetic tremor. Freezing of gait to tiptoe walking. Pyramidal signs. |
Parkinsonism. Blepharospasm. Pyramidal signs. Freezing of gait Falls. Tiptoe walking. |
Parkinsonism. Restless leg syndrome. Freezing of gait. |
Parkinsonism. Postural tremor. Pyramidal signs. |
Tremulous segmental dystonia. Parkinsonism. |
Parkinsonism. Action tremor. Blepharospasm. Hyperreflexia. Myoclonus. Frequent episodes of freezing. |
Parkinsonism. Pyramidal signs. Ataxia. |
MDD. Pyramidal signs. Ataxia. |
|
| Response to treatment | Mild-to-moderate response to LD. Early-onset LD-induced dyskinesia. |
Mild-to-moderate response to LD. Early-onset LD-induced dyskinesia. |
No response to LD. Mild response to DA. Early-onset dyskinesia. |
Initial good response to LD. Severe motor fluctuations and LD-induced dyskinesia. Initial good response to DBS. |
Initial good response to LD. Severe motor fluctuations and LD-induced dyskinesia. Initial good response to DBS. |
Moderate response to LD. LD-induced dyskinesia. |
Poor response to LD. | N/A | Good response to levodopa. | N/A | |
| Brain MRI (age) | Mild cerebral atrophy, faint T2 hyperintensity in the posterior periventricular WM (20–25 yrs) | Normal. | Mild atrophy of the inferior cerebellar vermis. | Normal. | Normal. | Mild-to-moderate cerebral atrophy, thin CC, small AC, small midbrain, mild atrophy of superior cerebellar vermis (25–30 yrs) | Mild cerebellar cortical atrophy. | Retrocerebellar cyst. | Atrophy of cerebral hemispheres, basal ganglia, thalami, brainstem and cerebellum, with mineralization of the lentiform nuclei. | Mild T2 signal alterations in the anterior midbrain bilaterally. | |
| Other instrumental investigations | DaTscan: abnormal. MIBG: normal. DEXA: osteopenia. |
EEG: normal. EMG/NCS: normal. |
EEG: normal. EMG/NCS: normal. Muscle biopsy: normal. |
F-DOPA-PET: abnormal. FDG-PET: temporal hypometabolism. |
- | US: no organomegaly. Ophthalmological assessment: normal. Normal CK, lactate, pyruvate, copper, caeruloplasmin and 24-hour urinary copper excretion. |
EEG/Video-EEG: normal. | Low testosterone level. EEG: cerebral dysrhythmia. VEP: prolonged latency. Brain MRS: normal. Muscle MRI: atrophy of gluteal muscles, with increased muscle fat. DEXA: severe osteopenia. |
DaTscan: abnormal. | Muscle biopsy and enzymology: normal. | |
| Genotype PSMF1 (NM_006814.5) | C-HET c.724C>G p.(Arg242Gly) c.282+2T>A | HOM c.724C>G p.(Arg242Gly) | HOM c.724C>G p.(Arg242Gly) | HOM c.724C>T p.(Arg242Cys) | HOM c.725G>A p.(Arg242His) | HOM c.157C>A p.(Leu53Met) | HOM c.682C>G p.(Leu228Val) | C-HET c.724C>G p.(Arg242Gly) c.691C>T p.(Arg231*) | HOM c.282+5G>A | HOM c.282+5G>A | |
| Pedigree | Pedigree I | Pedigree J | Pedigree K | Pedigree L | Pedigree M | Pedigree N | Pedigree O | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | I-II-3 | J-II-1 | J-II-3 | J-II-4 | K-II-1 | K-II-3 | K-II-6 | L-II-1 | M-II-4 | M-II-5 | N-II-3 | O-II-2 | |
| Sex/Current age | M/20–25 yrs | F/10–15 yrs | F/5–10 yrs | F/5–10 yrs | F/Deceased (15–20 yrs) | F/Deceased (15–20 yrs) | M/Deceased (15–20 yrs) | F/0–1 yr | M/Deceased (0–1 yr) | F/Deceased (0–1 yr) | M/Deceased (0–1 yr) | M/Feticide (GA third trimester) | |
| Parental consanguinity | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | No | |
| Family history | Unremarkable | Two siblings affected | Two siblings affected | Two siblings affected | Two siblings affected | Two siblings affected | Two siblings affected | Unremarkable | One pregnancy loss; one sibling affected. | One pregnancy loss; one sibling affected. | One miscarriage | Unremarkable | |
| Core phenotype | DD with CC agenesis | DD with CC hypoplasia | DD with CC hypoplasia | DD with CC hypoplasia | Parkinsonian-pyramidal syndrome with CC hypoplasia | Parkinsonian-pyramidal syndrome with CC hypoplasia | Parkinsonian-pyramidal syndrome with CC hypoplasia | Arthrogryposis with CC hypoplasia | Arthrogryposis | Arthrogryposis with CC agenesis | Arthrogryposis | Arthrogryposis with CC agenesis | |
| Age at onset | Early childhood | Early childhood | Early childhood | Early childhood | 10–15 yrs | 10–15 yrs | Infancy | Prenatal | Prenatal | Prenatal | Prenatal | Prenatal | |
| Symptom at onset Neurological manifestations | GDD | GDD | SLDD | GDD | Memory and attentional deficits | Memory and attentional deficits | Epilepsy | N/A | N/A* | N/A | N/A | N/A | |
| Motor features | Global hypokinesia. Clumsiness. Mild hypotonia. |
Hypokinesia. Broad-based gait. Talipes. |
Hypokinesia | Hypokinesia. Talipes. |
Hypokinesia. Pyramidal tract signs. |
Hypokinesia. Pyramidal tract signs. |
Hypokinesia. Pyramidal tract signs. |
Arthrogryposis. Limb spasticity. Increased jitteriness. |
N/A* | Myoclonus. Arthrogryposis. Limb spasticity. Truncal hypotonia. GDD. |
Arthrogryposis. | Arthrogryposis | |
| Response to treatment | N/A | Partial response to LEV. Good rresponse to CBZ and VPA | N/A | N/A | N/A | Partial response to CBZ and VPA | Unspecified response to CBZ | N/A | N/A* | Poor response to AED. | N/A | N/A | |
| Brain MRI (age) | CC agenesis, malformation of the left cerebellar hemisphere, cerebellar vermis hypoplasia. Arachnoid cyst. |
Moderate cerebral atrophy, thin CC (10–15 yrs) | N/A | N/A | Progressive mild-to-moderate cerebral atrophy, CC hypoplasia, small AC, faint periventricular T2 signal alterations, mild superior cerebellar vermis atrophy (10–15 yrs) | Moderate cerebral atrophy, CC hypoplasia, small AC, faint periventricular T2 signal alterations, mild superior cerebellar vermis atrophy (10–15 yrs) | CC hypoplasia, small ACC, faint posterior periventricular T2 signal (0–5 yrs) | CC hypoplasia. Absent myelination within the posterior portion of the internal capsule. Multiple foci of T2 hyperintensity within the lentiform nuclei (infancy). |
N/A* | CC hypoplasia. Hypomyelination. Progressive cerebral atrophy. Subdural hygromas (infancy). |
Cavum septum pellucidum and cavum vergae | N/A | |
| Other investigations | Prenatal US: hydrocephalus. DEXA: osteopenia (on bisphosphonates). |
EEG: multifocal epileptiform discharges | N/A | N/A | Sleep EEG (age 10–15 yrs): general disorganization of brain electrical activity without epileptiform discharges. | EEG: abnormal, with focus | EEG: initially normal, then showing epileptic activity mainly in the temporo-parietal regions. | EEG: normal. Postnatal abdominal US: normal. |
N/A* | CTG: pathological. EEG: intermittent epileptic activity. Prolonged ECG: intermittent bradycardia. Postnatal echocardiography: ASD type II with left-right shunt. Spine MRI: low-lying conus medullaris. |
Prenatal US: polyhydramnios, clenched fists, clubfeet. Neonatal head and spine US: reduced gyration, low-lying conus medullaris/tethered cord. EEG: pathological pattern. Postnatal echocardiography: unremarkable. Postnatal abdominal US: left hydronephrosis and hydroureter. |
Prenatal US (GA 20–25 wks): normal. Prenatal US (GA 30–35 wks): polyhydramnios, suspected CC agenesis, small stomach. Prenatal US (GA 35–40 wks): fetal growth restriction, abnormal fetal movements, CC agenesis, intracerebral cyst, arthrogryposis, thickened myocardium, small stomach, left hydronephrosis and hydroureter. QF-PCR: negative. CGH: negative. |
|
| Genotype PSMF1 (NM_006814.5) | HOM c.605+1G>A | HOM c.764+5G>A | HOM c.764+5G>A | HOM c.764+5G>A | N/A | HOM c.764+5G>A | HOM c.764+5G>A | HOM c.691C>T p.(Arg231*) | N/A | HOM c.1A>T | C-HET c.101del p.(Gly34Valfs*47) c.129+2T>C | C-HET c.365+2T>C c.1A>T | |
Legend: AC = anterior commissure; AED = antiepileptic drugs; ASD = atrial septal defect; CBZ = carbamazepine; C-HET = compound heterozygote; CC = corpus callosum; CGH = comparative genomic hybridization; CK = creatine kinase; CTG = cardiotocography; DBS = deep brain stimulation; DD = developmental delay; DEXA = bone density scan; ECG = electrocardiogram; EEG = electroencephalogram; F = female; GA = gestational age; GDD = global developmental delay; HOM = homozygote; LEV = levetiracetam; M = male; MDD = motor developmental delay; MIBG = 123I-metaiodobenzylguanidine scintigraphy; MRI = magnetic resonance imaging; MRS = magnetic resonance spectroscopy; N/A = not applicable or not available; PD = Parkinson’s disease; QF-PCR = quantitative fluorescence polymerase chain reaction; SLDD = speech and language developmental delay; US = ultrasound; VEP = visual evoked potentials; VPA = valproic acid; yr(s) = year(s); wks = weeks.
Clinical details not available (M-II-4 reported with a phenotype similar to his sister M-II-5).
Detailed clinical information is available upon request to the corresponding author.
The cohort comprised 11 males and 11 females. Parental consanguinity was reported in 9 of 15 pedigrees (60%). Probands’ parents were healthy. Based on the phenotypic features and severity, we distinguished three subgroups across subjects with PSMF1-related disorder and recognized clear genotype-phenotype correlation (Fig.1b).
In Pedigrees A-B-C-D-E-F-G, affected individuals manifested with early-onset PD or parkinsonism as the core phenotypic feature. After unremarkable neurodevelopmental milestones, motor symptoms began between the first and fifth decade of life, particularly during late adolescence or the third decade. In this subgroup, parkinsonism was most often accompanied by pyramidal tract signs, moderate-severe dysphagia occurring early in the disease course and psychiatric comorbidity. Brain MRI was normal or detected minor abnormalities, including mild-to-moderate cerebral or mild cerebellar atrophy (Fig.1c). Four probands (A-II-2, B-II-3, C-II-2, G-II-3) had markedly abnormal presynaptic dopaminergic imaging (Fig.1c). Affected individuals had different degrees of response to dopaminergic treatment and tended to develop early motor and non-motor fluctuations and levodopa-induced dyskinesia. Two probands (C-II-2, D-II-4) underwent bilateral subthalamic nucleus deep brain stimulation, with initial good outcome followed by rapid disease deterioration. All patients in this subgroup harbored PSMF1 missense variants, either in the homozygous state (Pedigrees B-C-D-E-F) or in compound heterozygosity with a predicted loss-of-function variant (Pedigrees A-G).
Affected individuals belonging to Pedigrees H-I-J-K presented with miscellaneous movement disorders, encompassing parkinsonism, spasticity, ataxia or a combination thereof, most often in the context of neurodevelopmental delay and associated with mild-to-moderate intellectual disability, epilepsy and cognitive decline. They exhibited facial or skeletal dysmorphic features. In this subgroup, two probands (H-II-3, I-II-3) had sensorineural hearing loss. Brain MRI almost invariably revealed hypoplasia of the corpus callosum and/or various degrees of cerebral or vermian cerebellar atrophy. Four patients (H-II-1, K-II-1, K-II-3, K-II-6; Fig.1c) died during adolescence because of rapidly progressive neurological deterioration starting early in the second decade of life. All affected individuals within this subgroup carried homozygous PSMF1 splice variants.
The third subgroup includes affected subjects from Pedigrees L-M-N-O, who presented with severe neurological manifestations since prenatal development, including arthrogryposis multiplex congenita, abnormal fetal movements (suspected for in utero seizures), epilepsy, profound developmental delay, limb spasticity, severe respiratory insufficiency and poor feeding, with death occurring within infancy. Agenesis or hypoplasia of the corpus callosum was detected in three probands (L-II-1, M-II-5, O-II-2; Fig.1c). A history of pregnancy losses (M-II-3, N-II-1) preceding the index case was reported in two of these pedigrees (Fig.1a). All affected subjects in this subgroup had biallelic PSMF1 loss-of-function variants, of which at least one predicted to cause complete loss of function.
Validation and functional analysis of 14 missense and loss-of-function variant in PSMF1
We detected 14 different PSMF1 mutant alleles, including five missense and nine loss-of-function variants (six splice, one nonsense, one start-loss and one frameshift; Fig.1a-2a). Variant pathogenicity predictions and computational analysis are detailed in Supplementary File 1. In all subjects with genomic DNA available, segregation analysis confirmed that biallelic PSMF1 variants co-segregated with neurological disease phenotypes. Moreover, parents of affected individuals harbored one heterozygous PSMF1 variant, and unaffected siblings of affected subjects were either heterozygotes for one PSMF1 mutant allele or homozygotes for the wild-type PSMF1 allele (Fig.1a). Among 20 affected subjects with genotype available, 17 were homozygotes and 4 were compound heterozygotes for PSMF1 variants (Fig.1a).
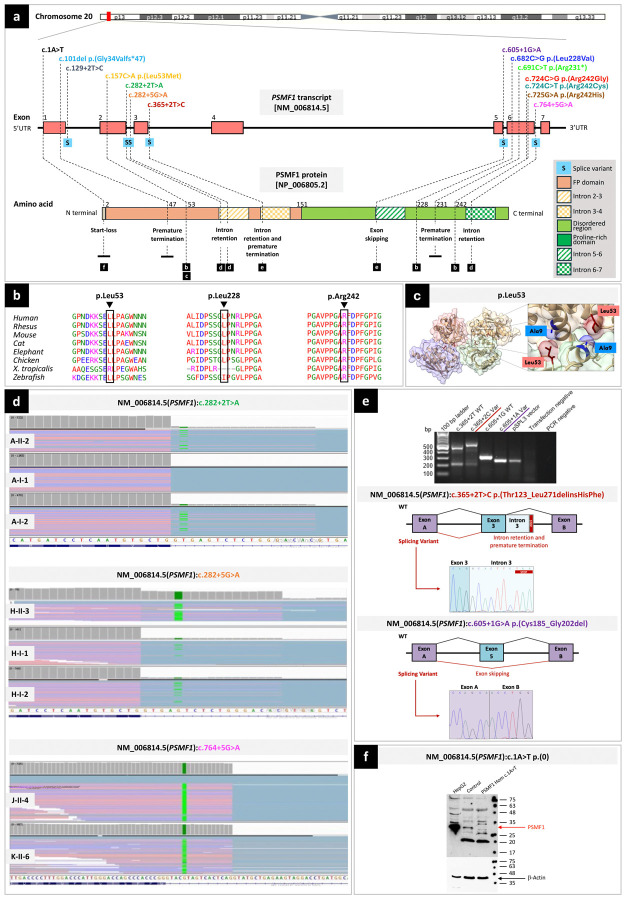
Figure 2. Location, computational analyses and functional characterization of PSMF1 variants.
(a) Schematic of PSMF1 and its protein product with location of 14 variants identified in this study and their effect. Upper part. Chromosome 20 showing position of PSMF1 on 20p13. Middle part. Schematic of PSMF1 with variants identified in the human disease gene discovery study cohort. Introns are not to scale. Exon numbers are according to the canonical transcript (NM_006814.5). Lower part. Schematic of PSMF1 protein with amino acid changes linked to disease (reference sequence NP_006805.2). (b) Interspecies alignment showing strong evolutionary conservation of the amino acids affected by the PSMF1 missense variants identified in this study across species, down to invertebrates. (c) Localization of the p.Leu53 residue within the PSMF1-PSMF1 homotetramer crystal structure, revealing a role for this residue in PSMF1 homodimerization. The residue p.Leu53 is involved in the PSMF1 variant detected in Pedigree E (Fig.1a; Supplementary File 4). Image derived from PDB 4OUH using ChimeraX. (d) RNA sequencing. Integrative Genomics Viewer screen captures showing results of RNA sequencing for Pedigree A (upper panel), Pedigree H (middle panel) and probands J-II-4 and K-II-6 (lower panel). For transcripts with PSMF1 splice variants c.282+2T>A (A-II-1, A-II-2), c.282+5G>A (H-II-3, H-I-1, H-I-2) and c.764+5G>A (J-II-4, K-II-6), IGV enables to visualize (partial) intron retention. (e) Minigene (splicing) assays. Upper panel. Agarose gel of the RT-PCR from the minigene cDNA. Medium panel. Splicing schematic of the NM_006814.5(PSMF1):c.365+2T>C variant showing intron 3 retention with stop codon sequence marked. Bottom panel. Splicing assay of the NM_006814.5(PSMF1):c.605+1G>A (bottom) variant shows exon skipping. (f) Western blot to functionally characterize the start-loss variant NM_006814.5(PSMF1):c.1A>T. Protein lysates (30μg) from HepG2 cells, and either control or patient fibroblasts (M-II-5) were analyzed by Western blotting using a polyclonal anti-PSMF1-Ab. HepG2 cells express PSMF1 at high levels, control but not patient fibroblasts express PSMF1. β-actin expression was used as loading control.
All PSMF1 variants identified in the families were absent or ultra-rare in the heterozygous state and not reported in the homozygous state in gnomAD,36 except for the missense variant c.725G>A, p.Arg242His (Supplementary File 1A), which had low frequency and four homozygous entries in gnomAD v4. Interestingly, this mutant allele was detected in the homozygous state in the proband (D-II-4) with the latest age at PD onset (45–50 years) in this study cohort, which could explain why this variant is not completely depleted from gnomAD v4.
PSMF1 missense variants identified in this study showed strong evolutionarily conservation across species down to invertebrates (Fig.2b; Supplementary File 1A).
All PSMF1 non-splice variants were predicted with a damaging effect according to most in silico prediction tools (Supplementary File 1A). All PSMF1 non-frameshift variants had a high Combined Annotation Dependent Depletion (CADD Phred range: 22.3–37). Pathogenicity prediction tools such as PolyPhen2, SIFT4G, PROVEAN and MutationTaster predicted the functional impact of all PSMF1 missense variants to be damaging/deleterious in most cases. In silico splice predictions of six PSMF1 splice variants using six computational tools (SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer, SpliceAI, and AbSplice) unanimously indicated reduction or complete abolishment of native splice donor sites according to most tools (Supplementary File 1B).
Four PSMF1 variants were detected in more than one family (Pedigrees A-B-G: c.724C>G, p.Arg242Gly; Pedigrees G-L: c.691C>T, p.Arg231*; Pedigrees J-K: c.764+5G>A; Pedigrees M-O: c.1A>T; Fig.1a). Specifically, the missense variant c.724C>G, p.(Arg242Gly) recurred in three families either in the compound heterozygous (Pedigrees A-G) or homozygous (Pedigree B) state. Furthermore, the missense variant c.724C>T, p.(Arg242Cys) detected in Pedigree C involved the same nucleotide as the previous mutant allele. Finally, the missense variant c.725G>A, p.(Arg242His) affected the same amino acid residue as the two previous mutant alleles. Intriguingly, although PSMF1 variants were distributed across the gene (Fig.2a), three different PSMF1 variants detected in a total of five families with PD/parkinsonism (Pedigrees A-B-C-D-G) involved the p.Arg242 residue, which could therefore represent a “hotspot codon”. Among splice variants, two were contiguous (c.282+2T>A in Pedigree A; c.282+5G>A in Pedigree H).
Autozygosity mapping.
Autozygosity mapping using AutoMap revealed that the PSMF1 locus lay within a region of homozygosity (ROH) in affected individuals carrying a homozygous PSMF1 variant who were born to consanguineous parents (Pedigrees B-C-E-F-J-K-L-M; raw ES data not available for proband I-II-3). On the contrary, PSMF1 did not map on a ROH in their unaffected family members or in affected individuals harboring homozygous PSMF1 variants who were born to nonconsanguineous parents (Pedigrees D-H; Supplementary File 2).
Haplotype analysis.
Haplotype analysis of the PSMF1 splice variant c.764+5G>A, which was detected in homozygosity in the proband of two families of different geographic provenance (J-II-1, K-II-6), revealed a different pattern of flanking variants, suggesting two independent mutational events rather than inheritance from a common ancestor (Supplementary File 3).
In silico modelling of PSMF1 missense variants.
To assess the functional consequences of putative pathogenic PSMF1 missense variants, modelling of the three-dimensional organization of the PSMF1/FBXO7 heterotetrameric complex was carried out using AlphaFold2 (Supplementary File 4). While the p.Leu228 and p.Arg242 residues are located in PSMF1 regions with low-confidence structural prediction, the p.Leu53 residue sits directly on the interface between PSMF1 monomers within the complex, forming hydrogen bonds with p.Val6 and p.Ala9 (Fig.2c). Substitution of methionine for the leucine at codon 53 likely disrupts these interactions, weakening the quaternary structure of the predicted heterotetramer. Hence, the PSMF1 p.Leu53Met variant detected in Pedigree E (Fig.1a) could perturb the function of the complex, acting as a loss-of-function variant. Further in vitro and cellular studies are required to test the impact of the p.Leu53Met variant, as well as to gain insight into the effect of coding variants in the C-terminal domain of PSMF1.
Functional characterization of 5 PSMF1 splice variants.
RNA sequencing (RNA-seq).
In Pedigree A, fibroblast RNA-seq with transcript visualization through Integrative Genomics Viewer (IGV) and Detection of RNA Outlier Pipeline (DROP) revealed partial intron retention in transcripts with the PSMF1 splice variant c.282+2T>A, which the proband (A-II-2) and his mother (A-I-2) harbored in the heterozygous state (Fig.1a; Fig.2d). PMSF1 was not detected as aberrant expression (AE) or aberrant splicing (AS) outlier in these individuals. In Pedigree H, fibroblast RNA-seq of the proband (H-II-3) and whole-blood RNA-seq of his parents (H-I-1, H-I-2) with transcript visualization by IGV showed intron retention in transcripts with the PSMF1 splice variant c.282+5G>A, which was detected in the homozygous state in the proband and in the heterozygous state in his parents (Fig.1a; Fig.2d). Here, DROP pipeline detected PSMF1 aberrant splicing of most transcript (OUTRIDER, PSMF1 as AE outlier, with fold change 0.13, z-score −9.84, FDR 6.29x10−14; FRASER2, PSMF1 as AS outlier, with |Δψ| 0.73, FDR 2.85x10−12). Finally, in probands J-II-4 and K-II-6, fibroblasts RNA-seq revealed that the PSMF1 splice variant c.764+5G>A caused intron retention (Fig.2d), and PSMF1 was detected as significant splicing outlier in the two probands (FRASER2, PSMF1 as AS outlier, with |Δψ| 0.88, FDR 4.06x10−5 in J-II-4 and with |Δψ| 0.89, FDR 9.05x10−6 in K-II-6; OUTRIDER, PSMF1 not detected as AE outlier).
Splicing (minigene) assay.
Minigene assay revealed that the PSMF1 splice variant c.365+2T>C (Pedigree O) led to intron retention (r.366_367inscacuucuaa) and premature termination (p.Thr123_Leu271delinsHisPhe; Fig.2e). Furthermore, the PSMF1 splice variant c.605+1G>A (Pedigree I) was proven to result in skipping of exon 5 (r.553_606del) and an in-frame deletion (p.Cys185_Gly202del; Fig.2e).
Functional characterization of the PSMF1 start-loss variant c.1A>T.
Western blotting of protein lysate obtained from fibroblasts of proband M-II-5 (Fig.1a), who was homozygote for the PSMF1 start-loss variant c.1A>T, revealed that PSMF1 expression was abolished (Fig.2f).
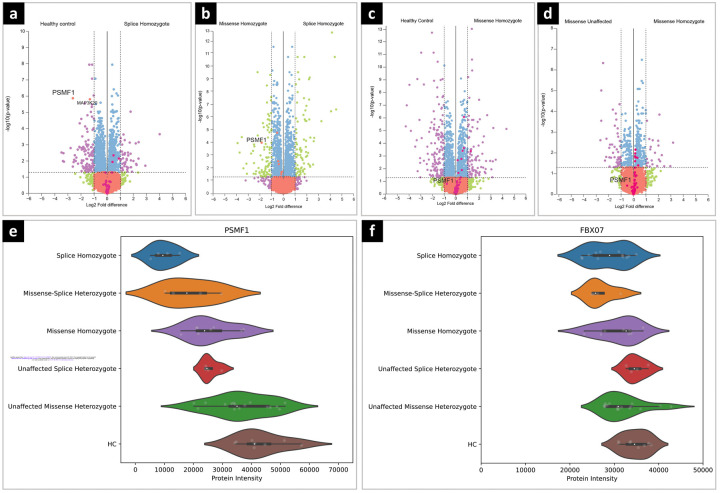
Proteomic analysis of human fibroblasts revealed a descending PSMF1 abundance profile from missense to loss-of function mutant alleles
Differential expression analysis of the proteomes of PSMF1 proband- and heterozygous carrier-derived fibroblasts was obtained by nano-liquid chromatography combined with mass spectrometry. This revealed statistically significant reduction in PSMF1 in probands carrying homozygous PSMF1 splice variants (H-II-3, J-II-4, K-II-6) compared to controls (Fig.3a). Statistically significant reduction in PSMF1 expression was also observed between probands harboring homozygous PSMF1 splice variants and those with homozygous PSMF1 missense mutations (B-II-3, E-II-3; Fig.3b). On the contrary, PSMF1 expression was not downregulated in probands carrying homozygous PSMF1 missense variants compared to controls or carriers of a single heterozygous PSMF1 missense variant (A-I-1, A-I-2, B-II-1, E-I-1, E-I-2; Fig.3c-d). Interestingly, other proteasome-associated proteins were significantly upregulated in probands with homozygous PSMF1 missense variants compared to homozygotes for PSMF1 splice variants, heterozygotes for a PSMF1 missense variants and controls (Fig.3b-c-d), although the majority of these do not pass the log2 fold-change threshold of 1. An upward trend was evident in PSMF1 levels from probands to carriers of single heterozygous PSMF1 splice or missense variants, approaching the highest PSMF1 levels recorded in controls (Fig.3e). We observed a trend for decreased FBXO7 expression in patients carrying homozygous and heterozygous PSMF1 splice variants (Fig.3f), which reached statistical significance in differential expression analysis for homozygotes having splice variants compared to controls, albeit with a log2 fold-change <1 (Fig.3f).
Figure 3. Quantitative proteomic analysis of PSMF1 patient- and carrier-derived dermal fibroblasts.
Volcano plots showing: (a) Statistically significant downregulation of PSMF1 in patients with homozygous PSMF1 splice variants compared to healthy controls; (b) Statistically significant reduction in PSMF1 expression in patients with homozygous PSMF1 splice variants compared to patients with homozygous PSMF1 missense variants; (c) PSMF1 expression was not downregulated in patients with homozygous missense variants compared to healthy controls or (d) patients having a heterozygous missense variant. Violin plots showing: (e) PSMF1 protein expression across patient groups; (f) a trend for decreased FBXO7 protein expression in patients with homozygous and heterozygous PSMF1 splice variants, which reached statistical significance in differential expression analysis for patients with homozygous PSMF1 splice variants versus healthy controls, albeit with a log2 fold-change <1. Violin plots showing: (e) PSMF1 protein expression across patient groups; (f) a trend for decreased FBXO7 protein expression in patients with homozygous and heterozygous PSMF1 splice variants, which reached statistical significance in differential expression analysis for splice variant homozygous patients versus healthy controls, albeit with a log2 fold-change <1.
Heterozygous rare and low-frequency PSMF1 variants are not associated with increased risk of sporadic Parkinson’s disease in the UKB and AMP-PD repositories
We looked for rare and low-frequency (minor allele frequency, MAF <0.005) PSMF1 variants in patients with sporadic (non-familial) PD and controls in the UKB and AMP-PD repositories. Gene-burden analysis revealed that single heterozygous PSMF1 variants do not contribute to the risk of developing sporadic PD (Supplementary File 5).
PSMF1 variants cause mitochondrial dysfunction and alteration of mitochondrial dynamics and mitophagy in human fibroblasts
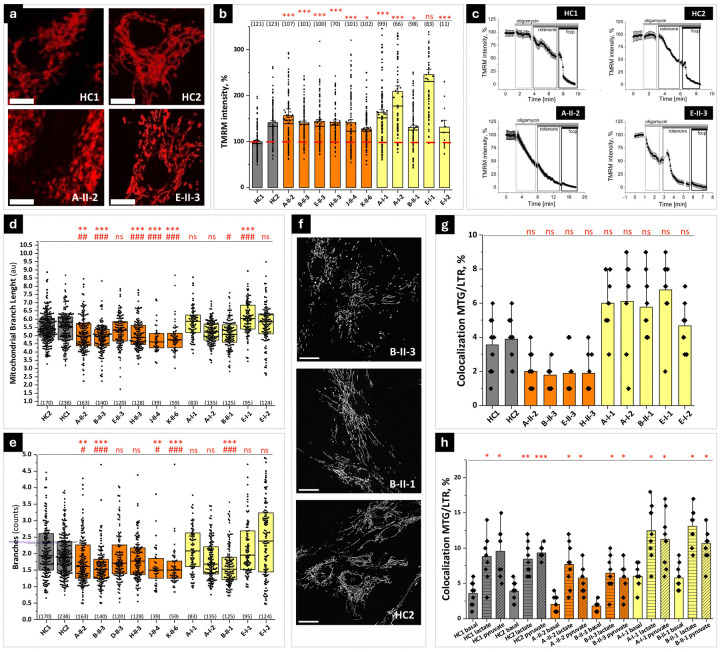
We explore the effects of PSMF1 variants on mitochondrial functions and mitophagy by live imaging of skin fibroblast of PSMF1 probands and carriers.
Mitochondrial membrane potential (Δψm).
Most processes in mitochondria depend on the Δψm, whose detection in live cells is an important indicator of healthy mitochondrial functioning. Using tetramethylrhodamine methyl ester (TMRM) as fluorescent probe for Δψm, we found that TMRM fluorescence in fibroblasts from PSMF1 probands was similar or higher and in unaffected PSMF1 carriers even higher than in controls (Fig.4a-b), which suggests mitochondrial hyperpolarization. Since Δψm is mainly maintained by the function of the mitochondrial electron transport chain (mETC), any perturbation of mitochondrial respiration switches ETC to alternative mechanisms to maintain Δψm in the physiological range, including reversal of the activity of the F0-F1-ATPase (complex V). Fibroblasts from controls or PSMF1 carriers showed no effect or small increase in TMRM fluorescence after application of the mitochondrial complex V inhibitor oligomycin (2μg/ml; Fig.4c, upper quadrants), indicating that that enzyme is working as ATP synthase. On the contrary, addition of oligomycin to PSMF1 proband-derived fibroblasts induced >50% depolarization (Fig.4c, lower quadrants), which suggests that complex V is working in ATPase mode due to pathological mETC functioning in these cells. Subsequent application of the inhibitor of mitochondrial complex I rotenone (5μM) induced decrease in TMRM fluorescence in control (by ~50%) or PSMF1 carriers’ fibroblasts (by ~60%; Fig.4c, upper quadrants), whereas in PSMF1 probands’ cells it induced complete mitochondrial depolarization, seen by the lack of effect of the mitochondrial uncoupler carbonyl cyanide p-trifluoro methoxyphenylhydrazone (FCCP; 1μM) on TMRM fluorescence after oligomycin and rotenone in these fibroblasts (Fig.4c, lower quadrants). In control or PSMF1 carriers’ fibroblasts, subsequent application of FCCP induced further drop of the TMRM signal (by 30–40%) to a complete depolarization (Fig.4c, upper quadrants). Overall, control fibroblasts maintain Δψm solely by the function of the mETC (including complexes I and II), while Δψm maintenance (~50%) in probands’ fibroblasts depends on ATP consumption by F0-F1-ATPase and the activity of complex I. Relevantly, we previously observed similar changes in Δψm maintenance in other cells with PD pathology, including PINK1-deficient cells, neurons with SNCA triplication and neurons with mitochondrial variants.37–39
Figure 4. PSMF1 deficiency is associated with defects in mitochondrial bioenergetics and dynamics. (a-c) Mitochondrial membrane potential status and maintenance:
(a) Representative images of tetramethylrhodamine methyl ester (TMRM)-loaded mitochondria in fibroblasts from healthy controls (HC1, HC2) and PSMF1 patients (A-II-2, E-II-3; see Fig. 1a). Scale bar = 20 μm. (b) Quantification bar graphs depict the mean mitochondrial membrane potential for healthy controls’ (gray bars; see also red dash line), probands’ (orange bars) and unaffected carriers’ (yellow boxes) mitochondria. (c) Inhibitor analysis of the mitochondrial membrane potential maintenance and electron transport chain function in healthy controls (upper quadrants) compared to probands’ (lower quadrants) mitochondria. (d-f) Mitochondrial dynamics: (d) Quantification box charts depicting average length of all mitochondrial rod and branches and (e) average number of branches analyzed in individual fibroblasts from healthy controls’ (gray boxes), probands’ (orange boxes) and unaffected relatives’ (yellow boxes) fibroblasts. (f) Representative images of mitochondrial shape and dynamics in one unaffected carrier (B-II-1), one proband (B-II-3) and one healthy control (HC2). Each dot represents a cell and total number of cells analyzed in 3–9 independent experiments is shown in brackets. Non-parametric Kruskal-Wallis ANOVA with post-hoc Dunn’s test for each group. *p < 0.05, **p < 0.01, ***p < 0.001 compared to control HC1; # p < 0.05, ## p < 0.01, ### p < 0.001 compared to control HC2. Scale bar = 20 μm. (g-h) Mitophagy: (g) Basal mitophagy rate assessed as colocalization of LysoTracker Red DND-99 with MitoTracker Green in healthy controls (gray bars), probands’ (orange bars) and unaffected carriers’ (yellow bars) fibroblasts. (h) Mitophagy rate after induction with either lactate or pyruvate. Data are presented as mean ± SEM. ***p < 0.0001.
Mitochondrial dynamics.
Mitochondrial dynamics is essential for proper mitochondrial metabolism and function. Mitochondria in PSMF1 proband- and control-derived fibroblasts differ not only by the TMRM signal intensity but also by their shape and size (Fig.4d-e-f). Further analysis of the mitochondrial shape showed that mitochondria in the probands’ fibroblasts appeared more fragmented and have shorter mitochondrial branch length (Fig.4d) and lower mean number of branches per network (Fig.4e) than control fibroblasts. Interestingly, fibroblasts of PSMF1 carriers had even longer mitochondrial branch length and mean number of branches per network than in control and probands’ fibroblasts (Fig.4d-e-f). Therefore, PSMF1 variants cause changes in mitochondrial dynamics, which could be secondary to an alteration of fission/fusion mechanisms or mitophagy.
Mitophagy.
Damaged or depolarized mitochondria are engulfed by lysosomes during mitophagy. To detect this translocation by live imaging of participant-derived fibroblasts, we analyzed the co-localization of mitochondria loaded with the green-fluorescent dye MitoTracker® Green FM with lysosomes loaded with the red-fluorescent dye LysoTracker Red DND-99.40 Although basal mitophagy levels in fibroblasts are relatively low and the difference between PD and control cells is difficult to detect,41 we found that the percentage of co-localization in PSMF1 probands’ fibroblasts was lower than in PSMF1 carriers (Fig.4g). Mitophagy activation by 30mM lactate or 30mM pyruvate41 increased the percentage of mitochondria-lysosome co-localization in all fibroblasts, but to a higher extent in control and PSMF1 carriers’ fibroblasts than in probands’ fibroblasts (Fig.4h).
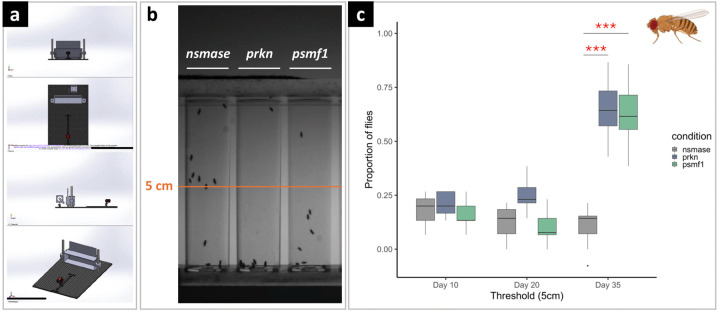
Psmf1-knockdown Drosophila melanogaster exhibits age-dependent locomotor dysfunction
In Drosophila melanogaster, we obtained pan-neuronal RNA interference (RNAi)-mediated targeted knockdown of psmf1 using the UAS/Gal4/Gal80 system of cell-type-specific transgene expression governed by nSyb-Gal4. Motor performance was assessed using the classic negative-geotaxis climbing test and measured at different ages in psmf1 knockdown flies versus positive (prkn) and negative (nsmase) controls (Fig.5a-b). At 10 and 20 days of age, we did not observe locomotor impairment in any of the three RNAi lines (Fig.5c). In 35-day-old flies, motor performance was impaired in the psmf1 and prkn RNAi strains, with the mean proportion of flies which did not reach the 5-cm threshold being significantly lower compared to nsmase knockdown (Fig.5c). Overall, these observations are consistent with Psmf1 loss of function causing age-dependent motor dysfunction in Drosophila.
Figure 5. Effect of psmf1 pan-neuronal knockdown on Drosophila melanogaster negative geotaxis measured by the climbing test.
Comparative climbing test (a) of three different Drosophila melanogaster RNA interference (RNAi) lines (prkn, psmf1, nsmase) with prkn used as positive control (significant motor impairment) and nsmase as a negative control (no motor impairment). (b) Climbing test performed at Days 10, 20 and 35 represented as percentage of flies that did not reach the 5-cm tube threshold. (c) Significant motor defect was observed at Day 35 for the psmf1 and prkn lines compared to the negative control nsmase. Significant comparisons are indicated by asterisks. ***p<0.0005. H. sapiens and D. melanogaster orthologues: PRKN (H. sapiens)/prkn (D. melanogaster); PSMF1 (H. sapiens)/psmf1 (D. melanogaster); SMPD2 (H. sapiens)/nsmase (D. melanogaster).
Conditional inactivation of Psmf1 in mice leads to motor dysfunction and diffuse gliosis
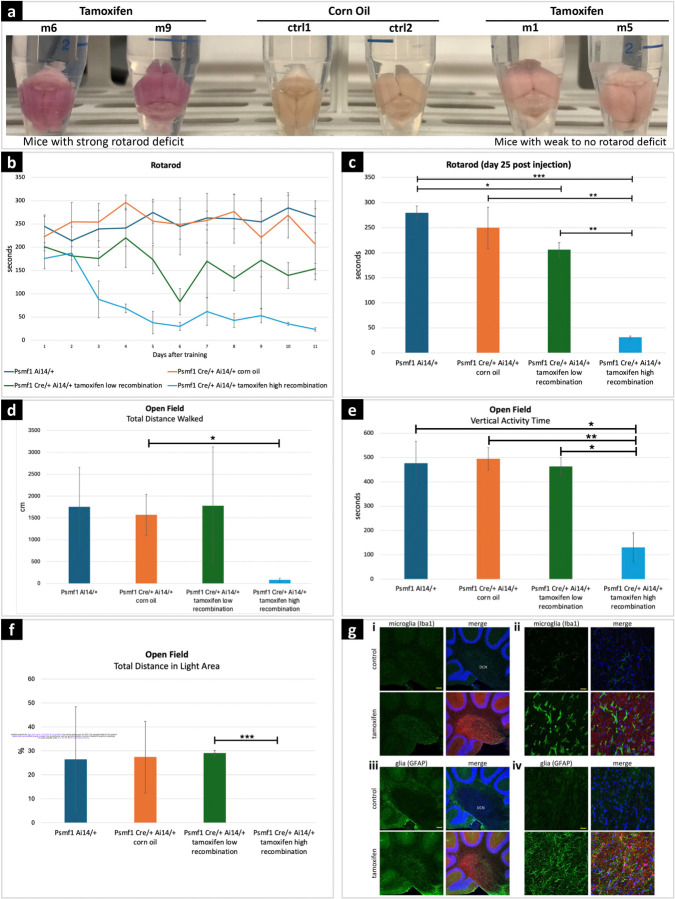
Mus musculus Psmf1 protein is 83% identical and 91% similar to human PSMF1. Members of our team previously reported that Psmf1 inactivation in mouse neurons causes age-progressive neurological phenotypes which culminate in neuronal cell death.22 To better examine the consequences of impaired Psmf1 function in the mouse, we bred mice with a tamoxifen-inducible Cre recombinase system with a Cre-ERT2 fusion gene under the control of the human ubiquitin C promoter (UBC-Cre-ERT2)42 with mice containing a loxP-flanked (floxed) third exon of Psmf1 (Psmf1fl),22 thus obtaining conditional knockout (CKO) of Psmf1. This strain enables conditional inactivation of Pmsf1 upon tamoxifen injection. To assess recombination efficiency, these mice also carried a transgenic Cre reporter, Ai14,43 which has a floxed STOP cassette preventing transcription of a CAG promoter-driven red fluorescent protein variant (tdTomato) inserted into the Gt(ROSA)26Sor locus. Upon successful Cre-mediated recombination, Ai14 mouse tissues express tdTomato red fluorescence. Depending on the amount of tamoxifen injected, we obtained recombination efficiency ranging from 30% to over 90% (Fig.6a; Supplementary File 6).
Figure 6. Behavioral tests and neuropathology in mice with conditional Psmf1 inactivation.
(a) Psmf1fl/fl UBC-Cre-ERT2/+ Ai14/+ brains post perfusion (28 days after injection of tamoxifen or corn oil as a control). Tamoxifen-induced recombination induced tdTomato (red) expression and marked cells in which Psmf1 was inactivated (Supplementary File 8). Successful tamoxifen-induced recombination was macroscopically assessed based on the color of dissected mouse brains, which appeared dark pink in mice with severe motor deficit in the rotarod assay (m6, m9) and light pink in mice with mild to no motor deficit in the rotarod assay (m1, m5) compared to controls (ctrl1, ctrl2), which reflected high (~90%) or low (30–50%) recombination rates, respectively. Psmf1fl/fl Ai14 UBC-Cre-ERT2 mice were injected with either corn oil (control) or tamoxifen to induce Cre recombination of floxed Psmf1 exon 3 for loss of Psmf1 and a floxed stop cassette in Ai14 for expression of tdTomato to evaluate recombination efficiency. Mice were sacrificed 28 days post injection. (b-c) Rotarod assay. In the rotarod assay, mice with high recombination rates (~90%) developed severe motor deficits (light blue line in (b) and light blue bar in (c)), whereas the motor performance of mice with low (30–50%) recombination rates was mildly or not impaired (green line in (b) and green bar in (c)). Panel (b) shows single rotarod trials for individual mice on each day. X-axis indicates days of testing after the initial four-day training period. Day 1 corresponds to day 14 after tamoxifen injection. In panel (c), each bar represents the average of three trials for a mouse on day 25 post tamoxifen injection (i.e., 12 days after the initial four-day training period). Error bars indicate standard deviations. (d-e-f) Open field assay. Mice with high recombination rates (~90%, fourth subgroup) strongly avoided walking in the open field, which reflects severe anxiety-related behavior. In contrast, the motor performance of mice with low recombination rates (30–50%, third subgroup) was comparable to controls. In panels c-d-e-f, error bars are standard deviations, p values derived by two-tailed t test, *p < 0.05, **p value < 0.01, *** p value < 0.001. (g) Sagittal brain sections stained with either Iba1 (green) for microglia or GFAP for glia (astrocytes) and with Hoechst 33342 for nuclei (blue). (i) In the deep cerebellar nuclei (DCN), reactive microglia in tamoxifen-induced mice indicated gliosis induction. Scale bar 100 μm. (ii) Higher magnification in the DCN shows morphologically distinct ramified microglia with more amoeboid appearance upon loss of Psmf1. Scale bar 20 μm. (iii) In the DCN of tamoxifen-induced mice we observe an increase of glia staining strongly for GFAP in the DCN, a marker for astrogliosis (of note, GFAP also stains the Bergmann glia in the molecular layer). Scale bar 100 μm. (iv) Higher magnification image of (C). Scale bar 20 μm.
Behavioral tests.
Rotarod assay.
We first assessed motor performance of Psmf1 CKO mice using the rotarod test (Fig.6b-c). Psmf1fl/fl Ai14 UBC-Cre-ERT2 mice were injected with tamoxifen or corn oil as control at 6–7 weeks of age, while Psmf1fl/fl Ai14 mice without Cre were injected with tamoxifen as an additional control for tamoxifen. Ten days after the last injection, mice were trained for four days on the rotarod and then tested once on each day after for 11 days (Fig.6b-c) and thrice on the subsequent day. Initially the performance of tamoxifen-injected Psmf1fl/fl Ai14 Ubc-Cre-ERT2 mice with a high recombination rate (>90% of neurons as assessed by Ai14) displayed a deficit by day 12, whereas tamoxifen-injected Psmf1fl/fl Ai14 Ubc-Cre-ERT2 mice with a low recombination rate (approximately 30% of neurons as assessed by Ai14) showed only a mild deficit (Fig.6b-c).
Open field assay.
Since individuals with PSMF1-related PD or parkinsonism present with severe anxiety, we also tested Psmf1fl/fl Ai14 UBC-Cre-ERT2 mice in the open field assay with dark insert. Anxiety-like behavior of rodents is frequently accompanied by reduced exploration. On day 26 after tamoxifen injection, Psmf1fl/fl Ai14 UBC-Cre-ERT2 mice displayed a clear difference to controls, consistent with increased anxiety (Fig.6d-e-f).
Brain immunohistochemistry.
We observed gliosis in various brain regions 28 days after tamoxifen injection in Psmf1fl/fl Ai14 UBC-Cre-ERT2 mice. Many neurodegenerative diseases are associated with gliosis, where glial cells become reactive and proliferate generally prior to the formation of aggregates, tangles and plaques, including PD.44–48 Significantly, we detected both microgliosis, as documented by increased staining of microglia with anti-Iba1 and a change in morphology of the microglia from ramified to more amoeboid, and astrogliosis, as demonstrated by elevated GFAP-staining of astrocytes (Fig.6g; Supplementary Files 7-8).
DISCUSSION
We provided extensive phenotypic, genetic and functional evidence to establish PSMF1 as a new human disease gene implicated in PD and childhood neurodegeneration. Specifically, we reported 15 unrelated pedigrees where biallelic PSMF1 missense and loss-of-function variants co-segregated with a clinical spectrum ranging from early-onset PD to perinatal lethality with neurological manifestations. Notably, we observed a clear genotype-phenotype correlation. Individuals carrying PSMF1 missense variants had normal neurodevelopmental milestones and manifested with parkinsonism between the first and fifth decade of life. Their clinical picture often encompassed pyramidal tract involvement, early dysphagia and psychiatric comorbidity, and abnormal presynaptic dopaminergic imaging was consistent with nigrostriatal degeneration. These individuals showed various response to antiparkinsonian agents and developed early motor and non-motor fluctuations and dyskinesia. Moving forward into the disease spectrum, patients harboring biallelic PSMF1 splice variants exhibited parkinsonism and/or other movement disorders and neurological manifestations since childhood or adolescence. Most often, these occurred in the context of neurodevelopmental delay and showed rapid progression leading to death within the second decade. Finally, subjects with two PSMF1 loss-of-function variants, of which at least one was predicted to abolish PSMF1 expression, presented with neurological manifestations and structural brain abnormalities since the prenatal period and died within infancy. Intriguingly, PSMF1 loss-of-function variants were almost invariably associated with agenesis or hypoplasia of the corpus callosum, which consists of millions of axons connecting the cerebral hemispheres. Since Psmf1 has been demonstrated to mediate neurosome-to-synapse fast transport and be crucial for synapse maintenance and neuronal survival in Drosophila and mice,20,22 it is tempting to speculate that loss of PSMF1 in humans affects axonal pathways leading to failed axonogenesis/synaptogenesis or premature degeneration of these neural structures.
PSMF1 encodes the evolutionarily highly conserved proteasome regulator PSMF1/PI31, which is ubiquitously expressed in human tissues, including the brain. PSMF1 shows strong protein expression in fetal brain tissue compared to non-brain tissues and elevated gene expression during the prenatal period across all brain regions.49 This could explain the widespread brain effects of its deficiency, not limited to the dopaminergic system. Notably, deep phenotyping of our study cohort revealed a tendency towards obesity and glucose intolerance, early osteoporosis/osteopenia and severe infections. Further delineation of PSMF1-related disorder will clarify whether PSMF1 deficiency might have multisystemic involvement in humans.
Our findings suggest that different PSMF1 mutant alleles drive disease via multiple loss-of-function mechanisms. In particular, the three missense variants p.Arg242Gly, p.Arg242Cys and p.Arg242His detected in five pedigrees with early-onset PD or parkinsonism affect a highly conserved residue within the natively disordered proline-rich C terminal of PSMF1. In vitro characterization of the complex human PSMF1/bovine 20S proteasome has demonstrated that PSMF1 Arg242 is inserted into the proteasome catalytic site, which results in an inhibitory mechanism.50 Considering the evidence of PSMF1 acting as a 20S proteasome inhibitor and a 26S proteasome upregulator in vitro as well as a proteasome enhancer in vivo, further studies are needed to establish whether variants at the Arg242 residue determine loss of function by converting PSMF1 from a proteasome inhibitor to a proteasome substrate, or by disrupting proteasome positive regulation, thus supporting the role of proteasome inhibition in the pathogenesis of PD. Similarly, the missense variant p.Leu53Met is located at the dimerization interface of PSMF1 and is likely to disrupt hydrophobic interactions within the PSMF1 homodimer, possibly influencing its activity on the proteasome and the stability of the PSMF1-FBXO7 heterocomplex. Finally, PSMF1 predicted and proven loss-of-function variants in our cohort are associated with moderate-to-severe phenotypes within the reported spectrum. Further analysis is needed to establish why biallelic PSMF1 splice variants lead to a milder, non-perinatal lethal phenotype compared to PSMF1 putative or confirmed complete loss-of-function variants, particularly for those splice variants without evidence of alternative splicing from RNA-seq data.
PSMF1 is a high-affinity binding partner to FBXO7 in Drosophila and mammalian cells. Phenotypic features of PSMF1 patients are reminiscent of those of FBXO7-related disorder, such as a clinical spectrum including juvenile-onset PD/parkinsonism, pyramidal tract signs only, or a combination thereof, initial response to levodopa and early occurrence of levodopa-induced dyskinesia.25 Human neuropathology is not available for PSMF1- and FBXO7-related disorders, but presynaptic dopaminergic imaging suggests dramatic reduction in the striatal dopaminergic innervation in both disorders.25 None of the PSMF1 variants identified in our cohort is predicted to affect amino acid residues at the interface between PSMF1 and FBXO7. Therefore, it remains elusive whether PSMF1 deficiency induces PD/parkinsonism per se, or by secondary disruption of FBXO7 functions in humans. Recently, ablation of the FBXO7-PSMF1 interaction by an FBXO7 missense variant has been demonstrated to affect expression and stabilization of these proteins as well as proteasomal and mitochondrial pathways in human fibroblasts.27 Importantly, Fbxo7 variants cause site-specific proteolytic cleavage and reduced Psmf1 activity in Drosophila and mouse.18,27,28 Moreover, transgenic Psmf1 expression in Fbxo7-null mice can robustly suppress neurological defects.21 This suggests that loss of Fbxo7 causes disease to a considerable extent by attenuating Psmf1 functions. Since FBXO7 acts synergically with PINK1 and PRKN to regulate basal mitochondrial functions and stress-induced mitophagy,51,52 we explored mitochondrial pathways by live imaging of PSMF1 patient-derived fibroblasts and documented mitochondrial dysfunction and alteration of mitochondrial dynamics and mitophagy. Extensive studies are needed to fully characterize the consequences of PSMF1 variants on mitochondrial homeostasis and, most importantly, to delineate their effects on proteasomal functions, whose disruption is likely to be the primary pathomechanism driving PSMF1-related pathology.
Comparison between Psmf1 knockout mouse models (previous data and current study) and humans with PSMF1-related disorder reveals striking parallelism. Similarly to Psmf1-null mice, which die at advanced embryonic stages,22 humans with biallelic complete or putative severe loss-of-function PSMF1 variants (Pedigrees L-M-N-O) exhibited neurological manifestations and brain structural abnormalities since prenatal development and died within infancy. On the contrary, conditional Psmf1 knockout mice develop age-dependent motor dysfunction,22 which is in keeping with motor phenotypes enabling survival into adolescence or adulthood observed in patients with biallelic PSMF1 splice and missense variants, which are predicted with decreasingly deleterious effects. Brain autopsy of our mouse model revealed diffuse micro- and astrogliosis with ameboid microglia, which likely reflects early neurodegeneration stages and the timing at which mice were sacrificed before neuropathology could be more informative. Further studies are needed, including expression of hypomorphic and knockin Psmf1 variants in mice, to provide detailed insights into the neuropathology of PSMF1-related disorder.
In conclusion, this study establishes a causal relationship between biallelic PSMF1 variants and early-onset PD and neurodegeneration in humans. Our data reveal impairment of mitochondrial membrane potential, dynamics and mitophagy in PSMF1 patient-derived fibroblasts, suggesting disruption of mitochondrial pathways as a contributing pathomechanism in PSMF1 deficiency. Additionally, we demonstrate that Psmf1 inactivation in Drosophila and mouse leads to age-dependent motor dysfunction, with diffuse gliosis in mice. Evidence herein presented and previous literature suggests that PSMF1 is a key factor in converging pathways of human neurodegeneration. Relevantly, Psmf1 deficiency in mice can be completely rescued with a Psmf1 transgene, and transgenic Psmf1 expression can also robustly suppress phenotypes of Fbxo7-knockout mice.21 Overall, this provides a proof of concept to explore PSMF1 gene therapy to treat neurodegenerative disorders in humans.
METHODS
Written informed consent for diagnostic and research activities and for publication of results was obtained from all study participants or their parents/guardians. Samples and data collection and analysis were not performed blind to the conditions of the experiments. No human fibroblasts, animals or data points have been excluded from the analyses.
Patient recruitment, phenotypic assessment, sampling and primary dermal fibroblast culture
Clinical and prenatal ultrasound assessments were conducted by experienced pediatric and adult neurologists and obstetrician-gynecologists, respectively. A detailed clinical proforma was completed by recruiting clinicians and clinical geneticists of affected individuals. Videos of neurological assessments were critically reviewed by experienced neurologists with expertise in movement disorders (F.M., K.P.B.). Neuroimaging was reviewed by an experienced paediatric neuroradiologist (M.S.). Genomic DNA was extracted from whole blood or amniotic fluid cells (proband O-II-2) and RNA was extracted from whole blood (parents H-I-1, H-I-2) according to standard procedures and protocols. In proband H-II-3, a muscle biopsy from the left anterior thigh was performed according to standard procedures and protocols. Punch skin biopsies of proband A-II-2 and his unaffected parents (A-I-1, A-I-2), proband B-II-3 and her unaffected brother (B-II-1), proband E-II-3 and his unaffected parents (E-I-1, E-I-2) and probands H-II-3, J-II-4 and K-II-6 (Fig.1a) were collected according to standard procedures and protocols for primary dermal fibroblast culture for RNA extraction and live cell imaging experiments. A punch skin biopsy was obtained from proband M-II-5 for primary dermal fibroblast culture for extracting protein lysate and performing Western blotting. Patient-derived primary skin fibroblasts were cultured in high glucose DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Thermo Fisher Scientific).
Genetic studies
Exome or genome sequencing and analysis.
ES of individuals A-I-1, A-I-2, A-II-2, C-II-2, G-II-3, J-II-1, K-II-6 was performed by Macrogen (Europe or South Korea) as detailed elsewhere.53 Briefly, target enrichment was performed with 2μg genomic DNA using the SureSelectXT Human All Exon Kit version 6 (Agilent) to generate barcoded ES libraries. Libraries were sequenced on the HiSeqX platform (Illumina) with 50x coverage. Quality assessment of the sequence reads was performed by generating QC statistics with FastQC. The bioinformatics filtering strategy included screening for only exonic and donor/acceptor splicing variants. In accordance with the pedigree and phenotype, priority was given to rare variants (<0.01% in public databases, including 1000 Genomes project, NHLBI Exome Variant Server, Complete Genomics, and Exome Aggregation Consortium [ExAC v0.2]) that were fitting a recessive (homozygous or compound heterozygous) or a de novo model and/or variants in genes previously linked to movement disorders, neurodevelopmental delay, and other neurological disorders. ES of individuals B-II-3, B-II-4 and F-II-3 was performed at the ICM IGenSeq core facility. Exons were captured using the Roche V.3 kit, followed by a massively parallel sequencing on NextSeq500 system (Illumina). Read alignment and variant calling were done using an in-house pipeline. Briefly, FastQC was used to check the quality of the reads and low-quality reads were removed using Trimmomatic. Sequencing data were then aligned to the human reference genome hg19 using the bwa suite36 and variant calling was performed using GATK HaplotypeCaller1. ES of individuals B-II-3, B-II-4 and F-II-3 was analyzed using VaraFT2 software. We filtered all homozygous variants shared by the two affected individuals that possibly affected the cDNA or localized in the splice site region (-8 +11bp from exon/intron junction), with a MAF <1% in the gnomAD public database and localized in region of loss of heterozygosity. Genome sequencing (GS) of D-II-4 was performed through the NIH facility. Read alignment and variant calling were done using Dragen version 07.021.624.3.10.4 (Illumina). Research for PSMF1 variants was done using an in-house R script (https://gitlab.com/icm-institute/corti-corvol/variants_tiers_gene). ES of probands B-II-3 and L-II-1 was carried out by CENTOGENE as previously described.54 GS was performed in individual C-II-2 by CENTOGENE as detailed elsewhere.55 ES of proband E-II-3 was performed using Agilent SureSelect Human All Exon V6 R2 kit with Illumina, NOVASeq6000 (Macrogen, Korea). Raw data obtained from ES were processed using the SEQ platform (Genomize, Istanbul, Turkey). Burrows-Wheeler Aligner (BWA) was used to align with the human reference genome, hg19. Freebayes was applied for variant calling, and the PCR duplicates were removed using the DeDup tool. ES of H-II-1 and H-II-3 was performed as described elsewhere.56 ES of proband I-II-3 was performed on a Novaseq 6000 platform (Illumina, San Diego, California) using IDT xGen Exome Research Panel V2 Kit (Integrated DNA Technologies, Coralville, Iowa). The average read depth was at least 100 reads, and the average percentage coverage of Refseq genes at 20X was at least 94%. The bioinformatic Analysis was performed using the Franklin variant interpretation platform (Genoox, Tel Aviv, Israel). Variants were filtered based on their effect on the protein (missense, nonsense, frameshift, splice-site) and a minor allele frequency <0.01 in general population databases (e.g. gnomAD), and the Rambam Genetics Institute internal database of over 3000 Israeli samples. ES of individual K-II-3 was carried out as reported previously.57 ES of subjects M-II-5, N-I-1, N-I-2 and N-II-3 was carried out as detailed elsewhere.58 ES of subjects O-I-1, O-I-2 and O-II-3 where performed as described elsewhere.59 Sanger sequencing was used to confirm next-generation sequencing findings and perform family segregation whenever possible. Recommendations of the Human Genome Variation Society were used to describe the cDNA and protein sequence variants using NM_006814.5 and NP_006805.2 as the reference. All next-generation sequencing findings were verified by Sanger sequencing while performing family segregation analysis in-house or in the referring centers.
Frequency, pathogenicity prediction and computational analysis of PSMF1 variants.
As a reference for potentially neutral variants and frequency of variants in the healthy population, gnomAD was inspected for all PSMF1 variants (Supplementary File 1). Pathogenicity prediction of PSMF1 non-splice variants was assessed using CADD, PolyPhen2 HVAR, SIFT4G, PROVEAN, MutationTaster and Human Splicing Finder (HSF) to predict the impact of variants on the protein structure and function (Supplementary File 1A). To examine the conservation of substituted amino acid positions, we used Genomic Evolutionary Rate Profiling (Supplementary File 1) and performed visual multiple sequence alignment of PSMF1 across multiple different species by inputting PSMF1 protein sequences for each species retrieved from UniProt in Clustal Omega (Fig.2b).60 For in silico splice predictions of six splice variants, we used the computational tools SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer, SpliceAI, and AbSplice (Supplementary File 1B).
Autozygosity mapping.
In all affected individuals having homozygous PSMF1 variants and their unaffected family members with raw ES or GS data available (Pedigrees B-C-D-E-F-H-J-K-L-M), regions of homozygosity larger than 2 Mb were identified using the AutoMap software (https://automap.iob.ch/) by inputting VCF files (Variant Call Format) files (Supplementary File 2).61 Whenever needed, we used UCSC LiftOver tool for conversion of hg19 VCF files to hg38.
Haplotype analysis.
Haplotype analysis was performed by plotting a color banding of variants flanking a homozygous PSMF1 variant detected in the proband of two pedigrees (J-II-1, K-II-6). By comparing the banding patterns, the status of being a recurrent or founder variant was investigated (Supplementary File 3).
RNA isolation and sequencing
Total RNA was extracted from fibroblasts of proband A-II-2 and his parents (A-I-1, A-I-2) and probands H-II-3, J-II-4 and K-II-6 using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. In H-I-1 and H-I-2, total RNA was isolated from peripheral whole blood treated with red blood cell lysis buffer (Qiagen) and white blood cells were collected for RNA extraction. RNA was extracted using TRIzol™ reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to manufacturer’s instruction. RNA samples with an RNA integrity number (RIN) ≥ 7 was used for RNA-seq. cDNA libraries were prepared by KAPA mRNA HyperPrep Kit (Roche Diagnostics, Tucson, AZ, USA) according to manufacturer protocol. Briefly, poly-A containing mRNA was collected and fragmented to 200–300 bp. Fragmented mRNA was then used as template for first-strand cDNA synthesis by random hexamer. In the second strand cDNA synthesis, blunt-end double stranded cDNAs were generated for 3’ adenylation and indexed adaptor ligation. The adaptor-ligated libraries enriched by polymerase chain reaction were sequenced as 151-bp paired-end runs on Illumina® NovaSeq 6000 platform (Illumina, San Diego, CA, USA) at the Center for PanorOmic Sciences (CPOS), University of Hong Kong. Data cleaning of the transcriptomics profile in fastq format was performed by AfterQC (v0.9.6). The sequence was aligned to the hg19/GRCh37 human reference genome and outputted as bam files using STAR (v2.5.2a, https://github.com/alexdobin/STAR) in two-pass mode. RNA sequencing data were processed with DROP v1.3.3 workflow,62 which integrates the statistical algorithm OUTRIDER63 and FRASER264,65 for detection of aberrant expression and aberrant splicing respectively by computing significant levels of extreme read count values. Both algorithms utilize a denoising autoencoder to control for the hidden confounder effects. In OUTRIDER for detection of aberrant expression, a false discovery rate (FDR) ≤ 0.05 was used as the cut-off for calling significant aberrant expression outliers.63 In FRASER2, split reads spanning the exon–exon junction and nonsplit reads spanning the splice sites were counted for splicing event calling. The Intron Jaccard Index is computed using split and non-split reads for capturing several types of aberrant splicing. Significant aberrant splicing events were defined with FDR ≤ 0.1.64,65
Minigene splicing assay
To functionally test the predicted impact of the c.365+2T>C (exon 3) and c.605+1G>A (exon 5) variants, an in vitro assay targeting each exonic region was performed through amplification from genomic DNA as previously described.66 Primers are shown in Supplementary File 9. Following amplification, clean-up and restriction enzyme digestion (XhoI, BamHI) of the PCR amplicon and pSPL3 exon trapping vector was performed prior to ligation between the vector-containing exons A and B of the linearized vector that was transformed into DH5α competent cells (NEB 5-alpha, New England Biolabs, Frankfurt, Germany), plated and incubated overnight. Colonies with the wild-type containing sequences were selected for correct size using colony PCR with an SD6 forward and the PSMF1 exon-specific reverse primer. Primers for site-directed mutagenesis were designed with NEBase Changer version 1.3.3 (New England Biolabs, https://nebasechanger.neb.com/). Site-directed mutagenesis followed the manufacturer’s protocol. Clones were selected for overnight LB-amp culture. Plasmids were isolated and Sanger sequence verified. The sequence-confirmed vectors with mutant and wild-type sequence were transfected into HEK 293T cells (ATCC, Manassas, VA, USA) with a density of 2x105 cells per mL. 2 µg of the respective pSPL3 vectors was transiently transfected using 6 µL of FuGENE 6 Transfection Reagent (Promega, Walldorf, Germany). Empty vector and transfection negative reactions were included as controls. The transfected cells were harvested 24 h after transfection. Total RNA was prepared using miRNAeasy Mini Kit (Qiagen, Hilden, Germany). Approximately 2 µg of RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s protocols. The cDNA was PCR amplified using vector-specific SD6 forward and SA2 reverse primers.
Proteomic studies
Protein structure modelling
The crystal structure for the PMSF1 homotetramer was accessed through the Protein databank repository, accession number 4OUH.67 Images were generated using ChimeraX.68 AlphaFold modelling69 of PSMF1 (UniProt: Q92530) and FBXO7 (Q9Y3I1–1) binding was performed using an HPC cluster with a local installation of ColabFold,70 using MMseqs2 sequence alignment and AlphaFold2-multimer-v3 model with subsequent AMBER structure relaxation. The predicted structures of the models obtained were visualized and analyzed using PyMOL 2.5.5.
Quantitative proteomics in human cultured dermal fibroblasts
Sample preparation.
Proband- and carrier-derived cultured dermal fibroblasts were harvested in lysis buffer containing 2% SDS (w/v), 100mM TEAB pH 8.5, with complete EDTA-free protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor cocktail tablets (Roche). Protein concentrations were determined via BCA assay (ThermoFisher) and 5µg protein aliquoted per sample for proteomic analysis. Samples were reduced with 10mM TCEP for 30 minutes at 60°C, followed by alkylation of cysteine residues with 40mM IAA for 30 minutes at room temperature in the dark. 20% SDS was then added to achieve a final concentration of 5% SDS in samples, which were then acidified by addition of trifluoracetic acid to a final concentration of 1%. Samples were subsequently processed for on-column tryptic digestion using S-Trap™ micro columns (Protifi, USA).71,72 Briefly, samples were diluted 6x with wash buffer (90% methanol, 10% 100mM TEABC) and loaded onto micro columns, with centrifugation at 1000g for 1 minute and flow-through discarded. After sample loading, the S-Trap™ micro columns were washed four times with 150μl wash buffer followed by centrifugation at 1000g, 1 minute. On-column digestion was performed by incubating 40μl (1μg) of Trypsin/Lys-C mix (MS grade, Promega, UK) in 50mM TEABC solution (pH 8) at 47°C for 1 hour and 20 minutes, followed by incubation at 22°C overnight. The samples were then eluted with addition of 40μl 50mM TEABC (pH 8), 40μl 0.15% (v/v) formic acid (FA), and 3x 40μl 80% acetonitrile (ACN), 0.15% FA. Peptides were then dried using a vacuum centrifuge at room temperature and stored at −20°C until mass spectrometry analysis.
Quantitative proteomic analysis.
Peptides were resuspended in 0.1% formic acid supplemented with 0.015% N-Dodecyl-B-D-Maltoside (DDM). 200ng of peptides were analyzed with a Vanquish Neo nano-liquid chromatography system in-line with an Orbitrap Astral mass spectrometer (ThermoScientific). Peptides were trapped and eluted using an Acclaim™ PepMap™ 100 C18 HPLC column (3μm particle size, 75μm diameter, 150mm length) and separated using an EASY-Spray™ PepMap™ Neo UHPLC column (2μm C18 particle, 75μm diameter,150mm length) with buffer A: 0.1% formic acid and buffer B: 80%ACN, 0.1% formic acid. Peptides were separated across a 13-minute gradient and the data was acquired using narrow window DIA mode. MS1 scans performed at a resolution of 240,000 across a m/z range of 380–980 and measured using ultra-high field orbitrap mass analyzer, with a normalized AGC target of 500% and maximum injection time of 3 milliseconds. DIA scans were performed of the precursor mass range with an isolation window of 4m/z, scan range of 150–2000m/z and a normalized collision energy of 25%.
Raw mass spectrometry data analysis.
Raw data were searched using DIA-NN (version 1.8.1) against a predicted spectral library generated from the reviewed human UniProt database (downloaded January 2023; 20383 entries with isoforms). Spectra were searched with strict Trypsin specificity (cleavage at K or R residues), allowing a maximum of two missed cleavages. Peptides with an amino acid length of 7–30 were considered. N terminal methionine excision and oxidation were specified as variable modifications and cysteine carbamidomethylation as a fixed modification, with a maximum of one variable modification permitted.
Statistical analysis and data visualization.
Statistical analysis and visualization of DIA-NN output files was performed with Python (version 3.11.5, Jupyter Notebook; Project Jupyter) using packages pandas (2.0.3), matplotlib (3.7.2), seaborn (0.12.2), numpy (1.26.1) and sci-kit learn (1.3.0); in conjunction with Perseus software (version 1.6.15.0) and uploaded to CURTAIN web tool (https://curtain.proteo.info/#/, Supplementary File 10). Proteins identified by a single peptide were excluded, with log2-transformed protein intensities then filtered by 100% detection in at least one sample condition. Missing values were imputed by random draws from a Gaussian distribution (width 0.3 and downshift 1.8). The Limma package (version 3.56.2; R, version 4.3.1) was used to fit the data to a linear model using the lmfit function, with eBayes correction to compute moderated t-statistics and false-discovery rate controlled by Benjamini-Hochberg correction.
Western blotting
Protein lysates were isolated from HepG2 cells, control-derived fibroblasts and proband M-II-5-derived fibroblasts (with homozygous PSMF1 variant c.1A>T) using RIPA buffer (Thermo Fisher) according to the manufacturer's protocol. 30μg protein lysates were separated by PAGE (18% gel), semi-dry blotted and incubated with a polyclonal anti-PSMF1 IgG Ab (Thermo Fisher cat. no. PA5–21603) at 4°C overnight. After washing and a 1-hour incubation with goat anti-rabbit IgG HRP, the blot was washed again before being developed with ECL-plus (Thermo Fisher) for 3 minutes. After stripping, the same blot was incubated with monoclonal β-actin antibody, washed and incubated with rabbit anti-mouse IgG HRP before being developed for 2 minutes with ECL-plus.
Frequency of PSMF1 variants in the UK Biobank and Accelerating Medicines Partnership-Parkinson’s Disease Initiative and gene burden
We searched for PSMF1 missense (nonsynonymous, nonframeshift indels) and loss-of-function (splice, nonsense, frameshift) variants in the ES data within the UK Biobank (UKB, https://www.ukbiobank.ac.uk/)73 and extracted exonic variants from GS data from the Accelerating Medicines Partnership-Parkinson’s Disease Initiative (AMP-PD, https://amp-pd.org/).74 Processing of these cohorts was detailed elsewhere.75 In brief, the UKB is a large-scale biomedical dataset with clinical and genetic information from ~500,000 participants.73 After filtering and removing those recruited in a genetic study, the database consists of a total of 45,857 individuals (22,040 males), of which 38,051 are controls, 1,105 cases, 6,033 individuals with a parent that is diagnosed with PD, and 668 individuals with a sibling that is diagnosed with PD (6,701 proxy cases). Among controls, individuals with age at recruitment <59 years, any reported neurological disorders (category 2406 and field codes: Dementia/42018, Vascular dementia/42022, FTD/42024, ALS/42028, Parkinsonism/42030, PD/42032, PSP/42034, MSA/42036), a parent with PD or dementia (field codes: 20107 and 20110) were filtered out.73,75
GS data from multiple datasets, including the Parkinson’s Progression Markers Initiative (PPMI), the Parkinson’s Disease Biomarkers Program (PDBP), the Harvard Biomarker Study (HBS), BioFIND, SURE-PD3, and STEADY-PD3, were obtained as part of the Accelerating Medicines Partnership in Parkinson’s Disease (AMP-PD) initiative. The AMP-PD cohorts (PPMI, PDBP, HBS, BioFIND, SURE-PD3, and STEADY-PD3) followed the GATK Best Practices guidelines established by the Broad Institute’s joint discovery pipeline, as well as additional details provided elsewhere.74 Data processing and quality control (QC) procedures have been described previously.74,76 All individuals included in the analysis were of European ancestry through principal component analysis using HapMap3 European ancestry populations.
The AMP-PD datasets consist of unrelated individuals and publicly available cohorts only. Similar filtering parameters were used for the AMP-PD databases resulting in a total of 4,007 individuals (2,197 males), with 2,556 being controls and 1,451 being PD cases.74,75 Variants were annotated using ANNOVAR, including the databases refGene, avsnp150, and clinvar_20220320.77 Variants were then selected/filtered for nonsynonymous (missense), stopgain, stoploss, nonframeshift, frame, splice (loss of function). For both analyses, we applied an upper MAF limit of 0.005, which captured most PSMF1 variants (Supplementary File 7). Raw genotype data files were generated to retrieve information on the allelic status (heterozygous, homozygous for reference allele, homozygous for non-reference allele). Following variant extraction, single variant association tests were performed using PLINK v1.90b4.4 (http://pngu.mgh.harvard.edu/purcell/plink/).78 To assess whether the average effect of genetic variants in this gene is associated with PD, burden testing was performed using Sequencing Kernel Association optimal unified test (Skat-O), Combined Multivariate and Collapsing (CMC) Wald test and CMC burden testing. This process, including variant annotation using SnpEff and SnpSift softwares, and the Loss-of-Function (LoF) Transcript Effect Estimator (LOFTEE) has been previously described.75
Mitochondrial studies in human cultured dermal fibroblasts
Live cell imaging of cultured dermal fibroblasts for studying mitochondrial bioenergetic state and function was performed in cell donors from Pedigrees A-B-E-H-J-K (Fig.1a). Images were acquired using Zeiss LSM 980 and LSM 710 confocal microscopes equipped with META detection system and 40× oil immersion objective. Images were analyzed using ZEN 2009 software and Fiji 2 software.
Measurement of mitochondrial membrane potential (Δψm) using TMRM.
For measurements of ΔΨm, dermal fibroblasts plated on 25-mm glass coverslips were loaded for 40 minutes at room temperature with 25 nM tetramethylrhodamine methylester (TMRM; Invitrogen), a cationic fluorescent dye that enters healthy mitochondria, in a HEPES buffered saline solution (HBSS) composed of 156mM NaCl, 3mM KCl, 2mM MgSO4, 1.25mM KH2PO4, 2mM CaCl2, 10mM glucose and 10mM HEPES; pH 7.35. The dye remained present in the media at the time of recording. Confocal images were obtained using a Zeiss 980 LSM Airy or 710 VIS CLSM equipped with a META detection system and a 40× oil immersion objective. TMRM was excited using the 561 nm laser line and fluorescence was measured above 580 nm. For basal ΔΨm measurements, Z-stack images were obtained by confocal microscopy and analyzed using ZEN 2009 software (Zeiss). TMRM values for control cases were set to 100% and the PMSF1 deficient and family members fibroblasts values were expressed relative to controls. For analysis of response to mitochondrial toxins, images were recorded in a time course-dependent manner from a single focal plane and analysed using ZEN Zeiss software (Zeiss).
For analysis of the maintenance mechanism of ΔΨm the response to mitochondrial toxins (oligomycin, rotenone fccp was studied. TMRM was used in the redistribution mode to assess ΔΨm, and therefore a reduction in TMRM fluorescence represents mitochondrial depolarization. Basal TMRM levels = 100%. TMRM fluorescence after complete depolarization by FCCP = 0%.
Dynamics.
We further studied the mitochondrial dynamics in the TMRM z-stacks by using the Mitochondrial Network Analysis (MiNA) toolset, a simplified pipeline to analyze the mitochondrial network morphology and fragmentation using fluorescence images and three-dimensional stacks using Fiji 2 software following the creators’ protocol,79 we studied the average length of all mitochondrial rods and branches in each cell (Fig.4d) and the number of branches per network in each cell (Fig.4e).
Mitophagy.
To study mitophagy via mitochondrial and lysosomal co-localization, cells were loaded with 200nM MitoTracker Green FM and 50nM LysoTracker Red in HBSS for 30 minutes before experiments. Confocal images were obtained using a Zeiss 710 confocal microscope equipped with 40X oil immersion objective. The 488nm argon laser line was used to excite MitoTracker Green fluorescence which was measured between 505 and 530nm. Illumination intensity was kept to a minimum (about 1% of laser output) to avoid phototoxicity and the pinhole set to give an optical slice of approximately 2μm. For LysoTracker Red, the 543nm Ne/He laser line was used with measurement above 650nm. All data presented were obtained from a minimum of five coverslips and three different cell preparations.
Drosophila melanogaster modelling and experiments
All Drosophila melanogaster experiments were carried out following published guidelines and approved by the local Sorbonne University ethics committee. In D. melanogaster, we obtained targeted knockdown of psmf1 expression in conjunction with the UAS/Gal4/Gal80 system of cell-type-specific transgene expression by RNA-interference (RNAi). Transgenic RNAi flies were generated according to a previously published protocol.80 Orthologues were identified through the “DIOPT ortholog finder”,81 with psmf1 (FBgn0033669), prkn (FBgn0041100) and nsmase (FBgn0035421) being considered as fly orthologues of human PSFM1, PRKN and SMPD2, respectively. Transgenic flies were obtained from VDRC Stock Center. We crossed the RNAi lines psmf1 (VDRC ID: 28858), prkn (VDRC ID: 47636) and nsmase (VDRC ID: 107062) with an nSyb-Gal4 strain to drive pan-neuronal RNAi expression. Growing flies were exposed to a standard 12h:12h light-dark schedule at 25°C and 60% relative humidity. Adult flies of the selected genotypes were tested for behavioral analyses at Days 10, 20 and 35. Based on previous studies and behavioral tests, prkn and nsmase were considered as positive and negative control, respectively. We assayed D. melanogaster locomotor activity using the climbing test based on negative-geotaxis analysis. Three experimental replicates were performed, each test including at least 15 flies. The test was performed on all lines five times at each time point after a 1-minute rest. Only females were tested. Flies failing to cross the tube at its midpoint (5cm) within 10 seconds from stimulation (repulsion) were interpreted as having a motor impairment. ImageJ software was used to automatically count flies.82
Mus musculus modelling and experiments
Mice.
Psmf1 knockout-first (KOF) mice were bred with a strain expressing FLP1 recombinase to generate a Psmf1 allele with the third exon flanked by 2 loxP sites (Psmf1fl and Psmf1tm1c(EUCOMM)Hmgu) as previously described.22 Psmf1fl/fl mice expressed normal levels of Psmf1, were viable and fertile, and appeared phenotypically normal. However, introduction of a Cre driver successfully inactivated Psmf1 and resulted in tissue-specific or time-specific loss of Psmf1 protein. UBC-Cre/ERT2 (007001) and Ai14 (007914) mice were purchased from Jackson Laboratories. All experiments in Mus musculus were performed in the US as required by the United States Animal Welfare Act and NIH’s policy to ensure appropriate care and use of laboratory animals for research, and according to established guidelines and supervision by the Institutional Animal Care and Use Committee of The Rockefeller University. Mice were housed in accredited facilities of the Association for Assessment of Laboratory Animal Care in accordance with NIH guidelines.
Tamoxifen injection.
Tamoxifen was dissolved in corn oil overnight at 37°C to a final concentration of 20mg/ml. Injection dose was determined by weight, using approximately 75 mg tamoxifen/kg body weight (about 94–100μl of tamoxifen solution for an adult mouse). Tamoxifen was administered by intraperitoneal injection into 6 to 7-week-old mice for 3 consecutive days. During the injection period and for one week after, the mice were monitored for any adverse reaction due to the treatment.
Behavioral tests assessing locomotion and anxiety-related behavior.
Rotarod test.
We used the rotarod test to assesses the motor performance of mice on a rotating accelerating rod (Ugo Basile model). The time-to-fall (rotarod score), i.e., the cumulative time (seconds) a mouse maintained its balance on the rotarod before falling off, was recorded. The maximum latency was set at 300 seconds per trial. A fall was recorded if the mouse held on to the rod for two consecutive revolutions. Mice treated with tamoxifen or corn oil (controls) started a four-day training period on the rotarod 10 days after the final injection. On day 14 post injection, the time-to-fall for one trial was recorded. The trial was repeated for 10 days to follow the progressive deficit in the tamoxifen treated Psmf1/Pi31 Ubc-CreERt2 mice. On day 25 post injection, three trials were conducted for each mouse and average latency to fall was determined.
Open field test.
We assessed motor activity as a reaction to an unknown environment (anxiety-related behavior). At 26 days post tamoxifen or corn oil injection mice were assayed for locomotion and anxiety in an open field chamber (42 × 42 × 30.5 cm) with a dark enclosure (half of open field chamber) for 10 minutes. Data was analyzed with Fusion 3.2 software (Open field chamber and software from AccuScan Instruments Inc.). Briefly, mice were moved to testing room 60 minutes prior to testing to acclimate. Then mice were placed in the dark enclosure and activity was recorded for 10 min.
Statistics.
Error bars are standard deviations and significance values (p values; Fig.6b-c-d-e-f) were derived by two-tailed T test with at least 2 mice per condition.
Immunofluorescence of mouse brain sections.
At Day 28 after final tamoxifen injection, mice were perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA) in PBS. Brains were removed and postfixed in 4% PFA PBS overnight at 4°C. They were then washed in PBS, soaked in 15% sucrose in PBS overnight at 4°C or until they sank, and then soaked in 30% sucrose in PBS overnight at 4°C. The brains were embedded in Richard-Allan Scientific Neg-50 Frozen Section Medium, and 50-μm sagittal sections were made using a Leica CM 3050S cryostat. The floating cryostat sections were permeabilized and blocked overnight in PBS with 0.3% Triton X-100 and 5% Normal Donkey Serum (NDS) at 4°C. The sections were then incubated overnight with primary antibodies diluted in PBS with 0.3% Triton X-100 and 5% NDS. Subsequently, the sections were washed 3 times for 20 min each in PBS with 0.1% Triton X-100 and incubated overnight with Alexa Fluor conjugated secondary antibodies diluted in PBS with 0.3% Triton X-100 and 5% NDS at 4°C. The sections were then washed for 30 minutes 4 times in PBS with 0.1% Triton X-100 and mounted with Molecular Probes ProLong Diamond antifade mounting medium. Images were captured on a Zeiss confocal LSM 780 with Zen software. Primary antibodies used were anti-Tyrosine Hydroxylase (TH) Rb pAb (1/500) Abcam ab112, anti-Tyrosine Hydroxylase (TH) Mouse mAb IgG1κ (clone LNC1) (1/500) Millipore MAB318, mCherry Chicken IgY (1/1000) Novus NBP-2–25158, GFAP chicken IgY (1/2000) EnCor CPCA-GFAP, GFAP rabbit IgG (1/500) Dako Cytomation Z0334, Iba1 rabbit IgG (1/1000) Wako 019–19741 and SQSTM1/p62 mouse IgG2a (1/500) Abcam ab56416.
Supplementary Material
Acknowledgments
The authors are indebted to the patients participating in this study as well as their families. For their kind support to this study, we are most grateful to Dr Andrew B. Singleton (National Institutes of Health, Bethesda, USA), Prof. Manju Kurian (University College London, London, United Kingdom), Dr Zane Jaunmuktane (University College London, London, United Kingdom), Prof. Olaf Riess and Dr German Demidov (University of Tübingen, Tübingen, Germany), Dr Julia Baptista (Exeter Genomics Laboratory, Exeter, United Kingdom), Dr Erin Torti (GeneDx), Dr Jill Anne Mokry (Baylor Genetics), Prof. Stephan Züchner (University of Miami, Miami, USA), Dr Rami Abou Jamra (University of Leipzig, Leipzig, Germany), Dr Nikola Kresojević (University Clinical Center of Serbia, Belgrade, Serbia), Dr Michele Matarazzo (HM CINAC, Madrid, Spain), Prof. Naci Karaağaç (Istanbul University, Istanbul, Turkey, Dr Bernd Wilken (Klinikum Kassel, Kassel, Germany), Dr Laura Mahoney Sanchez (The Francis Crick Institute, London, United Kingdom), Prof. Christine Klein, Dr Joanne Trinh and Dr Manu Sharma (University of Lübeck, Lübeck, Germany). Cr.Te., Th.Co., Cl.Pi, Ma.Ca, Ba.A.Ha., Gu.Co., Su.Le. and Al.Br. thank the DNA and cell bank of the ICM, Sorbonne Université, Paris, France, for sample preparation and acknowledge that part of this work was carried out on the iGenSeq and DAC core facilities of the ICM. Ay.N.Ba. and Tu.Gü. gratefully acknowledge the use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM), Istanbul, Turkey, funded by the Presidency of Turkey, Head of Strategy and Budget. Ch.Fa. is a member of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability ERN-ITHACA [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01–2016/739516]. Mi.Ze. is a member of the Medical and Scientific Advisory Council of the Dystonia Medical Research Foundation and a member of the Governance Council of the International Cerebral Palsy Genomics Consortium. We used data from the UK Biobank Resource under Application Number 33601. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://hpc.nih.gov). Data used in the preparation of this article were obtained from the AMP-PD Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. AMP-PD—a public–private partnership—is managed by the FNIH and funded by Celgene, GSK, the Michael J. Fox Foundation for Parkinson’s Research, the National Institute of Neurological Disorders and Stroke, Pfizer, and Verily. We would like to thank AMP-PD for the publicly available whole-genome sequencing data, including cohorts from the Fox Investigation for New Discovery of Biomarkers (BioFIND), the Parkinson’s Progression Markers Initiative (PPMI), and the Parkinson’s Disease Biomarkers Program (PDBP). The Parkinson’s Disease Biomarker Program (PDBP) consortium is supported by the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. Harvard Biomarker Study (HBS) is a collaboration of HBS investigators (full list of HBS investigators found at https://www.bwhparkinsoncenter.org/biobank) and funded through philanthropy and NIH and Non-NIH funding sources. The HBS Investigators have not participated in reviewing the data analysis or content of the manuscript.
Funding Statement
For their work on this study: Fr.Ma. was supported by the Edmond J. Safra Fellowship in Movement Disorders, the Edmond J. Safra Movement Disorders Research Career Development Award (Grant ID: MJFF-023893), the American Parkinson Disease Association and the David Pearlman Charitable Foundation; Cr.Te. received funding from the program "Investissements d'avenir" ANR-10-IAIHU-06; Ai.Sa. was supported by La Caixa Postgraduate Abroad fellowship; Ba.Vo. was funded by the German Research Foundation DFG VO 2138/7–1 grant 469177153; No.Es. was supported by a Ramón y Cajal Fellowship RYC2021–034267-I and a research grant PID2022–137011OA-I00 funded by the Spanish Ministry of Science and the European Union; Pa.A.Le. was supported by a Royal Society Industry Fellowship (IF\R2\222002) with LifeArc technologies; St.Ef. was supported by an MRC strategic award to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) MR/S005021/1'; Re.Bu. was supported by the German Academic Exchange Service (DAAD) as part of the German-Arab Transformation Program Line 4 (Project-ID 57166498); Gu.Co. was supported by the Global Parkinson's Genetics Program (GP2), which is funded by the Aligning Science Across Parkinson's (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson's Research (https://gp2.org; Grant ID: MJFF-024876); Ay.N.Ba. and Tu.Gü. were supported by Suna and İnan Kıraç Foundation and Koç University School of Medicine, Istanbul, Turkey. Mi.Ze. received research support from the German Research Foundation (DFG 458949627; ZE 1213/2–1). Mi.Ze. acknowledges grant support by the European Joint Programme on Rare Diseases (EJP RD Joint Transnational Call 2022), and the German Federal Ministry of Education and Research (BMBF, Bonn, Germany), awarded to the project PreDYT (PREdictive biomarkers in DYsTonia, 01GM2302), by the Federal Ministry of Education and Research (BMBF) and the Free State of Bavaria under the Excellence Strategy of the Federal Government and the Länder, as well as by the Technical University of Munich – Institute for Advanced Study. Mi.Ze.’s research is also supported by a “Schlüsselprojekt” grant from the Else Kröner-Fresenius-Stiftung (2022_EKSE.185). Vi.Bo. received financial support from the Stichting Parkinson Fonds, The Netherlands, Grant. n. 1880. Part of this study (Br.Ch., An.Kw., Ma.Ch., Ea.Ng., Sz.Fa.) was supported by the Society for the Relief of Disabled Children (SRDC) and the Commissioned Paediatric Research at Hong Kong Children Hospital (PR-HKU-4) under the Health and Medical Research Fund (HMRF) by the Hong Kong Food and Health Bureau. This research was supported in part by the Intramural research Program of the NIH, National institute on Aging. Part of this work was supported by grants from the Cure Alzheimer's Fund and The Pershing Square Foundation to He.St. Su.Le. received grants from Fondation pour la Recherche Médicale (FRM, MND202004011718), la Fondation de France, France-Parkinson Association, la Fédération pour la Recherche sur le Cerveau (FRC) and the French program “Investissements d’avenir” (ANR-10-IAIHU-06). Cr.Te., Th.Co., Cl.Pi, Ma.Ca, Ba.A.Ha., Gu.Co., Su.Le. and Al.Br. were supported by the RMF through the project “DBS - From genetic mutations to motor circuit dysfunctions & recovery”. He.Ho. was funded by the MRC (MR/S01165X/1, MR/S005021/1, G0601943, MR/V012177/1), The National Institute for Health Research, University College London Hospitals Biomedical Research Centre, Rosetrees Trust, Brain Research UK, Sparks GOSH Charity, the Rosetrees Trust PhD Plus award (PhD2022\100042) and Global Parkinson's Genetic Program (GP2; Grant ID: MJFF-022153).
Funding Statement
For their work on this study: Fr.Ma. was supported by the Edmond J. Safra Fellowship in Movement Disorders, the Edmond J. Safra Movement Disorders Research Career Development Award (Grant ID: MJFF-023893), the American Parkinson Disease Association and the David Pearlman Charitable Foundation; Cr.Te. received funding from the program "Investissements d'avenir" ANR-10-IAIHU-06; Ai.Sa. was supported by La Caixa Postgraduate Abroad fellowship; Ba.Vo. was funded by the German Research Foundation DFG VO 2138/7–1 grant 469177153; No.Es. was supported by a Ramón y Cajal Fellowship RYC2021–034267-I and a research grant PID2022–137011OA-I00 funded by the Spanish Ministry of Science and the European Union; Pa.A.Le. was supported by a Royal Society Industry Fellowship (IF\R2\222002) with LifeArc technologies; St.Ef. was supported by an MRC strategic award to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) MR/S005021/1'; Re.Bu. was supported by the German Academic Exchange Service (DAAD) as part of the German-Arab Transformation Program Line 4 (Project-ID 57166498); Gu.Co. was supported by the Global Parkinson's Genetics Program (GP2), which is funded by the Aligning Science Across Parkinson's (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson's Research (https://gp2.org; Grant ID: MJFF-024876); Ay.N.Ba. and Tu.Gü. were supported by Suna and İnan Kıraç Foundation and Koç University School of Medicine, Istanbul, Turkey. Mi.Ze. received research support from the German Research Foundation (DFG 458949627; ZE 1213/2–1). Mi.Ze. acknowledges grant support by the European Joint Programme on Rare Diseases (EJP RD Joint Transnational Call 2022), and the German Federal Ministry of Education and Research (BMBF, Bonn, Germany), awarded to the project PreDYT (PREdictive biomarkers in DYsTonia, 01GM2302), by the Federal Ministry of Education and Research (BMBF) and the Free State of Bavaria under the Excellence Strategy of the Federal Government and the Länder, as well as by the Technical University of Munich – Institute for Advanced Study. Mi.Ze.’s research is also supported by a “Schlüsselprojekt” grant from the Else Kröner-Fresenius-Stiftung (2022_EKSE.185). Vi.Bo. received financial support from the Stichting Parkinson Fonds, The Netherlands, Grant. n. 1880. Part of this study (Br.Ch., An.Kw., Ma.Ch., Ea.Ng., Sz.Fa.) was supported by the Society for the Relief of Disabled Children (SRDC) and the Commissioned Paediatric Research at Hong Kong Children Hospital (PR-HKU-4) under the Health and Medical Research Fund (HMRF) by the Hong Kong Food and Health Bureau. This research was supported in part by the Intramural research Program of the NIH, National institute on Aging. Part of this work was supported by grants from the Cure Alzheimer's Fund and The Pershing Square Foundation to He.St. Su.Le. received grants from Fondation pour la Recherche Médicale (FRM, MND202004011718), la Fondation de France, France-Parkinson Association, la Fédération pour la Recherche sur le Cerveau (FRC) and the French program “Investissements d’avenir” (ANR-10-IAIHU-06). Cr.Te., Th.Co., Cl.Pi, Ma.Ca, Ba.A.Ha., Gu.Co., Su.Le. and Al.Br. were supported by the RMF through the project “DBS - From genetic mutations to motor circuit dysfunctions & recovery”. He.Ho. was funded by the MRC (MR/S01165X/1, MR/S005021/1, G0601943, MR/V012177/1), The National Institute for Health Research, University College London Hospitals Biomedical Research Centre, Rosetrees Trust, Brain Research UK, Sparks GOSH Charity, the Rosetrees Trust PhD Plus award (PhD2022\100042) and Global Parkinson's Genetic Program (GP2; Grant ID: MJFF-022153).
References
- 1.Poewe W. et al. Parkinson disease. Nature Reviews Disease Primers 3, 17013 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Dorsey E.R., Sherer T., Okun M.S. & Bloem B.R. The Emerging Evidence of the Parkinson Pandemic. J Parkinsons Dis 8, S3–S8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trevisan L. et al. Genetics in Parkinson's disease, state-of-the-art and future perspectives. Br Med Bull 149, 60–71 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris H.R., Spillantini M.G., Sue C.M. & Williams-Gray C.H. The pathogenesis of Parkinson's disease. Lancet 403, 293–304 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Smolders S. & Van Broeckhoven C. Genetic perspective on the synergistic connection between vesicular transport, lysosomal and mitochondrial pathways associated with Parkinson’s disease pathogenesis. Acta Neuropathologica Communications 8, 63 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandres-Ciga S., Diez-Fairen M., Kim J.J. & Singleton A.B. Genetics of Parkinson's disease: An introspection of its journey towards precision medicine. Neurobiol Dis 137, 104782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasuhn J. & Bruggemann N. Genotype-driven therapeutic developments in Parkinson's disease. Mol Med 27, 42 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taymans J.M. et al. Perspective on the current state of the LRRK2 field. NPJ Parkinsons Dis 9, 104 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi M., Du X., Jiao Q., Chen X. & Jiang H. Expanding the role of proteasome homeostasis in Parkinson's disease: beyond protein breakdown. Cell Death Dis 12, 154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J.S., Davis R.L. & Sue C.M. Mitochondrial Dysfunction in Parkinson's Disease: New Mechanistic Insights and Therapeutic Perspectives. Curr Neurol Neurosci Rep 18, 21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borsche M., Pereira S.L., Klein C. & Grunewald A. Mitochondria and Parkinson's Disease: Clinical, Molecular, and Translational Aspects. J Parkinsons Dis 11, 45–60 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malpartida A.B., Williamson M., Narendra D.P., Wade-Martins R. & Ryan B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson's Disease: From Mechanism to Therapy. Trends Biochem Sci 46, 329–343 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Trinh D., Israwi A.R., Arathoon L.R., Gleave J.A. & Nash J.E. The multi-faceted role of mitochondria in the pathology of Parkinson's disease. J Neurochem 156, 715–752 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Chu-Ping M., Slaughter C.A. & DeMartino G.N. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain). Biochim Biophys Acta 1119, 303–11 (1992). [DOI] [PubMed] [Google Scholar]
- 15.McCutchen-Maloney S.L. et al. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J Biol Chem 275, 18557–65 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Uhlen M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zaiss D.M., Standera S., Holzhutter H., Kloetzel P. & Sijts A.J. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett 457, 333–8 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Bader M. et al. A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 145, 371–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho-Park P.F. & Steller H. Proteasome regulation by ADP-ribosylation. Cell 153, 614–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K. et al. PI31 Is an Adaptor Protein for Proteasome Transport in Axons and Required for Synaptic Development. Dev Cell 50, 509–524 e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minis A. & Steller H. PI31 expression prevents neuronal degeneration in a mouse Parkinson Disease model. bioRxiv, 2020.05.05.078832 (2020). [Google Scholar]
- 22.Minis A. et al. The proteasome regulator PI31 is required for protein homeostasis, synapse maintenance, and neuronal survival in mice. Proc Natl Acad Sci U S A 116, 24639–24650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yashiroda H. et al. N-terminal alpha7 deletion of the proteasome 20S core particle substitutes for yeast PI31 function. Mol Cell Biol 35, 141–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B.J. et al. Arabidopsis PROTEASOME REGULATOR1 is required for auxin-mediated suppression of proteasome activity and regulates auxin signalling. Nat Commun 7, 11388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Fonzo A. et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 72, 240–5 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Kirk R. et al. Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. J Biol Chem 283, 22325–35 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Al Rawi S. et al. Study of an FBXO7 patient mutation reveals Fbxo7 and PI31 co-regulate proteasomes and mitochondria. FEBS J (2024). [DOI] [PubMed] [Google Scholar]
- 28.Vingill S. et al. Loss of FBXO7 (PARK15) results in reduced proteasome activity and models a parkinsonism-like phenotype in mice. EMBO J 35, 2008–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemen C.S. et al. VCP and PSMF1: Antagonistic regulators of proteasome activity. Biochem Biophys Res Commun 463, 1210–7 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Scarian E. et al. The Role of VCP Mutations in the Spectrum of Amyotrophic Lateral Sclerosis-Frontotemporal Dementia. Front Neurol 13, 841394 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X. et al. VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington's disease. Nat Commun 7, 12646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherva R. et al. Identification of novel candidate genes for Alzheimer's disease by autozygosity mapping using genome wide SNP data. J Alzheimers Dis 23, 349–59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L. et al. A Rare Variant Nonparametric Linkage Method for Nuclear and Extended Pedigrees with Application to Late-Onset Alzheimer Disease via WGS Data. Am J Hum Genet 105, 822–835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesson C. et al. Identification of potential new genes involved in autosomal recessive forms of Parkinson’s disease [Abstract number 1113]. in International Congress of Parkinson's Disease and Movement Disorders (Nice, France, 2019). [Google Scholar]
- 35.Sobreira N., Schiettecatte F., Valle D. & Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 36, 928–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karczewski K.J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandhi S. et al. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell 33, 627–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludtmann M.H.R. et al. alpha-synuclein oligomers interact with ATP synthase and open the permeability transition pore in Parkinson's disease. Nat Commun 9, 2293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abramov A.Y. et al. Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain 133, 797–807 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezhnov A.V. et al. Intracellular pH Modulates Autophagy and Mitophagy. J Biol Chem 291, 8701–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komilova N.R. et al. Metabolically induced intracellular pH changes activate mitophagy, autophagy, and cell protection in familial forms of Parkinson's disease. FEBS J 289, 699–711 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Ruzankina Y. et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madisen L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kam T.I., Hinkle J.T., Dawson T.M. & Dawson V.L. Microglia and astrocyte dysfunction in parkinson's disease. Neurobiol Dis 144, 105028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacMahon Copas A.N., McComish S.F., Fletcher J.M. & Caldwell M.A. The Pathogenesis of Parkinson's Disease: A Complex Interplay Between Astrocytes, Microglia, and T Lymphocytes? Front Neurol 12, 666737 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian K. et al. Revisiting the critical roles of reactive astrocytes in neurodegeneration. Mol Psychiatry 28, 2697–2706 (2023). [DOI] [PubMed] [Google Scholar]
- 47.Wolf S.A., Boddeke H.W. & Kettenmann H. Microglia in Physiology and Disease. Annu Rev Physiol 79, 619–643 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Burda J.E. & Sofroniew M.V. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J. et al. Genome-Wide Association Analysis of Neonatal White Matter Microstructure. Cereb Cortex 31, 933–948 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu H.C., Wang J., Kjellgren A., Li H. & DeMartino G.N. High-resolution structure of mammalian PI31–20S proteasome complex reveals mechanism of proteasome inhibition. J Biol Chem 299, 104862 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Martinez A., Martinez A. & Whitworth A.J. FBXO7/ntc and USP30 antagonistically set the ubiquitination threshold for basal mitophagy and provide a target for Pink1 phosphorylation in vivo. PLoS Biol 21, e3002244 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kraus F. et al. PARK15/FBXO7 is dispensable for PINK1/Parkin mitophagy in iNeurons and HeLa cell systems. EMBO reports 24, e56399 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efthymiou S. et al. Biallelic mutations in neurofascin cause neurodevelopmental impairment and peripheral demyelination. Brain 142, 2948–2964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trujillano D. et al. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet 25, 176–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertoli-Avella A.M. et al. Successful application of genome sequencing in a diagnostic setting: 1007 index cases from a clinically heterogeneous cohort. Eur J Hum Genet 29, 141–153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwong A.K. et al. Exome sequencing in paediatric patients with movement disorders. Orphanet J Rare Dis 16, 32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han C. et al. Epileptic Encephalopathy Caused by Mutations in the Guanine Nucleotide Exchange Factor DENND5A. Am J Hum Genet 99, 1359–1367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brunet T. et al. De novo variants in neurodevelopmental disorders-experiences from a tertiary care center. Clin Genet 100, 14–28 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Chen W. et al. Clinical and molecular characterization of novel FARS2 variants causing neonatal mitochondrial disease. Mol Genet Metab 140, 107657 (2023). [DOI] [PubMed] [Google Scholar]
- 60.Madeira F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinodoz M. et al. AutoMap is a high performance homozygosity mapping tool using next-generation sequencing data. Nature Communications 12(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yepez V.A. et al. Detection of aberrant gene expression events in RNA sequencing data. Nat Protoc 16, 1276–1296 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Brechtmann F. et al. OUTRIDER: A Statistical Method for Detecting Aberrantly Expressed Genes in RNA Sequencing Data. Am J Hum Genet 103, 907–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mertes C. et al. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat Commun 12, 529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheller I.F., Lutz K., Mertes C., Yepez V.A. & Gagneur J. Improved detection of aberrant splicing with FRASER 2.0 and the intron Jaccard index. Am J Hum Genet 110, 2056–2067 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tompson S.W. & Young T.L. Assaying the Effects of Splice Site Variants by Exon Trapping in a Mammalian Cell Line. Bio Protoc 7(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shang J., Huang X. & Du Z. The FP domains of PI31 and Fbxo7 have the same protein fold but very different modes of protein-protein interaction. J Biomol Struct Dyn 33, 1528–38 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Meng E.C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci 32, e4792 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mirdita M. et al. ColabFold: making protein folding accessible to all. Nature Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nirujogi R.S., Fasimoye R., Phung T.K. & Alessi D.R. Protocol for Data Independent Acquisition - Mass spectrometry analysis – a DIA-based Organelle Proteomics. protocols.io (2022). [Google Scholar]
- 72.Fasimoye R. et al. Golgi-IP, a tool for multimodal analysis of Golgi molecular content. Proc Natl Acad Sci U S A 120, e2219953120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwaki H. et al. Accelerating Medicines Partnership: Parkinson's Disease. Genetic Resource. Mov Disord 36, 1795–1804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Makarious M.B. et al. Large-scale rare variant burden testing in Parkinson's disease. Brain (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bandres-Ciga S. et al. Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease. Acta Neuropathol 140, 341–358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valente A.J., Maddalena L.A., Robb E.L., Moradi F. & Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem 119, 315–326 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Qiao H.H. et al. An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila. Nat Commun 9, 4160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu Y. et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider C.A., Rasband W.S. & Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.