Abstract
Background:
Colorectal cancers (CRCs) from people with biallelic germline likely pathogenic/pathogenic variants in MUTYH or NTHL1 exhibit specific single base substitution (SBS) mutational signatures, namely combined SBS18 and SBS36 (SBS18+SBS36), and SBS30, respectively. The aim was to determine if adenomas from biallelic cases demonstrated these mutational signatures at diagnostic levels.
Methods:
Whole-exome sequencing of FFPE tissue and matched blood-derived DNA was performed on 9 adenomas and 15 CRCs from 13 biallelic MUTYH cases, on 7 adenomas and 2 CRCs from 5 biallelic NTHL1 cases and on 27 adenomas and 26 CRCs from 46 non-hereditary (sporadic) participants. All samples were assessed for COSMIC v3.2 SBS mutational signatures.
Results:
In biallelic MUTYH cases, SBS18+SBS36 signature proportions in adenomas (mean±standard deviation, 65.6%±29.6%) were not significantly different to those observed in CRCs (76.2%±20.5%, p-value=0.37), but were significantly higher compared with non-hereditary adenomas (7.6%±7.0%, p-value=3.4×10−4). Similarly, in biallelic NTHL1 cases, SBS30 signature proportions in adenomas (74.5%±9.4%) were similar to those in CRCs (78.8%±2.4%) but significantly higher compared with non-hereditary adenomas (2.8%±3.6%, p-value=5.1×10−7). Additionally, a compound heterozygote with the c.1187G>A p.(Gly396Asp) pathogenic variant and the c.533G>C p.(Gly178Ala) variant of unknown significance (VUS) in MUTYH demonstrated high levels of SBS18+SBS36 in four adenomas and one CRC, providing evidence for reclassification of the VUS to pathogenic.
Conclusions:
SBS18+SBS36 and SBS30 were enriched in adenomas at comparable proportions observed in CRCs from biallelic MUTYH and biallelic NTHL1 cases, respectively. Therefore, testing adenomas may improve the identification of biallelic cases and facilitate variant classification, ultimately enabling opportunities for CRC prevention.
Keywords: Colorectal cancer, mutational signature, adenoma, hereditary cancer predisposition, SBS18, SBS36, SBS30, MUTYH, NTHL1, variant of uncertain clinical significance
INTRODUCTION
Identifying people who have an increased risk of developing colorectal cancer (CRC), including people with a hereditary CRC or polyposis syndrome, provides important opportunities for cancer prevention. Individuals with homozygous or compound heterozygous likely pathogenic or pathogenic (LP/P) variants in the base excision repair genes MUTYH [1] and NTHL1 [2] (i.e., biallelic cases) predispose to the development of multiple pre-cancerous adenomas in the colon (adenomatous polyposis), CRC and a spectrum of extra-colonic cancers [2,3].
The application of tumour mutational signature profiling to identify hereditary cancer syndromes related to DNA repair defects has been highlighted [4]. Single base substitution (SBS) and insertion/deletion mutational signatures in CRC have been shown to be accurate predictors of Lynch syndrome and biallelic germline LP/P variants in MUTYH [5]. In particular, the combination of SBS18 and SBS36 (SBS18+SBS36) can accurately identify those with germline biallelic MUTYH LP/P variants [5,6], while for NTHL1, the SBS30 mutational signature has been identified in CRCs from those with biallelic NTHL1 LP/P variants [7]. Moreover, we previously identified two recurrent somatic mutations, namely the KRAS c.34G>T p.(Gly12Cys) and the PIK3CA c.1636C>A p.(Gln546Lys) mutations, that were strongly enriched in CRCs from biallelic MUTYH cases compared with CRCs from non-hereditary/sporadic cases (KRAS: p-value=1.4×10−6, PIK3CA: p-value=3.4×10−4) [6].
A further application of tumour mutational signature profiling is to aid variant classification. Previously, we have shown the presence of elevated levels of SBS18+SBS36 in CRCs provided evidence for an LP/P classification for the germline MUTYH variants c.1141G>T p.(Gly381Trp) and c.577–5A>G, where the second allele of MUTYH harboured an LP/P variant [6]. Alternatively, the absence of high levels of SBS18+SBS36 in CRCs supported a benign classification for MUTYH variants c.912C>G p.(Ser304Arg), c.821G>A p.(Arg274Gln), c.925C>T p.(Arg309Cys) and c.1431G>C p.(Thr477Thr) [6].
While these genomic features have been shown to be effective with CRC-derived data, there are important implications that could be facilitated by the ability to utilise mutational signature profiling in pre-cancerous adenomas namely: 1) identifying biallelic cases early before they develop cancer, including guiding surgical versus endoscopic management decision making, 2) enable pre-emptive genetic counselling and guide patient management strategies through risk assessment, 3) indicate if a second “unidentified” LP/P variant is present in monoallelic LP/P variant carriers, and 4) provide evidence for pathogenicity for variants of uncertain significance (VUS).
The aim of this study was to profile and compare the SBS18+SBS36 and SBS30 mutational signatures in adenomas and CRCs from biallelic MUTYH and biallelic NTHL1 cases, respectively, with sporadic adenomas and CRCs from participants without a hereditary CRC/polyposis syndrome to determine their discriminatory potential and ability to inform variant classification.
MATERIAL AND METHODS
Study cohort
Participants were men and women recruited to one of the following studies: 1) Applying Novel Genomic approaches to Early-onset and suspected Lynch Syndrome colorectal and endometrial cancers (ANGELS, n=4), 2) Colorectal Cancer Family Registry (CCFR, n=21) or 3) Genetics of Colonic Polyposis Study (GCPS, n=5) who were identified to have either germline biallelic MUTYH or germline biallelic NTHL1 LP/P variants from clinical diagnostic or research genetic testing. Formalin-fixed paraffin embedded (FFPE) tissue was collected for tumour mutational signature profiling comprising:
9 adenomas and 15 CRCs from 13 biallelic MUTYH cases;
4 CRCs from 4 monoallelic MUTYH cases;
7 adenomas, 1 hyperplastic polyp, 1 traditional serrated adenoma and 2 CRCs from 7 biallelic NTHL1 cases and
2 CRCs from 2 monoallelic NTHL1 cases.
A reference/control group of 46 participants from the CCFR who developed mismatch repair (MMR)-proficient adenomas (n=27) and/or MMR-proficient CRCs (n=26) and who were confirmed to not carry LP/P variants in 16 hereditary CRC/polyposis genes as defined in Seifert et al. [8] (i.e., non-hereditary/sporadic cases) were included in this study.
The mutational signature profiles of 12 CRCs from eight biallelic MUTYH cases and 4 CRCs from 4 monoallelic MUTYH cases described above have been reported previously [5,6]. Two CRCs from two biallelic and two monoallelic NTHL1 cases described above have been reported previously [7]. The studies were approved by the respective ethics committees and institutional review boards. All participants provided written informed consent for collection of tissue and peripheral blood samples.
Whole-exome sequencing and bioinformatic analysis
Adenoma and CRC tissue DNA and matched blood-derived DNA underwent whole-exome sequencing (WES) using the SureSelect Clinical Research Exome v.2 kit (Agilent Technologies, Santa Clara, CA, United States), to a median depth of 357.9 reads (interquartile range (IQR)=287.8–464.0) for FFPE tissue DNA samples and median depth of 179.1 reads (IQR=118.1–204.6) for blood-derived DNA samples. Somatic single-nucleotide variants and short insertion/deletions were determined using the intersection of calls from Strelka (v.2.9.2) [9] and Mutect2 (v.4.0) [10]. Tumour mutation burden (TMB) was calculated as the total number of all somatic single-nucleotide variants and short insertion/deletions observed in a sample divided by the size of the capture region (67Mb). A threshold for including variants was chosen based on a minimum depth (50 reads) and a minimum variant allele frequency of 10% as previously published [5]. Mutational signature profiles were calculated using the simulated annealing method previously described by SignatureEstimation [11] using a reduced set of 16 SBS signatures (Supplementary Table 1) as previously determined to be present in the colon/colorectal cancer tissue [5–7,12–19]. The following RefSeq transcripts were used: NM_001128425.1 (MUTYH), NM_002528.7 (NTHL1), NM_001369786.1 (KRAS) and NM_006218.4 (PIK3CA).
Statistical analysis
For each signature profile, we compared the biallelic cases with the corresponding CRCs or adenomas from the non-hereditary group. Statistical significance between two groups was determined using a two-sided t-test with a p-value<0.05 considered to be statistically significant. For group comparisons, one-way ANOVA was used. Additionally, we determined the Cohen’s d effect size to measure the difference between the means of two subgroups.
Source code
All data analysis was performed using Python v.3.11 [20], Numpy v.1.24 [21] and Scikit-Learn v.1.3 [22]. Data visualisation was done using the R programming language v.4.3.2 [23] and RStudio v.0.16.0 [24] using the following packages: ggplot2 v.3.5.1 [25], cowplot v.1.1.3 [26] and dplyr v.1.1.4 [27].
RESULTS
The clinicopathological characteristics of the participants and their adenomas and CRCs are shown in Table 1. The biallelic MUTYH and biallelic NTHL1 cases are presented in Supplementary Table 2. Of note, all adenomas and CRCs were MMR-proficient by immunohistochemistry except for two biallelic MUTYH cases; Pat_301 (2xCRCs at 50 years, one MMR-proficient and one MMR-deficient with MLH1/PMS2 loss), and Pat_315 (1xCRC at 39 years with MSH2/MSH6 loss). The SBS mutational signature profiles of each adenoma and CRC included in the study are presented in Figure 1.
Table 1.
The clinicopathological characteristics of the participants and their adenomas and CRCs from each of the biallelic MUTYH cases, biallelic NTHL1 cases and the adenomas and CRCs from the non-hereditary (control) groups included in this study. Overview of the phenotypes by sex, age at diagnosis (including mean and standard deviation), anatomical site, histological type, T stage, grade of tumour and study separated by adenoma and colorectal cancer tissue type and case subgroups.
| MUTYH cases (n=13) | NTHL1 cases (n=5) | Non-hereditary Controls (n=46) | Total (n=64) Individuals | ||||
|---|---|---|---|---|---|---|---|
| Adenoma biallelic MUTYH (n=9, 10.5%) | CRC biallelic MUTYH (n=15, 17.4%) | Adenoma biallelic NTHL1 (n=7, 8.1%) | CRC biallelic NTHL1 (n=2, 2.3%) | MMR-proficient Adenomas (n=27, 31.4%) | MMR-proficient CRCs (n=26, 30.2%) | Total (n=86, 100%) Tissues | |
| Sex, n (%) | |||||||
| Male | 5 (55.6%) | 11 (73.3%) | 3 (42.9%) | 0 (0.0%) | 12 (44.4%) | 12 (46.2%) | 43 (50.0%) |
| Female | 4 (44.4%) | 4 (26.7%) | 4 (57.1%) | 2 (100.0%) | 15 (55.6%) | 14 (53.8%) | 43 (50.0%) |
| Age at diagnosis, n (%) | |||||||
| Mean ± SD | 52.3 ± 14.4 | 52.2 ± 11.1 | 57.7 ± 5.7 | 68.5 ± 10.6 | 43.7 ± 10.6 | 42.8 ± 9.1 | 48.0 ± 12.1 |
| Min. - Max. | 33 – 73 | 33 – 64 | 51 – 66 | 61 – 76 | 27 – 61 | 21 – 59 | 21 – 76 |
| ≤50 years | 2 (22.2%) | 6 (40.0%) | 0 (0.0%) | 0 (0.0%) | 19 (70.4%) | 22 (84.6%) | 49 (57.0%) |
| >50 years | 7 (77.8%) | 9 (60.0%) | 7 (100.0%) | 2 (100.0%) | 8 (29.6%) | 4 (15.4%) | 37 (43.0%) |
| Ethnicity, n (%) | |||||||
| European | 9 (100.0%) | 15 (100.0%) | 7 (100.0%) | 2 (100.0%) | 27 (100.0%) | 24 (92.3%) | 84 (97.7%) |
| East Asian | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (3.8%) | 1 (1.2%) |
| South Asian | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (3.8%) | 1 (1.2%) |
| Anatomical site, n (%) | |||||||
| Proximal colon | 3 (33.3%) | 12 (80.0%) | 2 (28.6%) | 2 (100.0%) | 9 (33.3%) | 12 (46.2%) | 40 (46.5%) |
| Distal colon | 0 (0.0%) | 1 (6.7%) | 0 (0.0%) | 0 (0.0%) | 6 (22.2%) | 8 (30.8%) | 15 (17.4%) |
| Rectum | 0 (0.0%) | 2 (13.3%) | 1 (14.3%) | 0 (0.0%) | 6 (22.2%) | 6 (23.1%) | 15 (17.4%) |
| Unknown | 6 (66.7%) | 0 (0.0%) | 4 (57.1%) | 0 (0.0%) | 6 (22.2%) | 0 (0.0%) | 16 (18.6%) |
| Colorectal Adenoma Histological Type, n (%) | |||||||
| Tubular adenoma | 2 (22.2%) | - | 5 (71.4%) | - | 14 (51.9%) | - | - |
| Tubulovillous adenoma | 4 (44.4%) | - | 2 (28.6%) | - | 13 (48.1%) | - | - |
| Unknown | 3 (33.3%) | - | 0 (0.0%) | - | 0 (0.0%) | - | - |
| CRC Histological Type, n (%) | |||||||
| Adenocarcinoma | - | 15 (100.0%) | - | 2 (100.0%) | - | 24 (92.3%) | - |
| Mucinous adenocarcinoma | - | 0 (0.0%) | - | 0 (0.0%) | - | 0 (0.0%) | - |
| Signet ring adenocarcinoma | - | 0 (0.0%) | - | 0 (0.0%) | - | 1 (3.8%) | - |
| Undifferentiated (incl. medullary) | - | 0 (0.0%) | - | 0 (0.0%) | - | 1 (3.8%) | - |
| Grade of CRC, n (%) | |||||||
| Well differentiated | - | 1 (6.7%) | - | 0 (0.0%) | - | 3 (11.5%) | - |
| Moderately differentiated | - | 13 (86.7%) | - | 2 (100.0%) | - | 16 (61.5%) | - |
| Poorly differentiated | - | 0 (0.0%) | - | 0 (0.0%) | - | 4 (15.4%) | - |
| Unknown | - | 1 (6.7%) | - | 0 (0.0%) | - | 3 (11.5%) | - |
| Study, n (%) | |||||||
| ANGELS | 0 (0.0%) | 0 (0.0%) | 4 (57.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 4 (4.7%) |
| CCFR | 6 (66.7%) | 14 (93.3%) | 0 (0.0%) | 1 (50.0%) | 27 (100.0%) | 27 (103.8%) | 74 (86.0%) |
| GCPS | 3 (33.3%) | 1 (6.7%) | 3 (42.9%) | 1 (50.0%) | 0 (0.0%) | 0 (0.0%) | 8 (9.3%) |
Figure 1:
Mutational signatures observed across the cohort.
Abbreviations: MMR, DNA mismatch repair; SBS, single base substitution; CRC,colorectal cancer.
The SBS18+SBS36 mutational signature is elevated in both adenomas and CRCs from biallelic MUTYH cases
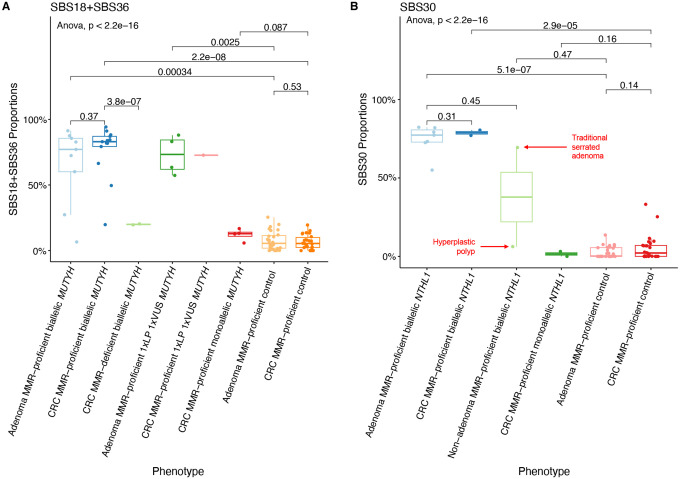
The mean (±standard deviation) proportion of SBS18+SBS36 in the adenomas (65.6%±29.6%) and MMR-proficient CRCs (76.2%±20.5%) from biallelic MUTYH cases were not significantly different (p-value=0.37) (Figure 2A, Table 2). This result is further highlighted when comparing the SBS18+SB36 proportions in adenomas and CRCs from the same participant (Figure 3). In contrast, the mean proportion of SBS18+SBS36 in adenomas and CRCs from biallelic MUTYH cases were significantly higher compared with the mean proportion in non-hereditary adenomas (65.6%±29.6% versus 7.6%±7.0%, p-value=3.4×10−4) and CRCs (76.2%±20.5% versus 6.5%±5.5%, p-value=2.2×10−8) (Figure 2A, Table 3).
Figure 2:
Boxplots of whole-exome sequencing derived genomic features for A) SBS18+SBS36 proportions in MUTYH cases and non-hereditary groups and B) SBS30 proportions in NTHL1 cases and non-hereditary groups.
Abbreviations: MMR, DNA mismatch repair; SBS, single base substitution; CRC,colorectal cancer; VUS, variant of uncertain significance; LP, likely pathogenic.
Table 2.
The mean, standard deviation, and range of five genomic features derived from whole-exome sequencing testing for their differences between tissue type and by MUTYH or NTHL1 case or non-hereditary status. Statistically significant p-values are highlighted in bold.
| Biallelic MUTYH cases | Biallelic NTHL1 cases | Proficient Controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adenom a biallelic MUTYH (n=9, 10.7%) | CRC biallelic MUTYH (n=13, 15.5%)1 | t-test (p-value)2 | Effect Size (Cohen’s d) | Adenoma biallelic NTHL1 (n=7, 8.3%) | CRC biallelic NTHL1 (n=2, 2.4%) | t-test (p-value)2 | Effect Size (Cohen’s d) | Adenoma proficient control (n=27, 32.1%) | CRC proficient control (n=26, 31.0%) | t-test (p-value)2 | Effect Size (Cohen’s d) | Total (n=84, 100%) | |
| SBS18+SBS36 | 0.37 | −0.4 | 0.30 | 0.5 | 0.53 | 0.2 | |||||||
| Mean | 65.6% | 76.2% | 1.0% | 0.4% | 7.6% | 6.5% | 23.4% | ||||||
| SD | 29.6% | 20.5% | 1.4% | 0.0% | 7.0% | 5.5% | 32.0% | ||||||
| Range | 6.6% – 91.3% | 19.8% – 94.2% | 0% – 3.2% | 0.4% – 0.4% | 0% – 25.4% | 0% – 19.5% | 0% – 94.2% | ||||||
| SBS30 | 0.38 | 0.4 | 0.31 | −0.5 | 0.14 | −0.4 | |||||||
| Mean | 6.0% | 2.0% | 74.5% | 78.8% | 2.8% | 5.4% | 11.6% | ||||||
| SD | 12.3% | 6.0% | 9.4% | 2.4% | 3.6% | 8.0% | 23.4% | ||||||
| Range | 0% – 32.4% | 0% – 21.4% | 55% – 82.3% | 77.1% – 80.5% | 0% – 13.6% | 0% – 33.2% | 0% – 82.3% | ||||||
| TMB | 0.12 | −0.8 | 0.87 | −0.1 | 7.4×10 −5 | −1.2 | |||||||
| Mean | 4.8 | 7.5 | 7.1 | 7.5 | 1.5 | 2.8 | 3.8 | ||||||
| SD | 4.3 | 2.9 | 4.2 | 2.0 | 0.9 | 1.3 | 3.3 | ||||||
| Range | 0.3 – 12.7 | 3.5 – 13.3 | 2.5 – 14.8 | 6.1 – 8.8 | 0.3 – 4.1 | 1.4 – 5.9 | 0.3 – 14.8 | ||||||
| INDEL count | 0.02 | −1.1 | 0.95 | 0.0 | 2.3×10 −3 | −0.9 | |||||||
| Mean | 4.4 | 9.7 | 10.7 | 10.5 | 7.1 | 11.3 | 8.9 | ||||||
| SD | 4.9 | 4.6 | 9.3 | 0.7 | 4.2 | 5.4 | 5.6 | ||||||
| Range | 1 – 17 | 2 – 16 | 2 – 25 | 10 – 11 | 1 – 17 | 3 – 20 | 1 – 25 | ||||||
| SNV count | 0.13 | −0.7 | 0.87 | −0.1 | 9.8×10 −5 | −1.2 | |||||||
| Mean | 318.6 | 494.3 | 466.9 | 491.0 | 91.1 | 178.1 | 245.6 | ||||||
| SD | 285.4 | 197.5 | 274.5 | 132.9 | 56.8 | 86.9 | 217.9 | ||||||
| Range | 19 – 851 | 231 – 895 | 168 – 974 | 397 – 585 | 16 – 262 | 84 – 381 | 16 – 974 | ||||||
Abbreviations: CRC, colorectal cancer; SBS, single base substitution; SD, standard deviation; TMB, tumour mutation burden; INDEL, large insertion/deletion; SNV, single nucleotide variant.
MMR-proficient CRCs only from biallelic MUTYH cases were included.
two-tailed t-test.
Figure 3.
Line plot displaying the comparison of SBS18+SBS36 signature proportions for adenomas and colorectal cancers related to each biallelic MUTYH case and for the participant with a pathogenic and variant of uncertain significance in MUTYH (Pat_763).
Abbreviations: SBS, single base substitution; CRC, colorectal cancer; VUS, variant of uncertain significance; LP, likely pathogenic; ID, identification; Pat, patient ID; Rel, relative ID.
Table 3.
The mean, standard deviation, and range of five genomic features derived from whole-exome sequencing testing assessed for their differences between MUTYH or NTHL1 case or non-hereditary status for colorectal adenomas and colorectal cancers separately. Statistically significant p-values are highlighted in bold.
| Colorectal Adenomas | Colorectal Cancers | Colorectal Adenomas | Colorectal Cancers | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biallelic MUTYH cases (n=9, 10.7 %) | Non-hereditary controls (n=27, 32.1%) | t-test (p-value)2 | Effect Size (Cohen's d) | Biallelic MUTYH cases (n=13 15.5 %)1 | Non-hereditary controls (n=26, 31.0%) | t-test (p-value)2 | Effect Size (Cohen's d) | Biallelic NTHL1 cases (n=7, 8.3%) | Non-hereditary controls (n=27, 32.1%) | t-test (p-value)2 | Effect Size (Cohen's d) | Biallelic NTHL1 cases (n=2, 2.4%) | Non-hereditary controls (n=26, 31.0%) | t-test (p-value)2 | Effect Size (Cohen's d) | Total (n=8 4, 100 %) | |
| SBS18+SBS36 | 3.4×10 −4 | 3.7 | 2.2×10 −8 | 5.6 | 8.0×10 −5 | −1.0 | 6.4×10 −6 | −1.1 | |||||||||
| Mean | 65.6% | 7.6% | 76.2% | 6.5% | 1.0% | 7.6% | 0.4% | 6.5% | 23.4% | ||||||||
| SD | 29.6% | 7.0% | 20.5% | 5.5% | 1.4% | 7.0% | 0.0% | 5.5% | 32.0% | ||||||||
| Range | 6.6% – 91.3% | 0% – 25.4% | 19.8 % – 94.2% | 0% – 19.5% | 0% – 3.2% | 0% – 25.4% | 0.4% – 0.4% | 0% – 19.5% | 0% – 94.2% | ||||||||
| SBS30 | 0.45 | 0.5 | 0.15 | −0.5 | 5.1×10 −7 | 13.8 | 2.9×10 −5 | 9.3 | |||||||||
| Mean | 6.0% | 2.8% | 2.0% | 5.4% | 74.5% | 2.8% | 78.8% | 5.4% | 11.6% | ||||||||
| SD | 12.3% | 3.6% | 6.0% | 8.0% | 9.4% | 3.6% | 2.4% | 8.0% | 23.4% | ||||||||
| Range | 0% – 32.4% | 0% – 13.6% | 0% – 21.4% | 0% – 33.2% | 55% – 82.3% | 0% – 13.6% | 77.1 % – 80.5% | 0% – 33.2% | 0% – 82.3% | ||||||||
| TMB | 4.7×10 −2 | 1.5 | 7.5×10 −5 | 2.3 | 1.2×10 −2 | 2.8 | 0.18 | 3.4 | |||||||||
| Mean | 4.8 | 1.5 | 7.5 | 2.8 | 7.1 | 1.5 | 7.5 | 2.8 | 3.8 | ||||||||
| SD | 4.3 | 0.9 | 2.9 | 1.3 | 4.2 | 0.9 | 2.0 | 1.3 | 3.3 | ||||||||
| Range | 0.3 – 12.7 | 0.3 – 4.1 | 3.5 – 13.3 | 1.4 – 5.9 | 2.5 – 14.8 | 0.3 – 4.1 | 6.1 – 8.8 | 1.4 – 5.9 | 0.3 – 14.8 | ||||||||
| INDEL count | 0.18 | −0.6 | 0.33 | −0.3 | 0.35 | 0.7 | 0.48 | −0.2 | |||||||||
| Mean | 4.4 | 7.1 | 9.7 | 11.3 | 10.7 | 7.1 | 10.5 | 11.3 | 8.9 | ||||||||
| SD | 4.9 | 4.2 | 4.6 | 5.4 | 9.3 | 4.2 | 0.7 | 5.4 | 5.6 | ||||||||
| Range | 1 – 17 | 1 – 17 | 2 – 16 | 3 – 20 | 2 – 25 | 1 – 17 | 10 – 11 | 3 – 20 | 1 – 25 | ||||||||
| SNV count | 4.4×10 −2 | 1.5 | 6.9×10 −5 | 2.4 | 1.1×10 −2 | 2.9 | 0.18 | 3.5 | |||||||||
| Mean | 318.6 | 91.1 | 494.3 | 178.1 | 466.9 | 91.1 | 491.0 | 178.1 | 245.6 | ||||||||
| SD | 285.4 | 56.8 | 197.5 | 86.9 | 274.5 | 56.8 | 132.9 | 86.9 | 217.9 | ||||||||
| Range | 19 – 851 | 16 – 262 | 231 – 895 | 84 – 381 | 168 – 974 | 16 – 262 | 397 – 585 | 84 – 381 | 16 – 974 | ||||||||
Abbreviations: CRC, colorectal cancer; SBS, single base substitution; SD, standard deviation; TMB, tumour mutation burden; INDEL, large insertion/deletion; SNV, single nucleotide variant.
MMR-proficient CRCs only from biallelic MUTYH cases were included
Two-tailed t-test
Co-occurrence of mutational processes related to defective MUTYH and defective DNA mismatch repair
In the two MMR-deficient CRCs from biallelic MUTYH cases (Pat_301 and Pat_315), the mean proportion of SBS18+SBS36, was significantly lower compared with the MMR-proficient CRCs from biallelic MUTYH cases (20.0%±0.5% versus 76.2%±20.5%, p-value=3.8×10−7) but they were still higher compared with non-hereditary CRCs (6.5%±5.5%, p-value=2.2×10−8) (Figure 2A, Table 3). Both these MMR-deficient CRCs also showed higher proportions of SBS15 and SBS44, which are mutational signatures associated with MMR-deficiency (Figure 1). In addition, the TMB of these two MMR-deficient CRCs (53.9 and 25.4 mutations/Mb, respectively) was higher compared with the mean TMB of the MMR-proficient CRCs from biallelic MUTYH cases (7.5±2.9 mutations/Mb) (Figure 4).
The cause of MMR-deficiency in Pat_301 and Pat_315 was not related to carrying a germline LP/P variant in one of the DNA MMR genes, but rather from two somatic MMR mutations causing biallelic inactivation in each CRC as determined from the WES data. The CRC showing loss of MLH1/PMS2 protein expression from Pat_301 had two somatic mutations in MLH1 (c.1813G>T p.(Glu605Ter) and c.1816G>T p.(Gly606Ter)) and no evidence of tumour MLH1 promoter hypermethylation. The CRC showing loss of MSH2/MSH6 protein expression from Pat_315 had a somatic mutation in MSH2 (c.394G>T p.(Glu132Ter)) and loss of heterozygosity indicating loss of the wildtype MSH2 allele. The somatic single nucleotide mutations observed in MLH1 and MSH2 matched the mutational contexts associated with SBS18 and SBS36 (Pat_301:TCT>TAT and TCC>TAC; Pat_315:TCA>TTA), suggesting the constitutionally defective MUTYH contributed to these somatic MMR mutational events and resulted in MMR-deficiency in these two CRCs. Interestingly, the synchronous MMR-proficient CRC from Pat_301 exhibited a high proportion of SBS18+SBS36 (94.2%) and a low TMB (9.4 mutations/Mb), further highlighting the impact of tumour MMR-deficiency on the SBS18+SBS36 signature proportions in biallelic MUTYH cases.
Somatic mutations as biomarkers of biallelic MUTYH status in adenomas
Previously, the KRAS c.34G>T p.(Gly12Cys) and PIK3CA c.1636C>A p.(Gln546Lys) somatic mutations were shown to be recurrent mutations significantly increased in CRCs from biallelic MUTYH pathogenic variant cases [6]. In adenomas, the KRAS c.34G>T mutation was present in 6/9 (66.7%) and 2/27 (7.4%) of the biallelic MUTYH and non-hereditary adenomas, respectively (p-value=3.1×10−2). The KRAS c.34G>T mutation had a positive predictive value of 75% and a negative predictive value of 89.3% in adenomas compared with a positive predictive value of 100% and negative predictive value of 86.7% in CRCs, indicating that the somatic KRAS mutation may not be as clinically useful in identifying biallelic MUTYH cases in adenomas as it is in CRCs. The PIK3CA c.1636C>A mutation was not observed in adenomas from biallelic MUTYH cases or in adenomas from the non-hereditary group (Supplementary Figure 1).
The SBS18+SBS36 mutational signature provides evidence for variant classification
We profiled four adenomas and a CRC from Pat_763 who carried a germline heterozygous pathogenic variant (c.1187G>A p.(Gly396Asp)) and a germline heterozygous VUS (c.533G>C p.(Gly178Ala)) in MUTYH. All four adenomas (mean proportion: 73.0%±14.9%, range: 57.3%-88.0%) and the CRC (72.7%) demonstrated high proportions of SBS18+SBS36 consistent with germline biallelic inactivation of MUTYH (Figure 2A, Figure 3). No somatic second hits in MUTYH were observed that may have accounted for the high SBS18+SBS36 signature proportions in the adenomas and CRC. These findings support a reclassification of the MUTYH c.533G>C p.(Gly178Ala) variant as likely pathogenic.
The SBS30 mutational signature is elevated in both adenomas and CRCs from biallelic NTHL1 cases
The mean proportion of SBS30 in adenomas (74.5%±9.4%) and CRCs (78.8%±2.4%) from biallelic NTHL1 cases were not significantly different (p-value=0.31) (Figure 2B, Table 2). The mean proportion of SBS30 in adenomas from biallelic NTHL1 cases was, however, significantly higher compared with the mean proportion in non-hereditary adenomas (74.5%±9.4% versus 2.8%±1.3%; p-value=5.1×10−7) (Figure 2B, Table 3). In addition to 7 adenomas and 2 CRCs, a hyperplastic polyp and a traditional serrated adenoma from two biallelic NTHL1 cases (Pat_005 and Pat_469) were tested. Of note, the traditional serrated adenoma showed high proportion of SBS30 at 69.4%, whereas the SBS30 proportion in the hyperplastic polyp was only 6.2% (Figure 2B).
DISCUSSION
In this study, we showed that the SBS18+SBS36 and SBS30 mutational signatures associated with biallelic MUTYH and biallelic NTHL1 deficiencies, were present in adenomas at similar proportions to those observed in CRCs and were significantly higher when compared with the proportions observed in non-hereditary adenomas and CRCs. Together, these results demonstrate the presence of these mutational processes and consequent mutational signatures, at diagnostic levels in the pre-malignant stage, thereby enabling the opportunity for early CRC detection by expanding the potential tissue available for profiling.
We identified two scenarios where SBS30 or SBS18+SBS36 may present with limitations. Firstly, although SBS30 was shown to be a predominant mutational signature in adenomas from biallelic NTHL1 cases, our results showed variable presence of SBS30 in two serrated polyp subtypes, 69.4% in the traditional serrated adenoma and only 6.2% in the hyperplastic polyp. As biallelic NTHL1 cases can present with mixed polyp types [28], further research is needed to determine the utility of testing serrated polyps for mutational signatures for NTHL1 and more broadly for other hereditary CRC/polyposis syndromes. Secondly, we tested two MMR-deficient CRCs from two biallelic MUTYH cases where the mutational signature profile showed defective MMR related to the presence of SBS15 and SBS44 and a hypermutated TMB that co-occurred with the SBS18+SBS36 signature, albeit at lower proportions than observed in MMR-proficient CRCs from biallelic MUTYH cases. These findings highlight MMR-deficiency as an important diagnostic caveat for utilising SBS18+SBS36 to identify biallelic MUTYH cases or for classifying variants.
This study extends on our previous work for applying SBS18+SBS36 in CRCs to reclassify VUSs in MUTYH [5]. We showed high levels of SBS18+SBS36 in the CRC and multiple adenomas from the same person provides high confidence that the MUTYH c.533G>C p.(Gly178Ala) variant is pathogenic. Additional evidence related to its absence in gnomAD and from in-silico predictions from REVEL, SIFT, PolyPhen-2 and Align-GVGD suggest this missense change affects protein function, further supporting pathogenicity (https://www.ncbi.nlm.nih.gov/clinvar/variation/481808/, last accessed date: August 1st, 2024). The ability to test multiple independent adenomas/CRCs provides high confidence for variant classification where all or none of the lesions have the signature. The clinical genetics community is increasingly challenged by VUS, where around half (47.8%, 1329/2782) of MUTYH variants in ClinVar are currently classified as VUS (stand: August 6th, 2024) [29]. Approaches to classify variants with existing and widely used infrastructure i.e., next generation sequencing and validated bioinformatic tools, will aid in reclassifying variants and optimising clinical management and cancer prevention for the patient and their relatives.
Limitations of this study include the lack of ethnic diversity within the case and non-hereditary groups which were predominantly white European. Similarly, there was a limited range of germline LP/P variants for both MUTYH and NTHL1. The consistency of mutational signature findings across a broader group of cases of different pathogenic variants and ethnic backgrounds would provide evidence of the robustness of this approach. All of the CRCs and adenomas tested in this study were from FFPE tissue, however we have previously shown that mutational signature profiling is effective in both FFPE and fresh frozen tissue DNA samples [5].
CONCLUSIONS
This study provides important findings demonstrating that testing adenomas for SBS18+SBS36 or SBS30 can be an equally effective alternative to identifying biallelic MUTYH or biallelic NTHL1 cases, respectively, if CRC has not yet developed or tissue is not available. This provides important opportunities for clinical management decision-making such as colectomy versus endoscopic polypectomy for CRC prevention given the established high CRC penetrance in biallelic cases. Furthermore, the specificity of these signatures enables the utility of mutational signature profiling to classify VUS. Our study identified potential caveats to using mutational signatures diagnostically, namely, the presence of MMR-deficiency which may diminish the SBS18+SBS36 signature, while for SBS30, testing of serrated polyps needs further investigation. This study adds to the growing evidence of the clinical utility of gene specific mutational signature profiling for identifying hereditary CRC/polyposis syndromes and further expands the opportunities to utilise mutational signatures as a supportive feature for variant classification.
AVAILABLILITY OF DATA AND MATERIALS
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Colorectal Oncogenomics Group and members from the Genomic Medicine and Family Cancer Clinic for their support of this manuscript. We thank the participants and staff from the ANGELS study, GCPS study, and from the Australasian and Ontario sites of the CCFR, in particular Maggie Angelakos, Samantha Fox, Cary Greenberg and Allyson Templeton for their support of this manuscript. The CCFR graciously thanks the generous contributions of their study participants, dedication of study staff, and the financial support from the U.S. National Cancer Institute, without which this important registry would not exist. The authors thank Melbourne Bioinformatics and the Australian Genome Research Facility for their collaboration on this project.
FUNDING
Funding by a National Health and Medical Research Council of Australia (NHMRC) Investigator grant GNT1194896 awarded to DDB and funding by a Cure Cancer Early Career Research Grant awarded to PG supported the design, analysis, and interpretation of data. DDB is supported by a University of Melbourne Dame Kate Campbell Fellowship. PG is supported by an NHMRC Investigator Grant (2026331). RW is supported by the University of Melbourne Early Career Researcher Grant. BJP is supported by a Victorian Health and Medical Research Fellowship from the Victorian Government. AKW is supported by an NHMRC Investigator grant (GNT1194392). JLH is supported by the University of Melbourne Dame Kate Campbell Fellowship. MAJ is supported by an NHMRC Investigator grant (GNT1195099).
The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). Support for case ascertainment was provided in part from the Surveillance, Epidemiology, and End Results (SEER) Program and the following U.S. state cancer registries: AZ, CO, MN, NC, NH; and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada). The content of this manuscript does not necessarily reflect the views or policies of the NIH or any of the collaborating centres in the CCFR, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government, any cancer registry, or the CCFR.
DECLARATION OF INTEREST STATEMENT
Robert C. Grant received a scholarship from Pfizer and provided consulting or advisory roles for Astrazeneca, Tempus, Eisai, Incyte, Knight Therapeutics, Guardant Health, and Ipsen. All other authors have no relevant financial or non-financial interests to disclose.
Funding Statement
Funding by a National Health and Medical Research Council of Australia (NHMRC) Investigator grant GNT1194896 awarded to DDB and funding by a Cure Cancer Early Career Research Grant awarded to PG supported the design, analysis, and interpretation of data. DDB is supported by a University of Melbourne Dame Kate Campbell Fellowship. PG is supported by an NHMRC Investigator Grant (2026331). RW is supported by the University of Melbourne Early Career Researcher Grant. BJP is supported by a Victorian Health and Medical Research Fellowship from the Victorian Government. AKW is supported by an NHMRC Investigator grant (GNT1194392). JLH is supported by the University of Melbourne Dame Kate Campbell Fellowship. MAJ is supported by an NHMRC Investigator grant (GNT1195099).
The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). Support for case ascertainment was provided in part from the Surveillance, Epidemiology, and End Results (SEER) Program and the following U.S. state cancer registries: AZ, CO, MN, NC, NH; and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada). The content of this manuscript does not necessarily reflect the views or policies of the NIH or any of the collaborating centres in the CCFR, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government, any cancer registry, or the CCFR.
REFERENCES
- [1].Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet 2002;30:227–32. 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- [2].Weren RDA, Ligtenberg MJL, Kets CM, de Voer RM, Verwiel ETP, Spruijt L, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet 2015;47:668–71. 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- [3].Win AK, Reece JC, Dowty JG, Buchanan DD, Clendenning M, Rosty C, et al. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. Int J Cancer 2016;139:1557–63. 10.1002/ijc.30197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Grolleman JE, Díaz-Gay M, Franch-Expósito S, Castellví-Bel S, de Voer RM. Somatic mutational signatures in polyposis and colorectal cancer. Mol Aspects Med 2019;69:62–72. 10.1016/j.mam.2019.05.002. [DOI] [PubMed] [Google Scholar]
- [5].Georgeson P, Pope BJ, Rosty C, Clendenning M, Mahmood K, Joo JE, et al. Evaluating the utility of tumour mutational signatures for identifying hereditary colorectal cancer and polyposis syndrome carriers. Gut 2021;70:2138–49. 10.1136/gutjnl-2019-320462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Georgeson P, Harrison TA, Pope BJ, Zaidi SH, Qu C, Steinfelder RS, et al. Identifying colorectal cancer caused by biallelic MUTYH pathogenic variants using tumor mutational signatures. Nat Commun 2022;13:3254. 10.1038/s41467-022-30916-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Grolleman JE, de Voer RM, Elsayed FA, Nielsen M, Weren RDA, Palles C, et al. Mutational Signature Analysis Reveals NTHL1 Deficiency to Cause a Multi-tumor Phenotype. Cancer Cell 2019;35:256–266.e5. 10.1016/j.ccell.2018.12.011. [DOI] [PubMed] [Google Scholar]
- [8].Seifert BA, McGlaughon JL, Jackson SA, Ritter DI, Roberts ME, Schmidt RJ, et al. Determining the clinical validity of hereditary colorectal cancer and polyposis susceptibility genes using the Clinical Genome Resource Clinical Validity Framework. Genet Med Off J Am Coll Med Genet 2019;21:1507–16. 10.1038/s41436-018-0373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 2012;28:1811–7. 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- [10].Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang X, Wojtowicz D, Przytycka TM. Detecting presence of mutational signatures in cancer with confidence. Bioinformatics 2018;34:330–7. 10.1093/bioinformatics/btx604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Crisafulli G, Sartore-Bianchi A, Lazzari L, Pietrantonio F, Amatu A, Macagno M, et al. Temozolomide Treatment Alters Mismatch Repair and Boosts Mutational Burden in Tumor and Blood of Colorectal Cancer Patients. Cancer Discov 2022;12:1656–75. 10.1158/2159-8290.CD-21-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941–7. 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Viel A, Bruselles A, Meccia E, Fornasarig M, Quaia M, Canzonieri V, et al. A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer. EBioMedicine 2017;20:39–49. 10.1016/j.ebiom.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Everall A, Tapinos A, Hawari A, Cornish A, Sud A, Chubb D, et al. Comprehensive repertoire of the chromosomal alteration and mutational signatures across 16 cancer types from 10,983 cancer patients 2023:2023.06.07.23290970. 10.1101/2023.06.07.23290970. [DOI] [PubMed] [Google Scholar]
- [17].Gurjao C, Zhong R, Haruki K, Li YY, Spurr LF, Lee-Six H, et al. Discovery and Features of an Alkylating Signature in Colorectal Cancer. Cancer Discov 2021;11:2446–55. 10.1158/2159-8290.CD-20-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020;580:269–73. 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walker R, Mahmood K, Joo JE, Clendenning M, Georgeson P, Como J, et al. A tumor focused approach to resolving the etiology of DNA mismatch repair deficient tumors classified as suspected Lynch syndrome. J Transl Med 2023;21:282. 10.1186/s12967-023-04143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Van Rossum G, Drake FL. Python 3 Reference Manual. Scotts Valley, CA: CreateSpace; 2009. [Google Scholar]
- [21].Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, et al. Array programming with NumPy. Nature 2020;585:357–62. 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- [23].R Core Team. R: a language and environment for statistical computing 2020. https://www.R-project.org/ (accessed October 5, 2022).
- [24].Allaire JJ. RStudio: Integrated Development Environment for R n.d.
- [25].Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York; 2016. [Google Scholar]
- [26].Wilke CO. cowplot: Streamlined Plot Theme and Plot Annotations for “ggplot2.” R package version 113 2024.
- [27].Wickham H, François R, Henry L, Müller K, Vaughan D, Software P, et al. dplyr: A Grammar of Data Manipulation 2023.
- [28].Weren RD, Ligtenberg MJ, Geurts van Kessel A, De Voer RM, Hoogerbrugge N, Kuiper RP. NTHL1 and MUTYH polyposis syndromes: two sides of the same coin? J Pathol 2018;244:135–42. 10.1002/path.5002. [DOI] [PubMed] [Google Scholar]
- [29].Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–7. 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.