Abstract
Background
Tuberculous meningitis (TBM) is difficult to diagnose. We investigated whether a 3-gene host response signature in blood can distinguish TBM from other brain infections.
Methods
The expression of 3 genes (dual specificity phosphatase 3 [DUSP3], guanylate-binding protein [GBP5], krupple-like factor 2 [KLF2]) was analyzed by RNA sequencing of archived whole blood from 4 cohorts of Vietnamese adults: 281 with TBM, 279 with pulmonary tuberculosis, 50 with other brain infections, and 30 healthy controls. Tuberculosis scores (combined 3-gene expression) were calculated following published methodology and discriminatory performance compared using area under a receiver operator characteristic curve (AUC).
Results
GBP5 was upregulated in TBM compared to other brain infections (P < .001), with no difference in DUSP3 and KLF2 expression. The diagnostic performance of GBP5 alone (AUC, 0.74; 95% confidence interval [CI], .67–.81) was slightly better than the 3-gene tuberculosis score (AUC, 0.66; 95% CI, .58–.73) in TBM. Both GBP5 expression and tuberculosis score were higher in participants with human immunodeficiency virus (HIV; P < .001), with good diagnostic performance of GBP5 alone (AUC, 0.86; 95% CI, .80–.93).
Conclusions
The 3-gene host signature in whole blood has the ability to discriminate TBM from other brain infections, including in individuals with HIV. Validation in large prospective diagnostic study is now required.
Keywords: 3-gene host response, tuberculous meningitis, diagnosis, HIV coinfection, brain infections
Host immune signatures to diagnose tuberculosis show promise, but have not been investigated in tuberculous meningitis. We demonstrate the ability of a whole-blood 3-gene host signature to discriminate tuberculous meningitis from other brain infections, including individuals with HIV.
An estimated 10.6 million people fell ill with tuberculosis in 2021 [1], an increase of 4.5% from 2020. True estimates of tuberculous meningitis (TBM), the most devastating form of tuberculosis, are unknown. Population-based estimates suggest that up to 1% (approximately 100 000) of total tuberculosis cases are TBM [2]. TBM has a mortality rate of 20%–50%, and neurodisability occurs in up to 50% of survivors [3, 4]. Poor patient outcomes are largely due to delayed diagnosis and treatment initiation [5]. Diagnosis is notoriously challenging, especially in settings where TBM is common [6]. Currently available diagnostic tests often fail to differentiate TBM from other brain infections, and the paucibacillary nature of TBM limits the utility of pathogen-based testing [7–9]. Mycobacterial culture, although considered the gold standard, is of restricted clinical value as results are too slow to impact clinical decision making. Xpert MTB/RIF Ultra is rapid, but has an inadequate negative predictive value to exclude TBM and is not widely available in tuberculosis endemic settings [10].
Poor outcomes from TBM are associated with an aberrant host inflammatory response to Mycobacterium tuberculosis [11]. Common inflammatory complications of TBM are characterized by neutrophil and inflammatory-mediated responses detected in whole blood, despite TBM compartmentalization to the brain [12, 13]. This signature has been detected weeks before the eventual onset of inflammatory neurological complications, suggesting that it might be clinically useful in predicting outcomes and defining therapeutic responses [14].
Whether whole-blood inflammatory signatures might aid in the diagnosis of TBM has not been investigated. Whole-blood analysis has major advantages over cerebrospinal fluid (CSF) collection, which can be contraindicated (eg, risk of cerebral herniation) or technically difficult. Conventional tuberculosis diagnostics for TBM are pathogen directed and all have considerable limitations, including variable sensitivities, high costs, and requirement for laboratory and technical expertise [11]. A simple and accurate whole-blood test for TBM would have major clinical advantages.
Host transcriptomic signatures have been developed as a novel diagnostic and/or triage test in pulmonary tuberculosis (PTB) [15, 16]. Guanylate-binding protein (GBP5) and dual specificity phosphatase 3 (DUSP3) mRNA in blood are differentially expressed whilst krupple-like factor 2 (KLF2) is similarly expressed in active PTB compared to other respiratory infections [15]. Both GBP5 and DUSP3 are upregulated in tuberculosis, whilst KLF2 is an anti-inflammatory gene and is downregulated in tuberculosis [17]. A combination score, known as the “tuberculosis score,” calculated from the gene expression of GBP5, DUSP3, and KLF2, has been developed to discriminate between PTB and other respiratory diseases with a diagnostic performance which met World Health Organization target product profile [15]. This 3-gene signature has been incorporated into an automated quantitative polymerase chain reaction (qPCR) test (Xpert mycobacterium host response [MTB-HR]; Cepheid), which uses a whole-blood sample taken via fingerprick, making it an attractive option as a point-of-care test.
The ability of host transcriptomic signatures to diagnose TBM has not been investigated. We therefore conducted an evaluation of the 3-gene host response to discriminate TBM from other brain infections. The secondary aim was to characterize the pattern of 3-gene host responses in TBM in comparison to PTB.
METHODS
Participant Cohorts
The study was conducted at the Hospital for Tropical Diseases and Pham Ngoc Thach Hospital in Ho Chi Minh City, Vietnam. Study participants with TBM or other brain infections were recruited from these 2 hospitals. Those with PTB were recruited from district tuberculosis units in Ho Chi Minh City. Healthy controls were office staff working in Ho Chi Minh City who volunteered to participate.
Four cohorts were assembled for the study. Cohort 1 comprised 281 Vietnamese adults with TBM enrolled in 2 previously described randomized controlled trials (LAST ACT and ACT HIV) [18, 19]. Participants had either definite, probable, or possible TBM, as defined by published criteria [20]. Participants were enrolled pretreatment or very early into treatment. Participants in ACT had < 4 days whilst participants in LAST ACT had <7 days of corticosteroids prior to enrolment. Cohort 2 comprised 279 adults with culture-confirmed PTB who had either drug-susceptible tuberculosis or multidrug-resistant tuberculosis as defined by molecular (Xpert MTB/RIF or Hain test) or drug susceptibility testing. All participants in this cohort had <7 days of antituberculosis drugs and were not suspected to have TBM. Cohort 3 comprised 50 adults with microbiologically confirmed brain infections other than TBM (other brain infections group) who had baseline CSF collected as part of standard of care for diagnosis. Standard laboratory testing included Gram stain, bacterial and fungal culture, viral PCR/serology, and antimicrobial susceptibility testing. Cohort 4 comprised 30 adults without signs or symptoms of tuberculosis or history of tuberculosis contact within the last 2 years as healthy controls. They were enrolled in a prospective study comparing epidemiology of tuberculosis household contacts, health care workers, and those with smear-positive PTB.
All the participants were prospectively identified and followed up for the acquisition of clinical and outcome data. Recruited participants were tested for human immunodeficiency virus (HIV) except in the other brain infections group where it was not mandatory and performed only when clinically indicated. The healthy controls were not tested for HIV infection.
Blood Sample Collection and RNA Sequence Preprocessing
Venous blood was collected from all participants at diagnosis in PAXgene Blood RNA tubes (PreAnalytiX) and stored at −80°C. RNA was manually extracted with the PAXgene Blood RNA Kit (Qiagen) according to the manufacturer's instructions at Oxford University Clinical Research Unit's laboratory in Ho Chi Minh City, Vietnam. DNA was digested on columns using the RNase-free DNase Set (Qiagen) before sending the sample to the Ramaciotti Centre for Genomics (Sydney, Australia) for RNA sequencing.
Quality control and alignment of RNA sequencing data were performed using an in-house pipeline derived from previously published RNA sequencing analysis [21, 22]. Gene expression counts were generated from sequencing fastq files. The counts were then entered into DESeq2 functions (DESeq2 package [23]) to generate log2 scale transformations of data, which were normalized for library size or other factors. To remove the batch effect caused by technical differences across batches, ComBat-seq function [24] was employed on transformed data. Data on GBP5, DUSP3, and KLF2 were extracted for the main analysis and expressed as the normalized gene expression in our study. The tuberculosis score was calculated as (GBP5 + DUSP3)/2 − KLF2 as defined in a previous study [25].
Statistical Analysis
Patient characteristics were compared between infection groups. For continuous variables, median and first and third quartiles are presented. For binary and categorical variables, number of cases and percentage are presented. No omnitest was used before pairwise testing. Pairwise testing was decided a priori. We used Mann-Whitney U test to compare gene expression between definite TBM versus other TBM; TBM versus PTB, other brain infection groups, and healthy controls; and bacterial versus viral brain infections. We evaluated the diagnostic performance of gene GBP5 and tuberculosis score in distinguishing all TBM, definite TBM from other brain infections based on the receiver operating characteristic (ROC) curves and the AUC, and stratified these results by HIV status. We examined the impact of sex, HIV status, age, PTB, CD4 count (Supplementary Figure 1), pretreatment variables (Supplementary Table 2), and PTB stratified by HIV status (Supplementary Figure 2) on GBP5 expression and tuberculosis score using the Mann-Whitney U test for 2-group comparisons and a linear regression model for association between 2 continuous variables. Covariates were gender, age, Glasgow coma score (GCS), and CSF characteristics, whilst tuberculosis score and GBP5 were outcomes. Association between HIV and PTB was examined based on a simple logistic regression model in which HIV was the outcome and PTB as covariate. Association between GBP5 and tuberculosis score with bacterial load was examined using hurdle Gaussian modeling (Supplementary Figure 3). All statistical analyses were conducted using R version 4.2.2 and we used the P value threshold of ≤ .05 as statistical significance.
Ethics
All participants, or their relatives if they were incapacitated, gave their written informed consent to take part in the study. The studies from which the cohorts were constructed had approval from the institutional review board at the Hospital for Tropical Diseases and Pham Ngoc Thach Hospital in Ho Chi Minh City, the ethics committee of the Ministry of Health, Vietnam, and the Oxford Tropical Research Ethics Committee, United Kingdom.
RESULTS
Cohort Characteristics
In the TBM cohort of 281 adults (> 18 years of age), the median age was 41 years (interquartile range [IQR], 31–54 years), 183 (65%) were male, and 74 (26%) were people with HIV (Table 1). Using the published uniform case definition for TBM clinical research [18], 134 (48%), 98 (35%), and 49 (17%) had definite, probable, and possible TBM, respectively. Of those with definite TBM, 91 of 134 (68%) were culture positive (mycobacterial growth indicator tube), 81 (60%) were Xpert or Xpert Ultra positive, and 83 (62%) were Ziehl-Neelson stain positive. The PTB cohort consisted of 296 adults with a median age of 44 years (IQR, 31–52 years). Of the 50 adults with other brain infections, 33 (66%) were male with a median age of 42 years (IQR, 30–60 years).
Table 1.
Baseline Characteristics in TBM, Definite TBM, PTB, Other Brain Infections, and Healthy Controls
| Characteristic | TBM (n = 281) | Definite TBM (n = 134) | PTB (n = 296) | Other Brain Infections (n = 50) | Healthy Controls (n = 30) |
|---|---|---|---|---|---|
| Age, y, median (IQR) | 41 (31–54) | 36 (29–47) | 44 (31–52) | 42 (30–60) | 33 (29–37) |
| Male, No. (%) | 183 (65) | 91 (68) | 228 (77) | 33 (66) | 11 (37) |
| HIV, No. (%) | |||||
| Negative | 207 (74) | 82 (61) | 295 (100) | 37 (100) | … |
| Positive | 74 (26) | 52 (39) | 0 (0) | 0 (0) | … |
| Unknown | 0 | 0 | 1 | 13 | … |
| Glasgow Coma Score, median (IQR) | 14 (12–15) | 14 (12–15) | … | 11 (9–13) | … |
| TBM diagnostic category, No. (%) | |||||
| Definite TBM | 134 (48) | 134 (100) | … | … | … |
| Probable TBM | 98 (35) | … | … | … | … |
| Possible TBM | 49 (17) | … | … | … | … |
| Microbiological tests, No. (%) | |||||
| MGIT culture positive | 91 (32) | 91 (68) | … | … | … |
| Xpert/Ultra positive | 81 (28.8) | 81 (60) | … | … | … |
| Microscopy positive | 83 (30) | 83 (62) | … | … | … |
| CSF parameters, median (IQR) | |||||
| CSF leukocyte count, × 103 cells/mL | 139 (17–350) | 206 (109–405) | … | 466 (114–2956) | … |
| CSF lymphocytes, % | 89 (70–100) | 78 (46–89) | … | 49 (10–86) | … |
| CSF neutrophils, % | 0 (0–23) | 15 (0–53) | … | 50 (14–90) | … |
| CSF protein level, g/L | 1.65 (0.97–2.41) | 1.88 (1.30–2.54) | … | 1.19 (0.67–3.91) | … |
| CSF glucose level, mmol/L | 2.57 (1.70–3.50) | 1.97 (1.30–2.67) | … | 2.96 (0.74–4.17) | … |
| CSF lactate level, mmol/L | 5.0 (3.8–7.2) | 5.6 (3.9–7.6) | … | 6.9 (3.0–14.2) | … |
| Blood parameters | |||||
| Blood leukocyte count, × 106 cells/mL | 8.4 (6.3–11.1) | 7.7 (6.1–10.5) | 9.2 (7.4–11.4) | 12.4 (8.8–17.8) | 6.4 (5.6–7.2) |
| Blood lymphocytes, % | 13 (8–20) | 11 (7–17) | 20 (16–26) | 7 (5–13) | 34 (32–38) |
| Blood monocytes, % | 7.0 (5.0–9.2) | 6.9 (5.0–9.0) | 8.7 (7.2–10.4) | … | 6.3 (5.3–7.9) |
| Blood neutrophils, % | 77 (67–84) | 80 (72–86) | 67 (60–74) | 85 (78–90) | 57 (52–60) |
| Blood eosinophils, % | 0.5 (0.1–2.0) | 0.3 (0.1–1.0) | 1.9 (1.1–3.4) | 0.2 (0.0–0.5) | 2.9 (1.3–3.6) |
| Blood platelets, × 103/μL | 289 (221–357) | 290 (222–354) | 378 (318–482) | … | 297 (259–321) |
Abbreviations: CSF, cerebrospinal fluid; HIV, human immunodeficiency; IQR, interquartile range; MGIT, mycobacterial growth indicator tube; PTB, pulmonary tuberculosis; TBM, tuberculous meningitis.
The other brain infections cohort had microbiologically confirmed bacterial meningitis (n = 25) or viral meningoencephalitis (n = 25). The bacterial species included Streptococcus sp (n = 21), Escherichia coli (n = 3), and Listeria monocytogenes (n = 1), with 1 participant having coinfection with Escherichia coli and Streptococcus sp. Viral meningoencephalitis was caused by herpes simplex virus (n = 11), varicella zoster (n = 6), Japanese encephalitis virus (n = 5), and dengue virus (n = 5). Two participants had immunoglobulin M (IgM) in CSF for both Japanese encephalitis and dengue viruses. Overall, the cohort had a low GCS with a median of 11 (IQR, 9–13) compared to the TBM group (median, 14; IQR, 12–15) at presentation (P < .001) (Table 1).
Three-gene Expression: All TBM Compared to Definite TBM
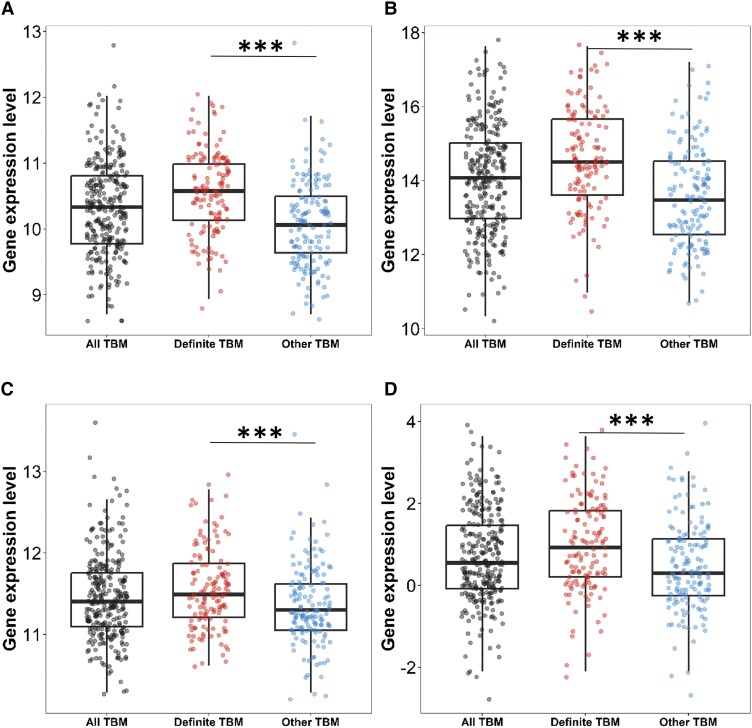
As no perfect reference standard for TBM exists, the 3-gene expression of the microbiological reference standard for TBM (definite TBM) was compared against other TBM (probable and possible TBM). When comparing definite TBM against other TBM, expression of all 3 genes (DUSP3, GBP5, and KLF2) were lower in the other TBM group P < .001 (Supplementary Table 1). Tuberculosis scores were higher in the definite TBM group. The clinical reference standard (definite, probable, and possible) for TBM did not deviate too much from the definite TBM (Figure 1).
Figure 1.
Three-gene expression and tuberculosis score in all tuberculous meningitis (TBM), definite TBM, and other TBM. All TBM comprises of definite, probable, and possible whilst other TBM comprises of probable and possible. A, DUSP3, (B) GBP5, (C) KLF2, and (D) tuberculosis score. Comparisons of gene expression in all TBM (n = 281, black dots), definite TBM, and other TBM group (n = 147, blue dots). Boxes indicate interquartile range, the line indicates the median, and dots indicate data in individuals. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Comparisons between definite TBM versus other TBM were performed using Mann-Whitney U test. ***P < .001.
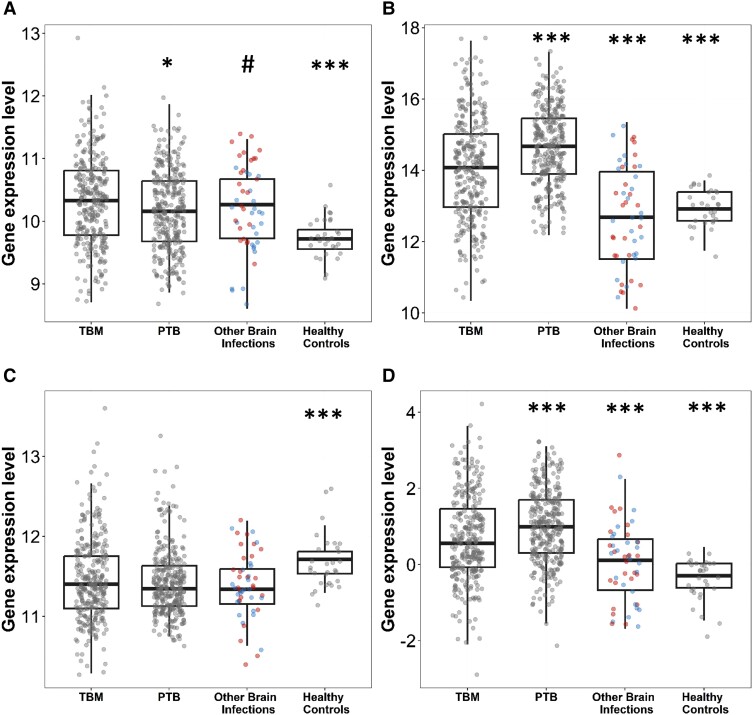
Three-gene Expression: All TBM Compared to the Other Cohorts
The mRNA expression of DUSP3, GBP5, and KLF2 was measured in the 4 cohorts and the combined tuberculosis score calculated for each (Figure 2). KLF2 was downregulated in TBM compared to healthy controls (P < .001), whilst the proinflammatory genes DUSP3 and GBP5 were upregulated (P < .001). When compared to the PTB group, DUSP3 expression in TBM was slightly higher (P = .013). However, GBP5 was significantly lower (P < .001), and KLF2 was comparable. GBP5 was the only gene that was significantly upregulated in TBM compared to other brain infections (P < .001). The tuberculosis score for TBM was significantly higher compared to the other brain infections group (P < .001) (Supplementary Table 1).
Figure 2.
Three-gene expression and tuberculosis score in tuberculous meningitis (TBM) compared to other groups: (A) DUSP3, (B) GBP5, (C) KLF2, and (D) tuberculosis score. Comparisons of gene expression in TBM (n = 281) and other groups: pulmonary tuberculosis (PTB; n = 296), other brain infections (n = 50), and healthy controls (n = 30). Other brain infections were classified as bacterial (n = 25, red dots) or viral infection (n = 25, blue dots). Boxes indicate interquartile range, the line indicates the median, and dots indicate data in individuals. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Comparisons between TBM versus PTB, TBM versus other brain infections, TBM versus healthy control, and bacterial infection versus viral infection were performed using Mann-Whitney U test. Unadjusted multiple testing was performed for exploratory analysis with *P < .05, ***P < .001. Difference in DUSP3 expression between bacterial versus viral infections, #P = .008.
When gene expression was analyzed according to bacterial or viral etiology in those with other brain infections, we found no difference in gene expression with the exception of DUSP3, where there was higher expression in bacterial meningitis (median, 10.58; IQR, 10.09–10.97) compared to viral meningoencephalitis (median, 9.97; IQR, 9.67–10.35; P = .008) (Figure 2).
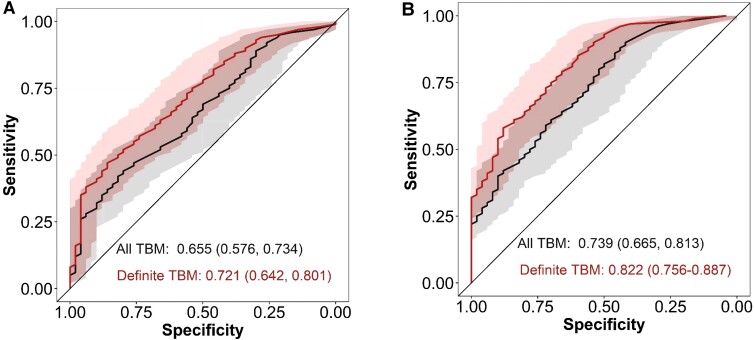
Diagnostic Performance
The diagnostic performance of the tuberculosis score and the most prominently expressed gene, GBP5, was assessed using both the clinical and definite TBM reference standards, as there is no single gold-standard diagnostic test for TBM. The diagnostic performance of GBP5 expression was slightly better compared to the diagnostic performance of the tuberculosis score in participants with all TBM (tuberculosis score AUC, 0.66; 95% confidence interval [CI], .58–.73 and GBP5 AUC, 0.74; 95% CI, .67–.81) and definite TBM (tuberculosis score AUC, 0.72; 95% CI, .64–.80 and GBP5 AUC, 0.82; 95% CI, .76–.89) (Figure 3).
Figure 3.
Diagnostic performance of tuberculosis score and GBP5 in tuberculous meningitis (TBM) and definite TBM. Receiver operating characteristics (ROC) curves for distinguishing all TBM (n = 281) and definite TBM (n = 134) against other central nervous system infections (n = 50) using 3-gene tuberculosis scores (A) or GBP5 (B). Shaded areas depict the 95% confidence intervals with values in parentheses.
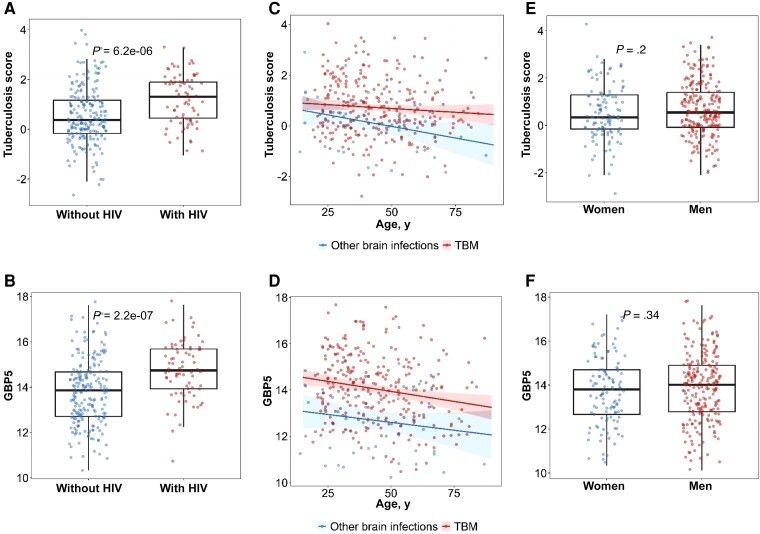
HIV Status, Age, and Gender
There was a significant difference between tuberculosis scores and GBP5 expression between HIV-negative and HIV-positive participants with TBM (tuberculosis score, P < .0001; GBP5, P < .0001; Figure 4A and 4B). Higher GBP5 and tuberculosis score gene expression was associated with HIV coinfection. However, the same association was not detected with CD4 count (median CD4 count = 67 cells/µL; 95% CI, 18.5–123.5; Supplementary Figure 1). In addition, increasing age was associated with lower GBP5 expression in the TBM group (estimate for 5-year age increase, −0.087; 95% CI, −.14 to −.032; P = .002; Figure 4C and 4D). Although the trend was similar, we did not observe evidence of an association between tuberculosis score and age (estimate for 5-year age increase, −0.030; 95% CI, −.072 to .012; P = .161). There was a trend to reduced GBP5 expression and tuberculosis score with increasing age in the other brain infections group; however, only the tuberculosis score reached statistical significance (GBP5 estimate for 5-year age increase, −0.068; 95% CI, −.18 to .040; P = .21 and tuberculosis score estimate for 5-year age increase, −0.091; 95% CI, −.18 to −.0077; P = .03). There was no evidence of a difference in GBP5 expression or tuberculosis score between women and men (Figure 4E and 4F).
Figure 4.
Association of human immunodeficiency virus (HIV) status, gender, and age with tuberculosis score and GBP5 gene expression. Each point represents individual data. A, Distribution of tuberculosis score expression between people with HIV (n = 207) and people without HIV (n = 74) in tuberculous meningitis (TBM) patients. B, Distribution of GPB5 expression between people with and without HIV. C, Regression line and shaded 95% confident interval band modeling linear trend between tuberculosis score and age. The red line corresponds to TBM patients (n = 281) and the blue line corresponds to other brain infections (n = 50). D, Regression line and shaded 95% confident interval band modeling linear trend between GBP5 and age. E, Distribution of tuberculosis score between women (n = 115) and men (n = 216). Boxes indicate interquartile range and the line indicates the median. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Comparisons were performed using Mann-Whitney U test. F, Distribution of GBP5 expression between women and men.
Diagnostic Performance Stratified by HIV Status
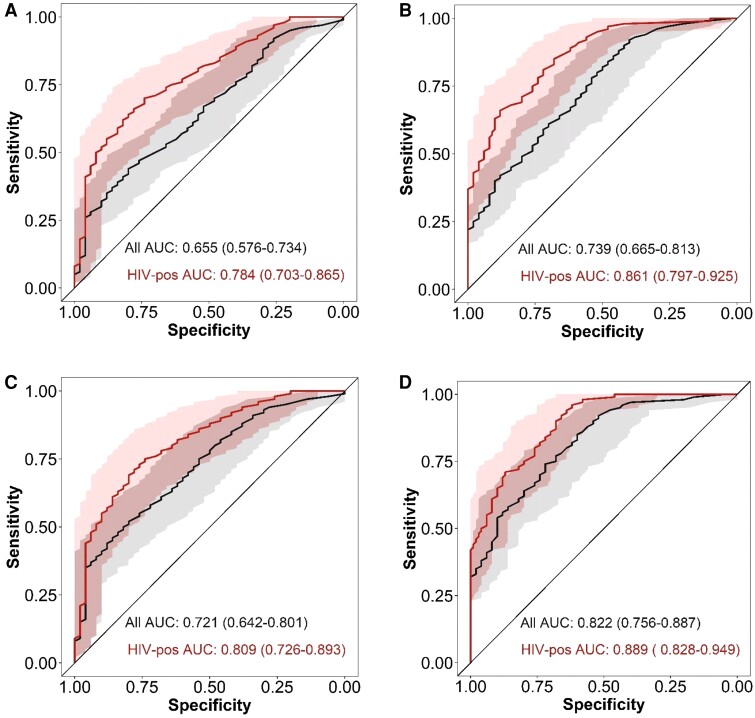
Given HIV status influenced GBP5 expression and tuberculosis score in participants with TBM, we stratified their diagnostic performance by HIV status. The performance of the tuberculosis score for diagnosing TBM was higher in participants with HIV (AUC, 0.78; 95% CI, .70–.87) compared to the combined group of participants with and without HIV (AUC, 0.66; 95% CI, .58–.73) (Figure 5). Similarly, GBP5 expression had a better diagnostic performance in the group of participants with HIV (AUC, 0.86; 95% CI, .80–.93) compared to all participants (AUC, 0.74; 95% CI, .67–.81).
Figure 5.
Diagnostic performance of tuberculosis score and GBP5 stratified by HIV status. ROC curves for distinguishing TBM and other brain infections using tuberculosis scores (A) or GBP5 (B). ROC curves for distinguishing definite TBM and other brain infections using tuberculosis scores (C) or GBP5 (D). Black lines are all participants in each comparison, red lines are participants with HIV only. Shaded areas depict the 95% confidence interval with values in parenthese. Abbreviations: AUC, area under the curve; HIV, human immunodeficiency virus; ROC, receiver operating characteristic; TBM, tuberculous meningitis.
Comparison Between TBM and TBM With Pulmonary Tuberculosis
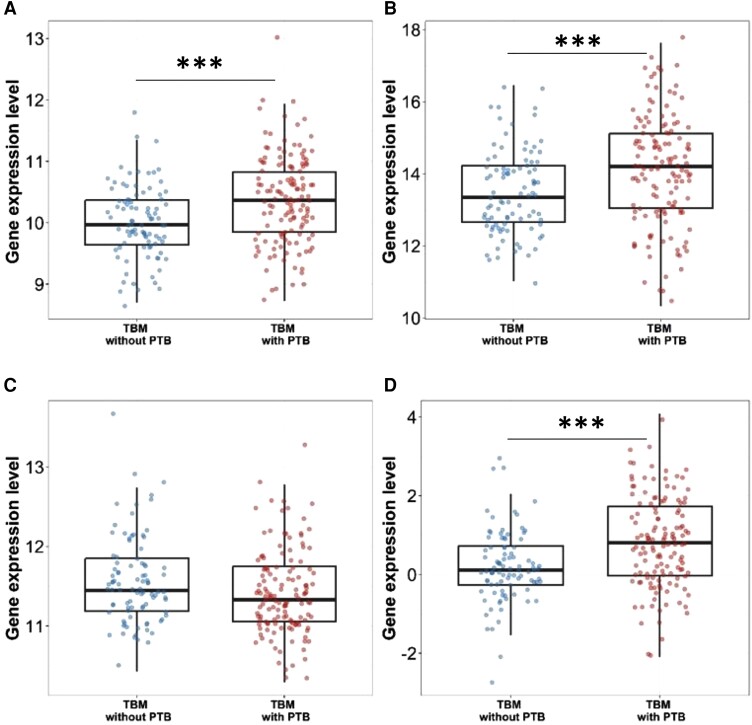
Given PTB was present in 59% (134 of 229) of participants with TBM in this study, we compared gene expression between TBM versus TBM with PTB. As no respiratory samples were collected from participants, PTB was defined as chest radiograph consistent with tuberculosis excluding miliary tuberculosis. Both DUSP3 and GBP5 expression were significantly upregulated in TBM-infected participants with PTB compared to those without (DUSP3 10.37 [IQR, 9.85–10.83] vs 9.97 [IQR, 9.64–10.37], P < .001 and GBP5 14.21 [IQR, 13.05–15.12] vs 13.35 [IQR, 12.66–14.23], P < .001). KLF2 expression was similar (P = .052) and the tuberculosis score was higher (P < .001) (Figure 6).
Figure 6.
Gene expression and tuberculosis score in tuberculous meningitis (TBM) and TBM with pulmonary tuberculosis (PTB): (A) DUSP 3, (B) GBP5, (C) KLF2, and (D) tuberculosis score. Comparisons of gene expression in TBM without PTB (n = 92, blue dots) and TBM with PTB (n = 134, red dots). Boxes indicate interquartile range, horizontal lines the median, and dots indicate individual patient data. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Comparisons were performed using Mann-Whitney U test. ***P < .001.
Gene Expression in TBM and TBM With Pulmonary Tuberculosis Stratified by HIV Status
Given gene expression was influenced by both HIV status and PTB, we explored for an association between HIV and PTB. TBM with PTB was significantly associated with HIV positive status (odds ratio, 3.79; 95% CI, 1.80–8.75; P < .001). To dissect whether this association influenced GBP5 and tuberculosis score we made comparisons of these in TBM and TBM with PTB and stratified by HIV status. GBP5 was upregulated and tuberculosis score higher in participants with HIV compared to those without regardless of whether PTB was present (Supplementary Figure 2).
Correlation With Pretreatment Variables
Associations between gender, age, GCS, and CSF characteristics with tuberculosis score and GBP5 expression were investigated. There was no association found between gene expression and tuberculosis score with GCS, CSF protein, and CSF lactate. A doubling in CSF lymphocyte count was associated with a lower tuberculosis score (P < .001) and GBP5 gene expression (P = .029) in the all-TBM group whilst a doubling of CSF neutrophil count was associated with an increase in GBP5 expression (P = .032). No associations were found in the definite TBM group (Supplementary Table 2). Whilst tuberculosis score and GBP5 were associated with the probability of M. tuberculosis detection there was no association between gene expression and mycobacterial loads (Supplementary Figure 3).
DISCUSSION
To the best of our knowledge, this is the first study to investigate whether whole-blood host 3-gene expression can be used to discriminate TBM from other brain infections and the first to explore differences in 3-gene host response between 2 tuberculosis phenotypes, TBM and PTB. The main findings were that there was differential gene expression between TBM and other brain infections. These differences were summarized by the tuberculosis scores but were particularly marked for GBP5. Our data suggest that either the tuberculosis score, or potentially GBP5 expression alone, could be a useful diagnostic tool for TBM. Secondly, gene expression profiles differed with higher DUSP3 expression and lower GBP5 expression in TBM compared to PTB, whilst KLF2 was similarly expressed. The pattern of 3-gene host response was similar when compared to healthy controls, although it differed in magnitude of expression. This suggests TBM and PTB share similar characteristics of the immune response.
The performance of the tuberculosis score to diagnose definite TBM had a higher AUC than in all TBM. GBP5 performed a little better with an AUC of 0.82 (95% CI, .76–.89) in definite TBM and 0.74 (95% CI, .67–.81) in all TBM, suggesting that a single gene (GBP5) instead of the 3-gene may be sufficiently discriminative. The diagnostic performance of GBP5 and the tuberculosis score in all TBM using the clinical reference standard was lower than in definite TBM. This is not unexpected as the clinical reference standard of TBM consisted of about 50% unconfirmed (probable and possible) cases, some of which may not have had the disease. We believe, however, that in the absence of a perfect reference standard, the clinical reference standard of TBM is a truer reflection of real-world medicine and thus more relevant for practicing clinicians.
Importantly, we found that the performance of tuberculosis score and GBP5 was influenced by HIV status and age, but not sex. In our study, HIV coinfection improved diagnostic performance in TBM; however, we are careful to generalize this to all patients with HIV as many in the other brain infections group had unknown HIV status. This is an important finding as HIV coinfection is universally associated with significantly higher mortality and morbidity [4]. A simple, accurate diagnostic tool to identify TBM therefore has the potential to improve clinical outcomes in this population. We have previously shown that the CSF cytokine concentrations in adults with HIV and TBM are higher than in those without HIV, suggesting a hyperinflammatory phenotype associated with poor outcomes [26]. A higher CSF mycobacterial load in HIV-positive adults with TBM was also positively correlated with a hyperinflammatory phenotype [27]. It is therefore plausible that expression of the proinflammatory GBP5 is more prominent in HIV-positive adults with TBM. Age also influenced GBP5, with reduced expression occurring with increasing age. The influence of age on 3-gene expression requires further investigation.
The 3-gene host response has been characterized in PTB. GBP5 was most prominently expressed followed by KLF2 and DUSP3 [15]. In our study, DUSP3 was more highly expressed and GBP5 less expressed when TBM was compared to PTB. KLF2 expression was similar between both tuberculosis phenotypes. In our TBM cohort, HIV coinfection influenced 3-gene expression whilst it did not in PTB [15]. This was evident even though our study suggested that concomitant PTB in those with TBM behaved as a mediator of the 3-gene host response. This suggests that regulation of DUSP3, GBP5, and KLF2 is different in TBM compared to PTB in HIV coinfection. GBP5 results in activation of proinflammatory responses through induction of COX-2 [28]. DUSP3 is an upregulator of innate immune responses and has an essential role in macrophage polarization and inflammation, whereas KLF2 inhibits monocyte proinflammatory gene expression. In contrast to GBP5 and DUSP3, KLF2 is considered anti-inflammatory and has been term a housekeeping gene due to its very stable expression [29, 30]. The role of GBP5, DUSP3, and KLF2 in TBM is unknown.
The limitations of this study are its single geographical setting, retrospective testing of archived blood samples, and the lack of a replication analysis. TBM diagnosis with the 3-gene host response would not give any information on drug resistance. Further investigation in other populations with high burden of TBM (eg, children) and a prospective evaluation of control groups that include other subacute meningitides (eg, cryptococcal and parasitic meningitis) are required. Whilst differing host genetics in various populations may impact 3-gene diagnostic performance, there was no evidence of this when tested in Southeast Asian and African populations with PTB [15].
A strength of our study is that the RNA sequencing data generated will allow us to explore other gene sets or combinations of genes as potential diagnostic signatures. We demonstrated proof-of-concept that using whole-blood RNA sequencing the 3-gene host response could be used to discriminate TBM from other brain infections. We also showed that GBP5 is the most discriminatory of the 3-gene host response and should be further explored. Whilst the sensitivity and specificity of GBP5 alone is not adequate to rule in or out TBM, it may be helpful when combined with pathogen-based diagnostic tools or CSF parameters in a diagnostic prediction model. We believe our findings are an important step to the discovery of a new diagnostic test for TBM that does not rely on CSF testing and therefore may improve the early diagnosis of a disease that continues to kill or disable nearly half of those infected.
CONCLUSION
The 3-gene host response has the ability to discriminate TBM from other brain infections. GBP5 expression is the most discriminatory, and may provide comparable or better TBM diagnostic performance than the tuberculosis score. HIV coinfection appears to improve the diagnostic performance of GBP5 and the tuberculosis score, which is an important finding given TBM is more common and more lethal in those with HIV. Large, prospective studies further defining the diagnostic utility of the 3-gene test are needed.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Julie Huynh, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, Oxford University, Oxford, United Kingdom.
Le Hoang Thanh Nhat, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Nguyen Le Hoai Bao, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Hoang Thanh Hai, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Do Dang Anh Thu, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Trinh Thi Bich Tram, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Vu Thi Mong Dung, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Do Dinh Vinh, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Nghiem My Ngoc, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Joseph Donovan, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, Oxford University, Oxford, United Kingdom.
Nguyen Hoan Phu, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; School of Medicine, Vietnam National University of Ho Chi Minh City, Ho Chi Minh City, Vietnam.
Dang Van Thanh, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Nguyen Thi Anh Thu, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Nguyen Duc Bang, Pham Ngoc Thach Hospital for Tuberculosis and Lung Disease, Ho Chi Minh City, Vietnam.
Dang Thi Minh Ha, Pham Ngoc Thach Hospital for Tuberculosis and Lung Disease, Ho Chi Minh City, Vietnam.
Ho Dang Trung Nghia, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Pham Ngoc Thach University of Medicine, Ho Chi Minh City, Vietnam; The Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam.
Le Van Tan, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, Oxford University, Oxford, United Kingdom.
Le Hong Van, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
Guy Thwaites, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, Oxford University, Oxford, United Kingdom.
Nguyen Thuy Thuong Thuong, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, Oxford University, Oxford, United Kingdom.
Notes
Acknowledgment. We acknowledge Dr Vu Thi Ty Hang and members of the emerging infections group for their help with data collection and performing laboratory diagnosis of the other brain infections group.
Author contributions. T. T. T. N. conceptualized the study, and reviewed and edited the full manuscript. J. H. drafted, reviewed, and edited the full manuscript. H. T. N. L. conceptualized and coordinated the data analysis and statistical methods. L. H. B. N. performed the data and statistical analysis. T. H. H. performed the quality control of RNA data. G. T., J. D., T. N. H. D., H. P. N., V. T. L., M. N. N., B. N., T. M. D., V. T. D., T. A. T. N., and H. V. L. were responsible for enrolling participants into studies from which whole blood was collected. A. T. D. D., T. B. T. T., M. D. V. T., and V. D. V. performed RNA extraction and tuberculosis diagnostics. All authors reviewed and approved the manuscript for submission.
Financial support. This work was supported by a Wellcome Trust Fellowship in Public Health and Tropical Medicine (grant numbers 206724/Z/17/Z to N. T. T. T. and 204904/Z/16/Z to L. V. T.); and a Wellcome Trust Investigator Award to G. T. Oxford University Clinical Research Unit is supported by the Wellcome Trust in the Vietnam Africa Asia Programme. Funding to pay the Open Access publication charges for this article was provided by Wellcome Trust.
References
- 1. World Health Organization . Global tuberculosis report 2022. Geneva, Switzerland: WHO, 2022. [Google Scholar]
- 2. Seddon JA, Tugume L, Solomons R, Prasad K, Bahr NC. Tuberculous meningitis international research consortium. The current global situation for tuberculous meningitis: epidemiology, diagnostics, treatment and outcomes. Wellcome Open Res 2019; 4:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiang S, Khan F, Milstein M, et al. . Treatment outcomes of childhood tuberculous meningitis: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:947–57. [DOI] [PubMed] [Google Scholar]
- 4. Stadelman AM, Ellis J, Samuels THA, et al. . Treatment outcomes in adult tuberculous meningitis: a systematic review and meta-analysis. Open Forum Infect Dis 2020; 7:ofaa257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thwaites GE, Tran TH. Tuberculous meningitis: many questions, too few answers. Lancet Neurol 2005; 4:160–70. [DOI] [PubMed] [Google Scholar]
- 6. Donovan J, Walker TM. Diagnosing tuberculous meningitis: a testing process. Int J Tuberc Lung Dis 2021; 25:605–6. [DOI] [PubMed] [Google Scholar]
- 7. Donovan J, Thu DDA, Phu NH, et al. . Xpert MTB/RIF ultra versus Xpert MTB/RIF for the diagnosis of tuberculous meningitis: a prospective, randomised, diagnostic accuracy study. Lancet Infect Dis 2020; 20:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cresswell FV, Tugume L, Bahr NC, et al. . Xpert MTB/RIF ultra for the diagnosis of HIV-associated tuberculous meningitis: a prospective validation study. Lancet Infect Dis 2020; 20:308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bahr NC, Nuwagira E, Evans EE, et al. . Diagnostic accuracy of Xpert MTB/RIF ultra for tuberculous meningitis in HIV-infected adults: a prospective cohort study. Lancet Infect Dis 2018; 18:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donovan J, Cresswell FV, Thuong NTT, et al. . Xpert MTB/RIF ultra for the diagnosis of tuberculous meningitis: a small step forward. Clin Infect Dis 2020; 71:2002–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huynh J, Donovan J, Phu NH, Nghia HDT, Thuong NTT, Thwaites GE. Tuberculous meningitis: progress and remaining questions. Lancet Neurol 2022; 21:450–64. [DOI] [PubMed] [Google Scholar]
- 12. Marais S, Lai RPJ, Wilkinson KA, Meintjes G, O'Garra A, Wilkinson RJ. Inflammasome activation underlying central nervous system deterioration in HIV-associated tuberculosis. J Infect Dis 2017; 215:677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rohlwink UK, Figaji A, Wilkinson KA, et al. . Tuberculous meningitis in children is characterized by compartmentalized immune responses and neural excitotoxicity. Nat Commun 2019; 10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thuong NTT, Thwaites GE. Treatment-associated inflammatory deterioration in tuberculous meningitis: unpicking the paradox. J Infect Dis 2017; 215:665–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sutherland JS, van der Spuy G, Gindeh A, et al. . Diagnostic accuracy of the cepheid 3-gene host response fingerstick blood test in a prospective, multi-site study: interim results. Clin Infect Dis 2022; 74:2136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warsinske H, Vashisht R, Khatri P. Host-response-based gene signatures for tuberculosis diagnosis: a systematic comparison of 16 signatures. PLoS Med 2019; 16:e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mendelsohn SC, Mbandi SK, Fiore-Gartland A, et al. . Prospective multicentre head-to-head validation of host blood transcriptomic biomarkers for pulmonary tuberculosis by real-time PCR. Commun Med (Lond) 2022; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donovan J, Phu NH, Thao LTP, et al. . Adjunctive dexamethasone for the treatment of HIV-uninfected adults with tuberculous meningitis stratified by leukotriene A4 hydrolase genotype (LAST ACT): study protocol for a randomised double blind placebo controlled non-inferiority trial. Wellcome Open Res 2018; 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donovan J, Phu NH, Mai NTH, et al. . Adjunctive dexamethasone for the treatment of HIV-infected adults with tuberculous meningitis (ACT HIV): study protocol for a randomised controlled trial. Wellcome Open Res 2018; 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marais S, Thwaites G, Schoeman JF, et al. . Tuberculous meningitis: a uniform case definition for use in clinical research. Lancet Infect Dis 2010; 10:803–12. [DOI] [PubMed] [Google Scholar]
- 21. Conesa A, Madrigal P, Tarazona S, et al. . A survey of best practices for RNA-Seq data analysis. Genome Biol 2016; 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yalamanchili HK, Wan YW, Liu Z. Data analysis pipeline for RNA-Seq experiments: from differential expression to cryptic splicing. Curr Protoc Bioinformatics 2017; 59:11.5.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-Seq count data. NAR Genom Bioinform 2020; 2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med 2016; 4:213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thuong NTT, Heemskerk D, Tram TTB, et al. . Leukotriene A4 hydrolase genotype and HIV infection influence intracerebral inflammation and survival from tuberculous meningitis. J Infect Dis 2017; 215:1020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thuong NTT, Vinh DN, Hai HT, et al. . Pretreatment cerebrospinal fluid bacterial load correlates with inflammatory response and predicts neurological events during tuberculous meningitis treatment. J Infect Dis 2019; 219:986–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jian CZ F, Li W, Wan Y, et al. . Inducible GBP5 mediates the antiviral response via interferon-related pathways during influenza A virus infection. J Innate Immun 2016; 9:419–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh P, Dejager L, Amand M, et al. . DUSP3 genetic deletion confers M2-like macrophage-dependent tolerance to septic shock. J Immunol 2015; 194:4951–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das H, Kumar A, Lin Z, et al. . Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci U S A 2006; 103:6653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.