Abstract
Diagnosing liver lesions is crucial for treatment choices and patient outcomes. This study develops an automatic diagnosis system for liver lesions using multiphase enhanced computed tomography (CT). A total of 4039 patients from six data centers are enrolled to develop Liver Lesion Network (LiLNet). LiLNet identifies focal liver lesions, including hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC), metastatic tumors (MET), focal nodular hyperplasia (FNH), hemangioma (HEM), and cysts (CYST). Validated in four external centers and clinically verified in two hospitals, LiLNet achieves an accuracy (ACC) of 94.7% and an area under the curve (AUC) of 97.2% for benign and malignant tumors. For HCC, ICC, and MET, the ACC is 88.7% with an AUC of 95.6%. For FNH, HEM, and CYST, the ACC is 88.6% with an AUC of 95.9%. LiLNet can aid in clinical diagnosis, especially in regions with a shortage of radiologists.
Subject terms: Cancer screening, Cancer imaging, Liver cancer, Machine learning
The distinction of liver lesions is critical for the accurate diagnosis and treatment of liver cancer. Here, the authors develop LiLNet, a deep learning-based system to identify focal liver lesions as well as benign and malignant liver tumours from CT images with high accuracy across multiple patient cohorts.
Introduction
Liver disease poses an ongoing, urgent challenge in the medical field, imperiling the lives and well-being of millions of people worldwide. These conditions, ranging from benign liver cysts to malignant hepatocellular carcinoma, present substantial threats to patients’ health. Notably, liver cancer ranks as the sixth most common cancer globally and the fourth leading cause of cancer-related fatalities1–3. The 5-year survival rate for liver cancer patients remains distressingly low, particularly among advanced cases, at approximately 10%4–6. While some similarities may exist among different liver lesions, their treatment strategies and prognoses differ markedly. Hence, precise classification plays a pivotal role in formulating optimal treatment and management strategies to effectively address the multifaceted challenges of liver diseases.
Liver lesion classification is a complex task that requires the differentiation of various tumor types, including malignant lesions such as hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC), and metastatic tumors (METs) and benign lesions such as focal nodular hyperplasia (FNH), hemangioma (HEM), and cysts. This necessitates both expert knowledge and meticulous analysis of medical imaging data. However, there are artifacts in hepatic images due to uneven intensity information, different shapes, and low contrast between the hepatic parenchyma and the focus7. Moreover, different acquisition protocols, contrast agents, scanner resolutions, and enhancement technologies make high-precision automatic classification of liver lesions more challenging.
In light of these challenges, AI-assisted methods have demonstrated remarkable potential in revolutionizing the precision and efficiency of liver lesion diagnosis. Previous research methods have utilized quantitative radiology8–11 or deep learning models12–14 to extract image features for liver lesion identification. In radiomics, researchers initially extract feature parameters from liver images and then use machine learning models to distinguish liver lesions based on important selected parameters. However, this feature extraction process relies on the subjective expertise and time-consuming efforts of researchers. The extracted features cannot be dynamically optimized as the dataset evolves. With advancements in deep learning algorithms, such as CNNs, AI-aided automatic analysis of liver lesion images has emerged as a possibility. CNNs can automatically learn and extract intricate patterns and features from medical images, enabling automated liver lesion classification with exceptional accuracy. However, previous studies12–16 have primarily concentrated on predicting HCC specifically, overlooking the classification and diagnosis of many other common liver lesions. Although the initial findings of these studies show promise, the clinical applicability of these methods remains uncertain due to low sensitivity, inadequate datasets for training, limited external validation, and insufficiently robust verification. To address these concerns and ensure more reliable and robust results, we collected a dataset of more than 4000 patients from six medical centers to develop our deep learning model. Additionally, we rigorously verified the robustness and generalizability of the model in four independent external validation centers.
Self-supervision, an unsupervised learning approach, has demonstrated significant potential across various domains, including medical applications17–20. Self-supervised learning harnesses pretext tasks to extract valuable supervision information from large, unlabeled medical datasets, enabling the training of neural networks based on this constructed supervision. In addition, self-supervised learning can also be utilized as a type of weak supervision by leveraging auxiliary tasks or data information to guide the training of models. However, most existing methods employ pretext tasks for pretraining models, followed by fine-tuning with weak labels to perform downstream tasks. Limited research has explored the direct use of pretextual tasks as target tasks to assist in classification tasks without separate pretraining.
In this study, we demonstrate the performance of our AI system, LiLNet, in distinguishing six common types of focal liver lesions. We develop the model using data from six centers and assess its generalization through extensive testing on a test set and four external validation centers. We compare LiLNet’s performance with radiologists’ interpretations of contrast-enhanced CT images in a reader study. To address real-world clinical implementation, we deploy LiLNet in two hospitals, integrating it into routine workflows across outpatient, emergency, and inpatient settings. This integration evaluates the system’s performance in various clinical environments, ensuring its robustness and reliability in practical use.
Results
Patient characteristics
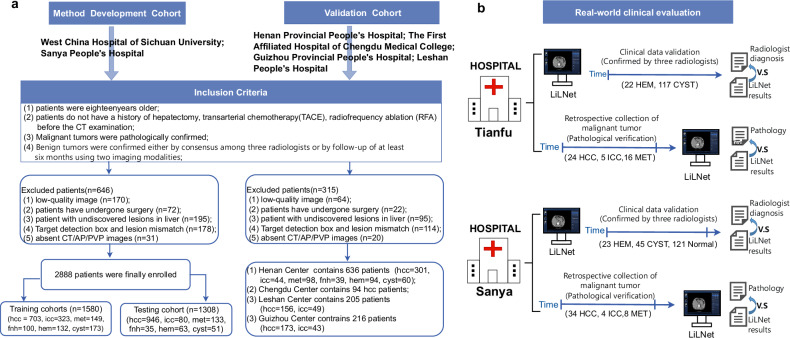
Reporting of the study adhered to the STARD guidelines. Between June 2012 and December 2022, multiphase contrast-enhanced CT images, including arterial phase (AP) and portal venous phase (PVP), were collected from a total of 4039 patients across six hospitals. The method used for retrospective data collection is depicted in Fig. 1a. Furthermore, clinical testing was conducted on two real-world clinical evaluation queues (Fig. 1b): West China Tianfu Center and Sanya People’s Hospital. At Tianfu Center, we examined 184 cases, while at Sanya People’s Hospital, 235 cases were assessed. Patient cohorts comprised an internal training set, internal test set, four external validation sets, and two real-world clinical datasets. Demographic details, including age and sex distributions, varied across cohorts: for instance, the training set showed a female-to-male ratio of 155:548 in HCC cases with an average age of 53.06 years, while the internal test set had 196 females and 750 males for HCC, averaging 52.34 years. Additional specifics can be found in Table 1.
Fig. 1. The Flowchart of the Cohort Setup.
a Patient recruitment process of the training, testing, and external validation cohorts. b The real-world clinical test datasets were obtained from two hospitals. HCC denotes Hepatocellular Carcinoma, ICC denotes Intrahepatic Cholangiocarcinoma, MET denotes Metastatic Cancer, FNH denotes Focal Nodular Hyperplasia, HEM denotes Hemangioma, and CYST denotes Cyst.
Table 1.
Baseline characteristics
| Tumor type | Internal Training (n = 1580) | Internal Test (n = 1308) | Validation HN (n = 636) | Validation CD (n = 94) | Validation GZ (n = 205) | Validation LS (n = 216) | |
|---|---|---|---|---|---|---|---|
|
Age, years (mean, std) |
HCC | 53.06 ± 11.90 | 52.34 ± 12.69 | 55.56 ± 10.34 | 59.79 ± 12.20 | 54.04 ± 11.00 | 57.5 ± 10.88 |
| ICC | 57.16 ± 12.10 | 57.27 ± 12.28 | 59.29 ± 10.38 | — | 59.18 ± 11.70 | 59.92 ± 12.14 | |
| MET | 55.07 ± 14.37 | 56.24 ± 13.44 | 58.61 ± 12.89 | — | — | — | |
| FNH | 35.12 ± 13.49 | 33.65 ± 13.21 | 35.46 ± 15.22 | — | — | — | |
| HEM | 50.13 ± 15.63 | 47.96 ± 11.06 | 50.77 ± 10.71 | — | — | — | |
| CYST | 58.53 ± 12.93 | 56.66 ± 12.36 | 59.13 ± 11.12 | — | — | — | |
|
Sex (Female/ Male) |
HCC | 155/548 | 196/750 | 42/259 | 20/74 | 31/142 | 21/135 |
| ICC | 157/166 | 43/37 | 18/26 | 0/0 | 23/20 | 31/18 | |
| MET | 70/79 | 60/73 | 39/59 | 0/0 | 0/0 | 0/0 | |
| FNH | 59/41 | 19/16 | 20/19 | 0/0 | 0/0 | 0/0 | |
| HEM | 84/48 | 33/30 | 63/31 | 0/0 | 0/0 | 0/0 | |
| CYST | 77/96 | 23/28 | 31/29 | 0/0 | 0/0 | 0/0 |
Note: Std denotes standard deviation, HN denotes Henan Provincial People’s Hospital, CD denotes the First Affiliated Hospital of Chengdu Medical College, GZ denotes Guizhou Provincial People’s Hospital, and LS denotes Leshan People’s Hospital. HCC denotes Hepatocellular Carcinoma, ICC denotes Intrahepatic Cholangiocarcinoma, MET denotes Metastatic Cancer, FNH denotes Focal Nodular Hyperplasia, HEM denotes Hemangioma, and CYST denotes Cyst.
Performance of Lesion Detection
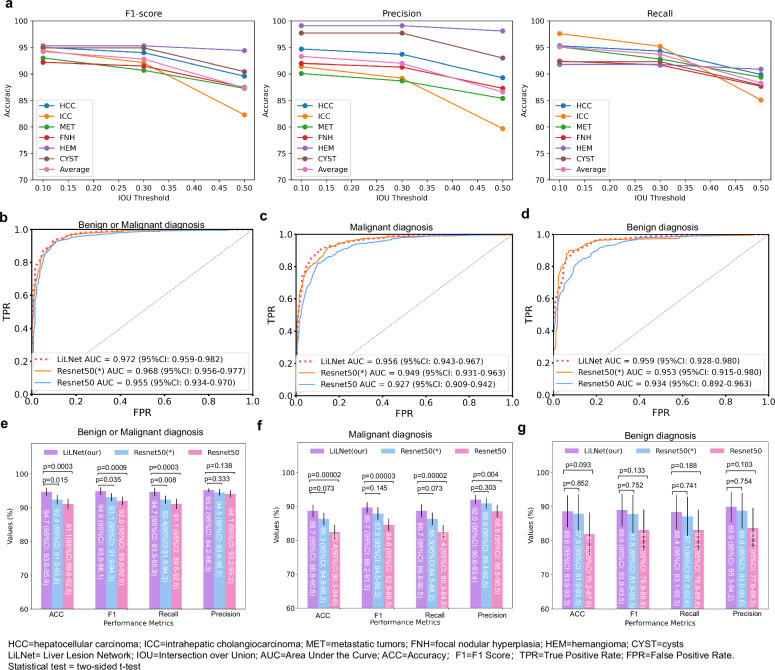
In the lesion detection task, we filtered out bounding boxes with a confidence level above 0.25 and compared them with the actual ground truth boxes. Boxes with an intersection over union (IoU) greater than the threshold are true positives, while those with a lower IoU or fewer repeats are false-positives. Undetected boxes are false-negatives. As shown in Fig. 2a, we analyzed F1, recall, and precision at different IoU thresholds. At an IoU of 0.1, we achieved an F1 of 94.2%, a recall of 95.1%, and a precision of 93.3%. At an IoU of 0.3, the F1 was 92.8%, the recall was 93.7%, and the precision was 91.3%. An IoU of 0.5 yielded an F1 of 87.4%, a recall of 88.3%, and a precision of 86.6%. These results demonstrate the robust performance of our model across different IOU thresholds. In the LiLNet system, we chose an IOU threshold of 0.1. Despite the minimal overlap between the detection box and the true bounding box at this IoU value, the subsequent classification images were extended to a 224 × 224 detection box, ensuring coverage of a portion of the lesion.
Fig. 2. Performance of the proposed model in the testing cohort.
a The outcome of lesion detection at various IOU thresholds. b, c, and d ROC curves for distinguishing benign and malignant tumors, malignant tumors (HCC, ICC, and METs), and benign tumors (FNH, HEM, and cysts), respectively. e ACC, F1, recall, and precision for benign and malignant tumor classification. f ACC, F1, recall, and precision for malignant tumor classification. g The same metrics for benign tumor classification. (*) denotes the use of pretrained parameters from ResNet trained on ImageNet. Source data are provided as a Source Data file Source_data_Figure_2.xlsx.
Performance of LiLNet
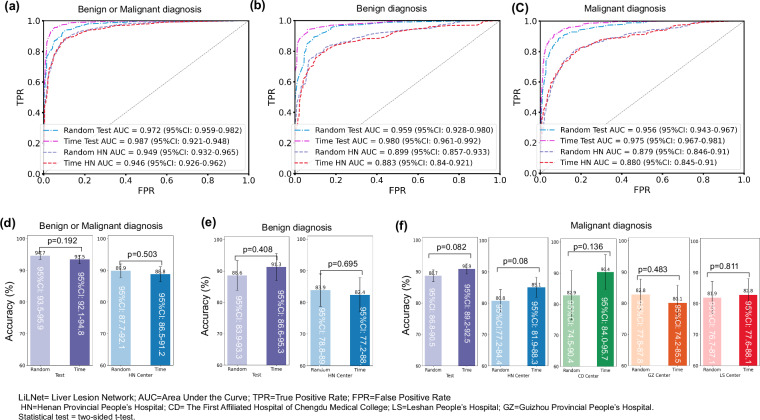
We trained three variations of the LiLNet model on the training set: LiLNet_BM was used to distinguish between benign and malignant liver lesions, LiLNet_M was used to distinguish between three types of malignant liver lesions, and LiLNet_B was used to distinguish between three types of benign liver lesions. On the test set, the LiLNet_BM model achieved the following performance metrics: an AUC of 97.2% (95% CI: 95.9–98.2), an ACC of 94.7% (95% CI: 93.5–95.9), an F1 of 94.9% (95% CI: 93.8–96.1), a recall of 94.7% (95% CI: 93.5-95.9), and a precision of 95.2% (95% CI: 94.2–96.3) (Fig. 2b, e). The LiLNet_M model achieved an AUC of 95.6% (95% CI: 94.3–96.7), an ACC of 88.7% (95% CI: 86.8-90.5), an F1 of 89.7% (95% CI: 88.2-91.3), a recall of 88.7% (95% CI: 86.8–90.5), and a precision of 92.0% (95% CI: 90.6–93.4) (Fig. 2c, f). Finally, the LiLNet_B model achieved an AUC of 95.9% (95% CI: 92.8–98.0), an ACC of 88.6% (95% CI: 83.9–93.3), an F1 of 89.0% (95% CI: 83.9–93.5), a recall of 88.4% (95% CI: 83.1–93.3), and a precision of 89.9% (95% CI: 85.3-94.2) (Fig. 2d, g). As a reference, we also constructed two benchmark models: a naive ResNet50 model and a ResNet50 model loaded with pretraining parameters. Upon a comprehensive analysis of the AUC, the LiLNet model exhibited a 1–2% improvement compared to the two baseline models, demonstrating enhanced discriminative capabilities between positive and negative samples.
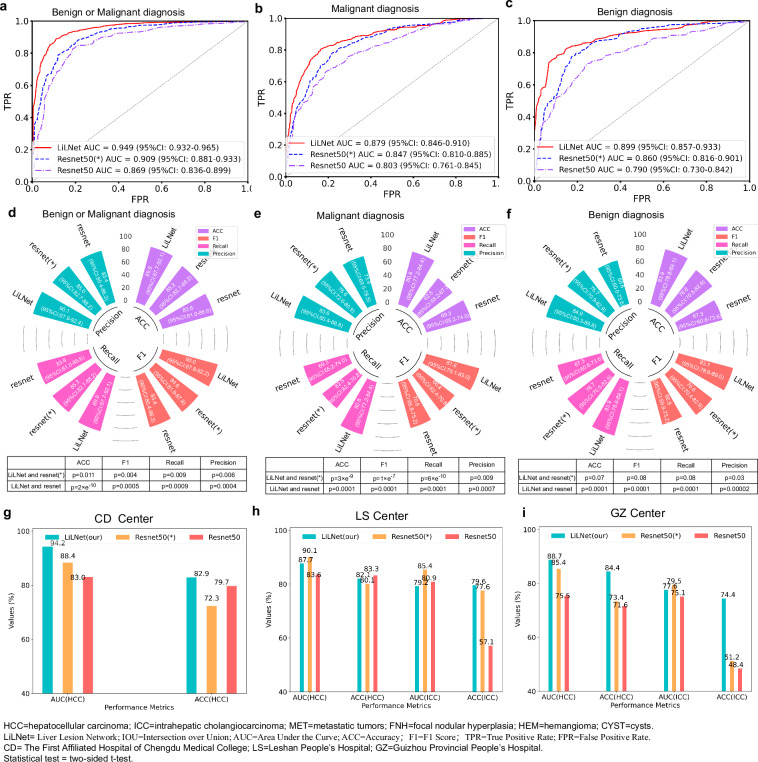
We evaluated our model’s performance using 1151 patients from four different centers. In the Henan Provincial People’s Hospital (HN Center), our model achieved an AUC of 94.9% (95% CI:93.2–96.5) for distinguishing benign and malignant tumors, with an 89.9% (95% CI:87.7–92.1) ACC, a 90.0% (95% CI:87.8–92.2) F1, an 89.9% (95%CI: 87.7–92.1) recall, and a 90.1% (95%CI: 87.9–92.4) precision (Fig. 3a, d). For malignant tumor diagnosis, it achieved an AUC of 87.9% (95% CI:84.6–91.0), with an 80.8% (95% CI:77.2-84.4) ACC, an 81.6% (95% CI: 78.1-85.0) F1, an 80.8% (95% CI: 77.2–84.4) recall, and an 83.6% (95% CI: 80.4-86.8) precision (Fig. 3b, e). For benign tumor diagnosis, it achieved an AUC of 89.9% (95% CI: 85.7-93.3), with an 83.9% (95% CI: 78.8–89.1) ACC, an 83.7% (95% CI: 78.6–89.0) F1, an 83.9% (95% CI: 78.8-89.1) recall, and an 84.9% (95% CI: 80.3-89.8) precision (Figs. 3c and 3f). Visual comparison of t-Distributed Stochastic Neighbor Embedding between the LiLNet, loaded with pre-trained ResNet50 (*) and the standard ResNet50 on the HN validation set can be found in Supplementary Fig. 1. In the First Affiliated Hospital of Chengdu Medical College (CD Center), an AUC of 94.2% and an ACC of 82.9% were achieved for diagnosing HCC (Fig. 3g). In the Leshan People’s Hospital (LS Center), the model achieved an AUC of 87.6% and an ACC of 82.1% for diagnosing HCC. Similarly, it maintained performance with an AUC of 79.2% and an ACC of 79.6% for ICC (Fig. 3h). Finally, at the Guizhou Provincial People’s Hospital (GZ Center), the ACC was 84.4% for HCC and 74.4% for ICC, as shown in Fig. 3i.
Fig. 3. Generalization performance of the LiLNet model on the external validation set.
a–c display ROC curves for differentiating benign and malignant tumors in the HN external validation set. d provides ACC, F1, Reacll and Precision for this distinction. e presents ACC, F1, recall, and precision for identifying malignant tumors, while f shows the same metrics for Benign tumors. g The model’s ACC and AUC for HCC in the CD validation set. h The model’s ACC and AUC for HCC and ICC in the LS validation sets. i The ACC and AUC for distinguishing HCC and ICC in the GZ validation sets. Source data are provided as a Source Data file Source_data_Figure_3.xlsx.
LiLNet Performance for Tumor Size
Table 2 presents the ACC for different tumor sizes in both the test set and the HN external validation set. Each cell shows the accuracy percentage for a tumor type within its size range, along with the total sample number. For instance, in the test set, tumors smaller than 1 cm achieved a 100% ACC for the HCC type, with a total of 4 samples. However, in the HN validation set, there were no samples in this size range, resulting in a 0% ACC. The ACC varies with size range, and specific tumor types show differing ACCs within these ranges. Hence, tumor size is not the sole factor influencing classification ACC. The results show varying accuracy levels for different tumor sizes, with no consistent trend. Some size ranges display high ACCs, while others show lower ACCs in both the test set and the HN validation set. The ACC also varies for specific tumor types within different size ranges, indicating that tumor size alone does not determine the ACC of classification. Other factors, such as tumor type, likely contribute to these variations.
Table 2.
Accuracy of classifying tumors with different sizes
| Size | HCC | ICC | MET | FNH | HEM | CYST | Average | |
|---|---|---|---|---|---|---|---|---|
| Accuracy of classifying tumors with different sizes on testing sets | ||||||||
| Acc/Num | <1 cm | 100%/4 | none/0 | 100%/7 | 0%/1 | 60.0%/5 | 100%/20 | 91.9%/37 |
| 1-3 cm | 83.4%/429 | 92.8%/28 | 85.8%/106 | 89.7%/29 | 89.6%/48 | 89.7%/29 | 85.2%/669 | |
| 3-5 cm | 91.9%/349 | 76.2%/42 | 78.9%/19 | 80.0%/5 | 90.0%/10 | 50.0%/2 | 89.4%/427 | |
| >5 cm | 97.5%/164 | 90.0%/10 | 100%/1 | none/0 | none/0 | none/0 | 97.1%/175 | |
| Accuracy of classifying tumors with different sizes on HN validation sets | ||||||||
| Acc/Num | <1 cm | none/0 | none/0 | 66.6%/3 | none/0 | 83.3%/6 | 100%/4 | 85.7%/13 |
| 1-3 cm | 69.7%/132 | 87.5%/8 | 94.3%/53 | 73.3%/30 | 91.3%/46 | 81.8%/44 | 79.5%/313 | |
| 3-5 cm | 89.1%/129 | 80.7%/26 | 73.5%/34 | 62.5%/8 | 97.3%/37 | 66.7%/9 | 85.6%/243 | |
| >5 cm | 92.5%/40 | 60%/10 | 28.6%/7 | 100%/1 | 100%/5 | 100%/3 | 81.8%/66 | |
Note: ACC denotes accuracy, Num denotes number. HCC denotes Hepatocellular Carcinoma, ICC denotes Intrahepatic Cholangiocarcinoma, MET denotes Metastatic Cancer, FNH denotes Focal Nodular Hyperplasia, HEM denotes Hemangioma, and CYST denotes Cyst. HN denotes Henan Provincial People’s Hospital.
The numerical values in each cell represent the accuracy of classifying tumor types within the corresponding size range, are presented as percentages, and are annotated with the total number of samples. Source data are provided as a Source Data file Source_data_Table_2.xlsx.
LiLNet Performance for Liver Background
To assess the potential impact of background liver conditions, such as fibrosis or inflammation, on the performance of our proposed system in analyzing CT images, we collected data from West Chian Tianfu Hospital, including 3 cases of HCC without hepatitis and liver fibrosis, 21 cases of HCC with hepatitis and liver fibrosis, 5 cases of ICC with similar liver conditions and 16 cases of MET without hepatitis or liver fibrosis. We observed that the system achieved an AUC of 88.1% and an ACC of 80.9% for HCC with liver fibrosis caused by hepatitis, while for ICC, the AUC was 96.4%, with an ACC of 80%. Our results show that the background liver condition has minimal impact on lesion extraction and imaging. This is because our data originate from real clinical events in which liver lesions often coexist with conditions such as cirrhosis, hepatitis, and liver fibrosis. During data collection, we did not exclude background liver diseases. The distinct imaging features of liver diseases, such as cirrhosis, fibrosis, or inflammation, on CT images typically differ from those of liver lesions, making it relatively straightforward for the model to differentiate between them.
LiLNet Performance for Different Phases
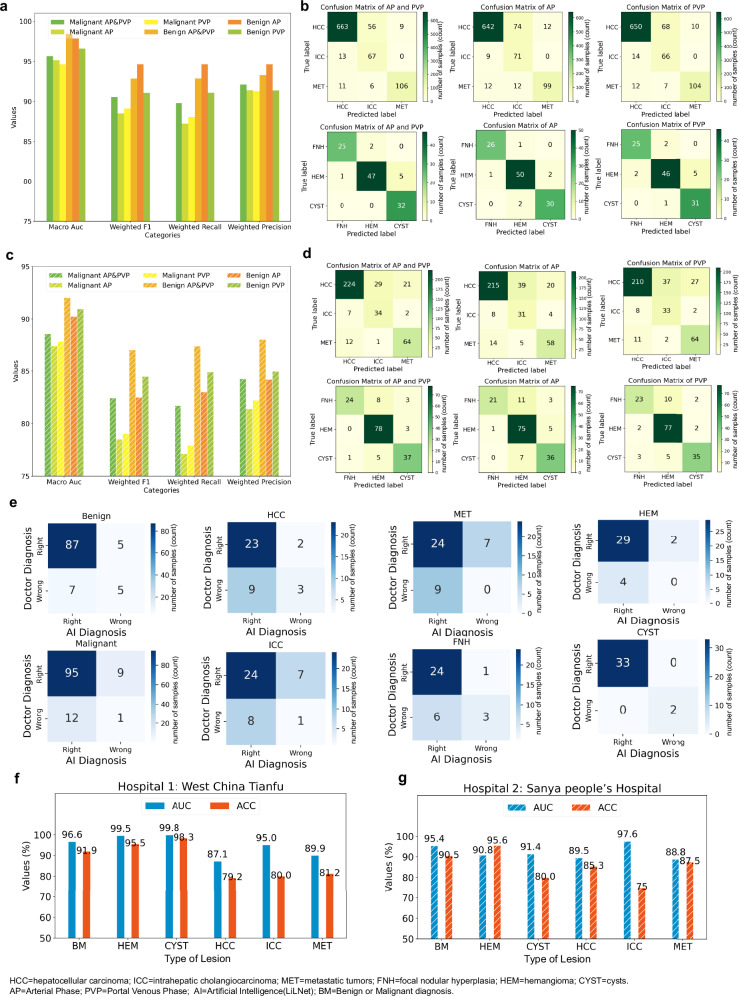
In clinical practice, lesions show different characteristics in various phases, each presenting unique features. Radiologists often use multiple phases for lesion diagnosis. Following this practice, our system simultaneously detects lesions in multiple phases, providing enhanced support for medical professionals. To evaluate the advantages of incorporating different phases, we conducted experiments on a dataset containing 1569 patients from both the test and HN external validation sets, covering data from multiple phases. The results are depicted in Figs. 4a–d. As depicted in Fig. 4a, for malignant triple classification in the test set, the diagnostic performance of using both AP and PVP was superior to that of using either AP or PVP alone, while the results for using AP or PVP alone were comparable. However, for benign triple classification, the AUC was optimal when utilizing both AP and PVP images simultaneously, followed by using AP alone and PVP alone; other performance indicators showed that AP outperforms AP and PVP, which outperforms PVP. As illustrated in Fig. 4c, in the validation set, the diagnostic performance of AP and PVP surpassed that of AP or PVP alone, regardless of malignant or benign classification. Analysis of the confusion matrices of the test set and external validation set (Fig. 4b, d) showed that employing images from both the AP and PVP phases simultaneously yielded superior results compared to using a single phase. Although the diagnostic outcomes of the two phases align in approximately 90% of cases, there are still instances where lesions exhibit better performance in the AP phase than in the PVP phase, and vice versa. This discrepancy may be attributed to the inherent characteristics of the data. In summary, integrating information from multiphase CT-enhanced images enables a comprehensive and accurate assessment of liver lesion characteristics and properties, thereby offering a more reliable basis for clinical diagnosis and treatment.
Fig. 4. LiLNet performance under different conditions.
a Comparison of the AUC, F1, recall, and precision for the classification of the three types of malignant lesions (HCC, ICC, and METs) and classification of the three types of benign lesions (FNH, HEM, and cysts) using different phases in the test set. “malignant AP&PVP” indicates the simultaneous use of AP and PVP for diagnosing malignant lesions, “malignant AP” indicates the use of only AP, and “Malignant PVP” indicates the use of only PVP for diagnosing malignant lesions. Similarly, “benign AP&PVP” indicates the simultaneous use of AP and PVP for diagnosing benign lesions, “benign AP” indicates the use of only AP, and “benign PVP” indicates the use of only PVP for diagnosing benign lesions. b Confusion matrices for classification of the three types of malignant lesions and classification of the three types of benign lesions using different phases in the test set. c Comparison of the AUC, F1, recall, and precision for classification of the three types of malignant lesions and classification of the three types of benign lesions using different phases in the HN external validation cohort. d Confusion matrices for classification of the three types of malignant lesions and classification of the three types of benign lesions using different phases in the validation set. e The confusion matrix is employed to depict the classification of lesions for patients, categorizing them into four groups based on the diagnoses provided by AI systems and radiologists. ‘Radiologist Right’ and ‘AI Right’ indicate instances where both the AI system and the doctor correctly diagnosed liver tumors. ‘Radiologist Right’ and ‘AI Wrong’ refer to cases where the AI system incorrectly diagnosed a liver tumor but the radiologist’s diagnosis was accurate. ‘Radiologist Wrong’ and ‘AI Right’ pertain to situations in which the AI system made a correct diagnosis of liver tumors but the radiologist’s diagnosis was incorrect. ‘Radiologist Wrong’ and ‘AI Wrong’ represent instances where neither the AI system nor the doctor diagnosed liver tumors correctly. f The results of clinical validation at West China Tianfu Hospital. g The results of clinical validation at Sanya People’s Hospital. Source data are provided as a Source Data file Source_data_Figure_4.xlsx.
Comparison with radiologists
We used a test set of 6743 images from 221 patients at West China Hospital of Sichuan University to compare the diagnostic ability of LiLNet with that of radiologists. The evaluation involved three radiologists with varying levels of experience. Radiologists independently labeled the 221 patients based on multiphase contrast-enhanced CT images. LiLNet demonstrated a diagnostic accuracy of 91.0% for distinguishing between benign and malignant tumors, 82.9% for distinguishing between malignant tumors, and 92.3% for distinguishing between benign tumors (Table 3). Compared to junior-level radiologists, LiLNet achieved 4.6% greater accuracy for benign and malignant diagnosis, 4.1% greater accuracy for middle-level radiologists, and 2.3% greater accuracy for senior-level radiologists. The diagnostic accuracy of radiologists for diagnosing malignant tumors was similar. Notably, compared with radiologists, LiLNet achieved a substantial 18% improvement in diagnostic accuracy. Additionally, in diagnosing benign tumors, LiLNet outperformed junior-level practitioners by 20%, middle-level practitioners by 10%, and senior-level practitioners by 6.7%. More information about the radiologists and their diagnostic results can be found in the supplementary information (Supplementary Table 1 and Table 2).
Table 3.
Comparison of diagnostic results between LiLNet and radiologists
| Diagnosis | Radiologist | Accuracy (%) | Recall (%) | Precision (%) |
|---|---|---|---|---|
| Benign and Malignant | LiLNet | 91.0 | 91.5 | 90.4 |
| Junior | 86.4 | 88.9 | 83.6 | |
| Middle | 86.9 | 88.0 | 85.6 | |
| Senior | 88.7 | 88.9 | 88.5 | |
| HCC, ICC, MET | LiLNet | 82.9 | 83.0 | 83.3 |
| Junior | 65.8 | 65.8 | 65.4 | |
| Middle | 64.1 | 63.9 | 64.1 | |
| Senior | 63.2 | 63.2 | 63.2 | |
| FNH, HEM, CYST | LiLNet | 92.3 | 92.3 | 92.4 |
| Junior | 71.9 | 71.4 | 71.7 | |
| Middle | 81.7 | 81.6 | 81.7 | |
| Senior | 85.6 | 85.5 | 85.8 |
Note: HCC denotes Hepatocellular Carcinoma, ICC denotes Intrahepatic Cholangiocarcinoma, MET denotes Metastatic Cancer, FNH denotes Focal Nodular Hyperplasia, HEM denotes Hemangioma, and CYST denotes Cyst.
Source data are provided as a Source Data file Source_data_Table_3.xlsx.
We calculated the Fleiss kappa coefficient between LiLNet and the radiologists to assess consistency. The Fleiss kappa values are 0.806 for benign and malignant cases and 0.848 for benign cases, surpassing the 0.8 threshold, indicating a very high level of agreement among evaluators in benign tumor diagnosis and 0.771 for malignant cases, which falls within the range of 0.6 to 0.8. This indicates a high level of agreement among the evaluators (details in Supplementary Table 3).
Figure 4e displays the comparison matrix of diagnoses between the AI system and radiologists (we selected the optimal diagnosis from the assessments provided by multiple radiologists) based on pathological diagnostic labels. This result indicates that 4% of cases are misdiagnosed by both the AI system and radiologists, while radiologists accurately diagnose 8% of cases where the AI system errs. In 16% of cases, the AI system was correct when radiologists made errors. Specifically, in the “benign” cases shown in Fig. 4e, the AI system and radiologists agreed on 92 cases. Among these, 87 cases were confirmed to be correct by pathology, while 5 cases were incorrect. Additionally, there were 12 cases of disagreement: 5 were incorrect AI judgments (false-negatives), 7 were correct (true positives), and 7 were incorrect radiologist diagnoses (false-negatives), with 5 being correct (true positives). Consequently, when the AI system and radiologists differed, the AI system achieved a 58.34% true positive rate for benign diagnoses, while the radiologists achieved a 41.67% true positive rate. For the “malignant” cases, the AI system and radiologists agreed on 96 cases. Among these, 95 cases were confirmed to be correct by pathology, while 1 case was incorrect. Additionally, there were 21 cases of disagreement: 9 were incorrect AI judgments (false negatives), 12 were correct (true positives), and 12 were incorrect radiologist diagnoses (false negatives), with 9 being correct (true positives). Consequently, when the AI system and radiologists differed, the AI achieved a 57.14% true positive rate for benign diagnoses, whereas the radiologists achieved a 42.85% true positive rate. Furthermore, additional diagnostic information for HCC, ICC, METs, FNH, HEM, and cysts can be found in Fig. 4e.
Figure 4e shows that AI and radiologists achieved congruent outcomes in cyst diagnosis, accurately identifying 32 cases while misdiagnosing 2 cases. Upon analyzing the misdiagnosed cyst images, we discovered one patient with a mixed lesion, showing characteristics of both HEM and a cyst. In this instance, the cyst was positioned near a blood vessel, resulting in misdiagnosis as HEM. Another misdiagnosis was due to the lesion size being less than 1 cm, presenting challenges in identification. However, there are some differences in diagnosis between AI and radiologists in other categories, indicating differences in diagnostic approach or focus. These findings highlight the potential for our AI-assisted software to collaborate with radiologists to enhance the diagnostic accuracy of liver lesions.
Real-world clinical evaluation
Our system (a simple web version is available in Supplementary Note 1) is currently suitable for routine clinical diagnoses, encompassing outpatient, emergency and inpatient scenarios with patients undergoing AP and PVP sequences. To authenticate the actual clinical efficacy of the system, we seamlessly integrated the system into the established clinical infrastructure and workflow at West China Tianfu Hospital and Sanya People’s Hospital in China, where we conducted a real-world clinical trial.
At West China Tianfu Hospital, we assessed outpatient and inpatient data from February 29th to March 7th, comprising 117 cysts, 22 HEMs, and 16 METs. To improve the evaluation of the model’s ability to diagnose malignancies, we included 24 HCC lesions and 5 ICC lesions from January 2022 to February 2024. All malignant tumors were pathologically confirmed, while benign tumors were diagnosed by three senior radiologists. As shown in Fig. 4f, the results of our system at the Tianfu Center indicated an AUC of 96.6% and an ACC of 91.9% for the diagnosis of benign and malignant lesions, respectively. For HEMs, the AUC was 99.54%, with an ACC of 95.45%, while for cysts the AUC was 99.8%, with an ACC of 98.3%. For HCCs, the AUC was 87.1%, with an ACC of 79.2%; for ICCs, the AUC was 95.0%, with an ACC of 80%; and for METs the AUC was 89.9%, with an ACC of 81.2%.
We assessed outpatient and inpatient data at Sanya People’s Hospital from March 15th to March 29th, comprising 45 cysts, 23 HEMs, 121 normal lesions, 1 ICC, and 3 METs. Additionally, we retrospectively collected data for 34 HCCs, 3 ICCs, and 5 METs from April 2020 to February 2024. All malignant tumors were pathologically confirmed, while benign tumors were diagnosed by three senior radiologists. As shown in Fig. 4g, the results of our system at Sanya Center indicated an AUC of 95.4% and an ACC of 90.5% for the diagnosis of benign and malignant lesions, respectively. For HEMs, the AUC was 90.8%, with an ACC of 95.6%, while for cysts, the AUC was 91.4% with an ACC of 80.0%. For HCCs, the AUC was 89.5%, with an ACC of 85.3%; for ICCs, the AUC was 97.6%, with an ACC of 75%; and for METs, the AUC was 88.8%, with an ACC of 87.5%.
Deep learning analysis
To better explain the deep learning model, we conducted two experiments: an analysis by professional radiologists on activation maps and gradient analysis.
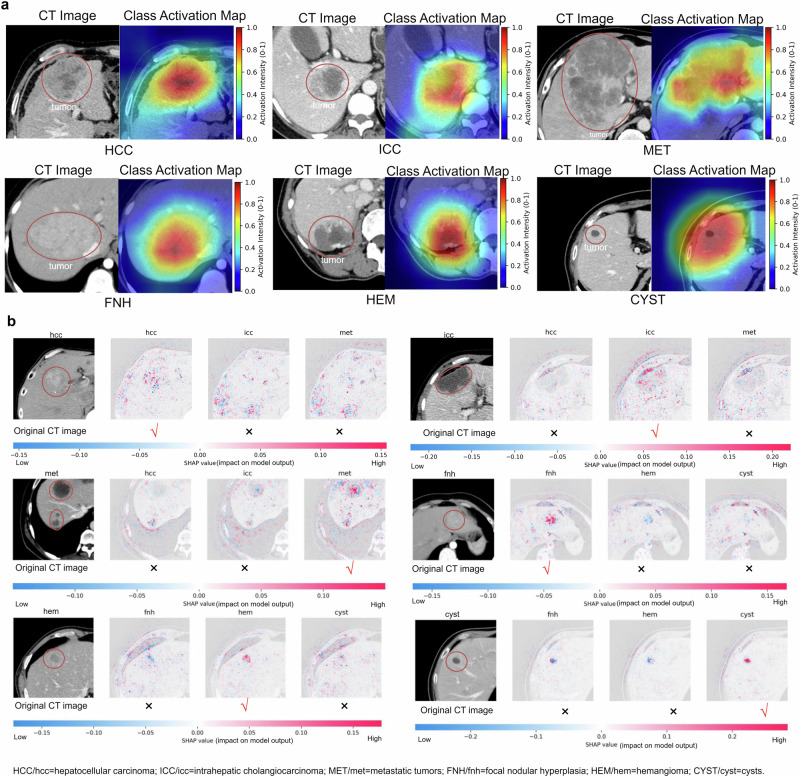
Class activation maps (CAMs) are generated by computing the activation level of each pixel in the image by the model, revealing the areas of focus within the image. Figure 5a shows that the model pays more attention to lesion areas relative to normal liver tissue to distinguish between different subtypes. HCC typically exhibits heterogeneity in internal structure and cellular composition, resulting in significant variation within the tumor. Rapid proliferation of tumor cells leads to increased cell density and richer vascularity in the central region, often manifested as arterial phase enhancement in imaging. Conversely, the surrounding area may display lower density and vascularity due to compression by normal hepatic tissue or the arrangement of tumor cells in a nest-like pattern, presenting as low density in imaging. Consequently, in this CAM image, the central region may exhibit deep activation, while the surrounding area may show secondary activation. Additionally, the irregular spiculated margins commonly observed in HCC are a critical feature, often encompassed within activated regions. ICC is characterized by tumor cells primarily distributed in peripheral regions, with fewer tumor cells and immune-related lymphocytes in the central area. Imaging typically reveals higher density and vascularity in the tumor periphery, contrasting with lower density and vascularity in the central region. These imaging features are reflected in the CAM image. Metastatic tumors, arising from either intrahepatic primary tumors or extrahepatic malignancies, often exhibit necrosis and uneven vascularity in tissue composition. This results in the characteristic imaging appearance of indistinct margins and multifocal lesions. CAM images frequently depict this process by demonstrating areas of diffuse and poorly defined activation, with uneven depth and distribution of activation regions.
Fig. 5. The visualization process of model decisions.
a The class activation map generated by the last convolution layer. We presented activation maps for liver lesions. The first line displays the original image, while the second line displays the corresponding activation map. Red denotes higher attention values, the color blue denotes lower values, and the red circle represents the tumor area. b SHAP plots revealing the influence of pixel on the model predictions for HCC, ICC, MET, FNH, HEM, and cyst lesions.
FNH typically arises from the abnormal arrangement of normal hepatic cells and contains abundant vascular tissue with high density. On images, it typically presents as homogeneous enhancement of focal lesions, while surrounding normal hepatic tissue appears relatively hypoenhanced due to compression. In CAM images, the lesion often exhibits uniform overall activation, while the compressed normal hepatic parenchyma demonstrates relatively lower activation. FNH is characterized by richer vascularity than other lesions, resulting in greater overall activation. HEM lesions usually contain abundant vascular tissue and manifest as focal lesions with significant enhancement during the contrast-enhanced phase of imaging. In CAM images, they typically appear as locally activated areas, exhibiting greater activation than other nonvascular lesions, with a more uniform distribution. Cysts typically consist of fluid or semisolid material, with uniform internal tissue distribution and clear borders. On images, they appear as circular or oval-shaped low-density areas with clear borders. In CAM images, cystic regions appear as circular areas with deep activation, and the activation intensity within the cyst is usually uniform, without significant differences. More class activation maps can be found in Supplementary Fig. 2.
Model interpretability refers to the process of explaining the outputs generated by a machine learning model, elucidating which features and how they influence the actual output of the model. In deep learning, particularly in computer vision classification tasks, where features are essentially pixels, model interpretability aids in identifying pixels that have either positive or negative impacts on predicting categories. To achieve this goal, we employ the SHapley Additive exPlanations (SHAP) library to interpret deep learning models. This process primarily involves analyzing the gradients within the model to gain a deeper understanding of how decisions are made. By inspecting gradients, we can determine which features contribute most significantly to the model’s predictions. In Fig. 5b, we present plots for HCC, ICC, MET, FNH, HEM, and CYST. Each SHAP plot comprises the original image alongside grayscale images corresponding to the number of output classes predicted by the model. Each grayscale image represents the model’s contribution to the output class. In these images, blue pixels indicate a negative effect, while red pixels indicate a positive effect. Conversely, white pixels denote areas where the model ignores input features. Below the images, there is a color scale ranging from negative to positive, illustrating the intensity of SHAP values assigned to each relevant pixel. For instance, in the case of correct HCC category prediction, the SHAP plot for HCC reveals that red activations are predominantly concentrated in the lesion area. However, in SHAP plots for other categories such as ICC and MET, although some red pixels are present, they are not concentrated in the lesion area. This suggests that the appearance of red activations outside the lesion area in other categories may indicate a misjudgment or confusion by the model during prediction. Meanwhile, the activation in the lesion area remains one of the key factors for accurate prediction.
Data Partitioning Strategy Experiments
We conducted a time-based data partitioning experiment to further validate the model’s generalization ability on the test set. We sorted the data used for model development chronologically, using early data for training and later data for testing (with the same test set size as random partitioning). We compared the results of random partitioning with those of time-based partitioning, as shown in Fig. 6. Using the time-based partitioning method, we achieved an AUC of 98.7% (95% CI: 92.1–94.8) and an ACC of 93.5% (95% CI: 92.1–94.8) for benign and malignant results. The diagnostic AUC for benign data was 98.0% (95% CI: 96.1–99.2), with an ACC of 91.3% (95% CI: 86.6–95.3), while for malignant diagnosis, the AUC was 97.5% (95% CI: 96.7–98.1) with an ACC of 90.9% (95% CI: 89.2–92.5). In HN external validation, the AUC for benign and malignant diagnosis was 94.6% (95% CI: 92.6–96.2), with an ACC of 88.8% (95% CI: 86.5–91.2). The AUC for benign data diagnosis was 88.3% (95% CI: 84.0-92.1), with an ACC of 82.4% (95% CI:77.2–88.1), while for malignant diagnosis, the AUC was 87.9% (95% CI: 84.6–91.0) with an ACC of 85.1% (95% CI: 81.9–88.3). In external validation for CD, the accuracy of malignant diagnosis was 90.4% (95% CI: 84.0–95.7). For GZ, the accuracy of malignant diagnosis was 80.1% (95% CI: 74.2–85.5), and for LS, it was 82.8% (95% CI: 77.6–88.1).
Fig. 6. Comparison of Results between randomly and time-divided data.
a displays ROC curves comparing the differentiation of benign and malignant tumors in the Test and HN external validation sets. b shows ROC curves comparing the differentiation of benign tumors in the Test and HN external validation sets. c presents ROC curves comparing the identification of malignant tumors. d displays ACC for distinguishing between benign and malignant tumors in the Test and HN external validation sets. e demonstrates ACC for distinguishing benign tumors in the Test and HN external validation sets. f provides ACC for identifying malignant tumors in the HN, CD, GZ, and LS validation sets. Source data are provided as a Source Data file Source_data_Figure_6.xlsx.
We conducted a statistical analysis of the accuracy of random and time-based data partitioning methods using a two-sided t-test. For the binary classification of benign and malignant lesions, the p-value is 0.192 on the test set and 0.503 on the HN external validation set. For the ternary classification of benign lesions, the p-value is 0.408 on the test set and 0.695 on the HN external validation set. For the ternary classification of malignant lesions, the p-values are 0.082 on the test set, 0.08 on the HN external validation set, 0.136 on the CD external validation set, 0.483 on the GZ external validation set, and 0.811 on the LS external validation set. The statistical results indicate that all p-values are greater than 0.05, suggesting no significant difference between the two methods.
Discussion
Liver lesions, encompassing a diverse range of malignancies and benign lesions, pose significant challenges in accurate diagnosis and representation. The advent of deep learning techniques ushered in an era in medical classification. We explored the potential of our proposed deep learning model for precise and efficient liver lesion classification, along with the challenges and future prospects in this evolving field.
Currently, there are several deep learning approaches for focal liver lesion diagnosis. However, most of these approaches primarily concentrate on HCC and ICC, often utilizing limited sample sizes from single or multiple centers14,16,21 and lacking external validation22 to confirm their general applicability. Furthermore, these approaches predominantly rely on convolutional networks or transfer learning techniques15,23,24. This study focused on developing a deep learning AI-assisted system for clinical liver diagnosis. Previous studies have indicated that deep learning algorithms outperform health care professionals with respect to some clinical outcomes14,25,26. We proposed a liver lesion diagnosis system based on deep learning, LiLNet, which can automate the analysis of radiation images and can rapidly screen and identify suspicious regions for further examination by radiologists. With the utilization of multicenter and large sample data, this system offers a relatively comprehensive diagnostic approach.
Our model exhibits robust performance in both the test set and external validation set, primarily owing to the integration of extensive datasets and advanced AI technology. The training data are comprehensive, encompassing a wide array of patterns, which include diverse imaging devices, variations in image window widths and levels, and adjustments in target area sizes. These factors are meticulously considered to accommodate differing background liver conditions, such as cirrhosis, fibrosis, inflammation, fatty liver, and abdominal fluid. Our model has shown excellent performance on test and external validation sets, primarily owing to the integration of big data and AI technology. Our training dataset is extensive and diverse, comprising images acquired from a variety of CT device models, each with unique specifications for window width and level settings. Additionally, the dataset includes samples representing a wide range of background liver conditions, including cirrhosis, fibrosis, inflammation, fatty liver, and the presence of abdominal fluid. Moreover, it encompasses target areas of varying sizes for comprehensive coverage. The richness of the data in our training set significantly contributed to enhancing the generalizability of the model. We adopted a two-stage approach, starting with detection-then-recognition technology. Initially, through object detection methodologies, we extracted ROIs to minimize irrelevant background information and direct the model’s attention to the tumor. Subsequently, we segmented the liver tumor classification task into benign and malignant stages and then performed subtype classification. This strategic classification approach not only reduces the complexity and difficulty of subsequent classifiers but also enhances the overall accuracy and stability of our classification system. Compared to the popular deep learning classification algorithms ResNet50 and pretrained ResNet50, our proposed model demonstrates better performance on both the test set and external validation. This is primarily attributed to several key enhancements. First, our model introduces an enhanced supervised signal, which selectively discards irrelevant regions in the feature maps and expands the original labels into joint labels during training. This additional supervision signal enables the model to better comprehend image content and learn more robust feature representations. Given the challenging nature of the liver tumor classification task, characterized by significant confusion or overlap between categories, our approach provides clearer supervisory signals to differentiate categories effectively, thereby reducing confusion and enabling the model to focus on recognizing detailed features. Additionally, leveraging self-distillation technology empowers our model to learn from its own generated responses, further improving its performance. This self-distillation process allows the model to refine its understanding and generalization ability over time, leading to enhanced performance in practical applications.
The LiLNet model outperforms clinical radiologists in third-tier cities due to its use of a vast dataset from a reputable comprehensive hospital in China for training, offering broader coverage and greater sample diversity. AI algorithms have undergone extensive standardization and optimization, ensuring consistent and accurate diagnoses. Conversely, radiologists in third-tier cities may face challenges such as limited medical resources and variations in personnel quality, hindering the level of professionalism and standardization in diagnosis. While there is high consistency between AI and radiologists in diagnosing straightforward cases such as cysts, discrepancies arise in easily confused cases such as HCC and ICC, HCC and FNH, and METs and HEM, suggesting differing diagnostic methods or focuses. The partnership between AI-assisted software and radiologists holds promise for enhancing the accuracy of liver disease diagnosis.
There are a few limitations. First, compared to other liver diseases, the higher incidence of HCC results in data imbalance, which may slightly affect the performance for diagnosing ICC. Comparative studies with clinical doctors have shown that the most accurate results are achieved when artificial intelligence collaborates with doctors to diagnose HCC and ICC. cHCC-CCA refers to combined hepatocellular-cholangiocarcinoma, a rare type of liver tumor that exhibits both hepatocellular carcinoma and cholangiocarcinoma characteristics. cHCC-CCA is a rare variant of liver cancer, with an incidence rate ranging from 0.4% to 14.2% compared to other primary liver cancers27–29. The performance of the LiLNet system in diagnosing cHCC-CCA is currently unclear due to the rarity of this tumor and limited research and data availability. Recognizing the importance of cHCC-CCA in the field of hepatocellular carcinoma, we plan to focus on this topic as a key area of future research. We aim to collaborate with pathology experts to collect relevant data and incorporate cHCC-CCA into our future studies, thereby expanding the scope of our research findings.
Methods
Ethical approval
Ethics committee approval was granted by the ethics review board of West China Hospital of Sichuan University (Ethical Approval No. 2024-424), and was carried out in adherence to the Declaration of Helsinki. Additionally, this study is officially registered with the Chinese Clinical Trial Registry, under the identifier ChiCTR2400081913 (accessible at https://www.chictr.org.cn/showproj.html?proj=212137). Recognizing the non-invasive nature of the methodology and the anonymization of data, the institutional review board granted a waiver for the informed consent requirement. Subject to data privacy and confidentiality, ethical review, and institutional policies, we use patient imaging data solely for system testing without requiring the patient to undergo additional testing, visits, or any activities directly related to the system. Under these conditions, generally, no additional compensation to the patient is necessary.
Data acquisition
Between June 2012 and December 2022, a total of 4039 patients’ multiphase (arterial phase and portal venous phase) contrast-enhanced CT images from six hospitals were included under the following inclusion criteria: Patients (1) were eighteen years or older; (2) did not have a history of hepatectomy, transarterial chemotherapy (TACE), or radiofrequency ablation (RFA) before CT imaging; (3) had pathologically confirmed malignant tumors; and (4) had benign tumors confirmed either by consensus among three radiologists or by follow-up of at least six months using two imaging modalities. The method used for retrospective data collection and basic patient information including sex and age are depicted in Fig. 1a and Table 1, respectively. Furthermore, clinical testing was conducted on two real-world clinical evaluation queues (Fig. 1b): West China Tianfu Center and Sanya People’s Hospital. At Tianfu Center, we examined 184 cases, while at Sanya People’s Hospital, 235 cases were assessed. Gender and Age assignment was based on government-issued IDs. No sex-based analysis was conducted as gender was unrelated to the model implementation or deployment. The primary reason for this is that the focus of our study was on evaluating the technical performance of the system, rather than examining potential differences based on gender or sex. Additionally, our primary objective was to ensure the system’s accuracy and efficiency in processing imaging data, regardless of the patient’s gender.
Triple-phase CT scans were performed on all participants, including non-contrast, arterial, and portal venous phases. Precontrast images were obtained before injecting the contrast agent (iodine concentration: 300–370 mg/mL; volume: 1.5–2.0 mL/kg; contrast type: iopromide injection, Bayer Pharma AG). The arterial phase and portal venous phase images were acquired at 25 s and 60–90 s after injection, respectively. Slice thickness was 5 mm for non-contrast images and 1–3 mm for arterial and portal venous phases. Specific details of the CT scanners used (GE Healthcare, Siemens Healthcare, Philips Healthcare, United Imaging Healthcare) are provided in the Supplementary Table 4.
We utilize the open-source libraries Pydicom (https://pydicom.github.io/, Version 2.2.2) and SimpleITK (https://simpleitk.org/, Version 2.0.2) to process the original DICOM files into BMP image format for convenient model training. The lesion areas are delineated using Jinglingbiaozhu (http://www.jinglingbiaozhu.com/, Version 2.0.4). All code is written in custom Python (Version 3.9.7).
Development of the LiLNet system
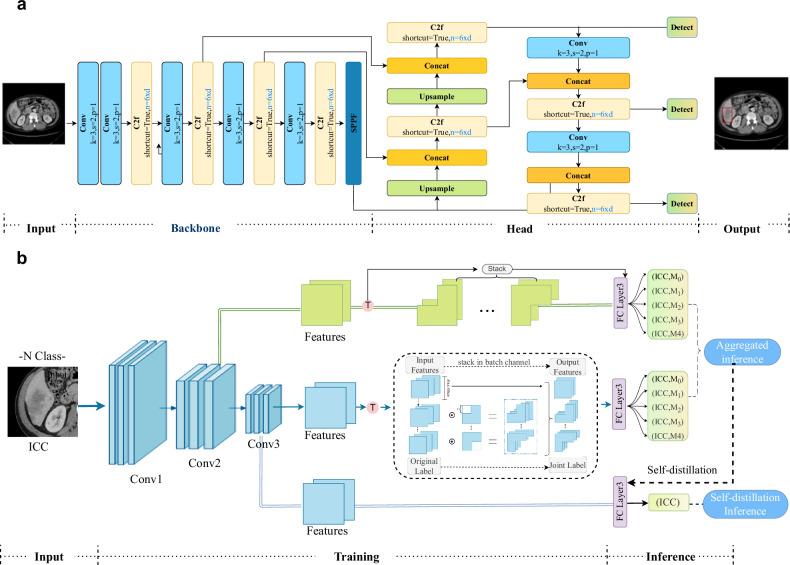
The ability of deep learning models to establish robust tumor classification frameworks depends on the extraction of discriminative features from input data. When excessive background information exists in the training image, the background overwhelms the foreground or hampers the visibility and identification of key features. To enhance the diagnostic ACC, we preprocessed the original CT images to extract regions of interest (ROIs) by removing redundant background information. Initially, a professional physician manually delineated the ROIs of the 500 patients used for training. Then, we developed a model based on YOLOv830 (Fig. 7a) to automatically obtain the ROI. To mitigate false positives, we implemented postprocessing on the target model using 3D liver segmentation technology31. This integration with liver segmentation results allowed us to effectively filter out false positives occurring outside the liver, leading to a notable enhancement in both the accuracy and reliability of the model.
Fig. 7. Workflow of the proposed LiLNet system.
a Lesion detection model. To enhance the classification algorithm’s focus on the relevant features of the image, we utilize an object detection model to locate the lesion and remove excessive background surrounding the lesion area. b Overview of our classification framework.
We established a classification model based on multiphase data that integrates image information using self-supervised tasks and joint labels, further improving diagnostic performance. Our model utilizes ResNet5032 as the backbone network, along with a feature-based pretext task that transforms different features for network recognition. To generate a transformed feature map, we applied binary masks to selectively remove spatially correlated information from a given feature map obtained from a random hidden representation layer. In this study, we utilized four distinct binary masks, which correspond to the covered regions of the upper left, upper right, lower left, and lower right portions. Additionally, we introduced joint labels to expand the original labels, facilitating the incorporation of the self-supervised pretext task into the model. We augmented the original label by including an additional self-supervised label that signifies the dropped region. After generating the joint labels, the number of input labels increased from the original N to 5 N. Subsequently, all feature maps generated by the classifiers were combined through aggregation inference for prediction.
We implemented multiclassifier techniques to enhance model performance. A classifier and a fully connected layer were introduced after the last two ResBlocks, specifically for training purposes, and could be omitted during inference. Recognizing the computational cost associated with the final convolutional layer, we also incorporated an extra classifier for the penultimate layer, mitigating additional convolutional computations while preserving accuracy improvements. In addition, we introduced a self-knowledge distillation module (SKD) to facilitate the interaction between deep-shallow features from different layers, thereby enhancing network learning and enabling adaptive information fusion. The SKD enhances the main network structure by incorporating a classifier that extracts deep features as teachers, which distills and learns the joint label layer of the source network. The network structure, depicted in Fig. 7b, selects the last feature layer from the residual network to build an additional classifier consisting of a fully connected layer and softmax activation. During the training process, these classifiers, as instructors, offer multidimensional guidance to the residual network to improve valuable information and enhance its efficiency. The tumor detection and training details are provided in the training strategy section.
Calculating Prediction Results
We compute the prediction probability for each image for all individuals, encompassing both AP and PVP images. Subsequently, we calculate the average prediction probability for each individual by averaging the prediction probabilities of all their images. If a patient has images with corresponding prediction probabilities , the average prediction probability is calculated as shown in Eq. (1):
| 1 |
Then, we apply softmax processing to these average prediction probabilities and determine the category with the highest probability as the final result. The softmax function is calculated as shown in Eq. (2):
| 2 |
where N is the total number of categories.
When different phase images of the same individual yield disparate results, we address this by averaging the prediction probabilities, thereby assigning higher weight to the image with the most confident prediction. This approach allows us to maximize the utilization of information from each image, rather than solely relying on the outcomes of a few images. By balancing the influence of each phase image, this method mitigates the impact of abnormal results from one phase image, thereby reducing misjudgments.
Training strategy
We utilized patients’ name-ID as the unique identifier to prevent duplicate IDs. Duplicate samples with the same name-ID were systematically removed, and patients were randomly assigned to either the training set or testing set to prevent data overlap. As shown in Fig. 1a, the training set comprised images from 1580 patients from West China Hospital of Sichuan University and Sanya People’s Hospital. The testing cohort consisted of 1308 patients from West China Hospital of Sichuan University, while external validation cohorts included 1151 patients from Henan Provincial People’s Hospital, The First Affiliated Hospital of Chengdu Medical College, Leshan People’s Hospital, and Guizhou Provincial People’s Hospital.
The model used for target detection is YOLOv8, and the code can be found at: https://github.com/ultralytics/ultralytics. For post-processing, we utilized 3D liver segmentation technology, which is available at: https://github.com/ellisdg/3DUnetCNN. Our classification network model is based on ResNet50, and the code can be referenced here: https://github.com/weiaicunzai/pytorch-cifar100/blob/master/models/resnet.py.
For the training strategy of the classification model, the images were cropped to 224 * 224, centered on the ROI. Data augmentation techniques such as random flipping and rotation were applied to further enhance the images for analysis. Additionally, to address sample imbalance, we performed resampling with transformations such as Gaussian noise and SaltPopperNoise. These techniques aimed to generate larger, more complex, and diverse datasets, ultimately improving the ACC and generalizability of the model. We initialized our network parameters by loading pretrained network layer parameters from the ImageNet dataset. The network was trained using random gradient descent and cross-entropy loss for weight adjustment and algorithm optimization. The initial learning rate was set to 0.01, which decreased by one tenth every 10 epochs until a final learning rate of 0.0001 was reached. To mitigate overfitting, batch normalization and a weight decay rate of 0.0001 were implemented during training. A batch size of 128 and a rectified linear unit (RLU) activation function were used. All codes were implemented using Python 3.9.7. The packages or softwares comprised Pytorch 1.12.1 for model training and testing, CUDA 11.4 and cuDNN 8.2.4 for GPU acceleration.
Statistical analysis
We evaluated the prediction model using statistical measures such as the ACC, sensitivity, specificity, precision, recall, F1-score (F1), area under the curve (AUC), and receiver operating characteristic (ROC) curve. For binary classification problems, we employed a default threshold of 0.5, a widely accepted standard. For multiclassification tasks, we chose the category with the highest probability as the prediction based on the softmax classifier output. By analyzing the ACC, AUC, F1, recall, and precision, we gained insights into the model’s ability to accurately classify instances, handle imbalanced datasets, and strike a balance between true positives, false-positives, true negatives, and false-negatives, depending on the specific requirements of the application. All statistical analyses employed two-tailed tests, with p-values of 0.05 or lower deemed significant. These analyses were conducted using Python, version 3.9.7 and scikit-learn version 1.1.3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by Municipal Government of Quzhou (Grant 2023D007 [ML] and 2023D033 [MHL]); National Natural Science Foundation of China (No.82202117 [YW]); 135 project for disciplines of excellence–Clinical Research Fund, West China Hospital, Sichuan University (23HXFH019 [YW]); Interdisciplinary Crossing and Integration of Medicine and Engineering for Talent Training Fund, West China Hospital, Sichuan University (HXDZ22010 [BS]); National health commission capacity building and continuing education center (YXFSC2022JJSJ007 [BS]) and Science and Technology Department of Hainan Province (ZDYF2024SHFZ052 [BS]).
We extend our gratitude to the radiologists at Quzhou People’s Hospital: Dr. Guozheng Zhang (Senior), Dr. Shufeng Xu (middle), and Dr. Zheying Zhan (Junior) for their unwavering support and commitment to the comparative experiments on AI.
Author contributions
Y.W. and M.Y. conceived and designed the study. Y.W., M.Z., F.G., N.Z., F.H., X.Z., S.Z. (Shasha Zhang), Z.H., S.Z. (Shaocheng Zhu), B.S. and M.L. did the investigation and acquired the data. M.Y., M.L. (Minghui Liu), J.D., X.C.,and T.X. did the statistical analyses. M.Y. and Y.W. developed, trained and applied the artificial neural network. M.Y., X.W., N.L. and H.G. implemented quality control of data and the algorithms. Y.W., L.X. and F.Z. accessed and verified the underlying raw data. Y.W. and M.Y., prepared the first draft of the manuscript. S.Z. (Shaocheng Zhu), B.S. and M.L. (Ming Liu) did the project administration and supervised and coordinated the work of the research team and provided senior oversight. M.Y. developed the web system, and all authors have validated the liver diagnosis system. All authors read and approved the final version of the manuscript. All authors were responsible for the final decision to submit the manuscript for publication.
Peer review
Peer review information
Nature Communications thanks Masahiko Kuroda, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper. All the analytical data underpinning the findings of this study are incorporated within this paper in the designated source data files (Source_data_Figure_2.xlsx to Source_data_Figure_6.xlsx and Source_data_Table_2.xlsx to Source_data_Table_3.xlsx). However, the original dataset used in this study is subject to access control, requiring licenses from multiple central institutions for access. The author of the article communicated with several hospital departments, including the Information Technology Department, Research Laboratory, and International Cooperation Office. Although these data do not involve blood or other biological samples, the hospital’s strict data management policy prohibits any external sharing of research data. These regulations aim to ensure that all data, regardless of its nature, are securely protected to prevent unauthorized use or disclosure. To promote further international cooperation and exchange, it is recommended to jointly apply for international multicenter cooperation projects. This approach will enable us to conduct further research within the hospital while ensuring compliance with hospital policies, data security, and patient privacy. For academic inquiries regarding the use and processing of raw data, please contact the corresponding author via email at cjr.songbin@vip.163.com or csmliu@uestc.edu.cn. Source data are provided with this paper.
Code availability
Our code is available at GitHub31 (https://github.com/yangmeiyi/Liver/tree/main). The trained model is available at Zenodo (https://zenodo.org/records/12646854). The trained model parameters can be accessed at Zenodo, including the detection model best.pt (https://zenodo.org/records/12646854/files/best.pt?download=1), the benign-malignant classification model Time_BM.pth.tar (https://zenodo.org/records/12646854/files/Time_BM.pth.tar?download=1), the benign classification model Time_B.pth.tar (https://zenodo.org/records/12646854/files/Time_B.pth.tar?download=1), and the malignant classification model Time_M.pth.tar (https://zenodo.org/records/12646854/files/Time_M.pth.tar?download=1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yi Wei, Meiyi Yang.
These authors jointly supervised this work: Shaocheng Zhu, Bin Song, Ming Liu.
Contributor Information
Shaocheng Zhu, Email: zsc2686@126.com.
Bin Song, Email: cjr.songbin@vip.163.com.
Ming Liu, Email: csmliu@uestc.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51260-6.
References
- 1.Sung, H. et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin.71, 209–249 (2021). 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Liu, Z. et al. The trends in incidence of primary liver cancer caused by specific etiologies: results from the global burden of disease study 2016 and implications for liver cancer prevention. J. Hepatol.70, 674–683 (2019). 10.1016/j.jhep.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 3.Anwanwan, D., Singh, S. K., Singh, S., Saikam, V. & Singh, R. Challenges in liver cancer and possible treatment approaches. Biochimica et. Biophysica Acta (BBA)-Rev. Cancer1873, 188314 (2020). 10.1016/j.bbcan.2019.188314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel, R. L., Miller, K. D. & Jemal, A. Global cancer statistics 2019. CA: a cancer J. clinicians69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Torre, L. A. et al. Global cancer statistics, 2012. CA: a cancer J. clinicians65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 6.El-Serag, H. B. & Hepatocellular, K. L. Rudolph carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology132, 2557–2576 (2007). 10.1053/j.gastro.2007.04.061 [DOI] [PubMed] [Google Scholar]
- 7.Survarachakan, S. & Prasad, P. J. R. Deep learning for image-based liver analysis-a comprehensive review focusing on malignant lesions. Artif. Intell. Med130, 102331 (2022). 10.1016/j.artmed.2022.102331 [DOI] [PubMed] [Google Scholar]
- 8.Kondo, S. et al. Computer-aided diagnosis of focal liver lesions using contrast-enhanced ultrasonography with perflubutane microbubbles. IEEE Trans. Med. imaging36, 1427–1437 (2017). 10.1109/TMI.2017.2659734 [DOI] [PubMed] [Google Scholar]
- 9.Gatos, I. et al. Focal liver lesions segmentation and classification in nonenhanced t2-weighted mri. Med. Phys.44, 3695–3705 (2017). 10.1002/mp.12291 [DOI] [PubMed] [Google Scholar]
- 10.Mao, B. et al. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur. Radiol.30, 6924–6932 (2020). 10.1007/s00330-020-07056-5 [DOI] [PubMed] [Google Scholar]
- 11.Mao, B. et al. Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur. Radiol.31, 4576–4586 (2021). 10.1007/s00330-020-07562-6 [DOI] [PubMed] [Google Scholar]
- 12.Wu, K., Chen, X. & Ding, M. Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik125, 4057–4063 (2014). 10.1016/j.ijleo.2014.01.114 [DOI] [Google Scholar]
- 13.Oestmann, P. M. et al. Deep learning–assisted differentiation of pathologically proven atypical and typical hepatocellular carcinoma (hcc) versus non-hcc on contrast-enhanced mri of the liver. Eur. Radiol.31, 4981–4990 (2021). 10.1007/s00330-020-07559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, R. et al. Deep learning for differential diagnosis of malignant hepatic tumors based on multi-phase contrast-enhanced ct and clinical data. J. Hematol. Oncol.14, 1–7 (2021). 10.1186/s13045-021-01167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasaka, A. O. K. S. & Akai, K. H. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced ct: A preliminary study. Radiology286, 887–896 (2018). 10.1148/radiol.2017170706 [DOI] [PubMed] [Google Scholar]
- 16.Wang, M. et al. Development of an ai system for accurately diagnose hepatocellular carcinoma from computed tomography imaging data. Br. J. Cancer125, 1111–1121 (2021). 10.1038/s41416-021-01511-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, H. et al. Case discrimination: self-supervised feature learning for the classification of focal liver lesions. in: Innovation in Medicine and Healthcare: Proceedings of 9th KES-InMed 2021, Springer, pp. 241–249 (2021).
- 18.Zhang, D., Chen, B., Chong, J. & Li, S. Weakly-supervised teacher-student network for liver tumor segmentation from non-enhanced images. Med. Image Anal.70, 102005.345 (2021). 10.1016/j.media.2021.102005 [DOI] [PubMed] [Google Scholar]
- 19.Hansen, S., Gautam, S., Jenssen, R. & Kampffmeyer, M. Anomaly detection-inspired few-shot medical image segmentation through self-supervision with supervoxels. Med. Image Anal.78, 102385 (2022). 10.1016/j.media.2022.102385 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, X. et al. Self-supervised tumor segmentation with sim2real adaptation. IEEE J. Biomed. Health Inform.27, 4373–4384 (2023). 10.1109/JBHI.2023.3240844 [DOI] [PubMed] [Google Scholar]
- 21.Zhou, J. et al. Automatic detection and classification of focal liver lesions based on deep convolutional neural networks: a preliminary study. Front. Oncol.10, 581210.365 (2021). 10.3389/fonc.2020.581210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu, X. et al. A knowledge-guided framework for fine-grained classification of liver lesions based on multi-phase ct images. IEEE J. Biomed. Health Inform.27, 386–396 (2022). 10.1109/JBHI.2022.3220788 [DOI] [PubMed] [Google Scholar]
- 23.W. Wang, et al. Classification of focal liver lesions using deep learning with fine-tuning, in: Proceedings of the 2018 International Conference on digital medicine and image processing, pp. 56–60, 2018.
- 24.Balagourouchetty, L., Pragatheeswaran, J. K., Pottakkat, B. & Ramkumar, G. Googlenet-based ensemble fcnet classifier for focal liver lesion diagnosis. IEEE J. Biomed. health Inform.24, 1686–1694 (2019). 10.1109/JBHI.2019.2942774 [DOI] [PubMed] [Google Scholar]
- 25.Peng, S. et al. Deep learning-based artificial intelligence model to assist thyroid nodule diagnosis and management: a multicentre diagnostic study. Lancet Digital Health3, e250–e259 (2021). 10.1016/S2589-7500(21)00041-8 [DOI] [PubMed] [Google Scholar]
- 26.Li, X. et al. Development and validation of a novel computed-tomography enterography radiomic approach for characterization of intestinal fibrosis in crohn’s disease. Gastroenterology160, 2303–2316 (2021). 10.1053/j.gastro.2021.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaufrère, Aurélie, Calderaro, Julien & Paradis, Valérie Combined hepatocellular-cholangiocarcinoma: an update. J. Hepatol.74, 1212–1224 (2021). 10.1016/j.jhep.2021.01.035 [DOI] [PubMed] [Google Scholar]
- 28.Ramai, D., Ofosu, A., Lai, J. K., Reddy, M. & Adler, D. G. Combined hepatocellular cholangiocarcinoma: a population-based retrospective study. J. Hepatol.114, 1496–1501 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Calderaro, J. et al. Deep learning-based phenotyping reclassifies combined hepatocellular-cholangiocarcinoma. Nat. Commun.14, 8290 (2023). 10.1038/s41467-023-43749-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis, D. G. & Aizenberg M. R. (2021) Trialing U-Net Training Modifications for Segmenting Gliomas Using Open Source Deep Learning Framework. In: Crimi A., Bakas S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science, 12659. Springer, Cham. 10.1007/978-3-030-72087-2_4.
- 31.M. Yang yangmeiyi/Liver: Focal Liver Lesion Diagnosis with Deep Learning and Multistage CT Imaging (v1.0.0). Zenodo. 10.5281/zenodo.12655750 (2024). [DOI] [PMC free article] [PubMed]
- 32.K. He, X. Zhang, S. Ren, J. Sun. Deep residual learning for image recognition. in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778, (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. All the analytical data underpinning the findings of this study are incorporated within this paper in the designated source data files (Source_data_Figure_2.xlsx to Source_data_Figure_6.xlsx and Source_data_Table_2.xlsx to Source_data_Table_3.xlsx). However, the original dataset used in this study is subject to access control, requiring licenses from multiple central institutions for access. The author of the article communicated with several hospital departments, including the Information Technology Department, Research Laboratory, and International Cooperation Office. Although these data do not involve blood or other biological samples, the hospital’s strict data management policy prohibits any external sharing of research data. These regulations aim to ensure that all data, regardless of its nature, are securely protected to prevent unauthorized use or disclosure. To promote further international cooperation and exchange, it is recommended to jointly apply for international multicenter cooperation projects. This approach will enable us to conduct further research within the hospital while ensuring compliance with hospital policies, data security, and patient privacy. For academic inquiries regarding the use and processing of raw data, please contact the corresponding author via email at cjr.songbin@vip.163.com or csmliu@uestc.edu.cn. Source data are provided with this paper.
Our code is available at GitHub31 (https://github.com/yangmeiyi/Liver/tree/main). The trained model is available at Zenodo (https://zenodo.org/records/12646854). The trained model parameters can be accessed at Zenodo, including the detection model best.pt (https://zenodo.org/records/12646854/files/best.pt?download=1), the benign-malignant classification model Time_BM.pth.tar (https://zenodo.org/records/12646854/files/Time_BM.pth.tar?download=1), the benign classification model Time_B.pth.tar (https://zenodo.org/records/12646854/files/Time_B.pth.tar?download=1), and the malignant classification model Time_M.pth.tar (https://zenodo.org/records/12646854/files/Time_M.pth.tar?download=1).