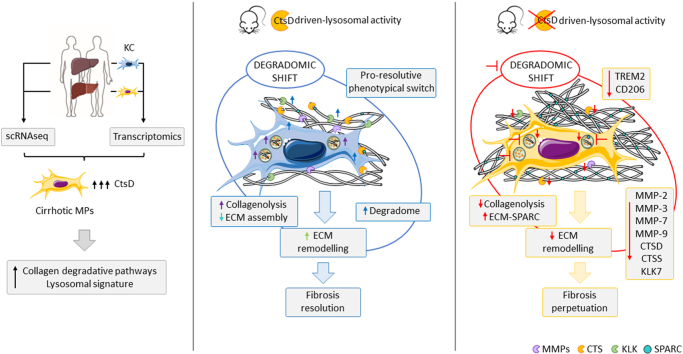
Abstract
Background and objectives
Fibrosis contributes to 45% of deaths in industrialized nations and is characterized by an abnormal accumulation of extracellular matrix (ECM). There are no specific anti-fibrotic treatments for liver fibrosis, and previous unsuccessful attempts at drug development have focused on preventing ECM deposition. Because liver fibrosis is largely acknowledged to be reversible, regulating fibrosis resolution could offer novel therapeutical options. However, little is known about the mechanisms controlling ECM remodeling during resolution. Changes in proteolytic activity are essential for ECM homeostasis and macrophages are an important source of proteases. Herein, in this study we evaluate the role of macrophage-derived cathepsin D (CtsD) during liver fibrosis.
Methods
CtsD expression and associated pathways were characterized in single-cell RNA sequencing and transcriptomic datasets in human cirrhosis. Liver fibrosis progression, reversion and functional characterization were assessed in novel myeloid-CtsD and hepatocyte-CtsD knock-out mice.
Results
Analysis of single-cell RNA sequencing datasets demonstrated CtsD was expressed in macrophages and hepatocytes in human cirrhosis. Liver fibrosis progression, reversion and functional characterization were assessed in novel myeloid-CtsD (CtsDΔMyel) and hepatocyte-CtsD knock-out mice. CtsD deletion in macrophages, but not in hepatocytes, resulted in enhanced liver fibrosis. Both inflammatory and matrisome proteomic signatures were enriched in fibrotic CtsDΔMyel livers. Besides, CtsDΔMyel liver macrophages displayed functional, phenotypical and secretomic changes, which resulted in a degradomic phenotypical shift, responsible for the defective proteolytic processing of collagen I in vitro and impaired collagen remodeling during fibrosis resolution in vivo. Finally, CtsD-expressing mononuclear phagocytes of cirrhotic human livers were enriched in lysosomal and ECM degradative signaling pathways.
Conclusions
Our work describes for the first-time CtsD-driven lysosomal activity as a central hub for restorative macrophage function during fibrosis resolution and opens new avenues to explore their degradome landscape to inform drug development.
Keywords: Fibrosis, Protease, Cathepsin, Resolution, Macrophage
Graphical abstract
Highlights
-
•
Human cirrhotic CtsD-expressing macrophages are enriched in ECM degradative pathways.
-
•
Myeloid-CtsD KO mice present enhanced liver fibrosis and impaired resolution.
-
•
CtsD contributes to functional, phenotypical and secretomic changes in macrophages.
-
•
CtsD-driven macrophage degradomic shift is essential for fibrosis resolution.
1. Introduction
Liver fibrosis is the result of a dysregulation of the normal wound-healing response due to repetitive damage or chronic injury. During liver fibrosis, replacement of the functional parenchyma by excessive electrodense extracellular matrix (ECM) results in architectural remodeling and a progressive stiffening of the liver, which can eventually lead to organ failure. It is currently estimated that fibrosis is a contributing factor to 45% of deaths in industrialized nations [1] and it exponentially increases liver-related mortality in nonalcoholic fatty liver disease patients [2]. Despite liver fibrosis being widely accepted to be reversible, little is known about the biological mechanisms controlling ECM remodeling and reabsorption. Changes in proteolytic activity are essential for ECM homeostasis, however, the proteolytic network involved in ECM deposition and reabsorption is far from understood. Considering that no specific treatments for liver fibrosis are currently available and that previous research efforts designing anti-fibrotic drugs focused on stopping ECM deposition have failed, increasing our biological understanding about the proteolytic pathways controlling ECM reabsorption can provide novel therapeutic approaches for the treatment of fibrotic diseases.
Cathepsins (Cts) are peptide-bond cleaving enzymes classically referred as lysosomal proteases. However, it is recognized that Cts can be found in other cellular locations and display activity outside their optimal acidic pH range [3]. They are classified into three families depending on their structure and catalytic sites: serine (CtsA and G), aspartate (CtsD and E) and cysteine (CtsB, C/DPP1, F, H, K, L, O, S, V, W and Z/X). Proteases finely tune several biological processes through efficient, highly selective, and limited cleavage of specific substrates, a process also known as proteolysis. Growing evidence suggests important roles for mainly cysteine Cts in the context of liver disease, participating in apoptosis, activation of hepatic stellate cells (HSC), autophagy, inflammation, matrix remodeling, lipid metabolism and cancer. However, our understanding of their specific targets and signaling networks remains very limited [3]. CtsB is one of the best studied Cts in the context of liver fibrosis, and it appears to have a dual role acting as a pro-apoptotic signal for hepatocytes [4] and a pro-fibrogenic signal for HSC [[5], [6], [7]]. Contrary to CtsB, the study of aspartyl CtsD has been challenging, partially due to the lack of an adult CtsD global knock-out mouse [8]. Recently, the generation of a CtsD floxed mouse [9] has opened new avenues to study the cell-specific role of CtsD during disease progression. Therefore, the current study was designed to evaluate CtsD cell-specific contribution to liver fibrosis. Our analysis of cirrhotic human tissues revealed high expression of CtsD in liver macrophages and hepatocytes, which was confirmed by the analysis of publicly available human single-cell RNA sequencing datasets. Hence, we generated two novel CtsD knock-out mice for myeloid cells (CtsDΔMyel) and hepatocytes (CtsDΔHep) to explore the contribution of CtsD expression in these cell types to liver fibrosis development. CtsD deletion in myeloid cells, but not in hepatocytes, resulted in increased liver fibrosis. In addition, proteomic analysis of fibrotic livers demonstrated an enrichment of the immune system and the matrisome signaling pathways in fibrotic livers from CtsDΔMyel mice. CtsDΔMyel liver macrophages displayed functional, phenotypical and secretomic changes that resulted in defective lysosomal proteolytic processing of collagen I in vitro and impaired collagen remodeling during fibrosis resolution in vivo. Analysis of CtsD mononuclear phagocyte (MP) subpopulations in cirrhotic human livers confirmed that CtsD-expressing subpopulations were differentially enriched in lysosomal and ECM containing and degradative signaling pathways. Our study demonstrates that CtsD is essential in regulating CtsD-driven lysosomal activity of macrophages, influencing their phenotype, secretomic profile and collagenolytic activity during liver fibrosis progression and resolution.
2. Methods
2.1. Generation of CtsD knock-out mouse strains for macrophages (CtsDΔMyel) and hepatocytes (CtsDΔHep)
To obtain the conditional KO mice for CtsD in myeloid cells or hepatocytes, LysMCre mice (Jackson Laboratory B6.129P2-Lyz2tm1(cre)Ifo/J) [10] or AlbCre (Jackson Laboratory B6.Cg-Tg(Alb-cre)21Mgn/J) [11] were bred with CtsD floxed mice [9]. Briefly, CtsDF/F LysMCre−/− or CtsDF/F AlbCre−/− females were bred with CtsDF/F LysMCre+/− or CtsDF/F AlbCre+/− males to generate CtsDF/F-LysMCre+/− (CtsDΔMyel) and CtsDF/F-LysMCre−/− (CtsDF/F) or CtsDF/F-AlbCre+/− (CtsDΔHep) and CtsDF/F-AlbCre−/− (CtsDF/F) littermates, respectively. CtsDΔMyel and CtsDΔHep were bred in heterozygosis and kept as separate strains. Genotyping was performed by PCR to detect the cre allele and floxed alleles in ear clipped biopsies and cre recombined allele in liver macrophages for the CtsDΔMyel or hepatocytes for the CtsDΔHep. Randomized littermates were used for the studies. Samples obtained from the animal studies were coded and histological analysis and scoring were performed blind. Males were preferentially used for the experimental models unless stated otherwise, as cathepsin D gene expression can be controlled by estrogens [12,13]. Females were included in all cell isolation experiments, as we did not detect any differences between male and female liver macrophages, and in the later in vivo experimental model of liver fibrosis reversion. All animal studies were done in accordance with the Comitè Ètic d'Experimentació Animal (CEEA-UB), under the approval of the Generalitat de Catalunya and according to Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. Primer sequences for genotyping are detailed in Table S1.
2.2. Liver fibrosis experimental animal models
8–12 weeks old males were preferentially used for the experimental models unless stated otherwise, as cathepsin D gene expression can be controlled by estrogens [12,13]. Hence, this can be translated into a higher variability of CtsD expression in females depending on their level of estrogens.
For the short-term chronic carbon tetrachloride (CCl4) model, CtsDΔMyel and CtsDF/F or CtsDΔHep and CtsDF/F littermates were injected intraperitoneally (IP) twice a week with 2 μL (CCl4/olive oil, 1:3 [v/v])/g body weight or olive oil for 8 weeks. For the bile duct ligation model, the bile duct was ligated for 7 or 14 days in CtsDΔMyel and CtsDF/F or for 14 days in CtsDΔHep and CtsDF/F mice. A sham operation was performed as control group. For the long-term chronic carbon tetrachloride (CCl4) model, CtsDΔMyel or CtsDF/F mice were injected intraperitoneally once a week with 2 μL (CCl4/olive oil, 1:10 [v/v])/g body weight or olive oil for 24 weeks. At least 6 animals were used per experimental group.
2.3. Liver reversion experimental animal model
Fibrosis was established for 4 weeks by intraperitoneal administration of 2 μL (CCl4/olive oil, 1:3 [v/v])/g body weight twice a week in CtsDΔMyel and CtsDF/F male and female mice. Endpoints were 24 h and 72 h after the last CCl4 dose for the peak and the resolution experimental groups, respectively.
2.4. Statistical analysis
Results are expressed as mean ± SEM unless otherwise stated in the figure legend. All p-values were calculated using 1 way ANOVA followed by Bonferroni's post-test, 2 way ANOVA followed by Turkey's or Bonferroni's post-test or two tailed unpaired student's t-test. ∗P ≤ 0.05; ∗∗P ≤ 0.01; ∗∗∗P ≤ 0.001 was considered statistically significant.
More details about the experimental procedures can be found in the supplementary material.
3. Results
3.1. CtsD expression increases in macrophages and serum from cirrhotic patients
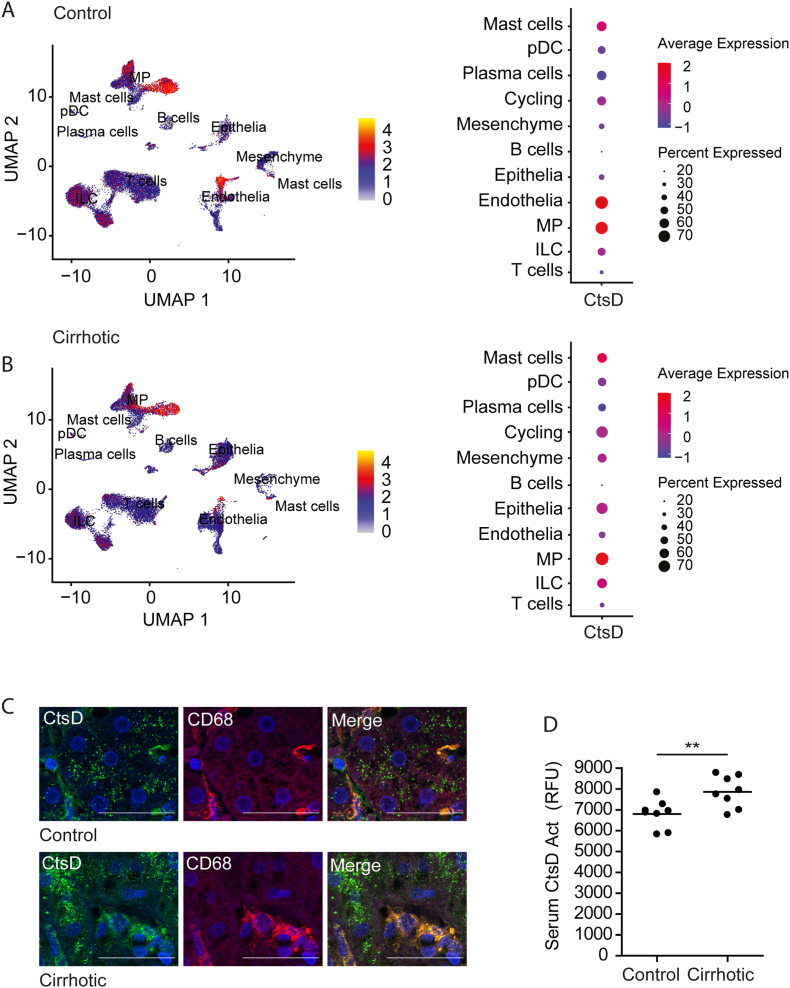
First, we investigated CtsD expression in publicly available single-cell RNA sequencing (scRNAseq) datasets from five normal and five cirrhotic livers [14]. Our analysis identified 11 common cell lineages, annotated according to the original publication (Fig. S1 and Table S2), across control and cirrhotic livers. From those, CtsD was predominantly expressed in the mononuclear phagocyte population in both control (Figure 1A) and cirrhotic (Figure 1B) livers, but it was also expressed to a lesser extend in endothelial and epithelial cells. Interestingly, while the percentage of CtsD-expressing cells increased in the cirrhotic epithelial cells, it decreased in cirrhotic endothelial cells. Next, we analyzed the expression of CtsD in tissue biopsies from control and cirrhotic patients. Immunofluorescent staining confirmed that CtsD was expressed in both macrophages (CD68+ve) and hepatocytes in control and cirrhotic tissues (Figure 1C), displaying a common cytosolic vesiculated pattern compatible with lysosomal expression, in agreement with classical studies performed by Yokota and co-workers in rat liver [15]. Further transcriptomic analysis of KCs isolated from control and cirrhotic human livers revealed a significant up-regulation of CtsD in human cirrhotic KCs (Log2 Fc = 2.63 p ≤ 0.001) [16]. Finally, we observed that CtsD activity was also significantly increased in serum from cirrhotic patients (Figure 1D).
Figure 1.
CtsD is elevated in human cirrhosis and is highly expressed in liver MPs.
UMAP projection depicting CtsD expression across cell types and dotplots representing percentage and average expression of CtsD within human (A) control and (B) cirrhotic livers (GSE136103). (C) Representative images of dual IF staining for CtsD (green) and CD68 (red) in human control and cirrhotic livers. (D) CtsD activity in serum from control and cirrhotic patients. Scale bars represent 20 μm. T-test analysis, ∗∗p ≤ 0.01.
3.2. Characterization of CtsDΔMyel and CtsDΔHep mice reveals normal basal phenotype
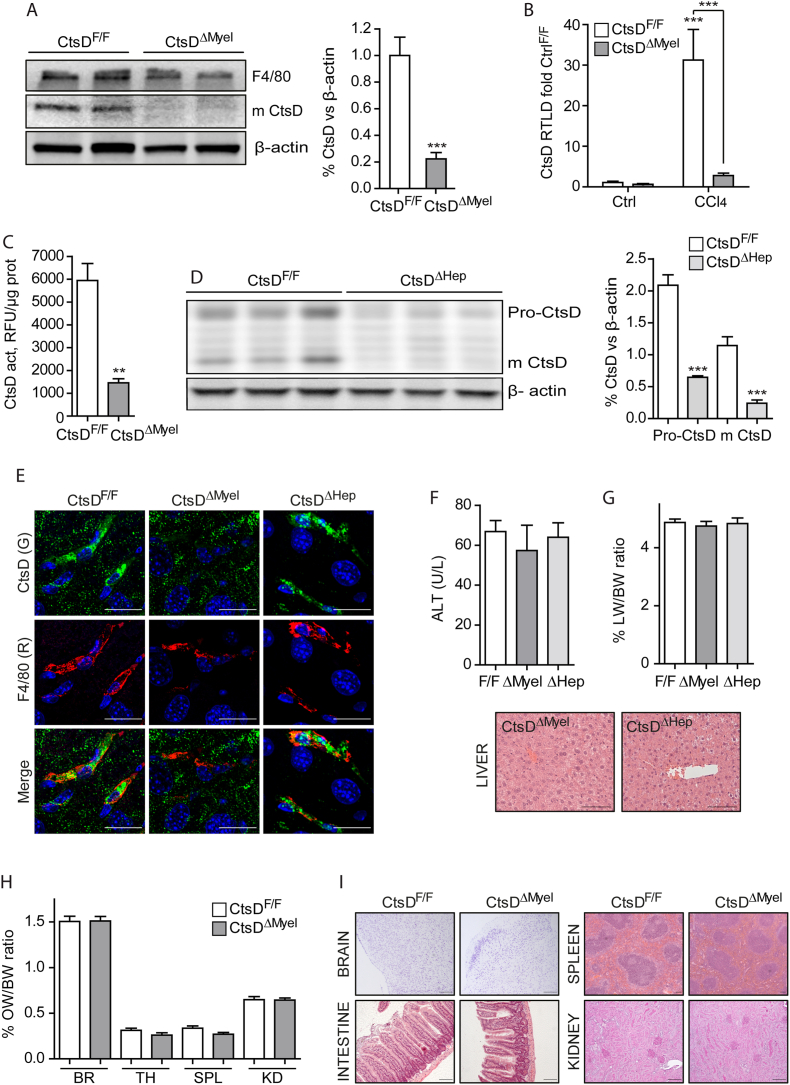
Our results demonstrated that macrophages and, to a lesser extent, hepatocytes, are the main cell types expressing CtsD in human cirrhosis. Thus, to better understand the role of CtsD in fibrosis development we generated two novel CtsD cell-specific knock-out mice for myeloid lineage cells including liver macrophages, both resident [17] and infiltrated [18], or for hepatocytes by breeding CtsD floxed mouse [9] with LysMCre [10] or AlbCre [11] mice respectively, from now on referred as CtsDΔMyel or CtsDΔHep. To validate CtsD floxed allele recombination in liver macrophages, we assessed CtsD expression by WB in liver macrophages isolated from CtsDF/F and CtsDΔMyel mice, demonstrating a significant decrease of CtsD in F4/80+ve cells from CtsDΔMyel mice (Figure 2A). In agreement with results obtained from the human cirrhotic liver macrophages, CtsDF/F liver macrophages isolated from CCl4 injured livers displayed a significant increase in CtsD gene expression (Figure 2B) and activity (Figure 2C) that was absent in CtsDΔMyel liver macrophages. Validation of CtsDΔHep mouse was carried out by WB in primary mouse hepatocytes isolated from CtsDF/F and CtsDΔHep mice (Figure 2D), demonstrating a lack in CtsD expression in hepatocytes from CtsDΔHep mice. Of note, no significant differences in the gene expression of other cathepsins (CtsA, B, C, L, S, X/Z) between phenotypes were observed in liver macrophages nor in hepatocytes (Figs. S2A–B). Similarly to what was observed in human cirrhotic livers, mouse CtsDF/F fibrotic livers displayed the strongest CtsD signal in macrophages (F4/80+ve cells) and a mild signal in hepatocytes with a cytosolic vesiculated pattern compatible with lysosomes. As expected, CtsD signal in macrophages or in hepatocytes from CtsDΔMyel or CtsDΔHep livers respectively was not detected (Figure 2E). Next, analysis of serum alanine aminotransferase (ALT) and liver histology revealed no liver damage and normal liver histology in both CtsDΔMyel and CtsDΔHep adult healthy mice (Figure 2F) and no differences in the liver to body weight ratios (Figure 2G). In addition, organ to body weight ratios and histology of extrahepatic tissues (brain, spleen, intestine and kidneys) from adult healthy CtsDΔMyel mouse, which are affected by the widespread deletion of CtsD in the global CtsD KO mouse [8], demonstrated no affectation in comparison with CtsDF/F mouse (Figure 2H–I).
Figure 2.
Characterization and validation of CtsDΔMyeland CtsDΔHepmice.
(A) F4/80, mature CtsD and β-actin expression by WB from liver macrophages of CtsDF/F and CtsDΔMyel mice and WB quantification. (B)CtsD gene expression of liver macrophages isolated from CtsDF/F and CtsDΔMyel mice treated ± CCl4 for 72 h and (C) CtsD activity in liver macrophages isolated from CtsDF/F and CtsDΔMyel mice treated with CCl4 for 72 h. (D) Pro-CtsD, mature CtsD and β-actin expression by WB of hepatocytes from CtsDF/F and CtsDΔHep mice and WB quantification. (E) CtsD (green) and F4/80 (red) dual IF in fibrotic liver tissue from CtsDF/F, CtsDΔMyel and CtsDΔHep mice. (F) Serum ALT and liver H&E representative images from CtsDF/F, CtsDΔMyel and CtsDΔHep mice. (G) Liver to body weight ratio in CtsDF/F, CtsDΔMyel and CtsDΔHep mice. (H) Organ to body weight ratios from brain (BR), thymus (TH), spleen, (SPL) and kidney (KD) and (I) representative images of brain, spleen, intestine and kidney tissues from 10-week-old CtsDF/F and CtsDΔMyel mice stained with Cresyl violet for the brain, H&E for the spleen and intestine and PAS for the kidney. Scales represent 20 μm in panel E and 50 μm in panel F and I. Data in A and E is expressed as percentage of CtsD signal versus β-actin. 1 way ANOVA or T-test analysis, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
3.3. Liver fibrosis is exacerbated in CtsDΔMyel mouse, but not in CtsDΔHep one
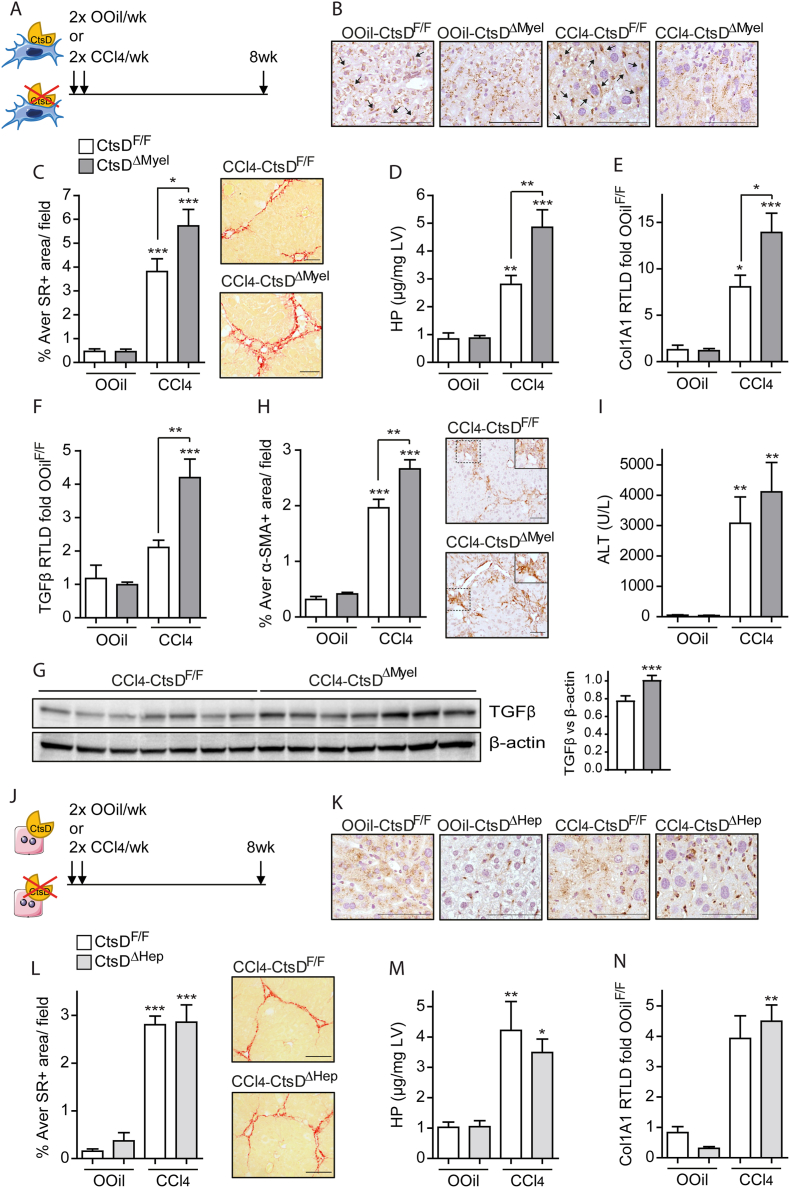
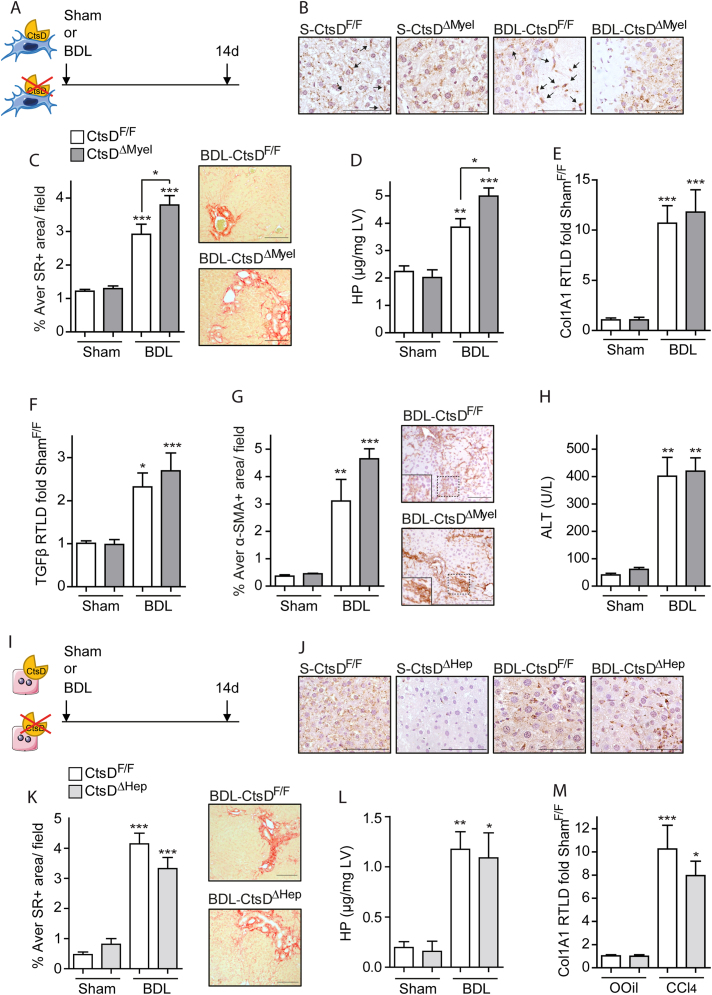
Next, we investigated liver fibrosis development in CtsDΔMyel or CtsDΔHep in comparison with CtsDF/F mice. To induce liver fibrosis, both CtsDF/F and CtsDΔMyel or CtsDF/F and CtsDΔHep littermates were injected with CCl4 or Olive Oil, as study controls, twice a week for 8 weeks as depicted in Figure 3A or Figure 3J. As expected, CtsD IHC demonstrated lack of CtsD in tissue macrophages from OOil-CtsDΔMyel and 8wkCCl4-CtsDΔMyel livers (Figure 3B) or in hepatocytes from OOil-CtsDΔHep and 8wkCCl4-CtsDΔHep livers (Figure 3K). When studying matrix deposition, liver collagen content was increased in all CCl4-treated groups, however, only 8wkCCl4CtsDΔMyel but not 8wkCCl4CtsDΔHep, displayed significantly enhanced collagen deposition as determined by Sirius Red morphometric analysis of tissue liver sections (Figure 3C,L) and liver hydroxyproline content (Figure 3D,M). Classical fibrogenic markers Col1A1 and TGF-β were assessed by RT-qPCR and WB, last one only for TGF-β (Figure 3E–G), and α-SMA expression was determined by IHC (Figure 3H) in liver tissue. All of them were significantly increased only in 8wkCCl4-CtsDΔMyel livers compared to 8wkCCl4-CtsDF/Fones, but not in 8wkCCl4-CtsDΔHep (Figures 3N and S3A-B). Of note, liver damage (serum ALT) was increased after CCl4 treatment but no significant differences were observed between phenotypes (Figures 3I and S3C) after CCl4 administration. Prolonged 6-month administration of CCl4, at a lower dose (Figure S3D), resulted in comparable results to those obtained with the 8 weeks CCl4 model, demonstrating increased liver collagen accumulation and α-SMA expression in 6MCCl4-CtsDΔMyel compared to 6MCCl4-CtsDF/F mice (Fig. S3E–H).
Figure 3.
CtsDΔMyelmice, but not CtsDΔHep, display enhanced liver fibrosis after 8-weeks CCl4administration.
(A) Experimental design of the 8-weeks CCl4 study in CtsDΔMyel mice. (B) Representative liver tissue images of CtsD IHC (black arrows indicate CtsD+ve macrophages), (C) morphometric analysis and representative images of Sirius Red (SR)+ve area/field (D) liver hydroxyproline quantification, liver (E)Col1A1 and (F)TGF-β gene expression from 8-weeks Olive Oil (OOil) or CCl4 treated CtsDF/F and CtsDΔMyel mice. (G) WB for TGF-β and β-actin in liver lysates from 8wkCCl4-CtsDF/F and 8wkCCl4-CtsDΔMyel mice and WB quantification. (H) Morphometric analysis of α-SMA+ve area/field with representative images with zoomed area and (I) serum alanine aminotransferase from 8-weeks Olive Oil (OOil) or CCl4 treated CtsDF/F and CtsDΔMyel mice. (J) Experimental design of the 8-weeks CCl4 study in CtsDΔHep mice. (K) Representative liver tissue images of CtsD IHC; (L) morphometric analysis and representative images of Sirius Red (SR)+ve area/field; (M) liver hydroxyproline quantification and (N) liver Col1A1 gene expression from 8 weeks Olive Oil (OOil) or CCl4 treated CtsDF/F and CtsDΔHep mice. Scale bars represent 50 μm. 1-way ANOVA, ∗p ≤ 0.05, ∗∗p ≤ 0.01 or ∗∗∗p ≤ 0.001.
To determine whether the effect of CtsD deletion in macrophages or hepatocytes on fibrosis was common to different types of injury, we performed bile duct ligation (BDL) or sham surgery in CtsDF/F and CtsDΔMyel mice (Figure 4A) or CtsDF/F and CtsDΔHep mice (Figure 4I) for 14 days. CtsD deletion in macrophages and hepatocytes was confirmed by IHC in liver tissue from CtsDΔMyel or CtsDΔHep mice, respectively (Figure 4B,J). In agreement with the 8wk-CCl4 model, BDL-CtsDΔMyel mice displayed increased liver collagen deposition (Figure 4C–D) but no differences were observed in BDL-CtsDΔHep mice when comparing with their BDL-CtsDF/F littermates (Figures 4K–M and S4A,B). Unlike the previous study, no significant differences were detected in Col1A1, TGF-β or α-SMA expression between CtsDΔMyel and CtsDF/F after 14 days of BDL (Figure 4E–G). Nevertheless, early changes in gene expression showing increased Col1A1, TGF-β and ACTA2 were demonstrated 7 days after BDL in CtsDΔMyel compared to CtsDF/F (Figs. S4D–G). Finally, as in the 8wk-CCl4 model, no significant differences were observed in liver damage after BDL amongst the different phenotypes studied (Figures 4H, S4C and S4H). Our results demonstrate that CtsD deletion in macrophages, but not in hepatocytes, contributes to an enhancement in liver fibrosis in two etiologically different models of experimental fibrosis without inducing changes in liver damage.
Figure 4.
CtsD deletion in macrophages, but not in hepatocytes, exacerbates liver fibrosis after BDL.
(A) Experimental design of the 14-day BDL study in CtsDΔMyel mice. (B) Representative liver tissue images of CtsD IHC (black arrows indicate CtsD+ve macrophages), (C) morphometric analysis and representative images of Sirius Red (SR)+ve area/field (D) liver hydroxyproline quantification, liver (E)Col1A1 and (F)TGF-β gene expression, (G) morphometric analysis of α-SMA+ve area/field and representative images with zoomed area and (H) serum alanine aminotransferase from 14 days Sham or BDL CtsDF/F and CtsDΔMyel mice. (I) Experimental design of the 14-day BDL study in CtsDΔHep mice. (J) Representative liver tissue images of CtsD IHC; (K) morphometric analysis and representative images of Sirius Red (SR)+ve area/field; (L) liver hydroxyproline quantification and (M) liver Col1A1 gene expression from 14 days Sham or BDL CtsDF/F and CtsDΔHep mice. Scale bars represent 50 μm. 1-way ANOVA, ∗p ≤ 0.05, ∗∗p ≤ 0.01 or ∗∗∗p ≤ 0.001.
3.4. Fibrotic CtsDΔMyel livers present enhanced inflammatory proteomic signature
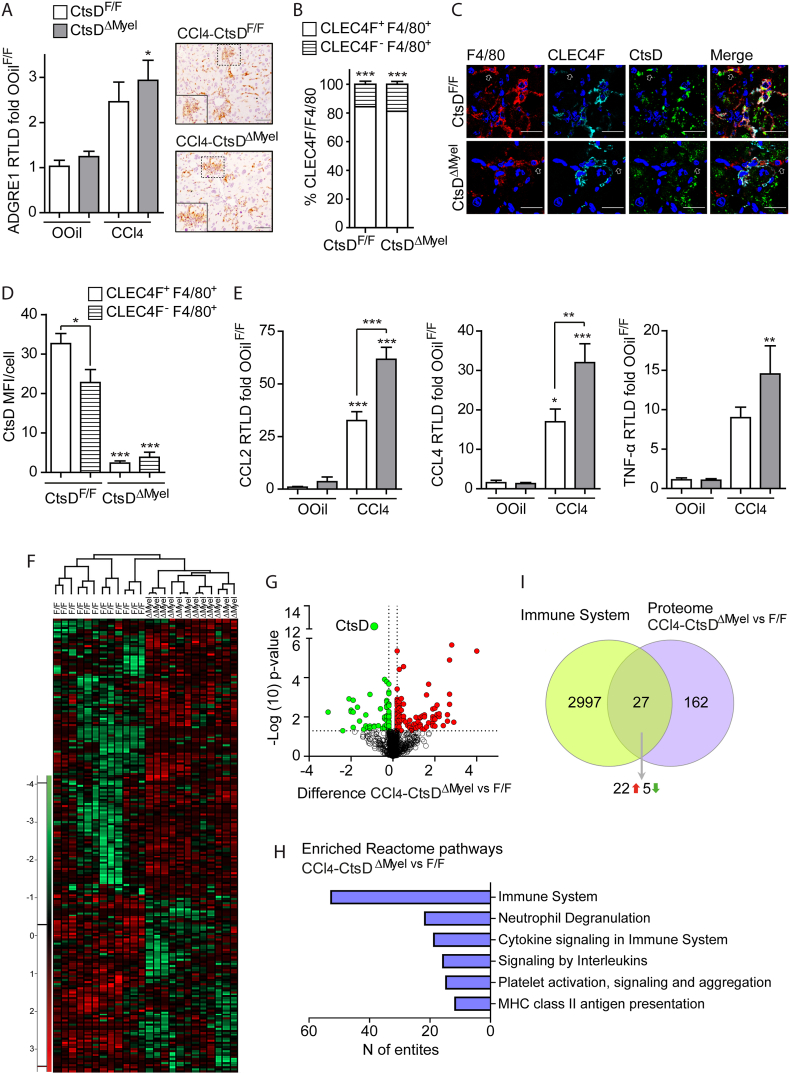
Both Kupffer cells (KCs) and infiltrated bone marrow monocyte-derived macrophages (MoMϕs) play an essential role during the immune response followed by any liver injury, which directly contributes to the initiation and development of the fibrotic response. For that reason, we evaluated the inflammatory response during liver fibrosis in CtsDF/F and CtsDΔMyel mice after both 8wk-CCl4 administration and BDL. As expected, fibrotic livers displayed an increase in macrophages (F4/80+ve) (Figures 5A and S5A), despite no differences between CtsDΔMyel and CtsDF/F mice were detected. Closer characterization of liver macrophages from fibrotic livers revealed that a significantly high proportion of macrophages (>80%) from 8wk-CCl4 livers were KCs (CLEC4F+ve-F4/80+ve) in both CtsDF/F and CtsDΔMyel mice. Besides, no differences in this proportion were observed between phenotypes (Figure 5B). Analysis of CtsD expression in CLEC4F+ve and CLEC4F-ve macrophages demonstrated that CtsD expression was significantly higher in KCs, which represented the main liver macrophage population in our model (Figure 5C–D). As expected, almost no CtsD expression was detected in CtsDΔMyel liver macrophages (Figures 5C–D and S5B). Of note, fibrotic livers also displayed an increase in granulocytes (NIMP+ve) and neutrophils (MPO+ve) (Fig. S5C–D), but no differences between phenotypes were detected. Similar results were obtained when analyzing neutrophil chemotactic proteins S100A8 and CXCL1 (Fig. S5E–F). However, significantly higher levels of classical inflammatory mediators CCL2, CCL4 and TNF-α were displayed by 8wkCCl4-CtsDΔMyel livers in comparison with 8wkCCl4-CtsDF/F ones (Figure 5E). WB for CCL2 in total liver also demonstrated an increase in 8wkCCl4-CtsDΔMyel livers despite not being statistically significant (Fig. S5G).
Figure 5.
CtsDΔMyelfibrotic livers present distinct and enriched immune and inflammatory proteomic profile.
(A) Liver ADGRE1 (F4/80) gene expression in liver from 8-weeks Olive Oil (OOil) or CCl4 treated CtsDF/F and CtsDΔMyel mice and representative images of F4/80 IHC with zoomed area in liver tissue sections; (B) percentage CLEC4F+ve-F4/80+ve or CLEC4F-ve-F4/80+ve cells/field; (C) representative images of triple IF staining for F4/80 (red), CLEC4F (cyan) and CtsD (green). White arrows point CLEC4F-ve-F4/80+ve cells; (D) CtsD MFI within CLEC4F+ve-F4/80+ve or CLEC4F-ve-F4/80+ve cells in liver tissue sections from 8-weeks CCl4 treated CtsDF/F and CtsDΔMyel mice. (E)CCL2, CCl4 and TNF-α gene expression in liver from 8-weeks Olive Oil (OOil) or CCl4 treated CtsDF/F and CtsDΔMyel mice. (F) Heatmap for the proteome distribution based on the Z-score-normalized LFQ abundances throughout all the samples. Green and red color ranges refer to lower and higher abundance, respectively; (G) Volcano plot of differentially regulated proteins. Green and red dots show the significant down- and up-regulated proteins, respectively, with highlight of CtsD protein; (H) Statistically significant enriched Reactome immune pathways in livers from CCl4-treated CtsDΔMyel versus CtsDF/F mice; (I) Eulero-Venn diagram with common hits between the differential proteome of CtsDΔMyel mice and the genes of the immune system from the MGI database. Scale bars represent 50 μm. 1-way ANOVA, ∗p ≤ 0.05, ∗∗p ≤ 0.01 or ∗∗∗p ≤ 0.001.
In agreement, early changes in gene expression were detected 7 days after BDL also demonstrating an increased expression of TNF-α, but not CCL2 and CCL4 (Figs. S5H–J). A label-free quantitative (LFQ) proteomic analysis was performed comparing fibrotic 8wkCCl4-CtsDΔMyel versus 8wkCCl4-CtsDF/F livers, showing differential protein expression clustering for both phenotypes (Figure 5F). The LFQ analysis quantified 1333 proteins (identified by a minimum of 2 unique peptides) and 189 proteins were detected as differentially expressed, of which 121 were up- and 68 down-regulated in the 8wkCCl4-CtsDΔMyel livers (Figure 5G). Pathway enrichment analysis revealed significantly enriched Reactome immune pathways (p < 0.01) in fibrotic CtsDΔMyel livers (Figure 5H). In addition, Eulero-Venn diagram intersecting the differential proteome of 8wkCCl4-CtsDΔMyel livers and the genes of the immune system from the MGI database demonstrated 27 common hits, 22 of which were up-regulated (Figure 5I). Of note, neutrophil degranulation pathway was also enriched in fibrotic CtsDΔMyel livers. Considering that CtsD deletion in CtsDΔMyel mouse might also affect neutrophils, we further investigated the degranulation capacity of neutrophils isolated from bone marrow of CtsDΔMyel mouse compared to CtsDF/F ones. Activity analysis of secreted neutrophil elastase (NE), one of the main proteases stored in neutrophil azurophilic granules, after PMA stimulation showed no significant differences in neutrophil degranulation between phenotypes (Fig. S5K).
Our observations demonstrate an enhanced inflammatory signature in fibrotic CtsDΔMyel livers with no differences in the total numbers of liver macrophages or neutrophils, pointing towards a differential phenotypical change of CtsDΔMyel inflammatory cells as responsible for the enriched inflammatory proteomic profile.
3.5. CtsD deficient liver macrophages exhibit functional, phenotypical and secretomic alterations compatible with impaired pro-resolutive shift
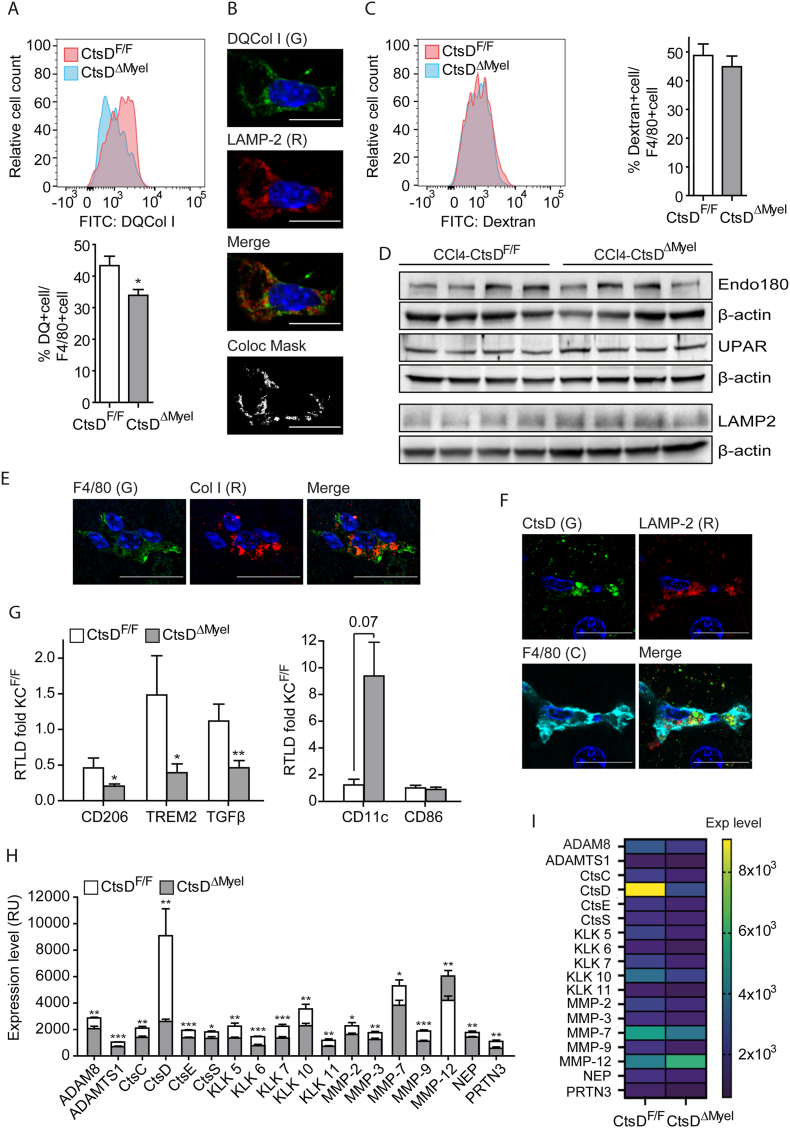
Macrophages not only play an important role as drivers of the inflammatory response during liver fibrosis but can also participate in the remodeling and recycling of the extracellular matrix. Thus, we decided to analyze whether phenotypical changes in CtsDΔMyel macrophages also affected their collagenolytic capacity. First, Eulero-Venn analysis was performed combining the differential proteome of 8wkCCl4-CtsDΔMyel livers and the databases for the matrisome, cytoskeleton and lysosome, revealing 9, 22 and 5 common hits, respectively. Most of the hits identified were up-regulated, confirming an increased abundance of proteins within these signaling pathways in fibrotic CtsDΔMyel livers (Fig. S6A). Next, collagenolytic activity of isolated liver macrophages was assessed using DQ Collagen I probe, which only emits fluorescence when is proteolytically cleaved. Proteolytic processing of DQ Collagen I was significantly decreased in CtsDΔMyel liver macrophages in comparison with CtsDF/F ones (Figure 6A) and partially colocalized with the lysosomal marker LAMP2, confirming DQ Collagen I processing was occurring within the lysosomes (Figure 6B). To prove whether decreased DQ Collagen I signal was caused by impaired endocytosis, dextran internalization assay was performed in liver macrophages isolated from CtsDF/F and CtsDΔMyel mice demonstrating similar levels of endocytosis independent of CtsD presence (Figure 6C). In addition, no differences were observed in the expression of the collagen internalization receptor, UPARAP/Endo 180, its co-receptor UPAR and the lysosomal marker LAMP2 in 8wkCCl4-CtsDF/F and 8wkCCl4-CtsDΔMyel fibrotic livers (Figure 6D). Supporting our results in liver macrophages, collagen I was detected within F4/80+ve macrophages in CCl4-treated livers (Figure 6E) and CtsD showed a lysosomal distribution (LAMP-2) in liver macrophages (Figure 6F). Pro-resolutive macrophages are responsible for ECM remodeling and recycling during fibrosis resolution. Hence, a decrease in the lysosomal collagenolytic activity of liver macrophages might be an indication of a wider defective pro-resolutive phenotype of macrophages. To study this, liver macrophages were isolated from CtsDF/F and CtsDΔMyel CCl4 treated mice. Indeed, CtsDΔMyel hepatic macrophages displayed a significant decrease in the expression of pro-resolutive markers such as CD206, TREM2 and TGF-β and an increase in the pro-inflammatory marker CD11c, with no changes in CD86 (Figure 6G). Despite some reports indicate that CtsD is able to directly process collagen I [19] and IV [20], it is unlikely to be the only protease responsible for the phenotype observed in the CtsDΔMyel mice, as proteases do not operate alone but in networks of several different proteases. Thus, we decided to investigate CtsDΔMyel liver macrophage secretory protease signature to further elucidate CtsD associated protease network. Conditioned medias from hepatic macrophages isolated after an acute dose of CCl4 were run in a protease array. Protease array detected 33 proteases, 18 of which were differentially secreted in CtsDΔMyel liver macrophages (Figures 6H–I and S6B) and could be organized in an interactome (Figs. S6C–D). Most of the proteases detected were down-regulated in CtsDΔMyel liver macrophages including MMP-2 [21,22] and MMP-9 [[21], [22], [23]], which are both well-described gelatinases, responsible for the degradation of collagen during liver fibrosis. In addition, other less well-known enzymes with ECM degrading activity, such as CtsS [24,25], KLK7 [26], MMP-3 [21,23] and MMP-7 [23] were also significantly less secreted by CtsDΔMyel liver macrophages. Surprisingly, MMP-12 was the only enzyme whose secretion was significantly up-regulated in CtsDΔMyel hepatic macrophages. Interestingly, CtsD secreted by hepatic macrophages was proteolytically active and significantly downregulated in CtsDΔMyel mice (Fig. S6E). Our results demonstrate CtsDΔMyel liver macrophages present deficient pro-resolutive characteristics, including defective lysosomal proteolytic processing of collagen, and altered secretomic proteolytic profile, which contributes to the enhanced liver fibrosis displayed by these mice.
Figure 6.
CtsDΔMyelliver macrophages exhibit phenotypical and secretomic alterations compatible with impaired pro-resolutive shift.
(A) Flow cytometry plot of relative cell counts and quantification of DQ collagen I expression in F4/80+ve liver macrophages isolated from CtsDF/F and CtsDΔMyel mice. (B) Representative confocal images and colocalization mask of CtsDF/F liver macrophages treated with DQCol I (green) and stained for LAMP-2 (red). (C) Flow cytometry plot of relative cell counts and quantification of Dextran internalization in F4/80+ve liver macrophages isolated from CtsDF/F and CtsDΔMyel mice. (D) WB for Endo180, UPAR, LAMP2 and their respective β-actin in liver lysates from 8wkCCl4-CtsDF/F and 8wkCCl4-CtsDΔMyel mice. Representative images of IF staining (E) for F4/80 (green) and Collagen I (red) and (F) for CtsD (green), LAMP-2 (red) and F4/80 (cyan) in liver tissue from 8wkCCl4-CtsDF/F mice. (G)CD206, TREM2, TGF-β, CD11c and CD86 gene expression in liver macrophages isolated from CtsDF/F and CtsDΔMyel mice treated with CCl4 for 72 h. (H) Average protein expression level and (I) heatmap of proteases differentially regulated in conditioned media of KC isolated from CtsDF/F or CtsDΔMyel mice treated 72 h with CCl4. Scale bars represent 10 μm for B and 20 μm for E-F. T-test analysis, ∗p ≤ 0.05, ∗∗p ≤ 0.01 or ∗∗∗p ≤ 0.001.
3.6. Changes in CtsDΔMyel liver macrophages result in impaired fibrosis resolution
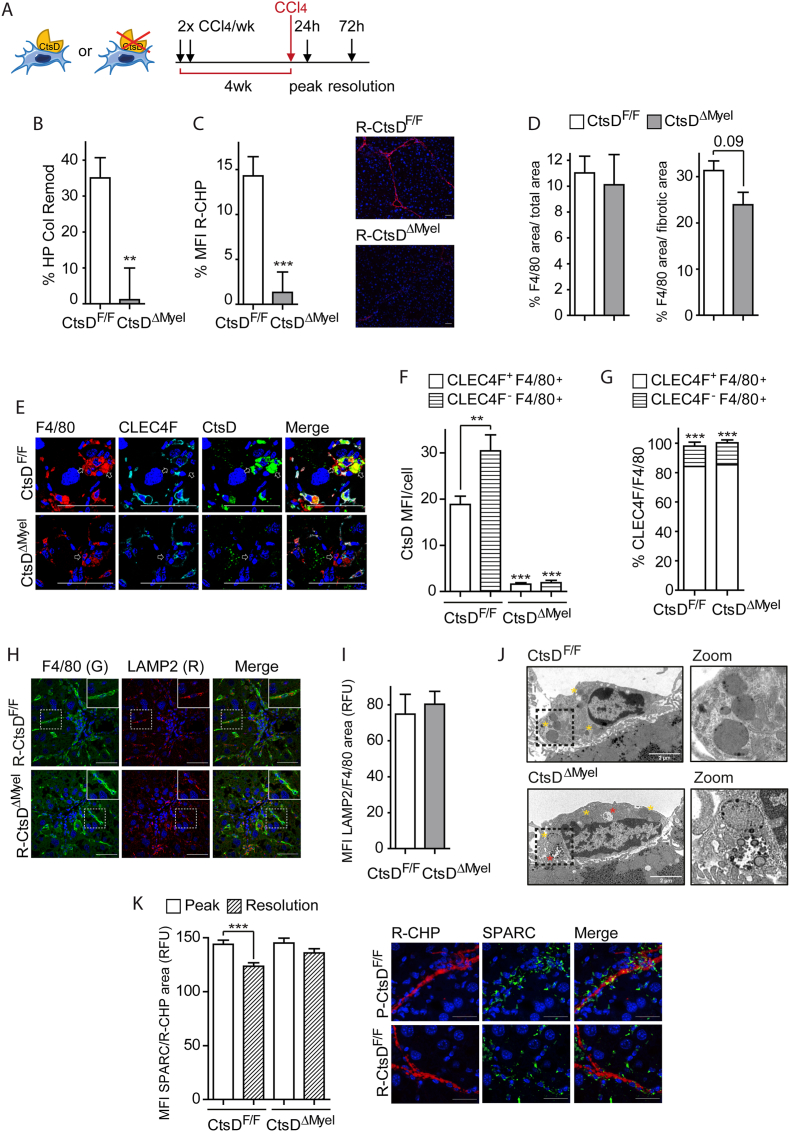
To analyze whether these phenotypical and secretomic alterations in CtsDΔMyel liver macrophages translated into changes in fibrosis resolution in vivo, we administered CCl4 for 4 weeks and established two endpoints after the last dose of CCl4, 1 day for the peak-CCl4 and 3 days for the resolution-CCl4 (Figure 7A). The percentage of liver collagen remodeling during resolution was assessed by hydroxyproline (Figure 7B) and liver staining of denatured collagen chains using fluorescently labelled Gly-X-Y collagen hybridizing peptides (Figure 7C) demonstrating that collagen remodeling and degradation was significantly decreased during resolution in CtsDΔMyel mice. Further study of liver tissues during the resolution phase showed that, despite no significant differences were observed in the total area occupied by F4/80+ve cells, the percentage of macrophages surrounding the fibrotic area was decreased in the CtsDΔMyel livers, supporting the idea that macrophages are not properly fulfilling their role during resolution (Figure 7D). In addition, and opposite to our observation in the 8wk-CCl4 livers, during the resolution phase CtsD expression was significantly higher in MoMϕs (CLECF4-ve-F4/80+ve) instead of in the KCs (Figure 7E–F), despite similar proportion of KCs and MoMϕs were maintained as in the 8wk-CCl4 model (Figure 7G). Closer analysis of the lysosomal content of liver macrophages during the resolution phase demonstrated similar levels between phenotypes (Figure 7H–I), but analysis by TEM revealed the presence of abnormal lysosomal structures in CtsDΔMyel liver macrophages from normal adult livers (Figure 7J). These findings might underpin an alteration on lysosomal function, but not lysosome numbers, as responsible for the secretomic and phenotypical changes observed in CtsDΔMyel mice. Besides the direct effect that CtsDΔMyel liver macrophage altered secretome (Figures 6H–I and S6C,D) can have over ECM remodeling, the imbalance in extracellular protease content might also exert an indirect effect over the function of proteins that participate in the ECM assembly and organization. SPARC (secreted protein acidic and rich in cysteine), which is a collagen-binding matricellular protein that participates in collagen I assembly, exhibits 13 cleaving sites for CtsD, spanning 18–303 residue range (Table S3) [27]. Analysis of SPARC expression within the collagen I area demonstrated that while in CtsDF/F mice there was a significant reduction in the expression of SPARC associated with collagen during resolution, no changes were observed in CtsDΔMyel mice (Figure 7K). These results might indicate that during fibrosis resolution active CtsD secreted by macrophages (Fig. S6E) could proteolytically process SPARC, facilitating ECM remodeling and degradation (Figure 7K). Lack of secreted active CtsD would result in sustained SPARC-collagen association, contributing to fibrosis perpetuation. Our results revealed that phenotypical and secretomic alterations of CtsDΔMyel macrophages caused suboptimal collagen remodeling in vivo, hampering fibrosis resolution and perpetuating liver fibrosis.
Figure 7.
Phenotypical and secretomic alterations of CtsDΔMyelliver macrophages causes deficient fibrosis resolution in vivo.
(A) Diagram displaying experimental setting for the fibrosis resolution model. (B) Percentage of HP collagen remodeling in liver tissue, (C) percentage of mean fluorescent intensity (MFI) of R-CHP staining in liver sections and representative R-CHP images from resolution-CCl4 CtsDF/F and CtsDΔMyel mice. (D) Percentage F4/80 area in total area or fibrotic area, (E) representative images of triple IF staining for F4/80 (red), CLEC4F (cyan) and CtsD (green). White arrows point CLEC4F-ve-F4/80+ve cells; (F) CtsD MFI within CLEC4F+ve-F4/80+ve or CLEC4F-ve-F4/80+ve cells; (G) percentage CLEC4F+ve-F4/80+ve or CLEC4F-ve-F4/80+ve cells/field in liver tissue sections from resolution-CCl4 CtsDF/F and CtsDΔMyel mice. (H) Representative images of dual IF of F4/80 (green) and LAMP2(R) with zoomed area and (I) LAMP2 MFI within F4/80 area in liver sections from resolution-CCl4 CtsDF/F and CtsDΔMyel mice. (J) Representative liver images of transmission electron microscopy displaying liver macrophages with zoom in lysosomal structures from CtsDF/F and CtsDΔMyel mice. (K) SPARC MFI within R-CHP area in liver sections from peak- and resolution-CCl4 CtsDF/F and CtsDΔMyel mice and representative images of dual IF staining for R-CHP (red) and SPARC (green) in liver sections from peak- and resolution-CCl4 CtsDF/F mice. Scale bars represent 50 μm. T-test analysis, ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001.
3.7. Human cirrhotic CtsD-expressing MPs subpopulations are enriched in ECM degradation pathways
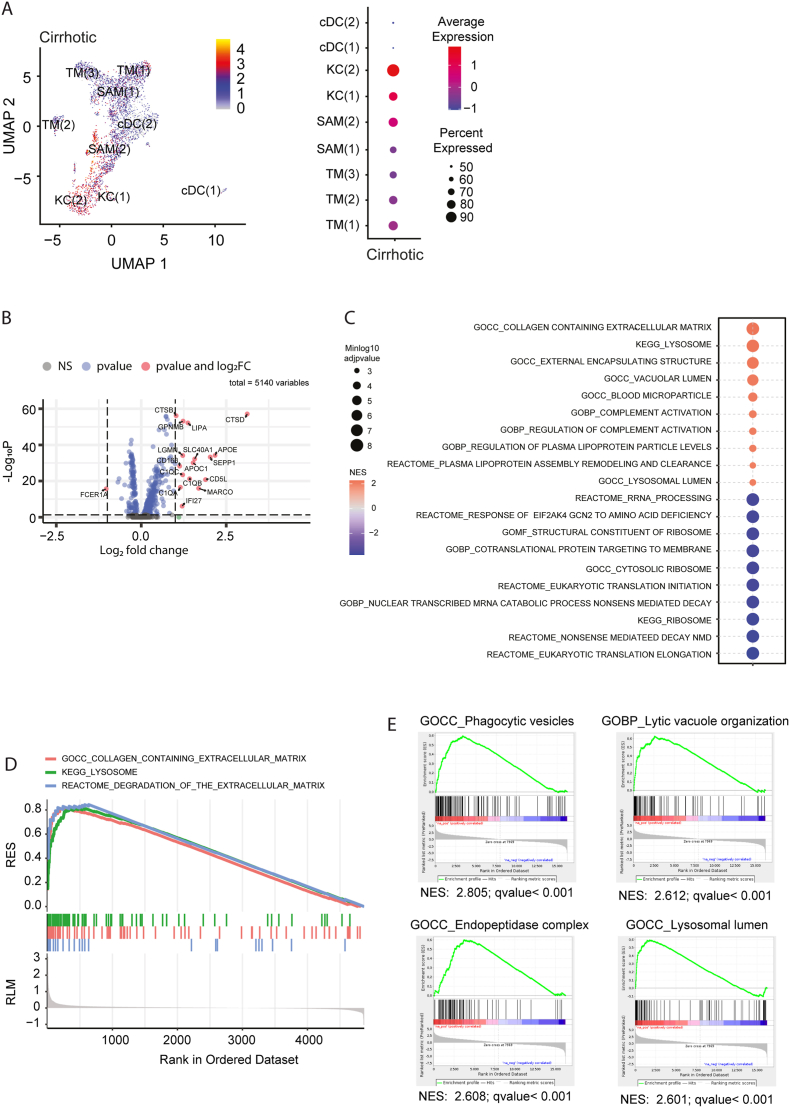
To investigate the potential biological translation of our results in mouse liver fibrosis into human cirrhosis, we performed further analysis of the single-cell transcriptomes [14]. Specific analysis of CtsD-expressing MPs identified KCs and SAM(2) as highly expressing CtsD (Figure 8A and Fig. S7A–G) in human cirrhotic livers. Analysis of differentially expressed transcripts between CtsD-expressing and non-expressing MPs in cirrhotic livers revealed 17 differentially expressed genes, 16 of which were upregulated and included the previously described pro-resolutive marker GPNMB [28,29] (Figure 8B). Additional heatmap analysis demonstrated a distinct gene expression pattern of the highest-CtsD expressing MPs subpopulations (KC(1), KC(2) and SAM(2)) in cirrhotic livers (Fig. S8A). Volcano plot comparing the differential expression of these three MPs subpopulations versus the other MPs confirmed these subpopulations as the ones highly expressing the macrophage pro-resolutive markers FABP5, GPNMB and TREM-2 [28,29] (Fig. S8B). Further analysis of differentially regulated pathways in CtsD-expressing versus non-expressing MPs revealed an enrichment in genes related to the ECM content and degradation and the lysosome (Figure 8C–D). In agreement with these results, gene set enrichment analysis of the transcriptome of human cirrhotic KCs [16] confirmed an enrichment in pathways related to the lytic organization, the endopeptidase complex and the lysosomal lumen (Figure 8E). Our results demonstrate that CtsD-expressing MPs in human cirrhosis display an enriched gene expression signature in the lysosome and the ECM degradation related pathways.
Figure 8.
CtsD-expressing MPs in cirrhotic patients are associated to ECM degradation in human liver cirrhosis.
(A) UMAP projection depicting CtsD expression and dotplot representing percentage CtsD expression within the different MPs subpopulations in human cirrhotic livers (GSE136103). (B) Volcano plots for differential expression of CtsD-expressing MP versus CtsD-non-expressing MPs in cirrhotic livers. (C) Top 10 up and down regulated signalling pathways and (D) selected pathways in CtsD-expressing MPs versus CtsD-non-expressing MPs in cirrhotic livers. (E) Selected gene set enrichment analysis within the 15 pathways with highest NES of human cirrhotic KCs (GSE123661).
4. Discussion
In this study we demonstrate that CtsD-driven lysosomal activity in macrophages is essential for liver fibrosis development and resolution. Liver fibrosis is caused by a dysregulation of the normal homeostatic wound healing response due to chronic injury and can lead to severe organ dysfunction or even failure, as a result of the replacement of functional parenchyma by fibrotic ECM. Liver fibrosis is currently widely accepted to be reversible, however, no specific treatments to slow-down or reverse fibrosis have been successfully developed so far. Little is still known about the biological mechanisms controlling ECM remodeling during liver fibrosis. Changes in proteolytic activity are essential to liver fibrosis development and resolution as ECM homeostasis is controlled, among others, by proteases. The contribution of cysteine cathepsins to the development of fibrotic diseases has been previously reported in heart (CtsL) [30], lung (CtsK) [31], and liver (CtsB) [3]. However, the role of aspartyl cathepsins, such as CtsD, has not yet been explored.
Analysis of CtsD expression in control and cirrhotic human scRNAseq datasets [14] and liver biopsies revealed CtsD expression in hepatocytes and macrophages (Figure 1A–C). Analysis of CtsD in KCs (Log2 Fc = 2.63 p ≤ 0.001) and in serum (Figure 1D) from cirrhotic patients revealed a significant increase of CtsD during cirrhosis. It is not surprising that CtsD major reservoir in the liver are macrophages, as they have been reported to display increased proteolytic activity during liver injury [32] and proteolytic enzymes are essential for processing phagocytic products and for macrophage mesenchymal migration [33]. To investigate the cell-specific role of CtsD in macrophages or in hepatocytes during liver fibrosis we generated two novel cell-specific KO mouse strains by breeding CtsD floxed [9] mice with LysMCre mice [10] (CtsDΔMyel) or AlbCre [11] (CtsDΔHep) mice and validated CtsD deletion in liver macrophages and hepatocytes, respectively (Figures 2 and S2). Liver fibrosis was induced by chronic CCl4 administration or BDL. Enhanced liver fibrosis was demonstrated by an increase in collagen deposition (Figure 3C–D, Fig. S3F and Figure 4C–D) and fibrogenic markers (Figure 3E–H, S3G-H and S4E-G) in liver tissue from CtsDΔMyel mice, but not in CtsDΔHep ones (Figures 3L-N, 4K-M, S3A-B and S4A-B), after both CCl4 and BDL induced liver fibrosis without affectation of the liver damage (Figurs 3I, S3C, 4H, S4C and S4H). When we assessed liver inflammation, we observed an increase in gene expression of inflammatory markers (CCL2, CCL4 and TNF-α) (Figures 5E and S5H-J) and a distinct proteomic signature (Figure 5F–G) with a significant enrichment in the Reactome immune pathways in 8wkCCl4-CtsDΔMyel fibrotic livers (Figure 5H–I). No significant differences were observed in the number of macrophages (Figures 5A and S5A) and the proportion of KCs and MoMϕs (Figure 5B) detected in fibrotic liver tissue between CtsDF/F and CtsDΔMyel mice. Interestingly, CtsD expression was significantly higher in KCs (CLECF4+ve-F4/80+ve) (Figure 5C–D), which represented the main liver macrophage population in our model. These results suggest that functional or phenotypical changes of the inflammatory cell population might be responsible for the enriched inflammatory signature displayed in the liver in the absence of CtsD, instead of changes in the number or proportion of liver macrophages. While the role of macrophages during initiation, progression and resolution of liver fibrosis is quite well defined, the role of neutrophils is still controversial [34]. Regarding our work, it is important to notice Cre recombination driven by lysozyme 2 gene promoter also occurs in granulocytes, including neutrophils. Our results demonstrated that neither the number of granulocytes or neutrophils (Figs. S5C–D), nor the expression of neutrophil chemotactic proteins (Figs. S5E–F), or their degranulation capacity (Fig. S5K) are different between phenotypes. Nevertheless, although several reports have shown that neutrophil depletion during chronic liver disease does not influence liver fibrosis [35], we still cannot discard that CtsD deletion in neutrophils might also partially contribute to the phenotype observed in CtsDΔMyel mice. Thus, further studies will be needed to clarify the full impact of CtsD deletion in neutrophils over liver fibrosis, as this was not the aim of our study.
During liver fibrosis, macrophage plasticity allows them to adapt their phenotype, playing different and even opposing roles in disease progression. Hence, macrophages are responsible for the immune and inflammatory mechanisms driving liver fibrosis but also promote tissue repair and remodeling, contributing to fibrosis resolution. To analyze the effect of CtsD deletion in macrophages over ECM remodeling, we intersected the 8wkCCl4-CtsDΔMyel differential proteome with the matrisome database, confirming increased abundance of ECM-related proteins in fibrotic CtsDΔMyel livers (Fig. S6A). Little is still known about the function and phenotypical characteristics of restorative macrophages. While their indirect role modulating ECM remodeling due to their ability to secrete proteolytic enzymes, such as MMPs, is been recognized [28], their role as fibrolytic cells, directly phagocytosing and degrading ECM proteins, is far less understood [36]. Our experiments demonstrated that CtsD is important for lysosomal proteolytic processing of DQ Collagen I in liver macrophages (Figure 6A–B). Impaired lysosomal proteolysis has also been reported due to the absence of CtsE in macrophages [37] supporting our findings. Receptor-mediated collagen endocytosis is mediated in macrophages by the uPAR-associated protein (uPARAP/Endo180) in cooperation with UPAR [38,39]. No differences in endocytosis in hepatic macrophages (Figure 6C) or the expression of collagen internalization receptor UPARAP/Endo180, and its partner UPAR (Figure 6D) were detected in CtsDF/F and CtsDΔMyel fibrotic livers, suggesting that changes in internalized DQ Collagen I signal were caused by defective proteolytic processing instead of alterations in the receptor-mediated endocytic pathway. Indeed, collagen I was visualized within macrophages in fibrotic livers demonstrating collagen I was being internalized by liver macrophages in vivo during liver fibrosis (Figure 6E). Furthermore, confirmation of CtsD lysosomal distribution in liver macrophages (Figure 6F) from fibrotic livers supported an intracellular role for CtsD in collagen remodeling.
Since pro-resolutive macrophages are responsible for ECM remodeling and recycling during fibrosis resolution, a decrease in the lysosomal collagenolytic activity of liver macrophages might be an indication of a wider defective pro-resolutive phenotype of macrophages. Analysis of CtsDΔMyel hepatic macrophages demonstrated decreased pro-resolutive markers such as CD206, TREM2 [29] and TGF-β and an increase in the pro-inflammatory marker CD11c (Figure 6G). In line with our findings, strong enrichment of CtsD has been previously reported in TREM+ve macrophage subclusters during murine NASH [29], displaying pro-resolutive markers resembling those previously described in pre-monocyte-derived KCs and MMP12+ve macrophages [29,40]. In agreement with our findings, combined pharmacological inhibition of cathepsins B, L and S in tumor associated macrophages from pancreatic cancer resulted in phenotypical shifting from M2 to M1 linked to distinct lysosomal alterations, however, the exact mechanism is still unknown [41].
Despite direct proteolytic processing of collagen I19 and IV [20] by CtsD has been previously reported, it is unlikely to be the only protease responsible for the defective collagen remodeling observed in CtsDΔMyel fibrotic livers. Indeed, protease array from hepatic macrophage conditioned medias revealed 18 out of 33 proteases differentially regulated in CtsDΔMyel liver macrophages, most of them downregulated, which could be built in an interactome providing novel insights on hepatic macrophage protease network (Figure 6H–I and S6C-D). Among the proteases with decreased secretion were well-described gelatinases MMP-2 [21,22] and MMP-9 [[21], [22], [23]] and less well-known enzymes with ECM degrading activity, such as CtsS [24,25], KLK7 [26], MMP-3 [21,23] and MMP-7 [23]. Interestingly, MMP-12 was the only enzyme whose secretion was significantly up-regulated in CtsDΔMyel liver macrophages, opposite to what could be expected from recent reports proposing MMP-12 as a maker for restorative macrophages in murine experimental models of liver fibrosis [28,29]. Since MMP-12 is synthetized as an inactive pro-form (zymogen), gene transcription is not the first regulatory step, but the proteolytic activation of the existing cellular pool. Hence, opposite observations at the transcript, protein and even activity level are not unusual when it comes to protease study, as changes in gene transcription not always encompass the same changes at the protein or activity level. Thus, further study is required to fully clarify the role of MMP-12 in restorative macrophages not only as a marker but also as part of their proteolytic landscape. Phenotypical and secretomic alterations of CtsDΔMyel liver macrophages resulted in deficient fibrosis resolution in vivo (Figure 7A–C) and decreased number of macrophages in the fibrotic areas (Figure 7D) supporting macrophages might not be fulfilling their role during resolution. Interestingly, a switch in CtsD expression was observed from KCs (CLECF4+ve-F4/80+ve) (Figure 5C–D) to MoMϕs (CLECF4-ve-F4/80+ve) (Figure 7E–F) during the resolution phase, despite similar proportion of KCs and MoMϕs (Figure 5, Figure 7G) being maintained. Despite no change in lysosomal content (Figure 7H–I) of macrophages was observed during liver fibrosis resolution, TEM analysis of normal mouse livers revealed already the presence of abnormal lysosomal structures in CtsDΔMyel liver macrophages (Figure 7J), underpinning an alteration on lysosomal function, but not lysosomal content, as responsible for the secretomic and phenotypical changes observed in CtsDΔMyel mice. Similarly, mice with lysosomal dysfunction due to the loss of glycosylated lysosomal membrane protein (GLMP) displayed spontaneous [42] and aged-dependent [43] liver fibrosis, pointing out correct lysosomal activity as an essential mechanism of control for liver fibrosis resolution. Complementary to its role within the lysosome, our experiments also demonstrated that CtsD can be actively secreted by liver macrophages (Fig. S6E). As an active secreted enzyme, CtsD can proteolytically process not only ECM proteins, but also proteins involved in ECM assembly and stabilization such as SPARC, which is one of CtsD substrates exhibiting 13 cleaving sites (Table S3). Considering its important role in fibril aggregation and growth, it is not surprising that SPARC knock-out mouse displays reduced collagen deposition and fibrosis in multiple organs including the liver [44]. Our experiments demonstrate that the amount of SPARC located within collagen I was significantly decreased during the resolution phase only in CtsDF/F mouse (Figure 7K). This result suggests that the lack of active secreted CtsD could be responsible for sustained SPARC-collagen association, which would also contribute to fibrosis perpetuation.
Analysis of human scRNAseq cirrhotic datasets identified KC(1), KC(2) and SAM(2) as the MPs subpopulations with the highest CtsD expression (Figure 8A), and with a distinct gene expression pattern compared to the other MPs subpopulations in cirrhotic livers (Figures 8B and S8A). Interestingly, FABP5, GPNMB and TREM-2, which have been proposed as markers of restorative macrophages [28,29], were up-regulated in these three MPs subpopulations (Fig. S8B). Analysis of differentially regulated pathways in CtsD-expressing versus non-expressing MPs revealed an enrichment in genes related to both the ECM content and degradation and the lysosome (Figure 8C–D). This was confirmed by gene set enrichment analysis of human cirrhotic KC transcriptome [16] (Figure 8E) in agreement with previous reports, restorative macrophages present enriched signature in lysosomal signaling pathways [28]. Thus, our results demonstrate that CtsD-expressing MPs in human cirrhosis display an enriched gene expression signature in the lysosome and the ECM degradation related pathways.
Our work provides the first description of how lysosomal function driven by CtsD modulates macrophage resolutive capacity and fibronolytic activity. Despite further work needs to be done to fully elucidate the multiple roles of the macrophage proteolytic network controlling ECM remodeling, our results put in the spotlight for the first time the importance of the lysosomal protease balance for the correct functioning of macrophages during fibrosis resolution. This knowledge is the first critical step to start exploring, designing and testing novel drugs based on proteases that will enhance ECM degradation and promote fibrosis resolution. In summary, our work demonstrates CtsD-driven lysosomal activity is essential for the degradomic shift, which comprises all the secretomic, phenotypical and functional changes required by macrophages to proteolytically process the ECM during liver fibrosis resolution. Our findings start to elucidate a novel and unknown degradome landscape for restorative macrophages in liver disease.
Financial support
This work was mainly funded by MCIN/AEI/10.13039/501100011033/FEDER, UE through the project grants PID2021-123652OB-I00 and RTI2018-097475-A-100 (AM); by MCIN/AEI/10.13039/501100011033 and El FSE invest in your future through the contract RYC-2016-19731 (AM); by Pfizer grant #77131383 (AM); by Ministerio de Universidades fellowships FPU19/05357 (PRB) and FPU20/01367 (MFF); by MCIN/AEI/10.13039/501100011033/FEDER, UE contract PRE2022-101676 (JCP) and CSIC contract JAEINT_23_01245 (AdCC). Additional funding sources can be found in the financial support and sponsorship section of the manuscript.
CRediT authorship contribution statement
Paloma Ruiz-Blázquez: Writing – review & editing, Methodology, Investigation, Formal analysis. María Fernández-Fernández: Writing – review & editing, Methodology, Investigation, Formal analysis. Valeria Pistorio: Methodology, Investigation, Formal analysis. Celia Martinez-Sanchez: Methodology, Investigation. Michele Costanzo: Writing – review & editing, Methodology, Investigation. Paula Iruzubieta: Writing – review & editing, Resources, Methodology. Ekaterina Zhuravleva: Methodology, Investigation. Júlia Cacho-Pujol: Methodology, Investigation. Silvia Ariño: Investigation, Methodology. Alejandro Del Castillo-Cruz: Investigation, Methodology. Susana Núñez: Methodology. Jesper B. Andersen: Resources. Margherita Ruoppolo: Writing – review & editing, Resources, Methodology. Javier Crespo: Resources. Carmen García-Ruiz: Resources. Luigi Michele Pavone: Writing – review & editing, Resources, Methodology. Thomas Reinheckel: Resources. Pau Sancho-Bru: Resources. Mar Coll: Resources, Methodology. José C. Fernández-Checa: Resources. Anna Moles: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Support and technical advice from Dr Carme Casal, head of the confocal microscopy platform IIBB-CSIC and the Animal House Facilities from the University of Barcelona were highly appreciated. We thank the TEM-SEM Electron Microscopy Unit from Scientific and Technological Centers (CCiTUB), Universitat de Barcelona, and all the staff for their support and advice on TEM.
This work was mainly funded by MCIN/AEI/10.13039/501100011033/FEDER, UE through the project grants PID2021-123652OB-I00 and RTI2018-097475-A-100 (AM); by MCIN/AEI/10.13039/501100011033 and El FSE invest in your future through the contract RYC-2016-19731 (AM); by Pfizer grant #77131383 (AM); by Ministerio de Universidades fellowships FPU19/05357 (PRB) and FPU20/01367 (MFF); by MCIN/AEI/10.13039/501100011033/FEDER, UE contract PRE2022-101676 (JCP) and by CSIC contract JAEINT_23_01245 (AdCC).
Additional funding was provided by:
Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy grant BIOSS.
EXC-294 (TR).
The Collaborative Research Centre 850 project B7 (TR).
GRK 2606 project ID 423813989 (TR).
German Cancer Consortium DKTK projects L627 and FR01-371 (TR).
FEDER/Ministerio de Ciencia e Innovación – Agencia Estatal de Investigación grant PID2019.
111669RB-100 (JCFC) and PID2020-115055RB-I00 (CGR).
AGAUR grant SGR-2017-1112 (JCFC).
Fundació Marató TV3 Project 201916/31 (JCFC).
European Cooperation in Science & Technology (COST) ACTION CA17112 (JCFC).
Red Nacional de Enfermedades Metabólicas y Cáncer project 2018-102799-T (JCFC).
European Horizon's research and innovation program Spanish National Research Council's.
Cancer Hub HORIZON-HLTH-2022-STAYHLTH-02 grant No 101095679 (JCFC).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101989.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Wynn T.A. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dulai P.S., Singh S., Patel J., Soni M., Prokop L.J., Younossi Z., et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. 2017;65:1557–1565. doi: 10.1002/hep.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz-Blázquez P., Pistorio V., Fernández-Fernández M., Moles A. The multifaceted role of cathepsins in liver disease. J Hepatol. 2021;75:1192–1202. doi: 10.1016/j.jhep.2021.06.031. [DOI] [PubMed] [Google Scholar]
- 4.Canbay A., Guicciardi M.E., Higuchi H., Feldstein A., Bronk S.F., Rydzewski R., et al. Cathepsin B inactivation attenuates hepatic injury and fibrosis during cholestasis. J Clin Invest. 2003;112:152–159. doi: 10.1172/JCI17740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moles A., Tarrats N., Fernández-Checa J.C., Marí M. Cathepsins B and D drive hepatic stellate cell proliferation and promote their fibrogenic potential. Hepatology. 2009;49:1297–1307. doi: 10.1002/hep.22753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moles A., Tarrats N., Fernández-Checa J.C., Marí M. Cathepsin B overexpression due to acid sphingomyelinase ablation promotes liver fibrosis in Niemann-Pick disease. J Biol Chem. 2012;287:1178–1188. doi: 10.1074/jbc.M111.272393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moles A., Tarrats N., Morales A., Domínguez M., Bataller R., Caballería J., et al. Acidic sphingomyelinase controls hepatic stellate cell activation and in vivo liver fibrogenesis. Am J Pathol. 2010;177:1214–1224. doi: 10.2353/ajpath.2010.091257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saftig P., Hetman M., Schmahl W., Weber K., Heine L., Mossmann H., et al. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995;14:3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ketscher A., Ketterer S., Dollwet-Mack S., Reif U., Reinheckel T. Neuroectoderm-specific deletion of cathepsin D in mice models human inherited neuronal ceroid lipofuscinosis type 10. Biochimie. 2016;122:219–226. doi: 10.1016/j.biochi.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 10.Clausen B.E., Burkhardt C., Reith W., Renkawitz R., Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 11.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 12.Augereau P., Miralles F., Cavaillès V., Gaudelet C., Parker M., Rochefort H. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol Endocrinol. 1994;8:693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- 13.Cavaillès V., Augereau P., Rochefort H. Cathepsin D gene is controlled by a mixed promoter, and estrogens stimulate only TATA-dependent transcription in breast cancer cells. Proc Natl Acad Sci USA. 1993;90:203–207. doi: 10.1073/pnas.90.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota S., Tsuji H., Kato K. Localization of cathepsin D in rat liver. Histochemistry. 1985;82:141–148. doi: 10.1007/BF00708198. [DOI] [PubMed] [Google Scholar]
- 16.Pose E., Coll M., Martínez-Sánchez C., Zeng Z., Surewaard B.G.J., Català C., et al. Programmed death ligand 1 is overexpressed in liver macrophages in chronic liver diseases, and its blockade improves the antibacterial activity against infections. Hepatology. 2021;74:296–311. doi: 10.1002/hep.31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai M., Troutman T.D., Seidman J.S., Ouyang Z., Spann N.J., Abe Y., et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain kupffer cell identity. Immunity. 2019;51:655–670.e8. doi: 10.1016/j.immuni.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beattie L., Sawtell A., Mann J., Frame T.C.M., Teal B., de Labastida Rivera F., et al. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J Hepatol. 2016;65:758–768. doi: 10.1016/j.jhep.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott P.G., Pearson H. Cathepsin D: specificity of peptide-bond cleavage in type-I collagen and effects on type-III collagen and procollagen. Eur J Biochem. 2005;114:59–62. doi: 10.1111/j.1432-1033.1981.tb06172.x. [DOI] [PubMed] [Google Scholar]
- 20.Zou J., Henderson L., Thomas V., Swan P., Turner A.N., Phelps R.G. Presentation of the goodpasture autoantigen requires proteolytic unlocking steps that destroy prominent T cell epitopes. J Am Soc Nephrol. 2007;18:771–779. doi: 10.1681/ASN.2006091056. [DOI] [PubMed] [Google Scholar]
- 21.Zhen E.Y., Brittain I.J., Laska D.A., Mitchell P.G., Sumer E.U., Karsdal M.A., et al. Characterization of metalloprotease cleavage products of human articular cartilage. Arthritis Rheum. 2008;58:2420–2431. doi: 10.1002/art.23654. [DOI] [PubMed] [Google Scholar]
- 22.Prudova A., auf dem Keller U., Butler G.S., Overall C.M. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol Cell Proteomics. 2010;9:894–911. doi: 10.1074/mcp.M000050-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukui N., McAlinden A., Zhu Y., Crouch E., Broekelmann T.J., Mecham R.P., et al. Processing of type II procollagen amino propeptide by matrix metalloproteinases. J Biol Chem. 2002;277:2193–2201. doi: 10.1074/jbc.M105485200. [DOI] [PubMed] [Google Scholar]
- 24.Vidmar R., Vizovišek M., Turk D., Turk B., Fonović M. Protease cleavage site fingerprinting by label-free in-gel degradomics reveals <scp>pH</scp> -dependent specificity switch of legumain. EMBO J. 2017;36:2455–2465. doi: 10.15252/embj.201796750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo T., Xie Q., Liu J., Yang J., Shi J., Kong D., et al. Macrophage-derived cathepsin S remodels the extracellular matrix to promote liver fibrogenesis. Gastroenterology. 2023 doi: 10.1053/j.gastro.2023.05.039. [DOI] [PubMed] [Google Scholar]
- 26.Silva L.M., Kryza T., Stoll T., Hoogland C., Dong Y., Stephens C.R., et al. Integration of two in-depth quantitative proteomics approaches determines the kallikrein-related peptidase 7 (KLK7) degradome in ovarian cancer cell secretome. Mol Cell Proteomics. 2019;18:818–836. doi: 10.1074/mcp.RA118.001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcaraz L.B., Mallavialle A., David T., Derocq D., Delolme F., Dieryckx C., et al. A 9-kDa matricellular SPARC fragment released by cathepsin D exhibits pro-tumor activity in the triple-negative breast cancer microenvironment. Theranostics. 2021;11:6173–6192. doi: 10.7150/thno.58254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran P., Pellicoro A., Vernon M.A., Boulter L., Aucott R.L., Ali A., et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA. 2012;109 doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrikx T., Porsch F., Kiss M.G., Rajcic D., Papac-Miličević N., Hoebinger C., et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2+ macrophages that localize to fibrotic areas and limit NASH. J Hepatol. 2022;77:1373–1385. doi: 10.1016/j.jhep.2022.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Petermann I., Mayer C., Stypmann J., Biniossek M.L., Tobin D.J., Engelen M.A., et al. Lysosomal, cytoskeletal, and metabolic alterations in cardiomyopathy of cathepsin L knockout mice. Faseb J. 2006;20:1266–1268. doi: 10.1096/fj.05-5517fje. [DOI] [PubMed] [Google Scholar]
- 31.Bühling F., Röcken C., Brasch F., Hartig R., Yasuda Y., Saftig P., et al. Pivotal role of cathepsin K in lung fibrosis. Am J Pathol. 2004;164:2203–2216. doi: 10.1016/S0002-9440(10)63777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanner A., Keyhani A., Reiner R., Holdstock G., Wright R. Proteolytic enzymes released by liver macrophages may promote hepatic injury in a rat model of hepatic damage. Gastroenterology. 1981;80:647–654. [PubMed] [Google Scholar]
- 33.Vérollet C., Charrière G.M., Labrousse A., Cougoule C., Le Cabec V., Maridonneau-Parini I. Extracellular proteolysis in macrophage migration: losing grip for a breakthrough. Eur J Immunol. 2011;41:2805–2813. doi: 10.1002/eji.201141538. [DOI] [PubMed] [Google Scholar]
- 34.Tang J., Yan Z., Feng Q., Yu L., Wang H. The roles of neutrophils in the pathogenesis of liver diseases. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.625472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moles A., Murphy L., Wilson C.L., Chakraborty J.B., Fox C., Park E.J., et al. A TLR2/S100A9/CXCL-2 signaling network is necessary for neutrophil recruitment in acute and chronic liver injury in the mouse. J Hepatol. 2014;60:782–791. doi: 10.1016/j.jhep.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKleroy W., Lee T.-H., Atabai K. Always cleave up your mess: targeting collagen degradation to treat tissue fibrosis. Am. J. Physiol. Cell. Mol. Physiol. 2013;304:L709–L721. doi: 10.1152/ajplung.00418.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsukuba T., Yanagawa M., Kadowaki T., Takii R., Okamoto Y., Sakai E., et al. Cathepsin E deficiency impairs autophagic proteolysis in macrophages. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen D.H., Leonard D., Masedunskas A., Moyer A., Jürgensen H.J., Peters D.E., et al. M2-like macrophages are responsible for collagen degradation through a mannose receptor–mediated pathway. J Cell Biol. 2013;202:951–966. doi: 10.1083/jcb.201301081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madsen D.H., Bugge T.H. Imaging collagen degradation in vivo highlights a key role for M2-polarized macrophages in extracellular matrix degradation. OncoImmunology. 2013;2 doi: 10.4161/onci.27127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remmerie A., Martens L., Thoné T., Castoldi A., Seurinck R., Pavie B., et al. Osteopontin expression identifies a subset of recruited macrophages distinct from kupffer cells in the fatty liver. Immunity. 2020;53:641–657.e14. doi: 10.1016/j.immuni.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oelschlaegel D., Weiss Sadan T., Salpeter S., Krug S., Blum G., Schmitz W., et al. Cathepsin inhibition modulates metabolism and polarization of tumor-associated macrophages. Cancers. 2020;12:2579. doi: 10.3390/cancers12092579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong X.Y., Nesset C.K., Damme M., Løberg E.-M., Lübke T., Mæhlen J., et al. Loss of lysosomal protein NCU-G1 results in spontaneous liver fibrosis with accumulation of lipofuscin and iron in Kupffer cells. Dis. Model. Mech. 2014 doi: 10.1242/dmm.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nesset C.K., Kong X.Y., Damme M., Schjalm C., Roos N., Løberg E.M., et al. Age-dependent development of liver fibrosis in Glmp gt/gt mice. Fibrogenesis Tissue Repair. 2016;9:5. doi: 10.1186/s13069-016-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atorrasagasti C., Peixoto E., Aquino J.B., Kippes N., Malvicini M., Alaniz L., et al. Lack of the matricellular protein SPARC (secreted protein, acidic and rich in cysteine) attenuates liver fibrogenesis in mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.