Abstract
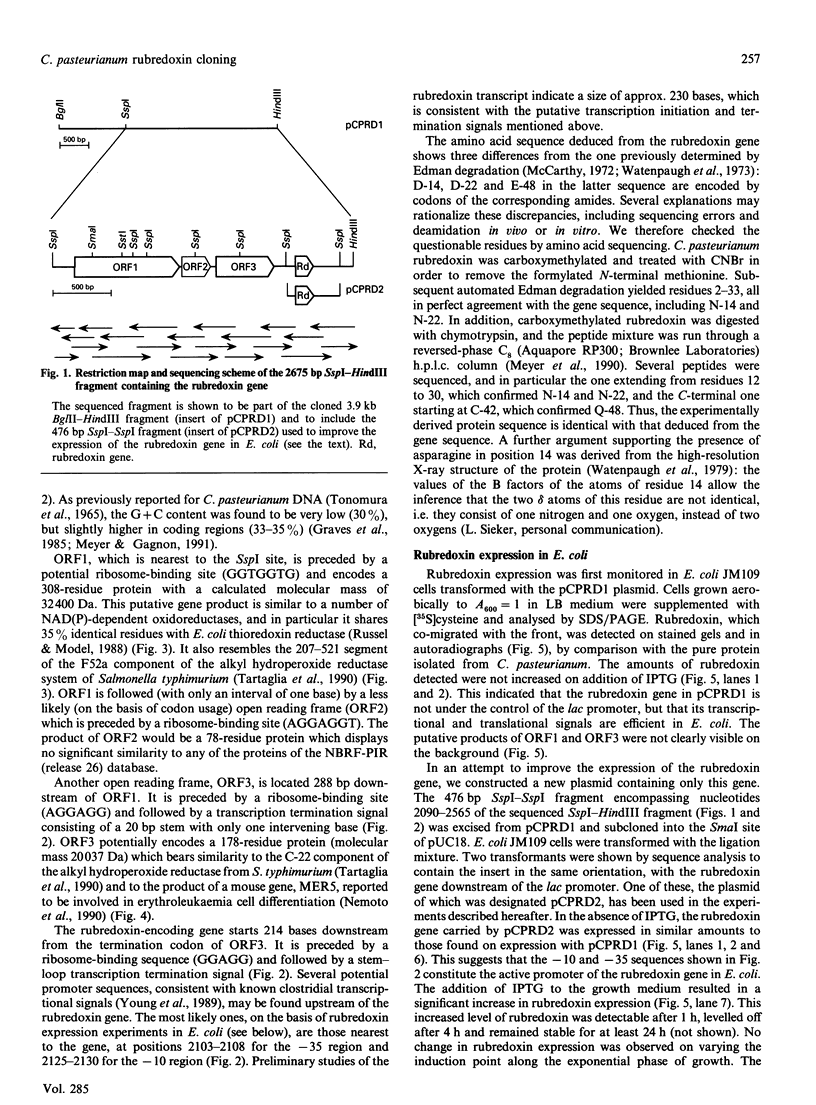
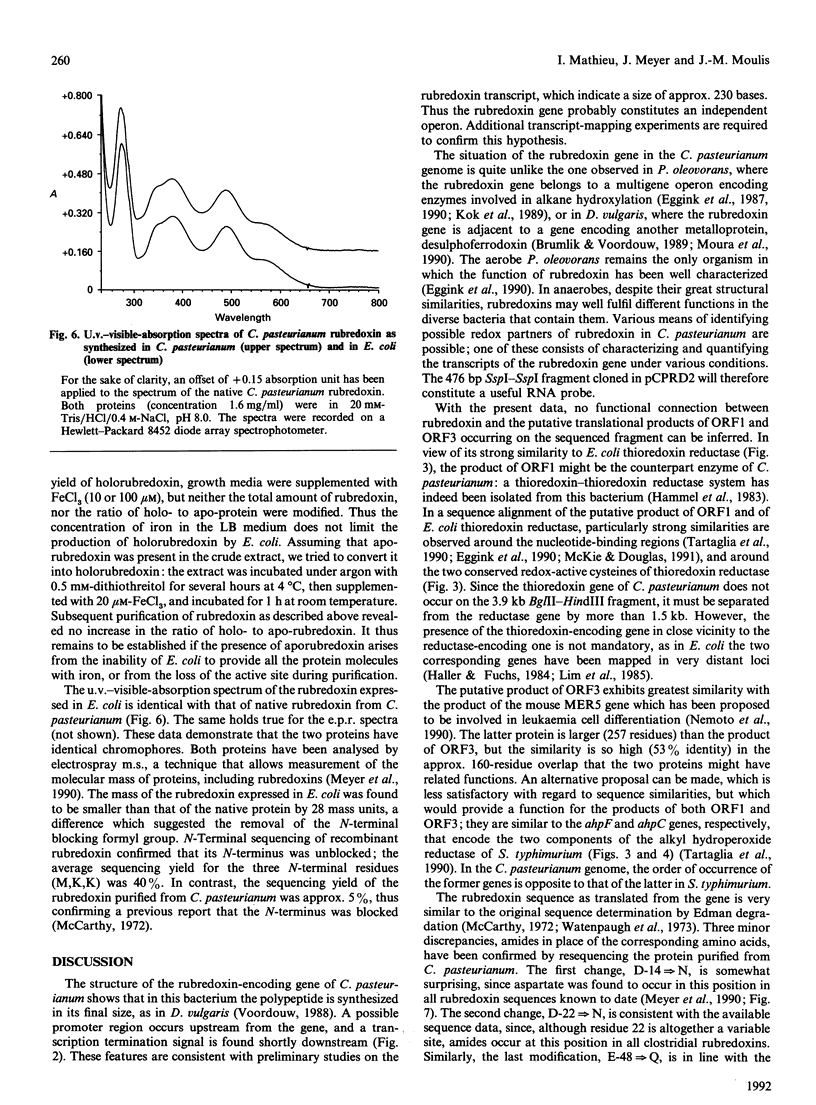
A 3.9 kb BglII-HindIII DNA fragment containing the rubredoxin gene from Clostridium pasteurianum has been cloned using oligonucleotide probes designed from the protein sequence. The 2675 bp SspI-HindIII portion of this fragment has been sequenced and found to contain three open reading frames in addition to the rubredoxin gene. The putative product of one of these open reading frames is similar to various thioredoxin reductases. The rubredoxin gene translates into a sequence that differs from the previously published protein sequence in three positions, D-14, D-22 and E-48 being replaced by the corresponding amides. These changes have been confirmed by partial resequencing of the protein. Promoter-like sequences and a transcription termination signal have been found near the sequence of the rubredoxin gene, which may therefore constitute an independent transcriptional unit. Expression of C. pasteurianum rubredoxin in Escherichia coli strain JM109 has been optimized by subcloning a 476 bp SspI-SspI fragment encompassing the rubredoxin gene. Under these conditions, the latter gene was partly under the control of the lac promoter of pUC18, and the level of rubredoxin production could be increased twofold on addition of a lactose analogue, thus reaching 2-3 mg of pure protein/l of culture. Recombinant rubredoxin was produced in E. coli cells as the holoprotein, and displayed a u.v.-visible-absorption spectrum identical with that of the rubredoxin purified from C. pasteurianum. M.s. and N-terminal sequencing showed that C. pasteurianum rubredoxin expressed in E. coli differs from its native counterpart by having an unblocked N-terminal methionine.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T., Sieker L. C., Jensen L. H. Structure of rubredoxin from Desulfovibrio vulgaris at 1.5 A resolution. J Mol Biol. 1991 Jan 20;217(2):337–352. doi: 10.1016/0022-2836(91)90547-j. [DOI] [PubMed] [Google Scholar]
- Ballongue J., Amine J., Masion E., Petitdemange H., Gay R. Rôle de l'acétate et du butyrate dans l'induction de la NADH: rubrédoxine oxydoréductase chez Clostridium acetobutylicum. Biochimie. 1986 Apr;68(4):575–580. doi: 10.1016/s0300-9084(86)80202-4. [DOI] [PubMed] [Google Scholar]
- Baur J. R., Graves M. C., Feinberg B. A., Ragsdale S. W. Characterization of the recombinant Clostridium pasteurianum ferredoxin and comparison of its properties with those of the native protein. Biofactors. 1990 Jul;2(3):197–203. [PubMed] [Google Scholar]
- Blake P. R., Park J. B., Bryant F. O., Aono S., Magnuson J. K., Eccleston E., Howard J. B., Summers M. F., Adams M. W. Determinants of protein hyperthermostability: purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry. 1991 Nov 12;30(45):10885–10895. doi: 10.1021/bi00109a012. [DOI] [PubMed] [Google Scholar]
- Brumlik M. J., Voordouw G. Analysis of the transcriptional unit encoding the genes for rubredoxin (rub) and a putative rubredoxin oxidoreductase (rbo) in Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1989 Sep;171(9):4996–5004. doi: 10.1128/jb.171.9.4996-5004.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. C., Chen J. S., Johnson J. L. Structural features of multiple nifH-like sequences and very biased codon usage in nitrogenase genes of Clostridium pasteurianum. J Bacteriol. 1986 Apr;166(1):162–172. doi: 10.1128/jb.166.1.162-172.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan V. M., Vickery L. E. Expression of human ferredoxin and assembly of the [2Fe-2S] center in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):835–839. doi: 10.1073/pnas.86.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink G., Engel H., Vriend G., Terpstra P., Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990 Mar 5;212(1):135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- Eggink G., Lageveen R. G., Altenburg B., Witholt B. Controlled and functional expression of the Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J Biol Chem. 1987 Dec 25;262(36):17712–17718. [PubMed] [Google Scholar]
- Frey M., Sieker L., Payan F., Haser R., Bruschi M., Pepe G., LeGall J. Rubredoxin from Desulfovibrio gigas. A molecular model of the oxidized form at 1.4 A resolution. J Mol Biol. 1987 Oct 5;197(3):525–541. doi: 10.1016/0022-2836(87)90562-6. [DOI] [PubMed] [Google Scholar]
- Grabau C., Schatt E., Jouanneau Y., Vignais P. M. A new [2Fe-2S] ferredoxin from Rhodobacter capsulatus. Coexpression with a 2[4Fe-4S] ferredoxin in Escherichia coli. J Biol Chem. 1991 Feb 15;266(5):3294–3299. [PubMed] [Google Scholar]
- Graves M. C., Mullenbach G. T., Rabinowitz J. C. Cloning and nucleotide sequence determination of the Clostridium pasteurianum ferredoxin gene. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1653–1657. doi: 10.1073/pnas.82.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller B. L., Fuchs J. A. Mapping of trxB, a mutation responsible for reduced thioredoxin reductase activity. J Bacteriol. 1984 Sep;159(3):1060–1062. doi: 10.1128/jb.159.3.1060-1062.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel K. E., Cornwell K. L., Buchanan B. B. Ferredoxin/flavoprotein-linked pathway for the reduction of thioredoxin. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3681–3685. doi: 10.1073/pnas.80.12.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton S. M., Slaughter C., Eisner W., Fisher T. The molybdenum-pterin binding protein is encoded by a multigene family in Clostridium pasteurianum. Gene. 1987;54(2-3):211–219. doi: 10.1016/0378-1119(87)90489-6. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Raatjes P., Kingma J., van Lelyveld P. H., Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J Biol Chem. 1989 Apr 5;264(10):5435–5441. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Gall J. Purification PARTIELLE ET 'ETUDE DE LA NAD: rubrédoxine oxydo-réductase de D. Gigas. Ann Inst Pasteur (Paris) 1968 Jan;114(1):109–115. [PubMed] [Google Scholar]
- Lim C. J., Haller B., Fuchs J. A. Thioredoxin is the bacterial protein encoded by fip that is required for filamentous bacteriophage f1 assembly. J Bacteriol. 1985 Feb;161(2):799–802. doi: 10.1128/jb.161.2.799-802.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovenberg W., Sobel B. E. Rubredoxin: a new electron transfer protein from Clostridium pasteurianum. Proc Natl Acad Sci U S A. 1965 Jul;54(1):193–199. doi: 10.1073/pnas.54.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makaroff C. A., Zalkin H., Switzer R. L., Vollmer S. J. Cloning of the Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase gene in Escherichia coli. Nucleotide sequence determination and properties of the plasmid-encoded enzyme. J Biol Chem. 1983 Sep 10;258(17):10586–10593. [PubMed] [Google Scholar]
- McKie J. H., Douglas K. T. Evidence for gene duplication forming similar binding folds for NAD(P)H and FAD in pyridine nucleotide-dependent flavoenzymes. FEBS Lett. 1991 Feb 11;279(1):5–8. doi: 10.1016/0014-5793(91)80236-v. [DOI] [PubMed] [Google Scholar]
- Meyer J., Gagnon J. Primary structure of hydrogenase I from Clostridium pasteurianum. Biochemistry. 1991 Oct 8;30(40):9697–9704. doi: 10.1021/bi00104a018. [DOI] [PubMed] [Google Scholar]
- Meyer J., Gagnon J., Sieker L. C., Van Dorsselaer A., Moulis J. M. Rubredoxin from Clostridium thermosaccharolyticum. Amino acid sequence, mass-spectrometric and preliminary crystallographic data. Biochem J. 1990 Nov 1;271(3):839–841. doi: 10.1042/bj2710839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulis J. M., Meyer J. Characterization of the selenium-substituted 2 [4Fe-4Se] ferredoxin from Clostridium pasteurianum. Biochemistry. 1982 Sep 14;21(19):4762–4771. doi: 10.1021/bi00262a037. [DOI] [PubMed] [Google Scholar]
- Moura I., Tavares P., Moura J. J., Ravi N., Huynh B. H., Liu M. Y., LeGall J. Purification and characterization of desulfoferrodoxin. A novel protein from Desulfovibrio desulfuricans (ATCC 27774) and from Desulfovibrio vulgaris (strain Hildenborough) that contains a distorted rubredoxin center and a mononuclear ferrous center. J Biol Chem. 1990 Dec 15;265(35):21596–21602. [PubMed] [Google Scholar]
- Nemoto Y., Yamamoto T., Takada S., Matsui Y., Obinata M. Antisense RNA of the latent period gene (MER5) inhibits the differentiation of murine erythroleukemia cells. Gene. 1990 Jul 16;91(2):261–265. doi: 10.1016/0378-1119(90)90097-b. [DOI] [PubMed] [Google Scholar]
- Rabinowitz J. Preparation and properties of clostridial ferredoxins. Methods Enzymol. 1972;24:431–446. doi: 10.1016/0076-6879(72)24089-7. [DOI] [PubMed] [Google Scholar]
- Russel M., Model P. Sequence of thioredoxin reductase from Escherichia coli. Relationship to other flavoprotein disulfide oxidoreductases. J Biol Chem. 1988 Jun 25;263(18):9015–9019. [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki S., Ikeda A., Ishimoto M. Rubredoxin as an intermediary electron carrier for nitrate reduction by NAD(P)H in Clostridium perfringens. J Biochem. 1988 Apr;103(4):583–584. doi: 10.1093/oxfordjournals.jbchem.a122310. [DOI] [PubMed] [Google Scholar]
- Shimizu F., Ogata M., Yagi T., Wakabayashi S., Matsubara H. Amino acid sequence and function of rubredoxin from Desulfovibrio vulgaris Miyazaki. Biochimie. 1989 Nov-Dec;71(11-12):1171–1177. doi: 10.1016/0300-9084(89)90020-5. [DOI] [PubMed] [Google Scholar]
- Stenkamp R. E., Sieker L. C., Jensen L. H. The structure of rubredoxin from Desulfovibrio desulfuricans strain 27774 at 1.5 A resolution. Proteins. 1990;8(4):352–364. doi: 10.1002/prot.340080409. [DOI] [PubMed] [Google Scholar]
- TONOMURA B., MALKIN R., RABINOWITZ J. C. DEOXYRIBONUCLEIC ACID BASE COMPOSITION OF CLOSTRIDIAL SPECIES. J Bacteriol. 1965 May;89:1438–1439. doi: 10.1128/jb.89.5.1438-1439.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia L. A., Storz G., Brodsky M. H., Lai A., Ames B. N. Alkyl hydroperoxide reductase from Salmonella typhimurium. Sequence and homology to thioredoxin reductase and other flavoprotein disulfide oxidoreductases. J Biol Chem. 1990 Jun 25;265(18):10535–10540. [PubMed] [Google Scholar]
- Voordouw G. Cloning of genes encoding redox proteins of known amino acid sequence from a library of the Desulfovibrio vulgaris (Hildenborough) genome. Gene. 1988 Jul 15;67(1):75–83. doi: 10.1016/0378-1119(88)90010-8. [DOI] [PubMed] [Google Scholar]
- Wang S. Z., Chen J. S., Johnson J. L. A nitrogen-fixation gene (nifC) in Clostridium pasteurianum with sequence similarity to chlJ of Escherichia coli. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1122–1128. doi: 10.1016/0006-291x(90)92012-o. [DOI] [PubMed] [Google Scholar]
- Wang S. Z., Chen J. S., Johnson J. L. The presence of five nifH-like sequences in Clostridium pasteurianum: sequence divergence and transcription properties. Nucleic Acids Res. 1988 Jan 25;16(2):439–454. doi: 10.1093/nar/16.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watenpaugh K. D., Sieker L. C., Jensen L. H. The structure of rubredoxin at 1.2 A resolution. J Mol Biol. 1979 Jul 5;131(3):509–522. doi: 10.1016/0022-2836(79)90005-6. [DOI] [PubMed] [Google Scholar]
- Young M., Minton N. P., Staudenbauer W. L. Recent advances in the genetics of the clostridia. FEMS Microbiol Rev. 1989 Dec;5(4):301–325. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]



