Abstract
Background
Understanding organic functions at a molecular level is important for scientists to unveil the disease mechanism and to develop diagnostic or therapeutic methods.
Aims
The present study tried to find genes selectively expressed in 11 rat organs, including the adrenal gland, brain, colon, duodenum, heart, ileum, kidney, liver, lung, spleen, and stomach.
Materials and Methods
Three normal male Sprague-Dawley (SD) rats were anesthetized, their organs mentioned above were harvested, and RNA in the fresh organs was extracted. Purified RNA was reversely transcribed and sequenced using the Solexa high-throughput sequencing technique. The abundance of a gene was measured by the expected value of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM). Genes in organs with the highest expression level were sought out and compared with their median value in organs. If a gene in the highest expressed organ was significantly different (p < 0.05) from that in the medianly expressed organ, accompanied by q value < 0.05, and accounted for more than 70% of the total abundance, the gene was assumed as the selective gene in the organ.
Results & Discussion
The Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO) pathways were enriched by the highest expressed genes. Based on the criterion, 1,406 selective genes were screened out, 1,283 of which were described in the gene bank and 123 of which were waiting to be described. KEGG and GO pathways in the organs were partly confirmed by the known understandings and a good portion of the pathways needed further investigation.
Conclusion
The novel selective genes and organic functional pathways are useful for scientists to unveil the mechanisms of the organs at the molecular level, and the selective genes’ products are candidate disease markers for organs.
Keywords: High-throughput sequencing, selective expression, organic markers, rat, genetic variations, DNA
1. INTRODUCTION
It was once believed that all somatic cells shared the same genome because all of a creature's cells and organs develop from a fertilized egg. The expression of an animal’s genome controls the animal’s functions, whose functions are executed by its cells. Therefore, cells have different functions depending on different gene expression profiles [1, 2], and so do different tissues and organs. The other gene expression profiles will doom cell differentiation [3], organ development [4], and its functions. Based on the understanding, it can be assumed that some genes as constructive ones must be universally expressed in all the cells with a nucleus, and some could be selectively expressed in cells, tissues, and organs at different developmental stages [5, 6]. At an animal’s adulthood, its gene expression profiles could be relatively stable to maintain its biological functions, and the gene expression profile would reflect its function. Therefore, the products (RNAs and proteins) from the gene selectively expressed in an organ suggest its function(s).
Health and disease are the eternal themes of humans, and are usually related to gene expression profiles. The mechanism study on human health and disease is generally carried on model animals at first, then on humans. Among them, adult rats and mice are model animals most frequently used by scientists, and no animals are studied more deeply than them. Therefore, it is a good strategy to understand humans by investigating gene expression profiles in rats. Identifying molecular targets and disease markers from rats and mice is usually the first step to understanding human health and disease, then to finding therapeutic strategies and methods. The selective gene products released into the blood can be used as damage markers. However, it is a big premise to understand the normal model animal’s biological features at the molecular level before scientists comprehensively understand human health and disease [7]. There were much data from animals suggesting that some genes selectively expressed in organs, e.g. NeuN (Rbfox3) in the brain or neuron [8] though with alternative opinions [9], troponin (Tnnc1, Tnni3) in the heart [10], glutamic pyruvic transaminase (GPT, Gpt) in the liver [11], and neutrophil gelatinase-associated lipocalin (NGAL) in the kidney [12]. The findings are very useful and even were adopted for clinic diagnosis and treatment. The gene products selectively and originally distributed can be used as molecular organic markers and then make disease diagnosis more accurate or earlier. Nevertheless, in the background of precision medicine [13], the selective gene products in organs are still insufficient for clinical practice, and it is still necessary to systematically screen the genes selectively expressed in organs.
Proteins and RNAs are the end products of genes and execute their functions. To identify the selective functions at the molecular level, all the selectively distributed proteins in organs should be screened out. However, among them, protein screening is a big economic burden because the study would consume plenty of antibodies. Since proteins and RNAs were transcribed and even then translated from genes, the present study would apply high-throughput sequencing technology to analyze gene expression profiles of 11 organs, including the adrenal gland, brain, colon, duodenum, heart, ileum, kidney, liver, lung, spleen, and stomach, at the RNA level, and then, based on the results, to find the likely organic markers and analyze the functional pathways the selective genes would be involved in.
2. MATERIALS AND METHODS
2.1. Materials
Adult male Sprague-Dawley (SD) rats (age, 45 days; body weight, 180-220 g) were obtained from Chengdu Dossy Experimental Animal Co. Ltd., Chengdu, China [Certification No. SCXK (Chuan) 2008–24]. TRIzol Plus RNA Purification kit was purchased from Invitrogen (Carlsbad, CA, USA). Ultra-pure water was produced with a Milli Q water purification system manufactured by EMD Millipore Group (Darmstadt, Germany). NanoDrop ND-1000 spectrophotometer was manufactured by PeqLab (Erlangen, Germany). The multimicroplate reader of Infinite 200pro was manufactured by Tecan Group (Mannedorf, Switzerland). Other instruments or reagents used in the present study were made in China if not mentioned.
2.2. Animal Treatment
Three rats were normally treated for three days. Then, the animals were intraperitoneally anesthetized with urethane (1.0 g/kg). The rats’ chests and abdomens were opened, and their organs were harvested, including the adrenal gland (Ad), brain (frontal cortex) (Br), colon (Co), duodenum (the first 5 cm) (Du), heart (left ventricle) (He), ileum (the end 5 cm) (Il), kidney (right) (Ki), liver (Li), lung (right) (Lu), spleen (Sp), and stomach (gastric antrum) (St). The tunica and mesentery of the organs were removed clearly. All the organs were frozen with liquid nitrogen and kept at -80°C by dry ice to keep them fresh, and then sent to Sangon Biotech Co. Ltd. (Shanghai China) (https://www.sangon.com/) immediately for high-throughput sequencing.
The animal experiments were approved by the Animal Care and Use Committee of Yunnan Provincial Key Laboratory of Molecular Biology for Sinomedicine (Approved No. LL-20171023-01), Yunnan University of Traditional Chinese Medicine.
2.3. High-throughput Sequencing of mRNA
The fresh organs were frozen with liquid nitrogen and ground to powder. The total RNA in the powder was extracted and purified using the TRIzol Plus RNA Purification kit (Invitrogen, Carlsbad, CA, USA). The quantity and quality of RNA were measured by the NanoDrop ND-1000 spectrophotometer. RNA integrity was assessed by three bands (28S, 18S, and 5S) using formaldehyde denaturing agarose gel electrophoresis RNA as previously described [14, 15].
Similar to the results of our previous study [16], double-stranded cDNA (ds-cDNA) was reversely transcribed from the total RNA using a SuperScript ds-cDNA synthesis kit (Invitrogen, Carlsbad, USA) in the presence of 100 pmol/L oligo dT primers. Solexa high-throughput sequencing technique was used to sequence the cDNA by Sangon Biotech Co. Ltd. (Shanghai, China). The raw data containing reads of 150 bases of nucleotide in fastq format was transformed to original sequences in fasta format by Seqkit software in the disc operation system (DOS) model [17]. The sequences that matched 27 bp or more to the rat’s reference mRNA sequences (https://www.ncbi.nlm.nih.gov/) were screened out by TBtools software (v0.664445552). The expected value of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) was used for the normalization of expression level [18].
2.4. Screening Genes Selectively Expressed
Values of gene’s FPKM in every organ were collected. The overall function of the organs at the gene expression level was analyzed by cluster analysis. The distance between organs was calculated by the Vegan package of Bray curtis method [19], and the cluster tree was established by Hcluster [20].
Based on the assumption that a gene is significantly overexpressed in an organ (statistical consideration), if its expression abundance accounts for the majority of that in all organs, say more than 70%, the gene is considered to be selectively expressed in that organ. The maximum FPKM value of a gene in any organs less than 5 was ignored because the expression level of the gene was supposed to be too low to analyze. Genes with FPKM above 5 were further analyzed. The means of a gene’s FPKM in all the organs were sorted. The organ with the median value and those with the biggest value were selected. Then, the expression level of the gene in the two organs (the highest and median organs) was compared with the Student t-test. The q-value, a false-discovery rate alternative to p-values, was also calculated as an adjustment for multiple comparisons [21]. If p-value and q-value were both less than 0.05, the gene was regarded as a candidate gene selectively expressed in the organ.
The means of the gene in all the organs were summed up as “Total”. The mean of the gene in the organ highest expressed it was regarded as “max mean”. Then, the MT ratio ((max mean)/total) was calculated. If the MT ratio was above 0.7, the gene was regarded as a selective gene in the organ. The gene’s product in the organ was regarded as an organic marker that may execute the selective function of the organ. The last reports on the relationship between the selective genes and the organs were searched at PubMed (www.pubmed.gov) on June 10, 2023.
The last report of the selective gene from the PubMed database was sought in the relative organ by searching the gene name and the organ both in the fields of title or abstract.
2.5. KEGG, and GO Analysis
The values of a gene in all the organs were sorted by its mean, and the organ that expressed the median value and that expressed the biggest value were selected. The expression abundance of the gene in the two organs was compared with the Student t-test. If there was significance (p < 0.05), the gene in the organ was regarded as an interesting gene. Interesting genes expressed in an organ were further analyzed to enrich the selective Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp/) and Gene Ontology (GO, http://www.geneontology.org/) pathways. KEGG enrichment [22] and KOG enrichment [23, 24] were performed by ClusterProfiler [25]. GO [26, 27] enrichment was performed by TopGO. The p-value and q-value were also calculated using the software mentioned above.
3. RESULTS
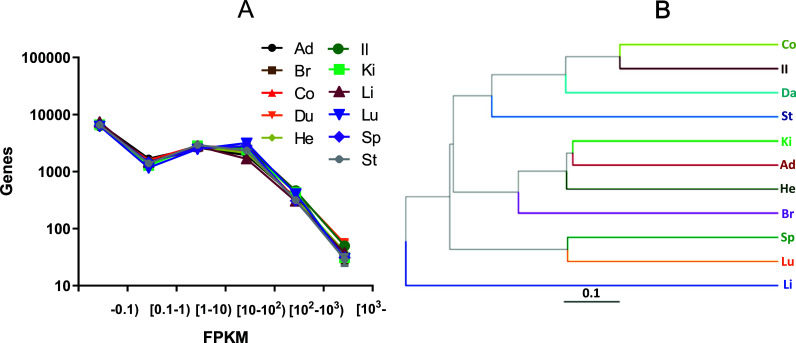
3.1. Total FPKM Distribution
In the normal rats, 32,623 genes’ transcripts were detected, and most genes were expressed at a very low level (FPKM < 1), only a small portion of genes expressed at a very high level (FPKM > 1000) (Fig. 1A). The overall FPKM distribution of every organ was similar. However, organs’ function is believed to be different, which suggests that the gene most highly expressed in one organ could be different from that in the other. According to the results of cluster analysis at the expression level (Fig. 1B), the function of the colon is near the ileum, then to the duodenum and stomach, which is easy to be understood. The function of the kidney is near to the adrenal gland, then to the heart and brain; and the spleen's function is near to the lung. To our surprise, the function of the liver was far from that of the other organs.
Fig. (1).
Distribution of gene expression and clustering analysis was made from 32,623 genes’ transcripts detected. The distribution of gene expression in different organs was similar (Mean ± SD, n = 3) (A). However, the function of the organs was different based on the clustering analysis of total gene expression from 11 organs (n = 3) (B). Abbreviations: Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Il, ileum; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; St, stomach.
3.2. Genes with Description Selectively Expressed in Different Organs
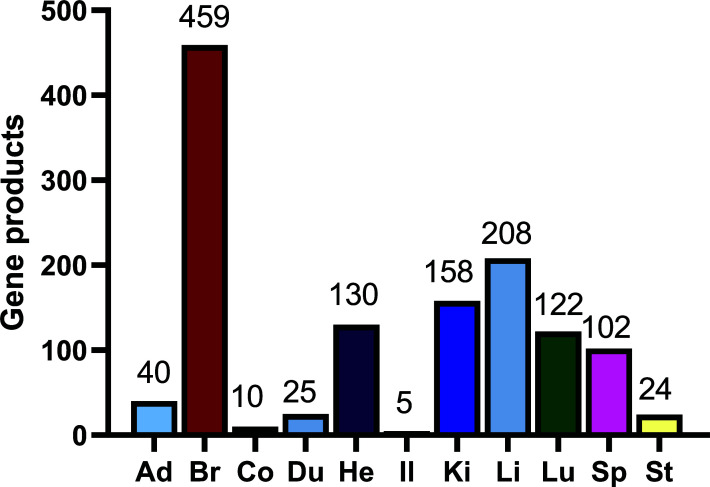
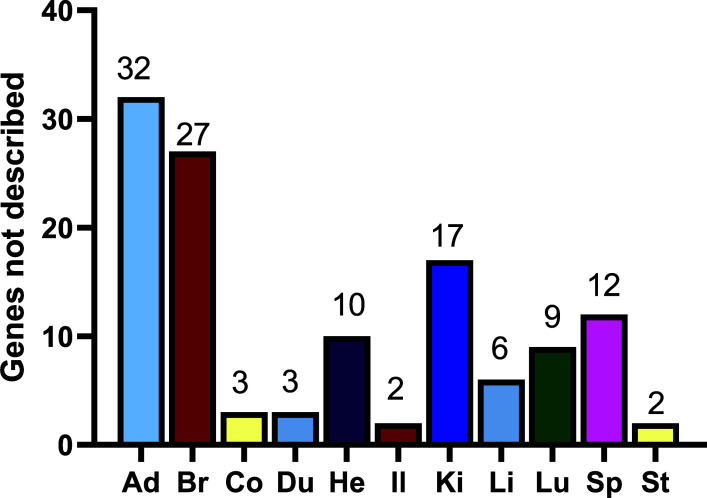
There were 15,922 genes with FPKM in any organ above 5, and 14,115 genes were significantly (p < 0.05) highly expressed in an organ. Among them, there were 12,617 genes accepted with q < 0.05. Apart from 123 genes without description, there were 1,283 genes with description selectively expressed in 11 organs (Fig. 2). From the results from Fig. (2), the brain (Br) was the organ with the most complex function because 459 genes were selectively expressed in it. Instead, the gastrointestinal tracts, including the stomach (St), duodenum (Du), ileum (Il), and colon (Co), selectively expressed fewer genes, suggesting that their functions could be relatively simple or similar to other organs.
Fig. (2).
Genes selectively expressed in different organs based on their abundance. Abbreviations: Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Il, ileum; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; St, stomach.
The total genes selectively expressed or the top 20 (if more) in 11 organs are listed in Tables 1-11. Their full lists can be seen in the supplementary data. According to the description of the gene name, most selective genes were associated with the known specific functions of the organ. For example, Mgarp (mitochondria-localized glutamic acid-rich protein) in the adrenal gland (Table 1) is associated with steroidogenesis [28]; Scg3 (secretogranin III) in the brain (Table 2) with neuroendocrine [29]; Reg3g (regenerating islet-derived 3 gamma) in the colon (Table 3) with intestinal bacterial translocation to the mesenteric lymph nodes [30]; Gip (gastric inhibitory polypeptide) in the duodenum (Table 4) with regulation of insulin secretion [31]; Klhl38 (kelch-like family member 38) in the heart (Table 5), though seldom reported, could be associated with the reversion of striated muscle atrophy [32]; Defa24 (defensin alpha 24) in the ileum (Table 6) with intestinal barrier [33]; Slc3a1 [solute carrier family 3 (amino acid transporter heavy chain), member 1] in the kidney (Table 7) with the transport of cystine and other amino acids across the membrane [34]; C5 (hemolytic complement) in the liver (Table 8) was early verified to execute innate immune [35]; Icam1 (intercellular adhesion molecule 1) in the lung (Table 9) with innate immune [36]; Coch (cochlin) used to highly expressed in the inner ear [37] also highly expressed in the spleen (Table 10); and Cxcl17 (chemokine (C-X-C motif) ligand 17) in the stomach (Table 11) with its innate immune [38]. Nevertheless, there were many genes that were not reported in the relative organs (supplementary data).
Table 1.
Top 20 of 40 genes with description selectively expressed in the adrenal gland (Ad) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | - | - | - | |||||
| 1 | Mgarp | Mitochondria-localized glutamic acid-rich protein | [39] | He | 469.5 | 473.5 | 1.018E-06 | 0.000 | 0.992 |
| 2 | Lrcol1 | Leucine rich colipase-like 1 | - | Ki | 7.6 | 7.8 | 1.165E-06 | 0.000 | 0.967 |
| 3 | Cyp21a1 | Cytochrome P450, family 21, subfamily a, polypeptide 1 | [40] | St | 9139.9 | 9148.9 | 2.277E-06 | 0.001 | 0.999 |
| 4 | Akr1b7 | Aldo-keto reductase family 1, member B7 | [41] | Ki | 2280.6 | 2281.5 | 5.124E-06 | 0.001 | 1.000 |
| 5 | Cyp11b2 | Cytochrome P450, family 11, subfamily b, polypeptide 2 | [42] | Lu | 327.8 | 337.2 | 5.121E-05 | 0.003 | 0.972 |
| 6 | Mir450a1 | MicroRNA 450a1 | - | St | 12.8 | 13.2 | 9.565E-05 | 0.004 | 0.967 |
| 7 | Star | Steroidogenic acute regulatory protein | [43] | St | 1438.9 | 1457.2 | 1.444E-04 | 0.005 | 0.987 |
| 8 | Ceacam16 | Carcinoembryonic antigen-related cell adhesion molecule 16 | - | Co | 73.6 | 74.6 | 1.595E-04 | 0.005 | 0.986 |
| 9 | Mrap | Melanocortin 2 receptor accessory protein | [44] | St | 413.8 | 448.9 | 1.809E-04 | 0.006 | 0.922 |
| 10 | Nkain3 | Na+/K+ transporting ATPase interacting 3 | - | St | 6.6 | 7.9 | 2.328E-04 | 0.006 | 0.837 |
| 11 | Nr0b1 | Nuclear receptor subfamily 0, group B, member 1 | [45] | Co | 41.1 | 42.0 | 2.828E-04 | 0.007 | 0.979 |
| 12 | Pbx4 | Pre-B-cell leukemia homeobox 4 | - | Du | 15.4 | 21.5 | 3.187E-04 | 0.007 | 0.715 |
| 13 | Slc27a3 | Solute carrier family 27 (fatty acid transporter), member 3 | - | St | 141.2 | 167.2 | 3.292E-04 | 0.007 | 0.844 |
| 14 | Mc2r | Melanocortin 2 receptor (adrenocorticotropic hormone) | [46] | St | 58.6 | 63.3 | 3.388E-04 | 0.008 | 0.925 |
| 15 | Eepd1 | Endonuclease/exonuclease/phosphatase family domain containing 1 | - | Co | 561.4 | 668.3 | 3.895E-04 | 0.008 | 0.840 |
| 16 | Nr5a1 | Nuclear receptor subfamily 5, group A, member 1 | [47] | Br | 51.8 | 61.4 | 3.918E-04 | 0.008 | 0.843 |
| 17 | Tmem200a | Transmembrane protein 200A | - | St | 23.9 | 30.3 | 4.488E-04 | 0.009 | 0.789 |
| 18 | LOC108348086 | Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 | - | Du | 557.5 | 558.7 | 4.900E-04 | 0.009 | 0.998 |
| 19 | Fdx1 | Ferredoxin 1 | [48] | Du | 2301.0 | 2657.4 | 5.368E-04 | 0.010 | 0.866 |
| 20 | Cyp11a1 | Cytochrome P450, family 11, subfamily a, polypeptide 1 | [49] | Co | 4795.7 | 4802.2 | 5.905E-04 | 0.010 | 0.999 |
Note: Sorted by q-value. Br, brain; Co, colon; Du, duodenum; He, heart; Ki, kidney; Lu, lung; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 11.
Top 20 of 24 genes with description selectively expressed in the stomach (St) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | Cxcl17 | Chemokine (C-X-C motif) ligand 17 | [152] | Br | 822.1 | 1042.0 | 1.41E-09 | 1.07E-05 | 0.789 |
| 2 | Kcnk16 | Potassium channel, two pore domain subfamily K, member 16 | - | Co | 7.8 | 9.2 | 4.69E-07 | 1.11E-04 | 0.855 |
| 3 | Anxa10 | Annexin A10 | [153] | Br | 946.4 | 954.5 | 1.83E-06 | 5.52E-04 | 0.991 |
| 4 | Fxyd3 | FXYD domain-containing ion transport regulator 3 | [154] | Li | 1153.4 | 1435.8 | 3.18E-05 | 2.04E-03 | 0.803 |
| 5 | Ptf1a | Pancreas-specific transcription factor, 1a | [155] | Ad | 10.1 | 12.0 | 1.99E-04 | 5.83E-03 | 0.849 |
| 6 | Slc9a4 | Solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 | [156] | Lu | 59.4 | 64.8 | 3.05E-04 | 7.22E-03 | 0.917 |
| 7 | Slc9b2 | Solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 | - | Sp | 18.4 | 22.4 | 4.37E-04 | 8.55E-03 | 0.820 |
| 8 | Adam28 | ADAM metallopeptidase domain 28 | [157] | Ad | 44.4 | 46.2 | 5.68E-04 | 9.90E-03 | 0.963 |
| 9 | Macc1 | Metastasis associated in colon cancer 1 | [158] | Li | 8.7 | 10.1 | 9.53E-04 | 1.29E-02 | 0.862 |
| 10 | Slc26a9 | Solute carrier family 26 (anion exchanger), member 9 | [159] | Ki | 98.8 | 116.7 | 9.95E-04 | 1.32E-02 | 0.847 |
| 11 | Psca | Prostate stem cell antigen | [160] | Co | 10716.9 | 10801.0 | 1.13E-03 | 1.41E-02 | 0.992 |
| 12 | Ghrl | Ghrelin/obestatin prepropeptide | [161] | Sp | 1965.4 | 2120.2 | 1.63E-03 | 1.70E-02 | 0.927 |
| 13 | Vsig1 | V-set and immunoglobulin domain containing 1 | [162] | Br | 270.6 | 274.9 | 2.01E-03 | 1.89E-02 | 0.984 |
| 14 | Pik3c2g | Phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma | - | Co | 19.2 | 25.9 | 2.39E-03 | 2.07E-02 | 0.741 |
| 15 | Atp4b | ATPase, H+/K+ exchanging, beta polypeptide | [163] | Du | 3191.2 | 3201.1 | 2.63E-03 | 2.18E-02 | 0.997 |
| 16 | Slc26a7 | Solute carrier family 26 (anion exchanger), member 7 | [164] | Co | 17.0 | 19.5 | 2.72E-03 | 2.21E-02 | 0.876 |
| 17 | Atp4a | ATPase, H+/K+ exchanging, alpha polypeptide | [163] | Ki | 1945.2 | 1952.4 | 3.51E-03 | 2.53E-02 | 0.996 |
| 18 | Clic6 | Chloride intracellular channel 6 | [165] | He | 230.4 | 241.4 | 5.96E-03 | 3.34E-02 | 0.954 |
| 19 | Gkn1 | Gastrokine 1 | [166] | Ad | 58685.7 | 59018.3 | 6.44E-03 | 3.48E-02 | 0.994 |
| 20 | Hdc | Histidine decarboxylase | [167] | Sp | 154.9 | 178.9 | 9.09E-03 | 4.19E-02 | 0.866 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Ki, kidney; Li, liver; Lu, lung; Sp, spleen.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 2.
Top 20 of 459 genes with description selectively expressed in the brain (Br) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | Dio2 | Preoptic regulatory factor 1 | [50] | Sp | 11.5 | 14.1 | 5.821E-08 | 3.324E-07 | 0.82 |
| 2 | Scg3 | Secretogranin III | [51] | Du | 197.4 | 215.7 | 4.057E-09 | 1.592E-05 | 0.91 |
| 3 | Gabbr1 | Gamma-aminobutyric acid (GABA) B receptor 1 | [52] | Ad | 448.3 | 627.1 | 2.797E-06 | 2.563E-05 | 0.71 |
| 4 | Asic2 | Acid-sensing (proton-gated) ion channel 2 | [53] | Co | 19.1 | 25.8 | 8.626E-08 | 9.567E-05 | 0.74 |
| 5 | Adcyap1r1 | Adenylate cyclase-activating polypeptide 1 receptor type 1 | [54] | Co | 36.9 | 44.8 | 1.593E-07 | 1.165E-04 | 0.82 |
| 6 | Chst10 | Carbohydrate sulfotransferase 10 | [55] | He | 37.1 | 47.6 | 3.197E-06 | 3.522E-04 | 0.78 |
| 7 | Larp6 | La ribonucleoprotein domain family, member 6 | - | St | 17.8 | 24.1 | 3.871E-06 | 3.944E-04 | 0.74 |
| 8 | Vsnl1 | Visinin-like 1 | [56] | Il | 406.5 | 450.9 | 1.252E-06 | 4.572E-04 | 0.90 |
| 9 | Snap91 | Synaptosomal-associated protein 91 | [57] | Co | 139.3 | 148.7 | 1.341E-06 | 4.692E-04 | 0.94 |
| 10 | Tceal3 | Transcription elongation factor A (SII)-like 6 | [58] | He | 132.8 | 141.2 | 2.179E-06 | 5.977E-04 | 0.94 |
| 11 | Pdzd4 | PDZ domain containing 4 | [59] | Du | 42.5 | 50.0 | 2.611E-06 | 6.401E-04 | 0.85 |
| 12 | LOC100911402 | Cell cycle exit and neuronal differentiation 1 | - | He | 231.8 | 236.2 | 3.063E-06 | 6.991E-04 | 0.98 |
| 13 | Acsbg1 | Acyl-CoA synthetase bubblegum family member 1 | - | Lu | 106.6 | 126.1 | 3.093E-06 | 7.091E-04 | 0.85 |
| 14 | Gdap1l1 | Ganglioside-induced differentiation-associated protein 1-like 1 | [60] | Du | 70.6 | 77.1 | 3.576E-06 | 7.453E-04 | 0.92 |
| 15 | Adgrb3 | Adhesion G protein-coupled receptor B3 | [61] | Du | 22.1 | 22.8 | 3.932E-06 | 7.981E-04 | 0.97 |
| 16 | Fam131b | Family with sequence similarity 131, member B | [62] | Lu | 56.2 | 58.1 | 3.942E-06 | 8.091E-04 | 0.97 |
| 17 | Plp1 | Proteolipid protein 1 | [63] | He | 1572.6 | 1599.8 | 4.805E-06 | 8.959E-04 | 0.98 |
| 18 | Nipal4 | NIPA-like domain containing 4 | - | Du | 6.3 | 7.9 | 1.199E-05 | 9.210E-04 | 0.79 |
| 19 | RragB | Ras-related GTP-binding protein B-like | [64] | He | 15.2 | 19.4 | 2.190E-05 | 1.143E-03 | 0.78 |
| 20 | Stmn3 | Stathmin-like 3 | [65] | Du | 887.3 | 918.2 | 8.338E-06 | 1.180E-03 | 0.97 |
Note: Sorted by q-value. Ad, adrenal gland; Co, colon; Du, duodenum; He, heart; Il, ileum; Ki, kidney; Lu, lung; Sp, spleen; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 3.
Top genes with description selectively expressed in the colon (Co) based on their abundance (n = 3).
| No | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | Q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | Reg3g | Regenerating islet-derived 3 gamma | [66] | Lu | 9161.7 | 12157.8 | 1.46E-08 | 4.93E-05 | 0.754 |
| 2 | Reg3b | Regenerating islet-derived 3 beta | [66] | Br | 6569.9 | 8784.4 | 6.70E-06 | 1.06E-03 | 0.748 |
| 3 | St6galnac1 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N- acetylgalactosaminide alpha-2,6-sialyltransferase 1 | [67] | Br | 286.1 | 294.8 | 7.56E-06 | 1.12E-03 | 0.971 |
| 4 | Ighg | Immunoglobulin heavy chain (gamma polypeptide) | [68] | Br | 78.0 | 106.8 | 7.38E-05 | 2.67E-03 | 0.730 |
| 5 | Hmcn2 | Hemicentin 2 | - | St | 30.5 | 31.2 | 1.55E-04 | 5.12E-03 | 0.977 |
| 6 | LOC290595 | Hypothetical gene supported by AF152002 | - | Ad | 103.0 | 146.0 | 1.75E-04 | 5.46E-03 | 0.706 |
| 7 | Ace | Angiotensin I converting enzyme | [69] | St | 51.5 | 59.7 | 6.53E-04 | 1.06E-02 | 0.861 |
| 8 | LOC691670 | Similar to natural killer cell protease 7 | - | Sp | 11.1 | 15.4 | 6.77E-03 | 3.56E-02 | 0.724 |
| 9 | Fgf19 | Fibroblast growth factor 19 | [70] | Ad | 41.2 | 43.2 | 9.63E-03 | 4.32E-02 | 0.953 |
| 10 | Mir192 | MicroRNA 192 | [71] | Ad | 6.9 | 6.9 | 1.17E-02 | 4.80E-02 | 1.000 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Lu, lung; Sp, spleen; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 4.
Top 20 of 25 genes with description selectively expressed in the duodenum (Du) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Refs.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | Gip | Gastric inhibitory polypeptide | [72] | Ad | 79.2 | 81.4 | 7.42E-06 | 1.11E-03 | 0.973 |
| 2 | LOC100910259 | Liver carboxylesterase-like | - | Sp | 498.7 | 699.5 | 5.32E-05 | 2.99E-03 | 0.713 |
| 3 | Prap1 | Proline-rich acidic protein 1 | - | Br | 4433.8 | 4622.2 | 9.65E-05 | 4.04E-03 | 0.959 |
| 4 | Papss2 | 3'-phosphoadenosine 5'-phosphosulfate synthase 2 | - | Sp | 649.9 | 779.5 | 1.20E-04 | 4.51E-03 | 0.834 |
| 5 | Tm4sf5 | Transmembrane 4 L six family member 5 | - | Ad | 940.9 | 1272.0 | 1.36E-04 | 4.81E-03 | 0.740 |
| 6 | RGD1311933 | Similar to RIKEN cDNA 2310057J18 | - | Ad | 221.2 | 221.9 | 2.62E-04 | 6.69E-03 | 0.997 |
| 7 | Cyp2c7 | Cytochrome P450, family 2, subfamily c, polypeptide 7 | - | Ad | 48.0 | 50.7 | 3.56E-04 | 7.82E-03 | 0.947 |
| 8 | Aadac | Arylacetamide deacetylase | - | St | 96.4 | 133.8 | 6.93E-04 | 1.10E-02 | 0.720 |
| 9 | Tmprss15 | Transmembrane protease, serine 15 | - | Sp | 138.8 | 139.2 | 7.87E-04 | 1.17E-02 | 0.997 |
| 10 | RGD1561551 | Similar to Hypothetical protein MGC75664 | - | Ad | 842.1 | 842.9 | 1.28E-03 | 1.50E-02 | 0.999 |
| 11 | Alppl2 | Alkaline phosphatase, placental-like 2 | - | Co | 60.3 | 71.1 | 1.40E-03 | 1.57E-02 | 0.848 |
| 12 | Akp3 | Alkaline phosphatase 3, intestine, not Mn requiring | [73] | Ad | 2279.3 | 2280.1 | 1.67E-03 | 1.72E-02 | 1.000 |
| 13 | Ada | Adenosine deaminase | [74] | Ki | 1461.9 | 2071.3 | 1.74E-03 | 1.76E-02 | 0.706 |
| 14 | Bco1 | Beta-carotene oxygenase 1 | [75] | Ki | 160.1 | 210.3 | 1.78E-03 | 1.78E-02 | 0.761 |
| 15 | Slc4a7 | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | [75, 76] | St | 108.5 | 137.6 | 1.90E-03 | 1.79E-02 | 0.789 |
| 16 | Alpi | Alkaline phosphatase, intestinal | [77] | Br | 1098.5 | 1193.8 | 1.84E-03 | 1.81E-02 | 0.920 |
| 17 | Treh | Trehalase (brush-border membrane glycoprotein) | [78] | Ki | 260.7 | 268.5 | 2.44E-03 | 2.09E-02 | 0.971 |
| 18 | Trpv6 | Transient receptor potential cation channel, subfamily V, member 6 | [79] | Sp | 24.2 | 32.7 | 2.45E-03 | 2.10E-02 | 0.741 |
| 19 | Otop3 | Otopetrin 3 | - | Co | 69.3 | 70.2 | 3.51E-03 | 2.53E-02 | 0.987 |
| 20 | Pdx1 | Pancreatic and duodenal homeobox 1 | [80] | Ad | 58.6 | 61.7 | 4.91E-03 | 3.02E-02 | 0.950 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Co, colon; Ki, kidney; Sp, spleen; St, stomach.
* Last Refs. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 5.
The top 20 of 130 genes with description are selectively expressed in the heart (He) based on their abundance(n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | Klhl38 | Kelch-like family member 38 | [81] | St | 10.4 | 13.4 | 4.25E-07 | 1.64E-04 | 0.776 |
| 2 | Rbm24 | RNA binding motif protein 24 | [82, 83] | Co | 45.0 | 58.7 | 7.37E-07 | 1.98E-04 | 0.768 |
| 3 | Ldb3 | LIM domain binding 3 | [84] | St | 541.1 | 590.9 | 5.15E-07 | 2.81E-04 | 0.916 |
| 4 | LOC100909784 | Leiomodin 2 (cardiac) | - | St | 92.6 | 93.6 | 5.36E-07 | 2.99E-04 | 0.989 |
| 5 | Hspb2 | Heat shock protein B2 | [85] | St | 183.4 | 212.0 | 2.62E-06 | 4.35E-04 | 0.865 |
| 6 | Itgb1bp2 | Integrin beta 1 binding protein 2 | [86] | Du | 121.2 | 140.6 | 1.30E-06 | 4.44E-04 | 0.862 |
| 7 | Klhl31 | Kelch-like family member 31 | [87] | Sp | 57.0 | 58.4 | 1.22E-06 | 4.49E-04 | 0.975 |
| 8 | Tnni3k | TNNI3 interacting kinase | [88] | Ad | 85.4 | 86.6 | 1.26E-06 | 4.58E-04 | 0.986 |
| 9 | Pla2g5 | Phospholipase A2, Group V | [89] | Sp | 54.0 | 60.0 | 2.19E-06 | 4.89E-04 | 0.899 |
| 10 | Fsd2 | Fibronectin type III and SPRY domain containing 2 | [90] | Du | 47.6 | 48.3 | 1.87E-06 | 5.56E-04 | 0.986 |
| 11 | Tmem182 | Transmembrane protein 182 | [91] | Ki | 79.9 | 84.0 | 2.18E-06 | 5.88E-04 | 0.951 |
| 12 | Rd3l | Retinal degeneration 3-like | - | Ad | 18.5 | 23.6 | 7.90E-06 | 1.10E-03 | 0.785 |
| 13 | Nkx2-5 | NK2 homeobox 5 | [92] | Lu | 75.9 | 84.4 | 7.41E-06 | 1.11E-03 | 0.899 |
| 14 | Sgcg | Sarcoglycan, gamma | [93] | Il | 93.4 | 110.5 | 1.83E-05 | 1.13E-03 | 0.845 |
| 15 | Hhatl | Hedgehog acyltransferase-like | [94] | Ki | 119.2 | 133.1 | 8.86E-06 | 1.22E-03 | 0.896 |
| 16 | Cav3 | Caveolin 3 | [95] | Sp | 116.1 | 123.7 | 9.50E-06 | 1.24E-03 | 0.939 |
| 17 | LOC691485 | Hypothetical protein LOC691485 | - | Br | 24.1 | 29.9 | 2.37E-05 | 1.26E-03 | 0.807 |
| 18 | Kbtbd12 | Kelch repeat and BTB (POZ) domain containing 12 | - | St | 16.2 | 18.2 | 1.23E-05 | 1.35E-03 | 0.891 |
| 19 | Txlnb | Taxilin beta | - | Co | 68.5 | 69.3 | 1.57E-05 | 1.62E-03 | 0.987 |
| 20 | Spink8 | Serine peptidase inhibitor, Kazal type 8 | - | Br | 166.1 | 187.3 | 2.10E-05 | 1.74E-03 | 0.887 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; Il, ileum; Ki, kidney; Lu, lung; Sp, spleen; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 20123.
Table 6.
Top genes with description selectively expressed in the ileum (Il) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | LOC100910656 | rCG60069-like | - | Sp | 244.0 | 341.0 | 0.001 | 0.011 | 0.715 |
| 2 | Defa24 | Defensin alpha 24 | - | Ad | 15591.7 | 19391.4 | 0.001 | 0.011 | 0.804 |
| 3 | Fabp6 | Fatty acid binding protein 6, ileal | [96] | Ki | 51493.4 | 56686.5 | 0.001 | 0.012 | 0.908 |
| 4 | Defal1 | Defensin alpha-like 1 | - | Ad | 29241.0 | 34877.4 | 0.001 | 0.015 | 0.838 |
| 5 | Pla2g4c | Phospholipase A2, group IVC-like 1 | - | St | 26.1 | 30.0 | 0.005 | 0.030 | 0.869 |
Note: Sorted by q-value. Ad, adrenal gland; Ki, kidney; Sp, spleen; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 7.
Top 20 of 158 genes with description selectively expressed in the kidney (Ki) based on their abundance(n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | C1qtnf3 | C1q and tumor necrosis factor-related protein 3 | - | He | 32.5 | 42.7 | 7.02E-07 | 7.13E-07 | 0.760 |
| 2 | Pter | Phosphotriesterase related | [97] | Co | 184.9 | 239.7 | 1.00E-08 | 2.91E-06 | 0.771 |
| 3 | Gclc | Glutamate-cysteine ligase, catalytic subunit | [98] | Ad | 1920.8 | 2266.6 | 1.91E-08 | 1.55E-05 | 0.847 |
| 4 | Slc3a1 | Solute carrier family 3 (amino acid transporter heavy chain), member 1 | [99] | He | 1569.4 | 1923.0 | 5.63E-09 | 3.01E-05 | 0.816 |
| 5 | Trpv4 | Transient receptor potential cation channel, subfamily V, member 4 | [100] | St | 30.2 | 37.6 | 1.65E-06 | 4.90E-05 | 0.803 |
| 6 | Skint10 | Selection and upkeep of intraepithelial T cells 10 | - | Ad | 5.3 | 5.5 | 2.71E-08 | 6.73E-05 | 0.965 |
| 7 | LOC688553 | Hypothetical protein LOC688553 | - | Du | 62.3 | 71.8 | 1.07E-06 | 1.09E-04 | 0.868 |
| 8 | Stra6 | Stimulated by retinoic acid 6 | [101] | Sp | 22.4 | 25.8 | 4.59E-07 | 1.45E-04 | 0.868 |
| 9 | RGD1310495 | Similar to KIAA1919 protein | - | Il | 71.5 | 82.3 | 1.64E-07 | 1.64E-04 | 0.869 |
| 10 | Wdr72 | WD repeat domain 72 | [102] | Il | 8.0 | 9.9 | 1.94E-07 | 1.80E-04 | 0.805 |
| 11 | Haao | 3-hydroxyanthranilate 3,4-dioxygenase | [103] | Il | 444.3 | 616.7 | 2.08E-07 | 1.86E-04 | 0.720 |
| 12 | Emx2 | Empty spiracles homeobox 2 | [104] | St | 13.9 | 16.2 | 2.79E-07 | 2.16E-04 | 0.857 |
| 13 | Gba3 | Glucosidase, beta, acid 3 | [105] | Ad | 172.9 | 173.3 | 3.61E-07 | 2.45E-04 | 0.998 |
| 14 | Car12 | Carbonic anyhydrase 12 | [106] | Ad | 352.9 | 454.9 | 2.45E-06 | 2.67E-04 | 0.776 |
| 15 | Pdzk1ip1 | PDZK1 interacting protein 1 | [107] | Du | 390.1 | 434.3 | 4.90E-07 | 2.77E-04 | 0.898 |
| 16 | Spo11 | SPO11 meiotic protein covalently bound to DSB | [108] | Br | 6.9 | 8.8 | 2.97E-06 | 2.93E-04 | 0.787 |
| 17 | Slc6a18 | Solute carrier family 6 (neutral amino acid transporter), member 18 | [109] | Ad | 273.3 | 274.3 | 7.07E-07 | 3.44E-04 | 0.996 |
| 18 | Glyat | Glycine-N-acyltransferase | [110] | Ad | 756.4 | 947.6 | 1.17E-06 | 4.42E-04 | 0.798 |
| 19 | Aspa | Aspartoacylase | [111] | Li | 140.1 | 199.1 | 4.18E-06 | 5.53E-04 | 0.703 |
| 20 | Cyp4a2 | Cytochrome P450, family 4, subfamily a, polypeptide 2 | [112] | Co | 561.2 | 737.0 | 2.15E-06 | 5.99E-04 | 0.761 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Il, ileum; Li, liver; Sp, spleen; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 8.
Top 20 of 208 genes with description selectively expressed in the liver (Li) based on their abundance (n = 3).
| No | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | C5 | Hemolytic complement | [113] | Sp | 118.2 | 142.0 | 9.584E-10 | 1.152E-05 | 0.833 |
| 2 | Serpind1 | Serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | [114] | Ad | 389.6 | 390.6 | 9.631E-08 | 1.267E-04 | 0.997 |
| 3 | Saa4 | Hermansky-Pudlak syndrome 5 | [115] | Ki | 691.8 | 743.6 | 1.417E-07 | 1.510E-04 | 0.930 |
| 4 | Crp | C-reactive protein, pentraxin-related | [116] | Ki | 5777.4 | 5787.5 | 1.605E-07 | 1.636E-04 | 0.998 |
| 5 | C8b | Complement component 8, beta polypeptide | [117] | Ad | 295.8 | 297.0 | 1.661E-07 | 1.664E-04 | 0.996 |
| 6 | C4bpa | Complement component 4 binding protein, alpha | [118] | He | 295.7 | 308.0 | 2.467E-07 | 2.024E-04 | 0.960 |
| 7 | Cfi | Complement factor I | [118] | Ki | 469.3 | 534.4 | 4.264E-07 | 2.665E-04 | 0.878 |
| 8 | C8g | Complement component 8, gamma polypeptide | [119] | Br | 180.5 | 214.3 | 6.792E-07 | 3.024E-04 | 0.842 |
| 9 | Slc13a4 | Solute carrier family 13 (sodium/sulfate symporter), member 4 | [120] | Il | 27.9 | 38.9 | 6.273E-07 | 3.033E-04 | 0.718 |
| 10 | Tmprss6 | Transmembrane protease, serine 6 | [121] | Il | 170.2 | 171.0 | 5.620E-07 | 3.060E-04 | 0.995 |
| 11 | Uroc1 | Urocanate hydratase 1 | - | Ki | 100.5 | 101.0 | 6.137E-07 | 3.200E-04 | 0.995 |
| 12 | Afm | Afamin | [122] | Br | 694.3 | 744.9 | 6.160E-07 | 3.206E-04 | 0.932 |
| 13 | Mug1 | Alpha-1-inhibitor III | [123] | Ad | 5659.4 | 5677.1 | 8.210E-07 | 3.702E-04 | 0.997 |
| 14 | Mbl1 | Mannose-binding lectin (protein A) 1 | [124] | Sp | 230.0 | 249.3 | 1.212E-06 | 4.497E-04 | 0.922 |
| 15 | F10 | Coagulation factor X | [125] | Il | 292.5 | 297.8 | 1.706E-06 | 5.317E-04 | 0.982 |
| 16 | LOC 100909524 |
Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 | - | Br | 95.6 | 98.0 | 1.825E-06 | 5.477E-04 | 0.975 |
| 17 | Slc38a4 | Solute carrier family 38, member 4 | [126] | St | 209.4 | 212.6 | 1.996E-06 | 5.758E-04 | 0.985 |
| 18 | Glyatl1 | Glycine-N-acyltransferase-like 1 | [127] | Il | 109.9 | 116.8 | 2.111E-06 | 5.936E-04 | 0.941 |
| 19 | C4bpb | Complement component 4 binding protein, beta | [118] | Ki | 305.3 | 313.2 | 2.177E-06 | 6.020E-04 | 0.975 |
| 20 | Pzp | Pregnancy-zone protein | [128] | Il | 2009.8 | 2053.1 | 2.243E-06 | 6.122E-04 | 0.979 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; He, heart; Il, ileum; Ki, kidney; Sp, spleen; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 9.
Top 20 of 122 genes with description selectively expressed in rat lung (Lu) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | St8sia2 | ST8 alpha-N-acetyl-neuraminide alpha-2,8- sialyltransferase 2 | [129] | Il | 7.0 | 8.1 | 3.25E-07 | 6.25E-07 | 0.872 |
| 2 | Ly6l | Lymphocyte antigen 6 family member L | [130] | Br | 77.2 | 103.7 | 5.49E-08 | 5.61E-05 | 0.745 |
| 3 | Icam1 | Intercellular adhesion molecule 1 | [131] | Il | 242.5 | 329.3 | 1.12E-06 | 7.80E-05 | 0.736 |
| 4 | LOC102546678 | Proline-rich Gla (G-carboxyglutamic acid) 3 (transmembrane) | - | Il | 18.0 | 20.5 | 1.97E-07 | 1.37E-04 | 0.879 |
| 5 | LOC102554838 | Stathmin domain-containing protein 1-like | - | Co | 6.2 | 8.6 | 2.53E-07 | 2.02E-04 | 0.726 |
| 6 | Thbd | Thrombomodulin | [132] | Il | 297.8 | 387.8 | 1.70E-06 | 2.35E-04 | 0.768 |
| 7 | Matn4 | Matrilin 4 | [133] | Ad | 36.4 | 46.0 | 2.30E-06 | 3.47E-04 | 0.791 |
| 8 | LOC681341 | Similar to paired immunoglobin-like type 2 receptor β | - | Co | 11.6 | 15.8 | 2.74E-06 | 3.79E-04 | 0.733 |
| 9 | Prrg3 | Proline-rich Gla (G-carboxyglutamic acid) 3 (transmembrane) | - | Co | 17.5 | 19.4 | 1.66E-06 | 5.23E-04 | 0.903 |
| 10 | Lhb | Luteinizing hormone beta polypeptide | [134] | Ki | 10.3 | 13.9 | 9.59E-06 | 5.31E-04 | 0.746 |
| 11 | Acvrl1 | Activin A receptor type II-like 1 | [135] | Ki | 238.0 | 336.0 | 2.12E-06 | 5.85E-04 | 0.708 |
| 12 | Pifo | Primary cilia formation | [136] | Ad | 6.3 | 7.9 | 2.64E-06 | 6.65E-04 | 0.803 |
| 13 | Scgb1a1 | Secretoglobin, family 1A, member 1 (uteroglobin) | [137] | Ad | 21465.0 | 21576.2 | 3.55E-06 | 7.70E-04 | 0.995 |
| 14 | Fhad1 | Forkhead-associated (FHA) phosphopeptide binding domain 1 | - | Ki | 9.2 | 10.7 | 5.42E-06 | 7.71E-04 | 0.854 |
| 15 | Nme9 | NME/NM23 family member 9 | - | Ad | 9.0 | 10.4 | 3.60E-06 | 7.76E-04 | 0.868 |
| 16 | RGD1561648 | RGD1561648 | - | Co | 7.6 | 10.6 | 9.11E-06 | 8.24E-04 | 0.718 |
| 17 | LOC108348266 | Cytochrome P450, family 2, subfamily b, polypeptide 1 | - | Br | 528.5 | 702.5 | 6.04E-06 | 1.00E-03 | 0.752 |
| 18 | Dram1 | DNA-damage regulated autophagy modulator 1 | [138] | Ad | 133.1 | 177.1 | 1.04E-05 | 1.04E-03 | 0.752 |
| 19 | Limch1 | LIM and calponin homology domains 1 | [139] | St | 174.7 | 216.9 | 8.99E-06 | 1.17E-03 | 0.805 |
| 20 | LOC680885 | Hypothetical protein LOC680885 | - | Ad | 14.2 | 15.3 | 1.09E-05 | 1.35E-03 | 0.928 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Co, colon; Il, ileum; Ki, kidney; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
Table 10.
Top 20 of 102 genes with description selectively expressed in the spleen (Sp) based on their abundance (n = 3).
| No. | Gene Name | Product (Description) | Last Ref.* | Median Organ | FPKM | p-value | q-value | Mean/total | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | Total | ||||||||
| 1 | Coch | Cochlin | [140] | Il | 318.4 | 345.0 | 6.96E-11 | 6.89E-08 | 0.923 |
| 2 | SNORD79 | Small nucleolar RNA, C/D box 79 | - | St | 13.3 | 17.8 | 6.87E-06 | 1.07E-03 | 0.747 |
| Tlx1 | T-cell leukemia, homeobox 1 | [141] | Br | 23.4 | 25.1 | 1.68E-05 | 1.66E-03 | 0.933 | |
| 3 | Erfe | Family with sequence similarity 132, member B | [142] | Du | 11.6 | 13.0 | 3.33E-05 | 2.17E-03 | 0.892 |
| 4 | Trim59 | Tripartite motif-containing 59 | [143] | Ki | 104.2 | 131.3 | 4.80E-05 | 2.60E-03 | 0.794 |
| 5 | Treml2 | Triggering receptor expressed on myeloid cells-like 2 | - | Du | 27.9 | 35.3 | 6.25E-05 | 3.24E-03 | 0.790 |
| SNORA4 | Small nucleolar RNA, H/ACA box 4 | - | St | 10.9 | 14.5 | 6.42E-05 | 3.29E-03 | 0.755 | |
| 6 | Spic | Spi-C transcription factor (Spi-1/PU.1 related) | [144] | Il | 38.4 | 43.6 | 7.65E-05 | 3.57E-03 | 0.880 |
| 7 | Adgre4 | EGF-like module containing mucin-like, hormone receptor-like sequence 4 | - | Du | 26.0 | 34.9 | 1.44E-04 | 4.25E-03 | 0.743 |
| 8 | Kel | Kell blood group, metallo-endopeptidase | [145] | Br | 140.8 | 146.6 | 1.10E-04 | 4.31E-03 | 0.961 |
| 9 | Tspo2 | Translocator protein 2 | - | Du | 45.0 | 47.3 | 1.18E-04 | 4.44E-03 | 0.950 |
| 10 | Defb36 | Defensin beta 36 | - | Ad | 6.1 | 7.3 | 1.70E-04 | 5.38E-03 | 0.833 |
| 11 | Icam4 | Intercellular adhesion molecule 4, Landsteiner-Wiener blood group | [146] | Ad | 20.1 | 22.7 | 1.93E-04 | 5.43E-03 | 0.884 |
| 12 | Mylk2 | Myosin light chain kinase 2 | - | Ad | 14.2 | 16.3 | 1.80E-04 | 5.49E-03 | 0.872 |
| 13 | Epb42 | Erythrocyte membrane protein band 4.2 | - | Ki | 88.5 | 91.6 | 1.95E-04 | 5.75E-03 | 0.966 |
| 14 | Tnn | Tenascin N | [147] | Ad | 8.0 | 9.3 | 2.22E-04 | 6.02E-03 | 0.862 |
| 15 | Grap2 | GRB2-related adaptor protein 2 | - | Br | 35.2 | 46.2 | 2.38E-04 | 6.32E-03 | 0.761 |
| 16 | Cxcl6 | Chemokine (C-X-C motif) ligand 6 | [148] | St | 6.3 | 8.0 | 3.06E-04 | 6.33E-03 | 0.791 |
| 17 | Clec4m | CD209b antigen | [149] | Ki | 45.6 | 46.6 | 2.35E-04 | 6.33E-03 | 0.978 |
| 18 | LOC681325 | Hypothetical protein LOC681325 | - | He | 17.2 | 20.7 | 2.59E-04 | 6.54E-03 | 0.830 |
| 19 | Ahsp | Alpha hemoglobin stabilizing protein | [150] | St | 2059.2 | 2118.7 | 2.61E-04 | 6.68E-03 | 0.972 |
| 20 | Rhag | Rh-associated glycoprotein | [151] | Il | 179.6 | 180.0 | 2.74E-04 | 6.84E-03 | 0.998 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Du, duodenum; He, heart; Il, ileum; Ki, kidney; St, stomach.
* Last Ref. was based on the reports documented in PubMed (www.pubmed.gov) before June 10, 2023.
3.3. KEGG and GO Pathway Enrichment
3.3.1. KEGG Pathway Enrichment
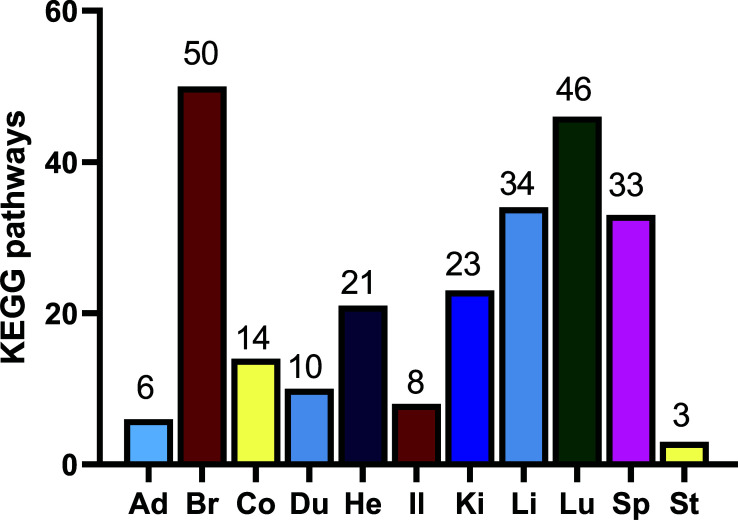
KEGG is a bioinformatics database resource for understanding high-level functions and utilities of the biological system, which includes the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies. The selective KEGG pathways were enriched based on the abundance of genes most highly expressed in organs. The number of the selective pathway is listed in Fig. (3) and the top 20 pathways are listed in Tables 12-22. Their full lists can be seen in the supplementary data. There were 179 “selective” pathways in 11 rat organs. Among them, 52 pathways were involved in two organs, 7 in three organs, and 1 in four organs. It should be noted that the “selective” pathways engaged in two or more organs were based on enrichment analysis. As can be seen from Fig. (3), organs with many selective pathways, like the brain, indicate that they undertake many complex functions. Conversely, organs with few selective pathways, like the adrenal glands and stomach, indicate their relatively simple functions. The results in Fig. (3), suggested that the lung could be the top 2 organs with the complex functions of the 11 organs.
Fig. (3).
Selective KEGG enrichment in different organs was based on the abundance of genes most highly expressed in organs. Abbreviations: Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Il, ileum; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; St, stomach.
Table 12.
Selective KEGG pathways in the adrenal gland.
| No | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko03010* | Ribosome | 21/283 | 133/5400 | 4.36E-06 | 0.001 |
| 2 | ko03050 | Proteasome | 10/283 | 39/5400 | 2.19E-05 | 0.002 |
| 3 | ko00061 | Fatty acid biosynthesis | 5/283 | 11/5400 | 1.36E-04 | 0.008 |
| 4 | ko03020 | RNA polymerase | 7/283 | 27/5400 | 3.61E-04 | 0.014 |
| 5 | ko04925* | Aldosterone synthesis and secretion | 9/283 | 44/5400 | 3.67E-04 | 0.014 |
| 6 | ko00240* | Pyrimidine metabolism | 12/283 | 78/5400 | 6.58E-04 | 0.020 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 22.
Selective KEGG pathways in the stomach.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04971* | Gastric acid secretion | 7/117 | 42/5400 | 2.70E-05 | 0.003 |
| 2 | ko04080* | Neuroactive ligand-receptor interaction | 14/117 | 218/5400 | 0.000 | 0.012 |
| 3 | ko04270* | Vascular smooth muscle contraction | 8/117 | 80/5400 | 0.0001 | 0.012 |
Note: * also significantly expressed in other organs. Sorted by q-value.
The function of some pathways was verified in relative organs based on common understandings, for example, ko04925 (Aldosterone synthesis and secretion) in the adrenal gland (Table 12), ko04721 (Synaptic vesicle cycle) in the brain (Table 13), ko04672 (Intestinal immune network for IgA production) in the colon (Table 14), ko04975 (Fat digestion and absorption) in the duodenum (Table 15), ko04260 (Cardiac muscle contraction) in the heart (Table 16), ko00520 (Amino sugar and nucleotide sugar metabolism) in the ileum (Table 17), ko04964 (Proximal tubule bicarbonate reclamation) in the kidney (Table 18), ko04976 (Bile secretion) in the liver (Table 19), ko04151 (PI3K-Akt signaling pathway) in the lung (Table 20), ko04640 (Hematopoietic cell lineage) in the spleen (Table 21), and ko04971 (Gastric acid secretion) in the stomach (Table 22).
Table 13.
Top 20 of 50 Selective KEGG pathways in the brain.
| No | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04721 | Synaptic vesicle cycle | 33/874 | 43/5400 | 0.000 | 0.000 |
| 2 | ko04724 | Glutamatergic synapse | 39/874 | 67/5400 | 0.000 | 0.000 |
| 3 | ko04723 | Retrograde endocannabinoid signaling | 38/874 | 65/5400 | 0.000 | 0.000 |
| 4 | ko04080* | Neuroactive ligand-receptor interaction | 77/874 | 218/5400 | 0.000 | 0.000 |
| 5 | ko04727 | GABAergic synapse | 31/874 | 55/5400 | 0.000 | 0.000 |
| 6 | ko04725 | Cholinergic synapse | 31/874 | 65/5400 | 0.000 | 0.000 |
| 7 | ko04728 | Dopaminergic synapse | 36/874 | 87/5400 | 0.000 | 0.000 |
| 8 | ko04713 | Circadian entrainment | 28/874 | 59/5400 | 0.000 | 0.000 |
| 9 | ko04360* | Axon guidance | 44/874 | 118/5400 | 0.000 | 0.000 |
| 10 | ko04020* | Calcium signaling pathway | 39/874 | 105/5400 | 0.000 | 0.000 |
| 11 | ko04726 | Serotonergic synapse | 30/874 | 73/5400 | 0.000 | 0.000 |
| 12 | ko04911 | Insulin secretion | 24/874 | 53/5400 | 0.000 | 0.000 |
| 13 | ko04921 | Oxytocin signaling pathway | 36/874 | 99/5400 | 0.000 | 0.000 |
| 14 | ko04024 | cAMP signaling pathway | 40/874 | 117/5400 | 0.000 | 0.000 |
| 15 | ko04540* | Gap junction | 26/874 | 63/5400 | 0.000 | 0.000 |
| 16 | ko04072* | Phospholipase D signaling pathway | 32/874 | 90/5400 | 0.000 | 0.000 |
| 17 | ko04261* | Adrenergic signaling in cardiomyocytes | 32/874 | 92/5400 | 0.000 | 0.000 |
| 18 | ko04114 | Oocyte meiosis | 29/874 | 80/5400 | 0.000 | 0.000 |
| 19 | ko04070 | Phosphatidylinositol signaling system | 23/874 | 58/5400 | 0.000 | 0.000 |
| 20 | ko04915 | Estrogen signaling pathway | 23/874 | 60/5400 | 0.000 | 0.000 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 14.
Selective KEGG pathways in the colon.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04630 | Jak-STAT signaling pathway | 19/218 | 94/5400 | 3.69E-09 | 5.12E-07 |
| 2 | ko04060* | Cytokine-cytokine receptor interaction | 23/218 | 163/5400 | 1.07E-07 | 7.46E-06 |
| 3 | ko04064* | NF-kappa B signaling pathway | 12/218 | 65/5400 | 8.39E-06 | 0.000 |
| 4 | ko04380* | Osteoclast differentiation | 13/218 | 87/5400 | 3.82E-05 | 0.001 |
| 5 | ko04210 | Apoptosis | 14/218 | 102/5400 | 5.09E-05 | 0.001 |
| 6 | ko04672* | Intestinal immune network for IgA production | 7/218 | 32/5400 | 2.25E-04 | 0.005 |
| 7 | ko04660* | T cell receptor signaling pathway | 11/218 | 78/5400 | 2.58E-04 | 0.005 |
| 8 | ko04071* | Sphingolipid signaling pathway | 11/218 | 85/5400 | 5.52E-04 | 0.010 |
| 9 | ko04214 | Apoptosis - fly | 7/218 | 43/5400 | 1.48E-03 | 0.021 |
| 10 | ko04620* | Toll-like receptor signaling pathway | 9/218 | 68/5400 | 1.49E-03 | 0.021 |
| 11 | ko04919* | Thyroid hormone signaling pathway | 9/218 | 69/5400 | 1.66E-03 | 0.021 |
| 12 | ko04621* | NOD-like receptor signaling pathway | 7/218 | 45/5400 | 1.94E-03 | 0.023 |
| 13 | ko04520* | Adherens junction | 7/218 | 46/5400 | 2.22E-03 | 0.024 |
| 14 | ko04068* | FoxO signaling pathway | 10/218 | 94/5400 | 4.34E-03 | 0.043 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 15.
Selective KEGG pathways in the duodenum.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko03010* | Ribosome | 29/264 | 133/5400 | 3.76E-12 | 6.37E-10 |
| 2 | ko04975 | Fat digestion and absorption | 10/264 | 26/5400 | 1.75E-07 | 1.48E-05 |
| 3 | ko04978* | Mineral absorption | 10/264 | 30/5400 | 8.30E-07 | 4.69E-05 |
| 4 | ko04974 | Protein digestion and absorption | 13/264 | 60/5400 | 4.44E-06 | 0.000 |
| 5 | ko04972 | Pancreatic secretion | 13/264 | 64/5400 | 9.47E-06 | 0.000 |
| 6 | ko00564 | Glycerophospholipid metabolism | 11/264 | 69/5400 | 0.000 | 0.013 |
| 7 | ko00450 | Selenocompound metabolism | 4/264 | 10/5400 | 0.001 | 0.021 |
| 8 | ko00561 | Glycerolipid metabolism | 8/264 | 44/5400 | 0.001 | 0.021 |
| 9 | ko04141* | Protein processing in the endoplasmic reticulum | 15/264 | 126/5400 | 0.001 | 0.021 |
| 10 | ko00051 | Fructose and mannose metabolism | 6/264 | 28/5400 | 0.002 | 0.033 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 16.
Top 20 of 21 Selective KEGG pathways in the heart.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko00190 | Oxidative phosphorylation | 74/331 | 108/5400 | 1.14E-66 | 2.01E-64 |
| 2 | ko04260 | Cardiac muscle contraction | 27/331 | 54/5400 | 2.79E-19 | 2.46E-17 |
| 3 | ko00020 | Citrate cycle (TCA cycle) | 16/331 | 24/5400 | 1.31E-14 | 7.70E-13 |
| 4 | ko01200* | Carbon metabolism | 28/331 | 94/5400 | 5.94E-13 | 2.61E-11 |
| 5 | ko00640* | Propanoate metabolism | 9/331 | 21/5400 | 1.67E-06 | 5.88E-05 |
| 6 | ko00620 | Pyruvate metabolism | 10/331 | 28/5400 | 3.18E-06 | 9.32E-05 |
| 7 | ko01210 | 2-Oxocarboxylic acid metabolism | 6/331 | 13/5400 | 6.02E-05 | 0.002 |
| 8 | ko00010 | Glycolysis / Gluconeogenesis | 11/331 | 48/5400 | 0.000 | 0.002 |
| 9 | ko02020 | Two-component system | 5/331 | 10/5400 | 0.000 | 0.003 |
| 10 | ko00280* | Valine, leucine and isoleucine degradation | 9/331 | 35/5400 | 0.000 | 0.003 |
| 11 | ko00720 | Carbon fixation pathways in prokaryotes | 5/331 | 11/5400 | 0.000 | 0.005 |
| 12 | ko04020* | Calcium signaling pathway | 16/331 | 105/5400 | 0.001 | 0.008 |
| 13 | ko04922* | Glucagon signaling pathway | 11/331 | 59/5400 | 0.001 | 0.010 |
| 14 | ko03010* | Ribosome | 18/331 | 133/5400 | 0.001 | 0.014 |
| 15 | ko04261 | Adrenergic signaling in cardiomyocytes | 14/331 | 92/5400 | 0.001 | 0.015 |
| 16 | ko00650* | Butanoate metabolism | 6/331 | 22/5400 | 0.002 | 0.017 |
| 17 | ko00710 | Carbon fixation in photosynthetic organisms | 6/331 | 22/5400 | 0.002 | 0.017 |
| 18 | ko00071* | Fatty acid degradation | 7/331 | 30/5400 | 0.002 | 0.018 |
| 19 | ko04022* | cGMP - PKG signaling pathway | 15/331 | 108/5400 | 0.002 | 0.021 |
| 20 | ko01230* | Biosynthesis of amino acids | 10/331 | 63/5400 | 0.005 | 0.040 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 17.
Selective KEGG pathways in the ileum.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04612 | Antigen processing and presentation | 20/333 | 63/5400 | 4.25E-10 | 7.87E-08 |
| 2 | ko04144* | Endocytosis | 35/333 | 189/5400 | 2.23E-09 | 2.06E-07 |
| 3 | ko04141* | Protein processing in endoplasmic reticulum | 22/333 | 126/5400 | 6.72E-06 | 0.000 |
| 4 | ko04145* | Phagosome | 21/333 | 121/5400 | 1.20E-05 | 0.001 |
| 5 | ko03010 | Ribosome | 22/333 | 133/5400 | 1.65E-05 | 0.001 |
| 6 | ko04672* | Intestinal immune network for IgA production | 9/333 | 32/5400 | 9.15E-05 | 0.003 |
| 7 | ko04514* | Cell adhesion molecules (CAMs) | 17/333 | 108/5400 | 0.000 | 0.007 |
| 8 | ko00520 | Amino sugar and nucleotide sugar metabolism | 9/333 | 37/5400 | 0.000 | 0.007 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 18.
Top 20 of 23 selective KEGG pathways in the kidney.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04146* | Peroxisome | 23/386 | 62/5400 | 1.04E-11 | 1.93E-09 |
| 2 | ko04961* | Endocrine and other factor-regulated calcium reabsorption | 13/386 | 35/5400 | 3.61E-07 | 3.35E-05 |
| 3 | ko04964 | Proximal tubule bicarbonate reclamation | 8/386 | 16/5400 | 4.91E-06 | 0.000 |
| 4 | ko00630* | Glyoxylate and dicarboxylate metabolism | 9/386 | 21/5400 | 6.03E-06 | 0.000 |
| 5 | ko00770 | Pantothenate and CoA biosynthesis | 7/386 | 13/5400 | 1.06E-05 | 0.000 |
| 6 | ko04142* | Lysosome | 18/386 | 87/5400 | 3.12E-05 | 0.001 |
| 7 | ko00280* | Valine, leucine and isoleucine degradation | 10/386 | 35/5400 | 0.000 | 0.003 |
| 8 | ko00260* | Glycine, serine and threonine metabolism | 9/386 | 29/5400 | 0.000 | 0.003 |
| 9 | ko00071* | Fatty acid degradation | 9/386 | 30/5400 | 0.000 | 0.003 |
| 10 | ko00480 | Glutathione metabolism | 9/386 | 33/5400 | 0.000 | 0.007 |
| 11 | ko00640* | Propanoate metabolism | 7/386 | 21/5400 | 0.000 | 0.007 |
| 12 | ko04614 | Renin-angiotensin system | 7/386 | 21/5400 | 0.000 | 0.007 |
| 13 | ko00040* | Pentose and glucuronate interconversions | 6/386 | 16/5400 | 0.001 | 0.008 |
| 14 | ko00790 | Folate biosynthesis | 4/386 | 7/5400 | 0.001 | 0.010 |
| 15 | ko00910* | Nitrogen metabolism | 6/386 | 17/5400 | 0.001 | 0.010 |
| 16 | ko01200* | Carbon metabolism | 16/386 | 94/5400 | 0.001 | 0.010 |
| 17 | ko04978* | Mineral absorption | 8/386 | 30/5400 | 0.001 | 0.010 |
| 18 | ko00330 | Arginine and proline metabolism | 8/386 | 35/5400 | 0.003 | 0.028 |
| 19 | ko00730 | Thiamine metabolism | 3/386 | 5/5400 | 0.003 | 0.032 |
| 20 | ko00650* | Butanoate metabolism | 6/386 | 22/5400 | 0.004 | 0.033 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 19.
Top 20 of 34 selective KEGG pathways in the liver.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04610 | Complement and coagulation cascades | 37/265 | 55/5400 | 1.98E-36 | 2.97E-34 |
| 2 | ko00140 | Steroid hormone biosynthesis | 15/265 | 33/5400 | 7.31E-12 | 5.46E-10 |
| 3 | ko00830 | Retinol metabolism | 14/265 | 38/5400 | 1.12E-09 | 5.59E-08 |
| 4 | ko00260* | Glycine, serine and threonine metabolism | 11/265 | 29/5400 | 5.09E-08 | 1.90E-06 |
| 5 | ko03320 | PPAR signaling pathway | 14/265 | 56/5400 | 3.03E-07 | 9.05E-06 |
| 6 | ko00120 | Primary bile acid biosynthesis | 6/265 | 10/5400 | 2.35E-06 | 5.85E-05 |
| 7 | ko04976 | Bile secretion | 12/265 | 51/5400 | 4.34E-06 | 9.27E-05 |
| 8 | ko00220 | Arginine biosynthesis | 6/265 | 12/5400 | 9.50E-06 | 0.000 |
| 9 | ko00980 | Metabolism of xenobiotics by cytochrome P450 | 9/265 | 32/5400 | 1.49E-05 | 0.000 |
| 10 | ko01230* | Biosynthesis of amino acids | 12/265 | 63/5400 | 4.30E-05 | 0.001 |
| 11 | ko00053 | Ascorbate and aldarate metabolism | 5/265 | 10/5400 | 5.64E-05 | 0.001 |
| 12 | ko00982 | Drug metabolism - cytochrome P450 | 8/265 | 31/5400 | 8.90E-05 | 0.001 |
| 13 | ko00340 | Histidine metabolism | 6/265 | 17/5400 | 0.000 | 0.001 |
| 14 | ko01040 | Biosynthesis of unsaturated fatty acids | 6/265 | 18/5400 | 0.000 | 0.002 |
| 15 | ko00591 | Linoleic acid metabolism | 6/265 | 22/5400 | 0.001 | 0.005 |
| 16 | ko01200* | Carbon metabolism | 13/265 | 94/5400 | 0.001 | 0.006 |
| 17 | ko00500 | Starch and sucrose metabolism | 6/265 | 24/5400 | 0.001 | 0.007 |
| 18 | ko00983 | Drug metabolism - other enzymes | 6/265 | 24/5400 | 0.001 | 0.007 |
| 19 | ko00100 | Steroid biosynthesis | 4/265 | 12/5400 | 0.002 | 0.016 |
| 20 | ko00430 | Taurine and hypotaurine metabolism | 3/265 | 6/5400 | 0.002 | 0.016 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 20.
Top 20 of 46 Selective KEGG pathways in the lung.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04510 | Focal adhesion | 45/703 | 126/5400 | 4.16E-11 | 4.01E-09 |
| 2 | ko04360* | Axon guidance | 43/703 | 118/5400 | 5.36E-11 | 4.01E-09 |
| 3 | ko04390 | Hippo signaling pathway | 40/703 | 110/5400 | 2.77E-10 | 1.38E-08 |
| 4 | ko04151 | PI3K-Akt signaling pathway | 59/703 | 223/5400 | 2.85E-08 | 1.06E-06 |
| 5 | ko04310 | Wnt signaling pathway | 34/703 | 100/5400 | 4.42E-08 | 1.32E-06 |
| 6 | ko04550 | Signaling pathways regulating pluripotency of stem cells | 32/703 | 93/5400 | 8.06E-08 | 2.01E-06 |
| 7 | ko04668* | TNF signaling pathway | 29/703 | 83/5400 | 2.26E-07 | 4.83E-06 |
| 8 | ko04392 | Hippo signaling pathway - multiple species | 12/703 | 19/5400 | 4.56E-07 | 7.60E-06 |
| 9 | ko04014* | Ras signaling pathway | 44/703 | 159/5400 | 4.57E-07 | 7.60E-06 |
| 10 | ko04010* | MAPK signaling pathway | 46/703 | 177/5400 | 1.76E-06 | 2.63E-05 |
| 11 | ko04060* | Cytokine-cytokine receptor interaction | 43/703 | 163/5400 | 2.52E-06 | 3.41E-05 |
| 12 | ko04015* | Rap1 signaling pathway | 37/703 | 132/5400 | 2.73E-06 | 3.41E-05 |
| 13 | ko04062* | Chemokine signaling pathway | 35/703 | 123/5400 | 3.50E-06 | 4.03E-05 |
| 14 | ko04916* | Melanogenesis | 22/703 | 63/5400 | 6.67E-06 | 7.12E-05 |
| 15 | ko04340 | Hedgehog signaling pathway | 13/703 | 27/5400 | 9.61E-06 | 9.57E-05 |
| 16 | ko04512 | ECM-receptor interaction | 17/703 | 46/5400 | 3.23E-05 | 0.000 |
| 17 | ko04341 | Hedgehog signaling pathway - Fly | 10/703 | 19/5400 | 4.01E-05 | 0.000 |
| 18 | ko04144* | Endocytosis | 44/703 | 189/5400 | 5.91E-05 | 0.000 |
| 19 | ko04650* | Natural killer cell mediated cytotoxicity | 25/703 | 86/5400 | 5.92E-05 | 0.000 |
| 20 | ko04810* | Regulation of actin cytoskeleton | 36/703 | 149/5400 | 0.000 | 0.001 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 21.
Top 20 of 33 selective KEGG pathways in the spleen.
| No. | ID | Description | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|
| 1 | ko04110 | Cell cycle | 48/667 | 95/5400 | 6.13E-20 | 9.87E-18 |
| 2 | ko04111 | Cell cycle - yeast | 32/667 | 57/5400 | 2.09E-15 | 1.68E-13 |
| 3 | ko03013 | RNA transport | 50/667 | 131/5400 | 2.02E-14 | 1.08E-12 |
| 4 | ko03040 | Spliceosome | 44/667 | 113/5400 | 3.47E-13 | 1.40E-11 |
| 5 | ko03030 | DNA replication | 20/667 | 29/5400 | 1.78E-12 | 5.73E-11 |
| 6 | ko04064* | NF-kappa B signaling pathway | 27/667 | 65/5400 | 2.64E-09 | 7.08E-08 |
| 7 | ko04113 | Meiosis - yeast | 22/667 | 49/5400 | 1.43E-08 | 3.28E-07 |
| 8 | ko03420 | Nucleotide excision repair | 18/667 | 37/5400 | 6.48E-08 | 1.30E-06 |
| 9 | ko04640 | Hematopoietic cell lineage | 21/667 | 49/5400 | 8.29E-08 | 1.48E-06 |
| 10 | ko03460 | Fanconi anemia pathway | 15/667 | 32/5400 | 1.52E-06 | 2.45E-05 |
| 11 | ko03430 | Mismatch repair | 9/667 | 14/5400 | 7.15E-06 | 0.000 |
| 12 | ko03015 | mRNA surveillance pathway | 23/667 | 73/5400 | 1.19E-05 | 0.000 |
| 13 | ko04662 | B cell receptor signaling pathway | 17/667 | 47/5400 | 2.23E-05 | 0.000 |
| 14 | ko04060* | Cytokine-cytokine receptor interaction | 39/667 | 163/5400 | 2.50E-05 | 0.000 |
| 15 | ko03008 | Ribosome biogenesis in eukaryotes | 20/667 | 62/5400 | 3.01E-05 | 0.000 |
| 16 | ko03410 | Base excision repair | 12/667 | 28/5400 | 5.29E-05 | 0.001 |
| 17 | ko04660* | T cell receptor signaling pathway | 22/667 | 78/5400 | 0.000 | 0.001 |
| 18 | ko04380* | Osteoclast differentiation | 23/667 | 87/5400 | 0.000 | 0.002 |
| 19 | ko03018 | RNA degradation | 18/667 | 61/5400 | 0.000 | 0.002 |
| 20 | ko04115 | p53 signaling pathway | 16/667 | 53/5400 | 0.000 | 0.004 |
Note: * also significantly expressed in other organs. Sorted by q-value.
3.3.2. GO Pathway Enrichment
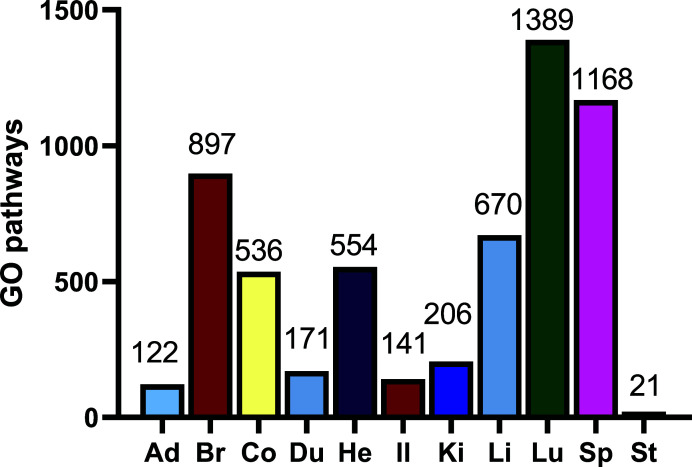
The GO database is the world’s largest source of bio-information on the functions of genes. This knowledge of the genes is a foundation for computational analysis of large-scale molecular biology and genetics experiments in biomedical research. Selective GO pathways were enriched based on the abundance of genes most highly expressed in organs. The number of the selective pathway is listed in Fig. (4) and the pathways of the adrenal gland, brain, colon, duodenum, heart, ileum, kidney, liver, lung, spleen, and stomach are listed in Tables 23-33, respectively. There were 4,432 relatively selective pathways in 11 rat organs. Among them, 971 pathways were involved in two organs, 357 in three organs, 86 in four organs, 21 in five organs, 7 in six organs, and 1 in seven organs. It should be noted that the “selective” pathways are involved in two or more organs based on the enrichment analysis.
Fig. (4).
Selective GO enrichment in different organs based on the abundance of genes most highly expressed in organs. Abbreviations: Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Il, ileum; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; St, stomach.
Table 23.
Top 20 of 122 selective GO pathways in the adrenal gland.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0043231* | Intracellular membrane-bounded organelle | cellular component | 621/998 | 8545/18378 | 8.00E-25 | 1.43E-21 |
| 2 | GO:0005739* | Mitochondrion | cellular component | 178/998 | 1536/18378 | 3.50E-23 | 3.12E-20 |
| 3 | GO:0044424* | Intracellular part | cellular component | 782/998 | 11898/18378 | 5.40E-22 | 2.85E-19 |
| 4 | GO:0043227* | Membrane-bounded organelle | cellular component | 686/998 | 9971/18378 | 6.40E-22 | 2.85E-19 |
| 5 | GO:0044429* | Mitochondrial part | cellular component | 107/998 | 727/18378 | 1.50E-21 | 5.35E-19 |
| 6 | GO:0043226* | Organelle | cellular component | 743/998 | 11246/18378 | 7.60E-20 | 2.26E-17 |
| 7 | GO:0005622* | Intracellular | cellular component | 800/998 | 12452/18378 | 1.90E-19 | 4.84E-17 |
| 8 | GO:0008152* | Metabolic process | biological process | 677/932 | 10277/17378 | 7.90E-19 | 1.18E-14 |
| 9 | GO:0043229* | Intracellular organelle | cellular component | 690/998 | 10283/18378 | 1.20E-18 | 2.67E-16 |
| 10 | GO:0005759* | Mitochondrial matrix | cellular component | 52/998 | 240/18378 | 4.70E-18 | 9.31E-16 |
| 11 | GO:0044237* | Cellular metabolic process | biological process | 611/932 | 9092/17378 | 3.00E-17 | 2.25E-13 |
| 12 | GO:0034660* | ncRNA metabolic process | biological process | 66/932 | 406/17378 | 4.50E-16 | 2.25E-12 |
| 13 | GO:0006807* | Nitrogen compound metabolic process | biological process | 417/932 | 5827/17378 | 1.80E-13 | 6.75E-10 |
| 14 | GO:0044422* | Organelle part | cellular component | 476/998 | 6775/18378 | 4.20E-13 | 7.27E-11 |
| 15 | GO:0071704* | Organic substance metabolic process | biological process | 621/932 | 9627/17378 | 4.60E-13 | 1.17E-09 |
| 16 | GO:0034641* | Cellular nitrogen compound metabolic process | biological process | 400/932 | 5563/17378 | 4.70E-13 | 1.17E-09 |
| 17 | GO:0005737* | Cytoplasm | cellular component | 593/998 | 8899/18378 | 5.10E-13 | 7.27E-11 |
| 18 | GO:0031974* | Membrane-enclosed lumen | cellular component | 264/998 | 3238/18378 | 5.30E-13 | 7.27E-11 |
| 19 | GO:0043233* | Organelle lumen | cellular component | 264/998 | 3238/18378 | 5.30E-13 | 7.27E-11 |
| 20 | GO:0070013* | Intracellular organelle lumen | cellular component | 263/998 | 3235/18378 | 8.40E-13 | 1.07E-10 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 33.
Top 20 of 21 selective GO pathways in the stomach.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0007586* | Digestion | Biological process | 20/490 | 111/17378 | 3.40E-11 | 5.10E-07 |
| 2 | GO:0001696 | Gastric acid secretion | Biological process | 9/490 | 17/17378 | 2.10E-10 | 1.57E-06 |
| 3 | GO:0055123* | Digestive system development | Biological process | 17/490 | 128/17378 | 1.20E-07 | 0.000 |
| 4 | GO:0022600 | Digestive system process | Biological process | 14/490 | 86/17378 | 1.20E-07 | 0.000 |
| 5 | GO:0031016 | Pancreas development | Biological process | 12/490 | 71/17378 | 6.20E-07 | 0.002 |
| 6 | GO:0004190 | Aspartic-type endopeptidase activity | Molecular function | 7/487 | 23/16814 | 2.70E-06 | 0.006 |
| 7 | GO:0070001 | Aspartic-type peptidase activity | Molecular function | 7/487 | 24/16814 | 3.70E-06 | 0.006 |
| 8 | GO:0001228* | Transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific | Molecular function | 25/487 | 315/16814 | 5.50E-06 | 0.006 |
| 9 | GO:0000981* | RNA polymerase II transcription factor activity, sequence-specific DNA binding | Molecular function | 38/487 | 601/16814 | 5.80E-06 | 0.006 |
| 10 | GO:0030855* | Epithelial cell differentiation | Biological process | 33/490 | 488/17378 | 3.50E-06 | 0.009 |
| 11 | GO:0046903* | Secretion | Biological process | 49/490 | 879/17378 | 4.20E-06 | 0.009 |
| 12 | GO:0046717* | Acid secretion | Biological process | 12/490 | 87/17378 | 5.70E-06 | 0.011 |
| 13 | GO:0031018 | Endocrine pancreas development | Biological process | 8/490 | 40/17378 | 1.30E-05 | 0.021 |
| 14 | GO:0044765* | Single-organism transport | Biological process | 99/490 | 2326/17378 | 1.40E-05 | 0.021 |
| 15 | GO:0009888* | Tissue development | Biological process | 69/490 | 1472/17378 | 1.80E-05 | 0.025 |
| 16 | GO:0048565* | Digestive tract development | Biological process | 13/490 | 117/17378 | 2.60E-05 | 0.032 |
| 17 | GO:0005882 | Intermediate filament | Cellular component | 15/533 | 144/18378 | 1.90E-05 | 0.034 |
| 18 | GO:0051050* | Positive regulation of transport | Biological process | 43/490 | 793/17378 | 3.20E-05 | 0.037 |
| 19 | GO:1903011 | Negative regulation of bone development | Biological process | 4/490 | 8/17378 | 4.00E-05 | 0.043 |
| 20 | GO:0060428 | Lung epithelium development | Biological process | 7/490 | 35/17378 | 4.60E-05 | 0.046 |
Note: * also significantly expressed in other organs. Sorted by q-value.
As can be seen from Fig. (4), organs with many selective pathways, like the lung, spleen and brain, indicate that they undertake many complex functions. Conversely, organs with few selective pathways, like the stomach and adrenal glands, indicate their relative sample functions. The results in Fig. (3), is similar to those in Fig. (4).
The top 20 GO pathways are shown in Tables 23-33, and their full lists can be seen in the supplementary data. As for the top 20 GO pathways, the adrenal gland (Table 23), colon (Table 25), and kidney (Table 29) had no real selective pathways, and the brain had the most selective pathways, suggesting that the brain has specific functions (Table 24). According to the results of GO enrichment, the adrenal gland is a hypermetabolic organ because mitochondria in the organ are very active (Table 23); the brain is a neural organ (Table 24), which is well-accepted by scientists; the colon is an immune and metabolic organ (Table 25); the duodenum is mainly an immune organ (Table 26); the heart is also a hypermetabolic organ (Table 27); the ileum is primarily an organ associated with protein synthesis, immune, and digestion (Table 28); the kidney (Table 29) and liver (Table 30) are mainly an organ associated with metabolism; the lung is an organ mainly associated with angiogenesis and blood circulation (Table 31); the spleen is an organ mainly associated with organelle metabolism (Table 32), and the stomach is an organ mainly associated with digestion and glandular secretion (Table 33).
Table 25.
Top 20 of 536 selective GO pathways in the colon.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0002376* | Immune system process | Biological process | 153/678 | 1949/17378 | 5.70E-18 | 8.55E-14 |
| 2 | GO:0031347* | Regulation of defense response | Biological process | 52/678 | 407/17378 | 4.10E-14 | 3.07E-10 |
| 3 | GO:0002682* | Regulation of immune system process | Biological process | 89/678 | 1014/17378 | 3.80E-13 | 1.31E-09 |
| 4 | GO:0019221* | Cytokine-mediated signaling pathway | Biological process | 44/678 | 322/17378 | 3.80E-13 | 1.31E-09 |
| 5 | GO:0045321* | Leukocyte activation | Biological process | 70/678 | 703/17378 | 4.60E-13 | 1.31E-09 |
| 6 | GO:0006952* | Defense response | Biological process | 93/678 | 1091/17378 | 5.70E-13 | 1.31E-09 |
| 7 | GO:0001775* | Cell activation | Biological process | 76/678 | 804/17378 | 6.10E-13 | 1.31E-09 |
| 8 | GO:0042110* | T cell activation | Biological process | 46/678 | 356/17378 | 8.50E-13 | 1.50E-09 |
| 9 | GO:0080134* | Regulation of response to stress | Biological process | 83/678 | 927/17378 | 9.00E-13 | 1.50E-09 |
| 10 | GO:0009607* | Response to biotic stimulus | Biological process | 74/678 | 797/17378 | 3.10E-12 | 4.63E-09 |
| 11 | GO:0009605* | Response to external stimulus | Biological process | 132/678 | 1856/17378 | 3.40E-12 | 4.63E-09 |
| 12 | GO:0006955* | Immune response | Biological process | 97/678 | 1208/17378 | 5.70E-12 | 7.12E-09 |
| 13 | GO:0048518* | Positive regulation of biological process | Biological process | 258/678 | 4587/17378 | 8.20E-12 | 9.46E-09 |
| 14 | GO:0002520* | Immune system development | Biological process | 67/678 | 706/17378 | 1.40E-11 | 1.32E-08 |
| 15 | GO:0035556* | Intracellular signal transduction | Biological process | 139/678 | 2034/17378 | 1.50E-11 | 1.32E-08 |
| 16 | GO:0071345* | Cellular response to cytokine stimulus | Biological process | 55/678 | 518/17378 | 1.50E-11 | 1.32E-08 |
| 17 | GO:0043207* | Response to external biotic stimulus | Biological process | 70/678 | 757/17378 | 1.50E-11 | 1.32E-08 |
| 18 | GO:0007159* | Leukocyte cell-cell adhesion | Biological process | 37/678 | 268/17378 | 2.20E-11 | 1.83E-08 |
| 19 | GO:0031349* | Positive regulation of defense response | Biological process | 34/678 | 231/17378 | 2.40E-11 | 1.89E-08 |
| 20 | GO:0046649* | Lymphocyte activation | Biological process | 60/678 | 604/17378 | 2.70E-11 | 1.91E-08 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 29.
Top 20 of 206 selective GO pathways in the kidney.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0003824* | Catalytic activity | Molecular function | 571/1203 | 5604/16814 | 4.10E-26 | 1.69E-22 |
| 2 | GO:0044281* | Small molecule metabolic process | Biological process | 218/1237 | 1566/17378 | 2.20E-23 | 3.30E-19 |
| 3 | GO:0005739* | Mitochondrion | Cellular component | 210/1275 | 1536/18378 | 8.60E-23 | 1.53E-19 |
| 4 | GO:0006082* | Organic acid metabolic process | Biological process | 136/1237 | 806/17378 | 6.90E-22 | 5.17E-18 |
| 5 | GO:0019752* | Carboxylic acid metabolic process | Biological process | 128/1237 | 740/17378 | 1.40E-21 | 7.00E-18 |
| 6 | GO:0044710* | Single-organism metabolic process | Biological process | 378/1237 | 3483/17378 | 4.80E-20 | 1.80E-16 |
| 7 | GO:0070062* | Extracellular exosome | Cellular component | 253/1275 | 2097/18378 | 7.70E-20 | 6.86E-17 |
| 8 | GO:0043436* | Oxoacid metabolic process | Biological process | 130/1237 | 793/17378 | 8.00E-20 | 2.40E-16 |
| 9 | GO:0055114* | Oxidation-reduction process | Biological process | 148/1237 | 967/17378 | 1.30E-19 | 3.25E-16 |
| 10 | GO:1903561* | Extracellular vesicle | Cellular component | 253/1275 | 2110/18378 | 1.80E-19 | 1.02E-16 |
| 11 | GO:0043230* | Extracellular organelle | Cellular component | 253/1275 | 2114/18378 | 2.30E-19 | 1.02E-16 |
| 12 | GO:1901605* | Alpha-amino acid metabolic process | Biological process | 50/1237 | 175/17378 | 5.30E-18 | 1.14E-14 |
| 13 | GO:0016491* | Oxidoreductase activity | Molecular function | 123/1203 | 775/16814 | 1.70E-17 | 3.50E-14 |
| 14 | GO:0006520* | Cellular amino acid metabolic process | Biological process | 59/1237 | 247/17378 | 6.30E-17 | 1.18E-13 |
| 15 | GO:0044282* | Small molecule catabolic process | Biological process | 54/1237 | 231/17378 | 3.60E-15 | 6.00E-12 |
| 16 | GO:0016054* | Organic acid catabolic process | Biological process | 45/1237 | 169/17378 | 4.50E-15 | 6.13E-12 |
| 17 | GO:0046395* | Carboxylic acid catabolic process | Biological process | 45/1237 | 169/17378 | 4.50E-15 | 6.13E-12 |
| 18 | GO:0031982* | Vesicle | Cellular component | 318/1275 | 3084/18378 | 9.50E-15 | 3.39E-12 |
| 19 | GO:0048037* | Cofactor binding | Molecular function | 59/1203 | 276/16814 | 1.70E-14 | 2.33E-11 |
| 20 | GO:1901565* | Organonitrogen compound catabolic process | Biological process | 47/1237 | 222/17378 | 9.80E-12 | 1.22E-08 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 24.
Top 20 of 897 selective GO pathways in the brain.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0097458 | Neuron part | Cellular component | 569/3717 | 1181/18378 | 1.00E-30 | 8.49E-29 |
| 2 | GO:0045202 | Synapse | Cellular component | 409/3717 | 718/18378 | 1.00E-30 | 8.49E-29 |
| 3 | GO:0044456 | Synapse part | Cellular component | 354/3717 | 593/18378 | 1.00E-30 | 8.49E-29 |
| 4 | GO:0043005 | Neuron projection | Cellular component | 428/3717 | 875/18378 | 1.00E-30 | 8.49E-29 |
| 5 | GO:0120025 | Plasma membrane-bounded cell projection | Cellular component | 565/3717 | 1477/18378 | 1.00E-30 | 8.49E-29 |
| 6 | GO:0098793 | Presynapse | Cellular component | 193/3717 | 302/18378 | 1.00E-30 | 8.49E-29 |
| 7 | GO:0036477 | Somatodendritic compartment | Cellular component | 311/3717 | 639/18378 | 1.00E-30 | 8.49E-29 |
| 8 | GO:0042995* | Cell projection | Cellular component | 581/3717 | 1558/18378 | 1.00E-30 | 8.49E-29 |
| 9 | GO:0097060 | Synaptic membrane | Cellular component | 146/3717 | 208/18378 | 1.00E-30 | 8.49E-29 |
| 10 | GO:0098794 | Postsynapse | Cellular component | 204/3717 | 354/18378 | 1.00E-30 | 8.49E-29 |
| 11 | GO:0030424 | Axon | Cellular component | 203/3717 | 360/18378 | 1.00E-30 | 8.49E-29 |
| 12 | GO:0030425 | Dendrite | Cellular component | 222/3717 | 436/18378 | 1.00E-30 | 8.49E-29 |
| 13 | GO:0044463* | Cell projection part | Cellular component | 349/3717 | 860/18378 | 1.00E-30 | 8.49E-29 |
| 14 | GO:0043025 | Neuronal cell body | Cellular component | 215/3717 | 437/18378 | 1.00E-30 | 8.49E-29 |
| 15 | GO:0045211 | Postsynaptic membrane | Cellular component | 108/3717 | 153/18378 | 1.00E-30 | 8.49E-29 |
| 16 | GO:0044297 | Cell body | Cellular component | 232/3717 | 497/18378 | 1.00E-30 | 8.49E-29 |
| 17 | GO:0098984 | Neuron to neuron synapse | Cellular component | 110/3717 | 181/18378 | 1.00E-30 | 8.49E-29 |
| 18 | GO:0014069 | Postsynaptic density | Cellular component | 107/3717 | 176/18378 | 1.00E-30 | 8.49E-29 |
| 19 | GO:0032279 | Asymmetric synapse | Cellular component | 108/3717 | 179/18378 | 1.00E-30 | 8.49E-29 |
| 20 | GO:0099572 | Postsynaptic specialization | Cellular component | 107/3717 | 177/18378 | 1.00E-30 | 8.49E-29 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 26.
Top 20 of 171 selective GO pathways in the duodenum.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0042571 | Immunoglobulin complex, circulating | Cellular component | 74/933 | 98/18378 | 1.00E-30 | 2.55E-28 |
| 2 | GO:0019814 | Immunoglobulin complex | Cellular component | 74/933 | 102/18378 | 1.00E-30 | 2.55E-28 |
| 3 | GO:0072562* | Blood microparticle | Cellular component | 78/933 | 173/18378 | 1.00E-30 | 2.55E-28 |
| 4 | GO:0005615* | Extracellular space | Cellular component | 214/933 | 1396/18378 | 1.00E-30 | 2.55E-28 |
| 5 | GO:0044421* | Extracellular region part | Cellular component | 335/933 | 3289/18378 | 1.00E-30 | 2.55E-28 |
| 6 | GO:0005576* | Extracellular region | Cellular component | 357/933 | 3681/18378 | 1.00E-30 | 2.55E-28 |
| 7 | GO:0009897* | External side of plasma membrane | Cellular component | 77/933 | 300/18378 | 1.00E-30 | 2.55E-28 |
| 8 | GO:0006910 | Phagocytosis, recognition | Biological process | 74/897 | 108/17378 | 1.00E-30 | 5.00E-28 |
| 9 | GO:0006958 | Complement activation, classical pathway | Biological process | 73/897 | 107/17378 | 1.00E-30 | 5.00E-28 |
| 10 | GO:0002455 | Humoral immune response mediated by circulating immunoglobulin | Biological process | 73/897 | 115/17378 | 1.00E-30 | 5.00E-28 |
| 11 | GO:0006911 | Phagocytosis, engulfment | Biological process | 74/897 | 120/17378 | 1.00E-30 | 5.00E-28 |
| 12 | GO:0099024 | Plasma membrane invagination | Biological process | 76/897 | 128/17378 | 1.00E-30 | 5.00E-28 |
| 13 | GO:0010324 | Membrane invagination | Biological process | 76/897 | 134/17378 | 1.00E-30 | 5.00E-28 |
| 14 | GO:0006956* | Complement activation | Biological process | 73/897 | 132/17378 | 1.00E-30 | 5.00E-28 |
| 15 | GO:0050853* | B cell receptor signaling pathway | Biological process | 73/897 | 132/17378 | 1.00E-30 | 5.00E-28 |
| 16 | GO:0072376 | Protein activation cascade | Biological process | 73/897 | 143/17378 | 1.00E-30 | 5.00E-28 |
| 17 | GO:0008037 | Cell recognition | Biological process | 80/897 | 182/17378 | 1.00E-30 | 5.00E-28 |
| 18 | GO:0050871* | Positive regulation of B cell activation | Biological process | 75/897 | 163/17378 | 1.00E-30 | 5.00E-28 |
| 19 | GO:0002377* | Immunoglobulin production | Biological process | 86/897 | 224/17378 | 1.00E-30 | 5.00E-28 |
| 20 | GO:0006959* | Humoral immune response | Biological process | 78/897 | 188/17378 | 1.00E-30 | 5.00E-28 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 27.
Top 20 of 554 selective GO pathways in the heart.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0005739* | Mitochondrion | Cellular component | 348/1048 | 1536/18378 | 1.00E-30 | 6.85E-29 |
| 2 | GO:0044429* | Mitochondrial part | Cellular component | 230/1048 | 727/18378 | 1.00E-30 | 6.85E-29 |
| 3 | GO:0005743* | Mitochondrial inner membrane | Cellular component | 141/1048 | 367/18378 | 1.00E-30 | 6.85E-29 |
| 4 | GO:0031966* | Mitochondrial membrane | Cellular component | 164/1048 | 508/18378 | 1.00E-30 | 6.85E-29 |
| 5 | GO:0005740* | Mitochondrial envelope | Cellular component | 167/1048 | 546/18378 | 1.00E-30 | 6.85E-29 |
| 6 | GO:0098800 | Inner mitochondrial membrane protein complex | Cellular component | 85/1048 | 125/18378 | 1.00E-30 | 6.85E-29 |
| 7 | GO:0019866* | Organelle inner membrane | Cellular component | 142/1048 | 409/18378 | 1.00E-30 | 6.85E-29 |
| 8 | GO:0098798 | Mitochondrial protein complex | Cellular component | 89/1048 | 144/18378 | 1.00E-30 | 6.85E-29 |
| 9 | GO:0044455* | Mitochondrial membrane part | Cellular component | 101/1048 | 195/18378 | 1.00E-30 | 6.85E-29 |
| 10 | GO:0031967* | Organelle envelope | Cellular component | 182/1048 | 867/18378 | 1.00E-30 | 6.85E-29 |
| 11 | GO:0031975* | Envelope | Cellular component | 182/1048 | 869/18378 | 1.00E-30 | 6.85E-29 |
| 12 | GO:0070469 | Respiratory chain | Cellular component | 65/1048 | 100/18378 | 1.00E-30 | 6.85E-29 |
| 13 | GO:0098803 | Respiratory chain complex | Cellular component | 59/1048 | 85/18378 | 1.00E-30 | 6.85E-29 |
| 14 | GO:0005746 | Mitochondrial respiratory chain | Cellular component | 58/1048 | 86/18378 | 1.00E-30 | 6.85E-29 |
| 15 | GO:0030016 | Myofibril | Cellular component | 76/1048 | 161/18378 | 1.00E-30 | 6.85E-29 |
| 16 | GO:0043292 | Contractile fiber | Cellular component | 76/1048 | 171/18378 | 1.00E-30 | 6.85E-29 |
| 17 | GO:0030017 | Sarcomere | Cellular component | 69/1048 | 142/18378 | 1.00E-30 | 6.85E-29 |
| 18 | GO:0044449 | Contractile fiber part | Cellular component | 71/1048 | 154/18378 | 1.00E-30 | 6.85E-29 |
| 19 | GO:1990204 | Oxidoreductase complex | Cellular component | 59/1048 | 105/18378 | 1.00E-30 | 6.85E-29 |
| 20 | GO:0005759* | Mitochondrial matrix | Cellular component | 79/1048 | 240/18378 | 1.00E-30 | 6.85E-29 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 28.
Top 20 of 141 selective GO pathways in the ileum.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:1990904* | Ribonucleoprotein complex | Cellular component | 144/1063 | 1129/18378 | 5.60E-20 | 9.98E-17 |
| 2 | GO:0030529* | Intracellular ribonucleoprotein complex | Cellular component | 143/1063 | 1128/18378 | 1.30E-19 | 1.16E-16 |
| 3 | GO:0003735* | Structural constituent of ribosome | Molecular function | 80/929 | 507/16814 | 9.90E-18 | 4.07E-14 |
| 4 | GO:0005840* | Ribosome | Cellular component | 86/1063 | 587/18378 | 1.10E-15 | 6.53E-13 |
| 5 | GO:0005198* | Structural molecule activity | Molecular function | 109/929 | 898/16814 | 3.30E-15 | 6.79E-12 |
| 6 | GO:0042611 | MHC protein complex | Cellular component | 16/1063 | 25/18378 | 1.80E-14 | 8.02E-12 |
| 7 | GO:0019882 | Antigen processing and presentation | Biological process | 27/976 | 91/17378 | 3.70E-13 | 5.55E-09 |
| 8 | GO:0043604* | Amide biosynthetic process | Biological process | 103/976 | 956/17378 | 9.30E-11 | 5.00E-07 |
| 9 | GO:0006412* | Translation | Biological process | 97/976 | 881/17378 | 1.00E-10 | 5.00E-07 |
| 10 | GO:0022626* | Cytosolic ribosome | Cellular component | 53/1063 | 350/18378 | 1.10E-10 | 3.92E-08 |
| 11 | GO:0043603* | Cellular amide metabolic process | Biological process | 113/976 | 1100/17378 | 1.90E-10 | 5.50E-07 |
| 12 | GO:0006518* | Peptide metabolic process | Biological process | 104/976 | 982/17378 | 2.00E-10 | 5.50E-07 |
| 13 | GO:0043043* | Peptide biosynthetic process | Biological process | 97/976 | 893/17378 | 2.20E-10 | 5.50E-07 |
| 14 | GO:0048002 | Antigen processing and presentation of peptide antigen | Biological process | 17/976 | 49/17378 | 5.70E-10 | 1.22E-06 |
| 15 | GO:0022627* | Cytosolic small ribosomal subunit | Cellular component | 26/1063 | 121/18378 | 4.70E-09 | 1.34E-06 |
| 16 | GO:0044391* | Ribosomal subunit | Cellular component | 58/1063 | 446/18378 | 5.80E-09 | 1.34E-06 |
| 17 | GO:0015935* | Small ribosomal subunit | Cellular component | 30/1063 | 158/18378 | 7.00E-09 | 1.34E-06 |
| 18 | GO:0044445* | Cytosolic part | Cellular component | 59/1063 | 460/18378 | 7.20E-09 | 1.34E-06 |
| 19 | GO:0005903* | Brush border | Cellular component | 23/1063 | 99/18378 | 7.50E-09 | 1.34E-06 |
| 20 | GO:0019538* | Protein metabolic process | Biological process | 372/976 | 5206/17378 | 1.20E-08 | 2.25E-05 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 30.
Top 20 of 670 selective GO pathways in the liver.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0044710* | Single-organism metabolic process | Biological process | 297/630 | 3483/17378 | 1.00E-30 | 1.50E-27 |
| 2 | GO:0043436* | Oxoacid metabolic process | Biological process | 131/630 | 793/17378 | 1.00E-30 | 1.50E-27 |
| 3 | GO:0006082* | Organic acid metabolic process | Biological process | 132/630 | 806/17378 | 1.00E-30 | 1.50E-27 |
| 4 | GO:0019752* | Carboxylic acid metabolic process | Biological process | 125/630 | 740/17378 | 1.00E-30 | 1.50E-27 |
| 5 | GO:0044281* | Small molecule metabolic process | Biological process | 181/630 | 1566/17378 | 1.00E-30 | 1.50E-27 |
| 6 | GO:0055114* | Oxidation-reduction process | Biological process | 125/630 | 967/17378 | 1.00E-30 | 1.50E-27 |
| 7 | GO:0006629* | Lipid metabolic process | Biological process | 128/630 | 1021/17378 | 1.00E-30 | 1.50E-27 |
| 8 | GO:0044712* | Single-organism catabolic process | Biological process | 102/630 | 695/17378 | 1.00E-30 | 1.50E-27 |
| 9 | GO:0044282* | Small molecule catabolic process | Biological process | 57/630 | 231/17378 | 1.00E-30 | 1.50E-27 |
| 10 | GO:0032787* | Monocarboxylic acid metabolic process | Biological process | 77/630 | 447/17378 | 1.00E-30 | 1.50E-27 |
| 11 | GO:0005615* | Extracellular space | Cellular component | 146/634 | 1396/18378 | 1.00E-30 | 1.78E-27 |
| 12 | GO:0016491* | Oxidoreductase activity | Molecular function | 99/614 | 775/16814 | 1.40E-28 | 5.76E-25 |
| 13 | GO:0003824* | Catalytic activity | Molecular function | 334/614 | 5604/16814 | 7.20E-28 | 1.48E-24 |
| 14 | GO:0008202 | Steroid metabolic process | Biological process | 50/630 | 204/17378 | 1.10E-27 | 1.50E-24 |
| 15 | GO:0016054* | Organic acid catabolic process | Biological process | 44/630 | 169/17378 | 1.20E-25 | 1.38E-22 |
| 16 | GO:0046395* | Carboxylic acid catabolic process | Biological process | 44/630 | 169/17378 | 1.20E-25 | 1.38E-22 |
| 17 | GO:0044255* | Cellular lipid metabolic process | Biological process | 92/630 | 774/17378 | 2.10E-24 | 2.25E-21 |
| 18 | GO:0005576* | Extracellular region | Cellular component | 234/634 | 3681/18378 | 8.70E-24 | 7.75E-21 |
| 19 | GO:0044421* | Extracellular region part | Cellular component | 214/634 | 3289/18378 | 1.20E-22 | 7.13E-20 |
| 20 | GO:1901605* | Alpha-amino acid metabolic process | Biological process | 41/630 | 175/17378 | 4.30E-22 | 4.30E-19 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 31.
Top 20 of 1389 selective GO pathways in the lung.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0072359* | Circulatory system development | Biological process | 300/2858 | 814/17378 | 1.00E-30 | 3.49E-28 |
| 2 | GO:0072358 | Cardiovascular system development | Biological process | 216/2858 | 515/17378 | 1.00E-30 | 3.49E-28 |
| 3 | GO:0001944 | Vasculature development | Biological process | 213/2858 | 506/17378 | 1.00E-30 | 3.49E-28 |
| 4 | GO:0048856* | Anatomical structure development | Biological process | 1053/2858 | 4553/17378 | 1.00E-30 | 3.49E-28 |
| 5 | GO:0044767* | Single-organism developmental process | Biological process | 1109/2858 | 4861/17378 | 1.00E-30 | 3.49E-28 |
| 6 | GO:0032502* | Developmental process | Biological process | 1117/2858 | 4909/17378 | 1.00E-30 | 3.49E-28 |
| 7 | GO:0001568 | Blood vessel development | Biological process | 204/2858 | 487/17378 | 1.00E-30 | 3.49E-28 |
| 8 | GO:0007275* | Multicellular organism development | Biological process | 969/2858 | 4155/17378 | 1.00E-30 | 3.49E-28 |
| 9 | GO:0009653* | Anatomical structure morphogenesis | Biological process | 550/2858 | 2009/17378 | 1.00E-30 | 3.49E-28 |
| 10 | GO:0010468* | Regulation of gene expression | Biological process | 828/2858 | 3414/17378 | 1.00E-30 | 3.49E-28 |
| 11 | GO:0051252* | Regulation of RNA metabolic process | Biological process | 725/2858 | 2910/17378 | 1.00E-30 | 3.49E-28 |
| 12 | GO:0048646 | Anatomical structure formation involved in morphogenesis | Biological process | 281/2858 | 814/17378 | 1.00E-30 | 3.49E-28 |
| 13 | GO:0031323* | Regulation of cellular metabolic process | Biological process | 1089/2858 | 4879/17378 | 1.00E-30 | 3.49E-28 |
| 14 | GO:0060255* | Regulation of macromolecule metabolic process | Biological process | 1074/2858 | 4798/17378 | 1.00E-30 | 3.49E-28 |
| 15 | GO:0044707* | Single-multicellular organism process | Biological process | 1102/2858 | 4954/17378 | 1.00E-30 | 3.49E-28 |
| 16 | GO:2001141* | Regulation of RNA biosynthetic process | Biological process | 699/2858 | 2797/17378 | 1.00E-30 | 3.49E-28 |
| 17 | GO:1903506* | Regulation of nucleic acid-templated transcription | Biological process | 698/2858 | 2792/17378 | 1.00E-30 | 3.49E-28 |
| 18 | GO:0019222* | Regulation of metabolic process | Biological process | 1141/2858 | 5184/17378 | 1.00E-30 | 3.49E-28 |
| 19 | GO:0006355* | Regulation of transcription, DNA-templated | Biological process | 690/2858 | 2762/17378 | 1.00E-30 | 3.49E-28 |
| 20 | GO:0019219* | Regulation of nucleobase-containing compound metabolic process | Biological process | 782/2858 | 3239/17378 | 1.00E-30 | 3.49E-28 |
Note: * also significantly expressed in other organs. Sorted by q-value.
Table 32.
Top 20 of 1168 selective GO pathways in the spleen.
| No. | GO.ID | Term | Ontology | Significant | Annotated | p-value | q-value |
|---|---|---|---|---|---|---|---|
| 1 | GO:0044428* | Nuclear part | Cellular component | 851/2479 | 3315/18378 | 1.00E-30 | 7.43E-29 |
| 2 | GO:0005634* | Nucleus | Cellular component | 1216/2479 | 5641/18378 | 1.00E-30 | 7.43E-29 |
| 3 | GO:0031981* | Nuclear lumen | Cellular component | 764/2479 | 2915/18378 | 1.00E-30 | 7.43E-29 |
| 4 | GO:0005694* | Chromosome | Cellular component | 327/2479 | 812/18378 | 1.00E-30 | 7.43E-29 |
| 5 | GO:0044427* | Chromosomal part | Cellular component | 297/2479 | 738/18378 | 1.00E-30 | 7.43E-29 |
| 6 | GO:0070013* | Intracellular organelle lumen | Cellular component | 781/2479 | 3235/18378 | 1.00E-30 | 7.43E-29 |
| 7 | GO:0031974* | Membrane-enclosed lumen | Cellular component | 781/2479 | 3238/18378 | 1.00E-30 | 7.43E-29 |
| 8 | GO:0043233* | Organelle lumen | Cellular component | 781/2479 | 3238/18378 | 1.00E-30 | 7.43E-29 |
| 9 | GO:0043228* | Non-membrane-bounded organelle | Cellular component | 814/2479 | 3656/18378 | 1.00E-30 | 7.43E-29 |
| 10 | GO:0043232* | Intracellular non-membrane-bounded organelle | Cellular component | 814/2479 | 3656/18378 | 1.00E-30 | 7.43E-29 |
| 11 | GO:0005654* | Nucleoplasm | Cellular component | 554/2479 | 2197/18378 | 1.00E-30 | 7.43E-29 |
| 12 | GO:0098687 | Chromosomal region | Cellular component | 136/2479 | 250/18378 | 1.00E-30 | 7.43E-29 |
| 13 | GO:0032991* | Macromolecular complex | Cellular component | 940/2479 | 4830/18378 | 1.00E-30 | 7.43E-29 |
| 14 | GO:0000228 | Nuclear chromosome | Cellular component | 177/2479 | 457/18378 | 1.00E-30 | 7.43E-29 |
| 15 | GO:0044446* | Intracellular organelle part | Cellular component | 1193/2479 | 6591/18378 | 1.00E-30 | 7.43E-29 |
| 16 | GO:0044454 | Nuclear chromosome part | Cellular component | 167/2479 | 429/18378 | 1.00E-30 | 7.43E-29 |
| 17 | GO:0044422* | Organelle part | Cellular component | 1204/2479 | 6775/18378 | 1.00E-30 | 7.43E-29 |
| 18 | GO:0000775 | Chromosome, centromeric region | Cellular component | 83/2479 | 142/18378 | 1.00E-30 | 7.43E-29 |
| 19 | GO:0005622* | Intracellular | Cellular component | 1939/2479 | 12452/18378 | 1.00E-30 | 7.43E-29 |
| 20 | GO:0000793 | Condensed chromosome | Cellular component | 82/2479 | 145/18378 | 1.00E-30 | 7.43E-29 |
Note: * also significantly expressed in other organs. Sorted by q-value.
3.4. Genes without Description but Selectively Expressed
Apart from the genes whose function is described, there were 123 genes without a clear description but selectively expressed in 11 organs (Fig. 5). From the results of Fig. (5), most genes without description were selectively expressed in the adrenal gland and brain. Instead, there were fewer genes without description in rat gastrointestinal tracts, including stomach, duodenum, ileum, and colon. The top 20 genes without description in the adrenal gland, brain, colon, duodenum, heart, ileum, kidney, liver, lung, spleen, and stomach were listed in Tables 34-44, respectively; and their full lists can be seen in the supplementary data. Because the genes were not described but selectively expressed in the organs, their products and functions need further investigation. Given the low number of genes selectively expressed in the adrenal gland, the high number of undescribed high expression of genes in this organ suggests that the organ may be less studied.
Fig. (5).
There were 123 Genes without description but selectively expressed in different organs based on their abundance. Abbreviations: Ad, adrenal gland; Br, brain; Co, colon; Du, duodenum; He, heart; Il, ileum; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; St, stomach.
Table 34.
The top 20/32 genes were not described but selectively expressed in the adrenal glands based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean /total | |
|---|---|---|---|---|---|---|---|---|
| Mean | Total | |||||||
| 1 | ENSRNOG00000041608 | AC123095.1 | St | 32.5 | 45.5 | 2.39E-05 | 2.00E-03 | 0.716 |
| 2 | ENSRNOG00000055956 | AABR07015078.1 | St | 103.8 | 141.0 | 3.62E-05 | 2.47E-03 | 0.736 |
| 3 | ENSRNOG00000030291 | Rn50_10_0698.6 | St | 871.4 | 1199.5 | 4.14E-05 | 2.57E-03 | 0.727 |
| 4 | ENSRNOG00000060657 | AABR07000404.1 | St | 14.2 | 19.1 | 1.05E-04 | 4.21E-03 | 0.742 |
| 5 | ENSRNOG00000029145 | AY172581.2 | St | 462.8 | 594.1 | 1.68E-04 | 5.34E-03 | 0.779 |
| 6 | ENSRNOG00000057514 | AABR07015080.1 | St | 26.2 | 35.7 | 1.79E-04 | 5.51E-03 | 0.734 |
| 7 | ENSRNOG00000057811 | AABR07015055.2 | St | 18.8 | 25.1 | 2.46E-04 | 6.47E-03 | 0.750 |
| 8 | ENSRNOG00000055836 | AABR07000402.1 | St | 30.7 | 42.9 | 3.34E-04 | 7.56E-03 | 0.717 |
| 9 | ENSRNOG00000046600 | AABR07015066.1 | St | 73.6 | 100.6 | 3.88E-04 | 8.16E-03 | 0.732 |
| 10 | ENSRNOG00000055323 | AABR07063421.1 | St | 33.9 | 47.6 | 4.49E-04 | 8.79E-03 | 0.712 |
| 11 | ENSRNOG00000046081 | AABR07015079.1 | St | 38.8 | 55.2 | 5.83E-04 | 1.00E-02 | 0.703 |
| 12 | ENSRNOG00000047991 | AABR07072283.1 | St | 125.3 | 143.0 | 1.02E-03 | 1.34E-02 | 0.876 |
| 13 | ENSRNOG00000053717 | Metazoa_SRP | Il | 121.7 | 171.4 | 1.69E-03 | 1.68E-02 | 0.710 |
| 14 | ENSRNOG00000046768 | AC135454.2 | St | 13.2 | 14.2 | 1.62E-03 | 1.70E-02 | 0.929 |
| 15 | ENSRNOG00000056945 | LOC102549408 | Sp | 22.1 | 26.9 | 1.97E-03 | 1.88E-02 | 0.820 |
| 16 | ENSRNOG00000049380 | Rn50_11_0375.8 | Du | 55.9 | 68.7 | 2.24E-03 | 1.99E-02 | 0.814 |
| 17 | ENSRNOG00000046106 | rno-mir-351-1 | St | 6.4 | 6.4 | 2.49E-03 | 2.11E-02 | 1.000 |
| 18 | ENSRNOG00000055947 | 7SK | Du | 24.6 | 30.8 | 2.63E-03 | 2.14E-02 | 0.800 |
| 19 | ENSRNOG00000048598 | AABR07037925.1 | St | 84.9 | 95.4 | 2.55E-03 | 2.14E-02 | 0.889 |
| 20 | ENSRNOG00000053888 | 5_8S_rRNA | St | 11.8 | 13.7 | 2.75E-03 | 2.23E-02 | 0.858 |
Note: Sorted by q-value. Du, duodenum; Il, ileum; Sp, spleen; St, stomach.
Table 44.
Genes were not described but selectively expressed in the stomach based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000060525 | AABR07007717.3 | Du | 8.9 | 12.7 | 5.70E-05 | 2.11E-03 | 0.703 |
| 2 | ENSRNOG00000062012 | Rn60_20_0037.1 | Ad | 35.3 | 35.3 | 6.37E-03 | 3.46E-02 | 1.000 |
Note: Sorted by q-value. Ad, adrenal gland; Du, duodenum.
4. DISCUSSION
Screening selectively expressed genes in organs is not only a tough task but also meaningful work because the results of the work will provide useful clues and even evidence for scientists to unveil the mechanism behind the overall dysfunction and symptoms. At least, we can obtain the putative organic markers for evaluating organic injury. There were good examples of some proteins selectively expressed in organs that were used as disease markers [8, 10-12] or used as therapeutic targets like trastuzumab on HER2 to treat breast cancer [167]. However, many selective genes have still not been revealed.
The present study screened out 1,406 genes selectively expressed in 11 rat organs, among which, 1,283 genes’ function was described, and 123 of which still need to be described in the near future. Some of the genes’ function was confirmed in the organs that were noted in Tables 1-11, but a good portion of them or the relationship between their function and the organs was not addressed. The new findings are useful to unveil the mechanism of their organic functions. Unfortunately, as for the selective genes in organs mentioned in the introduction, only troponin [10] was proved to be selective by the present study, and NeuN in the brain [8], GPT in the liver [11], and NGAL in the kidney [12] were not included in the present list of the selective genes. After consulting the FPKM values, it is exactly that the FPKM of NeuN in the brain was the highest, but not significant. The relative neuronal marker was further proved by recent work [9]. The highest GPT (GPT2) in the liver was significant, but the level of expression was not dominant (only about 45% of the total). Of course, if the criterion of selective genes was lowered, more genes would be included in the selective gene list, namely, in the list of putative organic markers. Phosphodiesterase 5 (PDE5a), an enzyme associated with angiectasis, is another similar example. PDE5a was verified to be the most highly expressed gene in the lung, but not included in the selective gene list (Table 9), supporting PDE5 inhibitors’ pharmacological effect on pulmonary arterial hypertension [168, 169].
The selective genes and their products can be used as physiological or disease markers. If a cell is injured, the selective gene’s product normally existing in its cytoplasm will be released to the blood. Based on the principle, some injury markers like serum Myl3 protein for heart injury [170] were screened out and verified by the present study. Theoretically, products from selective genes can be used as disease markers. However, it should be noted that because of some genes expressed in rats (e.g., Uox in the liver) [171], but not in humans, the fact that the products from the selective genes used as disease markers are only advisory, needing further verification.
The functional pathways of an organ enriched by the highest-expressed genes were largely supported by the known understanding. However, there are still some interesting functions that were not focused on. For example, KEGG pathways (Tables 12-22) like ko00061 (fatty acid biosynthesis) in the adrenal gland, ko04911 (insulin secretion) in the brain, ko00280 (Valine, leucine, and isoleucine degradation) in the heart and kidney, and ko04360 (axon guidance) in the lung were seldom paid attention to by scientists. Similar results would be obtained in the results of GO pathways (Table 23-33). The unpopular organic functional pathways enriched by the present study would open a new window to make insight into their mechanism. Especially the adrenal glands may be an organ with few basic researches.
Though the selective genes and the interesting genes only existed in one organ, the organic pathways including KEGG (Tables 12-22) and GO (Tables 23-33) pathways, enriched by them could exist in two or more organs. Since a pathway often involves many proteins, it is theoretically different for the real functions of the same selective pathway enriched by different selective genes. The same pathway is enriched in different organs with different profiles. Anyway, the functions are different from organ to organ, although they share some similarities at pathway levels.
CONCLUSION
In the end, because there were no standard criteria ready to evaluate a gene's selectivity, the present study used the dominant portion of FPKM value and statistical analysis. If the FPKM value of a gene in an organ accounted for 70% of the total values of all the organs concerned, the gene was assumed as the selective gene in the organ after excluding genes with low abundance. If the criterion were lowered, the list of the selective genes would be lengthened. On the other hand, the selective genes screened out by the present study were only based on the results of 11 organs in male rats, and some selective genes in other organs or female rats were neglected or missed. Moreover, the weights of the organs were not taken into account in the present study. Considering that the genome of rats has approximately 85% similarity with that of humans, this study provides a useful exploration of human organic markers and organ function, though the selective genes, the putative markers, and the functional pathways suggested are only advisory and worthy of further investigation.
Table 35.
The top 20/27 genes were not described but selectively expressed in the brain based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000047491 | AABR07037520.1 | St | 53.6 | 59.3 | 6.32E-07 | 3.22E-04 | 0.905 |
| 2 | ENSRNOG00000051341 | Rn50_X_0635.2 | Co | 28.1 | 35.5 | 3.05E-06 | 5.69E-04 | 0.792 |
| 3 | ENSRNOG00000054414 | AABR07043276.1 | Il | 23.4 | 28.2 | 4.13E-06 | 8.22E-04 | 0.831 |
| 4 | ENSRNOG00000060837 | AC132752.2 | Il | 58.1 | 76.0 | 1.22E-05 | 1.06E-03 | 0.764 |
| 5 | ENSRNOG00000003025 | Rn50_X_0749.3 | Ad | 48.1 | 52.7 | 4.53E-05 | 2.54E-03 | 0.914 |
| 6 | ENSRNOG00000060863 | AABR07017145.1 | Sp | 50.4 | 62.3 | 5.12E-05 | 2.92E-03 | 0.810 |
| 7 | ENSRNOG00000060211 | AABR07058699.2 | Co | 31.8 | 36.6 | 1.24E-04 | 4.36E-03 | 0.869 |
| 8 | ENSRNOG00000054809 | AABR07026032.1 | Lu | 8.5 | 8.9 | 1.34E-04 | 4.74E-03 | 0.954 |
| 9 | ENSRNOG00000038087 | AC110846.1 | Li | 7.7 | 10.3 | 4.35E-04 | 5.50E-03 | 0.746 |
| 10 | ENSRNOG00000058047 | AABR07000733.1 | Il | 10.3 | 11.2 | 2.19E-04 | 5.93E-03 | 0.918 |
| 11 | ENSRNOG00000022286 | Rn50_X_0746.6 | Ad | 16.9 | 20.2 | 2.85E-04 | 6.70E-03 | 0.834 |
| 12 | ENSRNOG00000022267 | Rn50_X_0747.1 | Il | 50.2 | 60.8 | 5.73E-04 | 9.89E-03 | 0.827 |
| 13 | ENSRNOG00000059081 | AABR07026032.3 | Co | 47.8 | 49.1 | 6.06E-04 | 1.02E-02 | 0.974 |
| 14 | ENSRNOG00000054155 | Rn50_5_1638.1 | Ad | 10.7 | 10.9 | 6.29E-04 | 1.04E-02 | 0.983 |
| 15 | ENSRNOG00000052831 | AABR07040629.1 | Co | 31.1 | 35.4 | 1.67E-03 | 1.72E-02 | 0.879 |
| 16 | ENSRNOG00000049802 | AABR07031533.1 | Ki | 36.4 | 37.0 | 2.35E-03 | 2.05E-02 | 0.984 |
| 17 | ENSRNOG00000002734 | AABR07042077.1 | Il | 8.2 | 8.4 | 2.83E-03 | 2.26E-02 | 0.984 |
| 18 | ENSRNOG00000054121 | AABR07061178.1 | Du | 21.5 | 22.4 | 4.28E-03 | 2.80E-02 | 0.960 |
| 19 | ENSRNOG00000060858 | AABR07043711.1 | Ki | 12.4 | 13.3 | 4.82E-03 | 2.95E-02 | 0.934 |
| 20 | ENSRNOG00000058276 | AABR07043200.1 | Sp | 7.5 | 8.5 | 5.24E-03 | 3.12E-02 | 0.887 |
Note: Sorted by q-value. Ad, adrenal gland; Co, colon; Du, duodenum; Il, ileum; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; St, stomach.
Table 36.
Genes were not described but selectively expressed in the colon based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000062185 | Rn60_20_0141.5 | Ad | 16.4 | 19.8 | 1.68E-05 | 1.68E-03 | 0.828 |
| 2 | ENSRNOG00000056727 | AABR07057353.2 | St | 11.5 | 11.7 | 4.92E-04 | 9.21E-03 | 0.979 |
| 3 | ENSRNOG00000038598 | AABR07032503.1 | Ad | 10.8 | 13.0 | 6.59E-03 | 3.52E-02 | 0.827 |
Note: Sorted by q-value. Ad, adrenal gland; St, stomach.
Table 37.
Genes were not described but selectively expressed in the duodenum based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000055064 | LOC102551636 | Ki | 415.4 | 557.4 | 5.88E-06 | 9.90E-04 | 0.745 |
| 2 | ENSRNOG00000056733 | AABR07004539.1 | Ad | 114.7 | 122.9 | 1.02E-03 | 1.33E-02 | 0.933 |
| 3 | ENSRNOG00000058562 | AABR07065651.7 | Br | 52.1 | 73.9 | 7.41E-03 | 3.74E-02 | 0.705 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; Ki, kidney.
Table 38.
Genes were not described but selectively expressed in the heart based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000023227 | AABR07052585.2 | Li | 745.4 | 754.6 | 6.97E-07 | 3.41E-04 | 0.988 |
| 2 | ENSRNOG00000043057 | AABR07025284.1 | Il | 18.5 | 20.0 | 1.58E-06 | 5.10E-04 | 0.924 |
| 3 | ENSRNOG00000052518 | AABR07025387.1 | Du | 26.7 | 33.3 | 2.28E-05 | 1.89E-03 | 0.801 |
| 4 | ENSRNOG00000048644 | AC115371.1 | St | 13.2 | 13.6 | 1.22E-04 | 4.56E-03 | 0.970 |
| 5 | ENSRNOG00000046133 | LOC102553613 | Du | 30.7 | 37.8 | 1.67E-04 | 4.59E-03 | 0.811 |
| 6 | ENSRNOG00000052389 | AABR07031489.1 | Co | 8.9 | 9.9 | 3.03E-04 | 7.12E-03 | 0.902 |
| 7 | ENSRNOG00000055328 | AABR07017268.1 | Ki | 10.3 | 11.7 | 8.42E-04 | 1.20E-02 | 0.881 |
| 8 | ENSRNOG00000060690 | AABR07052523.2 | Ad | 8.1 | 8.1 | 1.40E-03 | 1.57E-02 | 1.000 |
| 9 | ENSRNOG00000046229 | AC130940.1 | St | 11.9 | 12.7 | 2.82E-03 | 2.26E-02 | 0.935 |
| 10 | ENSRNOG00000058414 | LOC103690078 | Ad | 5.6 | 5.6 | 1.13E-02 | 4.71E-02 | 0.992 |
Note: Sorted by q-value. Ad, adrenal gland; Co, colon; Du, duodenum; Il, ileum; Ki, kidney; Li, liver; St, stomach.
Table 39.
Genes were not described but selectively expressed in the ileum based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000051194 | LOC108352134 | Ad | 22.7 | 30.7 | 4.35E-03 | 2.83E-02 | 0.739 |
| 2 | ENSRNOG00000051320 | Rn50_7_1164.3 | Lu | 22.0 | 25.6 | 4.74E-03 | 2.95E-02 | 0.861 |
Note: Sorted by q-value. Ad, adrenal gland; Lu, lung.
Table 40.
Genes were not described but selectively expressed in the kidney based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000056396 | AABR07006120.1 | Ad | 29.7 | 29.7 | 1.76E-06 | 5.42E-04 | 1.000 |
| 2 | ENSRNOG00000054801 | AABR07057997.1 | Br | 8.2 | 8.9 | 2.50E-06 | 6.28E-04 | 0.926 |
| 3 | ENSRNOG00000051964 | LOC103691699 | St | 67.9 | 75.7 | 6.31E-06 | 9.80E-04 | 0.897 |
| 4 | ENSRNOG00000054733 | LOC103690137 | He | 10.9 | 14.5 | 1.63E-05 | 1.11E-03 | 0.756 |
| 5 | ENSRNOG00000061754 | LOC102555924 | Sp | 5.5 | 7.2 | 1.78E-05 | 1.36E-03 | 0.762 |
| 6 | ENSRNOG00000057101 | AABR07050652.1 | Sp | 15.5 | 20.5 | 4.17E-05 | 2.47E-03 | 0.759 |
| 7 | ENSRNOG00000057904 | LOC102554608 | Ad | 64.2 | 64.2 | 2.14E-04 | 6.03E-03 | 1.000 |
| 8 | ENSRNOG00000061966 | Rn60_1_2220.2 | Ad | 15.2 | 17.1 | 2.46E-04 | 6.48E-03 | 0.891 |
| 9 | ENSRNOG00000061127 | AABR07057844.2 | He | 8.8 | 9.4 | 2.89E-04 | 6.98E-03 | 0.936 |
| 10 | ENSRNOG00000061436 | AABR07026778.1 | Ad | 27.7 | 27.8 | 4.18E-04 | 8.48E-03 | 0.997 |
| 11 | ENSRNOG00000059212 | AABR07025303.1 | Lu | 14.2 | 15.9 | 6.33E-04 | 1.03E-02 | 0.895 |
| 12 | ENSRNOG00000053953 | AABR07016672.1 | Ad | 6.9 | 6.9 | 9.28E-04 | 1.27E-02 | 1.000 |
| 13 | ENSRNOG00000057369 | AABR07027240.1 | Ad | 7.8 | 7.8 | 1.00E-03 | 1.32E-02 | 0.997 |
| 14 | ENSRNOG00000059314 | AABR07013477.2 | Ad | 16.7 | 16.8 | 1.14E-03 | 1.41E-02 | 0.994 |
| 15 | ENSRNOG00000046343 | - | Ad | 47.6 | 57.0 | 1.23E-03 | 1.47E-02 | 0.835 |
| 16 | ENSRNOG00000058847 | AABR07044001.4 | Br | 10.6 | 14.9 | 1.84E-03 | 1.80E-02 | 0.710 |
| 17 | ENSRNOG00000058611 | AABR07027137.1 | Lu | 14.2 | 19.6 | 9.08E-03 | 4.14E-02 | 0.723 |
Note: Sorted by q-value. Ad, adrenal gland; Br, brain; He, heart; Lu, lung; Sp, spleen; St, stomach.
Table 41.
Genes were not described but selectively expressed in the liver based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000054077 | AABR07024870.1 | Ad | 277.7 | 277.8 | 8.88E-05 | 3.87E-03 | 1.000 |
| 2 | ENSRNOG00000052176 | AC115255.1 | Du | 9.3 | 10.4 | 3.78E-04 | 7.88E-03 | 0.895 |
| 3 | ENSRNOG00000059330 | AABR07004549.1 | Ad | 802.9 | 803.1 | 1.56E-03 | 1.66E-02 | 1.000 |
| 4 | ENSRNOG00000062027 | Rn60_12_0107.3 | Ad | 89.4 | 89.5 | 1.73E-03 | 1.75E-02 | 0.999 |
| 5 | ENSRNOG00000021575 | AABR07021096.1 | Ad | 42.3 | 42.9 | 4.88E-03 | 3.01E-02 | 0.987 |
| 6 | ENSRNOG00000055973 | AABR07058498.1 | Ad | 14.5 | 14.9 | 5.54E-03 | 3.22E-02 | 0.975 |
Note: Sorted by q-value. Ad, adrenal gland; Du, duodenum
Table 42.
Genes were not described but selectively expressed in the lung based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000054709 | AABR07061382.2 | St | 14.2 | 18.3 | 2.75E-06 | 3.69E-04 | 0.776 |
| 2 | ENSRNOG00000053542 | AABR07067469.1 | Ad | 11.5 | 11.9 | 1.45E-05 | 1.56E-03 | 0.963 |
| 3 | ENSRNOG00000055889 | AABR07030901.1 | He | 5.7 | 7.2 | 6.63E-05 | 3.16E-03 | 0.792 |
| 4 | ENSRNOG00000036872 | AC119007.1 | St | 29.1 | 30.6 | 1.51E-04 | 5.07E-03 | 0.950 |
| 5 | ENSRNOG00000059588 | AC113785.2 | Ki | 1016.6 | 1365.0 | 2.14E-04 | 6.04E-03 | 0.745 |
| 6 | ENSRNOG00000046001 | AABR07030823.1 | He | 22.4 | 28.4 | 8.07E-04 | 1.16E-02 | 0.790 |
| 7 | ENSRNOG00000052597 | AABR07062477.2 | Ad | 7.0 | 7.0 | 8.03E-04 | 1.18E-02 | 0.995 |
| 8 | ENSRNOG00000050974 | AABR07030773.1 | St | 9.3 | 12.2 | 3.21E-03 | 2.39E-02 | 0.761 |
| 9 | ENSRNOG00000054935 | Rn50_7_1408.2 | St | 14.7 | 15.0 | 5.45E-03 | 3.19E-02 | 0.980 |
Note: Sorted by q-value. Ad, adrenal gland; He, heart; Ki, kidney; St, stomach.
Table 43.
Genes were not described but selectively expressed in the spleen based on their abundance (n = 3).
| No. | Gene ID | Gene Name | Median Organ | FPKM | p-value | q-value | Mean/ total | |
| Mean | Total | |||||||
| 1 | ENSRNOG00000062220 | LOC679342 | St | 9.1 | 12.0 | 3.88E-07 | 2.54E-04 | 0.764 |
| 2 | ENSRNOG00000062144 | AABR07035955.1 | St | 34.0 | 45.8 | 4.49E-06 | 8.67E-04 | 0.742 |
| 3 | ENSRNOG00000053879 | AABR07071821.1 | Ad | 8.8 | 8.9 | 5.70E-05 | 3.10E-03 | 0.988 |
| 4 | ENSRNOG00000060395 | AABR07025301.1 | St | 10.4 | 13.4 | 1.30E-04 | 4.70E-03 | 0.780 |
| 5 | ENSRNOG00000057558 | AC128792.2 | Ki | 1492.9 | 1879.3 | 1.89E-04 | 5.62E-03 | 0.794 |
| 6 | ENSRNOG00000053143 | Rn50_7_1407.3 | Du | 17.7 | 20.4 | 1.35E-03 | 1.41E-02 | 0.866 |
| 7 | ENSRNOG00000041826 | AABR07053152.1 | St | 14.6 | 19.8 | 1.66E-03 | 1.71E-02 | 0.736 |
| 8 | ENSRNOG00000041746 | AC095678.1 | St | 6.1 | 7.3 | 1.86E-03 | 1.82E-02 | 0.832 |
| 9 | ENSRNOG00000039025 | AABR07051947.1 | Lu | 24.8 | 34.8 | 4.04E-03 | 2.66E-02 | 0.713 |
| 10 | ENSRNOG00000052921 | AABR07021221.1 | Ki | 19.8 | 25.1 | 9.51E-03 | 4.25E-02 | 0.788 |
| 11 | ENSRNOG00000054411 | AABR07072897.1 | St | 6.4 | 8.9 | 1.07E-02 | 4.58E-02 | 0.714 |
| 12 | ENSRNOG00000062261 | Rn60_15_0518.2 | Ad | 6.5 | 6.7 | 1.18E-02 | 4.82E-02 | 0.977 |
Note: Sorted by q-value. Ad, adrenal gland; Du, duodenum; Ki, kidney; Lu, lung; St, stomach.
ACKNOWLEDGEMENTS
Declared none.
LIST OF ABBREVIATIONS
- DOS
Disc Operation System
- GO
Gene Ontology
- Icam1
Intercellular Adhesion Molecule 1
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NGAL
Neutrophil Gelatinase-associated Lipocalin
- PDE5a
Phosphodiesterase 5
- SD
Sprague-Dawley
AUTHORS’ CONTRIBUTIONS
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to its submission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The animal experiments were approved by the Animal Care and Use Committee of Yunnan Provincial Key Laboratory of Molecular Biology for Sinomedicine (Approved No. LL-20171023-01), Yunnan University of Traditional Chinese Medicine, Kunming, Yunnan, China.
HUMAN AND ANIMAL RIGHTS
All the animal experimentation was performed according to the Guide for the CARE and USE of Laboratory Animals and ARRIVE guidelines.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The raw data were uploaded as supplemental materials on the journal’s web.
FUNDING
This work was supported by the Foundation for Scien-tific Research provided by the National Natural Science Foundation of China (82260886), Yunnan Provincial Science and Technology Department–Applied Basic Research Joint Special Funds of Kunming Medical University (202101AY070001-007), and Yunnan Provincial Science and Technology Department–Applied Basic Research Joint Special Funds of Yunnan University of Traditional Chinese Medicine (202101AZ070001-010).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article. The raw data were uploaded on July 19, 2023 (Link: https://pan.baidu.com/s/1uOpvEIU_dRYgGmEIWc0SjA?pwd=DWG1 Password: DWG1)
REFERENCES
- 1.Robinson J.W., Martin R.M., Tsavachidis S., Howell A.E., Relton C.L., Armstrong G.N., Bondy M., Zheng J., Kurian K.M. Transcriptome-wide Mendelian randomization study prioritising novel tissue-dependent genes for glioma susceptibility. Sci. Rep. 2021;11(1):2329. doi: 10.1038/s41598-021-82169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y.M., Wu Z.K., Chai L.M., Zhang X.H., Li M., Chen Y.Y., Lv X.X., Zhu X.Y. [Effect on expression of mice alpha-hemoglobin stabilizing protein in different developmental stages treated with Yisui Shengxue granules]. Zhongguo Zhongyao Zazhi. 2007;32(7):609–612. [PubMed] [Google Scholar]
- 3.Girard T.J., Antunes L., Zhang N., Amrute J.M., Subramanian R., Eldem I., Remy K.E., Mazer M., Erlich E.C., Cruchaga C., Steed A.L., Randolph G.J., Di Paola J. Peripheral blood mononuclear cell tissue factor (F3 gene) transcript levels and circulating extracellular vesicles are elevated in severe coronavirus 2019 (COVID-19) disease. J. Thromb. Haemost. 2023;21(3):629–638. doi: 10.1016/j.jtha.2022.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W., Xia Z., Farré J.C., Subramani S. TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis. Autophagy. 2018;14(9):1574–1585. doi: 10.1080/15548627.2018.1463120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He C., Hua X., Sun S., Li S., Wang J., Huang X. Integrated bioinformatic analysis of SARS-CoV-2 infection related genes ACE2, BSG and TMPRSS2 in aerodigestive cancers. J. Inflamm. Res. 2021;14:791–802. doi: 10.2147/JIR.S300127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schubert T., Reisch N., Naumann R., Reichardt I., Landgraf D., Quitter F., Thirumalasetty S.R., Heninger A.K., Sarov M., Peitzsch M., Huebner A., Koehler K. CYP21A2 gene expression in a humanized 21-hydroxylase mouse model does not affect adrenocortical morphology and function. J. Endocr. Soc. 2022;6(6):bvac062. doi: 10.1210/jendso/bvac062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pras E., Arber N., Aksentijevich I., Katz G., Schapiro J.M., Prosen L., Gruberg L., Harel D., Liberman U., Weissenbach J., Pras M., Kastner D.L. Localization of a gene causing cystinuria to chromosome 2p. Nat. Genet. 1994;6(4):415–419. doi: 10.1038/ng0494-415. [DOI] [PubMed] [Google Scholar]
- 8.Tran V.D.T., Moretti S., Coste A.T., Amorim-Vaz S., Sanglard D., Pagni M. Condition-specific series of metabolic sub-networks and its application for gene set enrichment analysis. Bioinformatics. 2019;35(13):2258–2266. doi: 10.1093/bioinformatics/bty929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abou Nader N., Blais É., St-Jean G., Boerboom D., Zamberlam G., Boyer A. Effect of inactivation of Mst1 and Mst2 in the mouse adrenal cortex. J. Endocr. Soc. 2022;7(1):bvac143. doi: 10.1210/jendso/bvac143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartrampf P.E., Hüttmann T., Seitz A.K., Kübler H., Serfling S.E., Schlötelburg W., Michalski K., Rowe S.P., Pomper M.G., Buck A.K., Eberlein U., Werner R.A. SUVmean on baseline [18F]PSMA-1007 PET and clinical parameters are associated with survival in prostate cancer patients scheduled for [177Lu]Lu-PSMA I&T. Eur. J. Nucl. Med. Mol. Imaging. 2023;50(11):3465–3474. doi: 10.1007/s00259-023-06281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olivera J., Zhang V., Nemeth E., Ganz T. Erythroferrone exacerbates iron overload and ineffective extramedullary erythropoiesis in a mouse model of β-thalassemia. Blood Adv. 2023;7(14):3339–3349. doi: 10.1182/bloodadvances.2022009307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du R., Bai S., Zhao Y., Ma Y. Efficient generation of TBX3+ atrioventricular conduction-like cardiomyocytes from human pluripotent stem cells. Biochem. Biophys. Res. Commun. 2023;669:143–149. doi: 10.1016/j.bbrc.2023.05.104. [DOI] [PubMed] [Google Scholar]
- 13.Ihanus E., Uotila L.M., Toivanen A., Varis M., Gahmberg C.G. Red-cell ICAM-4 is a ligand for the monocyte/macrophage integrin CD11c/CD18: Characterization of the binding sites on ICAM-4. Blood. 2007;109(2):802–810. doi: 10.1182/blood-2006-04-014878. [DOI] [PubMed] [Google Scholar]
- 14.Abolbaghaei A., Turner M., Thibodeau J.F., Holterman C.E., Kennedy C.R.J., Burger D. The proteome of circulating large extracellular vesicles in diabetes and hypertension. Int. J. Mol. Sci. 2023;24(5):4930. doi: 10.3390/ijms24054930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong W., Xia Z., Chai Z., Qiu Z., Wang X., Yang Z., Wang J., Zhang T., Zhang Q., Jin J. Proteomic analysis of small extracellular vesicles from the plasma of patients with hepatocellular carcinoma. World J. Surg. Oncol. 2022;20(1):387. doi: 10.1186/s12957-022-02849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanczkowski W., Alexaki V.I., Tran N., Großklaus S., Zacharowski K., Martinez A., Popovics P., Block N.L., Chavakis T., Schally A.V., Bornstein S.R. Hypothalamo-pituitary and immune-dependent adrenal regulation during systemic inflammation. Proc. Natl. Acad. Sci. USA. 2013;110(36):14801–14806. doi: 10.1073/pnas.1313945110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., Wang F., Liu K., Long C., Chen Y., Li C., Li L., Liu F., Zhang X., Jing Y., Wang Y., Liang A., Yan H., Zhang H. αB‐crystallin/HSPB2 is critical for hyperactive mTOR‐induced cardiomyopathy. J. Cell. Physiol. 2021;236(12):8110–8121. doi: 10.1002/jcp.30465. [DOI] [PubMed] [Google Scholar]
- 18.Ballantyne C.M., Sligh J.E., Jr, Dai X.Y., Beaudet A.L. Characterization of the murine Icam-1 gene. Genomics. 1992;14(4):1076–1080. doi: 10.1016/S0888-7543(05)80132-6. [DOI] [PubMed] [Google Scholar]
- 19.Jiang H., Chen H., Wan P., Song S., Chen N. Downregulation of enhancer RNA EMX2OS is associated with poor prognosis in kidney renal clear cell carcinoma. Aging. 2020;12(24):25865–25877. doi: 10.18632/aging.202151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker F., Tang A.A.S., Rogers B., Carrington G., dos Remedios C., Li A., Tomlinson D., Peckham M. Affimers targeting proteins in the cardiomyocyte Z-disc: Novel tools that improve imaging of heart tissue. Front. Cardiovasc. Med. 2023;10:1094563. doi: 10.3389/fcvm.2023.1094563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., Cao J., Zhu L., Farrell K., Wang M., Guo L., Yang J., McKenzie A., Crary J.F., Cai D., Tu Z., Zhang B. Molecular differences in brain regional vulnerability to aging between males and females. Front. Aging Neurosci. 2023;15:1153251. doi: 10.3389/fnagi.2023.1153251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner C.A., Unwin R., Lopez-Garcia S.C., Kleta R., Bockenhauer D., Walsh S. The pathophysiology of distal renal tubular acidosis. Nat. Rev. Nephrol. 2023;19(6):384–400. doi: 10.1038/s41581-023-00699-9. [DOI] [PubMed] [Google Scholar]
- 23.Achom A., Das R., Pakray P. An improved Fuzzy based GWO algorithm for predicting the potential host receptor of COVID-19 infection. Comput Biol Med. 2022;151((Pt A)):106050. doi: 10.1016/j.compbiomed.2022.106050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bermúdez-Méndez E., Angelino P., van Keulen L., van de Water S., Rockx B., Pijlman G.P., Ciuffi A., Kortekaas J., Wichgers Schreur P.J. Transcriptomic profiling reveals intense host-pathogen dispute compromising homeostasis during acute rift valley fever virus infection. J. Virol. 2023;97(6):e00415–e00423. doi: 10.1128/jvi.00415-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvorjak M., Ahmed Y., Miller M.L., Sriram R., Coronnello C., Hashash J.G., Hartman D.J., Telmer C.A., Miskov-Zivanov N., Finn O.J., Cascio S. Cross-talk between colon cells and macrophages increases ST6GALNAC1 and MUC1-sTn expression in ulcerative colitis and colitis-associated colon cancer. Cancer Immunol. Res. 2020;8(2):167–178. doi: 10.1158/2326-6066.CIR-19-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng Z.H., Li W.C., Li Z.C., Wang Y.X., Han Z.W., Zhang Y.P. Neutrophil extracellular traps-associated modification patterns depict the tumor microenvironment, precision immunotherapy, and prognosis of clear cell renal cell carcinoma. Front. Oncol. 2022;12:1094248. doi: 10.3389/fonc.2022.1094248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunner H.I., Mueller M., Rutherford C., Passo M.H., Witte D., Grom A., Mishra J., Devarajan P. Urinary neutrophil gelatinase–associated lipocalin as a biomarker of nephritis in childhood‐onset systemic lupus erythematosus. Arthritis Rheum. 2006;54(8):2577–2584. doi: 10.1002/art.22008. [DOI] [PubMed] [Google Scholar]
- 28.Dong G., Wang M., Gu G., Li S., Sun X., Li Z., Cai H., Zhu Z. MACC1 and HGF are associated with survival in patients with gastric cancer. Oncol. Lett. 2017;15(3):3207–3213. doi: 10.3892/ol.2017.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ham M., Mizumori M., Watanabe C., Wang J.H., Inoue T., Nakano T., Guth P.H., Engel E., Kaunitz J.D., Akiba Y. Endogenous luminal surface adenosine signaling regulates duodenal bicarbonate secretion in rats. J. Pharmacol. Exp. Ther. 2010;335(3):607–613. doi: 10.1124/jpet.110.171520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekine K., Ikezono T., Matsumura T., Shindo S., Watanabe A., Li L., Pawankar R., Nishino T., Yagi T. Expression of cochlin mRNA splice variants in the inner ear. Audiol. Neurotol. 2010;15(2):88–96. doi: 10.1159/000231634. [DOI] [PubMed] [Google Scholar]
- 31.Naiki Y., Miyado M., Shindo M., Horikawa R., Hasegawa Y., Katsumata N., Takada S., Akutsu H., Onodera M., Fukami M. Adeno-associated virus-mediated gene therapy for patients’ fibroblasts, induced pluripotent stem cells, and a mouse model of congenital adrenal hyperplasia. Hum. Gene Ther. 2022;33(15-16):801–809. doi: 10.1089/hum.2022.005. [DOI] [PubMed] [Google Scholar]
- 32.Xu J., Song P., Nakamura S., Miller M., Barone S., Alper S.L., Riederer B., Bonhagen J., Arend L.J., Amlal H., Seidler U., Soleimani M. Deletion of the chloride transporter slc26a7 causes distal renal tubular acidosis and impairs gastric acid secretion. J. Biol. Chem. 2009;284(43):29470–29479. doi: 10.1074/jbc.M109.044396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachs G., Shin J.M., Vagin O., Lambrecht N., Yakubov I., Munson K. The gastric H,K ATPase as a drug target: Past, present, and future. J. Clin. Gastroenterol. 2007;141(2):S226–S242. doi: 10.1097/MCG.0b013e31803233b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y., Li Y. Comprehensive exploration of M2 macrophages and its related genes for predicting clinical outcomes and drug sensitivity in lung squamous cell carcinoma. J. Oncol. 2022;2022:1–12. doi: 10.1155/2022/1163924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou M., Wang Y., Qi S., Wang J., Zhang S. The expression of a mitochondria-localized glutamic acid-rich protein (MGARP/OSAP) is under the regulation of the HPG axis. Endocrinology. 2011;152(6):2311–2320. doi: 10.1210/en.2011-0050. [DOI] [PubMed] [Google Scholar]
- 36.Martínez-Saucedo M., Bárcenas-Gómez Y., Baeza-Capetillo P., Dedden M., Aguirre-Hernandez J., Téllez-Camacho S.A., Sánchez-Urbina R., Aquino-Jarquin G., Granados-Riveron J.T. Identification of human miR‐1839‐5p by small RNA‐seq, a miRNA enriched in neoplastic tissues. J. Gene Med. 2019;21(10):e3117. doi: 10.1002/jgm.3117. [DOI] [PubMed] [Google Scholar]
- 37.Sebrell T.A., Hashimi M., Sidar B., Wilkinson R.A., Kirpotina L., Quinn M.T., Malkoç Z., Taylor P.J., Wilking J.N., Bimczok D. A novel gastric spheroid co-culture model reveals chemokine-dependent recruitment of human dendritic cells to the gastric epithelium. Cell. Mol. Gastroenterol. Hepatol. 2019;8(1):157–171.e3. doi: 10.1016/j.jcmgh.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drobnis E.Z., Nangia A.K. Phosphodiesterase inhibitors (PDE Inhibitors) and male reproduction. Adv. Exp. Med. Biol. 2017;1034:29–38. doi: 10.1007/978-3-319-69535-8_5. [DOI] [PubMed] [Google Scholar]
- 40.Astudillo L., Therville N., Colacios C., Ségui B., Andrieu-Abadie N., Levade T. Glucosylceramidases and malignancies in mammals. Biochimie. 2016;125:267–280. doi: 10.1016/j.biochi.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Boncheva V., Linnebacher M., Kdimati S., Draper H., Orchard L., Mills K., O’Sullivan G., Tangney M., Guinn B. Identification of the antigens recognised by colorectal cancer patients using sera from patients who exhibit a crohn’s-like lymphoid reaction. Biomolecules. 2022;12(8):1058. doi: 10.3390/biom12081058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y., Hao Y., Zhuang Q., Ma X., Shi C. AKR1B10 regulates M2 macrophage polarization to promote the malignant phenotype of gastric cancer. Biosci. Rep. 2023;43(10):BSR20222007. doi: 10.1042/BSR20222007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prasad P., Tippana M. Morphogenic plasticity: The pathogenic attribute of Candida albicans. Curr. Genet. 2023;69(2-3):77–89. doi: 10.1007/s00294-023-01263-5. [DOI] [PubMed] [Google Scholar]
- 44.Weger M., Diotel N., Weger B.D., Beil T., Zaucker A., Eachus H.L., Oakes J.A., do Rego J.L., Storbeck K.H., Gut P., Strähle U., Rastegar S., Müller F., Krone N. Expression and activity profiling of the steroidogenic enzymes of glucocorticoid biosynthesis and the fdx1 co‐factors in zebrafish. J. Neuroendocrinol. 2018;30(4):e12586. doi: 10.1111/jne.12586. [DOI] [PubMed] [Google Scholar]
- 45.Swynghedauw B., Schwartz K., Léger J.J. Phylogenic and pathological changes. Basic Res. Cardiol. 1977;72(2-3):254–260. doi: 10.1007/BF01906370. [DOI] [PubMed] [Google Scholar]
- 46.Iwasa M., Yamagata T., Mizuguchi M., Itoh M., Matsumoto A., Hironaka M., Honda A., Momoi M.Y., Shimozawa N. ContiguousABCD1 DXS1357E deletion syndrome: Report of an autopsy case. Neuropathology. 2013;33(3):292–298. doi: 10.1111/j.1440-1789.2012.01348.x. [DOI] [PubMed] [Google Scholar]
- 47.Gawenis L.R., Greeb J.M., Prasad V., Grisham C., Sanford L.P., Doetschman T., Andringa A., Miller M.L., Shull G.E. Impaired gastric acid secretion in mice with a targeted disruption of the NHE4 Na+/H+ exchanger. J. Biol. Chem. 2005;280(13):12781–12789. doi: 10.1074/jbc.M414118200. [DOI] [PubMed] [Google Scholar]
- 48.Satala C.B., Jung I., Kovacs Z., Stefan-Van Staden R.I., Molnar C., Bara T., Patrichi A.I., Gurzu S. V-set and immunoglobulin domain containing 1 (VSIG1) as an emerging target for epithelial–mesenchymal transition of gastric cancer. Sci. Rep. 2022;12(1):16241. doi: 10.1038/s41598-022-19883-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langfelder P., Horvath S., Fast R. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012;46(11):i11. doi: 10.18637/jss.v046.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuehn F., Adiliaghdam F., Hamarneh S.R., Vasan R., Liu E., Liu Y., Ramirez J.M., Hoda R.S., Munoz A.R., Ko F.C., Armanini M., Brooks D.J., Bouxsein M.L., Demay M.B., Hodin R.A. Loss of intestinal alkaline phosphatase leads to distinct chronic changes in bone phenotype. J. Surg. Res. 2018;232:325–331. doi: 10.1016/j.jss.2018.06.061. [DOI] [PubMed] [Google Scholar]
- 51.Talaei M., Emmett P.M., Granell R., Tabatabaeian H., Northstone K., Bergström A., Shaheen S.O. Dietary patterns, lung function and asthma in childhood: A longitudinal study. Respir. Res. 2023;24(1):82. doi: 10.1186/s12931-023-02383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh H., Ha K., Hornick J.L., Madha S., Cejas P., Jajoo K., Singh P., Polak P., Lee H., Shivdasani R.A. Hybrid stomach-intestinal chromatin states underlie human barrett’s metaplasia. Gastroenterology. 2021;161(3):924–939.e11. doi: 10.1053/j.gastro.2021.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruppert V., Meyer T., Richter A., Maisch B., Pankuweit S. Identification of a missense mutation in the melusin-encoding ITGB1BP2 gene in a patient with dilated cardiomyopathy. Gene. 2013;512(2):206–210. doi: 10.1016/j.gene.2012.10.055. [DOI] [PubMed] [Google Scholar]
- 54.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou Z., Yang J., Wang G., Wang C., Zhang H. Bioinformatic analysis of gene expression profiles of pituitary gonadotroph adenomas. Oncol. Lett. 2017;15(2):1655–1663. doi: 10.3892/ol.2017.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H.S., Na M.J., Son K.H., Yang H.D., Kim S.Y., Shin E., Ha J.W., Jeon S., Kang K., Moon K., Park W.S., Nam S.W. ADAR1-dependent miR-3144-3p editing simultaneously induces MSI2 expression and suppresses SLC38A4 expression in liver cancer. Exp. Mol. Med. 2023;55(1):95–107. doi: 10.1038/s12276-022-00916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y., Smas C.M. Expression and regulation of transcript for the novel transmembrane protein Tmem182 in the adipocyte and muscle lineage. BMC Res. Notes. 2008;1(1):85. doi: 10.1186/1756-0500-1-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ago Y., Asano S., Hashimoto H., Waschek J.A. Probing the VIPR2 microduplication linkage to schizophrenia in animal and cellular models. Front. Neurosci. 2021;15:717490. doi: 10.3389/fnins.2021.717490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter M., Wang H., Lieber A. Role of fiber shaft length in tumor targeting with Ad5/3 vectors. Genes. 2022;13(11):2056. doi: 10.3390/genes13112056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L., Fouts D.E., Stärkel P., Hartmann P., Chen P., Llorente C., DePew J., Moncera K., Ho S.B., Brenner D.A., Hooper L.V., Schnabl B. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19(2):227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang H., Zhang Q., Zhang Y., Sun X., Liu C., Wang Q., Huang Y., Li Q., Wu Z., Pu C., Sun A. Identification of the level of exosomal protein by parallel reaction monitoring technology in HCC patients. Int. J. Gen. Med. 2022;15:7831–7842. doi: 10.2147/IJGM.S384140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaime-Cruz R., Sánchez-Gómez C., Villavicencio-Guzmán L., Lazzarini-Lechuga R., Patiño-Morales C.C., García-Lorenzana M., Ramírez-Fuentes T.C., Salazar-García M. Embryonic hyperglycemia disrupts myocardial growth, morphological development, and cellular organization: An in vivo experimental study. Life. 2023;13(3):768. doi: 10.3390/life13030768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oliver M.H., Jaquiery A.L., Connor K.L., Phua H.H., Harding J.E., Thorstensen E.B., Bloomfield F.H. Effect of maternal periconceptional undernutrition in sheep on cortisol regulation in offspring from mid-late gestation, through to adulthood. Front. Endocrinol. 2023;14:1122432. doi: 10.3389/fendo.2023.1122432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altrock E., Sens-Albert C., Hofmann F., Riabov V., Schmitt N., Xu Q., Jann J.C., Rapp F., Steiner L., Streuer A., Nowak V., Obländer J., Weimer N., Palme I., Göl M., Darwich A., Wuchter P., Metzgeroth G., Jawhar M., Hofmann W.K., Nowak D. Significant improvement of bone marrow-derived MSC expansion from MDS patients by defined xeno-free medium. Stem Cell Res. Ther. 2023;14(1):156. doi: 10.1186/s13287-023-03386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sampaio P., Waitzberg D.L., Machado N.M., de Miranda Torrinhas R.S.M., Fonseca D.C., Ferreira B.A.M., Marques M., Barcelos S., Ishida R.K., Guarda I., de Moura E.G.H., Sakai P., Santo M.A., Heymsfield S.B., Correa-Giannella M.L., Passadore M.D., Sala P. Gastrointestinal genetic reprogramming of vitamin A metabolic pathways in response of Roux-en-Y gastric bypass. Int. J. Vitam. Nutr. Res. 2024;94(1):27–36. doi: 10.1024/0300-9831/a000767. [DOI] [PubMed] [Google Scholar]
- 66.Hijazi H., Reis L.M., Pehlivan D., Bernstein J.A., Muriello M., Syverson E., Bonner D., Estiar M.A., Gan-Or Z., Rouleau G.A., Lyulcheva E., Greenhalgh L., Tessarech M., Colin E., Guichet A., Bonneau D., van Jaarsveld R.H., Lachmeijer A.M.A., Ruaud L., Levy J., Tabet A.C., Ploski R., Rydzanicz M., Kępczyński Ł., Połatyńska K., Li Y., Fatih J.M., Marafi D., Rosenfeld J.A., Coban-Akdemir Z., Bi W., Gibbs R.A., Hobson G.M., Hunter J.V., Carvalho C.M.B., Posey J.E., Semina E.V., Lupski J.R. TCEAL1 loss-of-function results in an X-linked dominant neurodevelopmental syndrome and drives the neurological disease trait in Xq22.2 deletions. Am. J. Hum. Genet. 2022;109(12):2270–2282. doi: 10.1016/j.ajhg.2022.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akhtar M.J., Khan S.A., Kumar B., Chawla P., Bhatia R., Singh K. Role of sodium dependent SLC13 transporter inhibitors in various metabolic disorders. Mol. Cell. Biochem. 2023;478(8):1669–1687. doi: 10.1007/s11010-022-04618-7. [DOI] [PubMed] [Google Scholar]
- 68.Gao Y., Yu Y., Qin W., Fan N., Qi Y., Chen H., Duan W. Uricase-deficient rats with similarly stable serum uric acid to human’s are sensitive model animals for studying hyperuricemia. PLoS One. 2022;17(3):e0264696. doi: 10.1371/journal.pone.0264696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camara-Clayette V., Rahuel C., Lopez C., Hattab C., Verkarre V., Bertrand O., Cartron J.P. Transcriptional regulation of the KEL gene and Kell protein expression in erythroid and non-erythroid cells. Biochem. J. 2001;356(1):171–180. doi: 10.1042/bj3560171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi X., Zhang Y., Gong Y., Chen M., Brand-Arzamendi K., Liu X., Wen X.Y. Zebrafish hhatla is involved in cardiac hypertrophy. J. Cell. Physiol. 2021;236(5):3700–3709. doi: 10.1002/jcp.30106. [DOI] [PubMed] [Google Scholar]
- 71.Bardy C., van den Hurk M., Kakaradov B., Erwin J.A., Jaeger B.N., Hernandez R.V., Eames T., Paucar A.A., Gorris M., Marchand C., Jappelli R., Barron J., Bryant A.K., Kellogg M., Lasken R.S., Rutten B.P.F., Steinbusch H.W.M., Yeo G.W., Gage F.H. Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Mol. Psychiatry. 2016;21(11):1573–1588. doi: 10.1038/mp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kondo T., Kitano S., Miyakawa N., Watanabe T., Goto R., Sato M., Hanatani S., Sakaguchi M., Igata M., Kawashima J., Motoshima H., Matsumura T., Araki E. The amount of residual incretin regulates the pancreatic β-cell function and glucose homeostasis. Intern. Med. 2021;60(9):1433–1442. doi: 10.2169/internalmedicine.6026-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cardoso-Moreira M., Sarropoulos I., Velten B., Mort M., Cooper D.N., Huber W., Kaessmann H. Developmental gene expression differences between humans and mammalian models. Cell Rep. 2020;33(4):108308. doi: 10.1016/j.celrep.2020.108308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koonin E.V., Fedorova N.D., Jackson J.D., Jacobs A.R., Krylov D.M., Makarova K.S., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Rao B.S., Rogozin I.B., Smirnov S., Sorokin A.V., Sverdlov A.V., Vasudevan S., Wolf Y.I., Yin J.J., Natale D.A. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siino V., Amato A., Di Salvo F., Caldara G.F., Filogamo M., James P., Vasto S. Impact of diet-induced obesity on the mouse brain phosphoproteome. J. Nutr. Biochem. 2018;58:102–109. doi: 10.1016/j.jnutbio.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 76.Kalisch-Smith J.I., Simmons D.G., Pantaleon M., Moritz K.M. Sex differences in rat placental development: From pre-implantation to late gestation. Biol. Sex Differ. 2017;8(1):17. doi: 10.1186/s13293-017-0138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calado J., Santos A.R., Aires I., Lebre F., Nolasco F., Rueff J., Ramalho J. The Na + ‐coupled glucose transporter SGLT 2 interacts with its accessory unit MAP 17 in vitro and their expressions overlap in the renal proximal tubule. FEBS Lett. 2018;592(19):3317–3326. doi: 10.1002/1873-3468.13233. [DOI] [PubMed] [Google Scholar]
- 78.Calvano J., Achanzar W., Murphy B., DiPiero J., Hixson C., Parrula C., Burr H., Mangipudy R., Tirmenstein M. Evaluation of microRNAs−208 and 133a/b as differential biomarkers of acute cardiac and skeletal muscle toxicity in rats. Toxicol. Appl. Pharmacol. 2016;312:53–60. doi: 10.1016/j.taap.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 79.Yang S., Wei Z., Wu J., Sun M., Ma Y., Liu G. Proteomic analysis of liver tissues in chicken embryo at Day 16 and Day 20 reveals antioxidant mechanisms. J. Proteomics. 2021;243:104258. doi: 10.1016/j.jprot.2021.104258. [DOI] [PubMed] [Google Scholar]
- 80.Mullen R.J., Buck C.R., Smith A.M. NeuN, a neuronal specific nuclear protein in vertebratesxs. Development. 1992;116(1):201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 81.Yang L., Wu Y., Su Y., Zhang X., Chakraborty T., Wang D., Zhou L. Cyp17a2 is involved in testicular development and fertility in male Nile tilapia, Oreochromis niloticus. Front. Endocrinol. 2022;13:1074921. doi: 10.3389/fendo.2022.1074921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Laere S., Van der Auwera I., Van den Eynden G., Van Hummelen P., van Dam P., Van Marck E., Vermeulen P.B., Dirix L. Distinct molecular phenotype of inflammatory breast cancer compared to non-inflammatory breast cancer using Affymetrix-based genome-wide gene-expression analysis. Br. J. Cancer. 2007;97(8):1165–1174. doi: 10.1038/sj.bjc.6603967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Yao E., Liu Y., Zhang Y., Ding M., Liu J., Chen X., Fan S. FUT2 facilitates autophagy and suppresses apoptosis via p53 and JNK signaling in lung adenocarcinoma cells. Cells. 2022;11(24):4031. doi: 10.3390/cells11244031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu C., Fu Y., Xia L., Li F., Huang K., Sun X. Expression profiles, prognosis, and ceRNA regulation of SRY-related HMG-Box genes in stomach adenocarcinoma. J. Environ. Pathol. Toxicol. Oncol. 2023;42(2):79–91. doi: 10.1615/JEnvironPatholToxicolOncol.2022044640. [DOI] [PubMed] [Google Scholar]
- 85.Mori H., Yoshino Y., Iga J., Ochi S., Funahashi Y., Yamazaki K., Kumon H., Ozaki Y., Ueno S. Aberrant expression of GABA-related genes in the hippocampus of 3xTg-AD model mice from the early to end stages of alzheimer’s disease. J. Alzheimers Dis. 2023;94(1):177–188. doi: 10.3233/JAD-230078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suga K., Kobayashi Y., Ochiai R. Impact of left heart bypass on arterial oxygenation during one-lung ventilation for thoracic aortic surgery. J. Cardiothorac. Vasc. Anesth. 2017;31(4):1197–1202. doi: 10.1053/j.jvca.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y., Guan Y., Xie Q., Gong W., Li J., Chen T., Xu Y., Xu N., Chen S., Chen M., Wang Z., Hao C.M. The metabolites of de novo NAD+ synthesis are a valuable predictor of acute kidney injury. Clin. Kidney J. 2023;16(4):711–721. doi: 10.1093/ckj/sfac262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yin H., Hou X., Tao T., Lv X., Zhang L., Duan W. Neurite outgrowth resistance to rho kinase inhibitors in PC12 Adh cell. Cell Biol. Int. 2015;39(5):563–576. doi: 10.1002/cbin.10423. [DOI] [PubMed] [Google Scholar]
- 89.Li Xu L., Wei Zhang H., Lin H., Mei Zhang X., Qi Wen Y., Long Zhao J., Xing Li Z., Gasset M. SWATH-MS-based proteomics reveals functional biomarkers of Th1/Th2 responses of tropomyosin allergy in mouse models. Food Chem. 2022;383:132474. doi: 10.1016/j.foodchem.2022.132474. [DOI] [PubMed] [Google Scholar]
- 90.Li T., di Stefano G., Raza G.S., Sommerer I., Riederer B., Römermann D., Tan X., Tan Q., Pallagi P., Hollenbach M., Herzig K.H., Seidler U. Hydrokinetic pancreatic function and insulin secretion are moduled by Cl − uniporter Slc26a9 in mice. Acta Physiol. 2022;234(1):e13729. doi: 10.1111/apha.13729. [DOI] [PubMed] [Google Scholar]
- 91.Nawata C.M., Hung C.C.Y., Tsui T.K.N., Wilson J.M., Wright P.A., Wood C.M. Ammonia excretion in rainbow trout (Oncorhynchus mykiss): evidence for Rh glycoprotein and H + -ATPase involvement. Physiol. Genomics. 2007;31(3):463–474. doi: 10.1152/physiolgenomics.00061.2007. [DOI] [PubMed] [Google Scholar]
- 92.Luo Y., Tian L., Liang C., Xu Y. KLHL38 facilitates staurosporine‐induced apoptosis in HL‐1 cells via myocardin degradation. IUBMB Life. 2022;74(5):446–462. doi: 10.1002/iub.2602. [DOI] [PubMed] [Google Scholar]
- 93.Wu Z., Liu X., Huang S., Li T., Zhang X., Pang J., Zhao J., Chen L., Zhang B., Wang J., Han D. Milk fat globule membrane attenuates acute colitis and secondary liver injury by improving the mucus barrier and regulating the gut microbiota. Front. Immunol. 2022;13:865273. doi: 10.3389/fimmu.2022.865273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Y., Gao J., Zhao D., Guan X., Morris S.C., Finkelman F.D., Huang H. The Hdc GC box is critical for Hdc gene transcription and histamine-mediated anaphylaxis. J. Allergy Clin. Immunol. 2023;152(1):195–204.e3. doi: 10.1016/j.jaci.2023.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hao J., Zeltz C., Pintilie M., Li Q., Sakashita S., Wang T., Cabanero M., Martins-Filho S.N., Wang D.Y., Pasko E., Venkat K., Joseph J., Raghavan V., Zhu C.Q., Wang Y.H., Moghal N., Tsao M.S., Navab R. Characterization of distinct populations of carcinoma-associated fibroblasts from non–small cell lung carcinoma reveals a role for ST8SIA2 in cancer cell invasion. Neoplasia. 2019;21(5):482–493. doi: 10.1016/j.neo.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alifanov V., Tashireva L., Zavyalova M., Perelmuter V. LIMCH1 as a new potential metastasis predictor in breast cancer. Asian Pac. J. Cancer Prev. 2022;23(11):3947–3952. doi: 10.31557/APJCP.2022.23.11.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Przygodzka P., Papiewska-Pająk I., Bogusz-Koziarska H., Sochacka E., Boncela J., Kowalska M.A. Regulation of miRNAs by Snail during epithelial-to-mesenchymal transition in HT29 colon cancer cells. Sci. Rep. 2019;9(1):2165. doi: 10.1038/s41598-019-39200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fregnan G.B., Frigerio L., Porta R., Prada M., Ruggieri F. Therapeutic properties of dihydroxy-dibutylether on sub-acute liver damage induced by several hepatotoxic agents in rats. Int. J. Tissue React. 1982;4(4):309–318. [PubMed] [Google Scholar]
- 99.Pegram M., Slamon D. Biological rationale for HER2/neu (c-erbB2) as a target for monoclonal antibody therapy. Semin. Oncol. 2000;27(5) Suppl. 9:13–19. [PubMed] [Google Scholar]
- 100.Wartenberg P., Lux F., Busch K., Fecher-Trost C., Flockerzi V., Krasteva-Christ G., Boehm U., Weissgerber P. A TRPV6 expression atlas for the mouse. Cell Calcium. 2021;100:102481. doi: 10.1016/j.ceca.2021.102481. [DOI] [PubMed] [Google Scholar]
- 101.Logantha S.J.R.J., Yamanushi T.T., Absi M., Temple I.P., Kabuto H., Hirakawa E., Quigley G., Zhang X., Gurney A.M., Hart G., Zhang H., Dobrzynski H., Boyett M.R., Yanni J. Remodelling and dysfunction of the sinus node in pulmonary arterial hypertension. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2023;378(1879):20220178. doi: 10.1098/rstb.2022.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gong S., Sun N., Meyer L.S., Tetti M., Koupourtidou C., Krebs S., Masserdotti G., Blum H., Rainey W.E., Reincke M., Walch A., Williams T.A. Primary aldosteronism: Spatial multiomics mapping of genotype-dependent heterogeneity and tumor expansion of aldosterone-producing adenomas. Hypertension. 2023;80(7):1555–1567. doi: 10.1161/HYPERTENSIONAHA.123.20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vir P., Kaur J., Mahmood A. Effect of chronic iron ingestion on the development of brush border enzymes in rat intestine. Toxicol. Mech. Methods. 2007;17(7):393–399. doi: 10.1080/15376510601102793. [DOI] [PubMed] [Google Scholar]
- 104.Gerges S.H., El-Kadi A.O.S. Sexual dimorphism in the expression of cytochrome P450 enzymes in rat heart, liver, kidney, lung, brain, and small intestine. Drug Metab. Dispos. 2023;51(1):81–94. doi: 10.1124/dmd.122.000915. [DOI] [PubMed] [Google Scholar]
- 105.Vasco C., Rizzo A., Cordiglieri C., Corsini E., Maderna E., Ciusani E., Salmaggi A. The role of adhesion molecules and extracellular vesicles in an in vitro model of the blood–brain barrier for metastatic disease. Cancers. 2023;15(11):3045. doi: 10.3390/cancers15113045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pan P., Leppilampi M., Pastorekova S., Pastorek J., Waheed A., Sly W.S., Parkkila S. Carbonic anhydrase gene expression in CA II‐deficient (Car2 −/−) and CA IX‐deficient (Car9 −/−) mice. J. Physiol. 2006;571(2):319–327. doi: 10.1113/jphysiol.2005.102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu D., Yun Y., Yang D., Hu X., Dong X., Zhang N., Zhang L., Yin H., Duan W. What is the biological function of uric acid? An antioxidant for neural protection or a biomarker for cell death. Dis. Markers. 2019;2019:1–9. doi: 10.1155/2019/4081962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhan X., Li F., Chu Q., Pang H. Secretogranin III may be an indicator of paraquat-induced astrocyte activation and affects the recruitment of BDNF during this process. Int. J. Mol. Med. 2018;42(6):3622–3630. doi: 10.3892/ijmm.2018.3909. [DOI] [PubMed] [Google Scholar]
- 109.Zhong Q., Yin J., Wang K., Chen X., Wang H., Hu X., Wang W., Wang L., Bei W., Guo J. FTZ promotes islet β-cell regeneration in T1DM mice via the regulation of nuclear proliferation factors. J. Ethnopharmacol. 2023;315:116564. doi: 10.1016/j.jep.2023.116564. [DOI] [PubMed] [Google Scholar]
- 110.Ding Y., Zhang Y., Wang Z., Zeng F., Zhen Q., Zhao H., Li J., Ma T., Huang C. Echinacoside from Cistanche tubulosa ameliorates alcohol‐induced liver injury and oxidative stress by targeting Nrf2. FASEB J. 2023;37(3):e22792. doi: 10.1096/fj.202201430R. [DOI] [PubMed] [Google Scholar]
- 111.Downs B.M., Ding W., Cope L.M., Umbricht C.B., Li W., He H., Ke X., Holdhoff M., Bettegowda C., Tao W., Sukumar S. Methylated markers accurately distinguish primary central nervous system lymphomas (PCNSL) from other CNS tumors. Clin. Epigenetics. 2021;13(1):104. doi: 10.1186/s13148-021-01091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abou-Elhamd A., Cooper O., Münsterberg A. Klhl31 is associated with skeletal myogenesis and its expression is regulated by myogenic signals and Myf-5. Mech. Dev. 2009;126(10):852–862. doi: 10.1016/j.mod.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 113.Kohane I.S. Ten things we have to do to achieve precision medicine. Science. 2015;349(6243):37–38. doi: 10.1126/science.aab1328. [DOI] [PubMed] [Google Scholar]
- 114.Shen W., Le S., Li Y., Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016;11(10):e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jin Y., Weberpals J.G., Wang S.V., Desai R.J., Merola D., Lin K.J. The impact of longitudinal DATA‐COMPLETENESS of electronic health record data on the prediction performance of clinical risk scores. Clin. Pharmacol. Ther. 2023;113(6):1359–1367. doi: 10.1002/cpt.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu J., Deng Y., Fan Z., Xu S., Wei L., Huang X., Xing X., Yang J. Construction and analysis of the abnormal lncRNA–miRNA–mRNA network in hypoxic pulmonary hypertension. Biosci. Rep. 2021;41(8):BSR20210021. doi: 10.1042/BSR20210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seo Y.E., Baine S.H., Kempton A.N., Rogers O.C., Lewis S., Adegboye K., Haile A., Griffin D.A., Peterson E.L., Pozsgai E.R., Potter R.A., Rodino-Klapac L.R. Systemic γ-sarcoglycan AAV gene transfer results in dose-dependent correction of muscle deficits in the LGMD 2C/R5 mouse model. Mol. Ther. Methods Clin. Dev. 2023;28:284–299. doi: 10.1016/j.omtm.2023.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kühn S., Williams M.E., Dercksen M., Sass J.O., van der Sluis R. The glycine N-acyltransferases, GLYAT and GLYATL1, contribute to the detoxification of isovaleryl-CoA - an in-silico and in vitro validation. Comput. Struct. Biotechnol. J. 2023;21:1236–1248. doi: 10.1016/j.csbj.2023.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo H., Liu R., He J., Yao W., Zheng W. Heat stress modulates a placental immune response associated with alterations in the development of the fetal intestine and its innate immune system in late pregnant mouse. Front. Physiol. 2022;13:841149. doi: 10.3389/fphys.2022.841149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fan J., Xia X., Fan Z. Hsa_circ_0129047 regulates the MIR ‐375/ACVRL1 axis to attenuate the progression of lung adenocarcinoma. J. Clin. Lab. Anal. 2022;36(9):e24591. doi: 10.1002/jcla.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.El-Gharbawi N., Shaheen I., Hamdy M., Elgawhary S., Samir M., Hanna B.M., Ali E.Y., Youssef E.A. Genetic variations of ferroportin-1(FPN1-8CG), TMPRSS6 (rs855791) and Hemojuvelin (I222N and G320V) among a cohort of egyptian β-thalassemia major patients. Indian J. Hematol. Blood Transfus. 2023;39(2):258–265. doi: 10.1007/s12288-022-01580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miyamae Y., Mochizuki S., Shimoda M., Ohara K., Abe H., Yamashita S., Kazuno S., Ohtsuka T., Ochiai H., Kitagawa Y., Okada Y. ADAM 28 is expressed by epithelial cells in human normal tissues and protects from C1q‐induced cell death. FEBS J. 2016;283(9):1574–1594. doi: 10.1111/febs.13693. [DOI] [PubMed] [Google Scholar]
- 124.Sinclair A., Park L., Shah M., Drotar M., Calaminus S., Hopcroft L.E.M., Kinstrie R., Guitart A.V., Dunn K., Abraham S.A., Sansom O., Michie A.M., Machesky L., Kranc K.R., Graham G.J., Pellicano F., Holyoake T.L. CXCR2 and CXCL4 regulate survival and self-renewal of hematopoietic stem/progenitor cells. Blood. 2016;128(3):371–383. doi: 10.1182/blood-2015-08-661785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brenner M., Gulko P.S. The arthritis severity locus Cia5a regulates the expression of inflammatory mediators including Syk pathway genes and proteases in pristane-induced arthritis. BMC Genomics. 2012;13(1):710. doi: 10.1186/1471-2164-13-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Montanes-Agudo P., Pinto Y.M., Creemers E.E. Splicing factors in the heart: Uncovering shared and unique targets. J. Mol. Cell. Cardiol. 2023;179:72–79. doi: 10.1016/j.yjmcc.2023.04.003. [DOI] [PubMed] [Google Scholar]
- 127.Hoang C.Q., Hale M.A., Azevedo-Pouly A.C., Elsässer H.P., Deering T.G., Willet S.G., Pan F.C., Magnuson M.A., Wright C.V.E., Swift G.H., MacDonald R.J. Transcriptional maintenance of pancreatic acinar identity, differentiation, and homeostasis by PTF1A. Mol. Cell. Biol. 2016;36(24):3033–3047. doi: 10.1128/MCB.00358-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang M., Wang X., Jiang B., Zhai Y., Zheng J., Yang L., Tai X., Li Y., Fu S., Xu J., Lei X., Kuang Z., Zhang C., Bai X., Li M., Zan T., Qu S., Li Q., Zhang C. Identification of MRAP protein family as broad‐spectrum GPCR modulators. Clin. Transl. Med. 2022;12(11):e1091. doi: 10.1002/ctm2.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Woodman A.G., Mah R.L., Kinney S., Holody C.D., Wiedemeyer A.R., Noble R.M.N., Clugston R.D., Bourque S.L. Perinatal iron deficiency causes sex-dependent alterations in renal retinoic acid signaling and nephrogenesis. J. Nutr. Biochem. 2023;112:109227. doi: 10.1016/j.jnutbio.2022.109227. [DOI] [PubMed] [Google Scholar]
- 130.Tan E., Kinoshita S., Suzuki Y., Ineno T., Tamaki K., Kera A., Muto K., Yada T., Kitamura S., Asakawa S., Watabe S. Different gene expression profiles between normal and thermally selected strains of rainbow trout, Oncorhynchus mykiss, as revealed by comprehensive transcriptome analysis. Gene. 2016;576(2):637–643. doi: 10.1016/j.gene.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 131.Liu Z., Liu H., Wang Y., Li Z. A 9-gene expression signature to predict stage development in resectable stomach adenocarcinoma. BMC Gastroenterol. 2022;22(1):435. doi: 10.1186/s12876-022-02510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Amaral-Silva L., Santin J.M. Molecular profiling of CO2/pH-sensitive neurons in the locus coeruleus of bullfrogs reveals overlapping noradrenergic and glutamatergic cell identity. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2023;283:111453. doi: 10.1016/j.cbpa.2023.111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tian Y., Jin Z., Zhu P., Liu S., Zhang D., Tang M., Wang Y., Li D., Yan D., Li G., Zhu X. TRIM59: A membrane protein expressed on Bacillus Calmette-Guérin-activated macrophages that induces apoptosis of fibrosarcoma cells by direct contact. Exp. Cell Res. 2019;384(1):111590. doi: 10.1016/j.yexcr.2019.111590. [DOI] [PubMed] [Google Scholar]
- 134.Alhajouj M.S., Alsharif G.S., Mirza A.A. Impact of sequential passaging on protein expression of E. coli using proteomics analysis. Int. J. Microbiol. 2020;2020:1–8. doi: 10.1155/2020/2716202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen L., Chen D.Q., Wang M., Liu D., Chen H., Dou F., Vaziri N.D., Zhao Y.Y. Role of RAS/Wnt/β-catenin axis activation in the pathogenesis of podocyte injury and tubulo-interstitial nephropathy. Chem. Biol. Interact. 2017;273:56–72. doi: 10.1016/j.cbi.2017.05.025. [DOI] [PubMed] [Google Scholar]
- 136.Navarro Garrido A., Kim Y.C., Oe Y., Zhang H., Crespo-Masip M., Goodluck H.A., Kanoo S., Sanders P.W., Bröer S., Vallon V. Aristolochic acid-induced nephropathy is attenuated in mice lacking the neutral amino acid transporter B 0 AT1 (Slc6a19). Am. J. Physiol. Renal Physiol. 2022;323(4):F455–F467. doi: 10.1152/ajprenal.00181.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vetrivel P., Nachimuthu S., Abuyaseer A., Bhosale P.B., Ha S.E., Kim H.H., Park M.Y., Kim G.S. Investigation on the cellular mechanism of Prunetin evidenced through next generation sequencing and bioinformatic approaches against gastric cancer. Sci. Rep. 2022;12(1):11852. doi: 10.1038/s41598-022-15826-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Muri L., Schubart A., Thorburn C., Zamurovic N., Holbro T., Kammüller M., Pluschke G., Ispasanie E. Inhibition of the different complement pathways has varying impacts on the serum bactericidal activity and opsonophagocytosis against Haemophilus influenzae type b. Front. Immunol. 2022;13:1020580. doi: 10.3389/fimmu.2022.1020580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen T.Y., Huang B.M., Tang T.K., Chao Y.Y., Xiao X.Y., Lee P.R., Yang L.Y., Wang C.Y. Genotoxic stress-activated DNA-PK-p53 cascade and autophagy cooperatively induce ciliogenesis to maintain the DNA damage response. Cell Death Differ. 2021;28(6):1865–1879. doi: 10.1038/s41418-020-00713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cuevas M., Terhune E., Wethey C., James M., Netsanet R., Grofova D., Monley A., Hadley Miller N. Cytoskeletal keratins are overexpressed in a zebrafish model of idiopathic scoliosis. Genes. 2023;14(5):1058. doi: 10.3390/genes14051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Busse T.M., Roth J.J., Wilmoth D., Wainwright L., Tooke L., Biegel J.A. Copy number alterations determined by single nucleotide polymorphism array testing in the clinical laboratory are indicative of gene fusions in pediatric cancer patients. Genes Chromosomes Cancer. 2017;56(10):730–749. doi: 10.1002/gcc.22477. [DOI] [PubMed] [Google Scholar]
- 142.Sumida S., Ichimura-Shimizu M., Miyakami Y., Kakimoto T., Kobayashi T., Saijo Y., Matsumoto M., Ogawa H., Oya T., Bando Y., Uehara H., Taira S., Shimada M., Tsuneyama K. Histological and immunohistochemical analysis of epithelial cells in epidermoid cysts in intrapancreatic accessory spleen. J Med Invest. 2023;70(1.2):251–259. doi: 10.2152/jmi.70.251. [DOI] [PubMed] [Google Scholar]
- 143.Wu C.L.S., Cioanca A.V., Gelmi M.C., Wen L., Di Girolamo N., Zhu L., Natoli R., Conway R.M., Petsoglou C., Jager M.J., McCluskey P.J., Madigan M.C. The multifunctional human ocular melanocortin system. Prog. Retin. Eye Res. 2023;95:101187. doi: 10.1016/j.preteyeres.2023.101187. [DOI] [PubMed] [Google Scholar]
- 144.Lawton M., Baig F., Toulson G., Morovat A., Evetts S.G., Ben-Shlomo Y., Hu M.T. Blood biomarkers with Parkinson’s disease clusters and prognosis: The oxford discovery cohort. Mov. Disord. 2020;35(2):279–287. doi: 10.1002/mds.27888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Neirijnck Y., Sararols P., Kühne F., Mayère C., Weerasinghe Arachchige L.C., Regard V., Nef S., Schedl A. Single-cell transcriptomic profiling redefines the origin and specification of early adrenogonadal progenitors. Cell Rep. 2023;42(3):112191. doi: 10.1016/j.celrep.2023.112191. [DOI] [PubMed] [Google Scholar]
- 146.Dhara M., Al Hoque A., Sen R., Dutta D., Mukherjee B., Paul B., Laha S. Phosphorothioated amino-AS1411 aptamer functionalized stealth nanoliposome accelerates bio-therapeutic threshold of apigenin in neoplastic rat liver: A mechanistic approach. J. Nanobiotechnology. 2023;21(1):28. doi: 10.1186/s12951-022-01764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu J., Kong X., Zhang M., Yang X., Xu X. RNA binding protein 24 deletion disrupts global alternative splicing and causes dilated cardiomyopathy. Protein Cell. 2019;10(6):405–416. doi: 10.1007/s13238-018-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Giblin S.P., Pease J.E. What defines a chemokine? – The curious case of CXCL17. Cytokine. 2023;168:156224. doi: 10.1016/j.cyto.2023.156224. [DOI] [PubMed] [Google Scholar]
- 149.Jin D., Li R., Mao D., Luo N., Wang Y., Chen S., Zhang S. Mitochondria-localized glutamic acid-rich protein (MGARP) gene transcription is regulated by Sp1. PLoS One. 2012;7(11):e50053. doi: 10.1371/journal.pone.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Noel J.G., Ramser S.W., Pitstick L., Bonamer J.P., Mackenzie B., Seu K.G., Kalfa T.A., Cancelas J.A., Gardner J.C. M-CSF supports medullary erythropoiesis and erythroid iron demand following burn injury through its activity on homeostatic iron recycling. Sci. Rep. 2022;12(1):1235. doi: 10.1038/s41598-022-05360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee S., Yang H.K., Lee H.J., Park D.J., Kong S.H., Park S.K. Systematic review of gastric cancer-associated genetic variants, gene-based meta-analysis, and gene-level functional analysis to identify candidate genes for drug development. Front. Genet. 2022;13:928783. doi: 10.3389/fgene.2022.928783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Goshima M., Sekiguchi R., Matsushita M., Nonaka M. The complement system of elasmobranches revealed by liver transcriptome analysis of a hammerhead shark, Sphyrna zygaena. Dev. Comp. Immunol. 2016;61:13–24. doi: 10.1016/j.dci.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 153.Nedvedova I., Kolar D., Neckar J., Kalous M., Pravenec M., Šilhavý J., Korenkova V., Kolar F., Zurmanova J.M. Cardioprotective regimen of adaptation to chronic hypoxia diversely alters myocardial gene expression in SHR and SHR-mtBN conplastic rat strains. Front. Endocrinol. 2019;9:809. doi: 10.3389/fendo.2018.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gonda X., Eszlari N., Torok D., Gal Z., Bokor J., Millinghoffer A., Baksa D., Petschner P., Antal P., Breen G., Juhasz G., Bagdy G. Genetic underpinnings of affective temperaments: A pilot GWAS investigation identifies a new genome-wide significant SNP for anxious temperament in ADGRB3 gene. Transl. Psychiatry. 2021;11(1):337. doi: 10.1038/s41398-021-01436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luo Z., Zhan Z., Qin X., Pan W., Liang M., Li C., Weng S., He J., Guo C. Interaction of teleost fish TRPV4 with DEAD box RNA helicase 1 regulates iridovirus replication. J. Virol. 2023;97(6):e00495–e23. doi: 10.1128/jvi.00495-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Y., Cai J., Lu W., Xu S., Qu M., Zhao S., Ding X. Comprehensive network-based analyses reveal novel renal function-related targets in acute kidney injury. Front. Genet. 2022;13:907145. doi: 10.3389/fgene.2022.907145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kähler A.K., Djurovic S., Rimol L.M., Brown A.A., Athanasiu L., Jönsson E.G., Hansen T., Gústafsson Ó., Hall H., Giegling I., Muglia P., Cichon S., Rietschel M., Pietiläinen O.P.H., Peltonen L., Bramon E., Collier D., Clair D.S., Sigurdsson E., Petursson H., Rujescu D., Melle I., Werge T., Steen V.M., Dale A.M., Matthews R.T., Agartz I., Andreassen O.A. Candidate gene analysis of the human natural killer-1 carbohydrate pathway and perineuronal nets in schizophrenia: B3GAT2 is associated with disease risk and cortical surface area. Biol. Psychiatry. 2011;69(1):90–96. doi: 10.1016/j.biopsych.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 158.Yuan Z., Li J., Li J., Gao X., Xu S. SNPs identification and its correlation analysis with milk somatic cell score in bovine MBL1 gene. Mol. Biol. Rep. 2013;40(1):7–12. doi: 10.1007/s11033-012-1934-z. [DOI] [PubMed] [Google Scholar]
- 159.Patyal P., Fil D., Wight P.A. Plp1 in the enteric nervous system is preferentially expressed during early postnatal development in mouse as DM20, whose expression appears reliant on an intronic enhancer. Front. Cell. Neurosci. 2023;17:1175614. doi: 10.3389/fncel.2023.1175614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sachetto A.T.A., Jensen J.R., Santoro M.L. Liver gene regulation of hemostasis-related factors is altered by experimental snake envenomation in mice. PLoS Negl. Trop. Dis. 2020;14(6):e0008379. doi: 10.1371/journal.pntd.0008379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Birchenough G.M.H., Johansson M.E.V., Stabler R.A., Dalgakiran F., Hansson G.C., Wren B.W., Luzio J.P., Taylor P.W. Altered innate defenses in the neonatal gastrointestinal tract in response to colonization by neuropathogenic Escherichia coli. Infect. Immun. 2013;81(9):3264–3275. doi: 10.1128/IAI.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen M., Praetorius J., Zheng W., Xiao F., Riederer B., Singh A.K., Stieger N., Wang J., Shull G.E., Aalkjaer C., Seidler U. The electroneutral Na +:HCO 3− cotransporter NBCn1 is a major pH i regulator in murine duodenum. J. Physiol. 2012;590(14):3317–3333. doi: 10.1113/jphysiol.2011.226506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.de O.C.P., Guarnier F.A., Figueiredo L.B. Identification of potential target genes associated with the reversion of androgen-dependent skeletal muscle atrophy. Arch. Biochem. Biophys. 2019;663:173–182. doi: 10.1016/j.abb.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 164.Sgro A., Cursons J., Waryah C., Woodward E.A., Foroutan M., Lyu R., Yeoh G.C.T., Leedman P.J., Blancafort P. Epigenetic reactivation of tumor suppressor genes with CRISPRa technologies as precision therapy for hepatocellular carcinoma. Clin. Epigenetics. 2023;15(1):73. doi: 10.1186/s13148-023-01482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Deng Y., Han Y., Gao S., Dong W., Yu Y. The physiological functions and polymorphisms of type II deiodinase. Endocrinol. Metab. 2023;38(2):190–202. doi: 10.3803/EnM.2022.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Polak J.M., Bloom S.R., Kuzio M., Brown J.C., Pearse A.G.E. Cellular localization of gastric inhibitory polypeptide in the duodenum and jejunum. Gut. 1973;14(4):284–288. doi: 10.1136/gut.14.4.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Trachoo O., Assanatham M., Jinawath N., Nongnuch A. Chromosome 20p inverted duplication deletion identified in a Thai female adult with mental retardation, obesity, chronic kidney disease and characteristic facial features. Eur. J. Med. Genet. 2013;56(6):319–324. doi: 10.1016/j.ejmg.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 168.Lei Z., Rong H., Yang Y., Yu S., Zhang T., Chen L., Nie Y., Song Q., Hu Q., Guo J. Loperamide induces excessive accumulation of bile acids in the liver of mice with different diets. Toxicology. 2022;477:153278. doi: 10.1016/j.tox.2022.153278. [DOI] [PubMed] [Google Scholar]
- 169.Felts S.K., Treanor L.L., Goodman J.S., Koenig M.G. Serum factors and the reticuloendothelial uptake of Staphylococcus aureus. II. Role of a zymosan-adsorbable serum opsonin. Infect. Immun. 1971;4(6):709–714. doi: 10.1128/iai.4.6.709-714.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hirade Y., Kubota M., Kitae K., Yamamoto H., Omori H., Shinoki S., Ohmura T., Tsujikawa K. A novel application of hectorite nanoclay for preparation of colorectal cancer spheroids with malignant potential. Lab Chip. 2023;23(4):609–623. doi: 10.1039/D2LC00750A. [DOI] [PubMed] [Google Scholar]
- 171.Yu Y., Wu M., Zhang N., Yin H., Shu B., Duan W. A pilot study on searching for peri-nuclear NeuN-positive cells. Peer J. 2020;8:e8254. doi: 10.7717/peerj.8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article. The raw data were uploaded on July 19, 2023 (Link: https://pan.baidu.com/s/1uOpvEIU_dRYgGmEIWc0SjA?pwd=DWG1 Password: DWG1)
Data Availability Statement
The raw data were uploaded as supplemental materials on the journal’s web.