Highlights
-
•
Three new nitrite-oxidizing bacteria (NOB) were recovered from full-scale WWTP.
-
•
Two of the NOBs were active showing high metabolic versatility in sludge and biofilms co-metabolizing with others.
-
•
A reductive glycine pathway (RGP) was transcribed by NOB02 likely for CO2 fixation.
-
•
Functional determination of the nitraspira and Ca. Nitrospira for potential gene transfer.
Keywords: Ammonia oxidation, Nitrite oxidation, Nitrogen removal, Wastewater treatment
Abstract
Nitrite-oxidizing bacteria (NOB) are undesirable in the anaerobic ammonium oxidation (anammox)-driven nitrogen removal technologies in the modern wastewater treatment plants (WWTPs). Diverse strategies have been developed to suppress NOB based on their physiological properties that we have understood. But our knowledge of the diversity and mechanisms employed by NOB for survival in the modern WWTPs remains limited. Here, Three NOB species (NOB01–03) were recovered from the metagenomic datasets of a full-scale WWTP treating duck breeding wastewater. Among them, NOB01 and NOB02 were classified as newly identified lineage VII, tentatively named Candidatus (Ca.) Nitrospira NOB01 and Ca. Nitrospira NOB02. Analyses of genomes and in situ transcriptomes revealed that these two novel NOB were active and showed a high metabolic versatility. The transcriptional activity of Ca. Nitrospira could be detected in all tanks with quite different dissolved oxygen (DO) (0.01–5.01 mg/L), illustrating Ca. Nitrospira can survive in fluctuating DO conditions. The much lower Ca. Nitrospira abundance on the anammox bacteria-enriched sponge carrier likely originated from the intensification substrate (NO2−) competition from anammox and denitrifying bacteria. In particular, a highlight is that Ca. Nitrospira encoded and treanscribed cyanate hydratase (CynS), amine oxidase, urease (UreC), and copper-containing nitrite reductase (NirK) related to ammonium and NO production, driving NOB to interact with the co-existed AOB and anammox bacteria. Ca. Nitrospira strains NOB01 and NOB02 showed quite different niche preference in the same aerobic tank, which dominanted the NOB communities in activated sludge and biofilm, respectively. In addition to the common rTCA cycle for CO2 fixation, a reductive glycine pathway (RGP) was encoded and transcribed by NOB02 likely for CO2 fixation purpose. Additionally, a 3b group hydrogenase and respiratory nitrate reductase were uniquely encoded and transcribed by NOB02, which likely confer a survival advantage to this strain in the fluctuant activated sludge niche. The discovery of this new genus significantly broadens our understanding of the ecophysiology of NOB. Furthermore, the impressive metabolic versatility of the novel NOB revealed in this study advances our understanding of the survival strategy of NOB and provides valuable insight for suppressing NOB in the anammox-based WWTP.
Graphical abstract
Introduction
Ammonia (NH3) oxidation, also named nitrification, is an essential process in various ecosystems, including marine and terrestrial environments for ammonia balance (Kuypers et al., 2018). Nitrification is driven by autotrophic microorganisms that are growing with NH3 or nitrite (NO2−) as energy-generating substrates. The nitrification process has long been believed to be carried out by two different communities, the conversion of ammonia to NO2− by autotrophic ammonia-oxidizing bacteria (AOB) and archaea (AOA) (Kuypers et al. 2018; Santoro et al. 2015), and the oxidation of NO2−to NO3− by chemolithoautotrophic nitrite-oxidizing bacteria (NOB). NOB were considered phylogenetically heterogeneous nitrifiers and widespread distribution in nature and man-made ecosystems (Bayer et al. 2021; Lücker et al. 2010, Leung et al. 2022). To date, characterized NOB are mainly affiliated with twelve genera, including Nitrobacter, Nitrococcus, Nitrospina, Nitrospira, Nitrospirota, Nitrotoga, Nitrolancea, Candidatus (Ca.) Nitromaritima, Ca. Nitrocaldera, Ca. Nitrotheca, Ca. Nitrohelix and Ca. Nitronauta (Daims et al. 2016; Daims and Wagner 2018; Su et al. 2023), suggesting an extensive phylogenetic pedigree of NOB. Comparatively, NOB Nitrospira in the family Nitrospiraceae is the most phylogenetically diverse and abundant NOB in various ecosystems, such as soil, oceans, freshwater, hot spring, and wastewater (Bayer et al. 2021; Daims et al. 2016; Liu et al. 2021; Ushiki et al. 2017b). Based on the phylogenetic analysis of the 16S rRNA gene sequence, Nitrospira was assigned to seven different phylogenetic lineages, in which lineages I, II and VII have been ubiquitously identified in wastewater treatment plants (WWTPs) (Gruber-Dorninger et al. 2015; Keuter et al. 2023; Ushiki et al. 2017a). Also, another NOB that is widely known is Nitrobacter, which belongs to the Alphaproteobacteria. Unlike Nitrospira, which prefers to survive in nitrite-limited and oxygen-minimum zones, resulting from the K-strategists with high affinity for nitrite and oxygen, NOB Nitrobacter is an r-strategists, that could tolerant high substrate concentrate (Nowka et al. 2015). Additionally, a recent study also indicated that Nitrospira moscoviensis could survive in nitrite-limited conditions because the presence of 2a [NiFe]-hydrogenase enabled it for aerobic growth using H2 as the sole energy source (Leung et al. 2022), and benefit from the mixotrophic lifestyle, including pathways for the transport, oxidation, and assimilation of simple organic compounds and the reverse tricarboxylic acid cycle, Nitrospira defluvii can survive in substrate-limited conditions (Lücker et al. 2010). Considering that the phylogenetic tree of functional classification has been constantly developed due to the expansion of available genomic data, such as bacteria involved in H2 oxidation (Koch et al. 2015, Leung et al. 2022), various carbon fixation pathways (Lücker et al. 2010; Starkenburg et al. 2006) and ammonia oxidation (Daims et al. 2015; van Kessel et al. 2015). An open question is still unclear whether the currently characterized NOB lineages can fully represent the diversity and metabolism of NOB. To better explain this question, it is imperative to recover more distinct branching lineages of NOB that will advance the elucidation of ecophysiological extension and evolutionary history.
A stable supply of nitrite is a prerequisite for the anammox-based nitrogen removal technologies, one of the major issue is to control the nitrite oxidation activity of NOB in WWTPs. AOB could survive in an extremely low DO condition (0.2–0.4 mg/L), which was much lower than that for NOB (1.5–12 mg/L) (Picioreanu et al. 1997). This provides a new inspiration that a low DO environment favors the growth of AOB and inhibits the activity of NOB. Whereas, following studies found that the oxygen affinity of various NOB is quite different. For example, Nitrospira-like nitrite oxidizer could survive in a much low DO concentration of 0.5 mg/L, a high DO concentration facilitated the growth of Nitrobacter (Daims et al. 2001; Regmi et al. 2014). It was suggested that, in order to achieve NOB suppression, the DO concentration should be controlled at a moderate level of 1.0 mg/L or a lower level of 0.5 mg/L (Cao et al. 2017). In addition, a previous study also indicated that NOB have a longer lag time than AOB during the transition from anoxic to oxic conditions due to the deactivation of hydroxylamine (Xu et al. 2012). As such, an intermittent aeration-based operation strategy had been proposed for suppressing NOB activity by creating a transient anoxic condition in SBR or continuous step-feed reactors (Gu et al. 2018; Miao et al. 2018). Most recently, a novel process of partial nitrification, anammox, and methane-dependent nitrite/nitrate reduction (PNAM) has been developed and applied in a single membrane biofilm reactor (MBfR) (Liu et al. 2021). In this system, NOB competes for nitrite with anammox bacteria, nitrite/nitrate-dependent anaerobic methane oxidation (n-DAMO) bacteria, and archaea, and thereby NOB can be maintained at a lower abundance by controlling the input of methane (Liu et al. 2021). This process, however, also encounters an unavoidable problem with the harsh oxygen availability, which directly affects the ammonia oxidation and overall nitrogen removal efficiencies. Above strategies have obtained a certain success in NOB suppression mostly in laboratorial bioreactors, but there are still many challenges for practical application in full-scale WWTPs. One of the crucial reasons is the extensive phylogenetic pedigree of NOB in WWTP with quite different physiologic properties, but our understanding on the ecophysiology of NOB remains limited. Therefore, more efforts are still needed to reveal the diversity and metabolic versatility of NOB in wastewater treatment systems, which will provide variluable insights on optimal WWTP configurations by suppressing NOB for efficient anammox-based nitrogen removal.
Here, metagenome and metatranscriptome analyses were performed on four distinct niches of a full-scale WWTP treating duck breeding wastewater, including two anaerobic tanks, one anoxic tank and one oxic tank. Two high-quality metagenome-assembled genomes (MAGs) representing a new NOB lineage were retrieved from the WWTP. These two novel NOB dominated the NOB communities in different reactors of the WWTP, but with the lowest abundances and transcriptional activities in the anoxic tank, reflecting the growth of NOB was significantly inhibited. By introducing the genomic characterization and transcriptional results, we revealed a high metabolic versatility and survival advantage of the novel NOB in wastewater treatment systems. Environmental conditions that inhibited the growth of NOB in the anoxic tank were further investigated.
Results and discussion
Recovery of novel NOB species from the full-scale WWTP
After shotgun sequencing and binning analysis, we retrieved a total of 27 NOB metagenome-assembled genomes (MAGs) from different reactors of the WWTP (Table S1). Based on the average nucleotide identity (ANI) and 43 concatenated markers phylogenetic analysis, the 27 MAGs were divided into four groups (Fig. S1, Table S1). After homologous gene prediction and comparison, 99.8–100 % identity was shared by MAGs within each group. So that four representative high-quality (Completeness > 91 %, Contamination < 7 %) draft genome assemblies (NOB01, NOB02, NOB03, and NOB04) from these four groups were selected for downstream analysis.
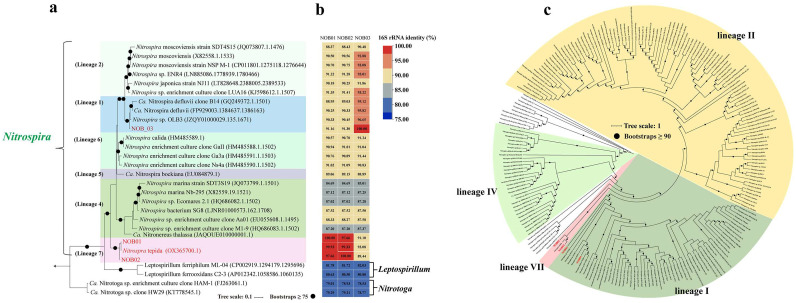
The preliminary classification carried out by the GTDB-tk could assigned NOB01, NOB02, NOB03 and NOB04 were classified as genus Nitrospira (Fig. S4). Consistently, phylogenetic analysis based on the 43 concatenated markers indicates that NOB03 and NOB04 had the closest lineage with “Nitrospira defluvii NOB2” (Fig. 1b). The highest ANI between NOB04 and the known NOB was 99.8 % (Table S2), larger than the recommended species cutoff (ANI > 95 %) (Konstantinidis et al. 2017), we thus named it “Ca. Nitrospira defluvii NOB04”. The highest ANI between NOB03 and the known NOB was 74.98 % (Fig. 1b, Table S2), lower than 95 %, but it shared a 98.81 % 16S rRNA gene identity with “Ca. Nitrospira sp. OLB3”, larger than the proposed species cutoff based on 16S rRNA gene (> 98.6%). Thereby, NOB03 was considered as a same species with “Ca. Nitrospira sp. OLB3” in the Nitrospira genus and named it “Ca. Nitrospira sp. NOB03”. Interestingly, the maximum ANI of NOB01 with the known Nitrospira genomes were 85.33 % than the recommended species cutoff (ANI > 95 %), while NOB02 share 99.90 % ANI with Nitrospira tebida (Table S2) (Keuter et al. 2023). Similarly, nearly full-length 16S rRNA gene sequences were successfully extracted from MAGs NOB01, NOB02 and a maximum likelihood tree based on 16S rRNA gene sequences was constructed (Fig. 1a). Comparable with the result of phylogenetic analysis based on the 43 concatenated markers, NOB01 and NOB02 formed a separate cluster with Nitrospira tebida (Fig. 1c). The nearly full-length 16S rRNA gene sequences of NOB01 and NOB02 were most similar (99.93 % and 99.33 %, respectively) to the 16S rRNA gene from Nitrospira tebida, which is larger than the genus cutoff based on 16S rRNA gene (94.5 %) (Yarza et al. 2014). Therefore, we proposed NOB01 and NOB02 belonge to the novel lineage VII, which were named as “Ca. Nitrospira NOB01” and “Ca. Nitrospira sp. NOB02”.
Fig. 1.
Phylogenetic analyses of 16S rRNA gene (a) and identity (b) show the affiliations of these three new NOB bacteria (red font) and known NOB strains (black font), Maximum likelihood tree of 43 concatenated markers gene (c) show the affiliations of retained for NOB bacteria (red font) and known NOB strains (black font). 16S rRNA gene tree was rooted by 16S rRNA genes of Nitrotoga, 43 markers phylogenetic tree was rooted by midpoint.
Active nitrogen metabolism of two lineage VII members in WWTP
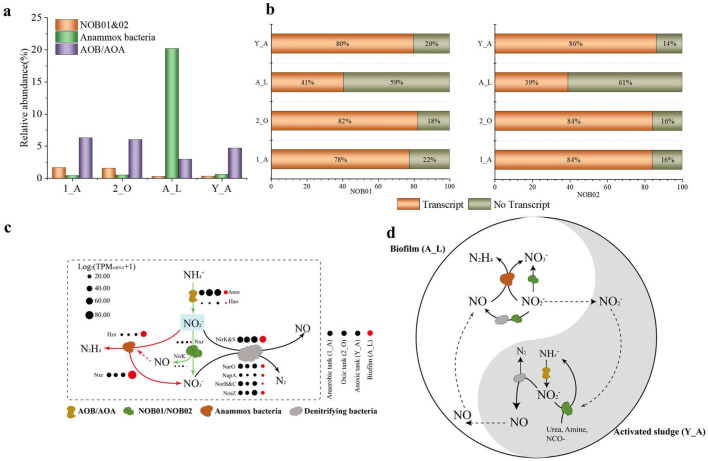
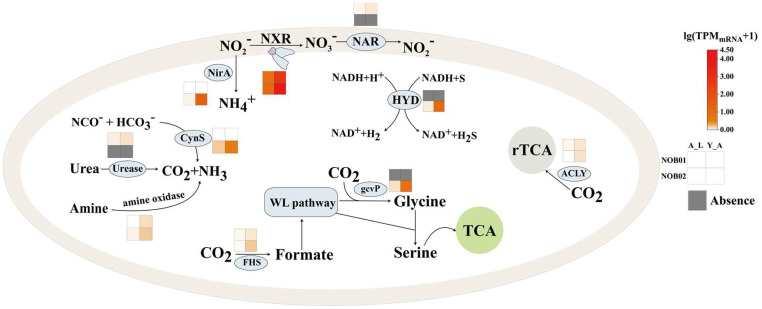
Around 80 % of the genes in both NOB01 and NOB02 in activated sludge niches (1_A, 2_A, and Y_A) were transcribed, but the proportion was dramaticlly decreased to 40 % in the biofilm niche in the anoxic tank (A_L) (Fig. 2b). NXR complex is an important part for nitrite-oxidizing bacteria associated with oxidizing nitrite to nitrate, which is composed by three subunits including NxrA, the main catalytic units, NxrB and NxrC responsible for converting electrons released by nitrite oxidation to cytochrome c (Lücker et al. 2010). As expected, genes conding for NXR complex were encoded and their transcripts were detected in the two novels Ca. Nitrospira MAGs (Table S3). In detail, one candidate nxrAB and two nxrC were identified in MAG of NOB01, while two copies of nxrA, one nxrB, and one nxrC were recovered from MAG of NOB02. NxrA of NOB01 and NOB02 shared a 98.98 % amino acid identity, while the two copies of NxrA in NOB02 had a 97.91 % amino acid identiy.
Fig. 2.
NOB Nitrospirae actively participated in nitrogen cycle in the full-scale WWTP with close cooperation with other organisms. (a) Relative abundances of nitrogen cycle related microorganisms in different niches. Relative abundance = TPM/total TPM *100 %. (b) Percentages of the in-situ transcribed genes of the two novel NOB. (c) The overall nitrogen metabolism networks in the four niches with the participation of the two novel NOB. (d) The constructed co-metabolism model between activated sludge (Y_A) and biofilm (A_L) niches in the anoxic tank (1_A) carried out by nitrogen cycle related microorganisms. Amo, ammonia oxidoreductase; Hao, Hydroxylamine oxidoreductase; Nxr, nitrite oxidoreductase; NirK, nitrite reductase (Cu-forming); NapA, periplasmic dissimilatory nitrate reductases; Nor, nitric oxide reductase; NosZ, nitrous-oxide reductase. Anaerobic tank (1_A), Anoxic tank (1_O), Anaerobic tank (2_A) and Oxic tank (2_O). Y_A and Y_B represent two separated parts in Anoxic tank (1_O).
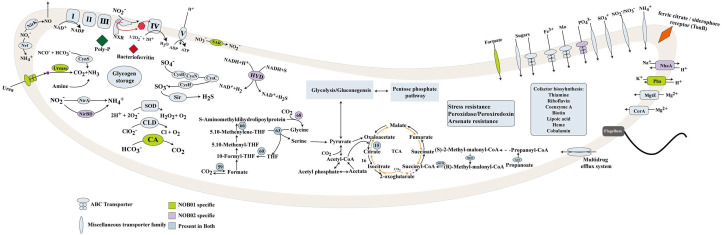
To better elaborate the function of NxrA, we select the nearly full-length nxrA as a representative sequence for further genetic function analysis. One [Fe-S] cluster and one molybdenum binding motif were identified in NxrA, likely suggesting NxrA of Ca. Nitrospira belongs to the Mo-co-binding enzymes in the dimethyl sulfoxide (DMSO) reductase family (Fig. S3a). Consistent with the NxrA in Nitrospira defluvii (Lücker et al. 2010), five potential substrate entry regulation channels or NO2−/NO3− catalytic active amino acid sites associated with substrate-binding during nitrite oxidation were identified, in which one threonine active binding site was replaced by asparagine (Fig. S3a). Additionally, four cystenine-rich binding motifs of [Fe-S] clusters related to intramolecular electron transfer were characterized in the NxrB of Ca. Nitrospira (Fig. S3b), which has been found in the NxrB of Nitrobacter and Nitrococcus, and the NarH of E. coli (Lücker et al. 2010; Ushiki et al. 2018). NxrC might act as membrane anchors and transfer electrons received from the β-subunit to the electron transport chain via one or two hemes (Rothery et al. 2008; Ushiki et al. 2018). Phylogenetic analysis suggests the nxrA of Ca. Nitrospira has high affiliation with the periplasmic nxrA from Nitrospira (Fig. S3a). Further analysis found that NxrA of Ca. Nitrospira contains an N-terminal signal peptide to export protein via the twin-arginine protein translocation (Tat) pathway (Kitzinger et al. 2018; Palmer and Berks 2012), and NxrC encode an N-terminal signal peptide for translocation via Sec pathway (Kitzinger et al. 2018), demonstrating that NxrA of Ca. Nitrospira was anchored in the periplasmic space and catalyzed an energetically advantageous nitrite oxidation reaction, and the produced protons can be directly released into the periplasm and contribute to proton motive force (PMF) (Kitzinger et al. 2018; Lücker et al. 2010). Ca. Nitrospira highly transcribed genes of NXR complex in all four samples for NO2− oxidation, and the released NO3− could be further utilized by the coexisting denitrifying bacteria (Fig. 2c).
These two VII Ca. Nitrospira strains encoded a copper-containing nitrite reductase (NirK) for nitrite reduction to NO and the transcription of nirK was detected (Fig. 2c, Table S3). The presence of NirK occurs in all sequenced Nitrobacter, Nitrococcus, Nitrospira, and Nitrospina genomes, suggesting that these NOB can produce NO (Daims et al. 2016; Koch et al. 2015). The produced NO can not be further reduced by Ca. Nitrospira because of the absence of nitric oxide reductase (NOR). NO is an essential intermediate of ammonia oxidation in AOA, AOB, and anammox bacteria (Caranto and Lancaster 2017; Hu et al. 2019; Kozlowski et al. 2016). Therefore, we proposed that the released NO by NOB01 and NOB02 in the aggregates could modulate the growth and the metabolism of the coexisted anammox bacteria and AOB/AOA and form a mutualistic interaction (Fig. 2c, Table S3). The two subunits of nrfAH were detected in NOB02, while only nrfH was found in the recovered part of NOB01 with slightly transcribed (Fig. 3), suggesting that nitrite could also be catalytically reduced to ammonium by periplasmic cytochrome c nitrite reductase (Simon et al. 2000; Simon et al. 2001; Ushiki et al. 2018). It has been suggested that nitrite reduction in Nitrospira was carried out by a cytoplasmic ferredoxin-nitrite reductase (NirA) for assimilation purpose rather than NrfAH (Daims et al. 2015; Koch et al. 2015; Lücker et al. 2010; Ushiki et al. 2018), and NirA also was identified in NOB01 and NOB02 genome. Thus, the pathway of assimilatory nitrite reduction to ammonium may operate as a result of the transcription of NirA (Fig. 3, Fig. 4).
Fig. 3.
Metabolic cartoon constructed from the annotation of the two nearly completely sequenced NOB01 and NOB02 genomes and the metatranscriptomic data. Numbers and gene abbreviations at partial pathway match the enzyme identifiers in Table S3.
Fig. 4.
Metabolic activitis of the two novel NOB in different niches.
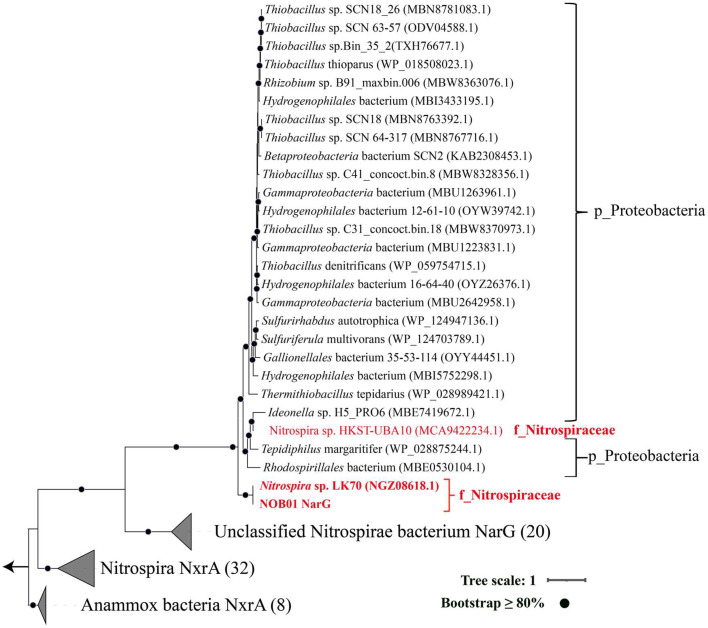
The retainment of a membrane-bound nitrate reductase complex (NAR, NarGHI) was recently reported in two comammox Nitrospira genomes, Ca. Nitrospira sp. LK70 and Ca. Nitrospira sp. HKST-UBA10, and inferred to be associated with nitrate reduction under anoxic conditions (Wang et al. 2021; Yang et al. 2020a). Interestingly, the NAR complex was encoded by NOB01 and exhibited transcriptional activity. This finding was unexpected, because NOB was proposed to use their NXR for catabolic nitrate reduction (Ehrich et al. 1995; Koch et al. 2015). From a bioenergetic perspective, the cytoplasmically oriented NAR could be a more efficient nitrate reductase than NXR. The presence of NAR likely confer a selective advantage to nitrate-reducing NOB01 over other NOB strains in anoxic conditions .
Another feature that makes Ca. Nitrospira attractive is that encoding of urease (UreABC) and cyanase (CynS), the two enzymes that catalyze the remineralization of organic matters to ammonium. Such enzymes also were discovered in Nitrospira and were considered to play an significant role. For example, urea transporters and cytoplasmic ureases were discovered in several Nitrospira genomes (Fig. 5), and the experimental results further directly confirm the ability of hydrolyses urea in N. moscoviensis (Daims et al. 2016; Koch et al. 2015). Also, NOB can better adapt to the scarce free ammonia condition by using urea as an energy source (Daims et al. 2016; Koch et al. 2015). Well-characterized cyanase also was identified in N. moscoviensi and catalyzes the reaction of cyanate with bicarbonateto produce ammonium for providing an alternative source of ammonium (Koch et al. 2015). Indeed, alternatively ammonium sources are not only an important survival strategy for NOB, but also allow it to form a mutual aid relationship with nitrifiers in the community (Daims et al. 2016; Koch et al. 2015). Studies have pointed out that urease-positive or/and cyanase-positive NOB can provide urease-negative or cyanase-lacking ammonium-oxidizing bacterium, such as Nitrosomonas europaea and Nitrosomonas nitrosa Nm90 with ammonia released from urea and cyanate degradation (Koch et al. 2015; Palatinszky et al. 2015). Again, this reciprocal feeding of NOB and AOM interaction also play a key role in co-existence system, because AOM produce nitrite, which is required assubstrate and also detoxified by NOB (Daims et al. 2016). Urease and cyanase were transcribed in retained Ca. Nitrospira (Fig. 4) demonstrated that they have the ability of urea and cyanate degradation, and likely forming a reciprocal feeding relationship with ammonia oxidizers, such as Nitrosomonas europaea, which was the prodomination AOM in our studied WWTP (Table S4), and the cleavage of urea and cyanase by Ca. Nitrospira likely illustrated the initial step that trigger nitrification by Ca. Nitrospira-AOM consortia.
Fig. 5.
The distribution of key marker genes related to different metabolisms in NOB and Comammox Nitrospira genomes.
Mixotrophic and alternative metabolism of Ca. Nitrospira
Pathways that related to organic substrates catabolic degradation and the assimilation of pyruvate, formate, acetate, and propanoate were encoded by the two Ca. Nitrospira genomes (Fig. 3). Since the gluconeogenesis and glycolysis (Embden-Meyerhof-Parnas, EMP) pathway is nearly complete (Table S3, Fig. 3), operation of the sugars catabolism in NOB01 and NOB02 was confirmed. The coding of mixotrophic metabolic pathway can help NOB01 and NOB02 survive in complex full-scale wastewater treatment systems and switch lifestyles with environmental changes (Lücker et al. 2010, Yang et al. 2020). Ca. Nitrospira contains the majority of genes related to oxidative tricarboxylic acid (oTCA) only except for the absence of 2-oxoglutarate dehydrogenase complex (ODH) (Table S3, Fig. 3), which was considered an alternative energy generation pathway in Nitrospira (Lücker et al. 2010). It was suggested that 2-oxoglutarate: ferredoxin oxidoreductase (ODOR) might replace ODH for 2-oxoglutarate dehydrogenase in Nitrospira (Lücker et al. 2010), and genes coding ODOR were identified in MAGs NOB01 and NOB02. However, such alternative strategy is only based on speculation of genomic information, because OGOR can replace the missing ODH in Helicobacter pylori (Lücker et al. 2010). Additionally, enzymes related to pentose phosphate pathway and galactose metabolism also were encoded by MAGs NOB01 and NOB02 for sugar catabolism (Table S3).
The existing genome data indicate that NOB Nitrobacter and Nitrococcususe encode the Calvin–Benson–Bassham (CBB) cycle for CO2 fixation (Starkenburg et al. 2006). Differently, NOB Nitrospira use CO2 as their carbon source through the reductive tricarboxylic acid (rTCA) cycle, and the key enzymes of the rTCA cycle are ATP citrate (pro-S)-lyase, 2-oxoglutarate: ferredoxin oxidoreductase (ODOR) and ferredoxin oxidoreductase (POR) (Lücker et al. 2010). Encoding of these genes suggesting NOB01 and NOB02 also fix CO2 via a typical rTCA cycle (Fig. 3). Interestingly, POR and OGOR are O2 sensitive enzymes, and in Nitrospira, a unique five subunit leads to better oxygen tolerance of these two enzymes (Lücker et al. 2010; Mueller et al. 2023), which helps Nitrospira adapt to the rTCA metabolic pathway in trace oxygen for survival. Recently, a metabolic pathway named the reductive glycine pathway (RGP) was constructed and found to be involved in CO2 fixation in Candidatus Phosphitivorax (Figueroa et al. 2018; Tashiro et al. 2018). Subsequently, an interconnected process Wood–Ljungdahl (WL) pathway and RPG has also been confirmed to fix CO2 in Clostridium drakei by 13C isotope-based metabolite-tracing experiments (Song et al. 2020). Indeed, the bottom line genes that are related to WL and RPG pathway, such as amino methyltransferase (gcvT), glycine cleavage system H protein (gcvH), hydroxymethyltransferase (glyA/ SHMT), glycine dehydrogenase (gcvP), formate dehydrogenase (NADP+) subunit (fdh), formate–tetrahydrofolate ligase (fhs), methylenetetrahydrofolate dehydrogenase (NADP+) (folD) and methylenetetrahydrofolate reductase (metF) were identified in NOB01, while NOB02 lack the key gene glycine dehydrogenase (gcvP) involved in the most critical step of the RPG pathway (Fig. 3, Fig. 5). Thus, NOB01 has the genetic potential to fix CO2 through the RPG and WL pathway (Table S3, Fig. 3). Previous research has found that the encoding of the RGP pathway enables Desulfovibrio deulfuricans to achieve autotrophic growth, and in the presence of sufficient ammonia, the growth rate of autotrophic Desulfovibrio deulfuricans increases with the increase of ammonia (Sánchez-Andrea et al. 2020). Therefore, it was speculated that the additional CO2 fixation pathway likely helps NOB01 survive unfavorable conditions and facilitate switches between lifestyles in fluctuating environments. The presence of RPG pathway regulation genes also were reported in some Nitrospira genomes (Fig. 5) (Lücker et al. 2010; Lawson et al. 2021; Ushiki et al. 2018; Yang et al. 2020a). Potential genes of polyhydroxyalkanoates (PHA) biosynthesis and degradation were identified in NOB01 and NOB02, including acetyl-CoA C-acetyltransferase (phaA), poly(R)-hydroxyalkanoic acid synthase subunit E (phaE) and poly(3-hydroxybutyrate) depolymerase like protein (phaZ) (Fig. 3, Fig. 5). Poly(R)-hydroxyalkanoic acid synthase subunit C (phaC) and acetoacetyl-CoA reductase (phaB) were absent probably due to the incomplete nature of genome of NOB01 and NOB02 (McCool and Cannon 2001; Parveez et al. 2015; Zher Neoh et al. 2022).
H2 and formate are two common products at oxic/anoxic interfaces, and putatively involved in alternative energy metabolisms under low O2 environment (Bayer et al. 2021). Recently researches suggested that some Nitrospira members possess the ability to utilize other than nitrite and CO2 as energy source. The first chemolithoautotrophic lifestyle independent of the nitrogen cycle was N. moscoviensis which encodes a group 2a [NiFe] hydrogenase and accessory proteins, and experimental result further comfirmed N. moscoviensis that can be achieved energy conservation via aerobic oxidizing H2, and can grow with H2 as the sole energy source and electron donor (Koch et al. 2015, Leung et al. 2022). [NiFe] hydrogenase also were identified in the genomes of Nitrospira (Bayer et al. 2021; Yang et al. 2020a) and Nitrospina gracilis (Luecker et al. 2013). Furthermore, N. moscoviensis also can couple formate oxidation to NO3− reduction to obtain energy under various O2 concentration condition, and formate dehydrogenase is the main function enzyme (Koch et al. 2015). A group 3b hydrogenases (HYD) and formate dehydrogenase were uniquely encoded with slightly transcription by Ca. Nitrospira (Fig. 5, Fig. S6), likely enabling Ca. Nitrospira to use H2 and formate as an alternative energy source in the biofilm and activated sludge niche.
Interaction of VII Ca. Nitrospira with other microbes
Suppressing NOB growth is a key element for removing nitrogen in anammox-based process, and many factors such as high temperature, high ammonium/ammonia concentration (Ganigué et al. 2007), high nitrite/ free nitrous acid concentration (Wang et al. 2014), and especially low dissolved oxygen (DO) concentration (Ma et al. 2015), have been identifed effectively inhibit or limit NOB growth. The relative abundance of Ca. Nitrospira and NXR activity, however, had no remarkable inhibition in the two anaerobic tanks (1_A and 2_A) with low DO concentrate (Fig. S2), Indeed, the recycled wastewater from the oxic tank (2_O, DO: 5.01 mg/L) might bring trace amounts of oxygen into the two anaerobic tanks (1_A and 2_A), which likely provided a promising opportunity for growth of Ca. Nitrospira. Interestingly, the abundance of Ca. Nitrospira was dramatically decreased in the anoxic tank (Y_A and A_L), in which anammox bacteria were highly enriched (Fig. 2a). The restricted oxygen diffusion leads to a relatively low DO microenvironment in biofilm (Cho et al. 2020; Ma et al. 2015). The low DO resulted in the low activity of NOB due to the unavailability of sufficient oxygen and securied a strong competitive edge for anammox bacteria. Furthermore, an obvious competitive relationship occurs in Ca. Nitrospira, anammox bacteria and denitrifers also seems to provide clues to the suppression of Ca. Nitrospira (Fig. 3c& d). Nitrite, as the growth substrate for Ca. Nitrospira, was heavily occupied by anammox bacteria and denitrifiers, because high transcriptional activities of nxr and nirK in anammox bacteria leading the mainstream NO2− was rapidly convert to NO3− and NO. Also, part of NO2− could be reduced to NO by denitrifiers due to the transcription of their nirK (Fig. 2).
Indeed, although the growth of Ca. Nitrospira was inhibited, it non-negligibly maintains the community's nitrogen transfer and construct a reciprocal feeding network. As mentioned above, encoding some ammonium generation enzymes, UreC CynS and amine layse, confer Ca. Nitrospira the ability to release ammonium into the community for feeding the AOM (Daims et al. 2016). Also, Ca. Nitrospira and denitrifying bacteria in the sludge (Y_A) also highly transcribed nirK (Fig. 3c), but most of them lack the nitric-oxide reductase (NOR) enzyme for further NO reduction, which likely resulted in a high amount of NO release from sludge. NO is a key intermediate in microbial interaction (Daims et al. 2016; Hu et al. 2019), thus, the released NO in the community might be directly used by anammox bacteria as the electron acceptor for anaerobic ammonium oxidation (Kartal et al. 2010) or could diffuse into the biofilm system to support the enrichment of anammox bacteria. In addition, NO released by Nitrospira-NOB may recruit AOB (such as in N. europaea) to form nitrifying aggregates, which helps to promote the formation of anaerobic cores and the enrichment of anammox bacteria. The detectable expression of NarG by NOB01 indicates that some NOB01 could respiration of organic substrates with nitrate as the terminal electron acceptor, and the released NO2− could be mainly utilized by denitrifiers and anammox bacteria (Fig. 3c).
NAR and NXR in Ca. Nitrospira
Membrane-anchored nitrate reductase (NAR), which belongs to the family of molybdenum (Mo)-containing DMSO reductases (Coelho and Romão 2015; González et al. 2006) and is often absent in Nitrospira-NOB, was annotated in the genome of NOB01. NAR complex has been discovered in many aerobic microorganisms, for example, in the model organism Escherichia coli. Unlike with NXR, the NAR complex is a trimeric enzyme, NarG and NarH are situated in the cytoplasm and NarI anchors to the membrane. Analogous cases are also discovered in some bacteria, such as Marinobacter hydrocarbonoclasticus, Paracoccus pantotrophus (Coelho and Romão 2015; Marangon et al. 2012). In all characterized Nitrospira genomes, only two Nitrospira (Nitrospira sp. LK70, and Nitrospira sp. HKST-UBA10) encode the NAR complex (Fig. 6) (Wang et al. 2021; Yang et al. 2020b), but the function was often elusive. Therefore, we first try to elucidate the potential function of the NAR complex in NOB01 based on a homologous modeling approach. Owing to the conserved structure of the NAP complex, thereby the three-dimensional structure of the NAR complex of NOB01 was modeled homologously based on existing models of the NAR complex (Fig. 6, default search for the best model). The best-matched model is 3EGW (PDB ID) (Gonzalez et al. 2017) and the quality related parameters such as GMQE (0.9) and QMEANDisCo Global (0.83 ± 0.05)were the expected result, suggesting model was constructed with good quality and high confidence (Studer et al. 2019; Waterhouse et al. 2018). The NAP complex of NOB01 also belongs to the DMSO reductases, in which the catalytic subunit NarG is the molybdopterin-dithiolenes (Mo-bisMGD) cofactor and an iron-sulfur center of the [4Fe-4S] type (FeS0) enzyme (Fig. S5b) which shares the same Mo-bisMGD and FeS0 binding sites with the NarG of E. coil (Fig. S5b) (Bertero et al. 2003). Five NO2−/NO3− catalytic active sites were encoded for biocatalytic nitrate reduction (Fig. S5). In E. coli, the NarH is the electron transfer subunit and contains three [4Fe-4S] (SF4) and a [3Fe-4S] (F3S) center, such as PDB ID: 1Q16, or two [4Fe-4S] (SF4) and two [3Fe-4S] (F3S) center like PDB ID: 3EGW, the small NarI subunit contains two b-type hemes cofactors and hooked NarGH at the cytoplasmatic side of the membrane, providing the binding site for the oxidation of electron quinol. As previously proposed operation process of nitrate reduction, menaquinone (MQH2) located on the cytoplasmic membrane is initially oxidized by NarI with the release of two electrons, which are subsequently transferred to NarG via NarH to activate the reduction of nitrate (Coelho and Romão 2015; González et al. 2006), suggesting the entire nitrate reduction process requires the collaboration of the three subunits. Comparably, the potential binding site of SF4, F3S, and heme were found in the NarHI sequence of NOB01 (Fig. S5c & d), but some amino acid residues are different with the NarI of E. coil. Intriguingly, although the binding sites of the two heme (the low-potential heme bL and the high-potential heme bH) of NarI in NOB01 differ from those of E. coil, the binding site of the histidine groups (heme bH coordinated with His56 and His205, and ferrous heme bL coordinated with His66 and His187) of the Fe atoms involved in the transfer of electrons is conserved (Fig. S5e). Thus, we concluded that the NAR complex of NOB01 should be capable of catalyzing the reduction of NO3− to NO2−. But the extent to which this affects the electron transfer efficiency and the biochemical processes mediated by NarHI subunits need further in-vitro verification.
Fig. 6.
Maximum likelihood tree highlights the affiliation of nitrate reductase (NarG) recovered from NOB01 in this study and other NarG and NxrA, the tree was rooted by NxrA from anammox bacteria.
Although both NXR and NAR belong to (Mo)-containing DMSO family enzymes, they seemly undergo different evolutionary histories. Previously studies suggested that the nxrA of Ca. N. defulvii has the closest affiliation to nxrA of Ca. Kuenenia stuttgartiensis (Daims et al. 2015; Lücker et al. 2010), and inferred the potential horizontal gene transfer (HGT) between anammox bacteria and Nitrospira. Herein, we also conducted the phylogenetic analysis of NxrA sequences of Ca. Nitrospira (Nitrospiraceae), Nitrospira, anammox bacteria, Nitrotoga, and Nitrobacter (Fig. S3b). The most adjacent branches of the nxrA cluster of Ca. Nitrospira and Nitrospira were anammox bacteria, which coincides with previous phylogenetic analysis (Daims et al. 2015; Kitzinger et al. 2018) and likely illustrates the nxrA of Ca. Nitrospira also evolved from anammox bacteria. In accordance with the result of nxrA, the closest homolog of Ca. Nitrospira nxrB was in anammox bacteria (Fig. S3d), and the closest homologous nxrB of Nitrospiraceae is Scalindua, which further proof of the speculation that NXR of the Nitrospiraceae was originated from anammox bacteria. Here, we speculated that the NXR of Ca. Nitrospira and Nitrospira might inherite from a common ancestor which acquired the NXR from anammox bacteria through HGT in the early evolution stage of these two lineages.
Intriguingly, comparing genomics revealed an unexpected evolutionary link between Nitrospirae and Proteobacteria phylum in the NAR complex. Phylogenetic analysis revealed that NarG of Ca. Nitrospira sp. NOB01 and Nitrospira sp. LK70 had higher affiliation with bacteria in Proteobacteria compared to other Nitrospirae (Fig. 6), which may indicate the quite different evolutionary sources of NarG in phylum Nitrospirae. NarG seuqences from NOB01 and Proteobacteria shared an over 60% amino acid sequence identity, which is much higher than with Nitrospirae (less than 49%), reflecting the HGT of NarG was occured between Ca. Nitrospira and Proteobacteria. Consistently, the high identities of NarH, NarI, and NarJ between NOB01 and Proteobacteria were also found (Table S4).
Stress defense
Many genes associated with coping with environmental adversities were detected in NOB01 and NOB02. Superoxide dismutase (SOD), thioredoxin-dependent peroxiredoxin (prx/tpx) (ref), and thioredoxin reductase (txrAB) (ref) related to oxygen resistance were found in NOB01 and NOB02 (Table S4). NOB01 has a more reactive oxygen species (ROS) scavenging system, which composed by superoxide dismutase (SOD), thioredoxin-dependent peroxiredoxin (prx/tpx) and thioredoxin reductase (txrAB), than NOB02, confers a survival advantage to NOB01 in the fluctuating wastewater systems (Y_A and Y_B). Resistance-nodulation-division (RND) system related genes acrAB and mtdABC were recovered in NOB01, while mtdABC was lacking in NOB02, likely suggesting NOB01 can be very effective in counteracting various toxic antibiotics, detergents, and biocide (Du et al. 2014; Hadchity et al. 2021; Nuonming et al. 2018). Additionally, heavy metals resistance genes, including cusABC efflux system and arsenite reductase (arsC) were encoded by NOB01 and NOB02, which potentially enhance their tolerance to heavy metals in sewage. Chlorite dismutase (CLD) often occurs in other NOB, including Nitrospira and Nitrobacter for chlorite detoxification (Daims et al. 2015; Lücker et al. 2010; Mlynek et al. 2011; Yang et al. 2020a), which also was found in NOB01 and NOB02. Interestingly, two identical locus coding for CLD were found in NOB02, whihc likely helps NOB02 to survive in the anoxic biofilm systems, since the producted oxygen by CLD could be used for aerobic respiration (Hofbauer et al. 2014; Kostan et al. 2010; Yang et al. 2020b). High-salinity concentrate is a common problem for many sewage, which may decrease the effectiveness of biological wastewater treatment via charging the microbial activity (Zhao et al. 2020). Na+/H+ antiporter (NhaA and NhaK), magnesium transporter (CorA), magnesium transporter (MgtE), and multicomponent K+:H+ antiporter complex (Pha) (Assaha et al. 2017; Sun et al. 2022) related to salt stress tolerance were found in the two MAGs. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs), together with CRISPR-associated (Cas) proteins, confer adaptive immunity to microbials in their defense against unwanted environment (Marraffini and Sontheimer 2010). Multiple families of Cas proteins also build unique CRISPR-Cas immune systems for microbes, for example, Csn-type systems have been shown to cleave invasive heterologous DNA (Garneau et al. 2010), and DNA silencing was mediated by Csm-type Cas proteins (Marraffini and Sontheimer 2010), and the Cmr-type complex are involved in the recognition and destruction of complementary RNAs in vivo (Hale et al. 2012). Some CRISPR-Cas immune systems regulation genes also were found in NOB01 and NOB02, especially in NOB01, which encode various Cas genes, this result suggests that NOB01 and NOB02 can effectively resist phage infestation.
Conclusions
After genomic recovery and characterization, we identified two novel VII Nitrospira species NOB01 and NOB02, and construct a substantial potential metabolic versatility. The identification of NXR consolidates the metabolic potential of Ca. Nitrospira for nitrite oxidation. In particular, the discovery of a urease and cyanase gene indicated Ca. Nitrospira can utilize urea and cyanate as substrates for ammonia production, and benefit from this, Ca. Nitrospira formed a reciprocal feeding relationship with the AOM in the community for performing nitrification. Our results also indicate that Ca. Nitrospira may interact with other members of the microbial communities, such as anammox bacteria and denitrifiers, through the supply of NO and NO3−, resulting from transcribed nirK and NarG in Ca. Nitrospira. In addition to classical r-TCA carbon fixation pathway, we also deduced that an interconnected process WL pathway and RPG for extra carbon fixation, and benefit from above carbon fixation pathway and H2 and formate metabolism would further increase the ecophysiological flexibility of Ca. Nitrospira in activated sludge and biofilm niche. In particular, nitrate reduction coupled with organic carbon oxidation might confer a selective advantage to NOB02 in the activated sludge niche. Our results indicate that coinciding with previous results, the closest relative of Ca. Nitrospira NXR is anammox bacteria, likely illustrating the occurrence of HGT. Importantly, the NAR complex only detected two Nitrospira genomes and NOB01, and its potential function profiling and evolution history also were illustrated by homology modeling and comparative genomic analysis. In summary, by introducing the novel genus of NOB Ca. Nitrospira and charactering their metabolic versality in WWTP system, our findings suggest that the diversity and abundance of NOB may be higher than previously thought. Moreover, there are also other prominent open questions pertaining to this genus of bacteria. Further studies should focus on the enrichment and physiological characterization of the novel NOB, such as the underlying reason for NAR complex coding and the real catalytic function of the NAR complex in Ca. Nitrospira.
Materials and methods
Sampling collection, sequencing, and binning
Procedures of sample collection, processing, and data generation and analysis are provided in our previous study (Hu et al. 2023). In brief, the activated sludge and biofilm were sampled in a full-scale wastewater treatment plant (WWTP) treating duck breeding wastewater, which was configured as an anaerobic-anoxic-anaerobic-oxic system. Samples were preserved in LifeGuard Soil Preservation (QIAGEN, Germany), and total DNA and RNA in all samples were extracted using the DNA and RNA extraction kits (QIAGEN, Germany) within 24 h. Shotgun metagenomics and metatranscriptomics sequencing were carried out by Novogene Co., Ltd. (Beijing). Raw shotgun metagenomic sequencing reads were quality controlled by MetaWRAP read_qc with default setting (Uritskiy et al. 2018), and retained clean reads were de novo assembled by MEGAHIT V1.1.4 (–k-min 23 –k-max 141 –k-step 20) (Li et al. 2015). Scaffolds larger than 1500 bp were binned by metabat2, maxbin2, concoct, and refined by MetaWRAP bin_refinement with the setting “-x 50 -c 50″ (Uritskiy et al. 2018). The quality of obtained MAGs was assessed using the module “lineage_wf” of CheckM v1.0.6 (Parks et al. 2015), and eligible MAGs (completeness > 50 % and contamination <10 %) were manually refined for downstream analysis. All refined MAGs were classified using GTDB-Tk v1.3.0 (Chaumeil et al. 2019) with the version Genome Database Taxonomy GTDB-r214 (Parks et al. 2020) as the database and default setting. Clean metagenomic reads were mapped to the scaffolds by BWA v0.7.17 (Li and Durbin 2009) with the default setting to calculate the genome and recorded gene abundance as TPM. For trimmed metatranscriptomic data, the non-rRNA dataset was constructed by removing the ribosomal RNA (rRNA) reads from raw data using SortMeRNA (version 2.1) (Kopylova et al. 2012) based on the SILVA 132 database. The transcription abundance (TPM) was calculated by mapping all non-rRNA datasets to all predicted ORFs by BWA v0.7.17 (Li and Durbin 2009) with a default setting.
Genome reassemble and annotation
In total three NOB MAGs were acquired and identified, including NOB01, NOB02, and NOB03. To improve the quality and exclude unexpected contaminations in binning, the two novel NOB MAGs NOB01 and NOB02 were subjected to highly iterated and rigorous reassembly (Yang et al. 2020a, Yang et al. 2021). Briefly, clean reads of the samples oxic tank (2_O) and biofilm (A_L) were mapped to the two NOB MAGs using BBmap (Bushnell 2014), and the successfully aligned reads were re-assembled using SPAdes v3.12.0 (Bankevich et al. 2012) with the k-mers of 21, 33, 55, 77, 99, 127, then, the quality of the resulting MAGs of two NOB was checked using CheckM v1.0.6 “lingage_wf” command again (Parks et al. 2015). The protein-coding genes in the reassembled MAGs NOB01 and NOB02 were predicated using Prodigal v2.6.3 using the “- p single” option (Hyatt et al. 2010). Genome annotation was carried out by GhostKOALA (Kanehisa et al. 2016) using the KEGG database, eggNOG-mapper (Huerta-Cepas et al. 2016). The interest function genes were also further confirmed using BLASTp against the NCBI non-redundant (nr) database. To compare the genome-wide gene expression levels of the two NOB MAGs in the targeted reactor and biofilm, gene transcription abundance showed by TPM was standardized by the genome abundance. Average Nucleotide Identity (ANI) was calculated between the newly recovered NOB MAGs and known Nitrospira genome downloaded from NCBI database by fastANI (Jain et al. 2018) with the default setting.
Phylogenetic analysis
The 43 concatenated marker genes were identified from the newly acquired NOB MAGs and known Nitrospira genomes download from NCBI and aligned using CheckM v1.0.6 “lineage_wf” module with default setting (Parks et al. 2015). In addition, a genome-wide de novo phylogenetic analysis based on the Genome Database Taxonomy GTDB-r214 database was also performed by GTDB-Tk v1.3.0 “de_novo_wf” command with “-outgroup_taxon P_Nitrospirae” (Chaumeil et al. 2019). The 16S rRNA gene sequences in MAGs NOB01, NOB02, and NOB03 were identified by BLASTn against the SILVA SSU 138 database and their close relatives in genus Nitrospira based on BLASTn results and 16S rRNA gene sequences from another characterized NOB genus Leptospirllum were downloaded from NCBI. All above 16S rRNA gene sequences were used for the phylogenetic tree built to highlight the phylogenetic placement of the Ca. Nitrospira genus in the family of Nitrospiraceae.
For the phylogenies of NxrA and NxrB (nitrite oxidoreductase alpha /beta subunit), the sequence of NOB01 and NOB02 were used as a query in BLASTp search (Mahram and Herbordt 2015) in the NCBI database with the requirements of E-value < 10e-10, amino acid identities > 60 %, and a minimum alignment length > 50 % to find its close relatives. All retained sequences were aligned using MAFFT v7.463 (Katoh et al. 2002) and gaps in alignment were removed using trimAl V2.0 with the setting ‘-gt 0.1′ (Capella-Gutiérrez et al. 2009). Maximum likelihood trees of NxrA and NxrB were constructed using IQ-TREE2 V1.6.12 (Minh et al. 2020) with the LG+R3 and LG+G4 substitution model and 1000 ultrafast bootstraps.
A maximum-likelihood phylogenetic tree of Ni/Fe hydrogenase, using online tools https://services.birc.au.dk/hyddb/ to determine the classification of the sequence, and then, additional Ni/Fe hydrogenase sequences of the family Nitrospiraceae and the genus Nitrospira deposited in the NCBI nr-database were also identified using BLASTp (Mahram and Herbordt 2015) using the Ni/Fe hydrogenase sequences of Ca. Nitrospira NOB02 as the query and the standard is E-value 〈 10e-10, amino acid identities 〉 60 %, and a minimum alignment length > 50 %. Sequences were aligned using MAFFT v7.463 (Katoh et al. 2002) and gaps in alignment were removed using trimAl V2.0 with the setting ‘-gt 0.1′ (Capella-Gutiérrez et al. 2009). Maximum likelihood trees were constructed using IQ-TREE2 V1.6.12 (Minh et al. 2020) with the Q.pfam+R8 substitution model and 1000 ultrafast bootstraps.
For the phylogeny of NarG (nitrate reductase alpha subunit), the sequence of Ca. Nitrospira NOB01 was used as the query in the BLASTp (Mahram and Herbordt 2015) search in the NCBI database (E-value 〈 10e-10, amino acid identities 〉 60 %, and a minimum alignment length > 50 %). To more accurately pinpoint the affiliation of NarG, additional NarG sequences of unclassified Nitrospirae bacterium, NxrA of Nitrosipra, and anammox bacteria deposited in the NCBI database were also identified using BLASTp using the NarG sequences of Ca. Nitrospira NOB01 as the query and the Evalue of 10e-6. All retained sequences were aligned using MAFFT v7.463 (Katoh et al. 2002) and gaps in alignment were removed using trimAl V2.0 with the setting ‘-gt 0.1’ (Capella-Gutiérrez et al. 2009). Maximum likelihood trees were constructed using IQ-TREE2 V1.6.12 (Minh et al. 2020) with the LG+R4 substitution model and 1000 ultrafast bootstraps. Finally, All the above-built trees were visualized and beautified using iTOL (Letunic and Bork 2021).
Homologous modeling of NAR complex
NAR complex structure homology modeling was performed by SWISS-MODEL workspace as described in (Bordoli et al. 2009). Briefly, the target NAR amino acid sequence, which is heteromeric and consists of three different protein chains (NarGHI) as subunits, was obtained from “Ca. Nitrospira NOB01”, then, the targeted sequence serve as a query to search for evolutionary-related protein structures against the SWISS-MODEL template library SMTL (https://swissmodel.expasy.org/templates/) (Biasini et al. 2014) using BLAST and HHblits (Bordoli et al. 2009; Remmert et al. 2012; Waterhouse et al. 2018). The three-dimensional NAR complex protein model was generated by aligning the first transfer ring conserving atoms to the template, and residue coordinates corresponding to insertions/deletions in the alignment also generated by loop modeling, the final three-dimensional NAR model was constructed by integrating the non-conserved amino acid side chains (Waterhouse et al. 2018). To quantify the modeling error and estimate the expected model accuracy, the expected quality of the resulting models was estimated by Global Model Quality Estimate (GMQE) and Quaternary Structure Quality Estimate (QSQE) (Bordoli et al. 2009; Waterhouse et al. 2018). Additionally, the NAR model also was further evaluated by SAVES v6.0 (Colovos and Yeates 1993; Pontius et al. 1996). Visualization and retouching of the model were carried out by PyMOL v 2.3.2 (Yuan et al. 2016). Cluster analysis of targeted NAR with the template was produced from SWISS-MODEL and PyMOL align module with default setting. Display for the alignment was generated using the ESPRIPT 3.0 tool (http://espript.ibcp.fr/).
CRediT authorship contribution statement
Pengfei Hu: Writing – original draft, Validation, Formal analysis, Data curation. Youfen Qian: Writing – original draft, Validation, Investigation. Yanbin Xu: Investigation, Data curation. Adi Radian: Writing – review & editing, Formal analysis. Yuchun Yang: Writing – review & editing, Validation, Formal analysis. Ji-Dong Gu: Writing – review & editing, Writing – original draft, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This research was supported by the National Key Research and Development Program of China (2021YFA0910300).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.wroa.2024.100237.
Appendix. Supplementary materials
Data availability
Raw metagenomic and metatranscriptomic data have been deposited into NCBI under the BioProject PRJNA815463. All NOB genome sequences from the current study have been deposited in the NCBI GenBank database, with accession numbers JANRMO000000000, JANRMP000000000, and JANRMQ000000000.
References
- Assaha D.V.M., Ueda A., Saneoka H., Al-Yahyai R., Yaish M.W. The Role of Na(+) and K(+) Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017;8:509. doi: 10.3389/fphys.2017.00509. -509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer B., Saito M.A., McIlvin M.R., Lücker S., Moran D.M., Lankiewicz T.S., Dupont C.L., Santoro A.E. Metabolic versatility of the nitrite-oxidizing bacterium Nitrospira marina and its proteomic response to oxygen-limited conditions. ISMe J. 2021;15(4):1025–1039. doi: 10.1038/s41396-020-00828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero M.G., Rothery R.A., Palak M., Hou C., Lim D., Blasco F., Weiner J.H., Strynadka N.C. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Mol. Biol. 2003;10(9):681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Ressearch. 2014;42:W252–W258. doi: 10.1093/nar/gku340. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009;4(1):1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Bushnell, B. (2014) BBMap: a Fast, Accurate, Splice-Aware Aligner. Conference: 9th Annual Genomics of Energy & Environment Meeting.
- Cao Y., van Loosdrecht M., Daigger G.T. Mainstream partial nitritation–anammox in municipal wastewater treatment: status, bottlenecks, and further studies. Appl. Microbiol. Biotechnol. 2017;101(4):1365–1383. doi: 10.1007/s00253-016-8058-7. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caranto J.D., Lancaster K.M. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proceedings of the National Academy of Sciences. 2017;114(31):8217–8222. doi: 10.1073/pnas.1704504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36(6):1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Kambey C., Nguyen V.K. Performance of Anammox Processes for Wastewater Treatment: a Critical Review on Effects of Operational Conditions and Environmental Stresses. Water. (Basel) 2020;12:1. [Google Scholar]
- Coelho C., Romão M.J. Structural and mechanistic insights on nitrate reductases. Protein science: a publication of the Protein Society. 2015;24(12):1901–1911. doi: 10.1002/pro.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein science: a publication of the Protein Society. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Lücker S., Wagner M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 2016;24(9):699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Lebedeva E.V., Pjevac P., Han P., Herbold C., Albertsen M., Jehmlich N., Palatinszky M., Vierheilig J., Bulaev A., Kirkegaard R.H., von Bergen M., Rattei T., Bendinger B., Nielsen P.H., Wagner M. Complete nitrification by Nitrospira bacteria. Nature. 2015;528(7583):504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Nielsen J.L., Nielsen P.H., Schleifer K.-H., Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 2001;67(11):5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Wagner M. Nitrospira. Trends in Microbiology. 2018;26(5):462–463. doi: 10.1016/j.tim.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T., Venter H., Chiu W., Luisi B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509(7501):512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich S., Behrens D., Lebedeva E., Ludwig W., Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol. 1995;164(1):16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- Figueroa I.A., Barnum T.P., Somasekhar P.Y., Carlström C.I., Engelbrektson A.L., Coates J.D. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proceedings of the National Academy of Sciences. 2018;115(1):E92–E101. doi: 10.1073/pnas.1715549114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganigué R., López H., Balaguer M.D., Colprim J. Partial ammonium oxidation to nitrite of high ammonium content urban landfill leachates. Water Res. 2007;41(15):3317–3326. doi: 10.1016/j.watres.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Garneau J.E., Dupuis M.-È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- González P., Correia C., Moura I., Brondino C.D., Moura J. Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 2006;100:1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Gonzalez P.J., Rivas M.G., Moura J.J.G. The Royal Society of Chemistry; 2017. Metalloenzymes in Denitrification: Applications and Environmental Impacts; pp. 39–58. [Google Scholar]
- Gruber-Dorninger C., Pester M., Kitzinger K., Savio D.F., Loy A., Rattei T., Wagner M., Daims H. Functionally relevant diversity of closely related Nitrospira in activated sludge. ISMe J. 2015;9(3):643–655. doi: 10.1038/ismej.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Yang Q., Liu Y. Mainstream anammox in a novel A-2B process for energy-efficient municipal wastewater treatment with minimized sludge production. Water Res. 2018;138:1–6. doi: 10.1016/j.watres.2018.02.051. [DOI] [PubMed] [Google Scholar]
- Hadchity L., Lanois A., Kiwan P., Nassar F., Givaudan A., Khattar Z.A. AcrAB, the major RND-type efflux pump of Photorhabdus laumondii, confers intrinsic multidrug-resistance and contributes to virulence in insects. Environ. Microbiol. Rep. 2021;13(5):637–648. doi: 10.1111/1758-2229.12974. [DOI] [PubMed] [Google Scholar]
- Hale Caryn R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch Alissa M., Glover Claiborne V.C., Graveley Brenton R., Terns Rebecca M., Terns Michael P. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45(3):292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer S., Gruber C., Pirker K., Schaffner I., Furtmüller P., Obinger C., Hagmüller A., Gysel K., Mlynek G., Kostan J., Djinović-Carugo K., Bellei M., Battistuzzi G., Daims H. 16th International Conference on Biological Inorganic Chemistry. 2014. Understanding chlorite dismutase from Candidatus nitrospira defluvii; p. 19. [Google Scholar]
- Hu P., Qian Y., Liu J., Gao L., Li Y., Xu Y., Wu J., Hong Y., Ford T., Radian A., Yang Y., Gu J.-D. Delineation of the complex microbial nitrogen-transformation network in an anammox-driven full-scale wastewater treatment plant. Water Res. 2023;235 doi: 10.1016/j.watres.2023.119799. [DOI] [PubMed] [Google Scholar]
- Hu Z., Wessels H., van Alen T., Jetten M.S.M., Kartal B. Nitric oxide-dependent anaerobic ammonium oxidation. Nature Communication. 2019;10(1):1244. doi: 10.1038/s41467-019-09268-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund S., Szklarczyk D., Jensen L., von Mering C., Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2016;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C., Rodriguez-R L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016;428(4):726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Kartal B., Tan N.C., Van de Biezen E., Kampschreur M.J., Van Loosdrecht M.C., Jetten M.S. Effect of nitric oxide on anammox bacteria. Appl. Environ. Microbiol. 2010;76(18):6304–6306. doi: 10.1128/AEM.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K.i., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuter S., Koch H., Nowka B., Lipski A., Kruse M., Lücker S., Spieck E. A novel Nitrospira lineage isolated from activated sludge using elevated temperatures. FEMS Microbiol. Lett. 2023;370 doi: 10.1093/femsle/fnad035. [DOI] [PubMed] [Google Scholar]
- Kitzinger K., Koch H., Lücker S., Sedlacek C.J., Herbold C., Schwarz J., Daebeler A., Mueller A.J., Lukumbuzya M., Romano S., Leisch N., Karst S.M., Kirkegaard R., Albertsen M., Nielsen P.H., Wagner M., Daims H. Characterization of the First "Candidatus Nitrotoga" Isolate Reveals Metabolic Versatility and Separate Evolution of Widespread Nitrite-Oxidizing Bacteria. mBio. 2018;9(4) doi: 10.1128/mBio.01186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H., Lücker S., Albertsen M., Kitzinger K., Herbold C., Spieck E., Nielsen P., Wagner M., Daims H. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proceedings of the National Academy of Sciences. 2015;112(36):11371–11376. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K.T., Rosselló-Móra R., Amann R. Uncultivated microbes in need of their own taxonomy. ISMe J. 2017;11(11):2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E., Noé L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Kostan J., Sjöblom B., Maixner F., Mlynek G., Furtmüller P.G., Obinger C., Wagner M., Daims H., Djinović-Carugo K. Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": identification of a catalytically important amino acid residue. J. Struct. Biol. 2010;172(3):331–342. doi: 10.1016/j.jsb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Kozlowski J.A., Stieglmeier M., Schleper C., Klotz M.G., Stein L.Y. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISMe J. 2016;10(8):1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers M.M.M., Marchant H.K., Kartal B. The microbial nitrogen-cycling network. Nature Reviews Microbiology. 2018;16(5):263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- Lücker S., Wagner M., Maixner F., Pelletier E., Koch H., Vacherie B., Rattei T., Damsté J.S.S., Spieck E., Paslier D.L., Daims H. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proceedings of the National Academy of Sciences. 2010;107(30):13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C.E., Mundinger A.B., Koch H., Jacobson T.B., Weathersby C.A., Jetten M.S.M., Pabst M., Amador-Noguez D., Noguera D.R., McMahon K., Lücker S. Investigating the Chemolithoautotrophic and Formate Metabolism of Nitrospira moscoviensis by Constraint-Based Metabolic Modeling and (13)C-Tracer Analysis. mSystems. 2021;6(4) doi: 10.1128/mSystems.00173-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung P.M., Daebeler A., Chiri E., Hanchapola I., Gillett D.L., Schittenhelm R.B., Daims H., Greening C. A nitrite-oxidising bacterium constitutively consumes atmospheric hydrogen. ISMe J. 2022;16:2213–2219. doi: 10.1038/s41396-022-01265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Lu Y., Zheng M., Hu S., Yuan Z., Guo J. Efficient nitrogen removal from mainstream wastewater through coupling Partial Nitritation, Anammox and Methane-dependent nitrite/nitrate reduction (PNAM) Water Res. 2021;206 doi: 10.1016/j.watres.2021.117723. [DOI] [PubMed] [Google Scholar]
- Luecker S., Nowka B., Rattei T., Spieck E., Daims H. The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer. Front. Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Bao P., Wei Y., Zhu G., Yuan Z., Peng Y. Suppressing Nitrite-oxidizing Bacteria Growth to Achieve Nitrogen Removal from Domestic Wastewater via Anammox Using Intermittent Aeration with Low Dissolved Oxygen. Sci. Rep. 2015;5:13048. doi: 10.1038/srep13048. -13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahram A., Herbordt M.C. NCBI BLASTP on High-Performance Reconfigurable Computing Systems. ACM. Trans. Reconfigurable Technol. Syst. 2015;7(4):33. doi: 10.1145/1862648.1862653. Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangon J., Paes de Sousa P.M., Moura I., Brondino C.D., Moura J.J., González P.J. Substrate-dependent modulation of the enzymatic catalytic activity: reduction of nitrate, chlorate and perchlorate by respiratory nitrate reductase from Marinobacter hydrocarbonoclasticus 617. Biochim. Biophys. Acta. 2012;1817(7):1072–1082. doi: 10.1016/j.bbabio.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Marraffini L.A., Sontheimer E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463(7280):568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool G.J., Cannon M.C. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol. 2001;183(14):4235–4243. doi: 10.1128/JB.183.14.4235-4243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Peng Y., Zhang L., Li B., Li X., Wu L., Wang S. Partial nitrification-anammox (PNA) treating sewage with intermittent aeration mode: effect of influent C/N ratios. Chemical Engineering Journal. 2018;334:664–672. [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynek G., Sjöblom B., Kostan J., Füreder S., Maixner F., Gysel K., Furtmüller P.G., Obinger C., Wagner M., Daims H., Djinović-Carugo K. Unexpected Diversity of Chlorite Dismutases: a Catalytically Efficient Dimeric Enzyme from Nitrobacter winogradskyi. J. Bacteriol. 2011;193(10):2408–2417. doi: 10.1128/JB.01262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.J., Daebeler A., Herbold C.W., Kirkegaard R.H., Daims H. Cultivation and genomic characterization of novel and ubiquitous marine nitrite-oxidizing bacteria from the Nitrospirales. ISMe J. 2023;17(11):2123–2133. doi: 10.1038/s41396-023-01518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowka B., Daims H., Spieck E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 2015;81(2):745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuonming P., Khemthong S., Dokpikul T., Sukchawalit R., Mongkolsuk S. Characterization and regulation of AcrABR, a RND-type multidrug efflux system, in Agrobacterium tumefaciens C58. Microbiol. Res. 2018;214:146–155. doi: 10.1016/j.micres.2018.06.014. [DOI] [PubMed] [Google Scholar]
- Palatinszky M., Herbold C., Jehmlich N., Pogoda M., Han P., von Bergen M., Lagkouvardos I., Karst S.M., Galushko A., Koch H., Berry D., Daims H., Wagner M. Cyanate as an energy source for nitrifiers. Nature. 2015;524(7563):105–108. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer T., Berks B.C. The twin-arginine translocation (Tat) protein export pathway. Nature Reviews Microbiology. 2012;10(7):483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- Parks D., Imelfort M., Skennerton C., Philip H., Tyson G. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015:25. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Chuvochina M., Chaumeil P.-A., Rinke C., Mussig A.J., Hugenholtz P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020;38(9):1079–1086. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- Parveez G.K.A., Bahariah B., Ayub N.H., Masani M.Y.A., Rasid O.A., Tarmizi A.H., Ishak Z. Production of polyhydroxybutyrate in oil palm (Elaeis guineensis Jacq.) mediated by microprojectile bombardment of PHB biosynthesis genes into embryogenic calli. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picioreanu C., van Loosdrecht M.C.M., Heijnen J.J. Modelling the effect of oxygen concentration on nitrite accumulation in a biofilm airlift suspension reactor. Water Science and Technology. 1997;36(1):147–156. [Google Scholar]
- Pontius J., Richelle J., Wodak S.J. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996;264(1):121–136. doi: 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
- Regmi P., Miller M.W., Holgate B., Bunce R., Park H., Chandran K., Wett B., Murthy S., Bott C.B. Control of aeration, aerobic SRT and COD input for mainstream nitritation/denitritation. Water Res. 2014;57:162–171. doi: 10.1016/j.watres.2014.03.035. [DOI] [PubMed] [Google Scholar]
- Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- Rothery R.A., Workun G.J., Weiner J.H. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta. 2008;1778(9):1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Santoro A., Dupont C., Richter A., Craig M., Carini P., McIlvin M., Yang Y., Orsi W., Moran D., Saito M. Genomic and proteomic characterization of “ Candidatus Nitrosopelagicus brevis”: an ammonia-oxidizing archaeon from the open ocean. Proc. Natl. Acad. Sci. U.S.A. 2015;112 doi: 10.1073/pnas.1416223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Andrea I., Guedes I.A., Hornung B., Boeren S., Lawson C.E., Sousa D.Z., Bar-Even A., Claassens N.J., Stams A.J.M. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nature Communication. 2020;11(1):5090. doi: 10.1038/s41467-020-18906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Gross R., Einsle O., Kroneck P.M., Kröger A., Klimmek O. A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol. Microbiol. 2000;35(3):686–696. doi: 10.1046/j.1365-2958.2000.01742.x. [DOI] [PubMed] [Google Scholar]
- Simon J., Pisa R., Stein T., Eichler R., Klimmek O., Gross R. The tetraheme cytochrome c NrfH is required to anchor the cytochrome c nitrite reductase (NrfA) in the membrane of Wolinella succinogenes. Eur. J. Biochem. 2001;268(22):5776–5782. doi: 10.1046/j.0014-2956.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- Song Y., Lee J., Shin J., Lee G., Jin S., Kang S., Lee J.-K., Kim D.R., Lee E., Kim S., Cho S., Kim D., Cho B.K. Functional cooperation of the glycine synthase-reductase and Wood–Ljungdahl pathways for autotrophic growth of Clostridium drakei. Proceedings of the National Academy of Sciences. 2020;117 doi: 10.1073/pnas.1912289117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg S.R., Chain P.S., Sayavedra-Soto L.A., Hauser L., Land M.L., Larimer F.W., Malfatti S.A., Klotz M.G., Bottomley P.J., Arp D.J., Hickey W.J. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 2006;72(3):2050–2063. doi: 10.1128/AEM.72.3.2050-2063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer G., Rempfer C., Waterhouse A., Gumienny R., Haas J., Schwede T. QMEANDisCo - distance constraints applied on model quality estimation. Bioinformatics (Oxford, England) 2019:36. doi: 10.1093/bioinformatics/btaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhao J., Zhou X., Bei Q., Xia W., Zhao B., Zhang J., Jia Z. Salt tolerance-based niche differentiation of soil ammonia oxidizers. ISMe J. 2022;16(2):412–422. doi: 10.1038/s41396-021-01079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z.C., Liu T., Guo J.H., Zheng M. Nitrite Oxidation in Wastewater Treatment: microbial Adaptation and Suppression Challenges. Environ. Sci. Technol. 2023;57(34):12557–12570. doi: 10.1021/acs.est.3c00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y., Hirano S., Matson M.M., Atsumi S., Kondo A. Electrical-biological hybrid system for CO2 reduction. Metab. Eng. 2018;47:211–218. doi: 10.1016/j.ymben.2018.03.015. [DOI] [PubMed] [Google Scholar]
- Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6(1):158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiki N., Fujitani H., Shimada Y., Morohoshi T., Sekiguchi Y., Tsuneda S. Genomic Analysis of Two Phylogenetically Distinct Nitrospira Species Reveals Their Genomic Plasticity and Functional Diversity. Front. Microbiol. 2017;8:2637. doi: 10.3389/fmicb.2017.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiki N., Fujitani H., Shimada Y., Morohoshi T., Sekiguchi Y., Tsuneda S. Genomic Analysis of Two Phylogenetically Distinct Nitrospira Species Reveals Their Genomic Plasticity and Functional Diversity. Front. Microbiol. 2018;8 doi: 10.3389/fmicb.2017.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiki N., Jinno M., Fujitani H., Suenaga T., Terada A., Tsuneda S. Nitrite oxidation kinetics of two Nitrospira strains: the quest for competition and ecological niche differentiation. Journal of Bioscience & Bioengineering. 2017;123(5):581–589. doi: 10.1016/j.jbiosc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- van Kessel M.A.H.J., Speth D.R., Albertsen M., Nielsen P.H., Op den Camp H.J.M., Kartal B., Jetten M.S.M., Lücker S. Complete nitrification by a single microorganism. Nature. 2015;528(7583):555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ye L., Jiang G., Hu S., Yuan Z. Side-stream sludge treatment using free nitrous acid selectively eliminates nitrite oxidizing bacteria and achieves the nitrite pathway. Water Res. 2014;55:245–255. doi: 10.1016/j.watres.2014.02.029. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ye J., Ju F., Liu L., Boyd J.A., Deng Y., Parks D.H., Jiang X., Yin X., Woodcroft B.J., Tyson G.W., Hugenholtz P., Polz M.F., Zhang T. Successional dynamics and alternative stable states in a saline activated sludge microbial community over 9 years. Microbiome. 2021;9(1):199. doi: 10.1186/s40168-021-01151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F., Beer T., Rempfer C., Bordoli L., Lepore A., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Xu X., Yang F., Liu S., Gao Y. Partial nitrification adjusted by hydroxylamine in aerobic granules under high DO and ambient temperature and subsequent Anammox for low C/N wastewater treatment. Chemical Engineering Journal. 2012;213:338–345. [Google Scholar]
- Yang Y., Daims H., Liu Y., Herbold C.W., Pjevac P., Lin J.-G., Li M., Gu J.-D., Santoro A.E., Capone D.G. Activity and Metabolic Versatility of Complete Ammonia Oxidizers in Full-Scale Wastewater Treatment Systems. mBio. 2020;11(2):e03175. doi: 10.1128/mBio.03175-19. -03119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Pan J., Zhou Z., Wu J., Liu Y., Lin J.-G., Hong Y., Li X., Li M., Gu J.-D. Complex microbial nitrogen-cycling networks in three distinct anammox-inoculated wastewater treatment systems. Water Res. 2020;168 doi: 10.1016/j.watres.2019.115142. [DOI] [PubMed] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E., Glöckner F.O., Ludwig W., Schleifer K.H., Whitman W.B., Euzéby J., Amann R., Rosselló-Móra R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014;12(9):635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- Yuan S., Chan H., Filipek S., Vogel H. PyMOL and Inkscape Bridge the Data and the Data Visualization. Structure. 2016;24 doi: 10.1016/j.str.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhuang X., Ahmad S., Sung S., Ni S.-Q. Biotreatment of high-salinity wastewater: current methods and future directions. World Journal of Microbiology and Biotechnology. 2020;36(3):37. doi: 10.1007/s11274-020-02815-4. [DOI] [PubMed] [Google Scholar]
- Zher Neoh S., Fey Chek M., Tiang Tan H., Linares-Pastén J.A., Nandakumar A., Hakoshima T., Sudesh K. Polyhydroxyalkanoate synthase (PhaC): the key enzyme for biopolyester synthesis. Curr. Res. Biotechnol. 2022;4:87–101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw metagenomic and metatranscriptomic data have been deposited into NCBI under the BioProject PRJNA815463. All NOB genome sequences from the current study have been deposited in the NCBI GenBank database, with accession numbers JANRMO000000000, JANRMP000000000, and JANRMQ000000000.