Abstract
Persistent inflammation driven by cytokines such as type-one interferon (IFN-I) can cause immunosuppression. We show that administration of the Janus kinase 1 (JAK1) inhibitor itacitinib after anti–PD-1 (programmed cell death protein 1) immunotherapy improves immune function and antitumor responses in mice and results in high response rates (67%) in a phase 2 clinical trial for metastatic non–small cell lung cancer. Patients who failed to respond to initial anti–PD-1 immunotherapy but responded after addition of itacitinib had multiple features of poor immune function to anti–PD-1 alone that improved after JAK inhibition. Itacitinib promoted CD8 T cell plasticity and therapeutic responses of exhausted and effector memory–like T cell clonotypes. Patients with persistent inflammation refractory to itacitinib showed progressive CD8 T cell terminal differentiation and progressive disease. Thus, JAK inhibition may improve the efficacy of anti–PD-1 immunotherapy by pivoting T cell differentiation dynamics.
Introduction
The administration of monoclonal antibodies to block the PD-1/PDL-1 inhibitory signaling axis and reactivate antitumor CD8 T cells has led to durable responses to many cancer types (PD-1, programmed cell death protein 1; PD-L1, programmed death ligand 1). In non–small cell lung cancer (NSCLC), the response rate to single-agent pembrolizumab (anti–PD-1) is ~45% in the first-line metastatic setting for patients with tumor PD-L1 expression ≥50% (1), making pembrolizumab the standard of care for this patient population. However, many patients fail to benefit from anti–PD-1 immunotherapy, and approximately two-thirds of NSCLC patients who do initially respond will relapse (2). Thus, developing new approaches to induce durable clinical responses is an important goal for immune checkpoint blockade (ICB) therapy. Interferon (IFN) signaling, which uses the Janus kinase (JAK) family, has well-recognized roles in immune stimulation and promoting antitumor immunity. However, IFN signaling can also have immunoregulatory effects. For example, in chronic lymphocytic choriomeningitis virus (LCMV) infection, high type-one interferon (IFN-I) signaling can inhibit ongoing immune responses and limit viral clearance (3). Blocking the IFN-I receptor (IFNAR1) improves viral control during chronic LCMV infection (4, 5), prevents antigen-specific CD8 T cells from becoming terminally exhausted, and preserves the CXCR5+ TCF1+ exhausted progenitor subset (6). In cancer, which is another disease characterized by chronic inflammation, high expression of a subset of interferon-stimulated genes (ISGs) in cancer cells is associated with immunotherapy resistance for multiple human tumor types, including in NSCLC after acquired resistance to anti–PD-1 (2, 7, 8). In CD8 T cells from patients with NSCLC and other cancer types, high expression of ISGs is also coupled to differentiation toward states that include terminal CD8 T cell exhaustion (9, 10). Moreover, tumor mutations of IFN pathway genes in NSCLC and other cancers can predict longer progression-free survival (PFS), and blocking IFN signaling in ISG-high mouse cancer models either genetically or by administration of JAK inhibitor (JAKi) can improve immune function and ICB response (7, 11, 12). Thus, persistent IFN signaling can have potent immunoregulatory effects. In cancer cells and immune cells, chronic IFN-I signaling is linked with ICB resistance in humans and impedes efficacy of immunotherapy in mice.
Delayed administration of a JAKi alters proliferating CD8 T cells and improves checkpoint blockade immunotherapy in mice
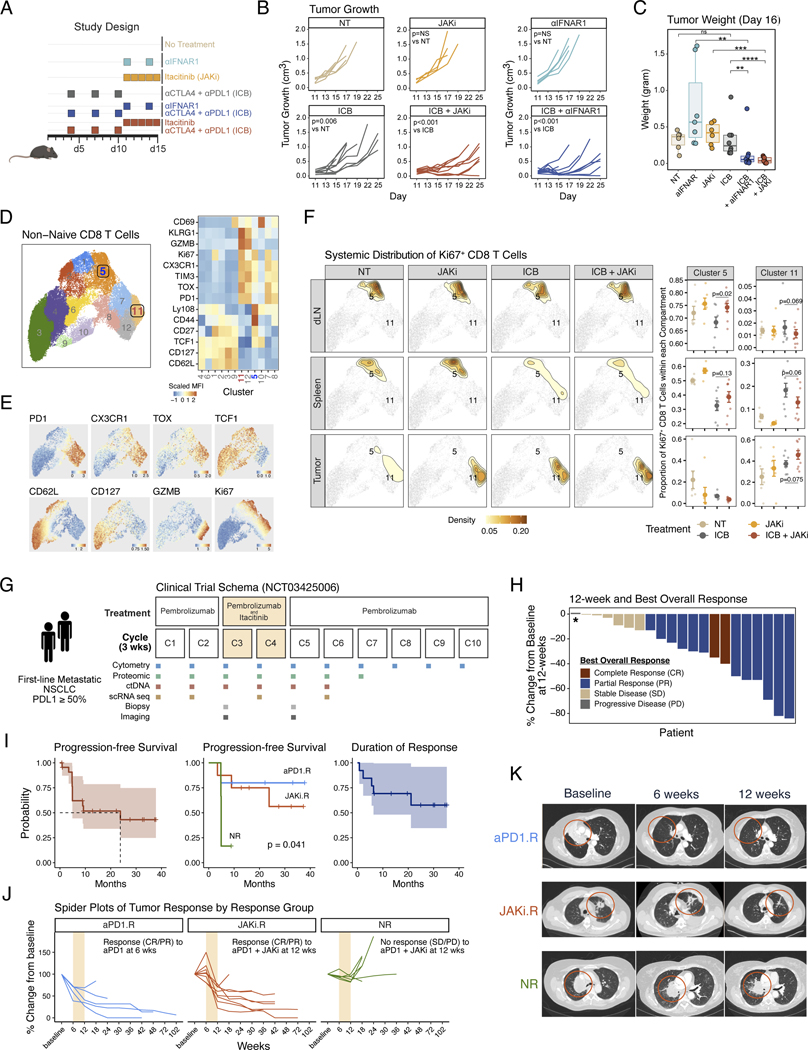
We previously demonstrated in mice that a JAK1 and 2 inhibitor (ruxolitinib) given after the start of ICB can resensitize ICB-resistant tumors from multiple cancer types (12). To examine whether the JAK1 selective inhibitor itacitinib can similarly improve ICB response, we used the Res 499 tumor model, a well-characterized ICB-resistant tumor derived from B16-F10 melanoma (13). Administration of itacitinib 7 days after the start of ICB by either anti–PD-L1 plus anti-CTLA4 (Fig. 1, A to C) or anti–PD-1 alone (fig. S1A) resulted in improved tumor responses. Because JAKi can inhibit numerous cytokine signaling pathways in addition to IFN, we also compared the effects of itacitinib with those of anti-IFNAR1 (Fig. 1B, right). This comparison demonstrated that anti-IFNAR1 largely phenocopied itacitinib, suggesting that inhibiting IFN-I signaling is an important property of—and sufficient for—the JAKi effect. Consistent with a role for IFN-I in dysfunction of CD8 T cells (14), CD8 T cells were among the leukocytes most significantly altered after addition of itacitinib or anti-IFNAR1 to anti–PD-L1 plus anti-CTLA4 (fig. S1, B and C). Therefore, we focused on non-naïve CD8 T cells and systemically evaluated changes in the tumor, draining lymph nodes (dLN), and spleen from treated mice. By flow cytometric analysis, non-naïve CD8 T cells were classified into 12 clusters (Fig. 1, D and E). To enrich for treatment-relevant CD8 T cells, we next restricted analysis to Ki67+-proliferating cells. These Ki67+-proliferating cells differentially distributed between dLN, spleen, and tumor (Fig. 1F, density plots; and fig. S1D). We focused on Ki67+ CD8 T cells belonging to clusters 5 and 11 because these were two clusters that significantly or near-significantly changed with the largest effect sizes in the dLN after JAKi plus ICB versus ICB alone (fig. S1E). Cluster 5 comprised CD8 T cells expressing TCF1, Ly108, low or intermediate PD-1 and TOX, and mixed expression of CX3CR1, resembling memory precursors and/or progenitor-like CD8 T cells (TPRE/PROG-like) (Fig. 1, D and E). Cluster 11 had high expression of PD-1 and TOX as well as CX3CR1, KLRG1, Ki67, and GZMB, consistent with an intermediate or circulatory subset of exhausted CD8 T cells with effector-like features (PD-1hi TEX-INT-like) (15–17). As a proportion of Ki67+ cells, TPRE/PROG-like cluster 5 cells increased in the dLN when JAKi was added to ICB, with similar trends in the spleen (Fig. 1F, top and middle). At the same time, PD-1hi TEX-INT-like cluster 11 cells trended to decrease in the periphery but increased in the tumor both proportionally and by absolute number of cells per gram of tumor (Fig. 1F, middle and bottom; and fig. S1F). The combination of JAKi and ICB also increased the total number of non-naïve CD8 T cells per gram of tumor (fig. S1G). In total, these preclinical data suggest that itacitinib improves ICB efficacy in resistant tumors characterized by high ISGs and functions through antagonizing IFN-I signaling. Compared with ICB alone, addition of JAKi increased both the proportion of proliferating cells in the periphery resembling precursor-like CD8 T cells and the number of intratumoral PD-1hi CD8 TEX-INT-like cells with effector-like features.
Fig. 1. Pre-clinical and phase 2 clinical trial results of anti-PD1 immunotherapy and a JAK1 inhibitor for non-small cell lung cancer.
(A) Pre-clinical treatment regimen using ICB plus either itacitinib, a JAK1 inhibitor (JAKi), or anti-IFNAR1 antibody for mice bearing resistant B16-derived Res 499 tumors. (B) Mouse tumor growth curves in response to treatment strategy outlined in (A). (C) Mouse tumor weights at day 16 after the indicated treatment. (D) Flow cytometry features from non-naive CD8 T cells from Res 499 mouse tumors, the draining lymph node (dLN), and spleen projected on UMAP space. Shown are 12 FlowSOM clusters (left) along with heat map of scaled MFI for all marker proteins arranged by hierarchical clustering (right). (E) MFI expression of select protein markers overlaid on cluster UMAP. (F) Systemic distribution of all Ki67+ CD8 T cells compared across dLN, spleen, and tumor (left density plots) overlaid on the UMAP from (D) (light grey dots). Locations for clusters 5 and 11 are labeled on the density plot overlay. The relative frequencies in each tissue compartment for cells belonging to cluster 5 or 11 are also shown (right dot plots). (G) Schema of phase 2 clinical trial for pembrolizumab and delayed administration of itacitinib for first-line metastatic NSCLC with tumor PDL1 ≥ 50%. Times of treatment, sample collection, and response assessment by imaging are shown relative to each 3-week treatment cycle. (H) Waterfall plot of 12-week tumor response for each patient. Patients are color-coded by best objective response (BOR) that includes response with additional follow-up beyond 12-weeks. Asterisk indicates a patient who clinically progressed prior to the 12-week assessment. (I) Survival curves for overall progression-free survival, progression-free survival by response group defined in (J), and overall duration of response. The 95% confidence intervals are shaded. (J) Spider plots indicating change in tumor measurements from baseline for patients in each response group. Patients were categorized as either an anti-PD1 responder (aPD1.R) if a complete response (CR) or partial response (PR) was observed at 6-weeks after pembrolizumab but prior to itacitinib, a JAKi responder (JAKi.R) if a CR or PR was not observed until 12-weeks after itacitinib, or a non-responders (NR) if a CR or PR was not observed at 12-weeks. Cycles when JAKi was added to anti-PD1 are highlighted in bisque. (K) Representative computerized tomography (CT) scan from aPD1.R, JAK1.R, and NR at baseline, 6 weeks, and 12 weeks. Significance for tumor growth was determined by a mixed-effect regression model. For pairwise comparisons, a two-sided Wilcox test or t-test was used for non-parametric or parametric data, respectively. Survival differences were determined by a log-rank test. Error bars represent SEM.
Clinical response to anti–PD-1 plus delayed JAKi for metastatic lung cancer
Motivated by our preclinical findings, we initiated a phase 2 clinical trial of pembrolizumab and delayed itacitinib for treatment-naïve metastatic NSCLC with tumor PD-L1 ≥50%. A total of 21 patients were treated and evaluated to examine the efficacy of adding itacitinib to pembrolizumab (Fig. 1G). Patients first received two cycles of pembrolizumab and then two cycles of itacitinib and pembrolizumab (start of cycles 3 and 4). At the start of cycle 5, patients were continued on pembrolizumab without itacitinib until disease progression. Imaging was performed after the first two cycles of pembrolizumab (at week 6, cycle 3) and then after itacitinib (at week 12, cycle 5). Objective response rate (ORR) was defined as the proportion of evaluable patients with a complete response (CR) or partial response (PR) on the week-12 scan, and the best overall response (BOR) was defined as the best response at any time, including with additional follow-up after 12 weeks. Clinicopathological characteristics of the 21 evaluable metastatic NSCLC patients were comparable to reported cohorts from other large academic institutions (18) and to the pembrolizumab arm of the KEYNOTE-24 multicountry randomized trial (1) (table S1). The 12-week ORR was 62%, and the BOR with additional follow up after 12 weeks was 67% (Fig. 1H). Only one patient had progression of disease as a BOR. After a median follow-up time of 27.6 months, the median PFS was 23.8 months [95% confidence interval (CI) 4.9 to not applicable (NA)], and the median duration of response (DOR) has not been reached (Fig. 1I, left and right; and fig. S1H). Although these results cannot be directly compared with other clinical trials, the ORR from randomized studies and select US academic centers has been reported to be ~44% with a median PFS of 6.5 to 10.3 months, and a median DOR of about 6 months (1, 18, 19). In addition to the 12-week response assessment, we also assessed response at 6 weeks. This analysis revealed an early (cycles 1 and 2) radiographic response to pembrolizumab before the addition of itacitinib in five patients. By contrast, eight patients failed to respond or had tumors that grew after initial pembrolizumab but responded at week 12 after itacitinib (cycles 3 and 4). Six patients remained nonresponders (failed to achieve PR or CR) at 12 weeks, and one responder did not have a 6-week scan, precluding assessment. On the basis of these response patterns (Fig. 1, J and K), we classified patients as either 6-week anti–PD-1 responders (aPD1.R), 12-week post-itacitinib responders (JAKi.R), or nonresponders (NR) at 12 weeks. These three response groups had similar clinicopathological features (table S2), but the PFS stratified by these groups was expectedly different (Fig. 1I, middle). In total, these findings suggested that the delayed administration of itacitinib after anti–PD-1 resulted in a high response rate and durable responses in NSCLC patients with tumor PD-L1 ≥50%. Of the patients with clinical benefit, some responded early after anti–PD-1, whereas others objectively responded only after addition of JAKi to anti–PD-1. Only one patient had progression of disease as a BOR.
JAK inhibition is associated with clinical response despite low initial CD8 T cell proliferative burst to anti–PD-1 immunotherapy
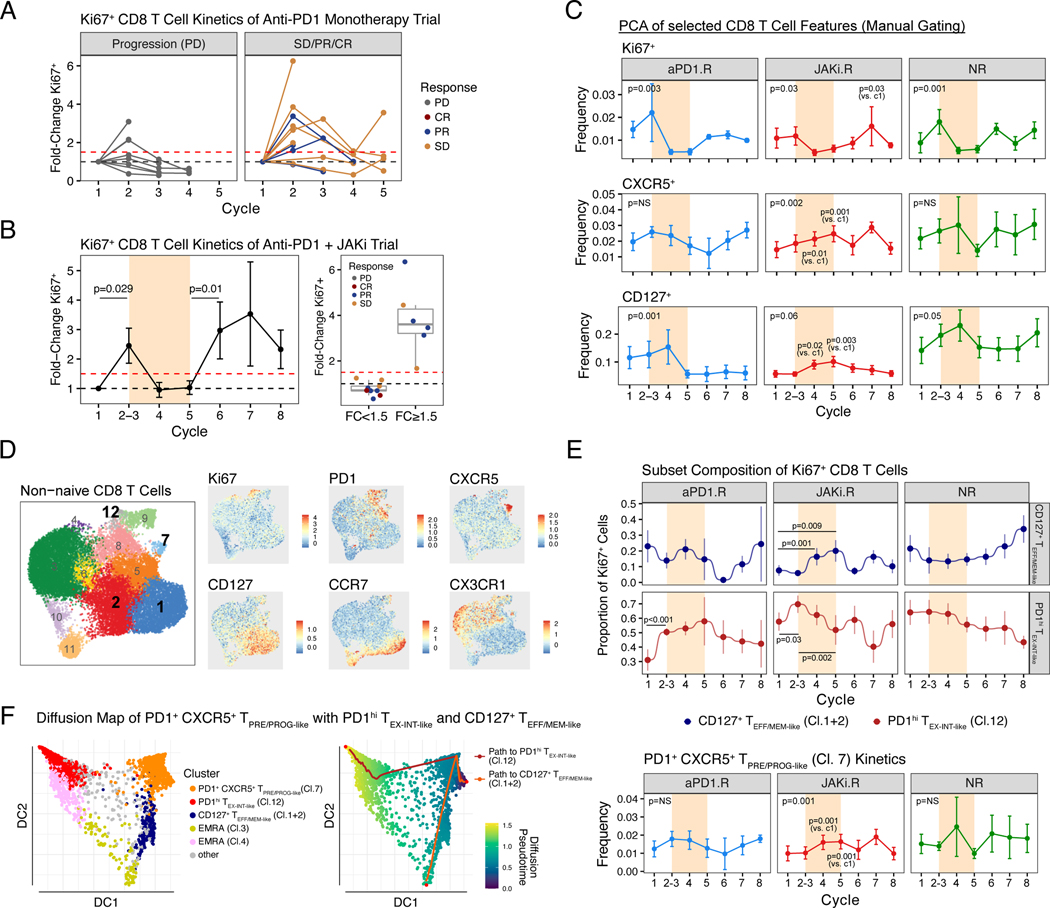
Although a single-arm study does not allow for a direct examination of the impact of JAKi on anti–PD-1 efficacy, we sought to gain insight into whether itacitinib may have altered the response to anti–PD-1 by examining discordance between clinical outcome and predictions from biomarkers for disease progression. In previous reports, approximately 60% of NSCLC patients who failed to achieve a significant increase in the percentage of peripheral Ki67+ CD8 T cells after the first one to two cycles of anti–PD-1 showed progression of disease (failed to achieve CR, PR, or SD) (20). To confirm that poor CD8 T cell proliferative responses to anti–PD-1 predicted disease progression, we first examined a cohort of NSCLC patients treated with anti–PD-1 monotherapy and used a previously defined 1.5-fold increase in Ki67+ CD8 T cells as a prediction threshold (20). Consistent with earlier reports, 71% (five out of seven) of patients with a CD8 T cell proliferative burst below this 1.5-fold threshold after one to two cycles of anti–PD-1 treatment had progression of disease (Fig. 2A). This early proliferative burst after one to two cycles of anti–PD-1 was also observed in our trial of anti–PD-1 plus JAKi; however, by contrast, none of the nine analyzable patients who fell below the 1.5-fold threshold showed progression of disease (Fig. 2B, P = 0.009 by Fisher’s exact test). As an average across all patients, a second CD8 T cell proliferative burst was observed after the period of JAKi treatment (Fig. 2B, left) but was rarely observed with continuous anti–PD-1 monotherapy (fig. S2A) (20–22). Thus, despite the absence of an early CD8 T cell response to anti–PD-1 in some patients, both clinical responses and a post-JAKi CD8 T cell proliferative burst could be observed, suggesting a possible clinical and immune effect when delayed JAKi was added to anti–PD-1 immunotherapy.
Fig. 2. Patient responses after anti-PD1 immunotherapy or JAK inhibition are associated with longitudinal changes in CD8 T cells.
(A) Fold-change in the percentage of Ki67+ CD8 T from a cohort of NSCLC patients treated with anti-PD1 monotherapy (MSKCC cohort). Patients are faceted by progression of disease (PD) or no progression (SD, PR, or CR) with dotted lines representing baseline (black) or a 1.5-fold increase over baseline (red). (B) Fold-change in the percentage of Ki67+ CD8 T from all analyzable patients treated on a clinical trial (this study) of anti-PD1 + JAKi (left). Cycles when JAKi was added to anti-PD1 are highlighted in bisque. Also shown are responses for patients grouped by the status of a 1.5-fold threshold change in Ki67+ CD8 T cells after cycle 1–2 of anti-PD1 (right). (C) Frequency changes of Ki67+, CXCR5+, and CD127+ CD8 T cells across treatment cycles and faceted by treatment response. Markers were selected based on PCA of manually gated flow cytometry features (fig. S2B-F). (D) UMAP and cluster assignment of peripheral non-naïve CD8 T cells analyzed by flow cytometry (left) along with an overlay of the MFI values for the indicated marker proteins (right). (E) Proportions of CD127+ TEFF/MEM-like clusters 1 and 2 and PDhi TEX-INT-like cluster 12 relative to all Ki67+ CD8 T cells (top) and the frequency of PD1+ CXCR5+ TPRE/PROG-like cluster 7 cells relative to non-naïve CD8 T cells (bottom). (F) Trajectory analysis by diffusion mapping for the indicated CD8 T cell subtypes (left) with pseudotime values and predicted paths overlaid (right). For longitudinal data, significance was determined by beta regression for frequency data and Dirichlet regression for compositional data. Error bars represent SEM.
JAKi modulates proliferating CD8 T cell subset composition
Because clinical responses occurred in multiple patients receiving JAKi despite a poor CD8 T cell proliferative burst to initial anti–PD-1, we reasoned that CD8 T cell markers besides Ki67 and/or the composition of treatment-responsive Ki67+ CD8 T cells might provide insight into the effects of JAKi. Thus, to identify CD8 T cell features that dynamically changed after initial administration of anti–PD-1 (samples collected at the start of cycles 2 and 3), during concurrent JAKi plus anti–PD-1 (start of cycles 4 and 5), or after return to anti–PD-1 monotherapy (start of cycles 6 and following), we used principal components analysis (PCA) of manually gated flow cytometry data from non-naïve peripheral CD8 T cells (fig. S2, B and C). This analysis revealed an association between initial anti–PD-1 treatment and changes in Ki67+ and PD-1+CD39+ CD8 T cells, between concurrent JAKi plus anti–PD-1 therapy and CD127+ and CXCR5+ CD8 T cell populations, and a correlation between post-JAKi anti–PD-1 monotherapy and CD127+ and PD-1+ CD8 T cell populations (fig. S2, D to F). Temporal changes in the frequency of Ki67+, CD127+, and CXCR5+ CD8 T cells during therapy also revealed notable differences by response groups (Fig. 2C). For example, after initial anti–PD-1, an increase in Ki67+ CD8 T cells was observed in all response groups except JAKi.R patients. Conversely, an increase in CXCR5+ and CD127+ CD8 T cells was only observed in JAKi.R patients and occurred during the window of concurrent JAKi and anti–PD-1 (cycle 4). This increase observed in JAKi.R patients was then followed by a post-JAKi increase in Ki67+ CD8 T cells. Thus, changes in CD8 T cells expressing Ki67, PD-1, CXCR5, and/or CD127 distinguished both effects of JAKi treatment and patient response. To quantify changes in the composition of Ki67+ CD8 T cells during treatment, we first defined T cell states from non-naïve peripheral CD8 T cells. Unbiased clustering analysis revealed 12 clusters of circulating CD8 T cells (Fig. 2D and fig. S3A). Cluster 12 comprised Ki67+ PD-1hi CD39+ CD8 T cells that resembled known TEX intermediate populations detectable in the blood (denoted as PD-1hi TEX-INT-like Cl.12) (21–23). CD127 expression was predominantly confined to clusters 1 and 2 and accompanied by variable expression of CX3CR1, TCF1, and EOMES, identifying these clusters as resembling effector-memory like (TEFF/MEM-like) CD8 T cells. The proportion of proliferating CD127+ TEFF/MEM-like Cl.1+2 cells and Ki67+ PD-1hi TEX-INT-like Cl.12 cells changed in opposite directions during treatment. In JAKi.R patients but not patients in other response groups, the proportion of proliferating CD127+ TEFF/MEM-like Cl.1+2 cells increased, whereas that of Ki67+ PD-1hi TEX-INT-like Cl.12 cells decreased during the window of JAKi (Fig. 2E, top plot), a pattern also not observed in a separate cohort of NSCLC patients treated with anti–PD-1 monotherapy (fig. S3D). Furthermore, this reciprocal compositional change in proliferating Ki67+ CD8 T cells during the JAKi window also correlated with an increase in the PD-1+ CXCR5+ TPRE/PROG-like Cl.7 population (Fig. 2E, bottom). Indeed, trajectory analysis predicted differentiation paths connecting PD-1+ CXCR5+ TPRE/PROG-like Cl.7 cells with both CD127+ TEFF/MEM-like Cl.1+2 and PD-1hi TEX-INT-like Cl.12 populations (Fig. 2F and fig. S3E). Thus, these findings suggest that in JAKi.R patients, addition of JAKi increased the frequency of PD-1+ CXCR5+ CD8 T cells resembling fate-flexible precursor cells predicted to be developmentally related to the CD127+ TEFF/MEM-like and/or PD-1hi TEX-INT-like populations. JAKi may have impacted this relationship by rebalancing the expansion of CD127+ CD8 TEFF/MEM-like and PD-1hi TEX-INT-like populations.
Progenitor-like, effector-memory, and exhausted CD8 T cell clonotypes coordinately change after JAK inhibition
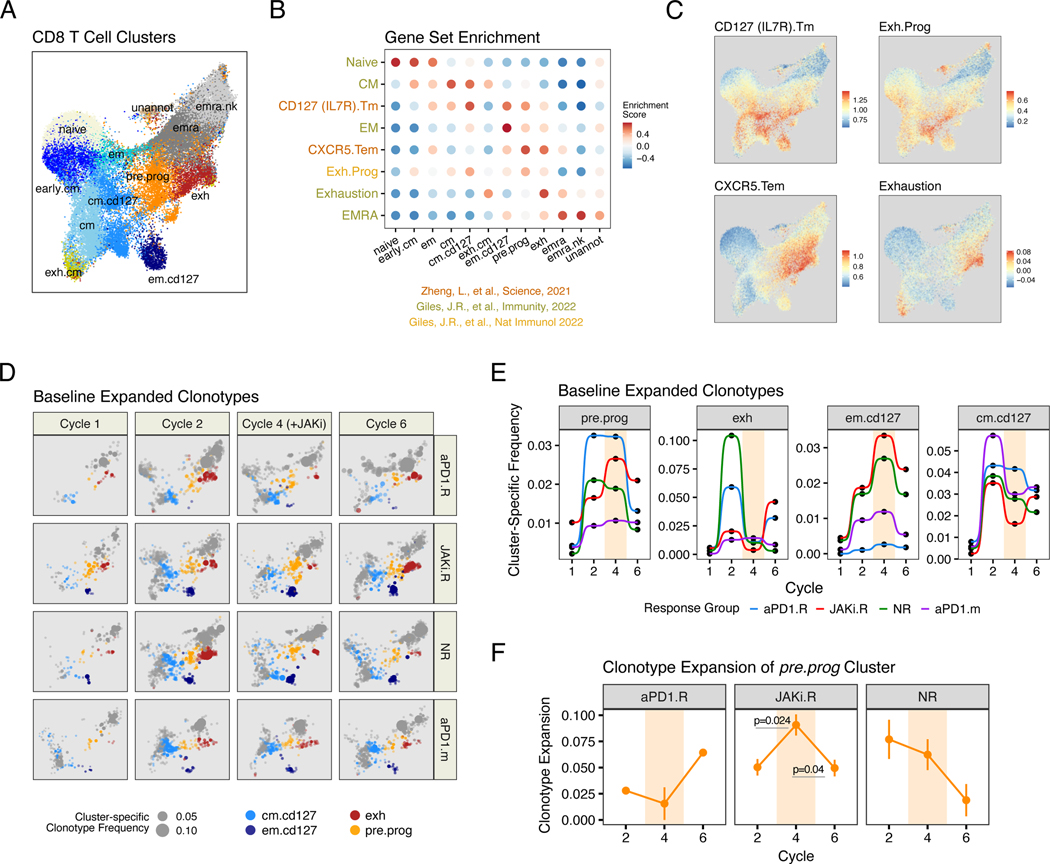
Because naïve T cells largely possess a distinctive T cell receptor (TCR), T cells that share the same TCR (clonotypes) are developmentally related. Thus, to more directly examine the impact of JAKi on T cell developmental relationships, we used single-cell RNA and TCR sequencing (scRNA/TCR-seq) on patients (n = 2 to 3) from each clinical response group at baseline (start of cycle 1), after anti–PD-1 (start of cycle 2), during concurrent JAKi and anti–PD-1 therapy (start of cycle 4), and after return to anti–PD-1 monotherapy (start of cycle 6). A reference CD8 T cell map for 40 samples was created and comprised 13 clusters annotated by expression of key genes and enrichment of CD8 T cell–atlas gene sets (10, 23, 29) (Fig. 3, A to C, and fig. S4A). Two clusters corresponding to naïve cells and a rare and poorly characterized population (unannotated) were not considered for downstream analysis. Using this annotated reference, we mapped CD8 T cells from all samples, keeping only T cells with a single TCR (a rearranged a and b chain) and a minimum annotation score [fig. S4B and supplementary materials (SM), materials and methods]. Using the clusters identified by scRNA/TCR-seq, we sought to approximate the relationships with clusters defined by flow cytometry—in particular, the PD-1hi TEX-INT-like Cl .12 cells, the CD127+ TEFF/MEM-like Cl.1+2 cells, and the PD-1+ CXCR5+ TPRE/PROG-like Cl.7 cells. A CD127 (IL7R) Tm gene set (10) enriched in several scRNA-seq clusters, including two memory and effector–memory-like clusters (cm.cd127 and em.cd127) likely approximating the CD127+ TEFF/MEM-like Cl.1+2 cells identified by flow cytometry. A cluster of TEX cells denoted exh was identified on the basis of enrichment for an Exhaustion gene set for CD8 T cell exhaustion (23), high expression of MKI67, and multiple TEX intermediate genes (e.g., GZMK, KLRG1, and CD38) that likely reflect recently proliferating PD-1hi TEX-INT-like Cl.12 cells identified by flow cytometry. A second rare cluster, denoted exh.cm, also modestly enriched for the Exhaustion gene set and for a CM gene set for central memory CD8 T cells. However, partly owing to sparsity of cells, low annotation scores (fig. S4B), and TCR sharing primarily with an unannotated cluster (fig. S5B), this cluster was not further examined. We next used two external gene sets to identify progenitor-like CD8 T cells: a CXCR5.Tem gene set for early effector-memory CD8 T cells and a Exh.Prog gene set for CD8 TEX progenitor cells. Indeed, both of these gene sets identified peripheral CD8 T cells from a previously described tumor T cell atlas (10) that are highly enriched for CXCR5 and TCF7 (fig. S4C). In this study, the CXCR5.Tem and Exh.Prog gene sets enriched in a cluster that we denoted pre.prog. This pre.prog cluster also had high expression of TCF7, CXCR5, GZMK, EOMES, IFIT1, and multiple genes that characterize previously described TEX progenitor cells (17, 25, 30, 31) (Fig. 3B and fig. S4A). This pre.prog cluster likely captures the fate-flexible PD-1+ CXCR5+ TPRE/PROG-like Cl.7 cells identified by flow cytometry, although cells that do not express CXCR5 transcript (fig. S4C) or protein (Fig. 2D) were also contained in this cluster. Other dysfunctional and/or terminal CD8 T cell subtypes were identified by using an EMRA gene set, including two EMRA subtypes (emra and emra.nk), with emra.nk expressing high levels of natural killer (NK) receptors. In our flow cytometry analysis, Ki67 was used to define CD8 T cells responding to anti–PD-1 and enriching for treatment-relevant T cells. Similarly, treatment-relevant T cell clonotypes were enriched by including only TCR clonotypes that expanded after therapy (fig. S4D and SM, materials and methods). This approach identified 5867 distinct clonotypes from 124,534 CD8 T cells. Changes in the cumulative frequencies of filtered clonotypes that belonged to pre.prog, exh, and the em.cd127 clusters approximated key patterns observed with PD-1+ CXCR5+ TPRE/PROG-like Cl.7, PD-1hi TEX-INT-like Cl.12, and CD127+ TEFF/MEM-like Cl.1+2 populations identified by flow cytometry (Fig. 3, D and E). Namely, JAKi.R patients had a blunted exh response at cycle 2 after anti–PD-1 and an increase in pre.prog and em.cd127 clonotypes during cycle 4 on JAKi. This pattern was followed by a post-JAKi increase in exh clonotypes that likely corresponded to the late CD8 T cell proliferative burst in JAKi.R patients (Fig. 2C). Neither aPD1.R patients nor patients from a separate NSCLC cohort treated with anti–PD-1 monotherapy (aPD-1.m) showed this pattern. Thus, as with the flow cytometry analysis, coordinated changes in expanded clonotypes from CD8 T cell populations that resemble PD-1+ CXCR5+ TPRE/PROG-like, CD127+ TEFF/MEM-like, and PD-1hi TEX-INT-like populations were linked to effects of adding JAKi to anti–PD-1 therapy.
Fig. 3. Evolution in CD8 T cell clonotypes after anti-PD1 immunotherapy and JAK inhibition correlates with treatment and response.
(A) UMAP of peripheral CD8 T cell subtypes analyzed by scRNA/TCR-seq from the start of cycles 1, 2, 4, and 6 from aPD1.R patients (n=2), JAKi.R patients (n=3), NR patients (n=3), and a separate cohort of patients treated with anti-PD1 monotherapy (aPD1.m) (n=2). (B) GSVA enrichment scores for each CD8 T cell cluster (x-axis) using the indicated CD8 T cell subtype gene set (y-axis). Source of the gene sets are indicated. (C) Enrichment scores for select gene sets overlaid on the UMAP from (A). (D) UMAP from (A) showing the frequency (expansion) of clonotypes belonging to the indicated color-coded CD8 T cell subtype (size of dot). Clonotypes that belonging to other subtypes are shown in grey. (E) Cumulative frequencies for expanded TCR clonotypes belonging to the indicated CD8 T cell subtype. Data for individual patients are pooled by treatment group. Cycles when JAKi was added to anti-PD1 are highlighted in beige.
(F) Clonotype expansion score (measuring degree of clonality) for pre.prog CD8 T cells from each indicated response group. Cycles when JAKi was added to anti-PD1 are highlighted. For longitudinal data, significance was determined by a repeated measures ANOVA using a mixed effect model and post-hoc interaction analysis. Error bars represent SEM.
Patient responses after JAK inhibition are associated with expanded CD8 T cell clonotypes with fate-flexible features
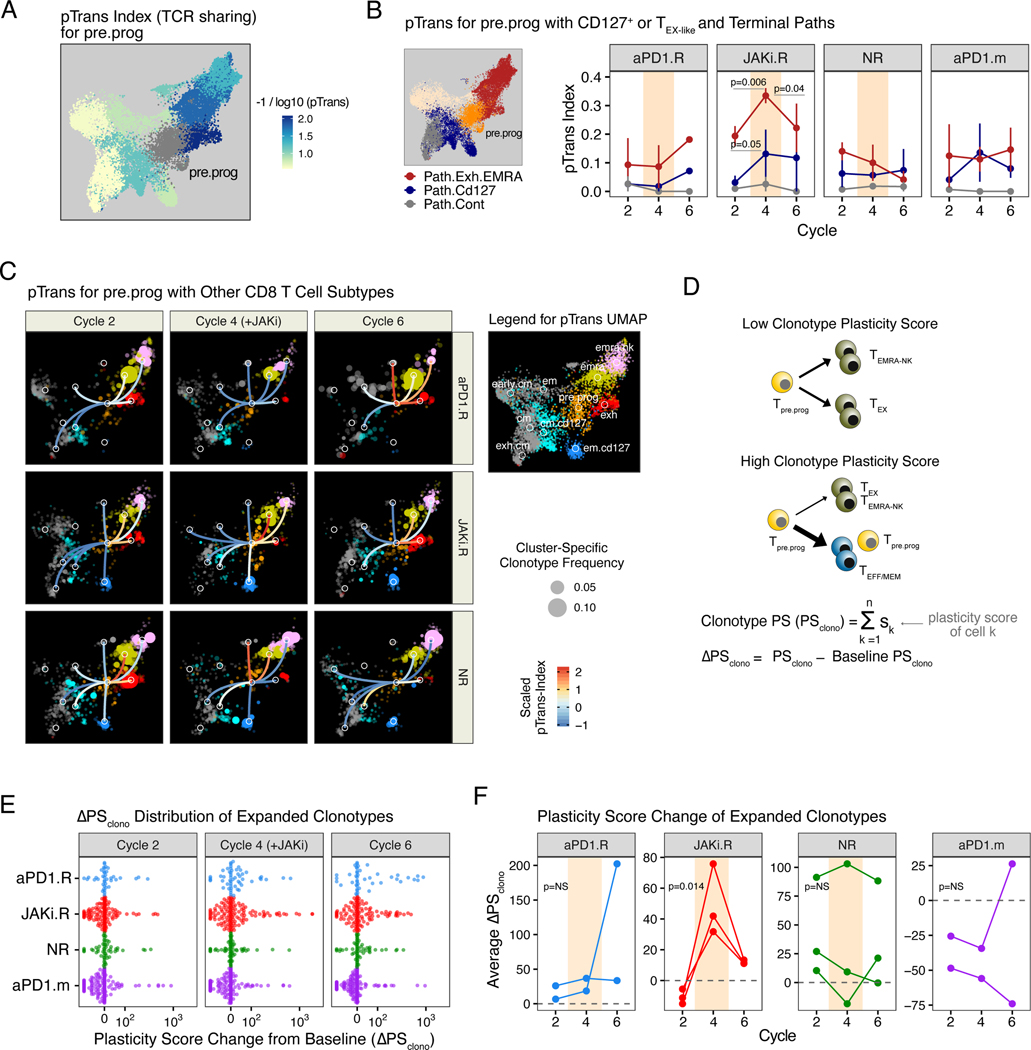
To begin assessing whether a developmental relationship between PD-1+ CXCR5+ TPRE/PROG-like, CD127+ TEFF/MEM-like, and PD-1hi TEX-INT-like CD8 T cells might be altered by JAKi, we examined clonotype expansion and TCR sharing between these populations. In JAKi.R patients who responded after the addition of JAKi to anti–PD-1, not only did JAKi result in an increase in the frequency of pre.prog clonotypes but also an increase in clonality (decreased clonal diversity) that was not observed in other patient response groups (Fig. 3F). This increase in clonality suggested that expansion of the pre.prog population may have been antigen-driven rather than a consequence of homeostatic proliferation or selective survival. To investigate whether this expansion of pre.prog clonotypes during JAKi that occurred together with an increased frequency in other clonotypes might be due to a developmental relationship, we used a previously described pairwise transition index (pTrans-index) (32) that measures the degree of TCR sharing between two T cell states. This analysis demonstrated that pre.prog CD8 T cells had a high degree of developmental relatedness with multiple different subtypes (Fig. 4A), which is consistent with pre.prog having fate-flexible properties. The degree of TCR sharing between pre.prog cells and either em.cd127 or cm.cd127 (Path.Cd127) increased after the addition of JAKi during cycle 4 in JAKi.R patients but not in other patient response groups (Figs. 4, B and C, middle rows). Simultaneously, the pTrans-index from pre.prog to exh or EMRA subtypes (Path.Exh.EMRA) also increased on JAKi but then decreased post-JAKi at cycle 6. No significant changes in the pTrans-index were observed with aPD1.R or NR patients or patients treated with anti–PD-1 monotherapy. Thus, these findings suggested that in JAKi.R patients, fate-flexible pre.prog CD8 T cells underwent specific expansion when JAKi was added, resulting in an increase in clonality and frequency. During JAKi treatment, these pre.prog clonotypes were developmentally linked to expansion of em.cd127 clones but also concomitantly gave rise to exh cells. These effects are consistent with JAKi impeding known functions of IFN-I signaling in driving progenitor differentiation toward downstream TEX subsets (9).
Fig. 4. Response to combined JAK inhibition and anti-PD1 immunotherapy is associated with alterations to CD8 T cell differentiation dynamics and clonotype plasticity.
(A) Pairwise transition (pTrans) index values (measuring TCR sharing) from the pre.prog subtype to other subtypes overlaid on the UMAP shown in Fig. 3A. (B) pTrans-index values between pre.prog CD8 T cells and either exh and terminal EMRA (emra, emra.nk) clusters (Path.Exh.EMRA, red), or em.cd127 and cm.cd127 clusters (Path.Cd127, blue). For comparison, results to the unrelated cm and exh.cm clusters (Path.Cont) are also shown. (C) pTrans-index values between the pre.prog subtype and other subtypes (legend in right margin) overlaid on a UMAP (from Fig. 3A) of expanded CD8 T cell clonotypes faceted by treatment cycle and response group. Edges connecting nodes from the pre.prog subtype to other subtypes are color-coded by the pTrans-index value (higher scores indicate greater TCR sharing and hence developmental relatedness, the absence of edges indicate no detectable sharing). Subtype-specific clonotype frequency is represented by dot size. (D) Schema and derivation of the plasticity scores (PS) using the pTrans-indices for each CD8 T cell subtype and the ΔPSclono using the difference of PS values from baseline. (E) ΔPSclono of all expanded clones colored by response group and faceted by treatment cycles. Positive ΔPSclono represents an altered clonotype subtype composition resulting from an increased plasticity (F) Mean ΔPSclono for patients in each of the indicated response groups. For longitudinal data, significance was determined by a repeated measures ANOVA using a mixed effect model and post-hoc interaction analysis. Error bars represent SEM.
Response after addition of a JAKi is linked to increased CD8 T cell plasticity
The ability of JAKi to impact the balance of progenitor CD8 T cell differentiation would be predicted to have broad consequences for the fate of individual CD8 T cell clonotypes after JAKi is added to anti–PD-1. Such an effect would result in changes in the subtype composition of individual clonotypes to include more nonterminal and nonexhausted T cell states. To test this hypothesis, we used the pTrans-index to derive a plasticity score (PS) for each CD8 T cell subtype. In this case, a subtype has a high PS if there is relatively even TCR sharing with other subtypes and T cell states, and it has a low PS if sharing is more restricted to only a few subtypes or states (Fig. 4D and fig. S5, A to C). By extension, the clonotype PS (PSclono) is the average of the PS values of all T cell subtypes that contribute to the composition of a clonotype. For each expanded clonotype from non-naïve CD8 T cells, we then determined the change in PSclono at the start of cycles 2, 4, and 6 compared with baseline (DPSclono). This analysis revealed an increase in DPSclono in multiple clonotypes during cycle 4 on JAKi and during cycle 6 post-JAKi in JAKi.R patients (Fig. 4, E and F). Neither aPD1.R patients, NR patients, nor the aPD-1.m patients from a separate cohort treated with anti–PD-1 monotherapy showed a significant difference in DPSclono across treatment cycles. For JAKi.R patients, restricting analysis to clonotypes shared with the pre.prog subtype provides specific examples of how changes in individual clonotype composition increased DPSclono after JAKi (fig. S5, D and E). Similarly, analysis of CD8 T cell clonotypes shared by the tumor and blood also provided examples of how JAKi possibly influenced the plasticity and evolution of tumor-relevant clonotypes (fig. S6). Thus, the ability of JAKi to potentially alter the balance of differentiation between PD-1+ CXCR5+ CD8 TPRE/PROG-like cells, CD127+ TEFF/MEM-like, and PD-1hi TEX-INT-like CD8 T cells may have broadly led to the evolution of CD8 T cells with less-committed and exhausted subtypes. This effect was particularly evident in JAKi.R patients, linking changes in CD8 T cell differentiation dynamics to clinical response after addition of JAKi to anti–PD-1.
Failure to respond to combined anti–PD-1 immunotherapy and JAK inhibition is coupled to persistent inflammation
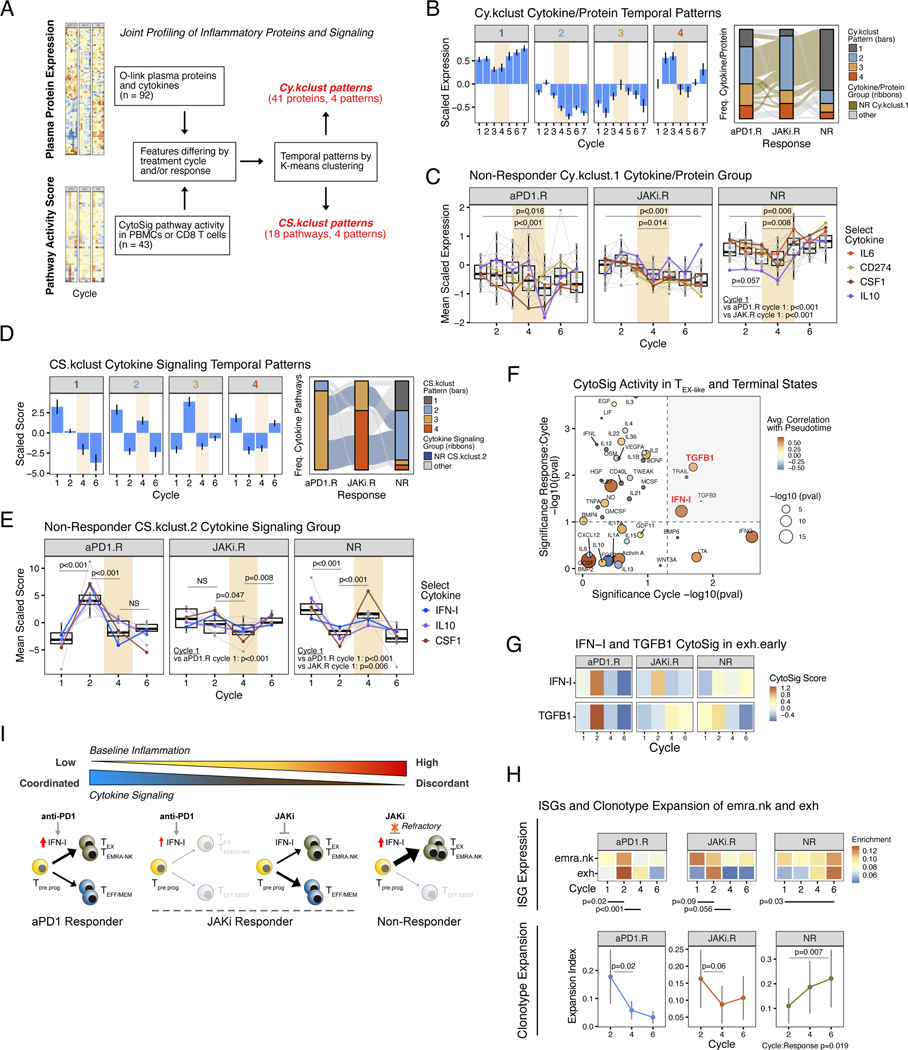
In contrast to patients who responded to anti–PD-1 and JAKi, NR patients showed marginal changes in features of CD8 T cell plasticity (Fig. 4, E and F), suggesting that NR patients may have been at least partially refractory to JAK inhibition. To investigate this notion, we profiled 92 plasma proteins by using Olink, together with 43 cytokine signaling pathways that we inferred from transcriptional changes in immune cells by using CytoSig (33) (Fig. 5A). For the plasma protein profiling, k-means clustering revealed four distinct temporal patterns (Cy.kclust) for 41 differentially expressed proteins (Fig. 5B, left bar plots; and fig. S7A). Compared with protein expression from aPD1.R and JAKi.R patients, that from NR patients, either group-averaged (fig. S7B) or examined across individual patients (fig. S7C), largely fell into pattern Cy.kclust.1 (Fig. 5B, gray stacked bar in right alluvial plot). Cy.kclust.1 proteins from NR patients included suppressive cytokines and proteins such as CD274, interleukin-6 (IL-6), IL-10, and CSF1, and they enriched for pathways such as IL-23 and IL-27 (fig. S7D) that have links to diseases of chronic inflammation (34). Expression of Cy.kclust.1 proteins from NR patients was higher at baseline than the other groups, modestly decreased on anti–PD-1, but then stably increased during the JAKi window (Fig. 5C). By contrast, most of these proteins took on a Cy.kclust.2 pattern in JAKi.R and aPD1.R patients (Fig. 5B, right-to-left path of tan-colored ribbons in right alluvial plot). Rather than having high baseline expression that transiently dipped and then increased during JAKi, these proteins in aPD1.R and JAKi.R patients had lower baseline expression that further decreased during and after JAKi (Fig. 5C). Thus, nonresponders displayed a specific pattern of high baseline levels of circulating inflammatory and immunoregulatory proteins that on average further increased during and after JAKi. To examine how persistently elevated cytokines might impact signaling in immune cells, we next assessed the activity of signaling pathways in peripheral blood mononuclear cells (PBMCs). As expected, this analysis revealed significant alterations in many of the signaling pathways known to use JAKs (JAK1, 2, and 3) after JAKi treatment (fig. S8A). Next, using only the signaling pathways that differed between response groups before or after JAKi (n = 18 CytoSig pathways), we again applied k-means clustering to determine temporal patterns (CS.kclust) (Fig. 5D, left bar plots; and fig. S8B). In NR patients, more than half of the signaling pathways belonged to CS.kclust.2 (Fig. 5D, blue stacked bar in right alluvial plot), which included IFN-I as well as suppressive cytokine pathways such as IL-10 and CSF1. Similarly to the expression of circulating inflammatory proteins, CS.kclust.2 signaling pathways from NR patients exhibited high relative baseline activity compared with their activity in aPD1.R and JAKi.R patients, decreased on anti–PD-1, but then increased during JAKi (Fig. 5E). By contrast, these pathways displayed a CS.kclust.3 pattern in aPD1.R patients and largely a CS.kclust.4 pattern in JAKi.R patients (Fig. 5D, right-to-left path of blue-colored ribbons in right alluvial plot). Specifically, in aPD1.R patients, NR CS.kclust.2 pathways had the lowest baseline activity that increased after anti–PD-1 and then decreased during JAKi. In JAKi.R patients, induction after anti–PD-1 was not apparent but instead observed post-JAKi (Fig. 5E). Thus, NR patients were characterized not only by high inflammation refractory to JAKi but also by immune signaling that appeared discordant and/or hyporesponsive to treatment.
Fig. 5. Refractoriness to JAK inhibition and persistent inflammation are associated with terminal CD8 T cell differentiation and therapy failure.
(A) Schema for joint profiling of plasma cytokines and immune signaling activity and their classification into temporal expression patterns. (B) Temporal expression patterns for plasma cytokines/proteins (Cy.kclust, left bar plots) along with their distribution in each response group (right alluvial plots). The ribbon in the alluvial plot is color-coded to track how Cy.kclust.1 cytokines/proteins from NR patients (tan-colored) change patterns in other response groups. (C) Expression of proteins/cytokines belonging to the Cy.kclust.1 pattern from NR patients (tan ribbon in alluvial plot from (B)) is shown for all response groups. Select suppressive cytokines are color-coded. P-values are for comparisons using all proteins/cytokines plotted (grey dots and lines). Cycles when JAKi was added to anti-PD1 are highlighted in bisque. (D) Temporal expression patterns for cytokine signaling activity in immune cells (CS.kclust, left bar plots) along with their distribution in each response groups (right alluvial plots). The ribbon in the alluvial plot is color-coded to track how CS.kclust.2 pathways from NR patients (blue-colored) change patterns in other response groups. (E) Activity score of cytokine pathways belonging to the CS.kclust.2 pattern from NR patients (blue ribbon in alluvial plot from (D)) is shown for all response groups. Select suppressive cytokines and IFN-I are color-coded. P-values are for comparisons using all pathways plotted (grey dots and lines). Cycles when JAKi was added to anti-PD1 are highlighted in bisque. (F) Cytokine pathway activity associated with CD8 T cell differentiation, treatment cycle, and response. Correlation of CD8 T cell pathway activity with pseudotime from trajectory analysis using all CD8 T cell subtypes are color-coded with circles size representing significance of the correlation (small grey dots are non-significant). The significance of changes in pathway activity in emra.nk and exh subtypes across treatment cycles (main effect) is shown on the x-axis, and the significance of whether changes across cycles differs by response group (interaction effect) is shown on the y-axis. Dotted lines represent significance levels (p=0.05 for main effect, p=0.10 for interaction effect) and grey upper-right quadrant show pathways that significantly differ by main and interaction effects. (G) Inferred CytoSig signaling activity for IFN-I and TGFB1 in exh CD8 T cells across treatment cycles. Shown are averages for each response group. Significance values are shown in (F). (H) Average ISG expression in terminal exh and emra.nk subtypes for each response group across treatment cycles (top). P-values for the indicated comparisons are shown. Also shown are the average expansion index (measure of clonality) for these subtypes (bottom). (I) Model summarizing relationship between inflammation, cytokine signaling in immune cells after anti-PD1, the impact of IFN-I on CD8 T cell differentiation toward either terminal or less committed states, and consequence of JAK inhibition. For longitudinal data, significance was determined by a repeated measures ANOVA using a mixed effect model and post-hoc interaction analysis. Error bars represent SEM.
IFN signaling that is refractory to JAK inhibition links progressive terminal CD8 T cell differentiation with treatment failure
Because IFN-I and other cytokines can promote differentiation of CD8 T cell progenitors (6) and because clinical response after JAKi was associated with enhanced features of CD8 T cell plasticity, the failure of JAKi to suppress inflammatory features in nonresponders might have contributed to progressive CD8 T cell exhaustion and/or terminal differentiation. Correlating all 43 CytoSig signaling pathway scores from CD8 T cells with pseudotime values from trajectory analysis (fig. S5A) revealed a strong positive correlation between IFN-I and IFN-g (IFNG) and the development of exh and emra.nk terminally differentiated CD8 T cell states (Fig. 5F, brown-hued dots). Indeed, IFN-I signaling (along with TGFB1) was among the few pathways that both significantly varied across treatment cycle and differed by response group (Fig. 5F, upper-right shaded quadrant; and Fig. 5G). Accordingly, ISGs were strongly induced on anti–PD-1 (cycle 2) in both emra.nk and exh CD8 T cell subtypes in aPD1.R patients, inconsistently increased in JAKi.R patients, but remained unchanged in NR patients (Fig. 5H, top; and fig. S9A). Only in NR patients did JAKi fail to decrease ISG expression (cycle 4), resulting in continuously high relative expression even post-JAKi. These differences in ISGs were corroborated by flow cytometry for ISG15 expression in PD-1hi and EMRA-like CD8 T cells (fig. S9B). In this analysis, NR patients had inconsistent blunting of ISG15 on JAKi and showed increasing expression post-JAKi, whereas patients belonging to the aPD1.R and JAKi.R groups showed significant decreases of ISG15 during JAKi. These persistently elevated ISG patterns in TEX-INT-like and EMRA with increased clonality in exh and emra.nk CD8 T cells (Fig. 5H, bottom), which is suggestive of progressive terminal differentiation in these populations. Moreover, the elevated ISG expression in such terminal CD8 T cells was associated with hyporesponsiveness to direct IFN-I stimulation in vitro compared with less-committed CD8 T cell subsets (fig. S9C), reminiscent of the overall immune signaling in NR patients. Thus, persistent IFN-I signaling and refractoriness to JAK inhibition may sustain progressive differentiation of CD8 T cells toward terminal and dysfunction states, contributing to therapy failure.
Discussion
The opposing functions of IFN rely on a temporal component that activates immune responses early but inhibits these responses at later times, especially in the setting of chronic inflammation. As shown here and in other preclinical studies (12), blocking the inhibitory function of IFNs to improve antitumor immunity is achievable in mice with delayed administration of JAKi or anti-IFNAR1 antibodies. We have now extended this concept to humans and demonstrated that in a phase 2 clinical trial for patients with metastatic NSCLC with tumor PD-L1 ≥50%, this therapeutic strategy in combination with anti–PD-1 is feasible and safe. Although proof of improved clinical efficacy from JAKi addition will require larger prospective randomized studies, the high response rates (67%) and long PFS (23.8 months) observed in this study encourage further clinical investigation. Additional goals for future clinical studies include investigating other settings amenable to improvement with JAKi (e.g., NSCLC with low PD-L1, other cancer types, and relapsed setting) and understanding properties of JAK inhibition required to complement immunotherapy (e.g., duration of treatment and JAKi selectivity). In this study, we identified immune features and immune cell types that distinguished the patients who had clinical response after addition of JAKi to anti–PD-1. These immune response characteristics include a subset of CD8 T cells that express CXCR5 and/or exhibit progenitor-like features. This population of cells is flexible in their developmental fate and can be modulated by JAKi. These CXCR5+ and progenitor-like CD8 T cells share similarities with memory precursor and exhausted progenitor CD8 T cells that can display a high degree of TCR sharing with multiple downstream subsets, express ISGs as part of an activation state, and circulate in the periphery (10, 24, 25). Blocking IFN-I signaling may impede the differentiation of such precursor or progenitor CD8 T cells toward terminal and exhausted states, promote fate flexibility during an immune response, and rebalance the proportion of nonexhausted to exhausted CD8 T cells to impact ICB efficacy. Besides the impact on CD8 T cells, JAK inhibition is also associated with the reprogramming of suppressive myeloid cells in anti–PD-1 refractory Hodgkin lymphoma patients who responded after the addition of the JAKi ruxolitinib to anti–PD-1, as shown in work published together with ours (35). Additional effects of JAKi on other CD8 T subsets—including more rare populations in the blood, or on other immune cells—cannot be excluded and will require further investigation. Furthermore, although the current findings together with previous studies (12) strongly implicate a role for impeding IFN signaling on the effects from JAK inhibition, JAKi can also impact signals downstream of many diverse cytokines. Indeed, the contribution of compensatory effects [e.g., the effect of IL-15 on CD8 T cell homeostatic proliferation (36)] and non-JAK cytokines [e.g., the effect of TGFB on CD8 T cell stemness (37)] will also be important to explore in future studies. Moreover, our previous work (7, 12) and recent genome-wide genetic screens in mice highlight a role for cancer-cell IFN signaling in ICB resistance (38), which is consistent with elevated levels of ISGs found in tumors from NSCLC patients that relapse after anti–PD-1 (2). Although detailed mechanisms are not yet clear, persistent IFN signaling in cancer cells can orchestrate immune suppression and impact CD8 T cell differentiation. Thus, some of the immune changes and improved ICB response evoked by JAK inhibition may indirectly result from inhibiting or preventing an IFN-driven resistant state in cancer cells. Whether JAKi can improve ICB may depend on levels and duration of baseline inflammation and how cytokine signaling pathways, including stimulatory and inhibitory pathways, are altered by chronic inflammation (Fig. 5I). The cellular consequences of chronic IFN and other cytokine pathways may involve changes in signaling thresholds, an altered balance between positive and negative regulators, changes in the composition of actively signaling cells, and/or inflexible epigenetic states leading to long-lasting changes in signaling behavior in tumor-specific T cells, other immune cells, or cancer cells (8, 39, 40). Indeed, immune cells from patients that did not respond in our trial appeared overtly refractory to JAKi or exhibited discordant signaling behavior that may have resulted from at least some of these features. A deeper understanding of how chronic inflammation alters cell behavior will provide insight into context-dependent differences in IFN and other cytokine pathways important for ICB response (41). This understanding may inform additional therapeutic strategies to “reset” signaling pathways in immune cells, cancer cells, or other cell types important for antitumor immunity.
Materials and methods
Clinical trial
The clinical trial (ClinicalTrials.gov identifier: NCT03425006) was approved by the institutional review board at the University of Pennsylvania and was completed in accordance with international standards of good clinical practice. All patients provided written informed consent at the time of enrollment. Patients with metastatic NSCLC who were treatment-naive with PD-L1 ≥50% by TPS PD-L1 IHC 22C3 pharmDx assay (Dako North America), eastern cooperative group performance status (ECOG PS) 0–1, RECIST v 1.1 measurable disease amenable to biopsy, and no untreated brain metastases were enrolled (n = 31 screened). Patients received pembrolizumab (200 mg every 21 days) and itacitinib 200 mg daily by mouth was started on cycle 3 day 1 of pembrolizumab and continued for 6 weeks. Of the 31 patients screened, 23 were enrolled between 16 October 2018 and 4 March 2021 and received at least 1 cycle of pembrolizumab. Primary endpoints were: (i) ORR determined by RECIST v1.1 PR and CR at 12 weeks, and (ii) toxicity of pembrolizumab and itacitinib by common terminology criteria for adverse events (CTCAE) v5.0 (table S3). Best ORR was defined as the best response determined by RECIST v1.1 over the study period. Secondary clinical objectives included PFS and overall survival (OS) from initiation of study therapy until disease progression or death due to any cause, respectively. DOR was defined as time from first response at 12 weeks until disease progression by RECIST v1.1. Response at 6 weeks was defined using a modified RECIST v1.1, allowing for changes of near −30% reduction to also be included as a PR. This resulted in a clear demarcation between responders (range: −27.3 to −61%) and nonresponders (range: −19.5% to 150%). Patients were censored at the date of last follow up if they came off trial for reasons other than progression or death and at data cutoff (1 December 2021) for those on trial without disease progression or death. Median PFS, DOR, OS were estimated using Kaplan-Meier methodology. Paired blood and tissue samples were collected for several translational and exploratory objectives.
Cell lines
The Res 499 melanoma cell line was derived from a B16-F10 melanoma tumor that relapsed after ICB-based immunotherapy and acquired elevated expression of a subset of IFS. This cell line was cultured as previously described (12) and is available upon request.
Mice
Mice were maintained in a specific-pathogen free facility at the University of Pennsylvania. Experiments and procedures were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania under protocol #803042. Five- to seven-weeks-old female C57BL/6 mice were obtained from Charles River Production. For ICB studies, 50,000 Res 499 cells were mixed 1:1 with reduced growth factor basement membrane extract (BME type 2) and injected into the hind limb of mice. Mice were randomized at tumor injection and then assigned to treatment groups with five or more mice per treatment group. Caliper measurements were started when palpable tumors were observed at approximately day 11. Mice with ulcerated tumors were censored. Endpoint for survival studies were a tumor size of 15 mm or greater in any dimension. Three independent experiments were performed.
In vivo mouse lymphocyte studies. Tumors, spleens, and dLNs were harvested at day 16 post-tumor implantation. For spleens and dLNs, single-cell suspensions were prepared after RBC lysis with ACK Lysis Buffer (Life Technologies). Tumors were weighed prior to enzymatic digestion with Type 4 collagenase and DNAse 1 at 1mg/ml. After enzymatic digestion or ACK Lysis, all tissues were filtered through 100 mm filters. Cells were stained with Fc Block and Zombie Live/Dead stain for 10 min prior to surface staining. Surface staining was done for 30 min at room temperature. Samples were fixed and permeabilized by incubating in 100 ml of Fix/Perm buffer at room temperature for 30 min and washed in Perm Buffer. Intracellular stains were performed overnight at 4°C. Cell counting beads were spiked into each sample prior to data acquisition. Data acquisition was done on a FACSymphony A5 and analyzed using OMIQ and/or R. See table S5 and table S7 for list of antibodies and buffers.
Human antibody panels and staining
Approximately 1×106 to 5×106 frozen PBMCs were used per patient per timepoint. PBMCs were thawed into 10% complete RPMI with DNase. Cells were stained with live/dead and Fc block for 10 min at room temperature. Chemokine receptors were stained at 37°C for 20 min followed immediately by surface staining for 30 min at room temperature. Samples were fixed and permeabilized by incubating in 100 ml of Fix/Perm buffer at room temperature for 30 min and washed in Perm Buffer. Intracellular stains were performed overnight at 4°C. Data acquisition was done on a FACSymphony A5 and analyzed using OMIQ and/or R. See table S4 and table S7 for list of antibodies and buffers.
Flow cytometry feature clustering
For quantification and statistical analysis of flow cytometry data, the flowCore and flow-Workspace R packages and custom R analysis pipelines were used. A down-sampled feature matrix was then created by equal random sampling of cells from each FCS file. This downsampled data were then used for dimensionality reduction by UMAP, as implemented in the umap R package, and clustering by self-organizing maps, as implemented in the FlowSOM R package. The number of clusters (K) was determined using random forest with stratified sampling, as implemented in the randomForestSRC R package, to estimate an out-of-bag overall prediction error rate for a range of K values. Then, a value for K was selected to maximize the number of clusters, while keeping the out-of-bag error rate to approximately 10% or below. Using this reference map, a random forest classifier was then trained and used to predict cluster membership for all cells for all samples.
Olink
Cytokines were measured from EDTA-plasma using the Olink Extension Assay (PEA) to measure 92 unique analytes. In brief, oligonucleotide-labeled capture antibodies were used to bind target cytokines and subsequently hybridized. The hybridized product was then amplified and measured by qPCR, allowing for parallel detection of 92 cytokines within the same sample.
pSTAT1 detection
Healthy donor PBMCs were plated overnight in RPMI at 1×106 cells per 100 ul. The next day, PBMCs were stimulated with 20ng/ml IFNa2 (Biolegend #592704) for 15 min. Reactions were stopped with a final concentration of 2% PFA and kept in methanol O/N at −80°C. The following day, cells were washed with PBS and stained with a pSTAT detection panel for 1 hour at room temperature. Data acquisition was done on a FACSymphony A5 and analyzed using OMIQ. See table S6 for list of all antibodies.
Single-cell RNA and TCR sequencing and processing
PBMCs from select patients were sorted for total live cells or live CD8+ cells on a BD FACs Aria II. Cells were sorted into GEMs using a 10x Chromium Controller and were made into libraries following the Chromium Next GEM Single Cell 5′ Reagent Kits v2 (Dual Index) Protocol. Libraries were sequenced using a NovaSeq 6000. Sequencing data was processed using the CellRanger pipeline v5 (10x Genomics). BCL files were converted to FASTQ and aligned to the human genome (GRCh38) to generate count matrices. For TCR libraries, BCL files were converted to FASTQ and the CellRanger VDJ pipeline was used for sequence assembly and clonotype calling. Cells with >10% mitochondrial DNA and/or <200 or >2500 RNA features were filtered out and conditions were integrated using Seurat v3.2.0. UMI barcodes were used to combine cell expression data with clonotype data.
Human CD8 T cell reference mapping and annotation
Using the Seurat objects for scRNA/TCR-sequencing data from patient PBMC-sorted CD8 T cells from all study timepoints (start of cycles 1, 2, 4, 6), 1000 cells with a rearranged TCR were randomly sampled and a separate Seurat object was created. Processing to filter out cells with high mitochondrial DNA and variations in UMI count were carried out as described. Data were then log normalized, variable features identified, scaled, and integrated using the RPCA reduction method from Seurat with k.anchors = 20. Predicted cell doublets were identified with the DoubletFinder R package (42). After dimensionality reduction by PCA and UMAP, cell clustering was performed by shared nearest-neighbor. Very sparse clusters or clusters consisting primarily of predicted doublets were removed along with any remaining predicted cell doublets. Clusters not considered to be independent CD8 T cell subtypes but rather activation states (10) (e.g., clusters characterized by high ISG expression) were merged with the subtype most resembling it by gene set enrichment and marker gene expression. This resulted in a CD8 T cell reference consisting of approximately 20,000 cells. After re-clustering, the reference was annotated using marker gene expression and enrichment of CD8 T cell subtype gensets by GSVA (described below). This annotated CD8 T cell reference was then used to map CD8 T cells from scRNA/TCR-sequencing of (i) CD8 T cells sorted from PBMCs, and (ii) matching PBMCs from the same patient and timepoint. To extract CD8 T cells from the latter sample type (the CD8 T cells from scRNA/TCR-sequenced PBMCs), the PBMC data were first mapped to the Azimuth Human PBMC reference. CD8 T cells identified from the level one predicted cell type annotation were extracted to create a separate Seurat object that was then merged with the Seurat object of sorted CD8 T cells from the same sample (i.e., same patient and same timepoint). All mapping of cells to reference maps was performed using MapQuery from Seurat.
Gene set enrichment and scores
Gene sets for human CD8 T cells from PBMCs and from a human pan-cancer T cell atlas were used to aid in cluster annotation and analysis (10, 23, 29). For the human PBMC CD8 T cell gene sets, the top 250 genes from each set were used. For the CD8 T cell gene sets from the human pan-cancer T cell atlas, the top 100 genes or all the genes in the gene set were used, whichever was smaller. The scRNA-sequencing data from all cells in a cluster were averaged to create pseudo-bulk data. Then, an enrichment score for each gene set of interest was determined using GSVA. To determine gene set scores for individual cells, the AddModuleScore function from Seurat was used.
Pseudotime trajectory analysis
Pseudotime trajectory analysis was carried out using Monocle 3. The integrated gene expression data from the CD8 T cell reference map were used as ordering genes to construct pseudotime trajectories. The naive CD8 T cell cluster was selected as a starting root state. Assigned pseudotime values for each cell were then used in downstream analysis.
CD8 T cell clonotype filtering and analysis
To enrich for well-annotated and treatment-relevant CD8 T cells and clonotypes, a set of filtering criteria were applied to each sample. These criteria included only keeping CD8 T cells with: (i) a single rearranged TCR comprised of a single TCR a and TCR b chain, (ii) predicted singlet by DoubletFinder (42), and (iii) reference mapping annotation score >0.50. CD8 T cells that met these criteria were then assigned to a clonotype using the amino acid sequences of the CDR3 region of TCR alpha and beta. Then, filtering criteria for clonotypes were used to keep only assigned clon otypes that: (i) were not exclusive to naive CD8 T cells, (ii) had a relative frequency in the blood of at least 0.0005, and (iii) increased in frequency above baseline by at least twofold. These filtering criteria were guided by examination of CD8 T cells from PBMCs from two healthy donors collected at two timepoints separated by 3 weeks. When these filtering criteria were applied to healthy donors, greater than 98% of clonotypes were excluded. To examine clonotypes shared by the tumor and the blood, the CDR3 of the TCR beta chain from Adaptive TCR sequencing was matched to the TCR beta chain from 10X Genomics sequencing with no requirement for a minimum frequency in the blood or increase above baseline.
TCR sharing, clonotype plasticity, and expansion
Assessment of clonotype sharing between two CD8 T cell states was calculated using Startrac (32) to determine the pairwise transition index (pTrans-index). The PS for each CD8 T cell state or cluster (representing a subtype or activation state) was then defined as the reciprocal of the variance of its pTrans-index with each of the other CD8 T cell states. A clonotype plasticity score (PSclono) was defined as the average of the PS for all CD8 T cell states comprising the clonotype. The ΔPSclono is the difference between the PSclono at a given seven-treatment cycle from the PSclono at baseline (start of cycle 1). To assess clonotype expansion, the expansion index from Startrac (32) was used.
Cytokine signaling activity score
The cytokine signaling activity of immune cells from scRNA-sequencing data was predicted using CytoSig (33). The scRNA-sequencing data from all cells in a cluster were averaged to create pseudo-bulk data. This was then used with the CytoSig Python package to return the predicted cytokine signaling activity scores for different immune cells or CD8 T cell subtypes.
Hierarchical and k-means clustering
Temporal patterns for plasma proteins and CytoSig activity values were examined using k-means clustering. We first rescaled the data matrix using the unitize function from the thresher R package. Then, we used k-means clustering as implemented in the stats R package. To choose K, or the number of clusters for k-means clustering, we plotted the within-cluster sum of squares as a function of K and selected K based on the “elbow” of the corresponding plot. For hierarchical clustering of gene or protein expression data, a Euclidean distance was used.
Statistical analysis
The significance of changes across treatment cycle for plasma protein levels (Olink), and cytokine signaling activity scores (CytoSig) were examined by a repeated measures ANOVA using a mixed-effect model implemented in the nlme R package. If the main effect was significant or near-significant, post-hoc interaction analysis was used to determine within-group or between-group differences using the phia R package. Z-scores, scaled, or normalized values were used for the models. For frequency data, such as CD8 T cell frequencies (flow cytometry) or clonotype frequencies (scRNA/TCR-seq), beta regression was used as implemented in the betareg R package. For compositional data, such as the proportional composition of Ki67+ CD8 T cells, Dirichlet regression was used as implemented in the DirichletReg R package. Differences in mouse tumor growth were determined using a mixed-effect regression model typically using a log-normal distribution (determined by inspection of data using Q-Q plot) using the MASS R package. Time points after significant death or study endpoint events had occurred were not considered. For survival analysis, the Kaplan-Meier estimate and a log-rank test from the survival R package were used. Median follow-up times were calculated using the prodlim R package and the reverse Kaplan-Meier method. For simple two-group comparisons, a two-sided Wilcoxon test or t test was used for nonparametric or parametric data, respectively. For multiple groups, a Kruskal-Wallis or ANOVA test was used along with Tukey HSD for post-hoc testing. Normality was assessed using a Shapiro’s test. The FactoMineR R package was used for PCA.
Supplementary Material
References
- 1.Reck M. et al. , Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 375, 1823–1833 (2016). doi: 10.1056/NEJMoa1606774; pmid: 27718847 [DOI] [PubMed] [Google Scholar]
- 2.Memon D. et al. , Clinical and molecular features of acquired resistance to immunotherapy in non-small cell lung cancer. Cancer Cell 42, 209–224.e9 (2024). doi: 10.1016/j.ccell.2023.12.013; pmid: 38215748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng CT et al. , Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe 17, 653–661 (2015). doi: 10.1016/j.chom.2015.04.005; pmid: 25974304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson EB et al. , Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207 (2013). doi: 10.1126/science.1235208; pmid: 23580528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teijaro JR et al. , Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211 (2013). doi: 10.1126/science.1235214; pmid: 23580529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu T. et al. , The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016). doi: 10.1126/sciimmunol.aai8593; pmid: 28018990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benci JL et al. , Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell 178, 933–948.e14 (2019). doi: 10.1016/j.cell.2019.07.019; pmid: 31398344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu J. et al. , Cancer cells resistant to immune checkpoint blockade acquire interferon-associated epigenetic memory to sustain T cell dysfunction. Nat. Cancer 4, 43–61 (2023). doi: 10.1038/s43018-022-00490-y; pmid: 36646856 [DOI] [PubMed] [Google Scholar]
- 9.Chen W. et al. , Chronic type I interferon signaling promotes lipid-peroxidation-driven terminal CD8+ T cell exhaustion and curtails anti-PD-1 efficacy. Cell Rep. 41, 111647 (2022). doi: 10.1016/j.celrep.2022.111647; pmid: 36384131 [DOI] [PubMed] [Google Scholar]
- 10.Zheng L. et al. , Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021). doi: 10.1126/science.abe6474; pmid: 34914499 [DOI] [PubMed] [Google Scholar]
- 11.Song E, Chow RD, Mutations in IFN-g signaling genes sensitize tumors to immune checkpoint blockade. Cancer Cell 41, 651–652 (2023). doi: 10.1016/j.ccell.2023.02.013; pmid: 36931275 [DOI] [PubMed] [Google Scholar]
- 12.Benci JL et al. , Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 167, 1540–1554.e12 (2016). doi: 10.1016/j.cell.2016.11.022; pmid: 27912061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twyman-Saint Victor C. et al. , Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377 (2015). doi: 10.1038/nature14292; pmid: 25754329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minn AJ, Wherry EJ, Combination Cancer Therapies with Immune Checkpoint Blockade: Convergence on Interferon Signaling. Cell 165, 272–275 (2016). doi: 10.1016/j.cell.2016.03.031; pmid: 27058661 [DOI] [PubMed] [Google Scholar]
- 15.Hudson WH et al. , Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity 51, 1043–1058.e4 (2019). doi: 10.1016/j.immuni.2019.11.002; pmid: 31810882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zander R. et al. , CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042.e4 (2019). doi: 10.1016/j.immuni.2019.10.009; pmid: 31810883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltra JC et al. , Developmental Relationships of Four Exhausted CD8+ T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841.e8 (2020). doi: 10.1016/j.immuni.2020.04.014; pmid: 32396847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilar EJ et al. , Outcomes to first-line pembrolizumab in patients with non-small-cell lung cancer and very high PD-L1 expression. Ann. Oncol. 30, 1653–1659 (2019). doi: 10.1093/annonc/mdz288; pmid: 31435660 [DOI] [PubMed] [Google Scholar]
- 19.Mok TSK et al. , Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised, open-label, controlled, phase 3 trial. Lancet 393, 1819–1830 (2019). doi: 10.1016/S0140-6736(18)32409-7; pmid: 30955977 [DOI] [PubMed] [Google Scholar]
- 20.Kamphorst AO et al. , Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl. Acad. Sci. U.S.A. 114, 4993–4998 (2017). doi: 10.1073/pnas.1705327114; pmid: 28446615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang AC et al. , A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med. 25, 454–461 (2019). doi: 10.1038/s41591-019-0357-y; pmid: 30804515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang AC et al. , T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). doi: 10.1038/nature22079; pmid: 28397821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giles JR et al. , Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 55, 557–574.e7 (2022). doi: 10.1016/j.immuni.2022.02.004; pmid: 35263570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He R. et al. , Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428 (2016). doi: 10.1038/nature19317; pmid: 27501245 [DOI] [PubMed] [Google Scholar]
- 25.Im SJ et al. , Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). doi: 10.1038/nature19330; pmid: 27501248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gueguen P. et al. , Contribution of resident and circulating precursors to tumor-infiltrating CD8+ T cell populations in lung cancer. Sci. Immunol. 6, eabd5778 (2021). doi: 10.1126/sciimmunol.abd5778; pmid: 33514641 [DOI] [PubMed] [Google Scholar]
- 27.Miller BC et al. , Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019). doi: 10.1038/s41590-019-0312-6; pmid: 30778252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnnidis JB et al. , Inhibitory signaling sustains a distinct early memory CD8+ T cell precursor that is resistant to DNA damage. Sci. Immunol. 6, eabe3702 (2021). doi: 10.1126/sciimmunol.abe3702; pmid: 33452106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giles JR et al. , Shared and distinct biological circuits in effector, memory and exhausted CD8+ T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat. Immunol. 23, 1600–1613 (2022). doi: 10.1038/s41590-022-01338-4; pmid: 36271148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jadhav RR et al. , Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl. Acad. Sci. U.S.A. 116, 14113–14118 (2019). doi: 10.1073/pnas.1903520116; pmid: 31227606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B. et al. , Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer 3, 108–121 (2022). doi: 10.1038/s43018-021-00292-8; pmid: 35121991 [DOI] [PubMed] [Google Scholar]
- 32.Zhang L. et al. , Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). doi: 10.1038/s41586-018-0694-x; pmid: 30479382 [DOI] [PubMed] [Google Scholar]
- 33.Jiang P. et al. , Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat. Methods 18, 1181–1191 (2021). doi: 10.1038/s41592-021-01274-5; pmid: 34594031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tait Wojno ED, Hunter CA, Stumhofer JS, The immunobiology of the Interleukin-12 family: Room for discovery. Immunity 50, 851–870 (2019). doi: 10.1016/j.immuni.2019.03.011; pmid: 30995503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zak J. et al. , JAK inhibition enhances checkpoint blockade immunotherapy in patients with Hodgkin lymphoma. Science 384, eade8520 (2024). doi: 10.1126/science.ade8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprent J, Zhang X, Sun S, Tough D, T-cell proliferation in vivo and the role of cytokines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 317–322 (2000). doi: 10.1098/rstb.2000.0568; pmid: 10794049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y. et al. , TGF-b regulates the stem-like state of PD-1+ TCF-1+ virus-specific CD8 T cells during chronic infection. J. Exp. Med. 219, e20211574 (2022). doi: 10.1084/jem.20211574; pmid: 35980386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubrot J. et al. , In vivo CRISPR screens reveal the landscape of immune evasion pathways across cancer. Nat. Immunol. 23, 1495–1506 (2022). doi: 10.1038/s41590-022-01315-x; pmid: 36151395 [DOI] [PubMed] [Google Scholar]
- 39.Philip M. et al. , Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017). doi: 10.1038/nature22367; pmid: 28514453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamada R. et al. , Interferon stimulation creates chromatin marks and establishes transcriptional memory. Proc. Natl. Acad. Sci. U.S.A. 115, E9162–E9171 (2018). doi: 10.1073/pnas.1720930115; pmid: 30201712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaretsky JM et al. , Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 375, 819–829 (2016). doi: 10.1056/NEJMoa1604958; pmid: 27433843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGinnis CS, Murrow LM, Gartner ZJ, DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 8, 329–337.e4 (2019). doi: 10.1016/j.cels.2019.03.003; pmid: 30954475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.