Summary
Abnormal epigenetics is the initial factor of the occurrence and development of osteoarthritis (OA), and abnormal mechanical load is a key pathogenic factor of OA. However, how abnormal mechanical load affects chondrocyte epigenetics is unclear. Chondrocytes reportedly respond to mechanics through the extracellular matrix (ECM), which has a role in regulating epigenetics in various diseases, and mitochondria are potential mediators of communication between mechanics and epigenetics. Therefore, it is hypothesized that the matrix mechanics of cartilage regulates their epigenetics through mitochondria and leads to OA. The matrix stiffness of OA cartilage on the stress-concentrated side increases, mitochondrial damage of chondrocyte is severe, and the chondrocyte H3K27me3 is demethylated. Moreover, mitochondrial permeability transition pore (mPTP) opens to increase the translocation of plant homeodomain finger protein 8 (Phf8) into the nucleus to catalyze H3K27me3 demethylation. This provides a new perspective for us to understand the mechanism of OA based on mechanobiology.
Subject areas: mechanobiology, epigenetics, cell biology
Graphical abstract

Highlights
-
•
Cartilage degeneration and mitochondrial damage correlated with matrix stiffness
-
•
Matrix stiffness affects epigenetics of chondrocytes by PHD finger protein 8 (Phf8)
-
•
H3K27me3 regulates the expression of matrix catabolism and apoptosis genes
Mechanobiology; Epigenetics; Cell biology.
Introduction
Osteoarthritis (OA) is a joint disease caused by abnormal mechanical stimulation.1,2 Abnormal mechanical loading leads to chondrocyte apoptosis or senescence.3 The mechanical microenvironment can activate Piezo1 and induce mitochondrial calcium uptake and oxidative phosphorylation (OXPHOS) by increasing cyclic AMP (cAMP) production.3,4 The mechanical properties of the extracellular matrix (ECM) affect chondrocyte metabolism through mechanotransduction.5,6,7,8 The mechanical environment of chondrocytes is highly dependent on the biomechanical properties of the ECM, and the stiffness of the ECM changes substantially during OA progression.9,10 The Young’s modulus of articular cartilage ECM in healthy mice ranges from 22.3 ± 1.5 kPa to 89.9 ± 2.8 kPa.11,12 The top-down calcification process begins with spherical mineral particle formation in the joint surface during the early stage of OA, followed by fiber formation and densely packed material transformation deep into the cartilage during advanced OA.12,13 As organic components of the cartilage matrix are replaced by inorganic minerals and the fibers that make up the ECM thicken and twist on the surface layer of cartilage,14 changes occur in its mechanical properties, making the articular cartilage harder and thinner; the matrix stiffness of OA cartilage is three times higher than that of normal cartilage.12
Mechanical stimulation plays an important role in intracellular biological reactions by affecting mitochondrial function.15 The shape and function of chondrocyte mitochondria become altered under mechanical stimulation.16 Moreover, excessive mechanical stimulation induces mitochondrial damage, subsequently leading to chondrocyte damage.15,16 Increasing evidence suggests that mitochondrial dysfunction promotes OA progression through myriad of mechanisms, including energy metabolism, oxidative stress, inflammatory response, cell senescence and death, and calcium homeostasis.16,17
Mitochondria communicate with the nucleus through metabolites or stress signals, leading to various epigenetic changes and causing diseases.18 Many OA risk factors are epigenetically related molecules and interact with noncoding regions of the genome and increase OA risk by regulating target gene expression.16,19 Moreover, various metabolites produced by mitochondria are associated with epigenetic modification of chromatin. For instance, s-adenosylmethionine is indispensable for histone methylation and alpha-ketoglutarate, which is the cofactor of the histone demethylase Jmjd family and is the key product in the tricarboxylic acid cycle (TCA) cycle.20 Mechanical stimulation triggers the release of Ca2+ in the endoplasmic reticulum (ER),16 thus causing mitochondrial calcium overload.17 Mitochondrial function is affected by its dynamics, which are regulated by dynamics-related proteins.21 Mitochondrial damage affects chondrocyte function through energy metabolism transformation and nuclear gene expression via epigenetic regulation.18,19 When mitochondrial membrane permeability increases, mitochondrial permeability transition pore (mPTP) opening and reactive oxygen species (ROS) accumulation lead to Phf8 upregulation, which leads to H3K27me3 demethylation.22
We hypothesize that matrix stiffness can promote OA progression through chondrocyte epigenetic modification (H3K27me3 demethylation), and mitochondria play an important role in mechanical conduction regulating chondrocyte epigenetics under different mechanical conditions. Matrix stiffness affects the mitochondrial function of chondrocytes and regulates epigenetics through mitochondria, so the potential mechanism of matrix stiffness as an epigenetic regulatory factor for OA should be explored. To this end, by simulating the response of ATDC5 chondrocytes to different matrix stiffnesses and tracking the fates and functions of mitochondria and mitochondrial-to-nuclear (mito-nuclear) communication, we propose a potential mechanism by which mechanical stimulation regulates H3K27me3 methylation after mitochondrial damage.
Results
ATDC5 chondrocytes respond to matrix stiffness through ER/Mito interactions
The Young’s modulus of articular cartilage ECM in healthy mice ranges from 22.3 ± 1.5 kPa to 89.9 ± 2.8 kPa.11,12 To simulate different ECM stiffnesses of chondrocytes, Young’s modulus of the three groups of Gelatin Methacryloyl (GelMA) was set to 60, 120, and 180 kPa through microindentation testing (Figure 1A). ATDC5 chondrocytes adhered to the surface without obvious cytotoxicity (Figure 1B). The effects of matrix stiffness on mechanotransduction of ATDC5 chondrocytes were further evaluated. Phosphorylation levels of focal adhesion kinase (FAK) and myosin light chain 2 (MLC2) were both upregulated with the increasing stiffness (Figures 1C–1E). At the same time, increased matrix stiffness led to increased Ca2+ inflow (Figure 1F). Intracellular Ca2+ is mainly stored in the ER, and mitochondria must contact the ER and accumulate high concentrations of Ca2+ in the local domain structure to promote Ca2+ uptake. Moreover, Ca2+ release from the ER follows the rule of “calcium release induced by calcium” in somatic cells.23 The expression of ER stress-related genes was increased, including transcription factor 6 (Atf6) and glucose-regulated protein 94 kDa (Grp94), while the Ca2+ release channels inositol-1,4,5-triphosphate receptor (Ip3r) and ryanodine receptor (Ryr), located on the ER membrane, were upregulated with increasing stiffness (Figure 1G). All RT-qPCR primers are in Table 1. ER and mito-trackers were used to explore the interaction of mitochondria and ER. Both colocalized with an increase in high stiffness (Figure 1H). Ip3r is the initiation factor of Ca2+ release from the ER, and 2-aminoethoxydiphenyl borate (2-APB) is a specific inhibitor of Ip3r. The level of Ca2+ was significantly reduced after the application of 2-APB at 180 kPa stiffness (Figure 1I). These results indicated that different stiffness surfaces induce ER/Mito interactions in ATDC5 chondrocytes in response to matrix mechanics.
Figure 1.
Matrix stiffness regulates mechanotransduction of ATDC5 chondrocytes
(A) Diagram of GelMA with different matrix stiffness and indentation testing results in 10%, 15%, and 20% GelMA groups (n = 5).
(B) The fluorescence images of cytotoxicity by Calcein-AM/PI staining in 60, 120, and 180 kPa and glass dish groups. Scale bars are 100 μm.
(C) The proteins expression of FAK, p-FAK, MLC, p-MLC, and Gapdh in 60, 120, and 180 kPa groups.
(D) Relative proteins expression of FAK, p-FAK, MLC, and p-MLC in 60, 120, and 180 kPa groups (n = 5).
(E) Immunofluorescence imaging of p-FAK and p-MLC in 60, 120, and 180 kPa groups. The nucleus is labeled by Hoechst. Scale bars are 20 μm.
(F) Fluorescent green Fluo4-AM staining imaging in 60, 120, and 180 kPa groups. Scale bars are 25 μm.
(G) Relative mRNAs expression levels of Grp94, Atf6, Ip3r, and Ryr in 60, 120, and 180 kPa groups (n = 5).
(H) Fluorescent imaging of MitoCMRox and ERGreen in 60, 120, and 180 kPa groups. Scale bars are 25 μm.
(I) Fluorescent green Fluo4-AM staining imaging in 180 kPa and 180 kPa+2-APB groups. Data are shown as means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Table 1.
Primers used in real-time PCR
| Target gene | Forward 5′-3′ | Reverse 5′-3′ |
|---|---|---|
| Col2a1 | GCTACACTCAAGTCACTGAACAACCA | TCAATCCAGTAGTCTCCGCTCTTCC |
| Mmp13 | TGGAGTAATCGCATTGTGAGAGTC | CCAGCCACGCATAGTCATATAGATAC |
| Bcl-2 | ATGCCTTTGTGGAACTATATGGC | GGTATGCACCCAGAGTGATGC |
| Bax | TGAAGACAGGGGCCTTTTTG | AATTCGCCGGAGACACTCG |
| Caspase3 | TGGTGATGAAGGGGTCATTTATG | TTCGGCTTTCCAGTCAGACTC |
| Mcu | ACTCACCAGATGGCGTTCG | CATGGCTTAGGAGGTCTCTCTT |
| Drp1 | CAAGGTTTTCTCGCCCAACG | CTGCCCTTACCATCTGGATCTA |
| Grp94 | TCGTCAGAGCTGATGATGAAGT | GCGTTTAACCCATCCAACTGAAT |
| Atf6 | GACTCACCCATCCGAGTTGTG | CTCCCAGTCTTCATCTGGTCC |
| IP3R | AGGCTTGGTTGATGACCGTTG | CGCCTACCTGGATCACGTT |
| Ryr | CCTTGGCTTCAGCCTTCTG | TCTGGGAGAGACACCTGTTGT |
| Phf8 | CATGGAGTCCTAAAGCCCGTG | GGTGTCAACTCTTACCTGCTG |
| Kdm1a | GTGGTGTTATGCTTTGACCGT | GCTGCCAAAAATCCCTTTGAGA |
| Kdm3a | CAGCAACTCCATCTAGCAAGG | TGTTCTCGGTACTTCAGGTTTTG |
| Kdm3b | GTGGTGCCAGTTGAATACCTT | TATAGGGACCTCGGCTGAAAA |
| Gapdh | ATGGTGAAGGTCGGTGTGAA | TGAGTGGAGTCATACTGGAACA |
| siNC | UUCUCCGAACGUGUCACGU | ACGUGACACGUUCGGAGAA |
| siPHF8 1 | GCUUCAUGAUCGAGUGUGA | UCACACUCGAUCAUGAAGC |
| siPHF8 2 | GCUACAAGUGUUCUGUGAA | UUCACAGAACACUUGUAGC |
| siPHF8 3 | GGUUGAAGACAUCUUCCAA | UUGGAAGAUGUCUUCAACC |
| ChIP-Mmp13 | ATTCTGCCAAGGATGGCACT | GCCAGTCACCTCTAAGCCAA |
| ChIP-Bax | GTCACCTGGTGGAAGCTCAC | CGCACACACCTGTAGCATCT |
Increased matrix stiffness induces ATDC5 chondrocyte catabolism and apoptosis in vitro
Chondrocytes require normal synthetic catabolism to maintain the articular cartilage matrix. The expression of the chondrocyte anabolic marker gene (Col2a1) decreased, whereas that of the catabolic marker gene (Mmp13) increased with increasing matrix stiffness (Figure 2A). Similarly, Col2a1 was down-regulated and Mmp13 was upregulated with increasing matrix stiffness (Figures 2B–2D). To evaluate the apoptosis of ATDC5 chondrocytes on different stiffness surfaces, ATDC5 chondrocytes were subjected to TUNEL, and it was found that the nuclei of ATDC5 chondrocytes exhibited obvious shrinkage and vacuoles with typical signs of apoptosis, as observed using transmission electron microscopy (TEM) (Figures 2E and 2F). The expression patterns of apoptosis-related proteins revealed that the expression of Bcl-2 decreased and that of Bax, caspase-9, Cleaved caspase-9, and Cleaved caspase-3 increased with increasing matrix stiffness (Figures 2G and 2H). These results indicated that the increased matrix stiffness promoted ATDC5 chondrocyte catabolism and apoptosis.
Figure 2.
Evaluation of ATDC5 chondrocytes’ physiological function on different matrix stiffness
(A) Relative mRNAs expression levels of Col2a1 and Mmp13 in 60, 120, and 180 kPa groups (n = 5).
(B) The proteins expression of Col2a1, Mmp13, and Gapdh in 60, 120, and 180 kPa groups.
(C) Relative proteins expression levels of Col2a1 and Mmp13 in 60, 120, and 180 kPa groups (n = 5).
(D) Immunofluorescence imaging of Col2a1 and Mmp13 in 60, 120, and 180 kPa groups. The nucleus is labeled by DAPI. Scale bars are 75 μm.
(E) TUNEL staining and TEM imaging in each group, and TUNEL-positive cells are shown by hollow arrow. Scale bars are 100 μm. Crumpled nuclear envelope is shown by white arrow and vacuole in cytoplasm is shown by asterisk in 60, 120, and 180 kPa groups.
(F) Ratio of TUNEL-positive cells in 60, 120, and 180 kPa groups (n = 5).
(G) The proteins expression of Bcl-2, Bax, Cleaved caspase-3, and Gapdh in 60, 120, and 180 kPa groups.
(H) Relative proteins expression levels of Bcl-2, Bax, Cleaved caspase-3, caspase-9, and Cleaved caspase-9 in 60, 120, and 180 kPa groups (n = 5). Data are shown as means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Excessive matrix stiffness induces mitochondrial damage in ATDC5 chondrocytes
Based on ER/Mito interactions caused by matrix stiffness, we hypothesized that increased matrix stiffness would further cause oxidative stress and mitochondrial damage in ATDC5 chondrocytes. To further explore ROS production induced by the increase in matrix stiffness, ROS generation in ATDC5 chondrocytes with different matrix stiffnesses was evaluated. ROS generation increased with increasing matrix stiffness (Figure 3A). Excessive accumulation of ROS leads to mitochondrial damage.16 Normal mitochondrial membrane potential depends on OXPHOS as electron transfer of mitochondrial respiration originates from OXPHOS, which is used to synthesize ATP. To evaluate the metabolic status of chondrocytes with different matrix stiffness, the results of total ATP content determination showed that increased matrix stiffness induced decreased ATP synthesis in ATDC5 chondrocytes (Figure 3B). Chondrocytes with different matrix stiffness were further treated with glycolysis and OXPHOS inhibitors, and the ATP content from glycolysis and mitochondrial synthesis was accurately determined. The level of OXPHOS represented by mitochondrial ATP is down-regulated with increasing matrix stiffness (Figure 3C). Normal mitochondrial membrane potential is necessary for maintaining OXPHOS. Flow cytometry analysis and JC-1 images revealed that the mitochondrial membrane potential showed a depolarization trend when the matrix stiffness increased from 60 to 180 kPa (Figures 3D and 3E). Under normal conditions, mitochondrial respiration and glycolysis work together to produce ATP to provide energy. However, when chondrocytes are stimulated by abnormal mechanical loading, they rely primarily on glycolysis to produce ATP.15
Figure 3.
Evaluation of mitochondrial function and dynamics on different matrix stiffness
(A) Fluorescent green DCFH-DA staining and bright-field imaging in 60, 120, 180 kPa and Rosup groups. Rosup was the positive control, and scale bars are 100 μm. And relative fluorescence intensity of DCFH-DA in 60, 120, 180 kPa and Rosup groups (n = 5).
(B) ATP content of ATDC5 chondrocytes in 60, 120, and 180 kPa groups (n = 5).
(C) Glycolytic ATP and mitochondrial ATP content and oxidative phosphorylation level of ATDC5 chondrocytes in 60, 120, and 180 kPa groups (n = 5).
(D) Flow cytometry analysis of JC-1 and aggregates ratio in 60, 120, 180 kPa, and Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) groups (n = 5). CCCP was the positive control.
(E) Fluorescence imaging of JC-1 in 60 , 120 , 180 kPa and CCCP groups. Scale bars are 25 μm.
(F) Fluorescent imaging of MitoGreen and mitochondrial length in 60, 120, and 180 kPa groups (n = 5). Scale bars are 10 μm.
(G) The proteins expression of Drp1, Fis1, Mfn1, Opa1, and Gapdh in 60, 120, and 180 kPa groups.
(H) Relative proteins expression levels of Drp1, Fis1, Mfn1, and Opa1 in 60, 120, and 180 kPa groups (n = 5).
(I) TEM imaging of mitochondrial morphology and cristae volume density in 60, 120, and 180 kPa groups (n = 5). Data are shown as means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Mitochondrial function was found to gradually decrease with increasing matrix stiffness. In the 60 kPa group, mitochondria showed a linear and network shape. When the matrix stiffness increased to 120 kPa, the mitochondrial network became discontinuous, and mitochondria appeared as rings and fragments in the 180 kPa group; the length of the mitochondrial network significantly shortened with increasing matrix stiffness (Figure 3F). The expression of mitochondrial division-related proteins (Drp1 and Fis1) significantly increased from 60 kPa to 180 kPa, while the expression of mitochondrial fusion-related proteins (Mfn1 and Opa1) was decreased (Figures 3G and 3H). TEM images showed morphological changes in the mitochondria of each group (Figure 3I). In summary, high matrix stiffness led to changes in mitochondrial dynamics and mitochondrial dysfunction in ATDC5 chondrocytes.
Intracellular Ca2+ mediates stiffness-induced apoptosis and mitochondrial damage
Based on the results of the previous experiment, we found that Ca2+ signal was significantly enhanced with increasing matrix stiffness, and there was a trend of strengthening with time in the 180 kPa group. To observe the effect of Ca2+ on ATDC5 chondrocyte apoptosis under different matrix stiffnesses, extracellular and intracellular Ca2+ were removed using BAPTA (Aminophenethane tetraacetic acid) and BAPTA-AM (1,2-bis (2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester), respectively. After extracellular Ca2+ was removed, high matrix stiffness still induced high expression of apoptosis-related proteins (Figure 4A), while the influence of different matrix stiffness on apoptosis was not significant after further using the BAPTA-AM to remove intracellular Ca2+ (Figure 4B). These results suggest that intracellular Ca2+ plays an important role in maintaining ATDC5 chondrocyte function in response to different matrix stiffness.
Figure 4.
Intracellular Ca2+ mediates stiffness-induced apoptosis and mitochondrial damage
(A) The proteins expression of Bcl-2, Bax, Cleaved caspase-3, and Gapdh in 60, 120, and 180 kPa groups after treatment with BAPTA (n = 5).
(B) The proteins expression of Bcl-2, Bax, Cleaved caspase-3, and Gapdh in 60, 120, and 180 kPa groups after treatment with BAPTA-AM (n = 5).
(C) Fluorescent green DCFH-DA staining and bright-field imaging and relative fluorescence intensity of DCFH-DA in 60, 120, 180 kPa, and Rosup groups after BAPTA treatment (n = 5). Scale bars are 250 μm.
(D) Fluorescent imaging of MitoGreen and mitochondrial length in 60, 120, and 180 kPa groups after BAPTA treatment (n = 5). Scale bars are 10 μm and 2 μm.
(E) The proteins expression of Mcu and Gapdh in 180 kPa and 180 kPa+2-APB groups (n = 5).
(F) Fluorescence imaging of JC-1 in 180 kPa, 180 kPa+2-APB, and CCCP groups. Scale bars are 50 μm.
(G) Glycolytic ATP and mitochondrial ATP content and oxidative phosphorylation level of ATDC5 chondrocytes in 180 kPa and 180 kPa+2-APB groups (n = 5). Data are shown as means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Similarly, a stiffer matrix did not cause obvious oxidative stress after removing intracellular Ca2+ using BAPTA-AM (Figure 4C). Further observation of mitochondrial dynamics did not show significant differences after BAPTA-AM treatment, and there was no significant difference in mitochondrial morphology (Figure 4D). The aforementioned results indicate that ER/Mito interactions are caused by increased matrix stiffness. After applying ER calcium release channel inhibitor 2-APB, we found that the expression of mitochondrial calcium inflow channel Mcu (mitochondrial Ca2+ uniporter) was significantly down-regulated (Figure 4E), and the downregulation of mitochondrial membrane potential was saved at high matrix stiffness (Figure 4F). The mitochondrial metabolism level of chondrocytes was further evaluated. The application of 2-APB to inhibit the release of calcium from the ER partially improved the mitochondrial ATP synthesis, i.e., OXPHOS level (Figure 4G).These results indicate that intracellular Ca2+ plays a major role in the response of ATDC5 chondrocytes to matrix stiffness.
The opening of mitochondrial mPTP upregulates Phf8 demethylation
Various intermediates of the TCA cycle in mitochondria also have epigenetic regulatory effects,16 and epigenetic alterations, such as histone methylation, have been found in OA. Besides mitochondrial dysfunction and dynamic changes, increased matrix stiffness affected mitochondrial membrane permeability and formed mPTP, accumulated Calcein AM in the cytoplasm, and only retained the Ca2+ signal in mitochondria. Results showed that, with increasing matrix stiffness, mitochondrial Ca2+ levels decreased, indicating that the degree of mPTP opening increased (Figure 5A). Phf8, which belongs to the Jmjd family, was regulated by ROS and the permeability of the mitochondrial membrane.21 The expression of Phf8 was higher among that of the demethylases (Figures 5B and 5C) and exhibited nuclear translocation to catalyze the demethylation of H3K27me3 (Figures 5D–5F), whereas other members showed no significant changes (Figures 5H and 5I). ATDC5 chondrocytes cultured in vitro also showed lower H3K27me3 expression induced by high matrix stiffness (Figure 5G). To determine the downstream target genes of Phf8 and H3K27me3, CUT&Tag was used to obtain chondrocyte DNA-chromatin complexes with low matrix stiffness (60 kPa) and high matrix stiffness (180 kPa) (Figure 5J). CUT&Tag-PCR analysis revealed that the promoters of Mmp13 and Bax in ATDC5 chondrocytes cultured on a soft matrix were more strongly bound to H3K27me3 (Figures 5K and 5L), indicating that demethylation of H3K27me3 activated Mmp13 and Bax transcription.
Figure 5.
Phf8 regulates H3K27me3 demethylation on the promoters of Mmp13 and Bax via mPTP opening
(A) Immunofluorescence imaging of mPTP by Calcein AM and mitochondrial tracker (MitoCMRox) staining in 60, 120, 180 kPa, and CCCP groups. Scale bars are 25 μm.
(B) The proteins expression of Phf8 and Gapdh in 60, 120, and 180 kPa groups.
(C) Relative protein expression level of Phf8 in 60, 120, and 180 kPa groups (n = 5).
(D) Immunofluorescence imaging of Phf8 in each group and the nucleus is labeled by Hoechst. Scale bars are 50 μm.
(E) The protein expression of H3K27me3 and Histone3 in 60, 120, and 180 kPa groups.
(F) Relative protein expression level of H3K27me3 in 60, 120, and 180 kPa groups (n = 5).
(G) Immunofluorescence imaging of H3K27me3 in 60, 120, and 180 kPa groups. The nucleus is labeled by Hoechst. Scale bars are 25 μm.
(H) The proteins expression of Kdm1, Kdm3, Kdm5, and Gapdh in 60, 120, and 180 kPa groups.
(I) Relative protein expression level of Kdm1, Kdm3, and Kdm5 in 60, 120, and 180 kPa groups (n = 5).
(J) Schematic diagram of Mmp13 and Bax promoters and corresponding H3K27me3 binding sites.
(K) CUT&Tag-PCR analysis of H3K27me3 on Mmp13 and Bax in promoters in soft (60 kPa) and hard (180 kPa) matrix (n = 5).
(L) Promoter occupancy analysis of H3K27me3 on Mmp13 and Bax promoters in soft (60 kPa) and hard (180 kPa) matrix. Data are shown as means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Phf8 catalyzes H3K27me3 demethylation on the promoters of Mmp13 and Bax
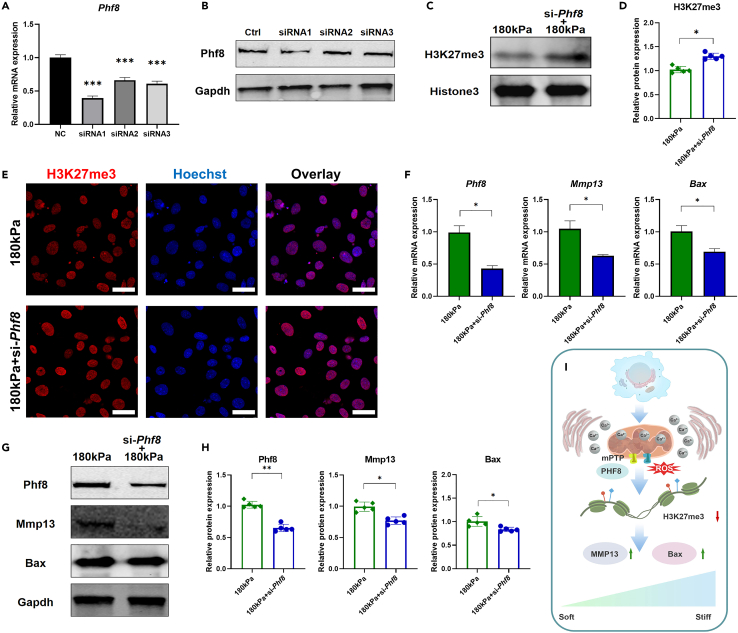
To verify whether the increase in mitochondrial mPTP-Phf8 regulated Mmp13 and Bax transcription through H3K27me3 demethylation, si-Phf8 was used to interfere with ATDC5 chondrocytes (Figures 6A and 6B). As a histone demethylase, knockdown of Phf8 rescued the demethylation of H3K27me3 on the high-stiffness matrix (Figures 6C–6E). Similarly, si-Phf8 inhibited the expression of Phf8 on the high-stiffness surface and the high expression of Mmp13 and Bax on a stiff matrix was inhibited (Figure 6F). The expression of the proteins corresponding to these genes was also inhibited (Figures 6G and 6H). These results showed that matrix stiffness promoted the demethylation of H3K27me3, which mainly affected the function of ATDC5 chondrocytes by regulating Mmp13 and Bax transcription. H3K27me3 has the function of silencing gene transcription, whereas its demethylation has the opposite effect. The Mmp13 and Bax promoters bound to H3K27me3 and matrix stiffening led to a decrease in H3K27me3 binding to both promoters, whereas transcriptional activation promoted chondrocyte apoptosis. Therefore, mitochondria not only have the role of mechanical conduction but also regulate epigenetics (Figure 6I).
Figure 6.
Deficiency of Phf8 inhibits the expression of Mmp13 and Bax caused by high matrix stiffness
(A) Phf8 knockdown efficiency by small interfering RNA (siRNA) was evaluated by RT-qPCR (n = 5).
(B) The protein expression of Phf8 and Gapdh in Ctrl, siRNA1, siRNA2, and siRNA3 groups.
(C) The proteins expression of H3K27m3 and Histone3 in 180 kPa and 180 kPa+si-Phf8 groups.
(D) Relative protein expression level of H3K27me3 in 180 kPa and 180 kPa+si-Phf8 groups (n = 5).
(E) Immunofluorescence imaging of H3K27me3 in 180 kPa and 180 kPa+si-Phf8 groups. The nucleus is labeled by Hoechst, and scale bars are 25 μm.
(F) Relative mRNAs expression levels of Phf8, Mmp13, and Bax in 180 kPa and 180 kPa+si-Phf8 groups (n = 5).
(G) The proteins expression of Phf8, Mmp13, Bax, and Gapdh in 180 kPa and 180 kPa+si-Phf8 groups.
(H) Relative proteins expression levels of Phf8, Mmp13, and Bax in 180 kPa and 180 kPa+si-PHF8 groups (n = 5).
(I) Schematic diagram.
Inhibition of Phf8 expression rescues OA cartilage in ACLT-induced mice
si-Phf8 was intra-articularly injected in the anterior cruciate ligament transection (ACLT) model to observe the change of cartilage status in OA. Osteoarthritis Research Society International (OARSI) scores decreased after si-Phf8 application (Figures 7A and 7B), while the expression of Col2a1 was significantly increased and the expression of Mmp13 was inhibited (Figures 7C and 7D). si-Phf8 rescued OA chondrocyte apoptosis (Figures 7E and 7F). Labeled multiple fluorescence of division and fusion proteins indicated that Opa1 and Mfn1 were mainly expressed in normal articular cartilage and Drp1 was mainly expressed in OA. The expression of fusion proteins (Mfn1 and Fis1) was increased after si-Phf8 application, while the expression of Drp1 and Opa1 decreased (Figures 7G and 7H). The mitochondrial morphology of mouse articular cartilage was further observed using TEM, and mitochondrial damage induced by si-Phf8 was milder (Figures 7I and 7J). Additionally, undamaged chondrocytes had a high expression of H3K27me3 and a low expression of Phf8. Moreover, si-Phf8 rescued the demethylation of H3K27me3 by inhibiting the expression of Phf8 (Figures 7K and 7L).
Figure 7.
The effect of si-Phf8 on OA cartilage in ACLT-induced mice
(A) H&E and Safranin-O/fast green staining of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups. Scale bars are 100 μm.
(B) Maximum and summed OARSI scores in Sham, OA vehicle, and OA si-Phf8 groups (n = 6).
(C) Immunofluorescence of Col2a1 and Mmp13 of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups. Scale bars are 50 and 100 μm, respectively.
(D) Relative fluorescence intensity of Col2a1 and Mmp13 of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups (n = 6).
(E) TUNEL staining of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups. Scale bars are 50 and 100 μm, respectively.
(F) TUNEL-positive cells in Sham, OA vehicle, and OA si-Phf8 groups (n = 6).
(G) Immunofluorescence of Drp1, Fis1, Mfn1, and Opa1 of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups. Scale bars are 20 and 40 μm, respectively.
(H) Relative fluorescence intensity of Drp1, Fis1, Mfn1, and Opa1 of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups (n = 6).
(I) TEM imaging of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups. Mitochondria of chondrocytes are as shown by hollow arrow.
(J) Mitochondrial area and cristae volume density of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups (n = 20).
(K) Immunofluorescence of H3K27me3 and Phf8 of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups. Scale bars are 20 μm. H3K27 methylated chondrocytes are shown by hollow arrow, and H3K27 demethylated chondrocytes are shown by asterisk in Sham, OA vehicle, and OA si-Phf8 groups.
(L) Relative fluorescence intensity of H3K27me3 and Phf8 of knee cartilage in Sham, OA vehicle, and OA si-Phf8 groups (n = 6). Data are shown as means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Increased matrix stiffness and mitochondrial damage are associated with OA progression
To investigate whether human OA cartilage still has similar mechanobiology characteristics, we obtained clinical specimens of OA cartilage. The demographic characteristics of clinical patients are shown in Table 2. Specimens were divided into mild, moderate, and severe OA groups (Figures 8A and 8B). The expression of COL2A1 and H3K27me3 decreased, whereas that of MMP13 and PHF8 increased significantly with the degradation of cartilage (Figures 8C and 8D). The mechanical properties of articular cartilage were analyzed using microindentation testing, and it was found that the stiffness of articular cartilage increased significantly with OA development (Figure 8E). The mitochondria of chondrocytes in severely wearied cartilage exhibited large volume and low cristae volume density. The mitochondria area was small in mild OA (<0.1 μm2) and increased significantly with OA progression (Figures 8F and 8G). In severe OA, the cristae volume density of chondrocyte mitochondria was lower than that in mild or moderate OA (Figure 8G). Next, TEM images of clinical specimens of OA cartilage showed that the mitochondrial damage was accompanied by increased contact with ER during OA progression (Figure 8H). The degree of mitochondrial damage increased with OA development, and the ratio of the area covered by ER in contact with the perimeter of mitochondria increased significantly, indicating that ER tended to wrap the damaged mitochondria in OA (Figure 8I). We further analyzed the correlation between OARSI scores, mitochondrial morphological parameters, and Young’s modulus of articular cartilage. The result showed that high OARSI scores were related to severe mitochondrial damage (Figure 8J) and increased stiffness was closely related to severe mitochondrial damage (Figure 8K).
Table 2.
The demographic data of all patients
| ID | Age | Sex | Kellgren-Lawrence (KL) grade |
|---|---|---|---|
| 1 | 66 | F | 3 |
| 2 | 69 | F | 3 |
| 3 | 78 | M | 4 |
| 4 | 82 | M | 3 |
| 5 | 67 | F | 4 |
| 6 | 72 | F | 4 |
Figure 8.
Correlation between OARSI scores, mitochondrial damage, and matrix stiffness at different stages of OA
(A) Macro, H&E, and Safranin O staining of OA cartilage sample at different stages. Scale bars are 500 μm.
(B) Maximum OARSI scores and summed OARSI scores of OA cartilage sample at different stages (n = 6).
(C) Immunohistochemistry of COL2A1, MMP13, PHF8, and H3K27me3. Scale bars are 100 and 200 μm, respectively.
(D) The positive proportions of COL2A1, MMP13, PHF8, and H3K27me3 (n = 6).
(E) Mechanical testing diagram of osteochondral unit and microindentation testing results of cartilage stiffness at different stages (n = 30).
(F) TEM imaging of OA cartilage sample at different stages. Mitochondria of chondrocytes are as shown by hollow arrow.
(G) Mitochondrial area and cristae volume density of OA cartilage sample at different stages (n = 60).
(H) TEM false-colored imaging of OA cartilage sample to observe the localization of ER-Mito at different OA stages. Red represents mitochondria, and green represents ER.
(I) quantitative statistics of the distance and the covered ratio of ER-Mito (n = 60).
(J) Correlation analysis of OARSI score and mitochondrial area and cristae volume density (n = 18).
(K) Correlation analysis of matrix stiffness and mitochondrial area and cristae volume density (n = 18). Data are represented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
In the current study, mitochondria were found to play a central role; mitochondria not only have a role in mechanotransduction but can also regulate epigenetics. These results indicate that Phf8 reduces the abundance of H3K27me3 on the promoters of Mmp13 and Bax, promoting the transcription of these genes, and a stiffer matrix promotes OA by damaging mitochondria. Mechanical loading affected mitochondrial function through intracellular Ca2+, which was impacted by ER stress.24,25,26 mPTP opening in damaged mitochondria upregulated Phf8, which induced H3K27me3 demethylation. These findings demonstrate that matrix stiffness promotes matrix degradation and that chondrocyte apoptosis aggravates OA by inhibiting H3K27me3 demethylation targeting matrix degradation and apoptosis.
ECM is a complex molecular network of chondrocytes. It is composed of many different biochemical components, including collagen, proteoglycans, and glycoproteins.5 ECM provides a physical scaffold for cells and regulates the processes of cell growth, differentiation, migration, homeostasis, and survival. The destruction of cartilage ECM is considered to be a sign of OA.11 During the development of OA, the integrity of the ECM network formed by type II collagen and proteoglycan is destroyed. The elastic storage capacity of OA cartilage is decreased, which further leads to collagen fiber cracks. Pathological calcification of cartilage surface occurs in early OA. As OA progresses, calcification invades the deeper layers of cartilage. Compared with normal cartilage, the ultrastructure and composition of OA cartilage have undergone great changes, and the crystal crystallinity and tissue stiffness have increased significantly.7,8 With the progression of OA, cartilage ECM gradually loses proteoglycans and glycosaminoglycan, and the cartilage layer becomes hard and thin, transferring most of the mechanical load to the underlying subchondral bone.8
Mitochondria are important for the maintenance of normal function of chondrocytes. Under the influence of mechanical load, chondrocytes rely on mitochondrial glycolysis and TCA to promote cell survival under stress.13 Abnormal metabolism of OA chondrocytes is a response to changes in mechanical environment and plays a key role in cartilage degeneration and OA progression. Elevated ROS levels in OA chondrocytes induce depolarization of mitochondrial membrane, thus causing oxidative stress and ultimately apoptosis of chondrocytes.14,15 During the occurrence and development of OA, chondrocytes are stimulated by abnormal mechanical load, which can affect chondrocyte function through mitochondria.13
Multiple forms of histone modifications exist during OA progression.27,28,29 H3K27 methylation modification is an important factor in promoting chondrogenesis. However, the change in H3K27 methylation in OA is still controversial.30 Mitochondria are closely related to histone methylation.20 The opening of the mPTP is a key event causing cell death and plays an important role in the regulation of cell survival and apoptosis.31 In healthy cells, the inner mitochondrial membrane maintains a normal gradient of mitochondrial membrane potential to ensure cellular respiration and energy supply.31 However, in the process of apoptosis or death, the opening of mPTP significantly changes mitochondrial permeability. Its continuous opening leads to overloading of Ca2+, mitochondrial oxidation of glutathione, and increased levels of ROS in mitochondria, which ultimately lead to the release of cytochrome c and the reduction of mitochondrial membrane potential.21
When chondrocytes are mechanically stimulated, Ca2+ enters cells through mechanically sensitive ion channels and causes intracellular Ca2+ oscillation, leading to changes in mitochondrial function and cell apoptosis.32 Before this, actin regulators specifically promote actin polymerization at the contact point between the ER and mitochondria, leading to mitochondrial division.33 Mitochondrial Ca2+ uptake depends on highly specialized regions formed by close contact between mitochondria and the ER. Ca2+ is released into the cytoplasm through IP3R, creating a cytosolic Ca2+ domain that is fully concentrated on the nanometer scale and sensed by the adjacent mitochondria.25
This study suggests that increased matrix stiffness induces mitochondrial calcium overload through ER/Mito interactions and that the opening of damaged mitochondrial mPTP activates Phf8 expression and nucleation and leads to demethylation of H3K27me3 on Mmp13 and Bax promoters, further aggravating OA. It provides a new mechanobiology mechanism for chondrocyte response to matrix stiffness and mitochondrial regulation of epigenetics.
Limitations of the study
Chondrocytes are surrounded by cartilage matrix, and their mechanical microenvironment is extremely complex.32 Studies have found that the volume of chondrocytes cultured in three-dimensional matrices with different Young’s moduli is significantly affected.34,35 The effects of three-dimensional and two-dimensional environments on chondrocyte function may be different, and more precise methods are needed to measure cartilage matrix mechanics under physiological or pathological conditions.36 Such studies will advance the current understanding regarding the potential mechanism of mitochondrial damage when chondrocytes respond to matrix stiffness as well as provide insights on how mechanical stimulation regulates the interaction between organelles and a new mechanobiology mechanism of OA.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Collagen Type II Polyclonal antibody | Proteintech | (Proteintech Cat# 28459-1-AP; RRID: AB_2881147) |
| MMP13 Polyclonal antibody | Proteintech | (Proteintech Cat# 18165-1-AP; RRID: AB_2144858) |
| Bcl2 Polyclonal antibody | Proteintech | (Proteintech Cat# 26593-1-AP; RRID: AB_2818996) |
| BAX Polyclonal antibody | Proteintech | (Proteintech Cat# 50599-2-Ig; RRID: AB_2061561) |
| Cleaved Caspase 3 Polyclonal antibody | Proteintech | (Proteintech Cat# 25128-1-AP; RRID: AB_3073913) |
| Caspase 9 Polyclonal antibody | Proteintech | (Proteintech Cat# 10380-1-AP; RRID: AB_2068632) |
| MCU Polyclonal antibody | Proteintech | (Proteintech Cat# 26312-1-AP; RRID: AB_2880474) |
| DRP1 Polyclonal antibody | Proteintech | (Proteintech Cat# 12957-1-AP; RRID: AB_2093525) |
| FIS1 Polyclonal antibody | Proteintech | (Proteintech Cat# 10956-1-AP; RRID: AB_2102532) |
| OPA1 Polyclonal antibody | Proteintech | (Proteintech Cat# 27733-1-AP; RRID: AB_2810292) |
| PHF8 Polyclonal antibody | Proteintech | (Proteintech Cat# 29516-1-AP; RRID: AB_2935473) |
| KDM1 Polyclonal antibody | Proteintech | (Proteintech Cat# 20813-1-AP; RRID: AB_10697809) |
| KDM3A Polyclonal antibody | Proteintech | (Proteintech Cat# 12835-1-AP; RRID: AB_1072935) |
| KDM5A Polyclonal antibody | Proteintech | (Proteintech Cat# 18825-1-AP; RRID: AB_10638436) |
| FAK Polyclonal antibody | Proteintech | (Proteintech Cat# 12636-1-AP; RRID: AB_2173668) |
| p-FAK Monoclonal antibody | Santa Cruz Biotechnology | (Santa Cruz Biotechnology Cat# sc-374668; RRID: AB_11150862) |
| MLC2 Polyclonal antibody | Proteintech | (Proteintech Cat# 10906-1-AP; RRID: AB_2147453) |
| p-MLC2 Polyclonal antibody | Cell Signaling Technology | (Cell Signaling Technology Cat# 3671; RRID: AB_330248) |
| GAPDH Monoclonal antibody | Proteintech | (Proteintech Cat# 60004-1-Ig; RRID: AB_2107436) |
| Histone3 Polyclonal antibody | Proteintech | (Proteintech Cat# 17168-1-AP; RRID: AB_2716755) |
| Histone H3K27me2 Monoclonal antibody | Active Motif | (Active Motif Cat# 61435; RRID: AB_2793635) |
| Histone H3K27me3 Polyclonal antibody | Active Motif | (Active Motif Cat# 39157; RRID: AB_2561020) |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum | Sigma | F8318 |
| DMEM/F12 medium | Gibco | 11320033 |
| Hoechst | Beyotime | P0133 |
| TUNEL Apoptosis Assay Kit | Beyotime | C1089 |
| Mitochondrial membrane potential assay kit | Beyotime | C2003S |
| MitoTracker Green | Beyotime | C1048 |
| Mito-Tracker Red CMXRos | Beyotime | C1049B |
| ER-Tracker Green | Beyotime | C1042S |
| Mitochondrial Permeability Transition Pore Assay Kit or MPTP Assay Kit | Beyotime | C2009S |
| Reactive Oxygen Species Assay Kit | Beyotime | S0033S |
| CUT&Tag-IT® Assay Kit, Anti-Mouse | Active Motif | 53165 |
| Experimental models: Cell lines | ||
| ATDC5 | Cell Bank of the Chinese Academy of Sciences | (RCB Cat# RCB0565, RRID:CVCL_3894) |
| Software and algorithms | ||
| ImageJ Version 1.53 | National Institutes of Health | https://www.nih.gov/ |
| GraphPad Prism Version 8 | GraphPad Software | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhifeng Yu (zfyu@outlook.com).
Materials availability
Relevant experimental materials and reagents can be obtained from the lead contact upon request.
Data and code availability
Experimental data reported in this manuscript will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Assesment of human OA cartilage samples
This study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital (No. SH9H-2021-T401-2). All patients were from Chinese Han population. Sample size requirements were calculated to detect a mean difference of 8 in total points of the Osteoarthritis Research Society International (OARSI) scores,12 and a sample size of n = 6 was chosen. All six patients provided written informed consent, and the demographic data of all patients are detailed in Table 2. Participants involved were diagnosed with primary symptomatic knee OA with varus deformity of lower limb, based on clinical and radiographic criteria. Those who had other forms of arthritis such as rheumatoid arthritis, or who were taking medications that could affect bone remodeling (including estrogen and bisphosphonates) were excluded. The tibial plateau of each OA patient was histologically stained and tanalyzed by TEM after microindentation testing. All OA cartilage specimens were divided into three group, mild, moderate and severe OA.
Establishment of anterior cruciate ligament transection (ACLT) mice
10-week-old male C57BL/6 mice were purchased from Shanghai SIPPR BK Laboratory Animals Ltd (Shanghai, China). Mice were housed under a 12 h light/-dark cycle with free access to food and water. All animals used in this study have been ethically approved and received care in compliance with the institutional guidelines established by the Committee of Ethics on Animal Experiments at the Shanghai Jiao Tong University School of Medicine (No. SH9H-2023-A3-1). OA mice were established by ACLT following the previous procedure. They were divided into sham, OA vehicle and OA si-Phf8 groups, and histological analysis were performed intra-articular injection of si-Phf8 for 14 days after operation.23
ATDC5 chondrocytes cells culture and chondrogenic differentiation
The ATDC5 chondrocytes cell line is a continuous, long-term cultured line derived from mouse teratocarcinoma cells and is commonly used as a model for in vitro chondrocyte research.21 ATDC5 chondrocytes chondrogenic cell line (Fuheng Center Cell Bank, China) was cultured in DMEM/F-12 (Hyclone, USA) containing 5% fetal bovine serum (Sigma, USA) and 1% penicillin/streptomycin (Gibco, USA) in 5% CO2 and 21% O2 at 37°C. To induce chondrogenic differentiation, ATDC5 chondrocytes cells were cultured in the differentiation medium supplemented with 1% insulin-transferrin-selenium (ITS) (Gibco, USA). The medium was changed every 2 days. And cells induced to differentiate for 14 days were used for subsequent experiments.
Establishment of a two-dimensional culture model
ATDC5 chondrocytes were cultured in 6-cm dishes with different GelMA stiffnesses at a density of 1 × 105/mL. In addition, ATDC5 chondrocytes were treated with inhibitors (BAPTA-AM and 2-APB) (Selleck Chemicals; China) or transfected with siRNA (Genomeditech; China) before two-dimensional cell culture. BAPTA-AM is a membrane-permeable Ca2+ chelator used to chelate intracellular Ca.2 2-APB is a Ca2+ inhibitor of the membrane permeability of inositol triphosphate (IP3) receptor, thereby inhibiting Ca2+ release from the ER. The culture medium was changed every 2 days.
Methods details
Hydrogel preparation
GelMA and lithium phenyl (2,4,6-trimethylbenzoyl) phosphinate (LAP) were purchased from Engineering for Life (Suzhou, China). GelMA was prepared from gelatin and methacrylic anhydride with an amino substitution of 60%. GelMA with the same degree of substitution is dissolved in LAP to produce GelMA solutions of 10%, 15% and 18% by mass volume ratio. To obtain hydrogels with different stiffnesses, 1 mL of GelMA at different concentrations was spread on the bottom of a 6-cm dish with a thickness of approximately 0.4 mm. Photocrosslinking was performed under a UV floodlight (EFL-LS-1601-405; Suzhou, China) for 1 min.
Microindentation
Microindentation testing were performed by using a displacement controlled PIUMA Nanoindenter (Optics11 B.V., Amsterdam, The Netherlands).37 A probe with 0.45 N/m cantilever stiffness (k) and a 48.5 μm radius (R) spherical tip was used in this study. Samples were treated as mechanically isotropic and tested by performing (for OA cartilage specimens, n = 30; for GelMA. n = 5) independent microindentation measurements on different (randomly selected) points to avoid any eventual effect due to repeated testing cycles on the same spot and sample pre-stress. GelMA values of 61.8 ± 7.5, 122.3 ± 15.8, and 180.2 ± 17.9 kPa were obtained (60-, 120-, and 180-kPa groups).
ROS analysis
After 5 days culture, the levels of ROS in different groups of ATDC5 chondrocytes were detected by using the fluorescence probe dichlorodihydrofluoresceindiacetate (DCFH-DA, Beyotime Biotechnology; China).38 Cells were observed with the fluorescence microscope (DM4000 B; Leica, Germany).
ATP content and metabolic level analysis
After 5 days culture, ATDC5 chondrocytes cultured on different stiffness were collected and homogenized. After centrifugation (12000 × g) for 10 min at 4°C, the supernatant was transferred into a 10 kDa MWCO spincolumn and centrifuged at 10000 × g for 20 min. The concentrations of arginine, fumarate, ornithine,and citrulline were, respectively, assessed by Arginine assay Kit, FumarateDetection Kit, Ornithine Assay Kit, and Citrulline Assay Kit according tothe provided protocols (Abcam, USA). For lactate and urea, the culture medium was collected and diluted by ultrapure waterto a suitable concentration. The final concentration was detected by a lactate assay kit (Dojindo, Japan).
Mitochondrial morphology analysis
After 5 days of two-dimensional culture of ATDC5 chondrocytes with/without 10 μM BAPTA-AM in the 60-, 120-, and 180-kPa groups, the morphology of mitochondria was observed and evaluated by the above-mentioned operating methods and reagents.38 Subsequently, MitoTracker Red CMXRos (1:1000) and MitoTracker Green working solution (1:5000) were added and incubated at 37°C for 30 min. The MitoTracker Red CMXRos working solution was removed, and fresh cell culture solution preheated at 37°C was added. The cells were observed with a laser confocal microscope (TCS SP8; Leica, Germany).
Mitochondrial membrane potential (JC-1) analysis
After 5 days of two-dimensional culture of ATDC5 chondrocytes with/without 10 μM BAPTA-AM in the 60-, 120-, and 180-kPa groups, 1 mL of JC-1 dye solution was added to each confocal culture dish. CCCP (Carbonyl cyanide 3-chlorophenylhydrazone) resulting in mitochondrial dysfunction was diluted to 10 μM, and cells were treated for 20 min as a positive control. The culture solution was removed, the cells were washed with PBS once, 1 mL of cell culture solution and 1 mL of JC-1 staining working solution were added, and the cells were incubated at 37°C for 20 min. The supernatant was removed and washed twice with JC-1 dye buffer. Two milliliters of cell culture medium were added, and the cells were observed under a laser confocal microscope. In addition, 100,000 cells from each group were suspended in 0.5 mL of cell culture solution, to which 0.5 mL of JC-1 staining working solution was added, incubated at 37°C for 20 min, centrifuged at 600 × g for 3 min at 4°C, washed twice with JC-1 staining buffer, and analyzed by flow cytometry (LSR Fortessa system; BD, USA).
Ca2+ imaging
ATDC5 chondrocytes were preincubated with/without 100 μM 2-aminoethoxydiphenyl borate (2-APB) (Selleck Chemicals) in the 180-kPa group after 5 days. Subsequently, the 60-kPa, 120-kPa, 180-kPa, and 180-kPa+2-APB groups were loaded with Fluo-4 a.m. (5 μM; Beyotime, China) at 37°C for ∼30 min. Calcium imaging was performed with the fluorescence microscope system MetaFluor software, and changes in fluorescence intensity were recorded within 480 s. Changes in intracellular Ca2+ concentration were estimated from Fluo-4 a.m. fluorescence images; 2-APB was used to block the release of Ca2+ from the ER.
ER-mito interaction analysis
After 5 days of two-dimensional culture, ER Tracker Green was diluted at a ratio of 1:1000. The cell culture solution was removed, and the cells growing on the culture dish were washed with an appropriate amount of solution, such as Hanks' balanced salt solution with Ca2+ and Mg2+. The washing solution was removed, and ER Tracker Green dye solution was preheated at 37°C and incubated at 37°C for 15 min. The cells were washed with cell culture solution 1–2 times. Subsequently, fluorescence microscopy or laser confocal microscopy was used for observation.
Mitochondrial permeablity transition pore (mPTP) analysis
After 5 days of two-dimensional culture of ATDC5 chondrocytes with different Ca2+-conditioned media, the culture medium was aspirated, and the cells were washed with PBS 2 times. Thereafter, 500 μL of calcein AM dye solution was added, and the samples were incubated in the dark at 37°C for 30 min. The dye solution was then replaced with fresh culture medium preheated at 37°C and incubated in the dark at 37°C for 30 min. The culture medium was removed, cells were washed with PBS 3 times, MitoTracker Red CMXRos was added to detect the activity of mitochondria, and the cells were observed under laser confocal fluorescence.
siRNA transfection and intra-articular injection
ATDC5 Cells were inoculated and cultured in six-well plates for 24 h to achieve a cell density of 60–70%. Next, 50 nM of si-NC or si-Phf8 duplexes were added according to the Lipofectamine 3000 siRNA transfection system (Thermo Fisher Scientific, Waltham, MA, USA). The gene knockdown effect was verified by qRT-PCR and western blot experiments. For an intra-articular injection, 10 μL of si-Phf8 or negative control labeled was weekly injected into knee joints of mice 14 days after ACLT surgery. The siRNAs sequences were listed in the Table 1.
Real-time PCR
Total RNA was isolated from the ATDC5 chondrocytes cultures using the Trizol reagent (Thermo Fisher) and converted to cDNA using PrimeScriptRT reagent Kit with gDNA Eraser (TaKaRa) according to the manufacturer’s instructions. RNA was reverse-transcribed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems). For real-time PCR, the PCR products were detected using LightCycler 480 SYBR Green I Master (Bimake) on a Q6 RT-PCR detection system (Life technologies). Primers for mouse genes in the Table 1. The relative expression levels for each target gene were calculated using the 2−ΔΔCt method.
Western blotting
The cells were lysed using RIPA lysis buffer supplemented with phosphatase and protease inhibitors (Roche Diagnostics, Basel, Switzerland). Total protein was quantified using a bicinchoninic acid assay (Thermo Fisher Scientific) and then equal quantities of extracted protein (20–30 μg) were separated via 4–20% sodium dodecyl sulfatepolyacrylamide gel electrophoresis and electroblotted onto 0.22 μm PVDF membranes (MilliporeSigma, Burlington, MA, USA). The membranes were blocked using 5% BSA-PBS (Beyotime Biotechnology) at room temperature (RT = 25°C) for 1 h, and followed by incubation at 4°C overnight with rabbit antibodies. The membranes were washed with Tris-buffered saline (TBS)-0.1% Tween 20 (TBST) and subsequently incubated with anti-rabbit IgG secondary antibody (cat. no. 5151; DyLight 800 4X PEG Conjugate; Cell Signaling Technology; 1:5000) for 1 h at RT in the dark. After washing in TBST, protein immunoreactivity was detected using the Odyssey Fluorescence Imaging system (LI-COR Biosciences, Lincoln, NE, USA). Semi-quantitative analysis of protein band intensity was conducted using the ImageJ v1.8.0 software (National Institutes of Health) and normalized to the intensity of the internal loading control.
Immunofluorescence
ATDC5 chondrocytes cells on the surface of GelMA with different matrix stiffness were cultured in 6 cm plates. The cells were treated with BAPTA, BAPTA-AM or 2-APB during the two-dimensional culture, or with or without siRNA transfection and cultured continuously for five days. The plates were then fixed using 4% paraformaldehyde at 25°C for 30 min, and then immersed in PBS (pH 7.4) and washed thrice for 5 min each. Blocking buffer (Cell Signaling Technology) was used to block for 60 min at 25°C The plates were subsequently incubated with primary antibodies in a wet box at 4°C overnight. The following day, the plates were washed using PBS and incubated with a recombinant Alexa Fluor 555 antimannose-6-phosphate receptor antibody for 60 min at 25°C in the dark. Subsequently, the slides were washed using PBS and incubated with Hoechst solution (Merck KGaA, Darmstadt, Germany) for 5 min at RT in the dark to stain cell nuclei. After a final PBS wash, the samples were airdried and sealed with anti-fluorescence quenching tablets. Digital fluorescence images were captured using a DM4000 B epifluorescence microscope (Leica Microsystems GmbH) at ×20 and ×40 magnifications, and interocular distance measurements were obtained using the Image Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
CUT&Tag-PCR
1 × 105 ATDC5 chondrocytes in the 60- and 180-kPa groups were harvested in NE buffer (20 mM HEPES-KOH, pH 7.5, 0.5 mM spermidine, 10 mM KCl, 0.1% Triton X-100, 10% glycerol, and 1 mM PMSF) and incubated on ice for 10 min. ConA beads were prewashed and resuspended in binding buffer (20 mM HEPES-KOH, pH 7.5, 10 mM KCl, 1 mM CaCl2, and 1 mM MnCl2). Beads (10 μL) were added to each sample and incubated at room temperature for 10 min. The beads were washed with washing buffer (20 mM HEPES-KOH, pH 7.5, 0.5 mM spermidine, 150 mM NaCl, and 0.1% BSA) and resuspended in blocking buffer (20 mM HEPES-KOH, pH 7.5, 0.5 mM spermidine, 150 mM NaCl, 0.1% BSA, and 2 mM EDTA) at 25°C for 5 min. Primary antibodies were added, and the samples were incubated at room temperature for 2 h. After washing with washing buffer, secondary antibodies were added, and the samples were incubated at room temperature for 30 min. Thereafter, 1.2 μL of PA-Tn5 transposomes was added to each sample and incubated at room temperature for 30 min. After washing with washing buffer, beads were treated with Pol II inhibitors or high salt (300 mM NaCl) in washing buffer for 10 min. The beads were resuspended in 30 μL of washing buffer with 10 mM MgCl2 and incubated at 37°C for 1 h. Reactions were stopped by adding 5.5 μL of stop buffer (2.25 μL of 0.5 M EDTA, 2.75 μL of 10% SDS, and 0.5 μL of 20 mg/mL Proteinase K) and incubated at 55°C for 30 min and then at 70°C for 20 min to inactivate Proteinase K. Clean DNA beads were added to each sample to extract the tagmentated DNA.39
DNA fragments were purified using DNA purification spin columns after reverse cross-linking of the DNA/protein complex. Enrichment of DNA was determined using PCR and quantitative real-time PCR. The primers used for CUT&Tag-PCR are listed in Table 1.
TEM analysis
The surface layer of cartilage tissue of the human OA tibial plateau and two-dimensional cultured ATDC5 chondrocytes were observed using TEM. Centrifuged adherent cell blocks and cartilage tissue blocks (1 mm3) were fixed and dehydrated. The sample was fixed with 2.5% glutaraldehyde for 2.5 h, washed in PBS (0.1 M, pH 7.0) three times (3 min each), and fixed with 1% osmic acid for 2 h. After washing three more times with 0.1 M PBS, the cells were continuously dehydrated in a graded ethanol series and dehydrated twice in 1:1 mixed ethanol+acetone:pure acetone. Each time, the cells were infiltrated with 3:1, 1:1, 1:1, 1:1, 1:1, and 1:1 acetone:resin for 1 h. The cells were then infiltrated with resin overnight and embedded in fresh resin for 3 h. After polymerization at 37°C for 8 h and 65°C for 48 h, the resin blocks were cut to ultra-thin slices (70–100 nm). Each ultra-thin slice was placed on a copper mesh with a carbon film, stained with uranyl acetate at 4°C for 7 min and lead citrate at 25°C for 3 min, and then observed and images were captured (Talos L120C, FEI; Thermo Fisher Scientific).
Quantification and statistical analysis
Statistical analysis
All data were collected from independent experiments or repeated measures, and the data of repeated experiments were averaged. Normal distribution and equal variance of the data were tested with the Shapiro-Wilk normality test and f test, respectively. Statistical significance was tested by Student’s paired t test. Based on normality repeated measures and one-way ANOVA followed by Tukey test for all pairwise multiple comparisons or by nonparametric Friedman test followed by Dunn’s test for all pairwise multiple comparisons. For evaluating linear relationships, Pearson correlation analysis method is used to test.33 The results are presented as mean ± standard deviation and statistical significance was set at p < 0.05. All statistical analyses were performed using GraphPad Prism 9.0 software.
Acknowledgments
T.K. and H.L. contributed equally to this work. This work was supported by the National Natural Science Foundation of China (12172223, 12272232, 82272997, and 81802679); Cross-disciplinary Research Fund of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (JYJC202201); and Shanghai Key Laboratory of Orthopaedic Implant (grant no. KFKT202203).
Author contributions
T.K. and H.L.: conceptualization, investigation, formal analysis, and writing – original draft. L.H.: investigation and formal analysis. J.C.: investigation and formal analysis. Y.W.: methodology and formal analysis. L.S.: investigation. L.W.: methodology and supervision. M.Y. and Z.Y.: conceptualization, supervision, funding acquisition, and writing – review and editing.
Declaration of interests
The authors declare no competing interests.
Published: July 17, 2024
Contributor Information
Mengning Yan, Email: yanmengning@163.com.
Zhifeng Yu, Email: zfyu@outlook.com.
References
- 1.Katz J.N., Arant K.R., Loeser R.F. Diagnosis and Treatment of Hip and Knee Osteoarthritis: A Review. JAMA. 2021;325:568–578. doi: 10.1001/jama.2020.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho K.K., Lee W.Y., Griffith J.F., Ong M.T., Li G. Randomized control trial of mesenchymal stem cells versus hyaluronic acid in patients with knee osteoarthritis - A Hong Kong pilot study. J. Orthop. Translat. 2022;37:69–77. doi: 10.1016/j.jot.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S.H., Mori D., Kobayashi H., Mori Y., Nakamoto H., Okada K., Taniguchi Y., Sugita S., Yano F., Chung U.I., et al. Excessive mechanical loading promotes osteoarthritis through the gremlin-1-NF-κB pathway. Nat. Commun. 2019;10:1442. doi: 10.1038/s41467-019-09491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Kordsmeier J., Nookaew I., Kim H.N., Xiong J. Piezo1 stimulates mitochondrial function via cAMP signaling. FASEB J. 2022;36 doi: 10.1096/fj.202200300R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng Z., Sun H., Bunpetch V., Koh Y., Wen Y., Wu D., Ouyang H. The regulation of cartilage extracellular matrix homeostasis in joint cartilage degeneration and regeneration. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120555. [DOI] [PubMed] [Google Scholar]
- 6.Mow V.C., Ratcliffe A., Poole A.R. Cartilage and diarthrodial joints as paradigms for hierarchical materials and structures. Biomaterials. 1992;13:67–97. doi: 10.1016/0142-9612(92)90001-5. [DOI] [PubMed] [Google Scholar]
- 7.Darling E.M., Wilusz R.E., Bolognesi M.P., Zauscher S., Guilak F. Spatial mapping of the biomechanical properties of the pericellular matrix of articular cartilage measured in situ via atomic force microscopy. Biophys. J. 2010;98:2848–2856. doi: 10.1016/j.bpj.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Wu Q., Zhang R., Fan Z., Li W., Mao R., Du Z., Yao X., Ma Y., Yan Y., et al. Stage-specific and location-specific cartilage calcification in osteoarthritis development. Ann. Rheum. Dis. 2023;82:393–402. doi: 10.1136/ard-2022-222944. [DOI] [PubMed] [Google Scholar]
- 9.Stolz M., Gottardi R., Raiteri R., Miot S., Martin I., Imer R., Staufer U., Raducanu A., Düggelin M., Baschong W., et al. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nat. Nanotechnol. 2009;4:186–192. doi: 10.1038/nnano.2008.410. [DOI] [PubMed] [Google Scholar]
- 10.Hodgkinson T., Kelly D.C., Curtin C.M., O'Brien F.J. Mechanosignalling in cartilage: an emerging target for the treatment of osteoarthritis. Nat. Rev. Rheumatol. 2022;18:67–84. doi: 10.1038/s41584-021-00724-w. [DOI] [PubMed] [Google Scholar]
- 11.Yan J., Shen M., Sui B., Lu W., Han X., Wan Q., Liu Y., Kang J., Qin W., Zhang Z., et al. Autophagic LC3+ calcified extracellular vesicles initiate cartilage calcification in osteoarthritis. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abn1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., Cai D., Zhou F., Yu J., Wu X., Yu D., Zou Y., Hong Y., Yuan C., Chen Y., et al. Targeting downstream subcellular YAP activity as a function of matrix stiffness with Verteporfin-encapsulated chitosan microsphere attenuates osteoarthritis. Biomaterials. 2020;232 doi: 10.1016/j.biomaterials.2019.119724. [DOI] [PubMed] [Google Scholar]
- 13.Jiang W., Liu H., Wan R., Wu Y., Shi Z., Huang W. Mechanisms linking mitochondrial mechanotransduction and chondrocyte biology in the pathogenesis of osteoarthritis. Ageing Res. Rev. 2021;67 doi: 10.1016/j.arr.2021.101315. [DOI] [PubMed] [Google Scholar]
- 14.Bolduc J.A., Collins J.A., Loeser R.F. Reactive oxygen species, aging and articular cartilage homeostasis. Free Radic. Biol. Med. 2019;132:73–82. doi: 10.1016/j.freeradbiomed.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco F.J., June R.K. Cartilage Metabolism, Mitochondria, and Osteoarthritis. J. Am. Acad. Orthop. Surg. 2020;28:242–244. doi: 10.5435/JAAOS-D-19-00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco F.J., Rego I., Ruiz-Romero C. The role of mitochondria in osteoarthritis. Nat. Rev. Rheumatol. 2011;7:161–169. doi: 10.1038/nrrheum.2010.213. [DOI] [PubMed] [Google Scholar]
- 17.Rice S.J., Beier F., Young D.A., Loughlin J. Interplay between genetics and epigenetics in osteoarthritis. Nat. Rev. Rheumatol. 2020;16:268–281. doi: 10.1038/s41584-020-0407-3. [DOI] [PubMed] [Google Scholar]
- 18.Matilainen O., Quirós P.M., Auwerx J. Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress. Trends Cell Biol. 2017;27:453–463. doi: 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Yan T., Zhao Y. Acetaldehyde induces phosphorylation of dynamin-related protein 1 and mitochondrial dysfunction via elevating intracellular ROS and Ca2+ levels. Redox Biol. 2020;28 doi: 10.1016/j.redox.2019.101381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying Z., Xiang G., Zheng L., Tang H., Duan L., Lin X., Zhao Q., Chen K., Wu Y., Xing G., et al. Short-Term Mitochondrial Permeability Transition Pore Opening Modulates Histone Lysine Methylation at the Early Phase of Somatic Cell Reprogramming. Cell Metab. 2019;29:502. doi: 10.1016/j.cmet.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Dong X., Xu X., Yang C., Luo Y., Wu Y., Wang J. USP7 regulates the proliferation and differentiation of ATDC5 cells through the Sox9-PTHrP-PTH1R axis. Bone. 2021;143 doi: 10.1016/j.bone.2020.115714. [DOI] [PubMed] [Google Scholar]
- 22.Peña-Oyarzun D., Rodriguez-Peña M., Burgos-Bravo F., Vergara A., Kretschmar C., Sotomayor-Flores C., Ramirez-Sarmiento C.A., De S.H., Reyes M., Perez W., et al. PKD2/polycystin-2 induces autophagy by forming a complex with BECN1. Autophagy. 2021;17:1714–1728. doi: 10.1080/15548627.2020.1782035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Z., Li H., Han X., Zhou F., Du J., Yang Y., Xu Q., Zhang S., Zhang S., Zhao N., et al. Irisin inhibits osteocyte apoptosis by activating the Erk signaling pathway in vitro and attenuates ALCT-induced osteoarthritis in mice. Bone. 2020;141 doi: 10.1016/j.bone.2020.115573. [DOI] [PubMed] [Google Scholar]
- 24.Huser C.A., Davies M.E. Calcium signaling leads to mitochondrial depolarization in impact-induced chondrocyte death in equine articular cartilage explants. Arthritis Rheum. 2007;56:2322–2334. doi: 10.1002/art.22717. [DOI] [PubMed] [Google Scholar]
- 25.Pathak T., Trebak M. Mitochondrial Ca2+ signaling. Pharmacol. Ther. 2018;192:112–123. doi: 10.1016/j.pharmthera.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habiballa L., Salmonowicz H., Passos J.F. Mitochondria and cellular senescence: Implications for musculoskeletal ageing. Free Radic. Biol. Med. 2019;132:3–10. doi: 10.1016/j.freeradbiomed.2018.10.417. [DOI] [PubMed] [Google Scholar]
- 27.Kim K.I., Park Y.S., Im G.I. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J. Bone Miner. Res. 2013;28:1050–1060. doi: 10.1002/jbmr.1843. [DOI] [PubMed] [Google Scholar]
- 28.Lian W.S., Ko J.Y., Wu R.W., Sun Y.C., Chen Y.S., Wu S.L., Weng L.H., Jahr H., Wang F.S. MicroRNA-128a represses chondrocyte autophagy and exacerbates knee osteoarthritis by disrupting Atg12. Cell Death Dis. 2018;9:919. doi: 10.1038/s41419-018-0994-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Y., Liu Z., Li Z., Li H., Zhu C., Li R., Zhou T., Fang B. Histone demethylase JMJD3 downregulation protects against aberrant force-induced osteoarthritis through epigenetic control of NR4A1. Int. J. Oral Sci. 2022;14:34. doi: 10.1038/s41368-022-00190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai J., Yu D., Wang Y., Chen Y., Sun H., Zhang X., Zhu S., Pan Z., Heng B.C., Zhang S., Ouyang H. Kdm6b regulates cartilage development and homeostasis through anabolic metabolism. Ann. Rheum. Dis. 2017;76:1295–1303. doi: 10.1136/annrheumdis-2016-210407. [DOI] [PubMed] [Google Scholar]
- 31.Yang F., Zhou S., Wang C., Huang Y., Li H., Wang Y., Zhu Z., Tang J., Yan M. Epigenetic modifications of interleukin-6 in synovial fibroblasts from osteoarthritis patients. Sci. Rep. 2017;7 doi: 10.1038/srep43592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lui J.C., Garrison P., Nguyen Q., Ad M., Keembiyehetty C., Chen W., Jee Y.H., Landman E., Nilsson O., Barnes K.M., Baron J. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat. Commun. 2016;7 doi: 10.1038/ncomms13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haberkamp S., Oláh T., Orth P., Cucchiarini M., Madry H. Analysis of spatial osteochondral heterogeneity in advanced knee osteoarthritis exposes influence of joint alignment. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aba9481. [DOI] [PubMed] [Google Scholar]
- 34.Major L.G., Holle A.W., Young J.L., Hepburn M.S., Jeong K., Chin I.L., Sanderson R.W., Jeong J.H., Aman Z.M., Kennedy B.F., et al. Volume Adaptation Controls Stem Cell Mechanotransduction. ACS Appl. Mater. Interfaces. 2019;11:45520–45530. doi: 10.1021/acsami.9b19770. [DOI] [PubMed] [Google Scholar]
- 35.Schwab A., Wesdorp M.A., Xu J., Abinzano F., Loebel C., Falandt M., Levato R., Eglin D., Narcisi R., Stoddart M.J., et al. Modulating design parameters to drive cell invasion into hydrogels for osteochondral tissue formation. J. Orthop. Translat. 2023;41:42–53. doi: 10.1016/j.jot.2023.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Y.X., Zhao J. Mechanobiology in cellular, molecular, and tissue adaptation. Mechanobiology in Medicine. 2023;1 doi: 10.1016/j.mbm.2023.100022. [DOI] [Google Scholar]
- 37.Meekel J.P., Mattei G., Costache V.S., Balm R., Blankensteijn J.D., Yeung K.K. A multilayer micromechanical elastic modulus measuring method in ex vivo human aneurysmal abdominal aortas. Acta Biomater. 2019;96:345–353. doi: 10.1016/j.actbio.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Kan T., Ran Z., Sun L., Jiang X., Hou L., Yang Y., Jia Z., Zhang W., Wang L., Yan M., Xie K. Cell-free fat extract-loaded microneedles attenuate inflammation-induced apoptosis and mitochondrial damage in tendinopathy. Mater. Today. Bio. 2023;22 doi: 10.1016/j.mtbio.2023.100738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Experimental data reported in this manuscript will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.