Abstract
Objective
Canine distemper virus (CDV) is a highly infectious virus that represents a threat for domestic dogs and several wild species. Despite recognized in several African countries, current knowledge of its molecular epidemiology is scarce and poorly updated.
Design
Twenty-two hemagglutinin sequences, obtained from symptomatic Namibian dogs from 2020 to 2023, were analysed through phylogenetic and phylodynamic analysis to characterize the local CDV epidemiology and contextualize it in the international scenario.
Results
Two unrelated clades were identified, including strains sampled in different Namibian towns, in the absence of a strong geographical clustering. The ancestors of the two clades were estimated to have originated from South America, likely Brazil, and South Africa, approximately in 2000 and 2006, respectively. While the introduction from South Africa was predictable, the introduction from Brazil was unexpected. The mediation of other African countries, particularly Angola, appears to be the most likely importation pathway.
Conclusions
The occurrence of multiple introduction events, likely originating from cross-border illegal animal trade between African countries, and the absence of any geographical clustering within Namibian regions, suggest a need for further investigation into its spreading pattern, as well as improved biosecurity measures to limit foreign viral introduction into the country.
Keywords: Canine distemper virus, Namibia, Phylogenesis, Phylogeography, International
Highlights
-
•
Phylogenetic and phylodynamic analysis of Namibian Distemper strains.
-
•
Strain migration from South Africa.
-
•
First description of distemper introduction from South America.
-
•
Role of cross-border illegal animal trade.
-
•
Threat for wild/endangered spercies
1. Introduction
Canine distemper virus (CDV) is a highly infectious virus affecting several carnivore species, thus displaying remarkable host flexibility. The host range of CDV primarily includes species from the order Carnivora, belonging to the families Canidae (dogs, dingoes, foxes, coyotes, jackals, and wolves), Procyonidae (raccoons and coatimundis), Mustelidae (weasels, ferrets, fishers, minks, skunks, badgers, martens, and otters), Ursidae (giant pandas), Ailuridae (red pandas), and a wide range of members from the family Felidae (lions, leopards, cheetahs, and tigers). To a lesser extent, it also affects other important families from different orders such as Artiodactyla, Primates, Rodentia, and Proboscidea [1,2].
CDV belongs to the species Morbillivirus canis, genus Morbillivirus, within the family Paramyxoviridae (https://ictv.global/taxonomy). It is an enveloped virus with a single-stranded, negative-sense RNA genome approximately 15 kb in length, encoding eight proteins. These include, from the 3′ to 5’ end: the nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin (H), and polymerase (L) proteins [3]. Additional V and C proteins are produced through RNA editing and leaky scanning of the P mRNA [4]. The H gene, which mediates viral attachment to cell receptors (SLAM, CD150, and Nectin-4), is the most investigated due to its implications for pathogenesis, and cellular and host tropism [4]. Certain sites have been reported to be under selective pressure and specific mutations can confer preferential adaptation to different hosts. Owing to this variability, the H gene is also used in molecular epidemiological studies to establish relationships among strains, cluster them based on genetic features, and estimate their dispersal over time and space [5]. To date, 18 lineages have been identified and are characterized by strong geographical clustering.
Clinically, CDV can present as catarrhal and nervous disease manifestations, either separately or combined, and as a chronic nervous form. Clinical disease is mainly the result of significant immunosuppression from viral infection of the bone marrow in the absence of a sufficient immune response, facilitating the occurrence of secondary infections. Catarrhal form symptoms are most common and may include pyrexia, mucopurulent conjunctivitis, cough, anorexia, gastrointestinal signs like vomiting or diarrhoea, and may be associated with enamel hypoplasia in permanent teeth. The disease becomes progressive once the neurological form manifests, and symptoms may encompass myoclonus, seizures, ataxia or paresis, postural reaction deficits, and may be associated with hyperkeratosis of the nasal planum and digital footpads [6].
Severe disease and mortality have been observed in several wild canids and carnivores, although great variations in the clinical disease were described, ranging from subclinical infection to death [7]. The disease's severity, combined with high infectivity and host plasticity, has made CDV a major threat to wildlife conservation, particularly among endangered species [1]. The introduction of vaccines has significantly reduced morbidity and mortality in domestic dogs and decreased spillover events to wildlife [8]. However, vaccination efficacy is never complete, particularly when not properly performed, and coverage varies significantly depending on the socio-economic context, potentially favouring infection circulation [9]. Moreover, CDV vaccination in wildlife is controversial and all CDV vaccine use in non-domestic animals is extra-label [7,10]. In low-income areas of several African countries vaccine administration poses a significant challenge and a high wildlife richness is often present. Dogs kept in a free-ranging lifestyle in such areas are often at an increased risk of interaction with wild, especially synanthropic, animals. Information on this topic in Africa is often sparse, not systematically collected, outdated, and sequence data is almost negligible, except for in South Africa [8,[11], [12], [13]].
CDV is a recognized disease in Namibia and its impact on domestic and wild carnivores has been reported [14]. However, updated survey and molecular epidemiological data are lacking. This study investigated CDV occurrence in clinically affected domestic dogs from three Namibian towns and characterized the detected strains by sequencing the H gene to investigate distribution and circulation patterns, within Namibia and in the international context.
2. Materials and methods
2.1. Sample collection
CDV-positive samples (blood, brain, and conjunctival swabs) from twenty-two dogs were collected by private veterinarians for routine diagnostic purpose between February 2020 and October 2023 from 3 Namibian towns (Windhoek, Gobabis, and Okahandja). The archived, secondary sample material was further evaluated in this study. All samples were stored at -80°C until processing. Metadata related to clinical presentation was recovered from patient records and reported for each patient. Windhoek is the capital located in the central Khomas region of Namibia. Okahandja and Gobabis are smaller towns located north of Windhoek, in the Otjozondjupa region, and east of Windhoek, in the Omaheke region, respectively.
2.2. RNA extraction, diagnosis, and sequencing
All brain and conjunctival samples were homogenized in 1 ml of sterile Phosphate Buffered Saline (PBS) using a TissueLyser LT (Qiagen, Germany). Total genomic RNA was extracted from the homogenized samples and blood using the High Pure Viral Nucleic Acid Kit (Hoffman-La Roche, Switzerland), with an elution volume of 100 μl, following the manufacturer's instructions.
RNA extracts were screened using a real-time PCR (qPCR) assay with specific primers and probe for CDV, as reported by Elia et al. (2006) [15]. Briefly, qPCR was performed on a C1000 Bio-Rad thermocycler (Bio-Rad, Hercules, CA, USA) with the SuperScript III Platinum One-Step Quantitative RT-PCR System (Invitrogen), under the following cycling conditions: activation at 95 °C for 10 min, followed by 45 cycles consisting of denaturation at 95 °C for 15 s, primer annealing at 48 °C for 1 min, and extension at 60 °C for 1 min.
Thereafter, the complete H gene was amplified in samples positive with a Cq ≤ 35 to the qRT-PCR reaction. For this purpose, four different primer pairs were used, and RT-PCR was conducted using a one-step RT-PCR kit (Qiagen, Hilden, Germany), as previously described by Trogu et al. (2022) [16]. All PCRs were conducted under the same conditions with the following thermocycler program: after reverse transcription, cDNA was denatured at 95 °C for 15 min, followed by 40 cycles of denaturation at 94 °C for 15 s, annealing at 51 °C for 30 s, extension at 68 °C for 1 min, and a final extension at 68 °C for 5 min.
Amplicons from positive samples were purified using the Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced commercially by LGC Genomics (Berlin, Germany). The sequences of the positive samples were edited and assembled using the Staden software package version 2.0.0b8. All obtained sequences were submitted to the GenBank database under accession numbers OR979445-OR979466.
2.3. Sequence dataset preparation
Besides the Namibian sequences obtained in the present study, all complete H sequences were downloaded from GenBank, provided they included the collection date and country. All sequences were aligned at the amino acid level and then back-translated to the nucleotide level using the MAFFT method implemented in TranslatorX [17]. After alignment, the obtained sequences were visually inspected to evaluate their quality. Sequences exhibiting poor alignment, frameshift mutations, and premature stop codons were removed from the analysis. Sequences of adequate quality were then merged with those obtained from Namibia. The presence of recombination in the considered region was evaluated using RDP4 [18], adjusting the parameters of the different methods based on the dataset features, according to the RDP4 manual's suggestions. Sequences were considered recombinant if detected by more than two methods with a significance level of p < 0.01, after Bonferroni correction, and were removed from further analysis.
2.4. Phylogenetic analysis
The relationships among Namibian sequences were investigated by reconstructing a maximum likelihood (ML) phylogenetic tree using IQ-Tree (http://iqtree.cibiv.univie.ac.at/). The best substitution model was selected using the Bayesian Information Criterion (BIC) calculated using the same software and the robustness of inferred clades was assessed by performing 10 000 ultrafast bootstrap replicates.
A comparable approach was selected to contextualize the Namibian sequences with the international scenario.
2.5. Phylodynamic analysis
The population history, evolution, and migration pattern of distemper strains were estimated to investigate the origin and timing of CDV introduction in Namibia, using the Bayesian serial coalescent approach implemented in BEAST 1.10.4 [19]. Several population parameters, including time to the most recent common ancestor (tMRCA), evolutionary rate, and viral population dynamics were jointly estimated. The nucleotide substitution model was selected based on the BIC calculated using JModelTest. The relaxed log-normal molecular clock and the Bayesian Skyline were preferred over alternative models based on Bayesian factor (BF) evaluation, calculated through marginal likelihood estimation through path sampling and stepping-stone methods, as suggested by Baele et al. [20]. A discrete state phylogeographic analysis was also performed as described by Lemey et al. [21] assuming the collection continent as the trait of interest and selecting the asymmetric migration model with Bayesian stochastic search variable selection (BSSVS). A 500 million generations Markov chain Monte Carlo run was performed sampling population parameters and trees every fifty-thousand generations. The log files were analysed using Tracer 1.7 and accepted only if the estimated sample size (ESS) was greater than 200 and the convergence and mixing were adequate. Parameter estimation was summarized in terms of mean and 95 % highest posterior density (HPD). Maximum clade credibility (MCC) trees were constructed and annotated using TreeAnnotator (BEAST package). The strain migration among countries was also reconstructed using the same approach. However, to reduce the computational complexity due to the higher feature number, a subset of sequences was selected by randomly subsampling a maximum of three sequences per country‐year. A total of six random datasets were generated and independently analysed to investigate the effect of sampling on the country-level analysis, but also on the complete dataset analysis. In fact, the differential sampling activity or sequence availability over time and geographic area could have biased the results.
3. Results
3.1. Sampled population features and CDV test results
Archived samples from a total of 12 male and 10 female dogs were enrolled in the study based on clinical features recorded, especially catarrhal and neurological symptoms, and confirmed to be CDV positive by qPCR. Patient metadata related to disease manifestation are presented in Supplemetary Table 1. Briefly, some of the CDV-infected dogs showed non-specific clinical features, including lethargy (N = 6), inappetence (N = 8), emaciation (N = 2), pyrexia (N = 5), and lymphadenomegaly (N = 1). Catarrhal signs were prevalent in most cases and included ocular and nasal discharge (N = 15), as well as associated gastrointestinal symptoms like diarrhoea (N = 7) and vomiting (N = 5). Neurological signs were present in many of the cases and included features like myoclonus (N = 8), ataxia (N = 5), nystagmus (N = 1), circling (N = 4), postural deficits (N = 3), salivation (N = 3), and seizures (N = 1). Hyperkeratosis of the nose and footpads (N = 2) were also recorded in selected cases. Neurological symptoms were concurrent in many of the patients where catarrhal signs were recorded, suggesting acute systemic infection. None of the animals recovered, and euthanasia was the course of treatment in all cases.
Table 1.
List of CDV positive samples and related metadata.
| ID | Accession | Age | Gender | Breed | Collection date | Sample type | Town | CDV vaccination history |
|---|---|---|---|---|---|---|---|---|
| D2 | OR979465 | 2 years | F | Mixed | 18/02/2020 | Blood (EDTA) | Windhoek | Vaccinated (2020) |
| D3 | OR979466 | 5 months | F | Mixed | 19/02/2020 | Blood (EDTA) | Gobabis | Partial vaccination (2020) |
| D4 | OR979456 | 3 months | M | Mixed | 19/02/2020 | Brain | Gobabis | No vaccination |
| D5 | OR979455 | 1 month | F | Mixed | 20/02/2020 | Blood (EDTA) | Windhoek | No vaccination |
| D6 | OR979454 | 1 month | F | Mixed | 24/02/2020 | Blood (EDTA) | Windhoek | Partial vaccination (2020) |
| D7 | OR979453 | 1 month | F | Mixed | 02/03/2020 | Brain | Windhoek | Partial vaccination (2020) |
| D8 | OR979452 | 7 years | M | Mixed | 23/03/2020 | Blood (EDTA) | Windhoek | No vaccination |
| D9 | OR979451 | 6 years | M | Mixed | 08/04/2020 | Brain | Windhoek | No vaccination |
| D10 | OR979450 | 6 months | M | Mixed | 30/05/2020 | Brain | Windhoek | No vaccination |
| D11 | OR979448 | 1 year | F | Mixed | 13/06/2020 | Blood (EDTA) | Okahandja | No vaccination |
| D12 | OR979449 | 2 years | F | Mixed | 13/06/2020 | Blood (EDTA) | Okahandja | No vaccination |
| D13 | OR979447 | 3 years | M | Mixed | 02/07/2020 | Brain | Windhoek | No vaccination |
| D14 | OR979446 | 6 months | M | Mixed | 10/07/2020 | Brain | Gobabis | No vaccination |
| D15 | OR979445 | 2 years | M | Mixed | 15/05/2023 | Blood (EDTA) | Windhoek | No vaccination |
| D16 | OR979462 | 1.5 years | F | Mixed | 01/08/2023 | Blood (EDTA) | Windhoek | No vaccination |
| D17 | OR979461 | 4 months | M | Mixed | 28/08/2023 | Blood (EDTA) | Windhoek | Partial vaccination (2023) |
| D18 | OR979460 | 7 months | M | Pure | 29/08/2023 | Blood (EDTA) | Windhoek | No vaccination |
| D19 | OR979459 | 10 years | M | Mixed | 24/08/2023 | Blood (EDTA) | Windhoek | Vaccinated (2020) |
| D20 | OR979458 | 1.8 years | M | Pure | 30/08/2023 | Ocular swab | Windhoek | Vaccinated (2023) |
| D21 | OR979457 | 9 months | F | Mixed | 04/09/2023 | Blood (EDTA) | Windhoek | No vaccination |
| D23 | OR979463 | 2 months | F | Pure | 19/09/2023 | Ocular swab | Windhoek | No vaccination |
| D24 | OR979464 | 9 months | M | Mixed | 02/10/2023 | Ocular swab | Gobabis | No vaccination |
Patient ranged between 1 month and ten years of age, and the majority were of mixed breed. Only seven animals had a history of CDV vaccination (Table 1). A total of 13 whole blood (EDTA), six brain, and three ocular swab archived samples were evaluated. All 22 dogs involved in the study tested positive for CDV and the H gene was successfully sequenced from all the positive samples.
3.2. Sequence, phylogenetic and phylogeographic analysis of Namibia strains in the national and international more surprising is the context
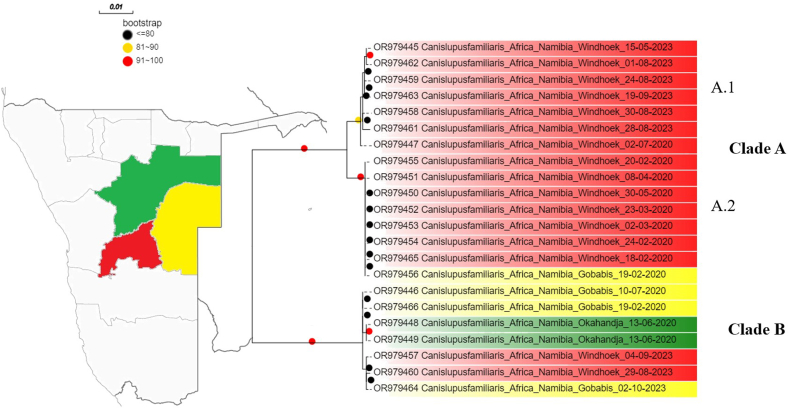
The average genetic distance among Namibian strains was 3.19 % [interval 0.00–6.63 %]. Two main clades with high bootstrap support were identified. The biggest one (Clade A) included only strains originating from Windhoek, with the only exception of one strain from Gobabis. This clade could be further divided in 2 closely related subclades (A.1 and A.2). The other clade (Clade B) included more geographically heterogeneous strains, 2 from Windhoek, 2 from Okahandja, and 3 from Gobabis. Strains collected from diseased dogs with a history of CDV vaccination were part of both Clades and subclades A.1 and A.2 (Fig. 1).
Fig. 1.
Maximum likelihood phylogenetic tree based on the H gene sequence analysis of Namibian strains. The labels have been color-coded depending on the region of origin, as depicted in the Namibian map (left image). The bootstrap support relative to each clade has also been color-coded.
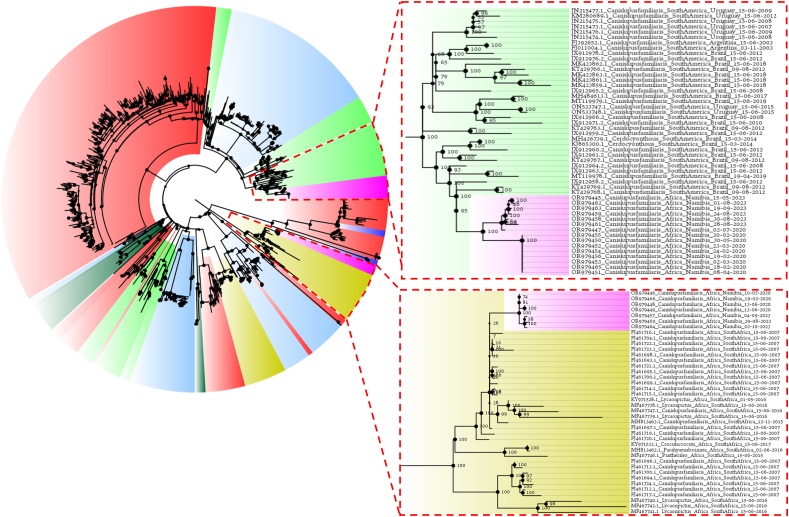
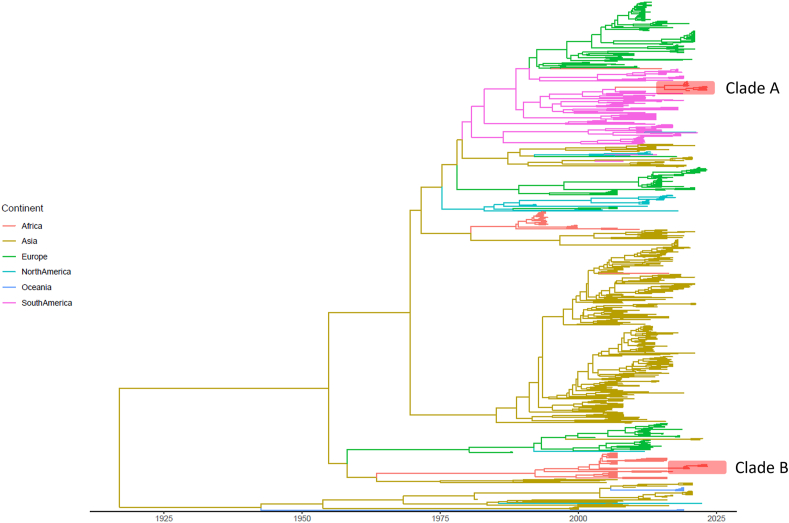
The phylogenetic analysis, performed on the international dataset, confirmed the presence of several CDV lineages with strong geographical clustering, with only limited exceptions. Clade B was part of the South African lineage, which included both domestic dogs and wild species. However, Namibian strains formed an independent cluster separated by a long branch. Clade A formed an independent cluster also, separated by a long branch with a high bootstrap support stemming from the Europe/South America-1 Lineage that included also Argentinian, Uruguayan, and Brazilian strains, with the latter showing the closest relationship with Namibian strains (Fig. 2). The phylodynamic and phylogeographic analyses estimated the CDV tMRCA in 1916.52 [95HPD: 1818.95–1955.32] and the evolutionary rate 9.10∙10−4 [95HPD: 8.18∙10−4-1.00∙10−3] and confirmed the strong phylogeographic clustering (Fig. 3), regardless of the considered randomly generated dataset.
Fig. 2.
Maximum likelihood phylogenetic tree based on the H gene sequence analysis of worldwide collected strains. The labels have been color-coded depending on the continent of origin (i.e. Africa in light brown, Asia in red, Europe in light blue, North America in dark green, Oceania in dark blue, South America in light green; Namibia is reported in Purple). The African and South American clades including the Namibian strain have been magnified.
Fig. 3.
Maximum Clade Credibility tree, reconstructing the migration of distemper strains among continents (color-coded) over time. The Namibian clades have been highlighted. Further details are available in Supplementary Fig. 1.
The origin of Namibian Clade A from South America and Clade B from South Africa were estimated around 2006 [95HPD: 2003.48–2009.48], with the two clades diverging around 2015 [95HPD: 2011.56–2018.04] and 2000 [95HPD: 1998.51–2003.88], respectively (Fig. 3 and Supplementary Fig. 1). The analysis of the randomly generated dataset confirmed the location origin and timing, even when countries rather than continents were included in the analysis (Supplementary Fig. 2). Brazil and South Africa were confirmed as the most likely introduction source of Clade A and B, respectively.
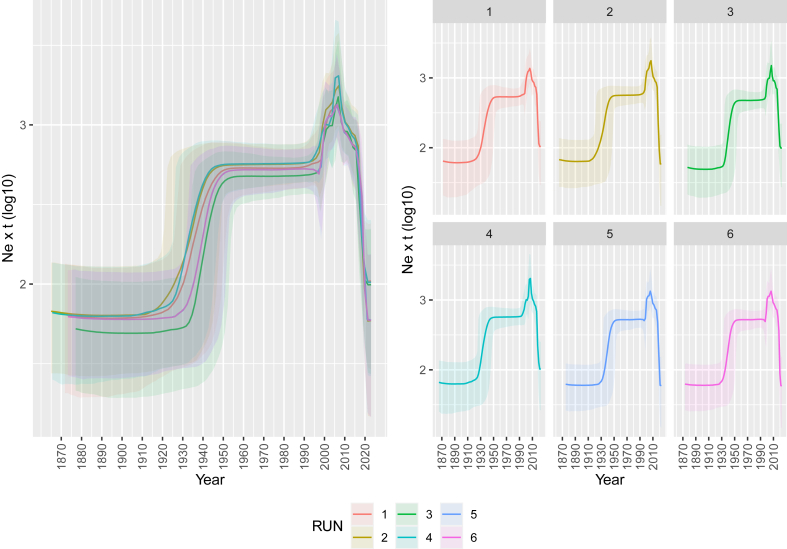
The evaluation of distemper population dynamics was characterized by an essentially constant population until approximately the 1990s, when a remarkable increase peaking around 2008 was observed, with a more stationary phase between 1995 and 2005. Thereafter, a decline, albeit with fluctuations characterized by alternating recoveries and stationary phases, marked the latter part of CDV history (Fig. 4).
Fig. 4.
Left figure: Mean relative genetic diversity (Ne x t) of the worldwide CDV population over time. The results of the 6 independent runs have been color-coded. Right figure: Mean, median and upper and lower 95HPD values are reported for each run.
4. Discussion
The occurrence of CDV has previously been reported in Namibia [14]; however, information is extremely limited, with no sequence data available to understand potential introduction, persistence, and spreading patterns. This study demonstrates significant genetic variability of CDV in Namibia, even within the relatively limited geographic area. Two main clades were identified, encompassing strains from different towns. Notably, Clade B included strains originating from various cities, indicating significant viral dispersal, and mixing through various mechanisms. Primarily, the movement of humans and their pets is common and Windhoek serves as a major centre where people from the countryside and smaller cities congregate for goods and services (Dr. Khaiseb, Namibian Deputy Chief Veterinary Officer, personal communication). This could explain the high number of diseased animals in this city/region and its potential role as a nucleus for viral dispersal. Furthermore, pet raising and trade in the poorer districts of Windhoek, intended for sale in other cities, is widespread. The lack of geographical clustering among canine infections such as canine parvovirus and canine circovirus was previously demonstrated in the same area [22,23] and the epidemiological significance of legal, and especially illegal, trade as a CDV spreading mechanism has been repeatedly demonstrated in other countries and continents [[24], [25], [26], [27]]. Therefore, common socio-economic factors can be considered as drivers shaping the epidemiology of several major canine pathogens. Informing and educating people about CDV disease remains vital to curbing the predictably significant implications disease spread has for companion animals' health and welfare.
The detection of identical or nearly identical strains in 2020 and 2023 from the same area suggests the effective establishment and maintenance of at least certain infectious clusters. However, Clade A.2 was detected only in 2020. Whether this scenario is due to limited sample size or conceals biological or epidemiological factors, such as increased population immunity, requires further investigation.
Vaccination has proven effective in preventing or limiting the disease and decreasing infection transmission [10]. Most animals in this study were not vaccinated, which could have facilitated viral persistence and spread. Several animals were only partially vaccinated at an age less than 6 months, suggesting interference of maternally derived antibodies (MDA) with vaccine efficacy. One animal had a CDV vaccination history outdating a 3-year cycle, suggestive of diminished immunocompetence and increased susceptibility to clinical disease [28]. Notably, some diseased animals had a current vaccination history, which could suggest a reduced cross-protection among distantly related strains. In the last few years, there have been reports of cases of CDV in vaccinated dogs at a worldwide level and differences in antigenic sites and variations in cross-protection have been proved [29]. In this study, vaccinated animals were affected by strains belonging to both Clade A and B, including both subtypes of Clade A (A.1 and A.2). This evidence might challenge the hypothesis of specific escape lineages. Nevertheless, a complex interaction between the host, circulating strains, and vaccine features might hinder the detection of a clear pattern. Despite the overall magnitude of cross-protection loss likely being limited, improper vaccine handling, competition with maternal antibodies, and decreasing immunity over time might contribute to outbreaks occurring in vaccinated populations [29,30]. Such phenomena cannot be excluded in the Namibian scenario also and may be contributing variables to disease emergence [3].
The comparison with the international scenario and the estimation of the most likely sources of introduction have yielded some of the most interesting findings. Our aim was not to describe the origin and overall dispersal patterns of CDV, since it has already been performed in Wang et al., 2023 [5]. Our estimation, although based on a broader dataset, overlaps Wang et al., 2023's study in terms of viral origin, evolutionary rate, population size dynamics, and main spreading patterns, thus supporting the goodness and reliability of both studies. Therefore, we refer to Wang et al., 2023 for a more extensive traction. Our study's focus was on the introduction pathways of CDV strains into Namibia. Clade B was likely introduced from South Africa around 2006. CDV has been characterized in domestic dogs and African wild dogs of South Africa from the Tswalu Kalahari Reserve in the Northern Cape province [8]. Strains from both domestic and wild animals from the Northern Cape providence, which neighbours Namibia's south-eastern borders, are closely related to those now identified in Namibia [8]. Therefore, this route of introduction was not unexpected, although the dynamics, mediated by domestic or wild species, remain elusive. The presence of the AA marker 530G/N and 549Y in Namibian viruses, commonly associated with dog infections [31,32], might suggest pet transportation as the most likely source of viral introduction. However, such an association, although commonly accepted, is not statistically significant and exceptions have been reported, despite being often justified as the consequence of potential contact with wildlife [32,33]. Therefore, other pathways and intermediate steps might have been involved. After the introduction, the considered clade underwent an independent evolution lasting for decades, leading to progressive diversification.
More surprising is the origin of Clade A, with an estimated origin from Brazil around 2000, supported by both phylogenetic and phylogeographic evidence. Previous studies reported well-supported migration routes from the USA to South Africa and from Europe to Gabon [5], but South America was never previously implicated in CDV's spread to Africa. Therefore, the connection between these two countries, which have negligible socio-economic relations, poses a significant challenge to explain.
Likely, intermediate steps were involved. One intriguing, albeit speculative, hypothesis suggests Angola as a potential intermediary in the viral migration. The historical and cultural connections between Brazil and Angola, both former colonies of Portugal, along with common trade and travel are relevant. Three factors may have facilitated the importation of CDV from Angola (Dr. Khaiseb, Namibian Deputy Chief Veterinary Officer, personal communication), supporting the hypothesis and explaining the presence of Brazilian-like strains in Namibia. Firstly, a significant community of wealthy Angolans, living in Namibia or frequently traveling between the two countries with their pets, has emerged and grown since 2000, coinciding with the estimated period of CDV introduction into Namibia. Additionally, the absence of barriers between Angola and Namibia allows for the exchange and trade of goods and animals among people of the same ethnic group (Owambo tribe) residing on both sides. Lastly, working dogs used in animal management could have crossed the border during transhumance. In past investigations of Foot-and-Mouth Disease outbreaks, Namibian cattle were found 250 km inside Angola, accompanied by dogs typically used to herd the animals. The potential for contact with CDV strains circulating in Angola cannot be ignored. Accordingly, the occurrence of multiple introduction events that likely originate from cross-border illegal animal trade between African countries was proposed for canine parvovirus also, highlighting the challenge of controlling illegal animal imports across land borders [22].
Alternative explanations might also be considered. For example, a strain previously identified in Gabon is part of the same lineage and thus shares a common ancestor with the Namibian ones. A within-Africa viral evolution and dispersal, from Gabon to Namibia, potentially mediated by Angola, might represent a plausible alternative. However, the long branches separating the considered strains, together with the presence of intermediate Brazilian strains estimated both through phylogenetic and phylodynamic analysis, seem to lessen this hypothesis. While more intricate and less parsimonious dispersal patterns are often regarded as less likely, their relevance must not be overlooked and will deserve further investigation if more sequences from the involved areas become available.
Although poorly documented, the history of distemper in Namibia is likely quite ancient. Nevertheless, it is fascinating to note that a major CDV outbreak, affecting both domestic dogs and jackals, occurred between 2002 and 2003. In early 2002, numerous distemper cases were reported in the Windhoek capital, thereafter signaled in the western region of Walvis Bay, and then in other southwestern coastal regions [14]. It would be tempting to speculate that the introduction of a new strain during those years might have led to this massive outbreak. Unfortunately, no adequate sequences were available for comparison, and while this theory is compelling, alternative pathways may exist. Further evidence is needed to corroborate this hypothesis, particularly through a more thorough understanding of CDV molecular epidemiology in Africa.
Overall, the present study highlighted the significance of CDV as a causative agent of pathogenic disease in Namibian dogs. Two main introduction events, followed by local persistence and diversification, were identified. While the introduction from South Africa could have been anticipated given the close ties between the countries, the spread from Brazil was unexpected. It was possible to hypothesize, though not confirm, that the virus may have been transmitted through other African nations like Angola. The detection of multiple instances of the virus's introduction, presumably through cross-border animal trade, including illegal ones, among African nations, coupled with the lack of geographical clustering of cases within Namibia, underscores the urgent need for more detailed epidemiological studies. Further investigations could map the spread and circulation of viral pathogens more accurately and help devise enhanced biosecurity strategies to prevent the ingress of foreign viruses into the country. Education of the general populace on early identification of CDV disease manifestation, the risks of disease spread, and the importance of vaccination should continue in the sustainment of companion animal health and the pursuit of limiting the well-known risks of disease spread at the domestic animal-wildlife interface.
Funding
This research was supported by EU funding within the NextGeneration EU-MUR PNRR Extended
Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT), by the Italian Ministry of Health through the Ricerca Corrente 2022, project “OneCoV: coronavirus animali emergenti e impatto nella Salute Pubblica”, recipient Alessio Lorusso, by the project “proDIACO” funded by the Tercas foundation, and supported by funds from the IAEA Peaceful Uses Initiative (PUI) VETLAB Network, Austria, and by the Department of Animal Medicine, Production and Health, University of Padua, Italy [grant number BIRD225455/22]
Disclosure statement
The authors declare no conflict of interest.
Ethical statement
Reviewer and/or approval by an ethics committee was not needed for this study since no experiments on live animals were performed; the study was carried out using remaining material obtained during routine clinical diagnostic activity.
Data availability
All obtained sequences were submitted to the GenBank database under accession numbers OR979445-OR979466.
CRediT authorship contribution statement
Giovanni Franzo: Writing – original draft, Software, Methodology, Formal analysis, Data curation, Conceptualization. Lourens de Villiers: Writing – review & editing, Formal analysis, Data curation. Lauren M. Coetzee: Visualization, Formal analysis, Data curation. Mari de Villiers: Investigation, Formal analysis. Francis N. Nyathi: Methodology, Formal analysis. Maya Garbade: Methodology, Investigation, Formal analysis, Data curation. Chantal Hansen: Formal analysis, Data curation. Shadia Berjaoui: Formal analysis, Data curation. Paola Ripà: Resources, Investigation. Alessio Lorusso: Writing – review & editing, Resources, Funding acquisition. Umberto Molini: Writing – original draft, Resources, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The study extends thanks to the following colleagues/entities, whether through direct or indirect involvement: Dr S. Khaiseb; Dr S. Phiri; Dr C. Van der Merwe; and Rhino Park Veterinary Clinic.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34805.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Maximum Clade Credibility tree, reconstructing the migration of distemper strains among continents (color-coded) over time. Metadata on strains included in the analysis are reported in the tip labels. The posterior probability associated to each ancestor state reconstruction support has been coded as circles of increasing size displayed over the corresponding node.
Maximum Clade Credibility tree, reconstructing the migration of distemper strains among countries (color-coded) over time. The results of six, randomly generated datasets are reported. Metadata on strains included in the analysis is reported in the tip labels. The posterior probability associated with each ancestor state reconstruction support has been coded as circles of increasing size displayed over the corresponding node.
References
- 1.Loots A.K., Mitchell E., Dalton D.L., Kotzé A., Venter E.H. Advances in canine distemper virus pathogenesis research: a wildlife perspective. J. Gen. Virol. 2017;98:311–321. doi: 10.1099/JGV.0.000666/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- 2.Beineke A., Baumgärtner W., Wohlsein P. Cross-species transmission of canine distemper virus—an update. One Health. 2015;1:49–59. doi: 10.1016/J.ONEHLT.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martella V., Elia G., Buonavoglia C. Canine distemper virus. Vet. Clin. Small Anim. Pract. 2008;38:787–797. doi: 10.1016/J.CVSM.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Rendon-Marin S., Da Fontoura Budaszewski R., Canal C.W., Ruiz-Saenz J. Tropism and molecular pathogenesis of canine distemper virus. Virol. J. 2019;16:1–15. doi: 10.1186/S12985-019-1136-6/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., Guo H., Hein V.G., Xu Y., Yu S., Wang X. The evolutionary dynamics history of canine distemper virus through analysis of the hemagglutinin gene during 1930–2020. Eur. J. Wildl. Res. 2023;69:1–12. doi: 10.1007/S10344-023-01685-Z/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sykes J.E. Elsevier Health Sciences; 2022. Greene's Infectious Diseases of the Dog and Cat-E-Book. [Google Scholar]
- 7.Wilkes R.P. Canine distemper virus in endangered species: species jump, clinical variations, and vaccination. Pathogens. 2023;12:57. doi: 10.3390/pathogens12010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loots A.K., Mokgokong P.S., Mitchell E., Venter E.H., Kotze A., Dalton D.L., et al. Phylogenetic analysis of canine distemper virus in South African wildlife. PLoS One. 2018;13:1–13. doi: 10.1371/journal.pone.0199993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zacarias J., Dimande A., Achá S., Dias P.T., Leonel E.M., Messa A., et al. Severe canine distemper outbreak in unvaccinated dogs in Mozambique. J. S. Afr. Vet. Assoc. 2016;87 doi: 10.4102/JSAVA.V87I1.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tizard I.R. Canine vaccines. Vaccines for Veterinarians. 2021;153 doi: 10.1016/B978-0-323-68299-2.00022-8. [DOI] [Google Scholar]
- 11.Roelke-Parker M.E., Munson L., Packer C., Kock R., Cleaveland S., Carpenter M., et al. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996;379:441–445. doi: 10.1038/379441A0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goller K.V., Fyumagwa R.D., Nikolin V., East M.L., Kilewo M., Speck S., et al. Fatal canine distemper infection in a pack of African wild dogs in the Serengeti ecosystem, Tanzania. Vet. Microbiol. 2010;146:245–252. doi: 10.1016/J.VETMIC.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Van De Bildt M.W.G., Kuiken T., Visee A.M., Lema S., Fitzjohn T.R., Osterhaus A.D.M.E. Distemper outbreak and its effect on african wild dog conservation. Emerg. Infect. Dis. 2002;8:212. doi: 10.3201/EID0802.010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gowtage-Sequelra S., Banyard A.C., Barrett T., Buczkowskl H., Funk S.M., Cleaveland S. Epidemiology, pathology, and genetic analysis of A canine distemper epidemic in Namibia. J. Wildl. Dis. 2009;45:1008–1020. doi: 10.7589/0090-3558-45.4.1008. [DOI] [PubMed] [Google Scholar]
- 15.Elia G., Decaro N., Martella V., Cirone F., Lucente M.S., Lorusso E., et al. Detection of canine distemper virus in dogs by real-time RT-PCR. J. Virol Methods. 2006;136:171–176. doi: 10.1016/J.JVIROMET.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Trogu T., Castelli A., Canziani S., Tolini C., Carrera M., Sozzi E., et al. Detection and molecular characterization of canine distemper virus in wildlife from northern Italy. Pathogens. 2022;11:1557. doi: 10.3390/PATHOGENS11121557/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abascal F., Zardoya R., Telford M.J. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:1–5. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4 doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baele G., Lemey P., Bedford T., Rambaut A., Suchard M.A., Alekseyenko A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzo G., De Villiers L., De Villiers M., Ravandi A., Gyani K., Van Zyl L., et al. Molecular epidemiology of canine parvovirus in Namibia: introduction pathways and local persistence. Prev. Vet. Med. 2022;209 doi: 10.1016/j.prevetmed.2022.105780. [DOI] [PubMed] [Google Scholar]
- 23.de Villiers L., Molini U., Coetzee L.M., Visser L., Spangenberg J., de Villiers M., et al. Molecular epidemiology of Canine circovirus in domestic dogs and wildlife in Namibia, Africa. Infect. Genet. Evol. 2023;112 doi: 10.1016/j.meegid.2023.105458. [DOI] [PubMed] [Google Scholar]
- 24.Willi B., Spiri A.M., Meli M.L., Grimm F., Beatrice L., Riond B., et al. Clinical and molecular investigation of a canine distemper outbreak and vector-borne infections in a group of rescue dogs imported from Hungary to Switzerland. BMC Vet. Res. 2015;11:1–15. doi: 10.1186/S12917-015-0471-0/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alfano F., Lanave G., Lucibelli M.G., Miletti G., D'Alessio N., Gallo A., et al. Canine distemper virus in autochtonous and imported dogs, southern Italy (2014–2021) Animals (Basel) 2022;12 doi: 10.3390/ANI12202852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrino M., Tassoni L., Campalto M., Cavicchio L., Mion M., Corrò M., et al. Molecular investigation of recent canine parvovirus-2 (CPV-2) in Italy revealed distinct clustering. Viruses. 2022;14 doi: 10.3390/V14050917/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campalto M., Carrino M., Tassoni L., Rizzo G., Rossmann M.C., Cocchi M., et al. Divergent minute virus of canines strains identified in illegally imported puppies in Italy. Arch. Virol. 2020;165:2945–2951. doi: 10.1007/S00705-020-04800-6/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day M.J., Horzinek M.C., Schultz R.D., Squires R.A. vol. 57. 2016. (GUIDELINES FOR THE VACCINATION OF DOGS AND CATS COMPILED BY THE VACCINATION GUIDELINES GROUP (VGG) OF THE WORLD SMALL ANIMAL VETERINARY ASSOCIATION (WSAVA)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anis E., Holford A.L., Galyon G.D., Wilkes R.P. Antigenic analysis of genetic variants of Canine distemper virus. Vet. Microbiol. 2018;219:154–160. doi: 10.1016/j.vetmic.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Bi Z., Xia X., Wang Y., Mei Y. Development and characterization of neutralizing monoclonal antibodies against canine distemper virus hemagglutinin protein. Microbiol. Immunol. 2015;59:202–208. doi: 10.1111/1348-0421.12238. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Gutierrez M., Ruiz-Saenz J. Diversity of susceptible hosts in canine distemper virus infection: a systematic review and data synthesis. BMC Vet. Res. 2016;12 doi: 10.1186/S12917-016-0702-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duque-Valencia J., Sarute N., Olarte-Castillo X.A., Ruíz-Sáenz J. Evolution and interspecies transmission of canine distemper virus—an outlook of the diverse evolutionary landscapes of a multi-host virus. Viruses. 2019;11:582. doi: 10.3390/V11070582. 2019;11:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolin V.M., Wibbelt G., Michler F.-U.F., Wolf P., East M.L. Susceptibility of carnivore hosts to strains of canine distemper virus from distinct genetic lineages. Vet. Microbiol. 2012;156:45–53. doi: 10.1016/j.vetmic.2011.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum Clade Credibility tree, reconstructing the migration of distemper strains among continents (color-coded) over time. Metadata on strains included in the analysis are reported in the tip labels. The posterior probability associated to each ancestor state reconstruction support has been coded as circles of increasing size displayed over the corresponding node.
Maximum Clade Credibility tree, reconstructing the migration of distemper strains among countries (color-coded) over time. The results of six, randomly generated datasets are reported. Metadata on strains included in the analysis is reported in the tip labels. The posterior probability associated with each ancestor state reconstruction support has been coded as circles of increasing size displayed over the corresponding node.
Data Availability Statement
All obtained sequences were submitted to the GenBank database under accession numbers OR979445-OR979466.