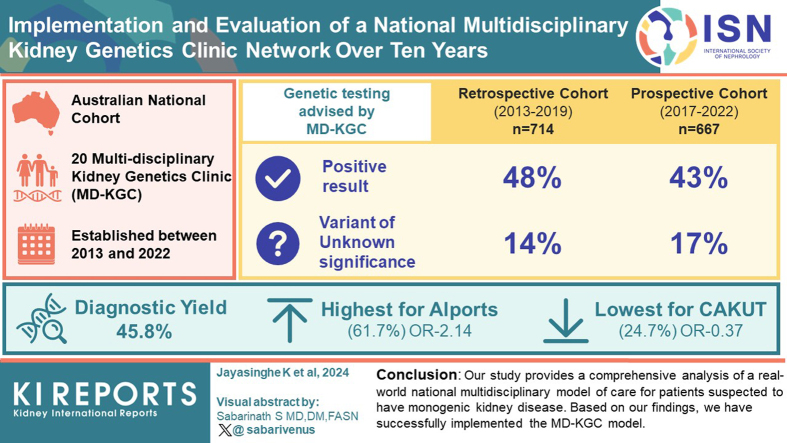
Abstract
Introduction
Diagnostic genomic sequencing is the emerging standard of care in nephrology. There is a growing need to scale up the implementation of genomic diagnostics nationally to improve patient outcomes.
Methods
This pragmatic study provided genomic or genetic testing to patients with suspected monogenic kidney disease through a national network of kidney genetics clinics (KGCs). We sought to evaluate the experiences of implementing genomic diagnostics across Australia and associated diagnostic outcomes between 2013 and 2022.
Results
We successfully established and expanded a nationwide network of 20 clinics as of 2022; concurrently developing laboratory, research, and education programs to scale the clinical application of genomics in nephrology. We report on an Australian cohort of 1506 kidney patients, of whom 1322 received their test results. We assessed barriers to implementation in the nephrology context, and where possible, applied real-time solutions to improve clinical processes over 10 years.
Conclusion
Developing a multidisciplinary kidney genetics model across multiple health services nationally was highly successful. This model supported optimal care of individuals with monogenic kidney disease in an economically responsible way. It has continued to evolve with technological and service developments and is now set to scale further as genomic testing for kidney patients transitions to health care system funding.
Keywords: genomic testing, implementation, kidney disease
Graphical abstract
Kidney diseases are a major global health priority, and monogenic causes account for 30% to 50% of childhood nephropathy1,2 and 10% to 30% of adult chronic kidney disease.3,4 A timely and accurate genetic diagnosis can provide valuable prognostic and predictive information, influence clinical decision-making, including treatment strategies and kidney transplant-related considerations, and inform reproductive decisions for affected individuals and their families.5,6 Furthermore, genomic sequencing can transform diagnostic pathways by facilitating the diagnosis of rare disorders and improving the understanding of disease mechanisms, paving the way for future therapies.
Although many studies have validated the diagnostic yield, clinical utility, and cost-effectiveness of genetic and genomic sequencing in kidney disease cohorts,7, 8, 9 most of these investigations have occurred within research settings, focusing on specific phenotypic subgroups.10 This context underscores the imperative to transition these findings into broader clinical implementation.11,12 Equitable and sustainable implementation in the clinical setting requires whole-system changes13 that overcome implementation challenges relating to workforce capacity, infrastructure, and funding, as well as the establishment of appropriate service delivery models.14,15
Multidisciplinary clinics are a well-recognized means of addressing such implementation challenges. This model of care has an established track record of supporting genomic implementation in kidney medicine and other specialties such as cardiology and oncology.16,17 For example, a 5-year review of a single KGC highlighted the benefits of including a multidisciplinary team (MDT) for kidney patients in the United Kingdom.16 The multidisciplinary KGC (MD-KGC) model allowed for a coordinated approach between clinical genetics and nephrology, and resulted in timely diagnosis and improved clinical management of genetic kidney disease.16 In the United States, an MD-KGC model consists of adult and pediatric nephrologists and genetic counselors and reported on only a relatively small number of patients.17 Support for genomic testing among nephrologists, with a strong preference for a multidisciplinary model involving a nephrologist, clinical geneticist, and genetic counselor, has also contributed to establishing a network of KGCs within the Australian public health care system.7 The first MD-KGC was established in Queensland (Australia) and provides services to pediatric and adult patients with suspected monogenic kidney disease.18 Subsequently, the MD-KGC started its implementation in other Australian jurisdictions, including a network of 4 academic centers in Victoria (Australia).19
The initial focus on demonstrating the clinical benefits of a multidisciplinary approach to genomic testing in kidney disease also precipitated the need to document practical experiences from its implementation into clinical practice.20 Furthermore, it is vital to scale up these models beyond individual tertiary centers to serve the needs of diverse and geographically dispersed populations. Studies that explore clinical testing at a broader scale, particularly in a nephrology context, are lacking. The implementation and contextual factors at organizational, provider, and individual levels (clinician and patient) that lead to the successful integration of genomic testing in nephrology are poorly defined. Nevertheless, some barriers have been identified that hinder the effective implementation of genomic testing into the routine clinical management of genetic kidney diseases. For example, a lack of funding and awareness among clinicians and their perceived lack of preparedness have been highlighted.16,17,21 A learning health care system has been suggested to address these challenges using continuous quality improvement strategies, which can keep up with technological advances in genetic medicine.17
We developed and implemented an iterative learning health care system approach to delivering kidney genomic services in Australia. Through this model, we established and used a national network of KGCs to continuously collect and aggregate sequencing data, including diagnostic and clinical outcomes. Analysis of these data informed implementation efforts to scale up genomic medicine in kidney disease at the national level. We actively collected feedback from clinical practices to inform our ongoing service model delivery. Here, we present a decade of extensive experience and proven results; our nationwide network of MD-KGCs has demonstrated significant improvements in the availability of genetic diagnosis for patients while optimizing the process for health care providers.
Methods
Ethics
This study was approved by the Human Research Ethics Committee at Melbourne Health (HREC/16/MH/251) as part of the Australian Genomics Health Alliance: preparing Australia for Genomic Medicine program. Governance site-specific approval for the project was obtained for all participating KGCs. All participants provided written informed consent for data collection and to undergo clinically indicated genomic/genetic testing.
Context
In 2013, the first MD-KGC in Australia was established in Brisbane, Queensland, with initial outcomes subsequently reported.18 Subsequently, a consortium of clinicians, counselors, scientists, and researchers (KidGen Collaborative) was formed to improve genetic kidney disease outcomes.22 These activities have occurred in the context of broader state and national genetic initiatives aimed at supporting the integration of genomics in clinical and research settings, including the Australian Genomics Health Alliance, the Melbourne Genomics Health Alliance, and the Queensland Genomics Health Alliance.14,23, 24, 25 These initiatives have focused on building the evidence base for using genomic testing in different clinical scenarios, including kidney disease while tackling some of the data infrastructure, policy, ethics, and workforce challenges that large-scale implementation poses. Some notable developments from the Australian Genomics Health Alliance include developing a national clinical consent form and the Shariant platform, which enables evidence sharing across Australian clinical genomic testing laboratories to support and promote variant interpretation consistency.20,26
Genomic test funding in the clinical setting is a pressing issue. One major enabler is that the KidGen Collaborative facilitated access to funding for genomic testing for all patients with suspected monogenic kidney disease. Funding sources varied, including local hospital departments, research projects, and state-based funding until 2022. From mid-2022, and partly due to the collaborative work undertaken by KidGen, genomic testing for kidney diseases has been funded through the Australian Federal Government's national Medicare health schemes (mbsonline.gov.au), which provides Australians with subsidized health care.
Assessment of Implementation
We conducted a formative evaluation to investigate the uptake of genomic testing while establishing a national MD-KGCs network. Clinicians delivering the MD-KGC service were invited via email to give written feedback on the perceived barriers to KGC service delivery at their site during 2021 and 2022. The rationale behind this approach was rooted in the clinicians' firsthand experiences with patients, which were crucial for shaping effective implementation strategies. For instance, these clinicians identified, nominated, and presented their cases at MDT meetings. Their active engagement ensured that MD-KGC operations were continuously refined to align with the evolving needs of both patients and health care providers, thereby enhancing the effectiveness and quality of care provided.
In addition, clinicians were encouraged to nominate any solutions they had implemented to address identified barriers, fostering a collaborative effort toward improvement. Three primary periods of interest along the patient journey were before referral (before the patient was referred to MD-KGC), pretesting (includes clinical assessments by MD-KGC and laboratory interactions), and posttesting (the return of results). We mapped the barriers and solutions to the Consolidated Framework for Implementation Research to generate findings that could be applied to other settings.27 The Consolidated Framework for Implementation Research is a versatile framework for analyzing and enhancing innovation and intervention implementation in complex organizations. It includes the following 5 key domains: (i) intervention characteristics (complexity and adaptability), (ii) outer setting (external factors like resources and culture), (iii) inner setting (organizational factors like leadership support and staff skills), (iv) characteristics of individuals (unique factors like motivation and beliefs), and (v) process (implementation activities like planning and training).27, 28, 29
Participants
Participants with kidney disease were recruited from our MD-KGCs network. Two distinct groups were enrolled. The first group consisted of a retrospective cohort comprising individuals undergoing clinical genomic testing directly or after being referred to MD-KGCs between 2013 and 2019. The second group comprised a prospective cohort of patients referred to MD-KGCs between 2017 and 2022, including individuals who did not undergo genomic testing. All patients were referred to a KGC, and advice was sought for clinical genomic testing by the MDT, which included representation from at least 1 of each discipline, such as a nephrologist, geneticist, and genetic counselor. Each patient was seen at least twice in the KGC, once before genomic testing and again for the return of the results. During each clinic visit, a team of 2 or more disciplines reviewed the patient's case. Patients with existing confirmed molecular genetic diagnoses were excluded from the study. In addition, patients without a clear clinical indication for genomic testing and those determined by the MDT to have a low likelihood of an underlying monogenic basis were excluded from this study.
Genetic/Genomic Testing and Variant Analysis
DNA was isolated from blood or saliva samples, and sequencing was undertaken at clinically accredited laboratories for both cohorts, as described elsewhere.19,30 Genomic and genetic testing in our cohort encompassed chromosomal microarray analysis, Sanger sequencing, whole exome sequencing, whole genome sequencing, and variable number of tandem repeats testing as clinically indicated. The earlier tests utilized targeted panels of genes and the newer technologies, such as whole genome sequencing and whole exome sequencing, were analyzed only for variants in a virtual panel of genes specific to the clinical phenotype using a curated list of several hundred genes associated with genetic kidney disease (PanelApp, Australia).31 In Supplementary Table S3, we provide a comprehensive list of all the genes included in the Kidneyome SuperPanel (Version 8.53). Detected variants were classified according to American College of Medical Genetics and Genomics guidelines for clinical sequencing interpretation.32 MDT meetings reviewed variants and phenotypic data when required to assist with interpretation at the case level before reporting.
Data Collection
Genomic and genetic test results and clinical and demographic data were collected and entered a research electronic data capture (REDCap) and data management platform33,34 hosted at the Murdoch Children's Research Institute, Melbourne, Australia. Data capture included age at the test, testing date, KGC location, sequencing approach, and diagnostic outcomes. The suspected clinical diagnosis before genomic testing was also collected and classified according to a broad aetiologic category.
Statistical Analysis
Baseline characteristics are expressed as counts (n, %) or medians with interquartile range, as appropriate. The Chi-square or Fisher exact tests were performed to compare the proportions of clinical characteristics between respective groups. The primary outcome was the diagnostic yield based on variants classified as “pathogenic” and “likely pathogenic” according to American College of Medical Genetics and Genomics guidelines,32 with consideration of appropriate inheritance patterns and phenotype consistency. Variants of uncertain significance (VUS)32 were not included. Accordingly, the diagnostic yield was calculated using the formula: Diagnostic Yield = positive result (PR / [PR + NR + VUS] × 100), where PR represents positive results, and NR denotes negative results. Diagnostic yield is expressed as percent (%). Data were analyzed using the Stata/MP 16.0 statistical package (StataCorp LP, College Station, TX). Statistical significance was determined using 2-sided P-values, with a significance level set at the conventional 5% threshold.
Results
Determinants of Implementation of Genomic Testing Within KGCs
As of 2022, 20 MD-KGCs have been established across all Australian state jurisdictions. Participating clinicians at these MD-KGCs identified contextual and individual provider issues that may impact future implementation programs. Barriers were identified by geneticists and nephrologists who were working in KGCs. Most were coded to the "inner setting" and "intervention characteristics" Consolidated Framework for Implementation Research domains (Table 1). Barriers relating to the "inner setting" domain dominated both the "before referral" and "post referral-pretesting periods," with barriers falling under the structural characteristics and implementation climate, knowledge, and available resources subdomains (Table 1). The "intervention characteristics" domain dominated the "post referral-pretesting" period, with responses relating to the complexity of whole genome sequencing or whole exome sequencing technology (Table 1). Key strategies to overcome barriers during the "before referral" period included presenting new evidence of genomic testing and highlighting the clinical utility of genomic testing in kidney disease through education and engagement activities such as conferences and workshops. Concerns about testing efficiency were noted. Strategies, including variant prioritization meetings and shared gene lists, aimed to enhance consistency in analysis. However, the MD-KGC model could not address all barriers, such as funding insecurity and patient remoteness (Table 1).
Table 1.
Barriers and suggested interventions to integrate routine kidney genomic testing into clinical practice
| Clinician reported barrier | CFIR construct | Description | Intervention applied in practice | Impact on the clinical setting |
|---|---|---|---|---|
| Before-referral period | ||||
| Delay in referrals and uptake by nephrologists not engaged with kidney genetics | Inner setting | Culture and implementation climate | Education initiatives | Earlier referrals to kidney genetics clinics |
| The inequity of referral for testing | Inner setting | Structural characteristics and underpinnings of inequity | Education initiatives | Increased referrals and reach of genomic testing |
| Variable access to testing between jurisdictions | Inner setting | Structural characteristics and readiness for implementation | Education initiatives | Equitable access to genomic testing in kidney disease |
| Nephrologists are unsure whom to refer to | Characteristics of individual | Knowledge and beliefs about the intervention | Education initiatives | Increased number of appropriate referrals |
| Nephrologists uncertain of the benefit of referral | Intervention characteristics | Evidence Strength & Quality | Education initiatives | Increased number of appropriate referrals |
| Poor community and consumer understanding of genomic testing | Process | Engaging | Engagement with communities and public engagement events | Increased utilization of genomic testing and application of findings |
| Patients and the general community are uncertain of the benefits of referral. | Intervention characteristics | Evidence strength & quality | Engagement with communities and public engagement events | Increased utilization of genomic testing and application of findings |
| Post referral-pretesting period | ||||
| The logistics of testing are too complicated to incorporate into clinics | Intervention characteristics | Complexity | Process simplification | Increased utilization of genomic testing |
| Nephrologists did not know how to order tests or prioritize patients for testing. | Inner setting | Knowledge and beliefs about the intervention, self-efficacy | Education initiatives | Increased utilization of genomic testing |
| Clinicians are unsure of which genes to test, which method to use | Inner setting and characteristics of individual | Access to knowledge and information | Developing standardized gene lists and recommended method | Increased utilization of genomic testing |
| Delay in sequencing | Process | Executing | Variant prioritization - meetings initiated | Earlier genomic testing |
| Variation in access to and cost of testing | Intervention characteristics | (Perceived) Cost | ||
| No consistent clinical genetics support | Inner setting | Available resources | Engagement with clinical genetics services and national consortia; Identification of clinical geneticist champions | Increased clinical genetics department engagement |
| The remote location of the clinical geneticist | Inner setting | Available resources | Telehealth clinics, remote/visiting clinics | Increased access to clinical geneticists |
| The remote location of patients who may lack literacy | Outer setting | The patient's needs and resources | ||
| Missed appointments to clinics reduce sustainability. | Outer setting | The patient's needs and resources | ||
| Unable to access clinical geneticist during clinic | Inner setting | Available resources | ||
| No secure funding for the multidisciplinary team (MDT) model of care | Inner setting | Available resources | Engagement with clinical services for the redesign of existing activities and demand; clinical champions engaged | Sustained operation of MDT models of care within existing resourcing |
| Variable hospital funding for tests | Outer setting | External policy and incentives | ||
| Post-testing period | ||||
| Long turn-around-time for sequencing and analysis | Intervention characteristics | Complexity | Variant prioritization - meetings initiated | Earlier genomic testing |
| Delay in return of results | Inner setting | Available resources | MDT results meetings and review systems | Improved accountable delivery of results to consumers |
| Results difficult for nephrologists without specific training to interpret/apply | Intervention characteristics | Complexity | Targeted and sustained multimodal education supported by the National Strategic Action Plan for Kidney Disease | Perceived improving clinician confidence |
| Results not received by the referring doctor | Process | Executing | Upfront information for doctors | Clinical translation of genomic testing |
| Unclear recommendations to referrer regarding the clinical application of findings | Intervention characteristics | Complexity | Facilitated interpretation and clinician guidance in interprofessional communication | Supported referrer actioning of result outcomes |
| Difficulty in understanding the implications of a variant of uncertain significance | Characteristics of individual | Knowledge and beliefs about the intervention and self-efficacy | ||
| Delay or failure of whom to clinically act on genetic findings | Characteristics of individual | Knowledge and beliefs about the intervention | Upfront information for doctors and patients | Clinical translation of genomic testing |
CFIR, Consolidated Framework for Implementation Research; MDT, multidisciplinary team.
In addition to establishing a publicly available nationwide network of MD-KGC, the KidGen Collaborative has also invested in many other resources to help improve equity and appropriate use of genomic testing for those with suspected monogenic kidney disease. The network has driven national consensus on gene panels for kidney genomic tests. These panels are hosted on PanelApp Australia, an openly accessible platform.31 The network developed an education program consisting of online modules and workshops to upskill nephrologists on various aspects of genomic testing, including patient selection, test ordering, and result interpretation. In addition, the KidGen Collaborative has recently hosted 5 educational events dedicated to genetic kidney diseases, marking the 10th anniversary of the first KGC in Australasia.18
Demographics
In total, 1506 participants were included in the study, of whom 125 did not proceed with genomic or genetic testing, leaving 1381 participants who underwent testing. Among the tested patients, results of 59 individuals (from the prospective cohort only) were not returned by the end of the audit cut-off date, bringing the total number of patients with genetic testing results to 1322 (Figure 1). The underlying reasons for the absence of results were not explicitly examined but included those that had not yet had results reported during the study period. The demographic characteristics of participants are shown in Table 2. Almost two-thirds (849/1381) of participants were adults, and the median age at testing was 29 (interquartile range: 10–45) years. Detailed patient recruitment data across various states, stratified by years, is presented in Supplementary Table S1. It is noteworthy that during the most severe period of the COVID-19 pandemic in 2020, recruitment activities nearly ceased across all states except Western Australia, where only 4 patients were successfully recruited.
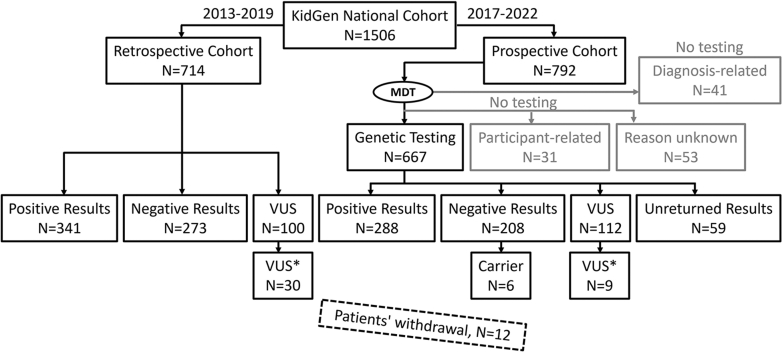
Figure 1.
Recruitment of the KidGen national cohort for genomic testing. Two participant nonoverlapping retrospective and prospective cohorts were recruited between 2013 and 2022, creating the KidGen national cohort. The dashed rectangle depicts the number of patients who withdrew and were excluded from the study. Carrier, an individual with an allele predisposing to disease; MDT, multidisciplinary team; VUS, a variant of uncertain significance; VUS×, VUS with suspected clinical relevance.
Table 2.
Demographic characteristics of participants who underwent genomic testing
| Characteristic | Retrospective cohort (2013–2019; n = 714) | Prospective cohort (2017–2022; n = 667) | All KidGen cohort (N = 1381) |
|---|---|---|---|
| Patient | |||
| Adult (≥18 yr) | 387 (54.20%) | 462 (69.27%) | 849 (61.48%) |
| Pediatric (<18 yr) | 327 (45.80%) | 205 (30.73%) | 532 (38.52%) |
| Total | 714 | 667 | 1381 |
| Median Age (IQR) | 22 (6–42) n = 707 (99%) | 33 (17–49) n = 594 (89%) | 29 (10–45) n = 1301 (94%) |
| Sexa | |||
| Male | 339 (47.48%) | 327 (49.02%) | 666 (48.23%) |
| Female | 374 (52.38%) | 289 (43.33%) | 663 (48.01%) |
| Unknown | 1 (0.14%) | 51 (7.65%) | 52 (3.76%) |
| Total | 714 | 667 | 1381 |
| Location | |||
| Queensland | 324 (45.38%) | 164 (24.59%) | 488 (35.34%) |
| New South Wales | 226 (31.65%) | 137 (20.54%) | 363 (26.29%) |
| Victoria | 121 (16.95%) | 244 (36.58%) | 365 (26.43%) |
| South Australia | 28 (3.92%) | 59 (8.85%) | 87 (6.30%) |
| Western Australia | 10 (1.40%) | 63 (9.45%) | 73 (5.29%) |
| Northern Territory | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| Tasmania | 5 (0.70%) | 0 (0.00%) | 5 (0.36%) |
| Total | 714 | 667 | 1381 |
Nominal variables are numbers with percent (%), while continuous variables are median with interquartile range (IQR).
Sex has here been defined as a set of biological attributes associated with physical and physiological features.
The retrospective cohort comprised 714 participants who all underwent genomic testing. The prospective cohort comprised 792 participants referred to KGCs, of whom 667 underwent genomic testing (Figure 1). Five percent (41/792) of participants within the prospective cohort were not recommended for testing by the MDT (greyed Figure 1) for the following reasons: testing would not impact the clinical management of either the participant or family members, an adequate prior diagnosis, or clinical presentation was not in keeping with monogenic kidney disease. In addition, 11% (84/792) of the prospective participants in the prospective cohort were not tested as the participant declined (n = 31) or for unknown reasons (n = 53) (Figure 1).
Within the retrospective cohort, 48% (341/714) received a positive (pathogenic/likely pathogenic) result (PR) from genomic testing. In addition, a VUS was detected in 14% (100/714). Of the patients with a VUS identified, 30 of 100 (30%) were VUS with suspected clinical relevance (VUS×). Within the prospective cohort, 43% (288/667) received a PR. In addition, a VUS was detected in 17% (112/667) participants, and of those, 8% (9/112) had a VUS with suspected clinical relevance (Figure 1).
Diagnostic Yield, Clinical Diagnoses, and Sequencing Method
The diagnostic yield for the national cohort between 2013 and 2022 was 45.8% (606/1322) (Table 3).
Table 3.
Diagnostic yield in the national KidGen cohort stratified by year of genomic testing
| Yr | Sequencing performed number of patients | Diagnostic variants present number of patients | Diagnostic yield percent |
|---|---|---|---|
| 2013 | 22 | 13 | 59.1 |
| 2014 | 78 | 41 | 52.6 |
| 2015 | 71 | 37 | 52.1 |
| 2016 | 157 | 68 | 43.3 |
| 2017 | 340 | 158 | 46.5 |
| 2018 | 295 | 125 | 42.4 |
| 2019 | 124 | 52 | 41.9 |
| 2020 | 4 | 2 | 50.0 |
| 2021 | 127 | 58 | 45.7 |
| 2022 | 60 | 30 | 50.0 |
| Unknown | 44 | 22 | 50.0 |
| Totala | 1322 | 606 | 45.8 |
NR, negative result; PR, positive result; VUS, a variant of uncertain significance.
Test results were unknown for 59 patients by the end of the 2022 cut-off. DY (diagnostic yield) was calculated using the formula: DY = (PR / [PR + NR + VUS] × 100) and is expressed in percent (%).
When the overall cohort is stratified according to the pretest clinical diagnosis, the diagnostic yield is highest for Alport syndrome (61.7%; 127/206) and lowest for congenital anomalies of the kidney and urinary tract (24.7%; 18/73), as shown in Table 4. The diagnostic odds ratio for Alport syndrome was 2.14, and for congenital anomalies of the kidney and urinary tract, it was 0.37 (Table 5). The evolution of sequencing methods throughout the study is presented in Table 6. In the earlier years (2013–2015), targeted panels of genes using massively parallel sequencing and Sanger sequencing were most commonly utilized. Since 2017, whole exome sequencing or whole genome sequencing-based approaches have been more widely used.
Table 4.
Diagnostic yield in the national KidGen cohort stratified by clinical diagnosis
| Clinical diagnosis | Sequencing performed number of patients | Positive results number of patients | Diagnostic yield percent |
|---|---|---|---|
| Alport syndrome | 206 | 127 | 61.7 |
| Tubular disease | 134 | 74 | 55.2 |
| Cystic kidney disease | 506 | 256 | 50.6 |
| Glomerular disease | 50 | 19 | 38.0 |
| Other | 122 | 45 | 36.9 |
| Complement-mediated | 89 | 26 | 29.2 |
| Nephrotic disease | 142 | 41 | 28.9 |
| CAKUT | 73 | 18 | 24.7 |
| Totala | 1322 | 606 | 45.8 |
CAKUT, congenital anomalies of the kidney and urinary tract-syndromic; DY, diagnostic yield (was calculated using the formula: DY = (PR / [PR + NR + VUS] × 100) and is expressed in percent (%); NR, negative result; PR, positive result; VUS, a variant of unknown significance.
Test results were unknown for 59 patients by the end of the 2022 cut-off.
Table 5.
Clinical predictors of genetic diagnosis
| Clinical diagnosis | Odds ratio | 95% confidence interval | P-value |
|---|---|---|---|
| Alport syndrome | 2.14 | 1.58-2.90 | <0.001 |
| Tubular disease | 1.52 | 1.06-2.18 | 0.022 |
| Cystic kidney disease | 1.36 | 1.09-1.70 | 0.006 |
| Glomerular disease | 0.72 | 0.40-1.28 | 0.259 |
| Other | 0.67 | 0.45-0.98 | 0.038 |
| Complement-mediated | 0.46 | 0.29-0.74 | 0.001 |
| Nephrotic disease | 0.44 | 0.30-0.65 | <0.001 |
| CAKUT | 0.37 | 0.21-0.63 | <0.001 |
CAKUT, congenital anomalies of the kidney and urinary tract-syndromic; P, 2-sided P-value.
Table 6.
The use of the testing method in the national KidGen cohort stratified by a year of testing
| Yr | Testing method number (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CMA | Targeted panel | Sanger | TE | VNTR | WES | WGS | Not specified | Total | |
| 2013 | 0 (0.0) | 6 (20.7) | 7 (24.1) | 4 (13.8) | 0 (0.0) | 4 (13.8) | 1 (3.4) | 7 (24.1) | 29 (100.0) |
| 2014 | 1 (1.2) | 6 (7.2) | 18 (21.7) | 24 (28.9) | 1 (1.2) | 2 (2.4) | 0 (0.0) | 31 (37.3) | 83 (100.0) |
| 2015 | 3 (3.9) | 7 (9.1) | 7 (9.1) | 27 (35.1) | 0 (0.0) | 2 (2.6) | 0 (0.0) | 31 (40.3) | 77 (100.0) |
| 2016 | 8 (4.5) | 15 (8.4) | 6 (3.4) | 53 (29.6) | 1 (0.6) | 12 (6.7) | 10 (5.6) | 74 (41.3) | 179 (100.0) |
| 2017 | 6 (2.0) | 9 (3.0) | 14 (4.7) | 78 (26.0) | 2 (0.7) | 67 (22.3) | 24 (8.0) | 100 (33.3) | 300 (100.0) |
| 2018 | 11 (3.7) | 8 (2.7) | 16 (5.4) | 40 (13.6) | 2 (0.7) | 63 (21.4) | 36 (12.2) | 118 (40.1) | 294 (100.0) |
| 2019 | 7 (5.6) | 6 (4.8) | 4 (3.2) | 31 (25.0) | 0 (0.0) | 4 (3.2) | 20 (16.1) | 52 (41.9) | 124 (100.0) |
| 2020 | 0 (0.0) | 0 (0.0) | 1 (25.0) | 2 (50.0) | 0 (0.0) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 4 (100.0) |
| 2021 | 1 (0.8) | 0 (0.0) | 8 (6.3) | 32 (25.2) | 2 (1.6) | 28 (22.0) | 51 (40.2) | 5 (3.9) | 127 (100.0) |
| 2022 | 4 (4.5) | 0 (0.0) | 11 (12.5) | 17 (19.3) | 2 (2.3) | 28 (31.8) | 21 (23.9) | 5 (5.7) | 88 (100.0) |
| Totala | 41 (3.1) | 57 (4.4) | 92 (7.0) | 308 (23.6) | 10 (0.8) | 211 (16.2) | 163 (12.5) | 423 (32.4) | 1305 (100.0) |
CMA, chromosome microarray analysis; Not specified, sequencing type not indicated; Sanger, Sanger sequencing, also known as the “chain termination method;” Targeted panel, using massively parallel sequencing (MPS); TE, targeted clinical exome sequencing with virtual panel; VNTR, variable number tandem repeat of the mucin 1 (MUC1) gene; WES whole exome sequencing (with virtual panel); WGS, whole-genome sequencing (with virtual panel).
A test was not performed, results were unreturned, or the year of testing was not specified in 201 patients.
Genes With Clinically Significant Variants
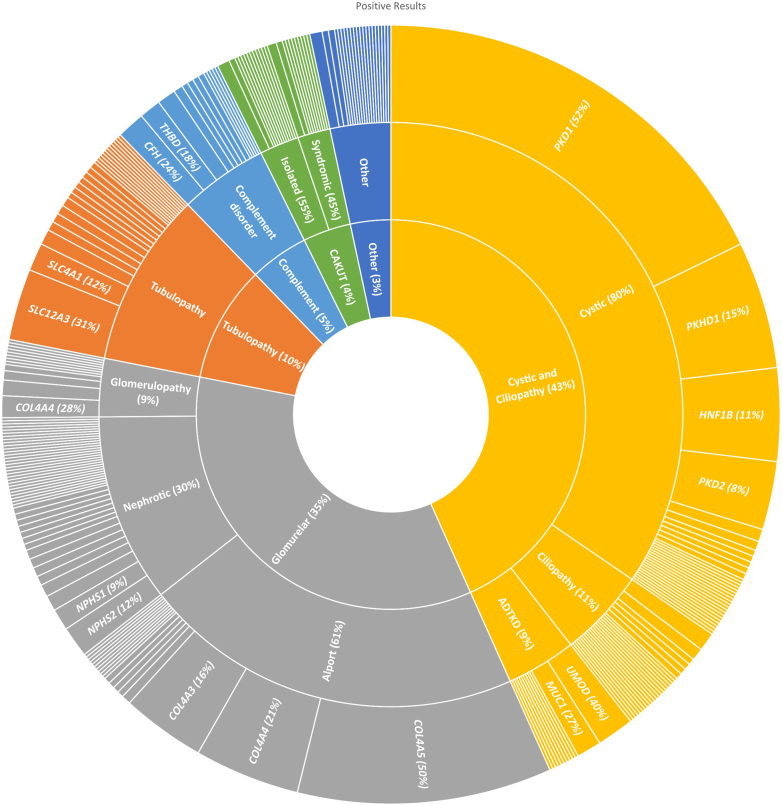
Pathogenic and likely pathogenic variants were identified in genes relating to cystic kidney disease, Alport syndrome, and renal tubular disorders in descending frequency, accounting for approximately two-thirds of the pathogenic and likely pathogenic variants identified (Figure 2). The most frequently affected gene in cystic kidney disease was PKD1(106 cases), followed by PKHD1 (56 cases) and HNF1B (28 cases). The SLC12A3 gene was the most frequent in tubular kidney disease (Figure 2), where we identified 18 compound heterozygotes and 12 homozygotes carrying pathogenic variants in this gene. All genes are listed in Supplementary Table S2. Variants of unknown significance (VUS or VUS with suspected clinical relevance) identified in the cohort are presented in Supplementary Figure S1.
Figure 2.
Genes with pathogenic/likely pathogenic variants in the national KidGen cohort. The genes are stratified by clinical diagnosis, with the width of the cut-outs indicating the number of times each gene appears. All genes are listed in Supplementary Table S2.
Discussion
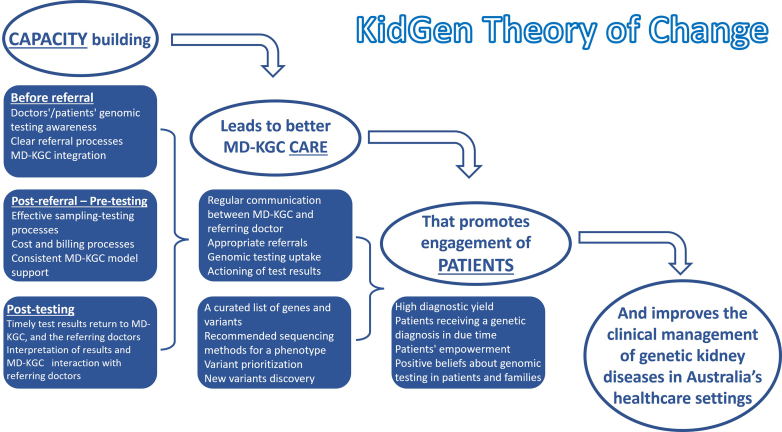
Our study delves into the complex process of implementing genomic testing within kidney medicine, emphasizing the successful role of a nationwide KGCs network embedded with a MDT. Focusing on real-world applicability, we explored the challenges and successes of integrating genetic testing seamlessly into clinical practice. We extended a theory-informed approach to determining barriers to implementing genomic testing in a nephrology context and, where possible, implementing real-time solutions to optimize respective processes.35 Our theory of change for the Australian MD-KGC model stands on 3 pillars: capacity, care, and patients (Figure 3). We hypothesized that building the capacity for genomic testing would improve KGC care, where patients would benefit immediately (given an outcome), and ultimately in the long term, improve the clinical management of chronic kidney diseases in Australia. Indeed, the program has succeeded in all 3 pillars of effective change. This accomplishment is evidenced by established adult and pediatric KGCs in 7 Australian states and territories, which have been operational for almost 10 years. Importantly, this model involved collaboration between geneticists, nephrologists, and genetic counselors at local sites, with support from the integrated national network, which created a practical, best-practice MDT approach to delivering genomic medicine in kidney disease. The effectiveness primarily stemmed from networking, because all KGCs were interconnected and actively exchanged ideas. This collaborative approach significantly eased the decision-making burden, particularly in handling complex cases and selecting the appropriate test based on the patient's phenotype. This collaborative network facilitated the rapid enhancement and spread of expertise, far outpacing what could be achieved in isolation. The collective clinical expertise, complemented by funding from various sources contingent on jurisdiction, was pivotal in establishing essential infrastructure to streamline the genomic testing process. The MD-KGC model, therefore, becomes the preferred service delivery model for kidney genetics in Australia among nephrologists7 and accounts for a relatively high diagnostic yield by selecting patients who were most likely to benefit from testing. The model also improves result interpretation and subsequent clinical actioning due to the availability of clinical genetics or genetic counselors and nephrology expertise within the clinic. Smaller clinics particularly benefitted from access to the highly specialized MD-KGC expertise. However, as demand for testing increases, this model may not be sustainable, and a “mainstreaming” model of care is already beginning to emerge in other specialties and some subgroups of genetic kidney disease, such as autosomal dominant polycystic kidney disease (ADPKD).21
Figure 3.
Visualizing the theory of change for the Australian MD-KGC model. The model is built upon 3 successive pillars: capacity, care, and patients. By addressing these areas through the MD-KGC approach, positive changes can be made in the clinical management of genetic kidney diseases. An essential aspect involves the implementation of test results by clinicians to guide patient care (actioning of results). MD-KGC, multidisciplinary kidney genetic clinic.
Concerning the implementation barriers identified in our clinics, we identified several challenges encountered by clinicians at local KGCs. Above all, the complexity of the entire process (outlined above) dominated as a significant barrier. Most of these concerns aligned with the “inner setting” of the Consolidated Framework for Implementation Research construct and could be addressed locally. For example, most confusion accumulated around referrals and sampling processes, selecting the right test, and using the appropriate sequencing method. Professional development educational activities address these challenges. In the global context, our identified barriers align with those reported worldwide; however, some were less pronounced in Australia. A US study with 150 nephrologists identified high laboratory or testing costs as a major barrier to genomic testing.36 Although testing costs were operationally not a primary hurdle in Australian practice, they were a perceived barrier for less-experienced clinicians owing to the complexity of accessing resourcing. Significant barriers included staff funding issues, such as genetic counselors and clinic infrastructure expenses. Another barrier identified in this study-the difficulty in interpreting genomic test findings-has already been acknowledged within an Australian context7 and was further reiterated during discussions at the Kidney Diseases: Improving Global Outcomes conference held in 2021.37 The MDT model addresses this barrier by integrating into kidney genetic clinics. Although this model of care results in high-quality health care due to subspecialty input, future efforts should further develop more capacity to overcome interpretation barriers while maintaining accessibility.
Our clinical findings, drawn from a cohort of over 1300 patients suspected of genetic kidney disease, represent the absolute number of 1 of the largest clinical cohorts in this domain. With a diagnostic yield of ∼46%, our results consistently match or surpass those of many published cohorts in research and clinical settings. However, the diagnostic yield in our study varied based on clinical phenotypes, with the highest yield observed in Alport syndrome (∼62%) rather than cystic kidney diseases, such as ADPKD (∼51%), which is the most prevalent monogenic kidney disease.38 This surprising outcome can be explained by several challenges that hindered the diagnosis of ADPKD using genomic testing for many patients. Key factors contributing to this discrepancy include constraints in local testing platforms, substantial expenses associated with overseas testing, and the absence of publicly funded genomic testing during the significant period of the study. These obstacles resulted in a focus on testing atypical cases and a period of underrepresentation of classic ADPKD patients. Consequently, the overall diagnostic yield for cystic kidney diseases was impacted, as the transient limited access to comprehensive genomic testing for ADPKD influenced the overall distribution of patients with various kidney diseases. Nevertheless, these findings are well within the range of previous reports where, for instance, diagnostic yield for cystic kidney disease is anywhere between 23% and 88%, depending on the clinical characteristics and sequencing approaches.1,3,10,39, 40, 41
North American data reinforced the diagnostic utility of clinical genetic and genomic testing in kidney medicine42 and reconfirmed previous findings in smaller cohorts at scale. Dahl et al.42 reported on 1623 patients with chronic kidney disease enrolled in a study evaluating the diagnostic and clinical utility of genomic testing in chronic kidney disease. They reported a yield of 20.8%, which altered management in 90% of patients with a positive result. This short-term clinical utility, as determined by clinician surveys, complements additional evidence from other cohorts43 and highlights the next gap in knowledge about implementation into practice. Our study addresses this information gap in implementing genetic testing in real-world clinical practice, adding generalizability, and confirming utility while identifying a higher diagnostic yield. However, comparing diagnostic yields in genetic kidney disease studies is challenging due to diverse study designs, cohort compositions, and test characteristics.10 In addition, in research studies, many patients never receive results, even if these are deemed clinically actionable.44 Our study, however, provides clinically accredited sequencing and interpretation. This design generates vital, policy-informing data on the impact of genetic diagnosis on patients with kidney disease.
Our study has some limitations. In addition to potential technical limitations inherent in the technology used, such as the possible omission of specific genetic causes such as MUC1 variants,45 it is essential to note that this was a pragmatic study. Therefore, we did not have predefined objective criteria for selecting patients for genomic testing. In addition, consumer engagement was outside the scope of this study; however, as a group, they are recognized as fundamental stakeholders in the development of service delivery. Within Australia and elsewhere, there is clear evidence of a need for the codevelopment of specialist genetic services regarding care delivery and providing resources that hurdle geographical and linguistic or cultural barriers.46,47 The authors group of this manuscript are currently undertaking a national research program into the experiences and needs of patients and their families to enable ongoing optimization of the service.
Nevertheless, our data reflect real-world practice. Every patient was assessed in the KGC using an MDT approach (based on relevant clinical indices) to ensure that genomic testing was only performed if clinically indicated and that the most suitable sequencing platform available was used. Second, it is important to note that our cohort does not encompass all patients who underwent sequencing during the 10 years. Although clinicians were requested to provide audit data for all patients seen, the inclusion of multiple health services and clinical laboratories, including international ones, posed challenges in determining the total number of patients sequenced nationwide and the number of patients who attended clinics but did not proceed to genomic testing. Consequently, despite encouraging clinicians to collect data on all sequenced patients, there remains a possibility of selection bias. In addition, because the data collection process relied on input from multiple clinicians across various renal genetics clinics, challenges arose in obtaining complete data, including instances of missing “year of recruitment” information when the relevant clinician did not provide it. In addition, the COVID-19 outbreak negatively impacted genomic testing and collection of data to this dataset, with data for only 4 patients recorded in 2020. Although our sample size may be considered relatively modest for a decade-long effort, it indicates the intrinsic challenges and complexities associated with establishing and expanding a national kidney genetics program.
In July 2022, federal funding for genomic testing for monogenic kidney disease was announced in Australia; however, data were only collected until October of the same year; and therefore, it is unlikely that this increase in test availability has impacted our cohort during the study timeline. Our study did not investigate the potential barriers and facilitators in implementing genomic testing outside MD-KGCs. This is especially important given the increasing availability of genomic testing for patients with kidney disease. Finally, given the longer-term relevance of the clinical utility of genomic testing, clinical utility information was not collected to reduce the burden on participants and clinicians. Nevertheless, the short-term and lifetime health and economic implications of genomic testing have been modeled to evaluate its relative cost-effectiveness compared to nongenetic diagnostic investigations in patients with suspected monogenic kidney disease.48 This economic evaluation established the cost-effectiveness of genomic testing for genetic kidney disease and demonstrated that it is cost-saving in patients with glomerular diseases.48 Further work is needed to evaluate the cost-effectiveness of this clinic model and identify ways to optimize its health and economic benefits to ensure a sustainable and equitable national service across Australia.
Conclusion
Our study provides a comprehensive analysis of a real-world national multidisciplinary model of care for patients suspected to have monogenic kidney disease. Through a decade-long iterative process, we gained insights into the strengths and weaknesses of this model. Based on our findings, we have successfully implemented the MD-KGC model. However, further research is necessary to evaluate its cost-effectiveness and long-term sustainability. Such studies would provide valuable insights into the feasibility and benefits of implementing this model more broadly.
Disclosure
All the authors declared no competing interests.
Acknowledgments
We want to thank Helen Healy, Julie McGaughran, Priyanka Sagar, Radhika Rajkumar, Sean Kennedy, Selma Torronen, William Majoni, Doris Chan, Neil Boudville, Suda Swaminathan, Germaine Wong, Nigel Toussaint, Sam Crafter, Andrew Talbot, and Joshua Kausman for their contributions to this project.
This work was supported by funding from the Australian Genomics Health Alliance (Australian Genomics), Melbourne Genomics Health Alliance (Melbourne Genomics), Australian Government Department of Health (Activity Reference Number: 4-E8LEZOZ), and Medical Research Future Fund (MRFF; Grant Reference Number: 2008249). Australian Genomics Health Alliance is funded by a National Health and Medical Research Council (NHMRC) grant (Grant Reference Number: 1113531) and the Australian Government's Medical Research Future Fund (MRFF). The research conducted at the Murdoch Children's Research Institute was supported by the Victorian Government's Operational Infrastructure Support Program. Clinical activities were supported by all Australian State and Territory Government Departments of Health and Local Hospital and Health Services. AJM was supported by a Queensland Government Advancing Clinical Research Fellowship. This work represents independent research, and the views expressed are those of the authors and not necessarily those of the NHMRC, MRFF, or Australian Government.
Data Availability Statement
The genomic data generated from this study involve patient information and are subject to confidentiality and privacy protections. Due to these legal and ethical considerations, the genomic data cannot be shared publicly or made available to other researchers. This restriction is in place to ensure the privacy and security of the patient's sensitive information. We are committed to upholding the highest data protection and privacy standards for the individuals involved in this study. Deidentified results supporting this study's findings are available from the corresponding author, AJM, upon reasonable request.
Footnotes
Figure S1. Genes with VUS/VUS× identified in the national KidGen cohort. The genes are stratified by clinical diagnosis, with the width of the cut-outs indicating the number of appearances. VUS is a variant of uncertain significance; VUS× is a VUS with suspected clinical relevance.
Table S1. Patient recruitment by state and year stratification.
Table S2. Occurrence of genes affected by pathogenic/likely pathogenic variants in patients with different kidney diagnoses.
Table S3. Genes listed in Kidneyome SuperPanel Version 8.53.
STROBE checklist.
Supplementary Material
Figure S1. Genes with VUS/VUS× identified in the national KidGen cohort. The genes are stratified by clinical diagnosis, with the width of the cut-outs indicating the number of appearances. VUS is a variant of uncertain significance; VUS× is a VUS with suspected clinical relevance. Table S1. Patient recruitment by state and year stratification. Table S2. Occurrence of genes affected by pathogenic/likely pathogenic variants in patients with different kidney diagnoses. Table S3. Genes listed in Kidneyome SuperPanel Version 8.53. STROBE checklist (PDF)
References
- 1.Bullich G., Domingo-Gallego A., Vargas I., et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 2018;94:363–371. doi: 10.1016/j.kint.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 2.Mann N., Braun D.A., Amann K., et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol. 2019;30:201–215. doi: 10.1681/ASN.2018060575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groopman E., Goldstein D., Gharavi A. Diagnostic utility of exome sequencing for kidney disease reply. N Engl J Med. 2019;380:2080–2081. doi: 10.1056/NEJMc1903250. [DOI] [PubMed] [Google Scholar]
- 4.Snoek R., van Jaarsveld R.H., Nguyen T.Q., et al. Genetics-first approach improves diagnostics of ESKD patients <50 years old. Nephrol Dial Transplant. 2020;37:349–357. doi: 10.1093/ndt/gfaa363. [DOI] [PubMed] [Google Scholar]
- 5.Jayasinghe K., Quinlan C., Stark Z., et al. Renal genetics in Australia: kidney medicine in the genomic age. Nephrol (Carlton) 2019;24:279–286. doi: 10.1111/nep.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knoers N., Antignac C., Bergmann C., et al. Genetic testing in the diagnosis of chronic kidney disease: recommendations for clinical practice. Nephrol Dial Transplant. 2022;37:239–254. doi: 10.1093/ndt/gfab218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayasinghe K., Quinlan C., Mallett A.J., et al. Attitudes and practices of Australian nephrologists toward implementation of clinical genomics. Kidney Int Rep. 2021;6:272–283. doi: 10.1016/j.ekir.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo-Gallego A., Pybus M., Bullich G., et al. Clinical utility of genetic testing in early-onset kidney disease: seven genes are the main players. Nephrol Dial Transplant. 2022;37:687–696. doi: 10.1093/ndt/gfab019. [DOI] [PubMed] [Google Scholar]
- 9.Jayasinghe K., Wu Y., Stark Z., et al. Cost-effectiveness of targeted exome analysis as a diagnostic test in glomerular diseases. Kidney Int Rep. 2021;6:2850–2861. doi: 10.1016/j.ekir.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claus L.R., Snoek R., Knoers N., van Eerde A.M. Review of genetic testing in kidney disease patients: diagnostic yield of single nucleotide variants and copy number variations evaluated across and within kidney phenotype groups. Am J Med Genet C Semin Med Genet. 2022;190:358–376. doi: 10.1002/ajmg.c.31995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shanks J., Butler G., Cheng D., Jayasinghe K., Quinlan C. Clinical and diagnostic utility of genomic sequencing for children referred to a Kidney Genomics Clinic with microscopic haematuria. Pediatr Nephrol. 2023;38:2623–2630. doi: 10.1007/s00467-022-05846-1. [DOI] [PubMed] [Google Scholar]
- 12.Morrow A., Chan P., Tucker K.M., Taylor N. The design, implementation, and effectiveness of intervention strategies aimed at improving genetic referral practices: a systematic review of the literature. Genet Med. 2021;23:2239–2249. doi: 10.1038/s41436-021-01272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark Z., Dolman L., Manolio T.A., et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark Z., Boughtwood T., Haas M., et al. Australian Genomics: outcomes of a 5-year national program to accelerate the integration of genomics in healthcare. Am J Hum Genet. 2023;110:419–426. doi: 10.1016/j.ajhg.2023.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooker G.W. Building an infrastructure to enable delivery of genomic medicine. Am J Med Genet C Semin Med Genet. 2021;187:95–99. doi: 10.1002/ajmg.c.31881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alkanderi S., Yates L.M., Johnson S.A., Sayer J.A. Lessons learned from a multidisciplinary renal genetics clinic. Q J M. 2017;110:453–457. doi: 10.1093/qjmed/hcx030. [DOI] [PubMed] [Google Scholar]
- 17.Thomas C.P., Freese M.E., Ounda A., et al. Initial experience from a renal genetics clinic demonstrates a distinct role in patient management. Genet Med. 2020;22:1025–1035. doi: 10.1038/s41436-020-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallett A., Fowles L.F., McGaughran J., Healy H., Patel C. A multidisciplinary renal genetics clinic improves patient diagnosis. Med J Aust. 2016;204:58–59. doi: 10.5694/mja15.01157. [DOI] [PubMed] [Google Scholar]
- 19.Jayasinghe K., Stark Z., Kerr P.G., et al. Clinical impact of genomic testing in patients with suspected monogenic kidney disease. Genet Med. 2021;23:183–191. doi: 10.1038/s41436-020-00963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanudisastro H.A., Holman K., Ho G., et al. Australia and New Zealand renal gene panel testing in routine clinical practice of 542 families. NPJ Genom Med. 2021;6:20. doi: 10.1038/s41525-021-00184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott M.D., James L.C., Simms E.L., et al. Mainstreaming genetic testing for adult patients with autosomal dominant polycystic kidney disease. Can J Kidney Health Dis. 2021;8 doi: 10.1177/20543581211055001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallett A., Patel C., Maier B., et al. A protocol for the identification and validation of novel genetic causes of kidney disease. BMC Nephrol. 2015;16:152. doi: 10.1186/s12882-015-0148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark Z., Boughtwood T., Phillips P., et al. Australian genomics: a federated model for integrating genomics into healthcare. Am J Hum Genet. 2019;105:7–14. doi: 10.1016/j.ajhg.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaff CL, M Winship I, M Forrest S, et al. Preparing for genomic medicine: a real world demonstration of health system change. 2017;2:16. https://doi.org/10.1038/s41525-017-0017-4 [DOI] [PMC free article] [PubMed]
- 25.Vidgen M.E., Williamson D., Cutler K., et al. Queensland Genomics: an adaptive approach for integrating genomics into a public healthcare system. NPJ Genom Med. 2021;6:71. doi: 10.1038/s41525-021-00234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tudini E., Andrews J., Lawrence D.M., et al. Shariant platform: enabling evidence sharing across Australian clinical genetic-testing laboratories to support variant interpretation. Am J Hum Genet. 2022;109:1960–1973. doi: 10.1016/j.ajhg.2022.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damschroder L.J., Reardon C.M., Widerquist M.A.O., Lowery J. The updated Consolidated Framework for Implementation Research based on user feedback. Implement Sci. 2022;17:75. doi: 10.1186/s13012-022-01245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damschroder L.J., Aron D.C., Keith R.E., Kirsh S.R., Alexander J.A., Lowery J.C. Fostering implementation of health services research findings into practice: a consolidated framework for advancing implementation science. Implement Sci. 2009;4:50. doi: 10.1186/1748-5908-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirk M.A., Kelley C., Yankey N., Birken S.A., Abadie B., Damschroder L. A systematic review of the use of the Consolidated Framework for Implementation Research. Implement Sci. 2016;11:72. doi: 10.1186/s13012-016-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayasinghe K., Stark Z., Patel C., et al. Comprehensive evaluation of a prospective Australian patient cohort with suspected genetic kidney disease undergoing clinical genomic testing: a study protocol. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2019-029541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiStefano M.T., Goehringer S., Babb L., et al. The Gene Curation Coalition: a global effort to harmonize gene-disease evidence resources. Genet Med. 2022;24:1732–1742. doi: 10.1016/j.gim.2022.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research Electronic Data Capture (REDCap)-a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris P.A., Taylor R., Minor B.L., et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95 doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kansal A., Quinlan C., Stark Z., et al. Theory designed strategies to support implementation of genomics in nephrology. Genes (Basel) 2022;13 doi: 10.3390/genes13101919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrug M., Bloom M.S., Seto C., et al. Genetic testing for chronic kidney diseases: clinical utility and barriers perceived by nephrologists. Kidney Med. 2021;3:1050–1056. doi: 10.1016/j.xkme.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.KDIGO Conference Participants Genetics in chronic kidney disease: conclusions from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 2022;101:1126–1141. doi: 10.1016/j.kint.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres V.E., Harris P.C., Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 39.Connaughton D.M., Kennedy C., Shril S., et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95:914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun D.A., Schueler M., Halbritter J., et al. Whole exome sequencing identifies causative mutations in the majority of consanguineous or familial cases with childhood-onset increased renal echogenicity. Kidney Int. 2016;89:468–475. doi: 10.1038/ki.2015.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallawaarachchi A.C., Lundie B., Hort Y., et al. Genomic diagnostics in polycystic kidney disease: an assessment of real-world use of whole-genome sequencing. Eur J Hum Genet. 2021;29:760–770. doi: 10.1038/s41431-020-00796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahl N., Bloom M.S., Chebib F.T., et al. The clinical utility of genetic testing in the diagnosis and management of adults with chronic kidney disease. J Am Soc Nephrol. 2023;34:2039–2050. doi: 10.1681/ASN.0000000000000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becherucci F., Landini S., Palazzo V., et al. A clinical workflow for cost-saving high-rate diagnosis of genetic kidney diseases. J Am Soc Nephrol. 2023;34:706–720. doi: 10.1681/ASN.0000000000000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nestor J.G., Marasa M., Milo-Rasouly H., et al. Pilot study of return of genetic results to patients in adult nephrology. Clin J Am Soc Nephrol. 2020;15:651–664. doi: 10.2215/CJN.12481019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirby A., Gnirke A., Jaffe D.B., et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet. 2013;45:299–303. doi: 10.1038/ng.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crellin E., Martyn M., McClaren B., Gaff C. What matters to parents? A scoping review of parents’ service experiences and needs regarding genetic testing for rare diseases. Eur J Hum Genet. 2023;31:869–878. doi: 10.1038/s41431-023-01376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevin S.M., McLoone J., Wakefield C.E., Kennedy S.E., McCarthy H.J. Genetic testing in the pediatric nephrology clinic: understanding families’ experiences. J Pediatr Genet. 2022;11:117–125. doi: 10.1055/s-0040-1721439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y., Jayasinghe K., Stark Z., et al. Genomic testing for suspected monogenic kidney disease in children and adults: a health economic evaluation. Genet Med. 2023;25 doi: 10.1016/j.gim.2023.100942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genes with VUS/VUS× identified in the national KidGen cohort. The genes are stratified by clinical diagnosis, with the width of the cut-outs indicating the number of appearances. VUS is a variant of uncertain significance; VUS× is a VUS with suspected clinical relevance. Table S1. Patient recruitment by state and year stratification. Table S2. Occurrence of genes affected by pathogenic/likely pathogenic variants in patients with different kidney diagnoses. Table S3. Genes listed in Kidneyome SuperPanel Version 8.53. STROBE checklist (PDF)
Data Availability Statement
The genomic data generated from this study involve patient information and are subject to confidentiality and privacy protections. Due to these legal and ethical considerations, the genomic data cannot be shared publicly or made available to other researchers. This restriction is in place to ensure the privacy and security of the patient's sensitive information. We are committed to upholding the highest data protection and privacy standards for the individuals involved in this study. Deidentified results supporting this study's findings are available from the corresponding author, AJM, upon reasonable request.