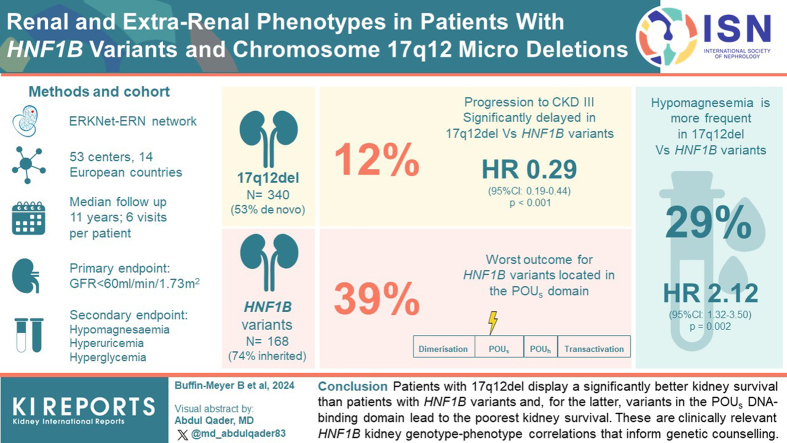
Abstract
Introduction
Hepatocyte nuclear factor 1-beta (HNF1B) gene variants or the chromosome 17q12 deletion (17q12del) represent the most common monogenic cause of developmental kidney disease. Although neurodevelopmental disorders have been associated with the 17q12del, specific genotype-phenotype associations with respect to kidney function evolution have not yet been fully defined. Here, we aimed to determine whether 17q12del or specific HNF1B variants were associated with kidney survival in a large patient population with HNF1B disease.
Methods
This was a retrospective observational study involving 521 patients with HNF1B disease from 14 countries using the European Reference Network for rare kidney diseases with detailed information on the HNF1B genotype (HNF1B variants or the 17q12del). Median follow-up time was 11 years with 6 visits per patient. The primary end point was progression to chronic kidney disease (CKD) stage 3 (estimated glomerular filtration rate [eGFR] < 60 ml/min per 1.73 m2). Secondary end points were the development of hypomagnesemia or extrarenal disorders, including hyperuricemia and hyperglycemia.
Results
Progression toward CKD stage 3 was significantly delayed in patients with the 17q12del compared to patients with HNF1B variants (hazard ratio [HR]: 0.29, 95% confidence interval [CI]: 0.19–0.44, P < 0.001). Progression toward CKD stage 3 was also significantly delayed when HNF1B variants involved the HNF1B Pit-1, Oct-1, and Unc-86 homeodomain (POUh) DNA-binding and transactivation domains rather than the POU-specific domain (POUs) DNA-binding domain (HR: 0.15 [95% CI: 0.06–0.37), P < 0.001 and HR: 0.25 (95% CI: 0.11–0.57), P = 0.001, respectively). Finally, the 17q12del was positively associated with hypomagnesemia and negatively associated with hyperuricemia, but not with hyperglycemia.
Conclusion
Patients with the 17q12del display a significantly better kidney survival than patients with other HNF1B variants; and for the latter, variants in the POUs DNA-binding domain lead to the poorest kidney survival. These are clinically relevant HNF1B kidney genotype-phenotype correlations that inform genetic counseling.
Keywords: chronic kidney disease, genotype-phenotype correlation, HNF1B disease, outcome
Graphical abstract
HNF1B-related disease is identified in 20% to 30% of fetuses with renal abnormalities. HNF1B disease, initially described as the renal cysts and diabetes syndrome (OMIM # 137920), has evolved to a much wider phenotype. Indeed, variants or whole gene deletions of HNF1B are the most common prenatal cause of hyperechogenic kidneys with or without cysts.1 When detected in the postnatal period, HNF1B disease is the most common cause of isolated renal hypodysplasia.2,3 Other renal manifestations associated with HNF1B disease include multicystic dysplastic kidneys, glomerulocystic kidney disease, oligomeganephronia, renal agenesis, renal hypoplasia, urinary tract defects, familial juvenile hyperuricemic nephropathy, and renal interstitial fibrosis.4, 5, 6 This wide variety of kidney phenotypes is probably due to the fact that the HNF1B protein is involved in the majority of the stages of kidney development, from the outgrowth of the uretic bud and its early branching7 to the elongation of the renal tubules,8 and is still expressed in the mature kidney. Individuals with HNF1B disease may also suffer from electrolyte disturbances such as hypomagnesemia and hyperuricemia, pancreatic hypoplasia, early-onset diabetes mellitus, as well as liver and genital defects.9
HNF1B is a transcription factor that controls key cystic disease genes during kidney development,10 where it controls the expression of genes required for kidney metabolism and solute transport by tubular epithelial cells in the adult kidney.11,12 HNF1B contains a dimerization domain located at the N-terminus of the protein which mediates the formation of HNF1B homodimers or heterodimers with the related protein HNF-1α.13 The protein also contains a homeo DNA-binding domain, consisting of a Pit-1, Oct-1, and Unc-86 (POU) homeodomain (POUh) and POU-specific domain (POUs). POUh is a classic homeodomain which recognizes DNA, whereas POUs cooperates with POUh to enhance the affinity and specificity of DNA binding.14 Finally, the C-terminal region of HNF1B contains a transactivation domain that is responsible for coactivator recruitment and transcriptional regulation.15
HNF1B disease transmission follows a dominant pattern, however, de novo variants are very common. Genetic alterations in HNF1B disease can be broadly divided into two categories.9 One category comprises base substitutions and small insertions and/or duplications and/or deletions, leading to missense, nonsense, frameshift, and splicing variants, most of which are described to be located in the DNA-binding and N-terminal dimerization domain of the protein.9 These HNF1B variants account for approximately 41% to 44% of patients with HNF1B disease.9 The other category is represented by the so-called 17q12del, which spans a region of approximately 1.5 Mb in which are located 14 genes, including HNF1B.9
The 17q12del compared to the HNF1B variants,16, 17, 18, 19 has already been shown to be associated with an increased risk of neurodevelopmental disorders in the pediatric population. Furthermore, in adults, the 17q12del has been suggested to be associated with improved kidney function, as demonstrated in a small subset of 169 adult patients.20 However, no clear kidney genotype-phenotype correlation has been established; though nowadays, HNF1B genetic testing is routinely obtained in most clinics for patients with developmental kidney abnormalities. Improved insight into this relationship would further inform the management and counseling of patients with HNF1B disease. We therefore aimed to investigate the association of HNF1B variants and the 17q12del with the development of CKD in a large multicenter European cohort of 521 individuals with genetically well-characterized HNF1B disease.
Methods
Patients and Data Collection
Observational anonymized data on patients with HNF1B disease were retrospectively collected from different European registries under the European Reference Network for rare kidney diseases (www.erknet.org) umbrella. Minimal data necessary for the study included the following: (i) the pathogenic HNF1B variant, specifically whether it was either the 17q12del or single nucleotide variants (missense, nonsense), small deletions or duplications (HNF1B variants were identified for 27% with Sanger sequencing, for 23% with multiplex ligation-dependent probe amplification, for 9% with quantitative multiplex polymerase chain reaction of short fluorescent fragments, for 8% with comparative genomic hybridization array and for 3% with fluorescence in situ hybridization); (ii) information on the inheritance of HNF1B genotype (the de novo status was determined by targeted analysis in parents, by trio-based exosome sequencing and by consulting family history or sonography of parents in 78%, 19%, and 3% of the cases, respectively); (iii) the prenatal or postnatal sonomorphologic kidney phenotype at diagnosis (multicystic dysplastic kidneys, cortical cystic kidneys, hyperechogenic kidneys, hypoplastic kidneys, agenesis, and “other”) and whether structural anomalies were unilateral or bilateral. Development of hypomagnesemia, hyperuricemia, and hyperglycemia was recorded. Maximum follow-up of patients was requested, preferably with new ultrasound (every 2–3 years) and biochemical-data (every 1–2 years, blood magnesium, potassium, uric acid, serum creatinine with associated method used [Jaffe or enzymatic]). We did not collect information on whether all patients were index patients or kindreds of index patients. Nonpaternity was not ruled out. Data were retrospectively collected from 53 centers in 14 European countries according to local standard-of-care. Given that HNF1B related disease is congenital, we considered the start of patient follow-up being birth.
Clinical Parameters
The CKD-Epidemiology Collaboration equation21 for eGFR calculation was used for patients aged ≥15 years. For patients aged <15 years, the eGFR was calculated using the Schwartz formula.22 In the particular case of eGFR estimation in the neonatal period (≤1 month), the Schwartz coefficient used was 0.31 and eGFR = 0.31 × (height/serum creatinine).23 CKD stage 3 was defined by an eGFR <60 ml/min per 1.73 m2 for patients aged ≥15 years. For patients aged 2 to 15 years, CKD stage 3 was defined by an eGFR <60 ml/min per 1.73 m2 in at least 2 consecutive visits. The thresholds for the definitions of hypomagnesemia, hyperuricemia, or hyperglycemia were <0.6 mmol/l, >320 μmol/l or >1.26 g/l (fasting), respectively, on at least 1 measurement.
Ethics
The data were fully anonymized. Approval was obtained from the medical ethical committees or institutional review boards for all participating countries and written informed consent was obtained from all participants or parents in adherence to the declaration of Helsinki.
Statistical Analysis
Patient characteristics were reported as number (percentage) or median (25%–75%) for qualitative and quantitative variables, respectively. They were compared according to the HNF1B genotype using a Chi-square test (N > 5) or a Fisher test (N ≤ 5) for categorical variables and a Wilcoxon rank sum test for continuous variables ((gtsummary package in R statistical software, V4.0.1)).
In the specific case of kidney ultrasound characteristics, where a global significant effect (P < 0.05) of the HNF1B genotype was observed, additional analysis was performed according to an approach based on calculating adjusted standardized residuals,24 in order identify features making the greatest contribution to the Chi-square test result: adjusted standardized residual ≥ 3 or ≤ −3 indicated that there were more or less patients, respectively, with the considered feature than would be expected by chance. Univariate and multivariate Cox proportional HR models were built to estimate the impact of HNF1B genotype on CKD stage 3 development (proportional hazard assumption was verified for each model using Schoenfeld residuals method) (survdiff package in R, V4.0.1). Progression to CKD was not evaluated in children aged <2 years, due to changes in eGFR in early life. HRs are reported with 95% CIs and P-value to assess whether the HR is statistically significantly different from 1 (survival package of R, V4.0.1). The equality of survival distributions was compared using log-rank test (survdiff package of R, V4.0.1). Considering that HNF1B disease is a congenital disease, the start of the follow-up was birth. Survival curves only used data from children aged ≥2 years. Odd ratios for hypomagnesemia, hyperuricemia, and hyperglycemia were obtained using logistic regression and reported with 95% CIs and P-value to assess whether the odd ratios is statistically significantly different from 1 (stats package of R, V4.0.1).
For analysis of eGFR evolution after birth, the first week was excluded, considering that creatinine levels reflected that of the mother. Changes in eGFR were assessed using a generalized linear mixed model with a negative binomial distribution, considering the HNF1B genotype (17q12del or HNF1B variant) as random effect (lme4 package in R, V4.0.1). For each period analyzed (1 week–2 years and 2–18 years) 4 models were analyzed as follows: (i) 1 model without random effect, (ii) a mixed-effect model with a random effect for the intercept and a fixed slope, (iii) a mixed-effect model with a random effect for the slope and a fixed intercept, and (iv) a mixed-effect model with both a random intercept and a random slope. These 4 models were compared among each other using Akaike's information criterion corrected for small samples.25 The model with the lowest Akaike's information criterion and with a value of at least 2 Akaike's information criterion units from the other models was considered the model best fitting the data. Patients with <3 eGFR measurements during the period of interest were excluded from the generalized linear mixed model analysis.
Results
Cohort Description
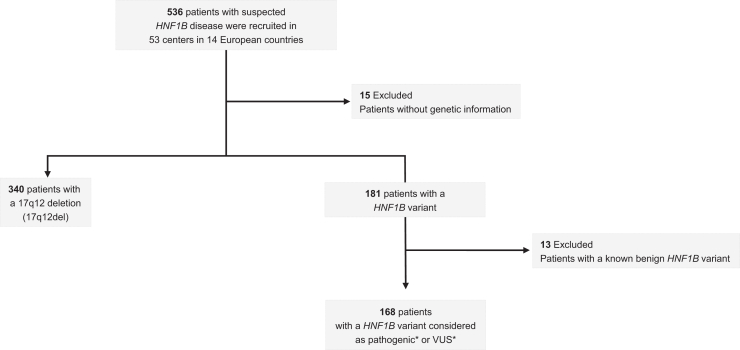
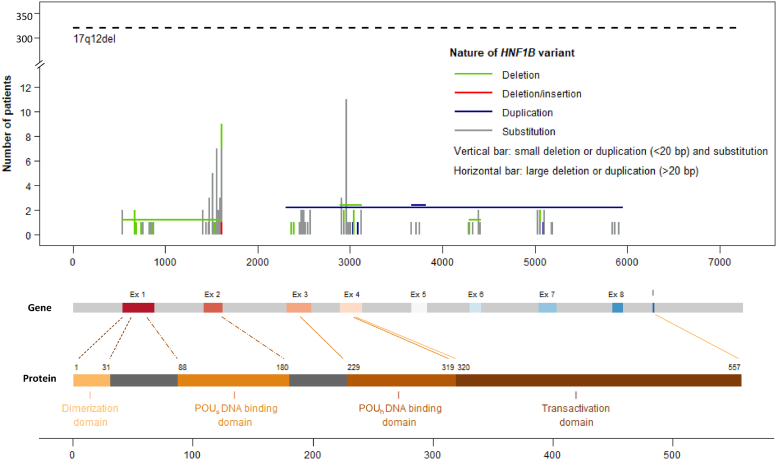
Retrospective data were initially collected from registries via ERKNet for 536 patients with suspected HNF1B disease from 53 centers in 14 European countries (Supplementary Figure S1). Fifteen patients were excluded because no clear information on the HNF1B variant was obtained leading to 340 patients with the 17q12del and 181 patients with HNF1B variants (Figure 1). Next, 13 additional patients were excluded in the HNF1B variants group because they were described as benign HNF1B variants (p.Val61Gly, p.Gly76Cys, p.Asp82Asn, p.Asn228Lys and p.His336Asp, Figure 1 and Supplementary Table S1 [lower grey section]). This led to a total of 168 patients with HNF1B variants (Supplementary Table S1). These HNF1B variants were mainly found in the DNA-binding domains POUs and POUh (67%, 88/132) and, to a lesser extent, in the transactivating domain (19%, 25/132) (Figure 2). The majority of the HNF1B variants were nucleotide substitutions (74%, Supplementary Figure S2a) leading to 47% of missense and 20% of nonsense variants at the protein level (Supplementary Figure S2b).
Figure 1.
Overview of patient recruitment and patient exclusion. ∗Definition of pathogenicity or variant of unknown significance (VUS) was based on the merger of the 3 following databases. ClinVar (https://www.ncbi.nlm.nih.gov/clinvar (db accessed July 11, 2023); LOVD (https://databases.lovd.nl/shared/variants/HNF1B#object_id=VariantOnTranscript%2CVariantOnGenome&id=HNF1B&order=VariantOnTranscript%2FDNA%2CASC&search_transcriptid=00009498&search_VariantOnTranscript/DNA=c.738G%3ET&page_size=100&page=1 (db accessed July 11, 2023)) and Leipzig_University (https://www.hnf1b.org (db accessed July 11, 2023).26 A variant was marked as pathogenic if in at least 1 database the variant was labelled “pathogenic”. In all other cases a variant was labelled “VUS.”
Figure 2.
Position of molecular modifications on the HNF1B gene and HNF1B protein in 508 patients with HNF1B disease. Domains in the HNF1B protein were positioned according to.27
Patients had a predominant antenatal diagnosis (58%, Table 1), a median of 6 visits and 11 years of follow-up with no difference between the 17q12del and the HNF1B variants (Supplementary Table S2). A 17q12del was identified in 67% (340/508) of the patients and occurred de novo in 53% of the cases (Table 1); 74% of the HNF1B variants were inherited.
Table 1.
Patients characteristics at inclusion
| Patient characteristics | n |
HNF1B |
P-valuea | Q-valueb | ||
|---|---|---|---|---|---|---|
| HNF1B variants | 17q12del | |||||
| All | 508 | 168 | 340 | |||
| Sex | 507 | 0.503 | 0.575 | |||
| Female | 208 (41%)c | 72 (43%) | 136 (40%) | |||
| Male | 299 (59%) | 95 (57%) | 204 (60%) | |||
| Origin | 286 | <0.001 | <0.001 | |||
| De novo | 123 (43%) | 28 (26%) | 95 (53%) | |||
| Inherited | 163 (57%) | 80 (74%) | 83 (47%) | |||
| Transmission mode (in case of inherited variants) | 156 | 0.036 | 0.097 | |||
| Mother | 100 (64%) | 44 (55%) | 56 (73%) | |||
| Father | 55 (35%) | 34 (43%) | 21 (27%) | |||
| Mother + father | 1 (0.6%) | 1 (1.3%) | 0 (0%) | |||
| Diagnosis | 309 | 0.023 | 0.090 | |||
| Antenatal | 180 (58%) | 49 (49%) | 131 (63%) | |||
| Postnatal | 129 (42%) | 51 (51%) | 78 (37%) | |||
| Age at antenatal diagnosis (wa) | 70 | 24 (20–30)d | 24 (20–27) | 24 (21–31) | 0.311 | 0.575 |
| Age at postnatal diagnosis (y) | 109 | 3 (0–17) | 4 (1–17) | 3 (0–16) | 0.454 | 0.575 |
| Number of kidneys with US lesions | 474 | 0.405 | 0.575 | |||
| No | 3 (0.6%) | 2 (1.3%) | 1 (0.3%) | |||
| 1 | 48 (10%) | 16 (10%) | 32 (10.0%) | |||
| 2 | 423 (89%) | 135 (88%) | 288 (90%) | |||
| Number of functional kidneys | 479 | 0.895 | 0.895 | |||
| 2 | 391 (82%) | 126 (81%) | 265 (82%) | |||
| 1 | 88 (18%) | 29 (19%) | 59 (18%) | |||
Pearson's Chi-square test; Fisher exact test; Wilcoxon rank sum test.
False discovery rate correction for multiple testing.
n (%)
Median (25%–75%).
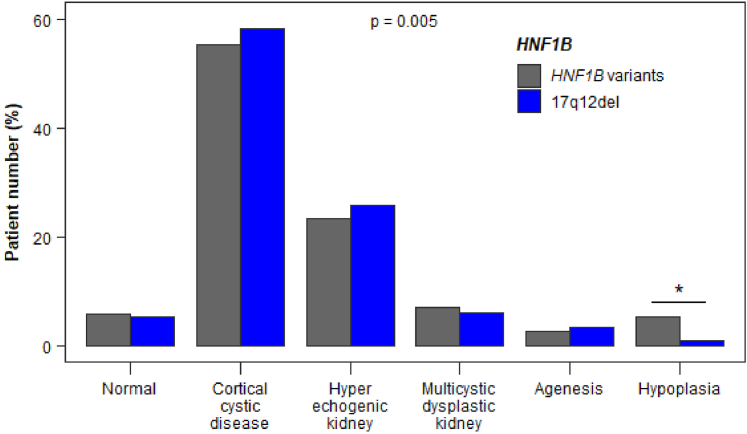
At diagnosis, main kidney malformations observed with ultrasound were cortical cystic disease or hyperechogenic kidneys, which together affected >80% of the patients (Figure 3). A single functional kidney due to unilateral multicystic dysplastic kidney or agenesis was observed in 18% of the patients (Table 1). The risk of having hypoplasia was significantly lower in patients with the 17q12del than in patients with HNF1B variants (1.2% vs. 5.5%, Figure 3).
Figure 3.
Kidney ultrasound characteristics at diagnosis in the 521 patients with HNF1B variants. A global significant effect (P = 0.005) of the HNF1B genotype was observed. The adjusted standardized residual was equal to 4.80 for hypoplasia in patients with HNF1B variants, thereby indicating (≥3, see statistical analysis) that there were more patients with hypoplasia than would be expected by chance. In contrast, the adjusted standardized residual was equal to −6.99 for hypoplasia in patients with the 17q12del, thereby indicating (≤ −3, see statistical analysis) that there were less patients with hypoplasia than would be expected by chance. This strongly suggests that the risk of having hypoplasia was significantly lower in patients with the 17q12del than in patients with HNF1B variants (∗).
Progression to Chronic Kidney Failure
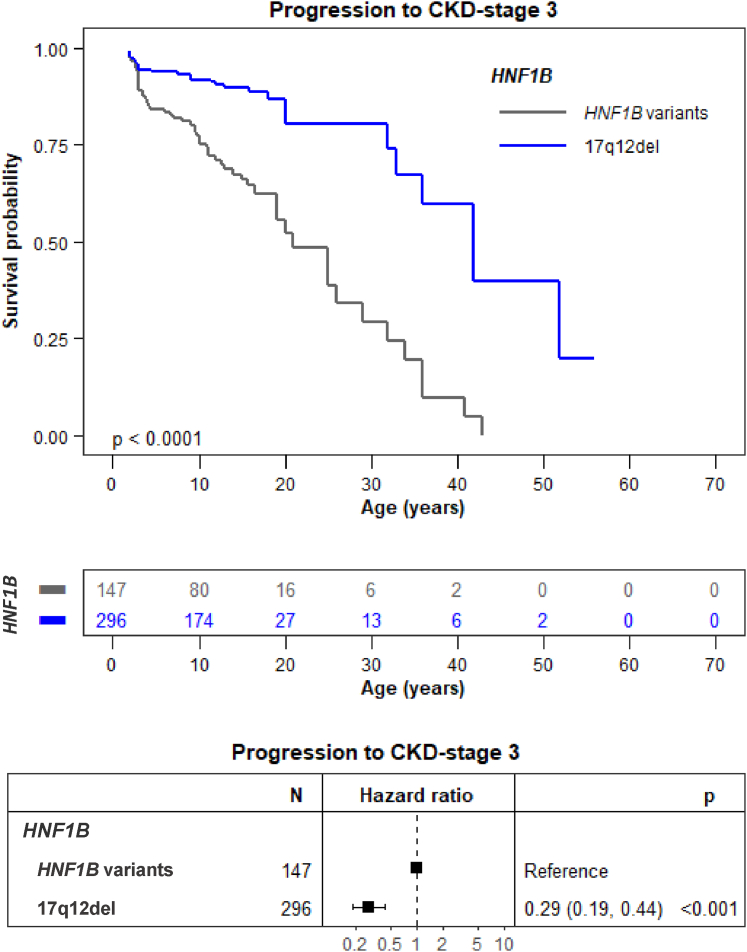
Twenty-one percent of the patients progressed toward the primary kidney end point (CKD stage 3, eGFR < 60 ml/min per 1.73 m2) during follow-up. This was less frequent in the population with the 17q12del than with HNF1B variants (12% vs. 39%, P < 0.001, Supplementary Table S2). In addition, progression toward CKD-stage 3 was significantly delayed in patients with the 17q12del compared to patients with HNF1B variants (Figure 4, HR: 0.29 [95% CI: 0.19–0.44], P < 0.001) with CKD-free survival of 87% (95% CI: 81–93) for the 17q12del versus 63% (95% CI: 54–73) for HNF1B variants at the age of 18 years. The association of HNF1B genotype with progression toward CKD stage 3 was still observed after adjustment for known CKD risk factors, including sex or hyperglycemia (HR: 0.30 [95% CI: 0.19–0.45], P < 0.001).
Figure 4.
Progression to CKD stage 3 of patients the 17q12del compared to HNF1B variants. Progression to CKD is significantly delayed in patients with the 17q12del compared to patients with HNF1B variants (HR: 0.29 [95% CI: 0.19–0.44], P < 0.001). The survival curve was generated using data from children aged ≥ 2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given the fact the HNF1B disease is a congenital nephropathy. The log-rank test for difference in survival yielded a P-value < 0.0001, indicating that the patients with 17q12del and HNF1B variants differed significantly in progression toward CKD stage 3. CI, confidence interval; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; HR, hazard ratio.
Patients with the 17q12del also developed end-stage kidney failure (ESKF) less frequently (Supplementary Table S2, P < 0.001) and displayed a delayed progression to ESKF (Supplementary Figure S3a, P < 0.001) with ESKF-free survival of 97% (95% CI: 95–100) for the 17q12del versus 86% (95% CI: 79–94) for HNF1B variants at the age of 18 years. Even after reaching CKD stage 3, a tendency for a slower progression to ESKF for the 17q12del was observed (Supplementary Figure S3b, P = 0.14).
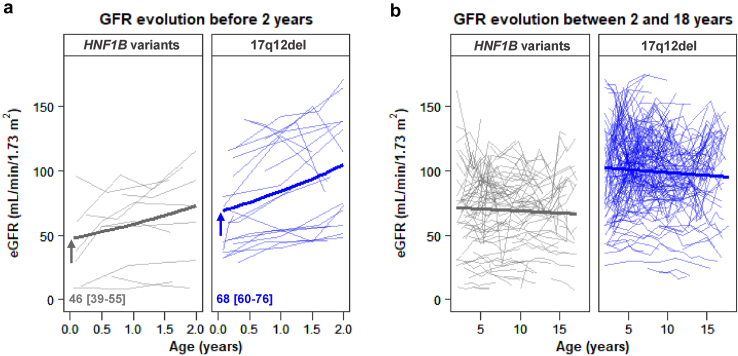
Individuals with the 17q12del had higher eGFR early in life (68 ml/min per 1.73 m2 [95% CI: 60–76] vs. 46 ml/min per 1.73 m2 [95% CI: 39–55] at 1 week, as defined by generalized linear mixed model analysis, Figure 5a). These higher eGFR values persisted throughout childhood (1 week–2 years, Figure 5a) and adolescence (2–18 years, Figure 5b). However, no difference between the 2 groups with respect to evolution of eGFR could be demonstrated (Figure 5a and b).
Figure 5.
Impact of the HNF1B genotype on eGFR trajectories in the pediatric period. Comparison of eGFR trajectories between the HNF1B variants and the 17q12del (a) before 2 years (10 and 21 patients for HNF1B variants and the 17q12del, respectively) and (b) between 2 and 18 years (91 and 175 patients for HNF1B variants and the 17q12del, respectively). Patients with <3 eGFR measurements during the period of interest were excluded. Individual and mean (bold) trajectories are plotted. The arrows and values indicate the mean (95% CI) eGFR at 1 week after birth. CI, confidence interval; eGFR, estimated glomerular filtration rate.
In case of the 17q12del, patients with a single functional kidney (due to 1 multicystic dysplastic kidney or unilateral agenesis) displayed accelerated CKD progression compared to patients with 2 functional kidneys (Supplementary Figure S4a, HR: 2.32 [95% CI: 1.04–5.17), P = 0.04). In contrast, in the group with HNF1B variants, the number of functional kidneys was not associated with CKD progression (HR: 0.92 [95% CI: 0.43–1.98), P = 0.836, Supplementary Figure S4b).
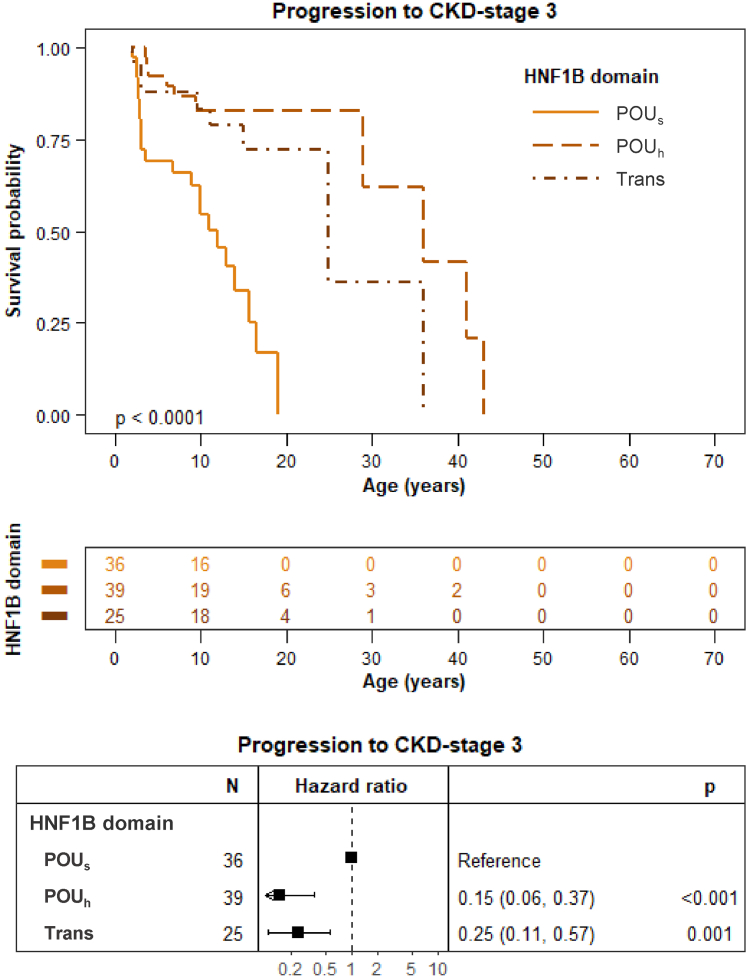
As observed in other studies,9 the majority of the patients had HNF1B variants located in the POU domains followed by variants in the transactivation domain. Patients with variants located in the POUh or transactivation domains progressed slower toward CKD stage 3 than patients with variants in the POUs domain (Figure 6, HR: 0.15 [95% CI: 0.06–0.37], P < 0.001 and HR: 0.25 [95% CI: 0.11–0.57], P = 0.001, respectively). Adjustment by sex or hyperglycemia did not impact progression (HR: 0.14 [95% CI: 0.05–0.35], P < 0.001 and HR 0.22 [95% CI: 0.08–0.58], P = 0.002, respectively).
Figure 6.
Impact of variants in the different HNF1B domains on the progression to CKD stage 3. Variants located in the POUh and transactivation domains displayed a significantly delayed progression toward CKD stage 3 compared to patients with variants in the POUs domain (HR: 0.15 [95% CI: 0.06–0.37], P < 0.001 and HR: 0.25 [95% CI: 0.11–0.57], P = 0.001, respectively). The survival curve was generated using data from children aged ≥2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. CI, confidence interval; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; HR, hazard ratio; Trans, transactivation domain. The log rank test for difference in survival yielded a P-value < 0.0001, indicating that the patients with variants in the 3 HNF1B domains differed significantly in progression toward CKD stage 3.
In contrast, the type of variants (missense, nonsense, splicing or frameshift) were not specifically associated with kidney survival (Supplementary Figure S5).
Hypomagnesemia or Extrarenal Disorders
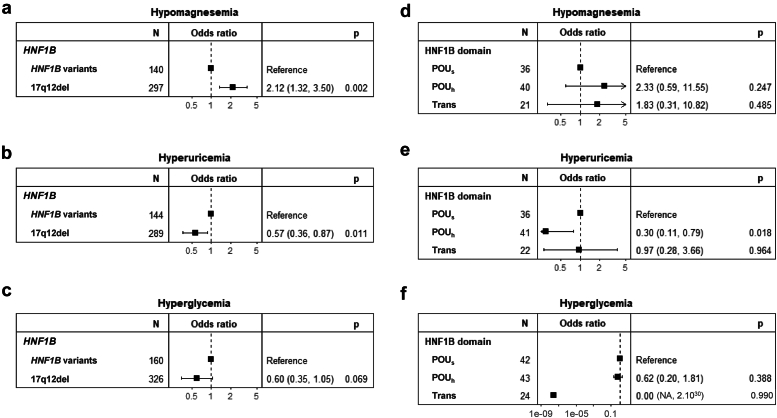
We next investigated whether there was a difference between the 17q12del and HNF1B variants in the development of hypomagnesemia or extrarenal disorders. Hypomagnesemia (defined as plasma magnesium <0.6 mmol/l) was observed in 29% of the patients during follow-up (Supplementary Table S2) and was significantly more frequent in patients with the 17q12del (HR: 2.12 [95% CI: 1.32–3.50], P = 0.002, Figure 7a). However, onset of hypomagnesemia was independent of the affected (POU or transactivation) protein domains (Figure 7d).
Figure 7.
The 17q12del and HNF1B variants in hypomagnesemia or extrarenal disorders. (a) Hypomagnesemia (magnesium < 0.6 mmol/l) was more frequent in patients with the 17q12del compared to patients with HNF1B variants. (b) Hyperuricemia (uric acid > 320 μmol/l) was less frequent in patients with the 17q12del compared to patients with HNF1B variants. (c) Hyperglycemia (fasting blood glucose > 1.26 g/l) was not different in the patient groups. (d) Hypomagnesemia was not different between patients with variants in the POUs, POUh, and transactivation domains. (e) Hyperuricemia was less frequent in patients with variants in the POUh than in the POUs and transactivation domains. (f) Hyperglycemia was not different between patients with variants in the POUs, POUh, and transactivation domains.
Overall, 65% of the patients developed hyperuricemia (defined by plasma uric acid >320 μmol/l, Supplementary Table S2) and was significantly less frequent in patients with the 17q12del compared to the HNF1B variants (HR: 0.57 [95% CI: 0.36–0.87), P = 0.011, Figure 7b). Among the HNF1B variants, patients with variants in the POUh domain, but not those with variants in the transactivation domain, developed significantly less frequent hyperuricemia than patients with variants in the POUs (HR: 0.30 [95% CI: 0.11–0.79], P = 0.018, Figure 7e). Finally, only 12% of the patients developed hyperglycemia (blood glucose > 1.26 g/l, Supplementary Table S2), which did not correlate with a specific HNF1B genotype even though a tendency to be less frequent in the population with the 17q12del was observed (HR: 0.60 [95% CI: 0.35–1.05), P = 0.069, Figure 7c). No significant difference for the development of hyperglycemia was observed between variants in the POU or transactivation domains (Figure 7f).
Discussion
Genotype-kidney survival phenotype correlations in patients with HNF1B disease have long been sought to improve genetic counseling. For a long time, there was no demonstrated correlation between genotype and kidney phenotype in HNF1B disease.9 However, a first small scale study in 2010, including 70 pediatric patients, showed a lower proportion of patients with renal failure in individuals with the 17q12del than in individuals with nonsense, splice, or frameshift HNF1B variants.4 Subsequently, a study in 2016 with 38 pediatric and adult individuals with HNF1B disease showed that patients with the 17q12del displayed higher eGFR compared to patients with HNF1B variants.18 Finally in 2018, Dubois-Lafforgue et al.20 showed in a larger population of 169 adult patients with HNF1B disease, primarily selected for HNF1B disease screening due the presence of maturity-onset diabetes of the young, that individuals with the 17q12del less often had CKD3–4/ESKF at diagnosis and at long-term follow-up (12–14.5 years). This present report clearly confirms in a large cohort of 521 patients with HNF1B disease that the 17q12del is associated with significantly better kidney survival than the HNF1B variants across all ages, including pediatric and adult patients. Moreover, this study identified for the first time that variants located in the POUs DNA binding domain of HNF1B had a significantly worse kidney survival than variants located in the POUh or transactivation domains.
The identification in childhood of a 17q12del and the presence of 2 functional kidneys will allow to reassure the parents with the information that their child has a low probability of developing CKD stage 3 before the age of 18 years. In contrast, it has been clearly documented that the 17q12del is associated with an increased risk of developing neurodevelopmental disorders.16, 17, 18, 19 Therefore, this important aspect should be considered and included in parental counseling. The presence of an HNF1B variant instead of the 17q12del would be an argument for early-in-life monitoring of signs of kidney failure and adopt conservative management. In addition, if the other HNF1B variant is located in the POUs DNA binding domain of the HNF1B protein, this monitoring should be further reinforced because variants in this domain compared to the POUh and transactivation domains led to a particularly high risk of progression to CKD stage 3 in our study.
The difference in terms of renal function between patients with the 17q12del and those with the HNF1B variants appears to be already present in the neonatal period, however, without differentially impacting eGFR evolution at least up to 18 years of age.
On a molecular level, the fact that the 17q12del resulted in a less severe kidney phenotype compared to the HNF1B variants is surprising. Such a difference has also been observed in mice. Hnf1b+/− mice were phenotypically normal28 with increased rather than decreased29 kidney HNF1B protein abundance, whereas mice carrying a heterozygous HNF1B splice variant lead presence of bilateral cystic kidneys with low kidney HNF1B protein levels.29 A possible explanation may be the fact that missense HNF1B variants might lead to a dominant negative effect because the variants are for the greater part located in the DNA-binding and N-terminal dimerization domain of the HNF1B protein9 and analysis of a Leu168Pro HNF1B variant located in the POUs domain clearly demonstrated a dominant effect on HNF1B activity when coexpressing the wild type and Leu168Pro HNF1B variant in vitro.30
The observation that variants in the POUs domain led to a more severe kidney outcome than variations in the POUh and transactivation domains is intriguing. It is thought that on the molecular level, the POUs domain cooperates with POUh to enhance the binding affinity and specificity of DNA binding and is not the initial DNA binding site.14 However, in vitro, missense variants in the POUs domain lead in general to a more pronounced reduction in HNF1B protein stability, transcriptional activity, and DNA binding than missense variants in the POUh domain.14
Hypomagnesemia developed in 29% of the patients with HNF1B disease. Hypomagnesemia is a common feature due to renal magnesium wasting in patients with HNF1B disease.9 However, we report for the first time a higher risk for this disorder in patients with the 17q12del. In contrast the risk of hyperuricemia, observed 65% of the patients during follow-up, was lower in patients with the 17q12del and in patients with a variant in the POUh compared to the POUs and transactivation domains.
The main strength of this study is the large number (>500) of patients enrolled for a rare disease. Patients were enrolled in a variety of >50 clinics, representing a diversity of health care systems across 14 European countries. In addition, the frequency of the 17q12del in our large cohort is close to what has been reported in the literature, 67% in the current study versus 56% to 59% in the literature.9 Therefore, the patient population studied is similar to routine clinical care.
Limitations of this study were the fact that this is a relatively young cohort with few patients aged over 20 years (59/521) at the end of the follow-up. This probably explains the low percentage (12%) of patients who developed hyperglycemia during follow-up in our cohort. It would therefore be interesting to reanalyze this cohort with an additional decade of follow-up. We also did not study the aforementioned development of psychiatric and autism spectrum disorders in our cohort16, 17, 18, 19 due to the trade-off of focusing on the kidney to maximize the number of patients included. Only 2 variants were located in the dimerization domain. Finally, this was a retrospective study.
In conclusion, our study has identified clinically relevant genotype-phenotype correlations in patients with HNF1B disease predicting kidney survival that inform genetic counseling.
Appendix
List of Additional Collaborators of the HNF1B Variant Study Group
Gema Ariceta, MD, PhD (Pediatric Nephrology, University Hospital Vall d’Hebron, Barcelona, Spain); Elisa Benetti, MD, PhD (Pediatric Nephrology, Dialysis and Transplant Unit, Department of Women's and Children's Health, Padua University Hospital, Padua, Italy); Marcus R Benz, MD (Pediatric Nephrology Dachau, Dachau, Germany); Anna Bjerre, MD, PhD (Division of Pediatric and Adolescent Medicine, Oslo University Hospital, Oslo, Norway); Bernard R Boudailliez, MD (Service de Néphrologie Pédiatrique, Département de Pédiatrie, CHU Amiens, Amiens, France); Antonia Bouts, MD, PhD (Emma Children's Hospital, Amsterdam University Medical Centers, Department of Pediatric Nephrology, Amsterdam Reproduction & Development, Amsterdam, the Netherlands); Jens Drube, (Department of Pediatric Kidney, Liver and Metabolic Diseases, Hannover Medical School Children's Hospital, Hannover, Germany); Ann Christin Gjerstad, MD, PhD (Division of Pediatric and Adolescent Medicine, Oslo University Hospital, Oslo, Norway); Augustina Jankauskiene, MD, PhD (Pediatric Center, Institute of Clinical Medicine, Vilnius University, Vilnius, Lithuania); Eszter Jávorszky, PhD (MTA -SE Lendület Nephrogenetic Laboratory, Budapest, Hungary); Nadine Jay, MD (Service de Pédiatrie, CHU de Brest, Brest, France); Martin Kirschstein, MD, PhD (Department of Pediatrics, General Hospital, Celle, Germany); Nataša Marčun Varda, MD, PhD (University Medical Centre Maribor, Department of Paediatrics, Maribor, Slovenia); Olivier Niel, MD, PhD (Pediatric Nephrology, Centre Hospitalier de Luxembourg, Luxembourg, Luxembourg); François Nobili, MD (Service de Pédiatrie 2, CHU Besancon, Besancon, France); Christine Pietrement, MD, PhD (Unité de Néphrologie Pédiatrique CHU Reims, Reims, France); Dovile Ruzgiene, MD (Pediatric Center, Institute of Clinical Medicine, Vilnius University, Vilnius, Lithuania); Raphael Schild, MD (University Children's Hospital, University Medical Center Hamburg Eppendorf, Hamburg, Germany); Hagen Staude, MD (Department of Pediatric Nephrology, University Children's Hospital, Rostock, Germany); Kálmán Tory, MD, PhD (MTA -SE Lendület Nephrogenetic Laboratory, Budapest, Hungary); Michel Tsimaratos, MD, PhD (Pédiatrie Multidisciplinaire Timone, Aix -Marseille Université, Marseille, France); Ulrike Walden, MD (Paediatric and Adolescent Medicine, University Medical Center, Augsburg, Germany); and Hildegard Zappel, MD (University Children's Hospital Göttingen, Göttingen, Germany).
Disclosure
All the authors declared no competing interests.
Acknowledgments
NEOCYST is funded by the German Federal Ministry of Education and Research–grant code 01GM1515A. This project has been supported by the European Reference Network for rare kidney diseases (ERKNet). ERKNet is funded by the European Union within the framework of the EU4Health Programme (grant No.HS g-23-49).
Data Availability Statement
All raw patient inclusion and follow-up data used in the study of the can be found in Supplementary Table S3.
Footnotes
Supplementary File (PDF and Excel)
Figure S1. An international multicenter study. (a) 14 European countries participated in the study. A total of (b) 22 centers from France and (c) 14 centers from Germany, the 2 major participating countries, were involved.
Figure S2. Distribution of HNF1B variants other than the 17q12del. (a) Nature of variants and (b) resulting variants.
Figure S3. Progression to ESKF of patients with HNF1B disease. (a) Progression to ESKF is significantly delayed in patients with the 17q12del compared to patients with HNF1B variants. (b) Progression to ESKF after developing CKD is not different between the 62 patients with HNF1B variants and 33 patients with the 17q12del. The survival curves were generated using data from children aged ≥2 years to dismiss changes in eGFR evolution in early life. The point in time of progression to ESKF was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. In (a), the log-rank test for difference in survival yielded a P-value < 0.0001, indicating that the patients with 17q12del and HNF1B variants differed significantly in progression toward ESKF. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate; ESKF, end-stage kidney failure.
Figure S4. Impact of 1 or 2 functional kidneys in patients with HNF1B disease on progression to CKD stage 3. (a) Kidney survival of patients with the 17q12del is worse in patients with 1 functional kidney. (b) Kidney survival of patients with HNF1B variants is similar irrespective of the number of functional kidneys. The survival curves were generated using data from children aged ≥2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. In (a) The log-rank test for difference in survival yielded a P-value of 0.034, indicating that the patients with 17q12del with 1 functional kidney differed significantly in progression toward CKD stage 3. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate.
Figure S5. Impact of the nature of the change at the HNF1B protein level on progression to CKD stage 3. Kidney survival of patients is not different between missense, nonsense, splicing, or frameshift HNF1B variants. The survival curves were generated using data from children aged >2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate.
Table S1. Characteristics of HNF1B gene variants.
Table S2. Follow-up data of patients with HNF1B disease.
Table S3. Individual patient data. (Excel)
STROBE Statement.
Contributor Information
Joost P. Schanstra, Email: Joost-peter.schanstra@inserm.fr.
Stéphane Decramer, Email: decramer.s@chu-toulouse.fr.
HNF1B variant study group:
Gema Ariceta, Elisa Benetti, Marcus R. Benz, Anna Bjerre, Bernard R. Boudailliez, Antonia Bouts, Jens Drube, Ann Christin Gjerstad, Augustina Jankauskiene, Eszter Jávorszky, Nadine Jay, Martin Kirschstein, Nataša Marčun Varda, Olivier Niel, François Nobili, Christine Pietrement, Dovile Ruzgiene, Raphael Schild, Hagen Staude, Kálmán Tory, Michel Tsimaratos, Ulrike Walden, and Hildegard Zappel
Supplementary Material
Figure S1. An international multicenter study. (a) 14 European countries participated in the study. A total of (b) 22 centers from France and (c) 14 centers from Germany, the 2 major participating countries, were involved. Figure S2. Distribution of HNF1B variants other than the 17q12del. (a) Nature of variants and (b) resulting variants. Figure S3. Progression to ESKF of patients with HNF1B disease. (a) Progression to ESKF is significantly delayed in patients with the 17q12del compared to patients with HNF1B variants. (b) Progression to ESKF after developing CKD is not different between the 62 patients with HNF1B variants and 33 patients with the 17q12del. The survival curves were generated using data from children aged ≥2 years to dismiss changes in eGFR evolution in early life. The point in time of progression to ESKF was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. In (a), the log-rank test for difference in survival yielded a P-value < 0.0001, indicating that the patients with 17q12del and HNF1B variants differed significantly in progression toward ESKF. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate; ESKF, end-stage kidney failure. Figure S4. Impact of 1 or 2 functional kidneys in patients with HNF1B disease on progression to CKD stage 3. (a) Kidney survival of patients with the 17q12del is worse in patients with 1 functional kidney. (b) Kidney survival of patients with HNF1B variants is similar irrespective of the number of functional kidneys. The survival curves were generated using data from children aged ≥2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. In (a) The log-rank test for difference in survival yielded a P-value of 0.034, indicating that the patients with 17q12del with 1 functional kidney differed significantly in progression toward CKD stage 3. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate. Figure S5. Impact of the nature of the change at the HNF1B protein level on progression to CKD stage 3. Kidney survival of patients is not different between missense, nonsense, splicing, or frameshift HNF1B variants. The survival curves were generated using data from children aged >2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate. Table S1. Characteristics of HNF1B gene variants. Table S2. Follow-up data of patients with HNF1B disease. Table S3. AAA. (Excel). STROBE Statement.
References
- 1.Decramer S., Parant O., Beaufils S., et al. Anomalies of the TCF2 gene are the main cause of fetal bilateral hyperechogenic kidneys. J Am Soc Nephrol. 2007;18:923–933. doi: 10.1681/ASN.2006091057. [DOI] [PubMed] [Google Scholar]
- 2.Weber S., Moriniere V., Knuppel T., et al. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the Escape study. J Am Soc Nephrol. 2006;17:2864–2870. doi: 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- 3.Thomas R., Sanna-Cherchi S., Warady B.A., Furth S.L., Kaskel F.J., Gharavi A.G. HNF1B and PAX2 mutations are a common cause of renal hypodysplasia in the CKiD cohort. Pediatr Nephrol. 2011;26:897–903. doi: 10.1007/s00467-011-1826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidet L., Decramer S., Pawtowski A., et al. Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin J Am Soc Nephrol. 2010;5:1079–1090. doi: 10.2215/CJN.06810909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellanné-Chantelot C., Chauveau D., Gautier J.F., et al. Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann Intern Med. 2004;140:510–517. doi: 10.7326/0003-4819-140-7-200404060-00009. [DOI] [PubMed] [Google Scholar]
- 6.Edghill E.L., Bingham C., Ellard S., Hattersley A.T. Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J Med Genet. 2006;43:84–90. doi: 10.1136/jmg.2005.032854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lokmane L., Heliot C., Garcia-Villalba P., Fabre M., Cereghini S. vHNF1 functions in distinct regulatory circuits to control ureteric bud branching and early nephrogenesis. Development. 2010;137:347–357. doi: 10.1242/dev.042226. [DOI] [PubMed] [Google Scholar]
- 8.Gresh L., Fischer E., Reimann A., et al. A transcriptional network in polycystic kidney disease. EMBO J. 2004;23:1657–1668. doi: 10.1038/sj.emboj.7600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clissold R.L., Hamilton A.J., Hattersley A.T., Ellard S., Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nat Rev Nephrol. 2015;11:102–112. doi: 10.1038/nrneph.2014.232. [DOI] [PubMed] [Google Scholar]
- 10.Coffinier C., Thépot D., Babinet C., Yaniv M., Barra J. Essential role for the homeoprotein vHNF1/HNF1beta in visceral endoderm differentiation. Development. 1999;126:4785–4794. doi: 10.1242/dev.126.21.4785. [DOI] [PubMed] [Google Scholar]
- 11.Ferrè S., Igarashi P. New insights into the role of HNF-1β in kidney (patho)physiology. Pediatr Nephrol. 2019;34:1325–1335. doi: 10.1007/s00467-018-3990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faguer S., Decramer S., Chassaing N., et al. Diagnosis, management, and prognosis of HNF1B nephropathy in adulthood. Kidney Int. 2011;80:768–776. doi: 10.1038/ki.2011.225. [DOI] [PubMed] [Google Scholar]
- 13.Mendel D.B., Khavari P.A., Conley P.B., et al. Characterization of a cofactor that regulates dimerization of a mammalian homeodomain protein. Science. 1991;254:1762–1767. doi: 10.1126/science.1763325. [DOI] [PubMed] [Google Scholar]
- 14.Lu P., Rha G.B., Chi Y.I. Structural basis of disease-causing mutations in hepatocyte nuclear factor 1beta. Biochemistry. 2007;46:12071–12080. doi: 10.1021/bi7010527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbacci E., Chalkiadaki A., Masdeu C., et al. HNF1beta/TCF2 mutations impair transactivation potential through altered co-regulator recruitment. Hum Mol Genet. 2004;13:3139–3149. doi: 10.1093/hmg/ddh338. [DOI] [PubMed] [Google Scholar]
- 16.Laliève F., Decramer S., Heidet L., et al. School Level of children carrying a HNF1B variant or a deletion. Eur J Hum Genet. 2020;28:56–63. doi: 10.1038/s41431-019-0490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno-De-Luca D., SGENE Consortium, Mulle J.G., et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clissold R.L., Shaw-Smith C., Turnpenny P., et al. Chromosome 17q12 microdeletions but not intragenic HNF1B mutations link developmental kidney disease and psychiatric disorder. Kidney Int. 2016;90:203–211. doi: 10.1016/j.kint.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois-Laforgue D., Bellanné-Chantelot C., Charles P., et al. Intellectual disability in patients with MODY due to hepatocyte nuclear factor 1B (HNF1B) molecular defects. Diabetes Metab. 2017;43:89–92. doi: 10.1016/j.diabet.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Dubois-Laforgue D., Cornu E., Saint-Martin C., et al. Diabetes, associated clinical spectrum, long-term prognosis, and genotype/phenotype correlations in 201 adult patients with hepatocyte nuclear factor 1B (HNF1B) molecular defects. Diabetes Care. 2017;40:1436–1443. doi: 10.2337/dc16-2462. [DOI] [PubMed] [Google Scholar]
- 21.Levey A.S., Stevens L.A., Schmid C.H., et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz G.J., Munoz A., Schneider M.F., et al. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20:629–637. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smeets N.J.L., IntHout J., van der Burgh M.J.P., Schwartz G.J., Schreuder M.F., de Wildt S.N. Maturation of GFR in term-born neonates: an individual participant data meta-analysis. J Am Soc Nephrol. 2022;33:1277–1292. doi: 10.1681/ASN.2021101326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharpe D. Chi-square test is statistically significant: now what? Pract Assess Res Eval. 20:8. doi: 10.7275/tbfa-x148 [DOI]
- 25.Burnham K.P., Anderson D.R., editors. Model Selection and Multimodel Inference. Springer; New York: 2004. Advanced issues and deeper insights; pp. 267–351. [DOI] [Google Scholar]
- 26.Vasileiou G., Hoyer J., Thiel C.T., et al. Prenatal diagnosis of HNF1B-associated renal cysts: is there a need to differentiate intragenic variants from 17q12 microdeletion syndrome? Prenat Diagn. 2019;39:1136–1147. doi: 10.1002/pd.5556. [DOI] [PubMed] [Google Scholar]
- 27.Hojny J., Bartu M., Krkavcova E., et al. Identification of novel HNF1B mRNA splicing variants and their qualitative and semi-quantitative profile in selected healthy and tumour tissues. Sci Rep. 2020;10:6958. doi: 10.1038/s41598-020-63733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbacci E., Reber M., Ott M.O., Breillat C., Huetz F., Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- 29.Niborski L.L., Paces-Fessy M., Ricci P., et al. Hnf1b haploinsufficiency differentially affects developmental target genes in a new renal cysts and diabetes mouse model. Dis Model Mech. 2021;14 doi: 10.1242/dmm.047498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida K., Mushimoto Y., Tanase-Nakao K., et al. A case report with functional characterization of a HNF1B mutation (p.Leu168Pro) causing MODY5. Clin Pediatr Endocrinol. 2021;30:179–185. doi: 10.1297/cpe.30.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. An international multicenter study. (a) 14 European countries participated in the study. A total of (b) 22 centers from France and (c) 14 centers from Germany, the 2 major participating countries, were involved. Figure S2. Distribution of HNF1B variants other than the 17q12del. (a) Nature of variants and (b) resulting variants. Figure S3. Progression to ESKF of patients with HNF1B disease. (a) Progression to ESKF is significantly delayed in patients with the 17q12del compared to patients with HNF1B variants. (b) Progression to ESKF after developing CKD is not different between the 62 patients with HNF1B variants and 33 patients with the 17q12del. The survival curves were generated using data from children aged ≥2 years to dismiss changes in eGFR evolution in early life. The point in time of progression to ESKF was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. In (a), the log-rank test for difference in survival yielded a P-value < 0.0001, indicating that the patients with 17q12del and HNF1B variants differed significantly in progression toward ESKF. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate; ESKF, end-stage kidney failure. Figure S4. Impact of 1 or 2 functional kidneys in patients with HNF1B disease on progression to CKD stage 3. (a) Kidney survival of patients with the 17q12del is worse in patients with 1 functional kidney. (b) Kidney survival of patients with HNF1B variants is similar irrespective of the number of functional kidneys. The survival curves were generated using data from children aged ≥2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. In (a) The log-rank test for difference in survival yielded a P-value of 0.034, indicating that the patients with 17q12del with 1 functional kidney differed significantly in progression toward CKD stage 3. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate. Figure S5. Impact of the nature of the change at the HNF1B protein level on progression to CKD stage 3. Kidney survival of patients is not different between missense, nonsense, splicing, or frameshift HNF1B variants. The survival curves were generated using data from children aged >2 years to dismiss changes in eGFR in early life. The point in time of progression to CKD stage 3 (eGFR < 60 ml/min per 1.73 m2) was entered as the chronological age of each patient. We considered that patients entered the study (baseline) at birth given that HNF1B disease is a congenital nephropathy. CKD, chronic kidney disease, eGFR, estimated glomerular filtration rate. Table S1. Characteristics of HNF1B gene variants. Table S2. Follow-up data of patients with HNF1B disease. Table S3. AAA. (Excel). STROBE Statement.
Data Availability Statement
All raw patient inclusion and follow-up data used in the study of the can be found in Supplementary Table S3.