Abstract
Precision medicine holds great promise for improving cancer outcomes. Yet, there are large inequities in the demographics of patients from whom genomic data and models, including patient-derived xenografts (PDX), are developed and for whom treatments are optimized. In this study, we developed a genetic ancestry pipeline for the Cancer Genomics Cloud, which we used to assess the diversity of models currently available in the National Cancer Institute–supported PDX Development and Trial Centers Research Network (PDXNet). We showed that there is an under-representation of models derived from patients of non-European ancestry, consistent with other cancer model resources. We discussed these findings in the context of disparities in cancer incidence and outcomes among demographic groups in the US, as well as power analyses for biomarker discovery, to highlight the immediate need for developing models from minority populations to address cancer health equity in precision medicine. Our analyses identified key priority disparity-associated cancer types for which new models should be developed.
Significance:
Understanding whether and how tumor genetic factors drive differences in outcomes among U.S. minority groups is critical to addressing cancer health disparities. Our findings suggest that many additional models will be necessary to understand the genome-driven sources of these disparities.
Introduction
Advances in our understanding of cancer genetics have led to the proliferation of improved precision treatments (1), yet there are disparities in the groups for which these treatments are most impactful. A major contributor to these disparities is an under-representation of donors with non-European ancestry in cancer cell lines, sequence data, and patient-derived models (2). Cancer health disparities are pervasive in US ethnic/racial minority communities and are driven by complex interactions between socioeconomic factors and potentially also by somatic and germline genetic variants differing in frequency between populations (3–5). Understanding whether and how genetic variants influence cancer health disparities will require a greater investment in developing models that better represent human genetic and epigenetic variation.
Genetic ancestry, which describes the relationships between individuals and populations based on shared genetic history, can provide insights into disease risk, prognosis, and therapy response. For example, studies in Latinos, who trace their ancestry to Europeans, African slaves, and Indigenous Americans, have shown that Indigenous ancestry has an inverse relationship with breast cancer risk (6, 7). This is consistent with data suggesting a low incidence of breast cancer in Latin American countries with large Indigenous American populations (8). Interestingly, we showed that Indigenous American ancestry is associated with breast tumor ERBB2 amplification (9), while other studies have shown similar associations between Indigenous ancestry and EGFR-mutated lung tumors (10). Studies in African Americans (AA) have also shown that African ancestry is associated with a higher risk of prostate cancer (11), and that genetic variants exclusively found in Africa are associated with the prevalence of triple-negative breast tumors in AA patients (12). Despite these associations between genetic ancestry and cancer phenotypes, a major limitation in the field has been the paucity of germline and somatic data from patients from diverse populations (13). Another major limitation has been the lack of minority patient-derived models. Such patient-derived models recapitulate their genome and epigenome and are needed to advance precision health equity in such populations (2). To address these research gaps, the National Cancer Institute (NCI) has supported two centers within the patient-derived xenograft (PDX) Development and Trial Centers Research Network (PDXNet) to work with minority populations to create models reflecting their genetic background and exposures, to characterize their genome, and to facilitate studies leading to minority-focused clinical trials that address cancer health disparities.
In this manuscript, we describe the development of an ancestry estimation pipeline on the Seven Bridges Genomics (SBG) Cancer Genomics Cloud (CGC) as part of the NCI-PDXNet project. We use this new pipeline to describe the diversity of samples currently in PDXNet in terms of the genetic ancestry of patients from whom the tumors were sampled. We further present power analyses to argue that additional PDX models are critically needed to address the cancer health disparities in U.S. minority groups.
Materials and Methods
Full method details are provided in the Supplementary Material. All patients who donated biospecimens for PDX model generation in PDXnet provided written informed consent. They were recruited using research protocols adhering to the Common Rule that were approved by IRBs at their corresponding institutions. In brief, we aggregated reference data from the 1,000 Genomes Project Phase III (14), GenomeAsia 100 K (15), and INMEGEN (16), filtered by minor allele frequency, Hardy-Weinberg equilibrium, linkage disequilibrium, and relatedness among individuals. We then used a principal component analysis to identify individuals with little admixture based on the continental ancestry group clustering. The result was a dataset of 264,153 SNPs from 1,990 individuals (Supplementary Table S1), which we used to prepare the weighting factors for ancestry inference with the program SNPweights v2.1 (17). We benchmarked the genetic ancestry estimates provided by this new reference panel using ADMIXTURE analyses. Finally, we developed an ancestry estimation workflow on the Cancer Genomics Cloud, which we used to quantify the diversity of PDX models in the PDXNet resource (Supplementary Table S2). To validate our ancestry estimation method and panel, we cross-validated our approach in a set of 1,000 Genome (14) individuals not used in the reference panel generation (Supplementary Data). The SNPweights panel estimates showed small differences for minority individuals compared to 1,000 Genomes (Supplementary Data; Supplementary Tables S3–S5). The largest difference was in Europeans, where the SNPweights panel showed a mean difference of −0.071 (SD: 0.023) compared to 1,000 Genomes estimates (Supplementary Table S5). The average differences of admixed AFR, AMR, EAS, and SAS individuals ranged from 0.000 (SD: 0.001) to 0.022 (SD: 0.040).
To contextualize the distribution of PDXnet genetic diversity we used epidemiological data from NCI to identify the top cancer outcome disparities for AAs and Latin Americans in the US (18, 19). We tabulated the number of relevant models currently available for understanding those disparities.
Data availability
The ancestry estimation models were trained using three publicly available datasets: (i) 1,000 Genomes Phase III (https://www.internationalgenome.org/); (ii) INMEGEN (http://www.inmegen.gob.mx); and (iii) GenomeAsia 100 K (https://browser.genomeasia100k.org/). The datasets supporting the findings of this study are available from multiple centers and repositories. The three main repositories for these PDX datasets are: (i) Baylor College of Medicine (BCM) PDX Portal (https://pdxportal.research.bcm.edu/), (ii) NCI Patient-Derived Models Repository (PDMR, https://pdxportal.research.bcm.edu/), and (iii) Cancer Data Service (CDS, https://dataservice.datacommons.cancer.gov/). The PIs listed in these repositories can be contacted directly with data access requests. Data contributors and their respective repositories are as follows: BCM, Fred Hutchinson Cancer Research Center (PDMR), Huntsman Cancer Institute: HCI (CDS), Jackson Laboratory (PDMR), Mayo Clinic (PDMR), MD Anderson Cancer Center (PDMR), National Cancer Institute (PDMR), UC Davis , University of Alabama (PDMR), Wistar Institute (CDS), and Washington University in St. Louis: WUSTL (CDS). Some of the PDX Sequence Datasets used in the study include: (i) BCM/HCI, available from NCBI SRA, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA756268); (ii) WUSTL, available from CDS (https://dataservice.datacommons.cancer.gov/#/study/phs002305); and (iii) Wistar Institute, available from CDS (https://dataservice.datacommons.cancer.gov/#/study/phs002432). Many of the PDXNet projects are ongoing, and dataset submissions to their respective repositories are in progress, but access to these datasets can be obtained upon request. Please contact the corresponding author, PDXNet, or the specific repository to obtain access to these datasets.
Results
Genetic ancestry among PDX models in PDXNet
After filtering reference data, the first three principal components explained 44.63%, 24.71%, and 17.6% of the variation of the filtered non-admixed ancestral reference genotype matrix, respectively. Samples clustered well by continental ancestry category among these three principal components (Supplementary Fig. S1), except for the Indigenous American and South Asian ancestry categories, which showed looser clustering, potentially due to population diversity or a lack of genome-wide references, particularly for Indigenous American populations.
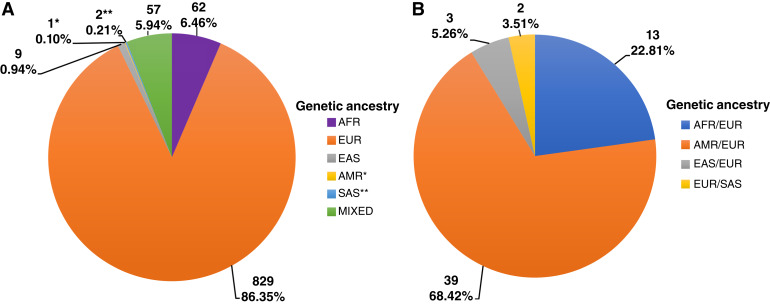
The models available in PDXNet as of September 2022 represented 960 unique patients, with 606 having self-reported race and ethnicity information. These models were developed by the six PDXnet centers and the NCI PDMR. Our genetic ancestry pipeline estimated 62 models with majority African ancestry and one model with majority Indigenous American ancestry (Fig. 1A). Thirteen models had mixed African and European ancestry and 39 models had mixed Indigenous American and European ancestry (Fig. 1B). The estimates of genetic ancestry proportions were highly concordant with self-reported race and ethnicity information (Supplementary Table S2).
Figure 1.
Diversity of genetic ancestry estimates from PDXNet models. A, Inferred genetic ancestry for 960 models across all cancer types. B, Top two categories for 57 “MIXED” samples with no category >70%. Numbers and percentages in each box reflect the models assigned to that category based on estimates from the SNPweights analysis.
Overall, most of the models in PDXNet originate from patients with a predominant European ancestry (Fig. 1). Certain cancer types, such as breast cancers (Supplementary Fig. S2A and S2B), have a greater representation of patients from non-European backgrounds due to the efforts of minority PDXNet centers at Baylor College of Medicine and University of California at Davis.
Power to detect drivers and develop new PDX models
In addition to estimating the PDX model genetic ancestry, we were interested in assessing whether their numbers were sufficiently large that we might discover alterations that were rare and potentially ethnicity- or race-specific. To do so, we carried out power analyses, which demonstrated that a study with 150 to 300 models in each of the two ancestry categories would have >80% to detect the presence of a driver mutation segregating between them (Supplementary Fig. S3). We found that for a driver mutation segregating at frequencies of 0.01 to 0.1 in a given population, sampling 50 to 100 individuals from that population should be sufficient to identify at least five patients from which PDX models could be developed (Supplementary Fig. S4). Although, to our knowledge, there are no guidelines or recommendations for the number of different models that should be used to obtain sufficiently robust preclinical data required for translation to clinical trials, our power estimation indicates that currently available models are insufficient for the study of rare and moderate frequency driver mutations.
Model race/ethnicity and cancer health disparities
After completing our genetic ancestry analyses, we wanted to assess whether the available minority models were appropriate to address existing cancer health mortality disparities in the US. Our goal was, first, to identify which cancer types resulted in disproportionally higher rates (we called these “priority cancer health disparity malignancies”) in minority groups and second, we used self-reported donor race and ethnicity data from PDXNet models to estimate the number of race/ethnicity-appropriate models for each one of the priority cancer health disparity malignancies. To identify the priority cancer health disparity malignancies for AAs and Latinos, we used SEER data to list the top 10 causes of cancer mortality for Non-Latino Whites (NLW), AAs, and Latinos (see Tables 1 and 2, ranked by age-adjusted mortality rates in NLW; refs. 18, 19). We then estimated the disparity ratio (DR, the ratio of age-adjusted mortality between NLW and minorities) for men and women. These analyses identified 11 different priority cancer health disparity malignancies, including nine cancer types in women and eight cancer types in men (Tables 1 and 2). In Table 3, we show the number of PDX models available in PDXNet derived from NLW, AA, and Latino patients (based on self-reported race and ethnicity) from the priority cancer health disparity malignancies shown in Tables 1 and 2. Our results show that of the 10 malignancies, only breast cancer has a relatively large number of race/ethnicity-appropriate models for both AAs and Latinos. Unfortunately, for the majority of cancers, the number of available models is dismal (with no models available for many cancer types), indicating that more work needs to be done to address this important cancer health disparity research gap.
Table 1.
Top 10 female age-adjusted mortality (per 100,000) and 5-year number of deaths (count) in NLW, AAs, and Latinos. Priority cancer health disparity malignancies have disparity ratio, DR, >1). Incidence-based mortality data from SEER (18 Registries, November 2019 Sub, 2000–2017)
| Non-Latino Whites | African Americans | Latinos | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Rank | Mortality rate | Count | Rank | Mortality rate | Count | DR | Rank | Mortality rate | Count | DR |
| Breast | 1 | 44.35 | 84,996 | 1 | 53.18 | 14,211 | 1.20 | 1 | 28.58 | 9,925 | 0.64 |
| Lung and bronchus | 2 | 38.73 | 71,424 | 2 | 36 | 9,726 | 0.93 | 2 | 16.83 | 5,451 | 0.43 |
| Colon and rectum | 3 | 19.92 | 39,257 | 3 | 25.82 | 6,793 | 1.30 | 3 | 14.77 | 4,932 | 0.74 |
| Corpus and uterus | 4 | 9.39 | 17,893 | 4 | 13.34 | 3,657 | 1.42 | 5 | 7.6 | 2,652 | 0.81 |
| Pancreas | 5 | 8.9 | 16,697 | 5 | 11.87 | 3,200 | 1.33 | 4 | 8.46 | 2,811 | 0.95 |
| Lymphoma | 6 | 8.34 | 16,126 | 7 | 6.63 | 1,735 | 0.79 | 6 | 7.5 | 2,426 | 0.90 |
| Ovary | 7 | 7.47 | 13,569 | 8 | 6.27 | 1,730 | 0.84 | 7 | 5.99 | 2,158 | 0.80 |
| Leukemia | 8 | 5.81 | 10,884 | 10 | 5.39 | 1,409 | 0.93 | 10 | 4.49 | 1,636 | 0.77 |
| Skina | 9 | 5.55 | 10,881 | 28 | 0.63 | 162 | 0.11 | 20 | 1.61 | 535 | 0.29 |
| Urinary bladder | 10 | 5.16 | 10,443 | 13 | 4.31 | 1,091 | 0.84 | 15 | 2.54 | 781 | 0.49 |
| Kidney and renal pelvis | 11 | 4.52 | 8,619 | 9 | 5.48 | 1,426 | 1.21 | 11 | 4.41 | 1,462 | 0.98 |
| Myeloma | 14 | 2.89 | 5,448 | 6 | 7.68 | 2,002 | 2.66 | 13 | 3.16 | 1,019 | 1.09 |
| Liver and bile duct | 15 | 2.86 | 5,265 | 14 | 4.23 | 1,217 | 1.48 | 8 | 5.97 | 2,004 | 2.09 |
| Cervix uteri | 16 | 2.73 | 4,208 | 11 | 5.18 | 1,427 | 1.90 | 12 | 3.84 | 1,497 | 1.41 |
| Stomach | 17 | 2.29 | 4,284 | 12 | 5.04 | 1,317 | 2.20 | 9 | 5.52 | 1,959 | 2.41 |
Excluding Basal and Squamous.
Table 2.
Top 10 male age-adjusted mortality rates (per 100,000) and 5-year number of deaths (count) in NLWs, AAs, and Latinos. Priority cancer health disparity malignancies have disparity ratio, DR, >1). Incidence-based mortality data from SEER (18 Registries, November 2019 Sub, 2000–2017)
| Non-Latino Whites | African Americans | Latinos | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Rank | Mortality rate | Count | Rank | Mortality rate | Count | DR | Rank | Mortality rate | Count | DR |
| Prostate | 1 | 66.63 | 92,170 | 1 | 125.6 | 19,303 | 1.89 | 1 | 51.2 | 10,477 | 0.77 |
| Lung and bronchus | 2 | 49.97 | 75,245 | 2 | 60.7 | 11,920 | 1.21 | 2 | 25.15 | 6,157 | 0.50 |
| Colon and rectum | 3 | 28.92 | 41,546 | 3 | 37.88 | 6,935 | 1.31 | 3 | 23.56 | 6,060 | 0.81 |
| Urinary bladder | 4 | 24.09 | 33,566 | 6 | 12.68 | 1,990 | 0.53 | 6 | 10.37 | 2,189 | 0.43 |
| Skina | 5 | 15.79 | 22,136 | 21 | 1.24 | 212 | 0.08 | 16 | 2.52 | 629 | 0.16 |
| Lymphoma | 6 | 14.09 | 20,030 | 8 | 10.92 | 2,181 | 0.78 | 5 | 11.55 | 3,041 | 0.82 |
| Pancreas | 7 | 11.46 | 17,479 | 5 | 13.78 | 2,840 | 1.20 | 7 | 9.6 | 2,669 | 0.84 |
| Leukemia | 8 | 10.91 | 15,398 | 11 | 8.24 | 1,529 | 0.76 | 10 | 7.13 | 2,143 | 0.65 |
| Kidney and renal pelvis | 9 | 10.39 | 15,294 | 7 | 12.31 | 2,321 | 1.18 | 8 | 9.53 | 2,566 | 0.92 |
| Liver and bile duct | 10 | 8.35 | 13,623 | 4 | 13.88 | 3,375 | 1.66 | 4 | 15.79 | 4,955 | 1.89 |
| Stomach | 13 | 5.61 | 8,325 | 10 | 10.29 | 1,956 | 1.83 | 9 | 9.42 | 2,624 | 1.68 |
| Myeloma | 14 | 5.14 | 7,496 | 9 | 10.54 | 1,903 | 2.05 | 11 | 4.96 | 1,239 | 0.96 |
aExcluding Basaling and Squamous.
Table 3.
Number of available models for priority cancer health disparity malignancies
| Average annual deaths (2013–2017)/available sex-matched models | ||||
|---|---|---|---|---|
| Cancer type | Disproportionally affected group | Non-Latino Whites | African Americans | Latinos |
| Lung and bronchus | African American, men | 29,334/44 | 4,329/2 | 2,322/0 |
| Prostate | African American, men | 18,434/7 | 3,861/0 | 2,095/0 |
| Breast | African Americans, women | 16,999/25 | 2,842/13 | 1,985/22 |
| Colon and rectum | African Americans, women and men | 16,161/109 | 2,746/4 | 2,198/3 |
| Pancreas | African Americans, women and men | 6,835/27 | 1,208/4 | 1,096/0 |
| Liver and Intrahepatic bile duct | African Americans and Latinos, women and men | 3,778/3 | 918/0 | 1,392/1 |
| Uterus | African Americans, women | 3,579/37 | 731/3 | 530/2 |
| Kidney and renal pelvis | Latinos, men | 3,059/22 | 464/1 | 513/0 |
| Multiple myeloma | African Americans, women and men | 2,589/0 | 781/0 | 1,096/0 |
| Stomach | African Americans and Latinos, women and men | 2,522/1 | 655/1 | 917/1 |
| Cervix | African Americans and Latinos, women | 842/5 | 285/0 | 299/1 |
Discussion
In this study, we developed a cloud-based genetic ancestry pipeline, which we used to assess the genetic ancestry diversity of PDXnet models. We found that our genetic ancestry estimates had a high correspondence with self-reported race and ethnicity and thus are useful for comparing the model diversity to cancer health disparities data. Patients with non-European genetic ancestry are highly under-represented in the PDXNet models, which reflects similar disparities in other cancer resources (2). Recent efforts of the PDXNet have yielded an increase in 15 models of breast cancers likely derived from patients with African genetic ancestry and 23 models likely derived from Latin American genetic ancestry, representing a 3-fold and 23-fold increase in models above those previously available, respectively. Furthermore, with the addition of two minority and disparity-focused centers in PDXNet in late 2018, several models from minority patients will soon be deposited in the PDMR. Yet, as our power analyses showed, there are still far fewer models than would be required to conduct a study with sufficient power to identify models with genetic variants with biologically relevant effects on the cancer health disparities between demographic groups. This highlights the critical need for further investment in model development to help reach health equity goals.
Health disparities are complex and involve the interaction of many factors, including structural inequities, social determinants of health, cultural factors, and variance in exposure to environmental harms (3–5). Our focus on genetics in this study is motivated by our belief that precision medicine holds great promise for improving patient outcomes and that this promise should be realized equitably. Cancer evolution leverages the germline and somatic background of patients in which tumorigenesis occurs and the progression of cancer depends on the interactions between somatic mutations and the normal tissue in the microenvironment on which it grows (20). It is therefore important to understand the diversity of genetic backgrounds in which cancers evolve to identify relevant biomarkers that could help to address treatment disparities. We want to emphasize that human genetic diversity is complex, there is large variance within human populations and we do not seek to naively assign risk factors for cancer incidence to broad, biologically dubious categories (21–23). Rather, we hope this work will help to motivate and facilitate an understanding of whether precision medicine approaches have the potential to help address current cancer health disparities by increasing the number and diversity of models available to identify whether and how segregating germline and somatic variants can help explain those disparities.
Categorization of models based on continental ancestry has several limitations. The concept of continental ancestry is premised on the concept of continental races, the biological and biomedical relevance of which is debated and controversial (21, 24). The history of human migration and gene flow is complex and cannot be circumscribed by continental borders (22, 23). Additionally, analyses based on the categorization of individuals by self-reported race or ancestry likely elide the complex interactions of demography, the environment, and socioeconomic factors (3, 4, 23). Our goal in this study was to help motivate the critical need for cancer models that better reflect the diversity of human genetic variation in order to help achieve equity in precision medicine. In this way, we seek to better understand the extent to which genetic variants that segregate among demographic groups impact cancer health disparities.
In a recent commentary, we argued for the need to diversify patient-derived models and the implications of the limited diversity of such models to advance precision medicine in minority populations (2). In Table 3, we showed that there remain large gaps for models available for model development from cancers with high burden in minorities; we referred to these cancers above as “priority cancer health disparity malignancies.” For stomach tumors, a malignancy that disproportionally affects both minority groups, only one appropriate PDX exists for AA and another one for Latinos. For liver cancer, another malignancy with a high burden in both groups, there are no race/ethnicity-appropriate models for AA and only one for Latinos in PDXNet. Kidney tumors also lack race/ethnic-appropriate models from Latinos. The number of models for malignancies with high burden in AA is also dismal, with no race/ethnicity-appropriate models for multiple myeloma or prostate cancer, four for pancreatic cancer, three for endometrial cancer, four for colorectal cancer, and two for lung cancer (Table 3). Only breast cancer is moderately represented for AAs, with 19 models from this minority group currently available in PDXNet. While rare cancers in NLW, such as multiple myeloma and stomach, liver, and kidney tumors, also have low numbers for this majority racial group, the disparities for common NLW cancers are striking. For example, there is only one colorectal cancer PDX from Latinos for every ∼36 NLW models. For each AA colorectal cancer PDX, there are 27 NLW models. The number of pancreas and lung cancer PDXs are also 7-fold and 22-fold higher for NLW than for AA, respectively. These analyses, therefore, not only identify disparities in existing models but also highlight that developing more diverse preclinical models is needed to address cancer mortality disparities through race/ethnicity-appropriate preclinical and translational research. Given the disproportionally high burden caused by liver and stomach tumors in AA and Latinos, by renal tumors in Latinos, by colorectal, pancreas, lung, prostate, breast, and uterine tumors, and multiple myeloma in AA, we suggest that they should represent priorities for model development in PDXNet and similar initiatives.
While acknowledging that PDX development requires specialized infrastructure and is time- and resource-intensive, we believe that a number of approaches can be taken to increase the number and diversity of models in the future. One approach is to support such efforts in cancer centers in historically under-served communities. In PDXNet, for example, the two centers contributing the most diverse models included the Baylor College of Medicine Cancer Center, which serves the largest safety net hospital in Houston, and the UC Davis Cancer Center, which recruits patients throughout the University of California System Comprehensive Cancer Center, which accept patients with MediCal (California’s Medicaid program) insurance, many of whom are ethnic/racial minorities. Another related approach is to increase model diversity to support such efforts in racially/ethnically diverse states. It is unsurprising that the centers contributing the largest number of models from ethnic/racial minorities were in California and Texas, two of the most diverse states in the nation. A third approach is to support collaborations and researchers based in cancer hospitals and research institutions in Latin America and Africa. Having the ability to make models from African and Latin American patients should also be encouraged as many of these individuals share genetic ancestry with U.S. minority populations.
In conclusion, we developed and implemented a pipeline for genetic ancestry evaluations in publicly available patient-derived models and highlight the fact that most of them have a predominantly European genetic ancestry. We estimated that to understand biological differences and responses to therapy between different genetic ancestries, hundreds of models are needed, highlighting the need to diversify the models. Furthermore, using self-reported race/ethnicity information available in ∼63% of the models and cancer health disparities data in the two largest U.S. minority populations, we showed that there very few or, in many cases, no models to develop therapies for the cancer types with the highest burden in minority patients. While ongoing efforts promise to diversify these available models, we encourage funders to support additional efforts aimed at developing and characterizing new models that equitable help realize the promise of cancer precision medicine to all Americans.
Supplementary Material
Figure S1 shows the Clustering of 1,990 non-admixed reference samples and 2,387 admixed samples(brown) individuals based on the first three principal components. Colors represent the continental ances
Figure S2 shows Diversity of genetic ancestry estimates from PDXNet breast cancer models. A: Inferred genetic ancestry for 115 breast cancer models in PDXnet. B: Top two categories for 26 "MIXED" samp
Figure S3 Shows the Power to detect a driver mutation that is absent in EUR but present in a non-EUR category at varying low frequencies.
Figure S4 Shows the Power to identify at least 5 patients with a known driver mutation that is present in populations at varying low frequencies.
The full list of collaborators for PDXNet Consortium and Supplementary Data and Methods
Populations of origin for unrelated individuals from the 1000 Genomes Phase III, GenomeAsia, and INMEGEN data used as reference populations to design a new SNPweights panel.
PDXnet metadata for samples for which genetic ancestry was estimated and samples were assigned into African (AFR), European (EUR), East Asian (EAS), American (AMR), and South Asian (SAS) continental ancestry.
1000Genomes K=10 Admixture Estimates downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/admixture_files/ALL.wgs.phase3_shapeit2_filtered.20141217.maf0.05.10.Q. Super-population designations were added to the partitions by sorting each partition and determining the corresponding designation for each partition.
Ancestral estimates 1000Genomes individuals (n=929) not used in reference panel generation. Estimates calculated from 1000Genomes K=10 admixture data, SNPWeights Panel, and the differences between these estimates for each individual.
Mean and Stdev of differences for 5 continental ancestral estimations between SNPWeights Panel and 1000Genomes K=10 Admixture Estimates by individual's super-population designation.
Acknowledgments
This work was primarily funded by grants from the Cancer Moonshots program (NCI U24CA224067, NCI U54CA224083, NCI U54CA224070, NCI U54CA224065, NCI U54CA224076, NCI U54CA233223, NCI U54CA233306). L.G. Carvajal-Carmona is grateful for funding received from The Auburn Community Cancer Endowed Chair in Basic Research, The Heart, BrEast, and BrAin HeaLth Equity Research (HEAL HER) program, a program made possible by residual class settlement funds in the matter of April Krueger v. Wyeth, Inc., Case No. 03-cv-2496 (US District Court, SD of Calif.), and from the National Cancer Institute (R01CA223978, R21CA199631, U54CA283766, P30CA093373 and U54CA280811) of the National Institutes of Health. J.E. Quino and A.P. Vang received diversity supplement support from the National Cancer Institute (R01CA223978-2S2 and R01CA223978-05S2). The full list of the collaborators for PDXNet Consortium can be found in the Supplementary Data. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
M.W. Lloyd reports grants from National Cancer Institute (NCI; HHS—NIH) during the conduct of the study; grants from National Cancer Institute (NCI; HHS—NIH) outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
P.C. Lott: Software, formal analysis. K. Chiu: Resources, data curation, software, investigation, writing-review and editing. J.E. Quino: Data curation, writing-review and editing. A.P. Vang: Data curation, software, formal analysis, writing-review and editing. M.W. Lloyd: Data curation, software. A. Srivastava: Conceptualization, data curation, software, supervision, methodology, writing-review and editing. J.H. Chuang: Conceptualization, resources, supervision, funding acquisition, writing-review and editing. L.G. Carvajal-Carmona: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing-original draft, writing-review and editing.
References
- 1. Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 2014;4:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Halmai NB, Carvajal-Carmona LG. Diversifying preclinical research tools: expanding patient-derived models to address cancer health disparities. Trends Cancer 2022;8:291–4. [DOI] [PubMed] [Google Scholar]
- 3. Adler N, Bush NR, Pantell MS. Rigor, vigor, and the study of health disparities. Proc Natl Acad Sci U S A 2012;109 Suppl 2(Suppl 2):17154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams DR, Mohammed SA, Leavell J, Collins C. Race, socioeconomic status, and health: complexities, ongoing challenges, and research opportunities. Ann N Y Acad Sci 2010;1186:69–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zavala VA, Bracci PM, Carethers JM, Carvajal-Carmona L, Coggins NB, Cruz-Correa MR, et al. Cancer health disparities in racial/ethnic minorities in the United States. Br J Cancer 2021;124:315–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fejerman L, John EM, Huntsman S, Beckman K, Choudhry S, Perez-Stable E, et al. Genetic ancestry and risk of breast cancer among US latinas. Cancer Res 2008;68:9723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shieh Y, Fejerman L, Lott PC, Marker K, Sawyer SD, Hu D, et al. A polygenic risk score for breast cancer in US latinas and Latin American women. J Natl Cancer Inst 2020;112:590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 9. Marker KM, Zavala VA, Vidaurre T, Lott PC, Vásquez JN, Casavilca-Zambrano S, et al. Human epidermal growth factor receptor 2-positive breast cancer is associated with indigenous American ancestry in Latin American women. Cancer Res 2020;80:1893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carrot-Zhang J, Soca-Chafre G, Patterson N, Thorner AR, Nag A, Watson J, et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed Latin American populations. Cancer Discov 2021;11:591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, Chou A, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 2021;53:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martini R, Chen Y, Jenkins BD, Elhussin IA, Cheng E, Hoda SA, et al. Investigation of triple-negative breast cancer risk alleles in an International African-enriched cohort. Sci Rep 2021;11:9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spratt DE, Chan T, Waldron L, Speers C, Feng FY, Ogunwobi OO, et al. Racial/ethnic disparities in genomic sequencing. JAMA Oncol 2016;2:1070–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. GenomeAsia100K Consortium . The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 2019;576:106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreno-Estrada A, Gignoux CR, Fernández-López JC, Zakharia F, Sikora M, Contreras AV, et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 2014;344:1280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen C-Y, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics 2013;29:1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller KD, Ortiz AP, Pinheiro PS, Bandi P, Minihan A, Fuchs HE, et al. Cancer statistics for the US Hispanic/Latino population, 2021. CA Cancer J Clin 2021;71:466–87. [DOI] [PubMed] [Google Scholar]
- 19. Giaquinto AN, Miller KD, Tossas KY, Winn RA, Jemal A, Siegel RL. Cancer statistics for African American/black people 2022. CA Cancer J Clin 2022;72:202–29. [DOI] [PubMed] [Google Scholar]
- 20. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res 2019;79:4557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keita SOY, Kittles RA, Royal CDM, Bonney GE, Furbert-Harris P, Dunston GM, et al. Conceptualizing human variation. Nat Genet 2004;36(11 Suppl):S17–20. [DOI] [PubMed] [Google Scholar]
- 22. Weiss KM, Long JC. Non-darwinian estimation: my ancestors, my genes’ ancestors. Genome Res 2009;19:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cerdeña JP, Grubbs V, Non AL. Genomic supremacy: the harm of conflating genetic ancestry and race. Hum Genomics 2022;16:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maglo KN, Mersha TB, Martin LJ. Population genomics and the statistical values of race: an interdisciplinary perspective on the biological classification of human populations and implications for clinical genetic epidemiological research. Front Genet 2016;7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 shows the Clustering of 1,990 non-admixed reference samples and 2,387 admixed samples(brown) individuals based on the first three principal components. Colors represent the continental ances
Figure S2 shows Diversity of genetic ancestry estimates from PDXNet breast cancer models. A: Inferred genetic ancestry for 115 breast cancer models in PDXnet. B: Top two categories for 26 "MIXED" samp
Figure S3 Shows the Power to detect a driver mutation that is absent in EUR but present in a non-EUR category at varying low frequencies.
Figure S4 Shows the Power to identify at least 5 patients with a known driver mutation that is present in populations at varying low frequencies.
The full list of collaborators for PDXNet Consortium and Supplementary Data and Methods
Populations of origin for unrelated individuals from the 1000 Genomes Phase III, GenomeAsia, and INMEGEN data used as reference populations to design a new SNPweights panel.
PDXnet metadata for samples for which genetic ancestry was estimated and samples were assigned into African (AFR), European (EUR), East Asian (EAS), American (AMR), and South Asian (SAS) continental ancestry.
1000Genomes K=10 Admixture Estimates downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/admixture_files/ALL.wgs.phase3_shapeit2_filtered.20141217.maf0.05.10.Q. Super-population designations were added to the partitions by sorting each partition and determining the corresponding designation for each partition.
Ancestral estimates 1000Genomes individuals (n=929) not used in reference panel generation. Estimates calculated from 1000Genomes K=10 admixture data, SNPWeights Panel, and the differences between these estimates for each individual.
Mean and Stdev of differences for 5 continental ancestral estimations between SNPWeights Panel and 1000Genomes K=10 Admixture Estimates by individual's super-population designation.
Data Availability Statement
The ancestry estimation models were trained using three publicly available datasets: (i) 1,000 Genomes Phase III (https://www.internationalgenome.org/); (ii) INMEGEN (http://www.inmegen.gob.mx); and (iii) GenomeAsia 100 K (https://browser.genomeasia100k.org/). The datasets supporting the findings of this study are available from multiple centers and repositories. The three main repositories for these PDX datasets are: (i) Baylor College of Medicine (BCM) PDX Portal (https://pdxportal.research.bcm.edu/), (ii) NCI Patient-Derived Models Repository (PDMR, https://pdxportal.research.bcm.edu/), and (iii) Cancer Data Service (CDS, https://dataservice.datacommons.cancer.gov/). The PIs listed in these repositories can be contacted directly with data access requests. Data contributors and their respective repositories are as follows: BCM, Fred Hutchinson Cancer Research Center (PDMR), Huntsman Cancer Institute: HCI (CDS), Jackson Laboratory (PDMR), Mayo Clinic (PDMR), MD Anderson Cancer Center (PDMR), National Cancer Institute (PDMR), UC Davis , University of Alabama (PDMR), Wistar Institute (CDS), and Washington University in St. Louis: WUSTL (CDS). Some of the PDX Sequence Datasets used in the study include: (i) BCM/HCI, available from NCBI SRA, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA756268); (ii) WUSTL, available from CDS (https://dataservice.datacommons.cancer.gov/#/study/phs002305); and (iii) Wistar Institute, available from CDS (https://dataservice.datacommons.cancer.gov/#/study/phs002432). Many of the PDXNet projects are ongoing, and dataset submissions to their respective repositories are in progress, but access to these datasets can be obtained upon request. Please contact the corresponding author, PDXNet, or the specific repository to obtain access to these datasets.