To the Editor:
Patients with relapsed/refractory high-risk neuroblastoma (HRNBL) have poor prognoses. Recent studies found that BARD1 harbored the most significantly enriched pathogenic/likely-pathogenic germline mutations in neuroblastoma patients1. Such mutations, when engineered to neuroblastoma cell-lines, can cause homologous recombination repair (HRR) deficiency, conferring sensitivity to PARP inhibitors (PARPi)2.
We report the first response of a child with neuroblastoma and a BARD1 germline mutation to a PARPi. The patient was diagnosed at 22 months with metastatic HRNBL. After induction therapy (chemotherapy, surgery), she received salvage therapy (cyclophosphamide/topotecan, irinotecan/temozolomide) for persistent bone marrow (BM) disease, followed by autologous stem cell transplantation, radiation, and isotretinoin. At the end of therapy, her disease progressed with 30% BM involvement.
The patient transferred to our institution and achieved stable disease (by INRC criteria)3 following 2 cycles of irinotecan, temozolomide, and dinutuximab (anti-GD2 antibody) and 2 cycles of cyclophosphamide, topotecan, and dinutuximab. Paired tumor-normal whole-exome analysis identified a pathogenic germline heterozygous frameshift in BARD1, but no other known driver variants (Fig. S1 and S2). Based on this finding and supporting pre-clinical data1,2, treatment was changed to talazoparib (PARPi) plus irinotecan (as per BMNIRN4). The patient had a complete response (INRC criteria) in the BM compartment (Cycle 2) and continued therapy. Irinotecan was decreased (Cycle 5) then eliminated (Cycle 6) due to thrombocytopenia. BM remained negative for tumor. Focal radiation therapy was administered, concurrently with talazoparib, to 3 minimally avid bone lesions (FDG-PET). Single agent talazoparib was discontinued following Cycle 26. The patient is 32 months off therapy with no clinical evidence of disease (treatment course: Fig. 1a).
FIGURE 1. Treatment history and molecular features of a high-risk neuroblastoma patient with sustained response to PARP inhibitor (talazoparib).
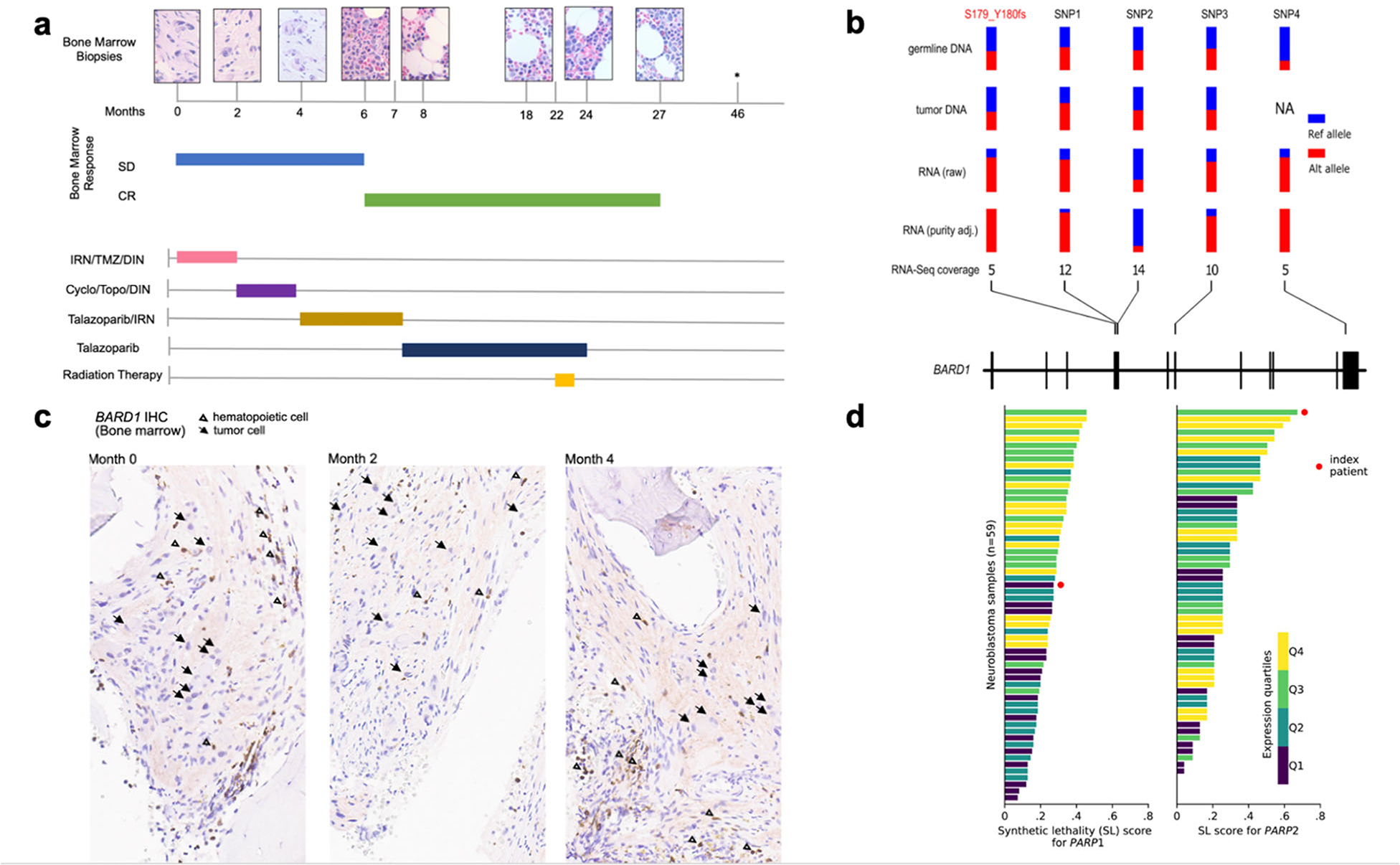
a) Patient’s (patient ID SJNBL031647) bone marrow response over time in months, depicted by bone marrow biopsy histology (top row) and response per Revised INRC Criteria3 (middle rows), in correlation with treatments received over time (bottom rows), demonstrated by colored horizontal bars. Month 0 indicates the start of the patient’s treatment for progressive neuroblastoma at our institution. The patient received PARPi (talazoparib 400 mcg/m2 IV, days 1–7) between months 4 and 24 as indicated by gold and dark blue bars; *: asymptomatic, clinically well with negative FDG-PET scan; bone marrow not assessed at this time point based on clinical judgment. Abbreviations: SD: stable disease, CR: complete response, IRN: irinotecan (initially given at 40 mg/m2 IV, days 1–5; reduced to 20 mg/m2 following cycle 5 and eliminated following cycle 6), TMZ: temozolomide, DIN: dinutuximab, Cyclo: cyclophosphamide, Topo: topotecan.
b) Allele-specific expression of BARD1 in tumor RNA-seq of the index patient (SJNBL031647). In addition to the S179_Y180fs frameshift (highlighted in red), four heterozygous SNPs (labeled SNP1‒4) identified from the germline exome were used for this analysis; each site had ≥5X coverage in RNA-seq. Their variant allele fraction (VAF) values in germline DNA, tumor DNA, tumor RNA-seq and purity-adjusted tumor RNA-seq are shown in parallel, indicating allele specific expression (ASE) in tumor RNA (p-value = 0.015 based on simulation analysis, see Supplementary Appendix.). c) BARD1 protein expression in index patient’s tumor cells at months 0, 2 and 4. Immunohistochemical staining (IHC) was performed against BARD1 on the bone marrow biopsies. Arrows indicate BARD1-negative tumor cells with definite ganglion cell differentiation. Open triangles indicate BARD1-positive background hematopoietic cells. Additional BARD1-negative cells correspond to tumor Schwann cells. d) Synthetic lethality (SL) score was estimated by a pediatric cancer-based gene network model for PARP1 and PARP2 inhibition (Supplementary Appendix). The index patient (red circle) is compared with 58 other neuroblastoma patient samples. Bars are color-coded corresponding to the quartiles of expression, measured by transcript per million (TPM), for each gene.
The patient’s response to talazoparib is likely attributed to bi-allelic loss of BARD1 in the tumor, based on mono-allelic expression (p = 0.015) of the frameshift mutation in RNA-seq5 (Fig. 1b). Immunohistochemical staining of serial BM samples, obtained before talazoparib administration, confirmed somatic BARD1 protein loss (Fig. 1c and S3). When analyzed by a synthetic lethality (SL) network model trained on pediatric cancer data, the patient’s tumor had the highest SL score for PARP2 among 59 profiled neuroblastomas, indicating that the expected SL response might have been primarily mediated by PARP2 inhibition, given its high PARP2 and low PARP1 expression rank (Fig. 1d).
The sustained clinical response in our patient demonstrates the potential for exploiting HRR deficiencies in pediatric cancer patients, supporting further evaluation in an expansion cohort of recurrent/refractory solid tumors with germline or somatic alterations in HRR genes in a clinical trial (NCT04901702).
Supplementary Material
Acknowledgments
This work was supported in part by National Cancer Institute (NCI) grant R01CA216391 to JZ and a Cancer Center Support (CORE) Grant (P30 CA21765) to St. Jude Children’s Research Hospital. All authors received support from American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Contributor Information
Margaret Cupit-Link, St. Jude Children’s Research Hospital, Memphis, TN.
Kohei Hagiwara, St. Jude Children’s Research Hospital, Memphis, TN.
Matthew Nagy, National Cancer Institute, Bethesda, MD.
Selene C. Koo, St. Jude Children’s Research Hospital, Memphis, TN.
Brent A. Orr, St. Jude Children’s Research Hospital, Memphis, TN
Eytan Ruppin, St. Jude Children’s Research Hospital, Memphis, TN
John Easton, St. Jude Children’s Research Hospital, Memphis, TN
Jinghui Zhang, St. Jude Children’s Research Hospital, Memphis, TN.
Sara M. Federico, St. Jude Children’s Research Hospital, Memphis, TN.
REFERENCES
- 1.Kim J, Vaksman Z, Egolf LE, et al. Germline pathogenic variants in neuroblastoma patients are enriched in BARD1 and predict worse survival. J Natl Cancer Inst. 2024;116:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randall MP, Egolf LE, Vaksman Z, et al. BARD1 germline variants induce haploinsufficiency and DNA repair defects in neuroblastoma. J Natl Cancer Inst. 2024;116:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JR, Bagatell R, Cohn SL, et al. Revisions to the International Neuroblastoma Response Criteria: A Consensus Statement From the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol 2017;35:2580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Federico SM, Pappo AS, Sahr N, et al. A phase I trial of talazoparib and irinotecan with and without temozolomide in children and young adults with recurrent or refractory solid malignancies. European Journal of Cancer 2020;137:204–13. [DOI] [PubMed] [Google Scholar]
- 5.Hagiwara K, Edmonson MN, Wheeler DA, Zhang J. indelPost: harmonizing ambiguities in simple and complex indel alignments. Bioinformatics 2022;38:549–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


