Abstract
Metastatic cancer presents with many, sometimes hundreds of metastatic lesions through the body, which often respond heterogeneously to treatment. Therefore, lesion-level assessment is necessary for a complete understanding of disease response. Lesion-level assessment typically requires manual matching of corresponding lesions, which is a tedious, subjective, and error-prone task. This study introduces a fully automated algorithm for matching of metastatic lesions in longitudinal medical images. The algorithm entails four steps: (1) image registration, (2) lesion dilation, (3) lesion clustering, and (4) linear assignment. In step (1), 3D deformable registration is used to register the scans. In step (2), lesion contours are conformally dilated. In step (3), lesion clustering is evaluated based on local metrics. In step (4), matching is assigned based on non-greedy cost minimization. The algorithm was optimized (e.g., choice of deformable registration algorithm, dilatation size) and validated on 140 scan-pairs of 32 metastatic cancer patients from two independent clinical trials, who received longitudinal PET/CT scans as part of their treatment response assessment. Registration error was evaluated using landmark distance. A sensitivity study was performed to evaluate the optimal lesion dilation magnitude. Lesion matching performance accuracy was evaluated for all patients and for a subset with high disease burden. Two investigated deformable registration approaches (whole body deformable (WDR) and articulated deformable (ADR) registrations) led to similar performance with the overall registration accuracy between 2.3 and 2.6 mm. The optimal dilation magnitude of 25 mm yielded almost a perfect matching accuracy of 0.98. No significant matching accuracy decrease was observed in the subset of patients with high lesion disease burden. In summary, lesion matching using our new algorithm was highly accurate and a significant improvement, when compared to previously established methods. The proposed method enables accurate automated metastatic lesion matching in whole-body longitudinal scans.
1. Introduction
Metastasis is the leading cause of cancer-related mortality (Chaffer and Weinberg, 2011). Although cancer patients have treatment options, the treatment response among lesions can be substantially different. This heterogeneous response poses a great challenge to cancer management in metastatic patients because the behaviour of one lesion (or even a few) can be unrepresentative of the global response to treatment. If response can be assessed at the lesion-level, inter-lesion heterogeneity can be quantified, providing valuable information for improved cancer management via early prediction of outcomes (Gayed et al., 2004; Schwarz et al., 2005) or via identification and subsequent selective targeting of resistant lesions (Lubner et al., 2013; Roth et al., 2020).
Molecular imaging is an essential tool for detecting metastatic lesions and monitoring their progression and therapeutic response (Baum et al., 2010; Scarpelli et al., 2018; Vanderhoek et al., 2013; Weissleder, 2006). Imaging can capture the heterogeneity in response because it is able to spatially resolve different lesion sites and to capture different quantitative and qualitative characteristics of each disease site. However, late-stage cancer patients can present with tens of metastatic lesions spread throughout their whole-body. Therefore, even though imaging can resolve different lesions, lesion-level disease analysis is impractical in daily clinical practice because it is overwhelming to manually assess individual lesion response. Therefore, response assessment employed by physicians often reduces the rich heterogeneous lesion-level image information to patient-level information either by summarizing all the lesions into a single score (Gallamini et al., 2014; Larson et al., 1999) or by evaluating a subset of lesions (Eisenhauer et al., 2009; Wahl et al., 2009) (e.g. RECIST or PERCIST). One way to overcome this information reduction is to achieve automatic assessment of individual lesion response via lesion matching.
Matching individual lesions across multiple time points has been investigated via rigid registration for identification of new cancerous lesions (Moltz et al., 2009) and for treatment response assessment of multiple sclerosis brain lesions (Bosc et al., 2003; Köhler et al., 2019; Shahar and Greenspan, 2005). Going beyond rigid registration, Yip et al introduced a hierarchical articulated registration that enabled treatment response assessment of late-stage prostate cancer bone lesions (Yip et al., 2014; Yip and Jeraj, 2014). Finally, two dimensional (2D) deformable registration was previously explored as a means of refining the rigid registration-based lesion matching of lymphoma lesions (Xu et al., 2011).
The aforementioned studies demonstrated that rigid, articulated, and deformable image registration enable lesion matching in specific organs (e.g. skeleton, brain, liver). However, a comprehensive algorithm that enables matching of metastatic lesions spread across the entire patient anatomy remains unavailable. Rigid registration applicability is limited to certain body-parts (e.g. brain) and fails to account for skeletal movement and soft-tissue deformations. Deformable registration showed promising results to aid in lesion matching, but the 2D nature of its previous application fell short on exploring the full potential of the deformations.
In this work we introduce a novel algorithm for lesion matching of metastatic lesions spread through the whole-body that is based on several distinctive steps, including 3D deformable registration, lesion dilation, clustering, and linear assignment of lesions. The lesion matching algorithm was tested in a population of cancer patients that received longitudinal PET/CT scans as part of their treatment response assessment.
2. Algorithm Description
We pose the lesion matching as a linear assignment problem. Following image registration (Step 1), the matching algorithm includes lesion dilation (Step 2), clustering (Step 3), and linear assignment (Step 4). A schematic description of the algorithm’s structure is presented in Figure 1.
Figure 1 -.
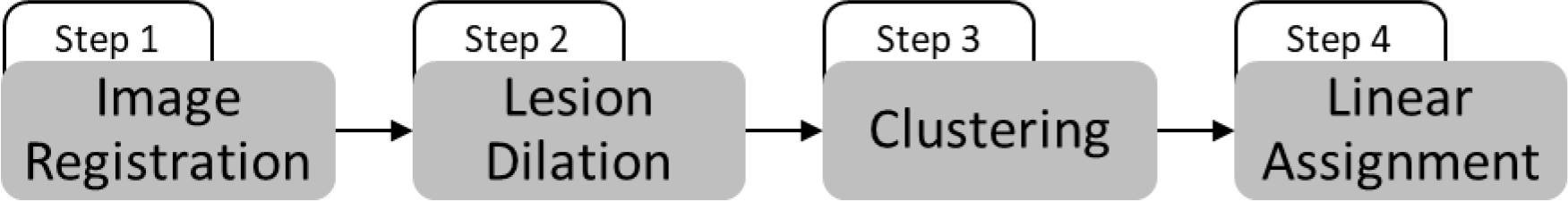
schematic description of the lesion matching algorithm
The algorithm acts on a scan-pair of anatomical medical images, each one having an associated binary lesion mask. The goal of the algorithm is to automatically perform lesion matching decisions based on lesion superposition. A matching decision consists of identifying if a lesion present in one scan has an associated lesion (homologous lesion) in the paired scan; if so, the longitudinal pair of lesions is called single corresponding lesion (CL). Lesions that have no corresponding lesion in the paired scan are either new lesions (NL) or disappearing lesion (DL). In each scan-pair, one matching decision had to be made for each unique lesion and the number of unique lesions per scan-pair was defined as the sum of CLs, NLs, and DLs. The concepts of scan-pairs, unique lesions, matching decisions, corresponding lesions, new lesions, and disappearing lesions are illustrated in Figure 2.
Figure 2 –
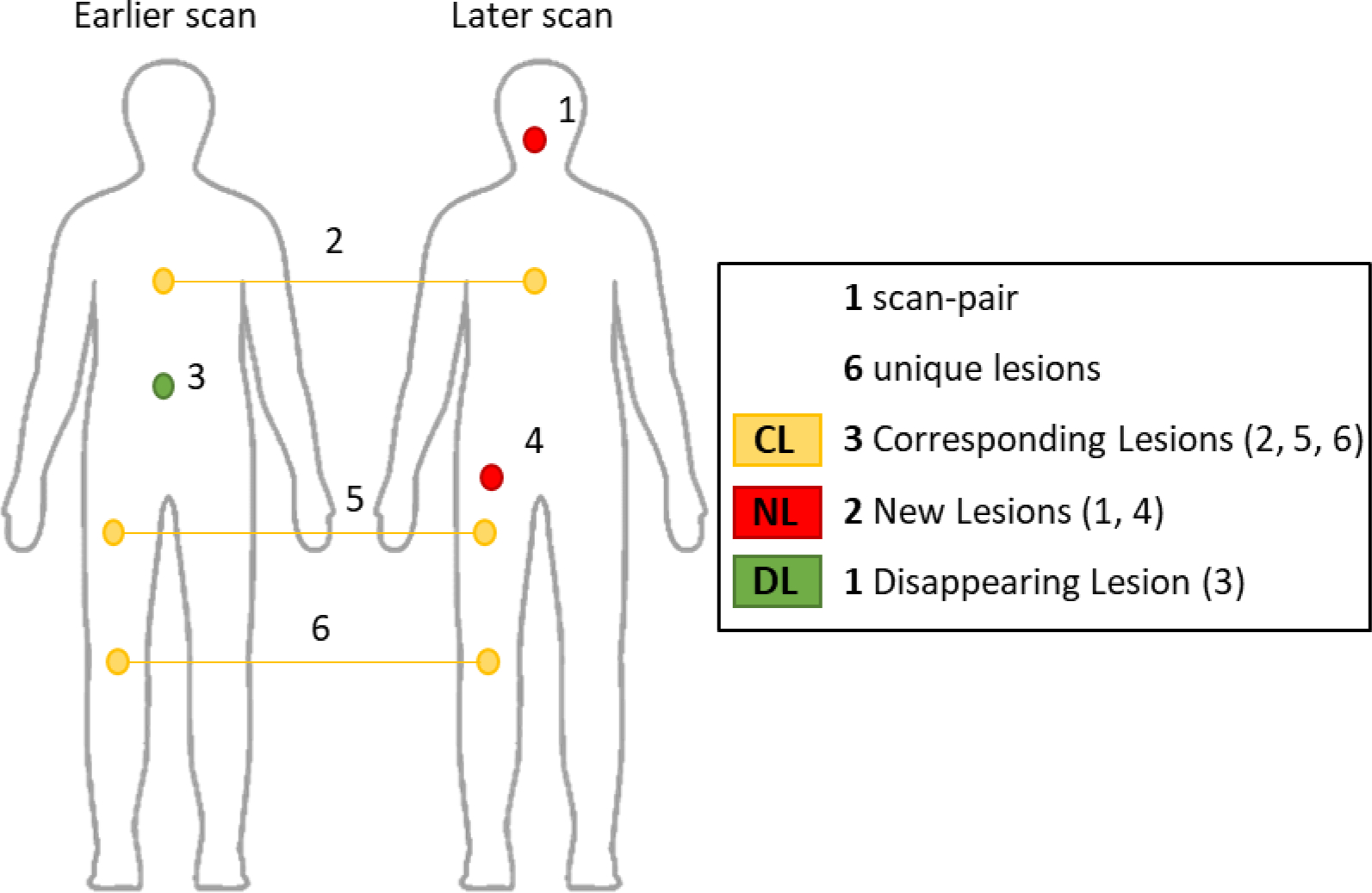
Illustration of types of unique lesions per scan-pair: corresponding lesions (CL), new lesions (NL), and disappearing lesions (DL).
In the first step, 3D deformable image registration is used to superimpose anatomically homologous regions of a scan-pair. The deformation vector field is estimated by minimization of the sum of square differences between voxel intensities in the moving and in the fixed anatomical medical images, respectively the later and the earlier scans. Deformable registration is done using free form deformation with 3rd order B-splines interpolation and hierarchical control grid (Rueckert et al., 1999). This technique was chosen for its wide acceptance in the medical image registration field, its local support, and for requiring relatively simple and fast calculations (Sotiras et al., 2014). The hierarchical control grid approach is also well established and is known to help in preserving the smoothness of the registration (Lee et al., 1996; Mattes et al., 2003; Rueckert et al., 1999).
The calculated transformation field is then applied to the binary lesion mask of the later scan in the scan-pair.
| (1) |
where is the set of coordinates of the lesions in the binary mask of the later scan and is this set of coordinates after transformation.
In the second step, a morphologic conformal lesion dilation operation is applied to the binary lesion masks (on both time points in the scan-pair) to increase the lesion superposition probabilities (Figure 3).
| (2) |
| (3) |
where is the set of coordinates of the lesions in the binary mask of the earlier scan, is the conformal dilation of the set , such that , and is the conformal dilation of the set , such that . Subsequently, the individual lesion components are numbered based on connected components analysis, the superposition of the lesions is evaluated and a matrix of intersections is automatically constructed as
| (4) |
where is the subset of the coordinates of the jth lesion in the later scan, is the subset of the coordinates of the ith lesion in the earlier scan, and is the cardinality of the intersection of these subsets. The matrix is a square matrix with elements where is the maximal integer between and , which are the number of lesions identified in the earlier and in the later scans.
Figure 3 –
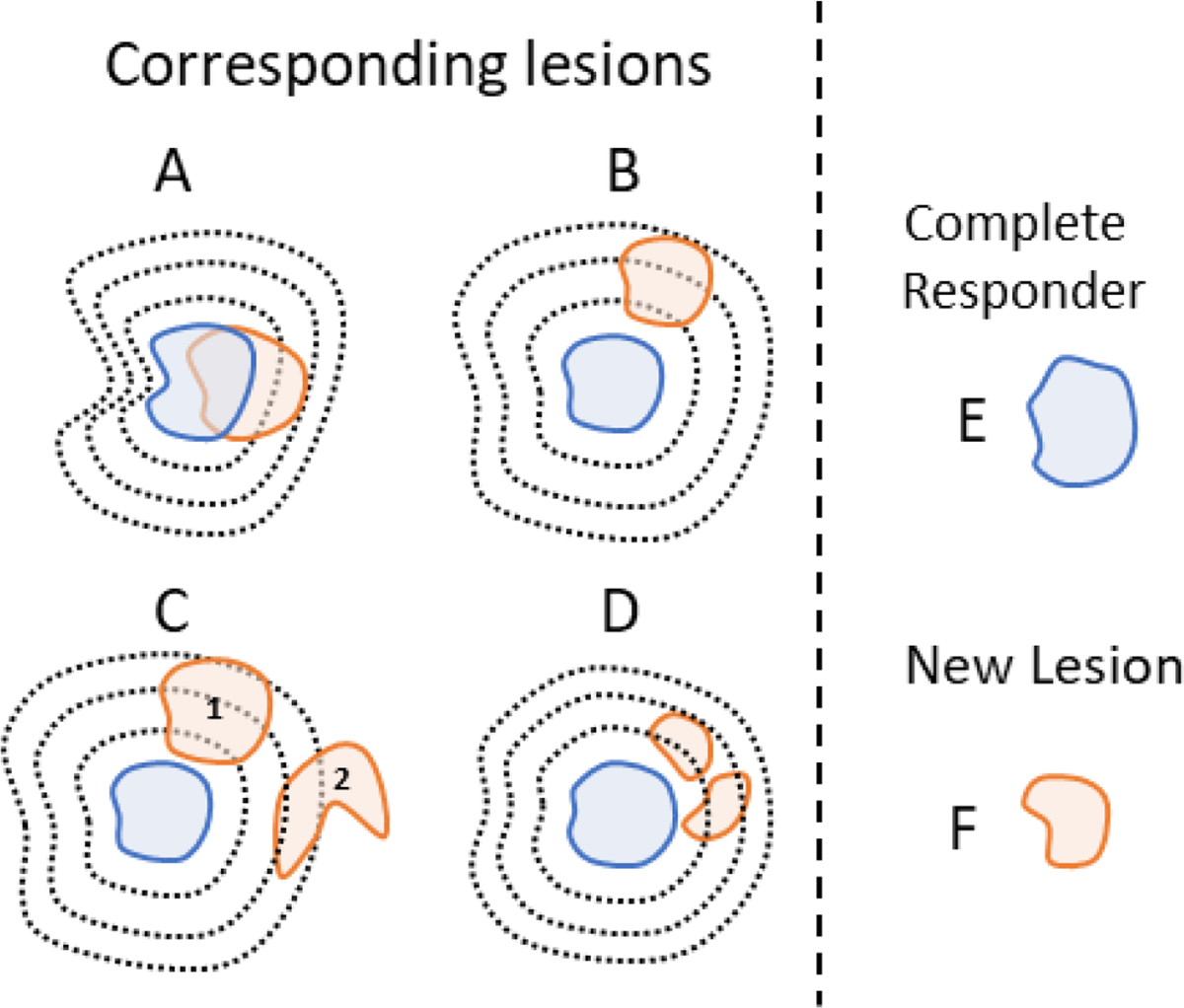
Illustration of the conformal dilation role in lesion matching. For clarity, lesion dilation is represented only on one scan of the scan-pair (blue). Lesions (solid contours) and lesions’ conformal dilations (black dotted lines), colors indicate lesions from different scans in a scan-pair. (A) Corresponding lesions are matched based on direct superposition. (B) Despite no direct superposition, match is still possible because of the conformal dilation. (C) lesion dilation yields a correct (1) and one incorrect (2) superposition, however, the Munkres algorithm selects the correct match. (D) Two lesions are superimposed to the dilation and are merged and matched. (E) A single disappearing lesion. (F) A single new lesion.
The algorithm’s third step is clustering of lesions, an important consideration for treatment response assessment, which can occur due to merging or splitting of multiple lesions from one to another imaging time point (Shafiei et al., 2021). Merging is usually a result of disease progression in adjacent lesions. Conversely, lesion splitting is usually caused by one lesion responding to treatment and separating into two or more smaller disjoint lesion components. After the second step, a lesion may intersect multiple lesions on the paired scan. This may be a spurious intersection with a neighboring lesion (Figure 3C), or may be due to merging or splitting (Figure 3D). We developed a novel lesion clustering algorithm designed specifically to distinguish between these two instances, so that the overall matching algorithm considers sets of components originating from splitting, and sets of components that eventually merge, as individual lesions. A set of lesion components is clustered when:
The components intersect to at least one common lesion on the other scan (Figure 4B), considering the dilation introduced on the second step
The distance between the centroids of the components (Figure 4C) is smaller than a distance (Figure 4D)
Figure 4 –
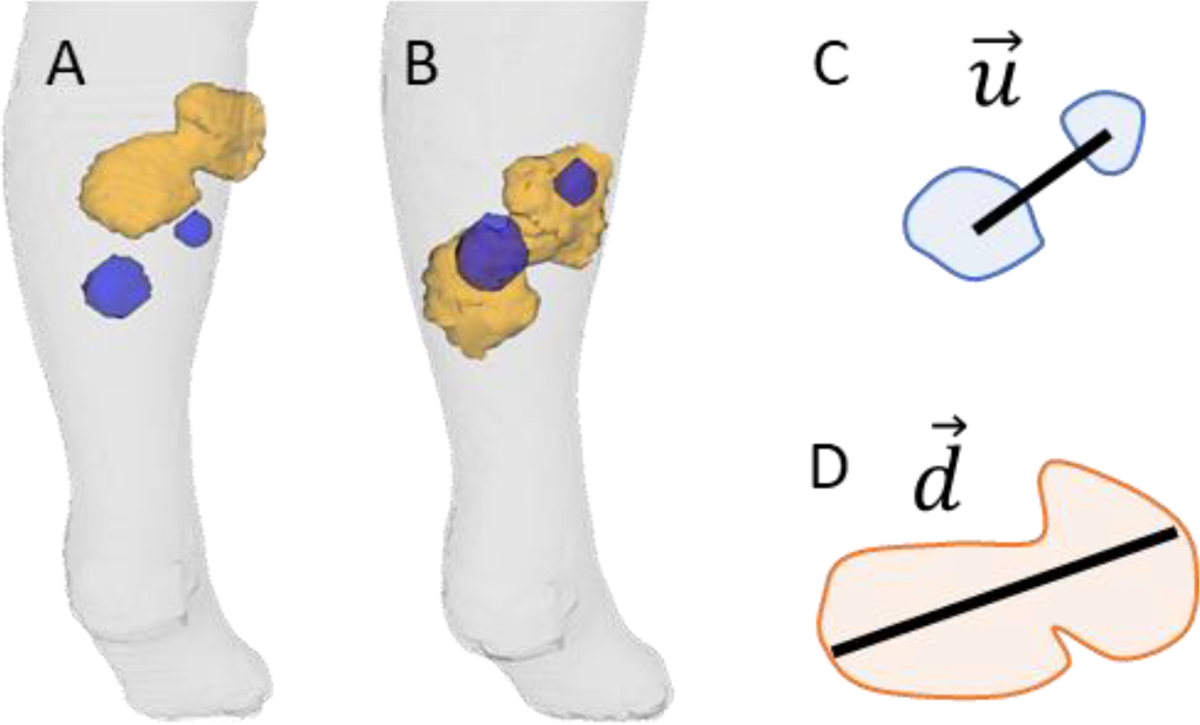
Situation illustrating clustering as lesion merging caused by disease progression. The blue masses are from an earlier scan and the orange mass is from a later scan. (A) Before registration, the lesions are not superimposed. (B) After registration, lesions are superimposed. (C) illustrates the vector between centroids and (D) the vector . After the clustering, the two blue masses correspond to one single lesion (orange).
The distance is defined as the longest chord in the common matching lesion that makes a smaller than 5° angle with the vector connecting the centroids of the components to be clustered (Figure 4), the angular constraint is expressed as
| (5) |
Assuming perfect registration, if a set of components originate from a homologous lesion in the paired scan (either merging or splitting), the components should all be spatially contained in the homologous lesion, in which case the condition will be satisfied. If registration is not perfect, but the dilation step guarantees that all the components overlap to the homologous lesion, the relationship between the vectors and will still hold. The angular constraint of equation (5) is imposed to consider that a chord in a direction in which the homologous lesion is long might be an overly lenient threshold for evaluating a vector connecting components separated along a different direction, in which the homologous lesion is short. The necessity for an angular constraint is illustrated in Figure 5. The 5° tolerance in equation (5) is used to consider that and need not be exactly parallel, but approximately so. The 5° value is an algorithm hyperparameter and different values could be explored depending on the application.
Figure 5 -.
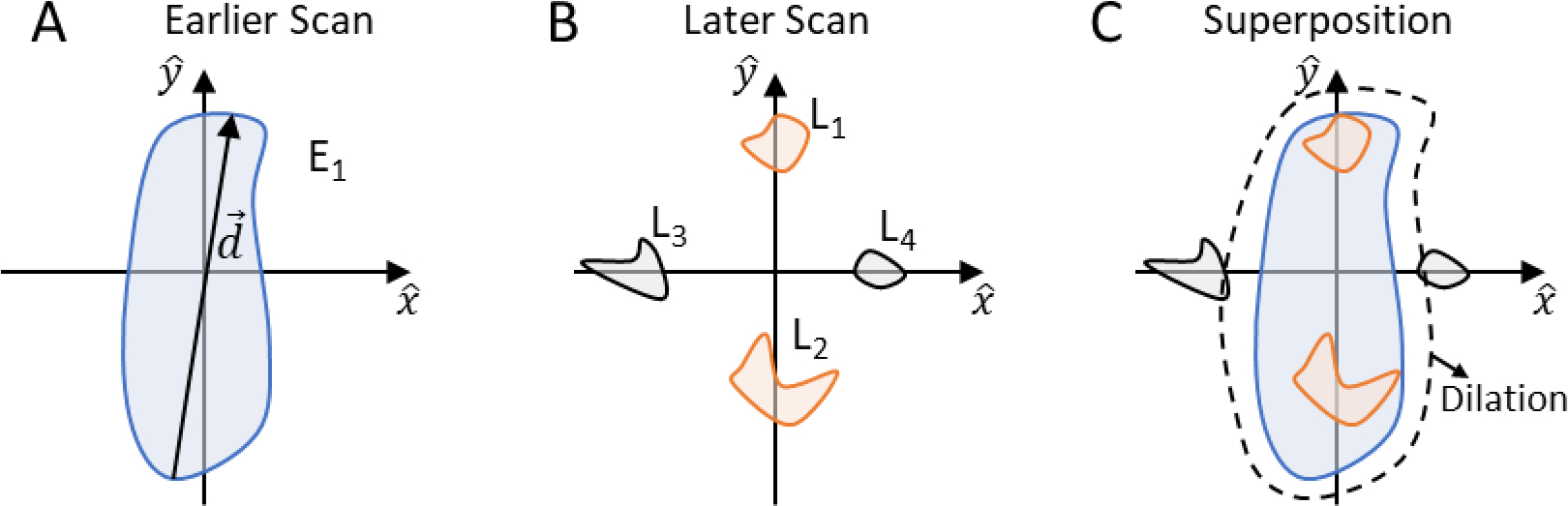
Illustration of the rationale for using a small angle constraint in the lesion clustering step. If lesion overlap were considered alone, lesions , and would cluster since all of them overlap to lesion (considering dilation). Using the constraint without an angular constraint would still be insufficient and imply in wrongful clustering of , and . Using the angular constraint, and are correctly excluded from the cluster.
In the fourth step, the lesions are renumbered and voxels that pertain to a given lesion are reevaluated based on the clustering decision. A new intersection matrix is calculated under the new conditions. A cost matrix is then defined as . The lesion matching problem is thus reduced to a linear assignment problem (Jaqaman et al., 2008), which consists of finding an optimal permutation matrix
| (6) |
within the constraints to the permutation matrix
| (7) |
where summations might be zero if a lesion is superimposed to no other (NLs or DLs). The Munkres assignment algorithm (Munkres, 1957) is used to solve the linear assignment problem. One of the possible mathematical interpretations of the Munkres algorithm is that it finds the optimal permutation matrix A that minimizes the global sum of costs defined by equation (6) within the constraints imposed by equation (7). Briefly, the Munkres algorithm performs a non-greedy minimization of , which is equivalent to a maximization of the volume of intersection between all lesions in a scan-pair.
3. Algorithm Validation
The developed algorithm was evaluated in 172 PET/CT images of 32 patients with metastatic cancer, all images were acquired under the institutional standard procedures. PET/CT images provide anatomical structure and functional information of the location of diseased tissue, which makes them suitable for validating the algorithm. The PET/CT images were retrospectively acquired from two independent clinical trials (Table 1). One of the trials contained 19 metastatic melanoma patients, for whom lesion-level disease assessment is particularly challenging because of the plurality of metastatic sites and the elevated number of lesions, which in the most extreme case was of 64 unique lesions spanning from the legs to the head. A second dataset containing 10 different types of cancers was included to evaluate the algorithm for its robustness regarding disease type.
Table 1 -.
Patient population description
| # of Patients | Tracer | Scanner | Disease | Treatment | Design |
|---|---|---|---|---|---|
|
| |||||
| 19 | 18F-FDG | GE Discovery IQ GE Discovery 710 GE Discovery STE |
Metastatic Melanoma | immune checkpoint inhibitor and antiangiogenic drugs | Retrospective |
| 13 | 18F-FLT | GE Discovery VCT | Upper tract urothelial, rectal, breast, lung (non-small cell), prostate, colon, ovarian, appendiceal, adenoid, and squamous cell | antiangiogenic drug | Prospective (Scarpelli et al., 2016) |
In the validation dataset, a scan-pair was defined as two longitudinal and sequential PET/CT scans of the same patient. Therefore, if is the number of scans that a patient received, is the number of scan-pairs; e.g. a patient that received four scans , will have three scan-pairs to to , and to . The dataset contained 140 scan-pairs.
In the validation, CT images served as the anatomical information, while the binary lesion masks were derived from the PET image. Using radiology reports as guidance, cancerous lesions, indicated by elevated PET SUV uptake relative to background, were manually contoured by an imaging scientist, defining thus the binary lesion masks associated to each PET/CT image. The manual contouring started by using an active-contours algorithm to automatically conform the contours to the lesions (Yang et al., 2009), then the contours were reviewed by a nuclear medicine physician and manually corrected when necessary. Reference matching decisions were made by visually analyzing the scan-pairs’ images for each patient and preparing a reference lesion matching decision matrix for each scan-pair. The merging or splitting of lesions over time was accounted for in the reference matching decisions. Clustering decisions were made when two or more lesions occupied the same anatomical position within the patient body as a larger lesion in the corresponding scan (Figure 4B).
The main metric used to assess the algorithm performance on the validation dataset was the overall accuracy of lesion matching, determined as the number of correct lesion matching decisions made divided by the total number of matching decisions. Additionally, the accuracy of matching corresponding lesions and the accuracy of identifying vanishing lesions and new lesions were estimated separately.
3.1. Image Registration Sensitivity Analysis
To fully understand the impact of image registration on lesion matching, we studied the algorithm performance under four distinct registration methods. Two deformable registration approaches were investigated: (1) Articulated Deformable Registration (ADR) which was preceded by Whole-body Rigid Registration (WRR) and Articulated Rigid Registration (ARR) and (2) Whole-body Deformable Registration (WDR), which was preceded only by WRR (Figure 6). Both deformable registration approaches (WDR and ADR) were done using free form deformation with 3rd order B-splines interpolation and hierarchical control grid (Rueckert et al., 1999). Additionally, lesion matching was assessed after WRR and ARR alone.
Figure 6 –
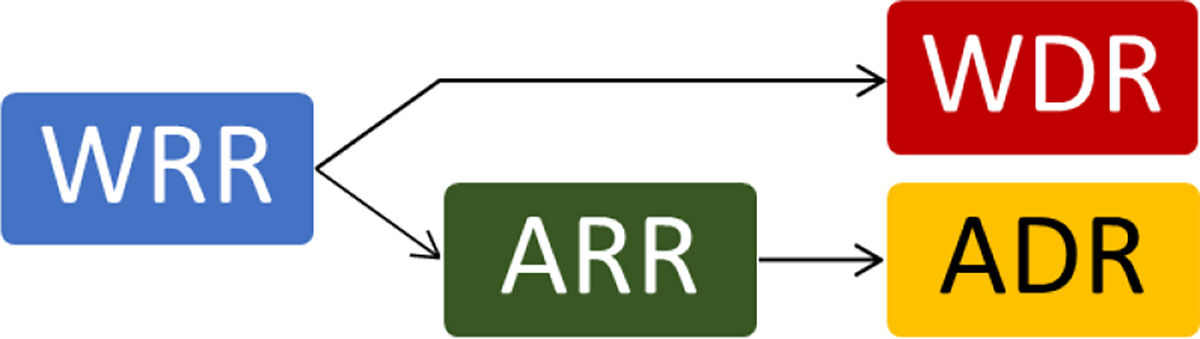
Diagram illustrating the approaches to arrive at whole-body and articulated deformable registrations.
The centroids of corresponding lesions were used as landmarks to analyze the performance of image registration, only lesions whose volume varied less than 50% between scans were used because high volume variation can lead to displacements of the lesion centroids which are unrelated to registration performance. The Wilcoxon signed-rank test was used to compare the centroid distance distributions. Additionally, the impact of the registration methods on lesion matching performance was assessed using the McNemar test to intercompare all the registration methods (0.05 significance level).
3.2. Lesion Dilation Sensitivity Analysis
A sensitivity study was conducted to assess dependence of the lesion matching accuracy on the magnitude of the conformal dilation (Step 2) by varying the dilation from 0 to 55 mm in 5 mm increments.
3.3. Anatomic Distribution Sensitivity Analysis
The overall lesion matching accuracy evaluation was stratified for the whole body and per individual body parts. The individual body parts considered were head and neck, chest+abdomen, pelvis, and left and right upper arms, forearms, hands, thighs, legs, and feet.
3.4. Lesion Burden Sensitivity Analysis
The lesion matching accuracy was analyzed on a subset of high burden scan-pairs to investigate if an accuracy drop would be observed in those cases. High burden scan-pairs were defined as those containing 10 or more unique lesions, twice as much as what is recommended for visual human evaluation. In a clinical setting and without automation, individual comprehensive lesion-level response assessment of these scan-pairs is prohibitive. Therefore, satisfactory performance of the algorithm on these scan-pairs is paramount for its possibility of translation into the clinic.
4. Results
In this study, 736 matching decisions were necessary to analyze all 140 scan-pairs. Based on the reference matching decisions, 298 of the 736 matching decisions (40%) were corresponding lesions and 438 (60%) were new or disappearing lesions. The accuracy of lesion matching is shown in Table 2. These results represent the accuracy with no dilation.
Table 2 -.
Accuracy of lesion matching when no dilation is used.
| Registration | Accuracy |
||
|---|---|---|---|
| Corresponding Lesions N = 298 | New or disappearing lesions N = 438 | Overall N = 736 | |
|
| |||
| Whole-body deformable (WDR) | 0.75 | 1.00 | 0.90 |
| Articulated deformable (ADR) | 0.72 | 0.99 | 0.88 |
|
| |||
| Whole-body rigid (WRR) | 0.58 | 0.99 | 0.82 |
| Articulated rigid (ARR) | 0.68 | 1.00 | 0.87 |
4.1. Image Registration Sensitivity Analysis
The centroids of 186 corresponding lesions were used as landmark. The landmark distance distributions are shown in Figure 7. All distributions were significantly different (). A visual understanding of how registration influences landmark distance, and thus proximity of corresponding lesions, is shown in Figure 8.
Figure 7 –
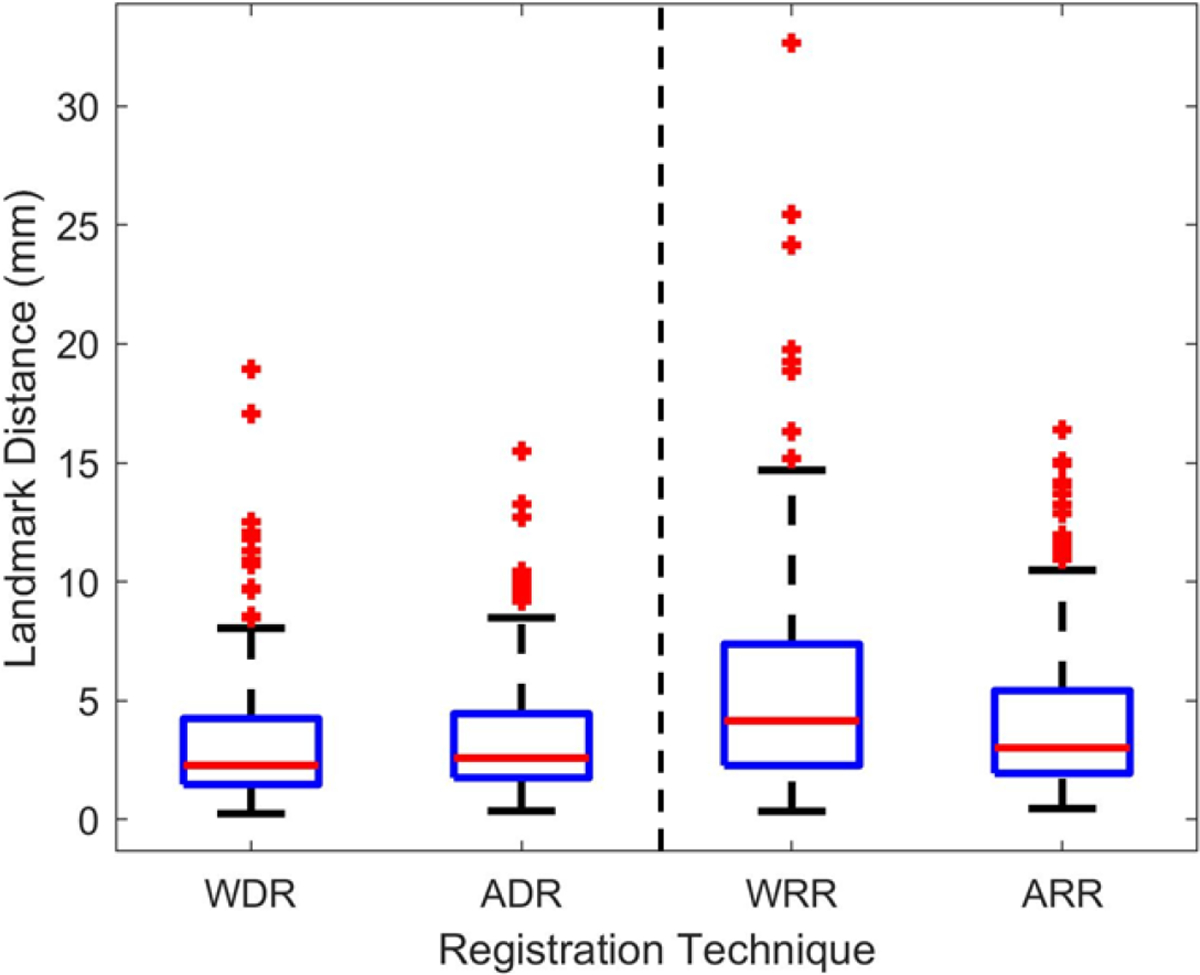
Landmark distance distribution of the 186 landmark lesions. Results on the left are for the 3D deformable registration methods: whole-body deformable (WDR) and articulated deformable (ADR). Results on the right are for previous explored registration methods: whole-body rigid (WRR) and articulated rigid (ARR).
Figure 8 -.
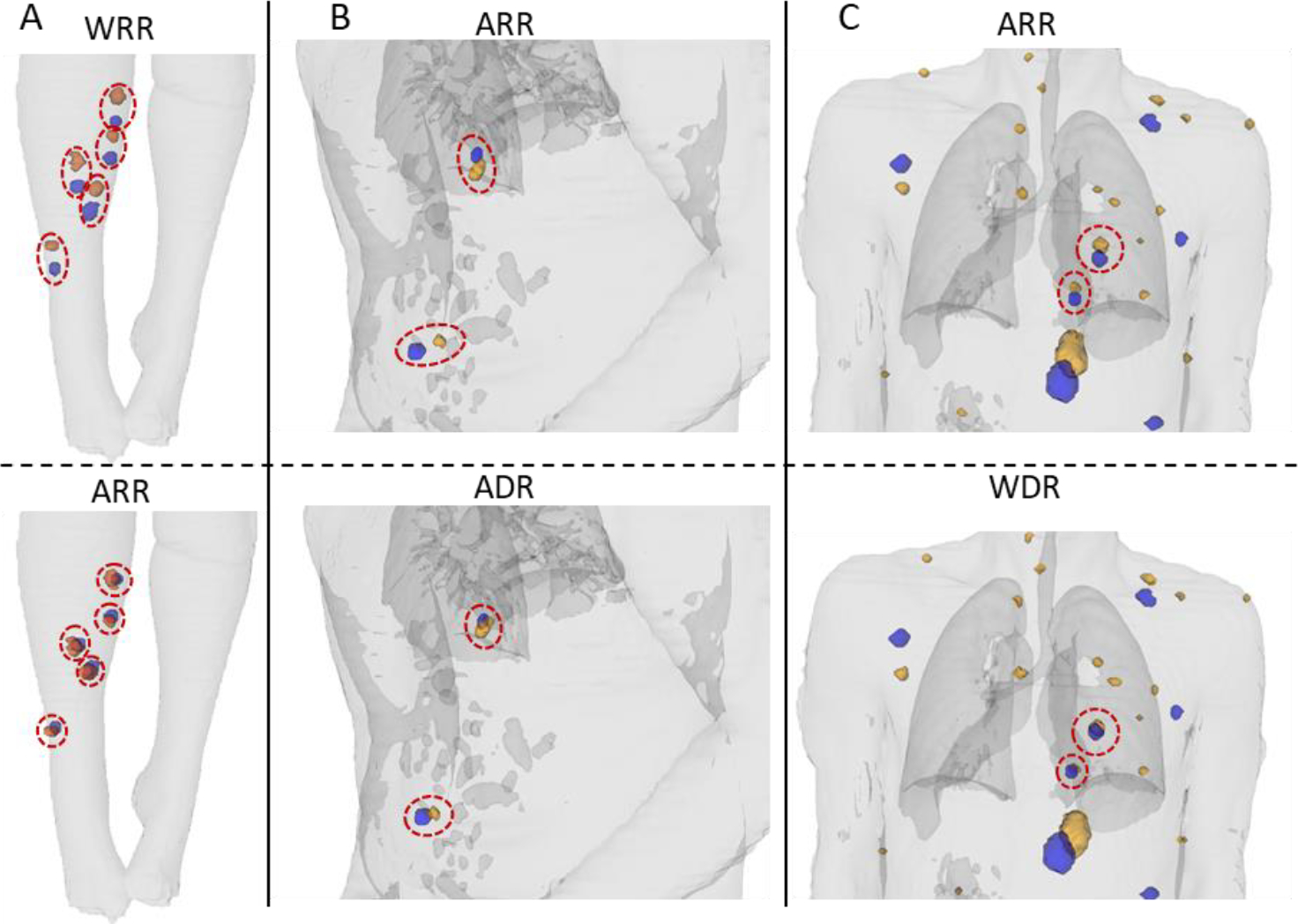
Visualization of the impact of different registrations on proximity of corresponding lesions. The CT contour is shown in light gray. Earlier scan lesions are shown in blue and later scan lesions in orange. Red ellipses highlight the corresponding lesions that had visible approximation. Images are shown for whole-body rigid (WRR), articulated rigid (ARR), articulated deformable (ADR), and whole-body deformable (WDR) registrations. Column (A) compares WRR to ARR. Column (B) compares ARR to ADR. And column (C) compares ARR to WDR.
The McNemar test was used to intercompare the deformable registration approaches’ impact on the lesion matching performance. Lesion matching performance using WDR was better than ADR, nevertheless with marginal statistical significance . We also used the McNemar test to compare both deformable approaches to the rigid approaches, WRR and ARR. Comparison of both approaches with WRR yielded statistically significant differences . WDR significantly improved lesion matching when compared to ARR . Improvement observed with ADR was close on the verge of significance .
4.2. Lesion Dilation Sensitivity Analysis
An increase in matching accuracy was observed for corresponding lesions across the dilation range (Figure 9A). By contrast, the accuracy for new or disappearing lesions decreased with lesion dilation, (Figure 9B). The overall accuracy was observed to have an initial increase associated with the CL accuracy trend, followed by a decrease associated with the NL/DL accuracy trend (Figure 9C). The optimal dilation magnitude was determined by the peaks in the overall accuracy curves. The optimal dilations were of 25 mm for all registration methods besides WRR, which had an optimal dilation of 30 mm.
Figure 9 –
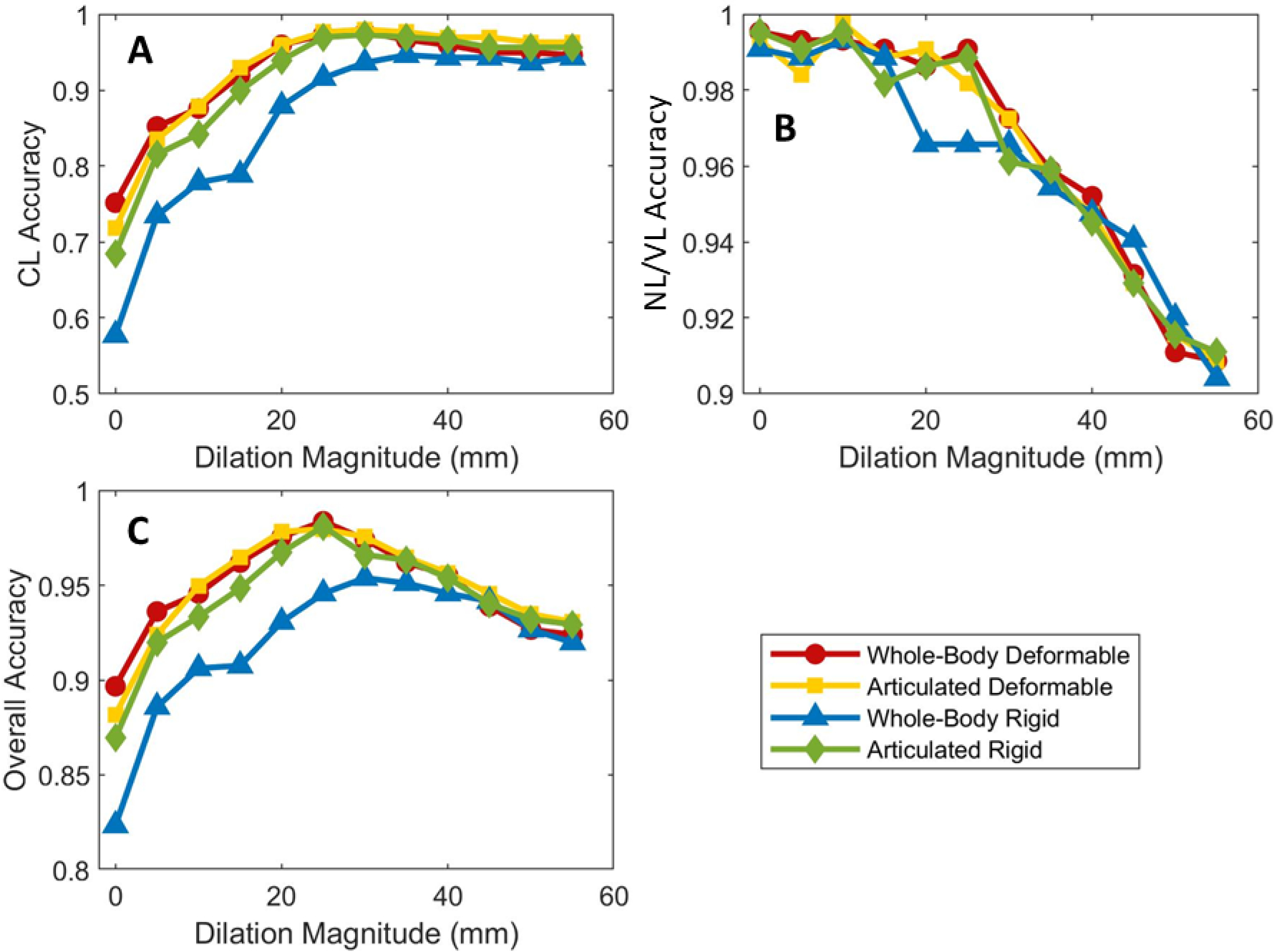
Accuracy as a function of dilation magnitude for (A) corresponding lesions (CL), (B) new or disappearing lesions (NL/DL), and (C) all lesions. The markers corresponding to 0 mm of dilation represent the accuracy of the method without lesion dilation.
4.3. Anatomic Distribution Sensitivity Analysis
The data in Table 3 are derived from our observation of the 736 lesions present in the patients in our datasets, the location of all unique lesions are shown in Table 3, most of which were the chest+abdomen (52% - 380/736) and in the pelvis (21% - 156/736). The thighs (13% - 99/736) and the legs (8% - 60/736) also presented with relatively high disease densities.
Table 3 -.
Number of lesions per body-part
| Body-part | # of lesions | % | Body-part | # of lesions | % |
|---|---|---|---|---|---|
|
| |||||
| Chest+abdomen | 380 | 52% | Feet | 9 | 1.2% |
| Pelvis | 156 | 21% | Forearms | 5 | 0.7% |
| Thighs | 99 | 13% | Arms | 5 | 0.7% |
| Legs | 60 | 8% | Hands | 3 | 0.4% |
| Head & neck | 19 | 3% | |||
For the four body-parts with highest number of lesions (chest+abdomen, pelvis, thighs, and legs), assessment of the overall accuracy was performed separately. The overall accuracies per body-part, evaluated at the optimal dilations (Figure 10) saw minimal variation across registration methods and body-parts. No accuracy result was smaller than 0.90 (WRR in the Thigs).
Figure 10 –
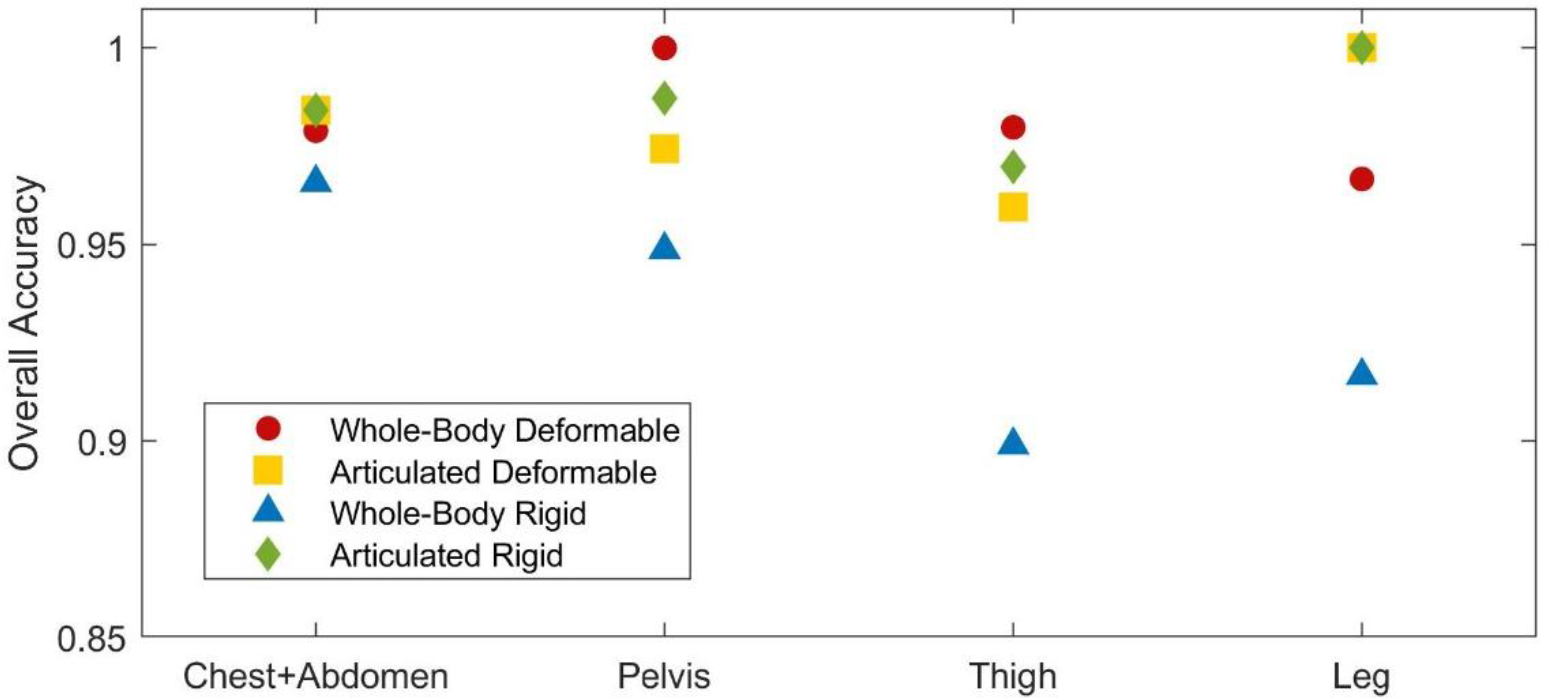
Overall accuracy, using each registration method in the four body-parts with highest number of lesions at the optimal dilations.
4.4. Lesion Burden Sensitivity Analysis
In the complete patient population, containing all 140 scan-pairs, the median number of lesion matching decisions was 3, with minimum of 1 and maximum of 68. In the high lesion burden subset, the median number of lesion matching decisions was 19, minimum of 11 and maximum of 68. The high lesion burden subset accounted for 345 of the 736 matching decisions (47% of the total) and for 12 of the 140 scan-pairs (8.5% of the total). Accuracy results were evaluated for the optimal dilation magnitudes (Figure 11).
Figure 11-.
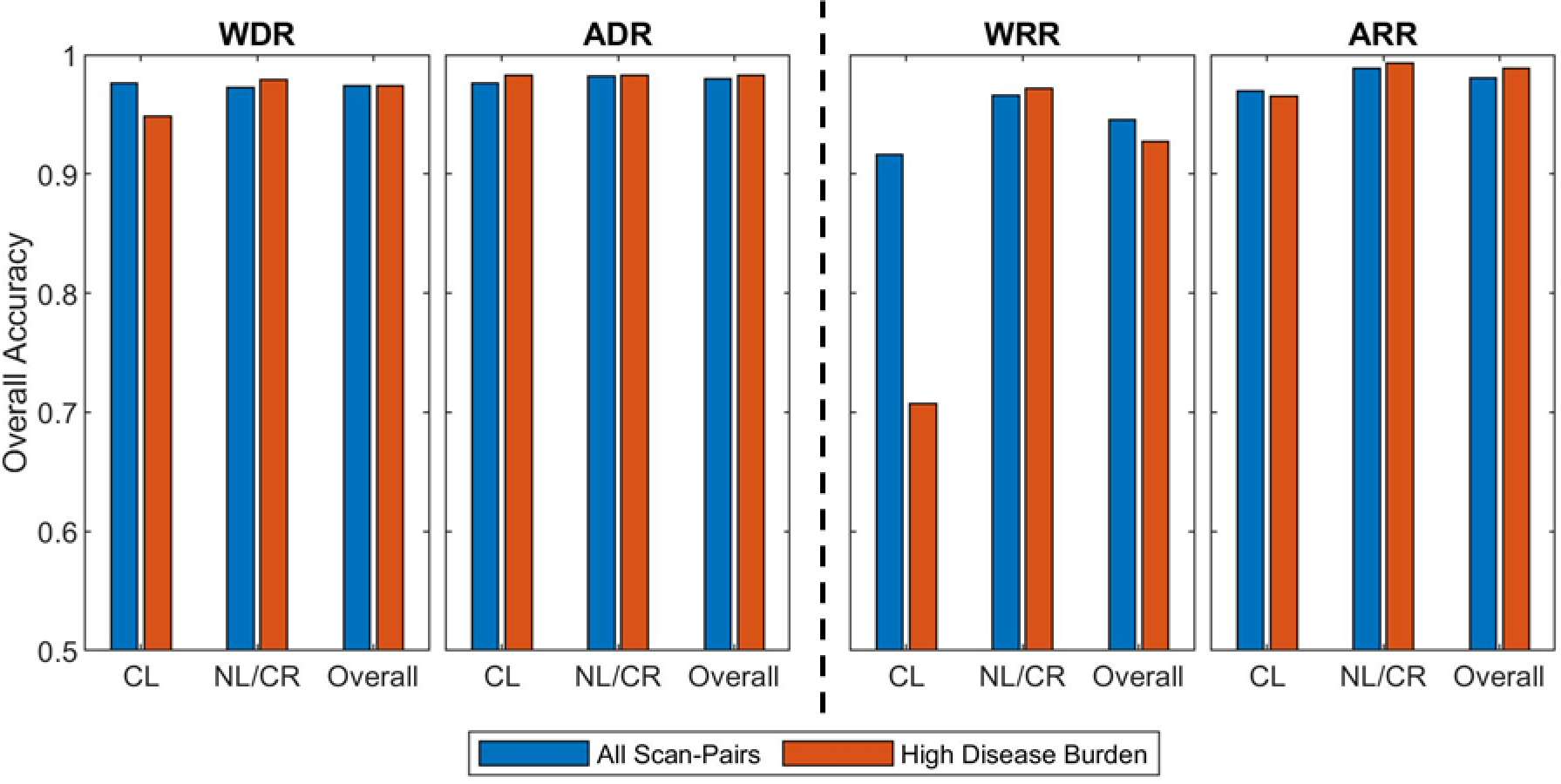
Comparison between overall accuracies obtained with the whole dataset and with the subset of high disease burden scan-pairs (more than 10 matching decisions per scan-pair). Results on the left are for the 3D deformable registration methods: whole-body deformable (WDR) and articulated deformable (ADR). Results on the right are for previous explored registration methods: whole-body rigid (WRR) and articulated rigid (ARR). The optimal lesion dilation was used, namely 25 mm for ARR, ADR, and WDR; and 30 mm for WRR. In addition to overall accuracy, the analysis was separated into corresponding lesions (CL) and new or disappearing lesions (NL/DL).
5. Discussion
In this work we presented a lesion matching algorithm based on 4 steps: image registration, lesion dilation, lesion clustering, and linear assignment (Figure 1). The algorithm was validated on two independent patient populations and the lesion matching accuracy obtained was excellent (0.98 overall accuracy). No trend in accuracy was observed across patient populations, overall accuracies were 0.98 for both. We explored the lesion matching accuracy sensitivity regarding image registration methods, lesion dilation magnitude, anatomic distribution, and lesion burden. The deformable registration techniques (WDR and ADR) showed superior registration results, when compared to rigid techniques (WRR and ARR) as shown by the landmark distance (Figure 7), which led to a significant improvement in the lesion matching accuracy. Furthermore, for WDR and ADR, residual registration error was in accordance with literature for image intensity based methods (Yin et al., 2011). Use of lesion dilation magnitude was an important factor in achieving the excellent accuracy level. For the best performing registration approach (WDR), the use of dilation caused overall accuracy to increase by 0.08, from 0.90 with no dilation to 0.98 with the optimal dilation of 25 mm (Table 2, Figure 9). In addition, the excellent accuracy levels were minimally dependent on lesion body-part location (Figure 10) and were sustained on the analysis of high burden scan-pairs (Figure 11). Lastly, the matching criteria used by our methodology are positional and make no assumption regarding lesion shape, instead, the shape is taken into consideration by the methodology via conformal dilation (step 2) and via the use of lesion derived distance metrics for clustering (step 3).
Previous works have explored rigid registration-based lesion matching (Bosc et al., 2003; Köhler et al., 2019; Moltz et al., 2009; Shahar and Greenspan, 2005; Xu et al., 2011). Moltz et al. suggested that rigid registration suffices for lesion matching applications; however, we hypothesize that this conclusion was based on their focus on liver and lung cancer lesions. These organs are located in the chest+abdomen body-part, where we observed WRR-based matching to have its best performance (0.96 overall accuracy - Figure 10). However, for the purpose of whole-body soft tissue lesion matching, the rigid registration method (WRR) resulted in the worst overall matching accuracy; matching accuracy of corresponding lesions was of only 0.58 without dilation (Table 2). When compared the ARR method (Yip et al., 2014), use of 3D deformable registration resulted in a remarkable lesion matching improvement, 29%, from 0.58 to 0.75 (Table 2). Deformable registration has been previously employed for the task of lesion matching (Xu et al., 2011). However, it was limited to 2D deformation of a few image slices. The use of 3D deformable registration for lesion matching purposes is one of the novelties presented in this work. Other novelties of our work are the use of conformal dilation for lesion matching, the development of a novel lesion clustering methodology that is specific for longitudinal monitoring of merging and splitting lesions, and the use of a non-greedy optimizer to solve a linear assignment problem in the setting of whole-body longitudinal disease matching using in-vivo imaging of human patients. Most importantly, the main novelty of our work consists in combining several image analysis techniques to solve the problem of whole-body soft tissue lesion matching, which had not been done previously.
One challenge of our work was defining the reference dataset for the lesion matching decisions, which is subject to human error and bias in its nature. However, these same limitations would be encountered in manual lesion matching performed for clinical decisions. In other words, some arbitrariness is unavoidably associated with visual interpretation of medical images. We observed, however, that this limitation is likely to have little impact in the validation of the algorithm because the correspondence between the vast majority of lesion pairs was conspicuous.
The 12 incorrect matching decisions made by the algorithm for the best performer parameters (WDR with dilation of 25 mm) had three causes. First, lesion dilation caused 2 incorrect decisions where a new and a disappearing lesion were registered close enough that dilation led to incorrect superposition. Second, misregistration caused 4 incorrect decisions, all in the same scan-pair and located in the patient’s legs. Largely different legs positioning resulted in a large deformation vector field that failed to accurately approximate the corresponding lesions; use of ADR for matching lesions in the legs is one way of overcoming this limitation. Finally, Incorrect clustering caused 6 incorrect decisions (Figure 12), where two or more lesions (blue) that should not cluster were superimposed to a common lesion (orange). Due to the particular geometric positioning, the threshold distance 𝑑 (Figure 4D) and the angle restriction failed to prevent the incorrect clustering. This limitation could be overcome by use of an adaptive dilation magnitude parameterized by local lesion density, which will be pursued in future developments. The impact of incorrect lesion matching decisions on possible applications of the herein presented algorithm, such as automated treatment response assessment, will be subject of future investigations.
Figure 12 -.
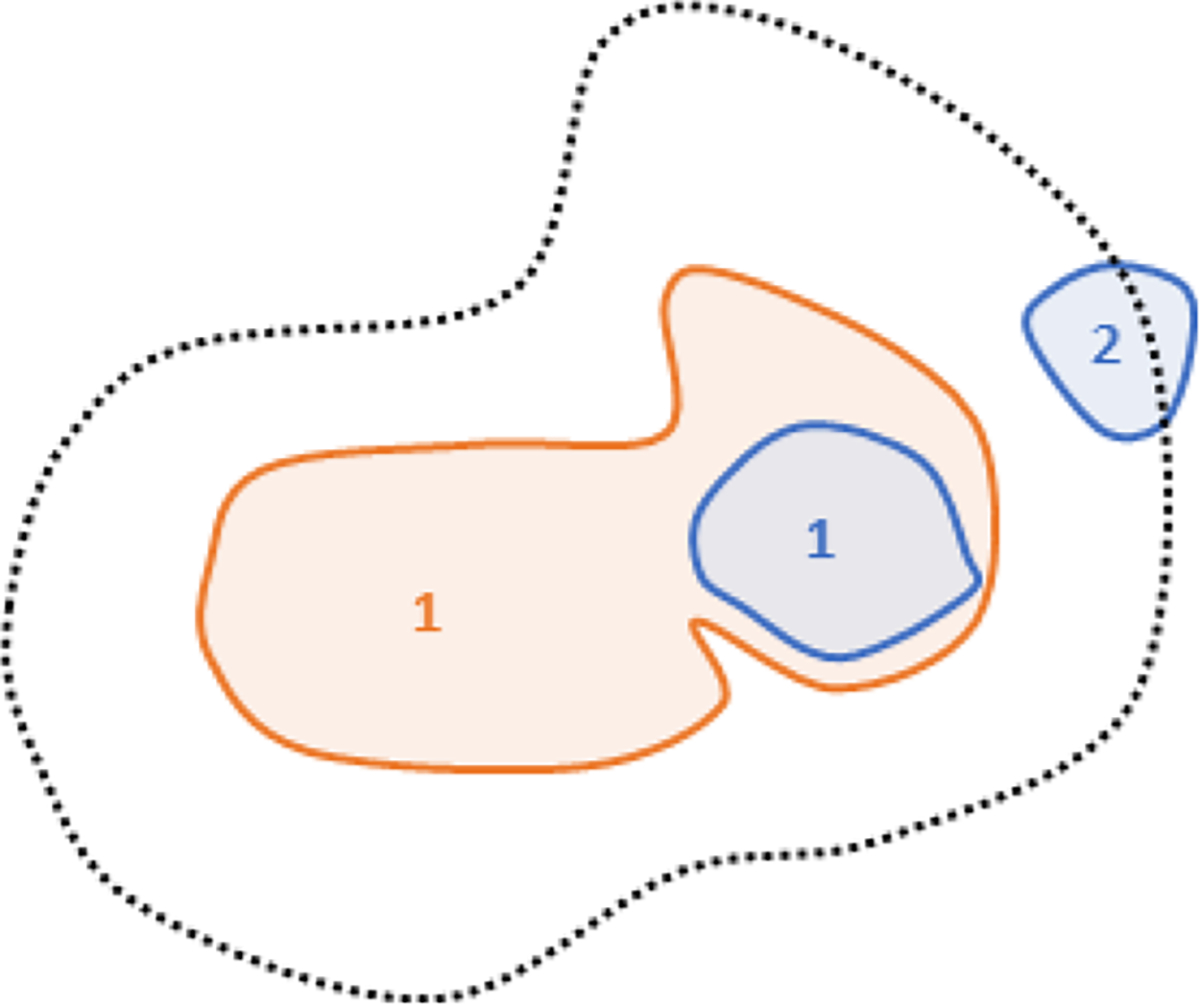
illustration of incorrect cluster decision
While the herein presented algorithm was validated using PET/CT images, we hypothesize that it is generalizable to other types of medical imaging modalities. The requirements for application of the algorithm are anatomical information that is used to guide the registration step and the existence of lesion conspicuity, which in our validation dataset was obtained through PET scans. We note, however, that the implementation of the method to other imaging modalities would require sensible adaptation of hyperparameters involved in all four steps of the algorithm.
6. Conclusion
In this work we introduced an algorithm that enables automated matching of metastatic lesion. We validated the algorithm for whole-body longitudinal PET/CT scans of metastatic cancer patients and showed it has high overall accuracy. One novelty of our algorithm is the leveraging of 3D deformable image registration, which outperformed the existing methods for every studied scenario and was a significant improvement, when compared to previously established methods. Our study suggests that whole-body lesion matching can be done automatically, possibly facilitating lesion-level disease assessment of metastatic cancer patients in clinical practice and in clinical trials, ultimately leading to enriched information for treatment response assessment and facilitating the development of novel treatment combinations.
References
- Baum RP, Prasad V, Müller D, Schuchardt C, Orlova A, Wennborg A, Tolmachev V, Feldwisch J, 2010. Molecular imaging of HER2-expressing malignant tumors in breast cancer patients using synthetic 111In- or 68Ga-labeled Affibody molecules. J. Nucl. Med. 10.2967/jnumed.109.073239 [DOI] [PubMed] [Google Scholar]
- Bosc M, Heitz F, Armspach JP, Namer I, Gounot D, Rumbach L, 2003. Automatic change detection in multimodal serial MRI: Application to multiple sclerosis lesion evolution. Neuroimage 20, 643–656. 10.1016/S1053-8119(03)00406-3 [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA, 2011. A perspective on cancer cell metastasis. Science (80-. ). 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J, 2009. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- Gallamini A, Zwarthoed C, Borra A, 2014. Positron emission tomography (PET) in oncology. Cancers (Basel). 6, 1821–1889. 10.3390/cancers6041821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayed I, Vu T, Iyer R, Johnson M, Macapinlac H, Swanston N, Podoloff D, 2004. The role of 18F-FDG PET in staging and early prediction of response to therapy of recurrent gastrointestinal stromal tumors. J. Nucl. Med. 45, 17–21. [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G, 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Wahl H, Ziemssen T, Linn J, Kitzler HH, 2019. Exploring individual multiple sclerosis lesion volume change over time: Development of an algorithm for the analyses of longitudinal quantitative MRI measures. NeuroImage Clin. 21, 101623. 10.1016/j.nicl.2018.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SM, Erdi Y, Akhurst T, Mazumdar M, Macapinlac HA, Finn RD, Casilla C, Fazzari M, Srivastava N, Yeung HWD, Humm JL, Guillem J, Downey R, Karpeh M, Cohen AE, Ginsberg R, 1999. Tumor treatment response based on visual and quantitative changes in global tumor glycolysis using PET-FDG imaging. The visual response score and the change in total lesion glycolysis. Clin. Positron Imaging. 10.1016/S1095-0397(99)00016-3 [DOI] [PubMed] [Google Scholar]
- Lubner MG, Lee FT, Brace CL, Ziemlewicz TJ, Hinshaw JL, 2013. Microwave Ablation of Hepatic Malignancy 1, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltz JH, Schwier M, Peitgen HO, 2009. A general framework for automatic detection of matching lesions in follow-up ct. Proc. - 2009 IEEE Int. Symp. Biomed. Imaging From Nano to Macro, ISBI 2009 843–846. 10.1109/ISBI.2009.5193184 [DOI] [Google Scholar]
- Munkres J, 1957. Algorithms for the assignment and transportation problems. Journal of the society for industrial and applied mathematics. J Soc Indust Appl Math 5, 32–38. [Google Scholar]
- Roth A, Cooley G, Smilowitz J, Ferjancic P, Liu G, Jeraj R, 2020. Treatment of Oligoresistant and Oligoprogressive Disease in Metastatic Prostate Cancer Patients with Radiation Therapy, in: AAPM Meeting. [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ, 1999. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18, 18:712–21. [DOI] [PubMed] [Google Scholar]
- Scarpelli M, Bruce JY, Carmichael L, Eickhoff J, Kolesar J, Perlman S, Jeraj R, Liu G, 2016. 18F-FLT PET/CT imaging in patients with advanced solid malignancies treated with axitinib on an intermittent dosing regimen. Cancer Chemother. Pharmacol. 78, 1245–1252. 10.1007/s00280-016-3183-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpelli M, Simoncic U, Perlman S, Liu G, Jeraj R, 2018. Dynamic 18F-FLT PET imaging of spatiotemporal changes in tumor cell proliferation and vasculature reveals the mechanistic actions of anti-angiogenic therapy. Phys. Med. Biol. 63. 10.1088/1361-6560/aad1be [DOI] [PubMed] [Google Scholar]
- Schwarz JD, Bader M, Jenicke L, Hemminger G, Jänicke F, Avril N, 2005. Early prediction of response to chemotherapy in metastatic breast cancer using sequential18F-FDG PET. J. Nucl. Med. 46, 1144–1150. [PubMed] [Google Scholar]
- Shafiei A, Bagheri M, Farhadi F, Apolo AB, Biassou NM, Folio LR, Summers RM, 2021. CT Evaluation of Lymph Nodes That Merge or Split during the Course of a Clinical Trial: Limitations of RECIST 1.1. Radiol. Imaging Cancer 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahar A, Greenspan H, 2005. A probabilistic framework for the detection and tracking in time of multiple sclerosis lesions. 2004 2nd IEEE Int. Symp. Biomed. Imaging Nano to Macro (IEEE Cat No. 04EX821) 440–443. 10.1109/isbi.2004.1398569 [DOI] [Google Scholar]
- Sotiras A, Davatzikos C, Paragios N, 2014. Deformable Medical Image Registration: A Survey. IEEE Trans Med Imaging 32, 1153–1190. 10.1109/TMI.2013.2265603.Deformable [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhoek M, Perlman SB, Jeraj R, 2013. PET-Based Quantification of Treatment Response 54, 1188–1195. 10.2967/jnumed.112.113332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl RL, Jacene H, Kasamon Y, Lodge MA, 2009. From RECIST to PERCIST: Evolving Considerations for PET Response Criteria in Solid Tumors. J. Nucl. Med. 50, 122S–150S. 10.2967/jnumed.108.057307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R, 2006. Molecular imaging in cancer. Science (80-. ). 312, 1168–1171. 10.1126/science.1125949 [DOI] [PubMed] [Google Scholar]
- Xu J, Greenspan H, Napel S, Rubin DL, 2011. Automated temporal tracking and segmentation of lymphoma on serial CT examinations. Med. Phys. 38, 5879–5886. 10.1118/1.3643027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Zheng J, Nofal A, Deasy J, Naqa I. El, 2009. Techniques and software tool for 3D multimodality medical image segmentation. J. Radiat. Oncol. Informatics 1, 1–22. https://doi.org/jroi-1-1-4 [Google Scholar]
- Yin Y, Hoffman EA, Ding K, Reinhardt JM, Lin CL, 2011. A cubic B-spline-based hybrid registration of lung CT images for a dynamic airway geometric model with large deformation. Phys. Med. Biol. 56, 203–218. 10.1088/0031-9155/56/1/013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S, Jeraj R, 2014. Use of articulated registration for response assessment of individual metastatic bone lesions. Phys. Med. Biol. 59, 1501–1514. 10.1088/0031-9155/59/6/1501 [DOI] [PubMed] [Google Scholar]
- Yip S, Perk T, Jeraj R, 2014. Development and evaluation of an articulated registration algorithm for human skeleton registration. Phys. Med. Biol. 59, 1485–1499. 10.1088/0031-9155/59/6/1485 [DOI] [PubMed] [Google Scholar]


