Abstract
Brain microstructural changes already occur in the earliest phases of Alzheimer’s disease (AD) as evidenced in diffusion magnetic resonance imaging (dMRI) literature. This study investigates the potential of the novel dMRI Apparent Measures Using Reduced Acquisitions (AMURA) as imaging markers for capturing such tissue modifications.Tract-based spatial statistics (TBSS) and support vector machines (SVMs) based on different measures were exploited to distinguish between amyloid-beta/tau negative (A
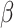 -/tau-) and A
-/tau-) and A
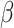 +/tau+ or A
+/tau+ or A
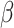 +/tau- subjects. Moreover, eXplainable Artificial Intelligence (XAI) was used to highlight the most influential features in the SVMs classifications and to validate the results by seeing the explanations’ recurrence across different methods.TBSS analysis revealed significant differences between A
+/tau- subjects. Moreover, eXplainable Artificial Intelligence (XAI) was used to highlight the most influential features in the SVMs classifications and to validate the results by seeing the explanations’ recurrence across different methods.TBSS analysis revealed significant differences between A
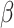 -/tau- and other groups in line with the literature. The best SVM classification performance reached an accuracy of 0.73 by using advanced measures compared to more standard ones. Moreover, the explainability analysis suggested the results’ stability and the central role of the cingulum to show early sign of AD.By relying on SVM classification and XAI interpretation of the outcomes, AMURA indices can be considered viable markers for amyloid and tau pathology. Clinical impact: This pre-clinical research revealed AMURA indices as viable imaging markers for timely AD diagnosis by acquiring clinically feasible dMR images, with advantages compared to more invasive methods employed nowadays.
-/tau- and other groups in line with the literature. The best SVM classification performance reached an accuracy of 0.73 by using advanced measures compared to more standard ones. Moreover, the explainability analysis suggested the results’ stability and the central role of the cingulum to show early sign of AD.By relying on SVM classification and XAI interpretation of the outcomes, AMURA indices can be considered viable markers for amyloid and tau pathology. Clinical impact: This pre-clinical research revealed AMURA indices as viable imaging markers for timely AD diagnosis by acquiring clinically feasible dMR images, with advantages compared to more invasive methods employed nowadays.
Keywords: Amiloyd-beta, tau, AMURA, tract-based spatial statistics, eXplainable Artificial Intelligence
I. Introduction
Amyloid-beta (A
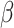 ) accumulation and neurofibrillary tangles due to phosphorylated tau protein define the Alzheimer’s disease (AD) molecular pathology [1]. Recent studies showed that both can occur from a pre-clinical and asymptomatic condition to the appearance of symptoms such as mild cognitive impairment (MCI) and diagnosed dementia [2]. For this reason, markers reflecting these two targets’ variations in the earliest disease phase are currently researched to develop potential therapies able to slow or stop its progression. At present, A
) accumulation and neurofibrillary tangles due to phosphorylated tau protein define the Alzheimer’s disease (AD) molecular pathology [1]. Recent studies showed that both can occur from a pre-clinical and asymptomatic condition to the appearance of symptoms such as mild cognitive impairment (MCI) and diagnosed dementia [2]. For this reason, markers reflecting these two targets’ variations in the earliest disease phase are currently researched to develop potential therapies able to slow or stop its progression. At present, A
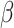 and tau can be revealed by positron emission tomography and cerebrospinal fluid via lumbar puncture [3]. However, more advantageous methods are required since these techniques are used at the cost of radioactive tracers, high spending and invasiveness [3]. Recent pathology studies employing diffusion magnetic resonance imaging (dMRI) reported brain microstructural abnormalities in the earliest phases of AD in both gray (GM) [4] and white matter (WM) [3], [5], [6], [7]. In particular, such abnormalities can be detected before the appearance of brain atrophy typically evidenced by classical T1-weighted (T1w) MRI [4], [5].
and tau can be revealed by positron emission tomography and cerebrospinal fluid via lumbar puncture [3]. However, more advantageous methods are required since these techniques are used at the cost of radioactive tracers, high spending and invasiveness [3]. Recent pathology studies employing diffusion magnetic resonance imaging (dMRI) reported brain microstructural abnormalities in the earliest phases of AD in both gray (GM) [4] and white matter (WM) [3], [5], [6], [7]. In particular, such abnormalities can be detected before the appearance of brain atrophy typically evidenced by classical T1-weighted (T1w) MRI [4], [5].
The diffusion tensor imaging (DTI) [8] is the most popular dMRI model used in clinics to quantitatively measure microstructural brain tissue properties. It was recently exploited by Chen et al. [3] to characterize WM differences among A
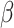 -negative/tau-negative (A
-negative/tau-negative (A
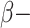 /tau−), A
/tau−), A
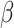 -positive/tau-negative (A
-positive/tau-negative (A
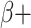 /tau−), and A
/tau−), and A
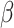 -positive/tau-positive (A
-positive/tau-positive (A
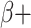 /tau+) cognitively normal controls (CN), as well as A
/tau+) cognitively normal controls (CN), as well as A
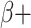 /tau+ MCI and AD subjects. They found widespread WM alterations in the whole AD continuum, but such a finding occurred especially early and correlated with tau pathology in the hippocampal cingulum revealing the potential of dMRI in the early AD detection challenge. The DTI popularity is mainly due to its simplicity and feasibility with data acquired in clinical conditions (i.e., low number of diffusion gradients in addition to their poor strength and timing). Nevertheless, it assumes a Gaussian trend of the dMRI signal with the intrinsic limitation of failing the reconstruction of complex WM architectural configurations where diffusion is anisotropic (e.g., fibers’ crossings, kissings, etc.). Recent overcomes of this limitation are represented by multi-shell dMRI and new modelling techniques like the Neurite Orientation Dispersion and Density Imaging (NODDI) [9], though not standard for clinical purposes. NODDI is a compartmental model which assumes that the brain tissue is divided in isotropic, intracellular diffusion, and extracellular microstructural compartments. Vogt et al. [6] showed that the NODDI-derived neurite density index was sensitive to GM modifications in A
/tau+ MCI and AD subjects. They found widespread WM alterations in the whole AD continuum, but such a finding occurred especially early and correlated with tau pathology in the hippocampal cingulum revealing the potential of dMRI in the early AD detection challenge. The DTI popularity is mainly due to its simplicity and feasibility with data acquired in clinical conditions (i.e., low number of diffusion gradients in addition to their poor strength and timing). Nevertheless, it assumes a Gaussian trend of the dMRI signal with the intrinsic limitation of failing the reconstruction of complex WM architectural configurations where diffusion is anisotropic (e.g., fibers’ crossings, kissings, etc.). Recent overcomes of this limitation are represented by multi-shell dMRI and new modelling techniques like the Neurite Orientation Dispersion and Density Imaging (NODDI) [9], though not standard for clinical purposes. NODDI is a compartmental model which assumes that the brain tissue is divided in isotropic, intracellular diffusion, and extracellular microstructural compartments. Vogt et al. [6] showed that the NODDI-derived neurite density index was sensitive to GM modifications in A
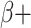 /tau+ CN before the onset of brain atrophy and cognitive impairment. Spotorno et al. [4] too demonstrated the higher potential of GM microstructural alterations in revealing the astrocytic response to A
/tau+ CN before the onset of brain atrophy and cognitive impairment. Spotorno et al. [4] too demonstrated the higher potential of GM microstructural alterations in revealing the astrocytic response to A
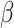 aggregation compared to macrostructural measurements as those derived from T1w-MRI. In particular, they relied on a multi-shell acquisition as in [6], but they employed the Mean Apparent Propagator (MAP)MRI model [10] which, differently from NODDI, does not assume any tissue composition. Indeed, avoiding assumptions on tissue composition is preferable especially in disease state because the underlying biophysical theoretical assumptions at the bases of the compartmental models probably do not hold in such conditions. In line with this consideration, Moody et al. [7] used MAPMRI to detect AD-related early neurodegenerative changes in WM, finding more spatially diffuse associations with A
aggregation compared to macrostructural measurements as those derived from T1w-MRI. In particular, they relied on a multi-shell acquisition as in [6], but they employed the Mean Apparent Propagator (MAP)MRI model [10] which, differently from NODDI, does not assume any tissue composition. Indeed, avoiding assumptions on tissue composition is preferable especially in disease state because the underlying biophysical theoretical assumptions at the bases of the compartmental models probably do not hold in such conditions. In line with this consideration, Moody et al. [7] used MAPMRI to detect AD-related early neurodegenerative changes in WM, finding more spatially diffuse associations with A
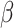 and tau cerebrospinal fluid markers compared to DTI and NODDI.
and tau cerebrospinal fluid markers compared to DTI and NODDI.
In the present study, we further investigated the potential of dMRI in highlighting WM microstructural alterations in the earliest phases of AD with the twofold goal of exploiting typical clinical acquisition protocols while enabling finer microstructural characterization. To this end, we relied on a recently proposed method called Apparent Measures Using Reduced Acquisitions (AMURA) [11], allowing to exploit single-shell acquisition protocols while maintaining the descriptive power of MAPMRI indices under certain conditions. AMURA applied to high b-value acquisitions provides microstructural indices with similar sensitivity compared to MAPMRI-derived ones [11], hence allowing a highly specific microstructural characterization at a finer granularity compared to DTI while bringing DTI-like advantages such as clinical feasibility, i.e. a reduced number of samples and low computational complexity. In this preliminary study, AMURA capability in the characterization of different A
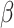 and tau status in the AD continuum was compared with the classical DTI in exquisitely clinical acquisitions (i.e. low b-value) allowing to test its suitability also in such a scenario. Of course, the complete characterization of the method would require to contrast it with the MAPMRI outcomes when relaxing the constraint of single-shell acquisitions. However, this is out of the scope of this contribution which focuses on classical single-shell acquisitions for which DTI is the de-facto benchmark. Though the applied method is not new, in our opinion its application to a hard and open problem at the state of the art also holding high translational potential deserves an in-depth analysis, and the post-hoc assessment of the outcomes would mark a step in the direction of the neurophysiological plausibility of the results, and thus on the relevance and usefulness of the method in the translational perspective highlighting its ability to capturing actual tissue alterations without being invasive.
and tau status in the AD continuum was compared with the classical DTI in exquisitely clinical acquisitions (i.e. low b-value) allowing to test its suitability also in such a scenario. Of course, the complete characterization of the method would require to contrast it with the MAPMRI outcomes when relaxing the constraint of single-shell acquisitions. However, this is out of the scope of this contribution which focuses on classical single-shell acquisitions for which DTI is the de-facto benchmark. Though the applied method is not new, in our opinion its application to a hard and open problem at the state of the art also holding high translational potential deserves an in-depth analysis, and the post-hoc assessment of the outcomes would mark a step in the direction of the neurophysiological plausibility of the results, and thus on the relevance and usefulness of the method in the translational perspective highlighting its ability to capturing actual tissue alterations without being invasive.
In the context of post-hoc assessments, the application of eXplainable Artificial Intelligence (XAI) methods is gaining increasing importance due to the outbreak of AI applied to the biomedical field in the last years. Indeed, XAI has the potential of offering a key for the interpretation and strengthening of the AI-derived results themselves by bringing to light some aspects of the internal thinking of complex models, deep networks on the top of the list, or providing ordered lists of input features leading the algorithm’s outcomes. Many examples can be found in the literature [12], [13], though a real awareness of the intrinsic limitations and related risks of such methods is rarely acknowledged and faced. Otherwise stated, the validation of the XAI method is most often overlooked, that is a serious risk especially in the biomedical field. In this work, in addition to employ AI and XAI to assess the early AD classification when based on different microstructural properties of the tissue, we faced the validation issue by comparing two different interpretability methods and through a more simple framework that is the relying on the prior knowledge derived from the literature. The last is a qualitative validation of the outcomes limited to the neurophysiological plausibility. Other validation strategies like post-hoc association studies as well as the analysis of other attributes of XAI methods are left for future investigation. In particular, in this work we rely on SHAP [14] and LIME [15] methods, that are among the most widespread for their conceptual simplicity and understandability, to derive an ordered list of features, i.e. brain WM tracts, allowing to discriminate across the A
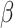 and tau spectra. To the best of our knowledge, only one work which specifically investigated the pre-clinical AD attempted to give an interpretation of the results by exploiting the XAI. In the mentioned study, Hwang et al. [16] classified A
and tau spectra. To the best of our knowledge, only one work which specifically investigated the pre-clinical AD attempted to give an interpretation of the results by exploiting the XAI. In the mentioned study, Hwang et al. [16] classified A
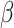 + and A
+ and A
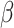 - CN with a deep generative model relying on T1w scans and many other features like demographics and cognitive scores. They subsequently used the integrated gradients XAI method to explain their outcomes. Hence, the main novelty represented by our current investigation consists of the information used at the basis of the classification. Indeed, the dMRI targets the microstructural properties of the tissue instead of the macrostructural ones as the T1w-MRI does. In this respect, this work constitutes a step forward in the comprehension of the mechanisms at the basis of AD, also thanks to the translational power of the XAI that provides reliable explanations of easy interpretation for the clinicians.
- CN with a deep generative model relying on T1w scans and many other features like demographics and cognitive scores. They subsequently used the integrated gradients XAI method to explain their outcomes. Hence, the main novelty represented by our current investigation consists of the information used at the basis of the classification. Indeed, the dMRI targets the microstructural properties of the tissue instead of the macrostructural ones as the T1w-MRI does. In this respect, this work constitutes a step forward in the comprehension of the mechanisms at the basis of AD, also thanks to the translational power of the XAI that provides reliable explanations of easy interpretation for the clinicians.
II. Methodology
A. Microstructural Brain Description Through AMURA
AMURA is an innovative method for studying cerebral microstructure [11]. The innovation is represented by the capability of computing ensemble average propagator (EAP)-based indices such as the return to origin/axis/plane probability (RTOP/RTAP/RTPP) with lower computational complexity and number of diffusion gradients compared to current state-of-the-art approaches like MAPMRI [10]. These three indices represent the probability that protons do not move during the dMRI acquisition, thus reflecting barriers’ restriction and consequently cell bodies’ size measures. In particular, RTAP and RTPP can be considered as the RTOP projections on the perpendicular plan and parallel direction to the maximum diffusion. The simpler computation with AMURA is possible because it treats the diffusion anisotropy as independent on the b-value (i.e., the factor indicating the acquisition diffusion gradients’ strength and timing). Given
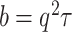 , the normalised signal
, the normalised signal
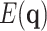 in the q-space can be formalized as:
in the q-space can be formalized as:
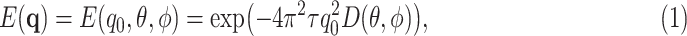 |
where
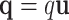 (
(
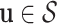 is a unit direction in space, and
is a unit direction in space, and
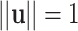 ),
),
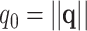 ,
,
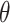 and
and
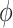 are the angular coordinates in the spherical system,
are the angular coordinates in the spherical system,
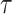 is the diffusion time, and
is the diffusion time, and
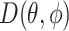 is the apparent diffusion coefficient (ADC) on a single-shell acquisition [17]. In light of the assumptions described above, a numerical implementation of the indices based on spherical harmonics (SH) expansions has been proposed in [11]:
is the apparent diffusion coefficient (ADC) on a single-shell acquisition [17]. In light of the assumptions described above, a numerical implementation of the indices based on spherical harmonics (SH) expansions has been proposed in [11]:
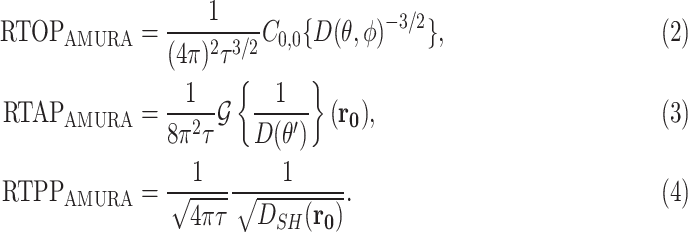 |
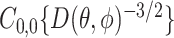 is the
is the
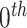 order coefficient of the SH series expansion,
order coefficient of the SH series expansion,
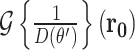 is the Funk-Radon transform of the inverse of the diffusion signal
is the Funk-Radon transform of the inverse of the diffusion signal
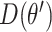 , i.e. the diffusion signal at the equator normal to
, i.e. the diffusion signal at the equator normal to
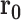 (the direction of maximum diffusion), parameterized by the angle
(the direction of maximum diffusion), parameterized by the angle
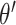 , and
, and
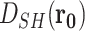 is the SH regularized version of the ADC evaluated at
is the SH regularized version of the ADC evaluated at
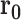 .
.
B. Dataset
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Images were collected from different centers with scanners from GE, Philips, and Siemens vendors. Please refer to https://adni.loni.usc.edu/ for up-to-date information. 442 subjects being CN or MCI were selected from the ADNI phase 3 (ADNI3) database. Although both basic and advanced dMRI acquisitions were performed in this phase of the study consisting of single or multiple shells acquisitions, only single-shell ones were considered for the current study [18].
For each subject, the 3D T1w-MRI volume (sagittal accelerated MPRAGE, TR/TE = shortest, TI =900 ms, FOV
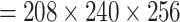 mm3, flip angle =9°, resolution
mm3, flip angle =9°, resolution
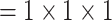 mm3) and the single-shell dMR image (acquisition through 3T MRI scanner, TR/TE =7200/56 ms, FOV
mm3) and the single-shell dMR image (acquisition through 3T MRI scanner, TR/TE =7200/56 ms, FOV
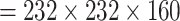 mm3, resolution
mm3, resolution
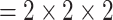 mm3,
mm3,
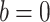 and 1000 s/mm2, diffusion time
and 1000 s/mm2, diffusion time
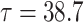 ms) were collected along with concentration values of A
ms) were collected along with concentration values of A
 and tau protein in the cerebrospinal fluid. These concentration values were used to stratify the cohort into 3 classes. In particular, subjects were classified as A
and tau protein in the cerebrospinal fluid. These concentration values were used to stratify the cohort into 3 classes. In particular, subjects were classified as A
 if [A
if [A
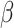 -protein
-protein
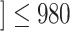 pg/mL, and tau+ if [tau-protein
pg/mL, and tau+ if [tau-protein
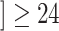 pg/mL (see Table 1).
pg/mL (see Table 1).
TABLE 1. Demographic Summary of the Study Cohort.
 tau− tau− |
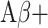 tau− tau− |
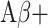 tau+ tau+ |
|
|---|---|---|---|
| N (%female) | 168 (58.3%) | 128 (44.5%) | 146 (50%) |
| Age |
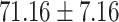 |
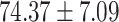 |
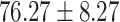 |
C. Preprocessing
A minimal preprocessing including the bias-field correction and the linear registration to the 2-mm MNI space was applied to T1w images by employing the fsl_anat tool (FSL, version 6.0, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) [19].
The dMRI data were preprocessed by extracting the brain and performing Eddy currents correction still using FSL [20]. Subsequently, data were denoised using the Python dipy library (https://dipy.org/) to apply a principal component analysis (PCA)-based denoising algorithm with automatic PCs classification grounding on the Marcenko-Pastur distribution [21] (the radius of the 3D sliding window was set equal to 2). The b0 volumes were averaged and registered to the T1w image of the subject through the epi_reg routine in FSL [22], and then linearly registered to the MNI space by applying the transformation obtained through fsl_anat. The same linear transformations applied to the average b0 were also applied to all the other volumes of the dMR image, and the result obtained was further non-linearly registered to the MNI space through ANTs software (http://stnava.github.io/ANTs/) [23] to correct for EPI-induced currents [24]. The dMRI gradients’ direction was rotated accordingly.
D. Microstructural Indices Extraction
RTOP/RTAP/RTPP
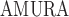 microstructural descriptors were derived following the numerical implementation described in Section II-A. A SH spherical order of 6 and a cost parameter for the Laplace-Beltrami regularization of
microstructural descriptors were derived following the numerical implementation described in Section II-A. A SH spherical order of 6 and a cost parameter for the Laplace-Beltrami regularization of
 were selected for the computations. In addition, relying on the dipy library, the diffusion tensor model [8] was used to obtain DTI-based versions of RTOP/RTAP/RTPP as follows [11]:
were selected for the computations. In addition, relying on the dipy library, the diffusion tensor model [8] was used to obtain DTI-based versions of RTOP/RTAP/RTPP as follows [11]:
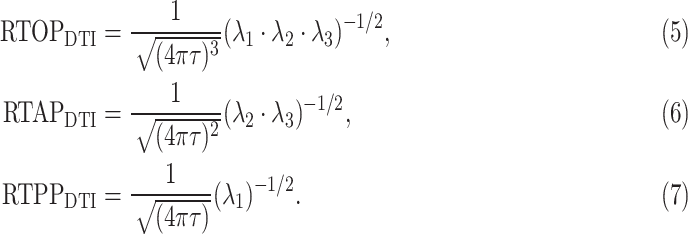 |
Specifically,
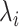 is the
is the
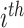 eigenvalue of the diffusion tensor. Standard Mean Diffusivity (MD) and Fractional Anisotropy (FA) microstructural descriptors were also derived [8].
eigenvalue of the diffusion tensor. Standard Mean Diffusivity (MD) and Fractional Anisotropy (FA) microstructural descriptors were also derived [8].
E. Tract-Based Spatial Statistics
The tract-based spatial statistics (TBSS) pipeline from FSL was performed on the aforementioned FA images. More in detail, all images were registered to the FA image of the JHU DTI-based WM atlas [25] through a non-linear transformation, and the resulting WM skeleton was obtained using a threshold of 0.2 on the calculated average volume. The same obtained registrations were subsequently applied to the images of all microstructural indices. The pipeline ended with a two-sample unpaired t-test performed through the FSL tool randomise
[26]. Both contrasts (CN > patients, and patients > CN) were investigated for each index by comparing A
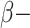 /tau− with A
/tau− with A
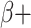 /tau−, and A
/tau−, and A
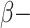 /tau− with A
/tau− with A
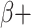 /tau+. For each test, the number of permutations performed was 1000. Images representing the threshold-free cluster enhanced p-value corrected for multiple comparisons across space were obtained.
/tau+. For each test, the number of permutations performed was 1000. Images representing the threshold-free cluster enhanced p-value corrected for multiple comparisons across space were obtained.
F. Support Vector Machine-Based Classification
A
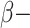 /tau− with A
/tau− with A
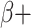 /tau−, and A
/tau−, and A
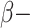 /tau− with A
/tau− with A
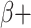 /tau+ were further investigated by performing their classifications through Support Vector Machines (SVMs) relying on Scikit-Learn library (https://scikit-learn.org/stable/) in Python. SVMs are notoriously able to perform classification tasks in a very versatile way (e.g., linearly or not linearly) [27], and they are commonly used for biomedical applications with good performance [28], [29]. Besides, a deep learning strategy was not affordable in this work because of the limited number of subjects available.
/tau+ were further investigated by performing their classifications through Support Vector Machines (SVMs) relying on Scikit-Learn library (https://scikit-learn.org/stable/) in Python. SVMs are notoriously able to perform classification tasks in a very versatile way (e.g., linearly or not linearly) [27], and they are commonly used for biomedical applications with good performance [28], [29]. Besides, a deep learning strategy was not affordable in this work because of the limited number of subjects available.
Considering only voxels belonging to the WM skeleton, for all subjects, the average value of each index was extracted from 48 Regions of Interest (ROIs) (i.e., WM tracts) based on the JHU DTI-based WM atlas previously introduced [25]. Thus, for each of the two classification tasks, we trained and validated 8 SVMs (one per microstructural index), each one based on a different initial data matrix of dimensions
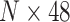 . N was the total number of subjects depending on the two classes of the classification task to handle, while 48 was the number of WM tracts.
. N was the total number of subjects depending on the two classes of the classification task to handle, while 48 was the number of WM tracts.
The optimal hyperparameters were found through an exhaustive search performed with a cross-validation strategy over all possible kernels and [0.01, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 10, 20] regularization parameters C, resulting in the linear kernel and
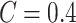 . The final classification tasks were carried out with a stratified 10-folds cross-validation, and the performance was assessed by calculating the mean accuracy, precision, sensitivity and specificity over the folds. Due to the limited cohort numerosity, we did not retain also a test set in addition to those used for training and validating the model.
. The final classification tasks were carried out with a stratified 10-folds cross-validation, and the performance was assessed by calculating the mean accuracy, precision, sensitivity and specificity over the folds. Due to the limited cohort numerosity, we did not retain also a test set in addition to those used for training and validating the model.
G. Explainable Artificial Intelligence Analysis
SHAP and LIME were used for identifying the features that contributed most to the SVM outcome. To this end, we focused on the classification task generally obtaining the best performance based on validation accuracy in order to maximise the generalizability, reliability and robustness of the results [30]. Indeed, poor classification performance would be an indication of the difficulty of the model in discriminating the classes relying on the available features, casting shadows on the actual relevance of the SHAP/LIME attribution values.
Given the lack of an independent test set, for each microstructural index, the fold obtaining the best accuracy among the ten was selected in order to maximize both predictive and descriptive accuracy [30]. The former is the classifier’s accuracy, while the latter is the objective capability of the interpretability method to capture the relationships learned by the classifier itself. Both predictive and descriptive accuracies should be high to obtain a trustworthy explanation, but the former constrains the latter. For this reason such a selection was done as in [31], [32], [33], and [34]. Thus, the SHAP and LIME values indicating the relevance of each feature to the classification of every subject in the validation set were calculated.
1). Shapley Additive Explanations
SHapley Additive exPlanation (SHAP) [14] is a model-agnostic and perturbation-based method for estimating the input feature importance. Basically, it is a method from coalitional game theory where a prediction is explained by assuming that each feature is a “player” in a game where the prediction is the payout. The SHAP value of a feature is calculated as the average marginal contribution of that feature across all possible coalitions. Calculating Shapley feature importance values thus becomes computationally expensive for complex models and high number of features. However, this is not the case for the problem at hand where both the number of features and data samples (subjects) is limited. SHAP has demonstrated its efficacy in the medical domain to explain clinical decision-making both from image [35], [36], [37] and non-image [38], [39], [40], [41] inputs.
In this work, the SHAP library (https://github.com/shap/ shap) in Python was used with a kernel explainer using a weighted linear regression to compute the importance of each feature. For each index and feature, the mean SHAP value over the validation set was derived.
2). Local Interpretable Model-Agnostic Explanations
The Local Interpretable Model-agnostic Explanations (LIME) [15] is a model-agnostic method based on perturbation like SHAP, with the main difference of focusing on explaining individual predictions instead of providing a global interpretation based on the whole dataset. Given an individual prediction, the approach starts with the creation of a fictitious dataset produced by perturbing the corresponding input features within a proximity usually defined by an exponential kernel based on the Euclidian distance. The local fidelity is ensured by assigning to each new data point a weight that is the higher the closer it is to the original one. The artificial dataset is thus used to train an interpretable surrogate simple model like the linear regression instead of the original complex one. With reference to the linear model, the relevance of each feature on the initial individual prediction is thus defined by the coefficients found by solving the fitting. Together with SHAP, LIME is the most commonly used method to evaluate the impact of every single feature to a AI-derived result [42]. In this work, we used the Python implementation of LIME (https://github.com/marcotcr/lime) for tabular data. The surrogate model chosen was the linear regression. Despite the local nature of LIME, a global explanation for each index was provided by finding the LIME values for each subject of the validation set and thus calculating the average across all of them [43].
III. Results
A. Qualitative Assessment
The maps for each dMRI index for a representative subject of each group (i.e., A
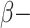 /tau−, A
/tau−, A
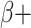 /tau−, and A
/tau−, and A
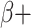 /tau+) are shown in Fig. 1. The cubic-root of RTOP, and the square-root of RTAP were calculated and reported to easily compare the three restriction indices (i.e., RTOP, RTAP, and RTPP).
/tau+) are shown in Fig. 1. The cubic-root of RTOP, and the square-root of RTAP were calculated and reported to easily compare the three restriction indices (i.e., RTOP, RTAP, and RTPP).
FIGURE 1.
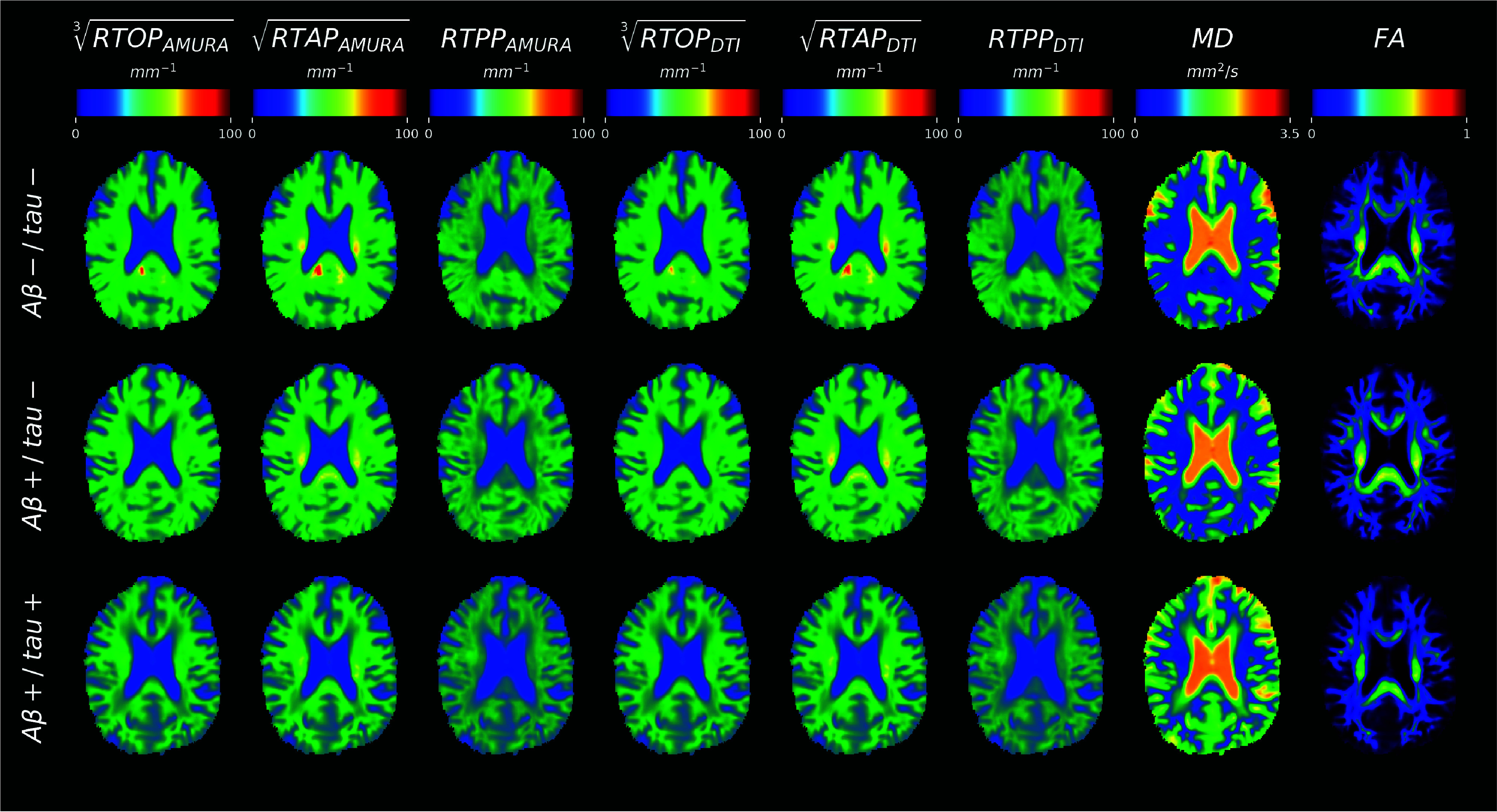
Microstructural maps of values of one representative subject for A
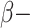 /tau−, A
/tau−, A
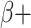 /tau−, and A
/tau−, and A
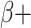 /tau+ groups (rows), respectively. The representative axial slices reported are in MNI space and are displayed in radiological convention. RTOP = return to the origin probability; RTAP = return to the axis probability; RTPP = return to the plane probability; MD = mean diffusivity; FA = fractional anisotropy.
/tau+ groups (rows), respectively. The representative axial slices reported are in MNI space and are displayed in radiological convention. RTOP = return to the origin probability; RTAP = return to the axis probability; RTPP = return to the plane probability; MD = mean diffusivity; FA = fractional anisotropy.
As expected, RTOP/RTAP/RTPP
 had similar contrast to FA, appearing hyperintense in regions where diffusion takes place preferentially along a single direction (e.g., corpus callosum). MD showed an opposite trend, reaching the highest values where diffusion is unrestricted. No evident differences across groups could be appreciated by qualitative assessment.
had similar contrast to FA, appearing hyperintense in regions where diffusion takes place preferentially along a single direction (e.g., corpus callosum). MD showed an opposite trend, reaching the highest values where diffusion is unrestricted. No evident differences across groups could be appreciated by qualitative assessment.
B. TBSS Analysis
The TBSS analysis results are shown in Fig. 2. The significant voxels for each of the considered indices are overlaid to the JHU-FA atlas. Results unveiled widespread statistically significant differences corrected for multiple comparisons (p-value
 ) between A
) between A
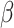 -/tau- and both A
-/tau- and both A
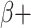 /tau− and A
/tau− and A
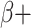 /tau+ groups for all indices. More in detail, RTOP/RTAP/RTPP
/tau+ groups for all indices. More in detail, RTOP/RTAP/RTPP
 and FA exhibited these significant differences only in the contrast A
and FA exhibited these significant differences only in the contrast A
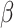 -/tau- > A
-/tau- > A
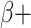 /tau− or A
/tau− or A
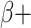 /tau+, while MD index displayed significance only in the opposite contrast.
/tau+, while MD index displayed significance only in the opposite contrast.
FIGURE 2.
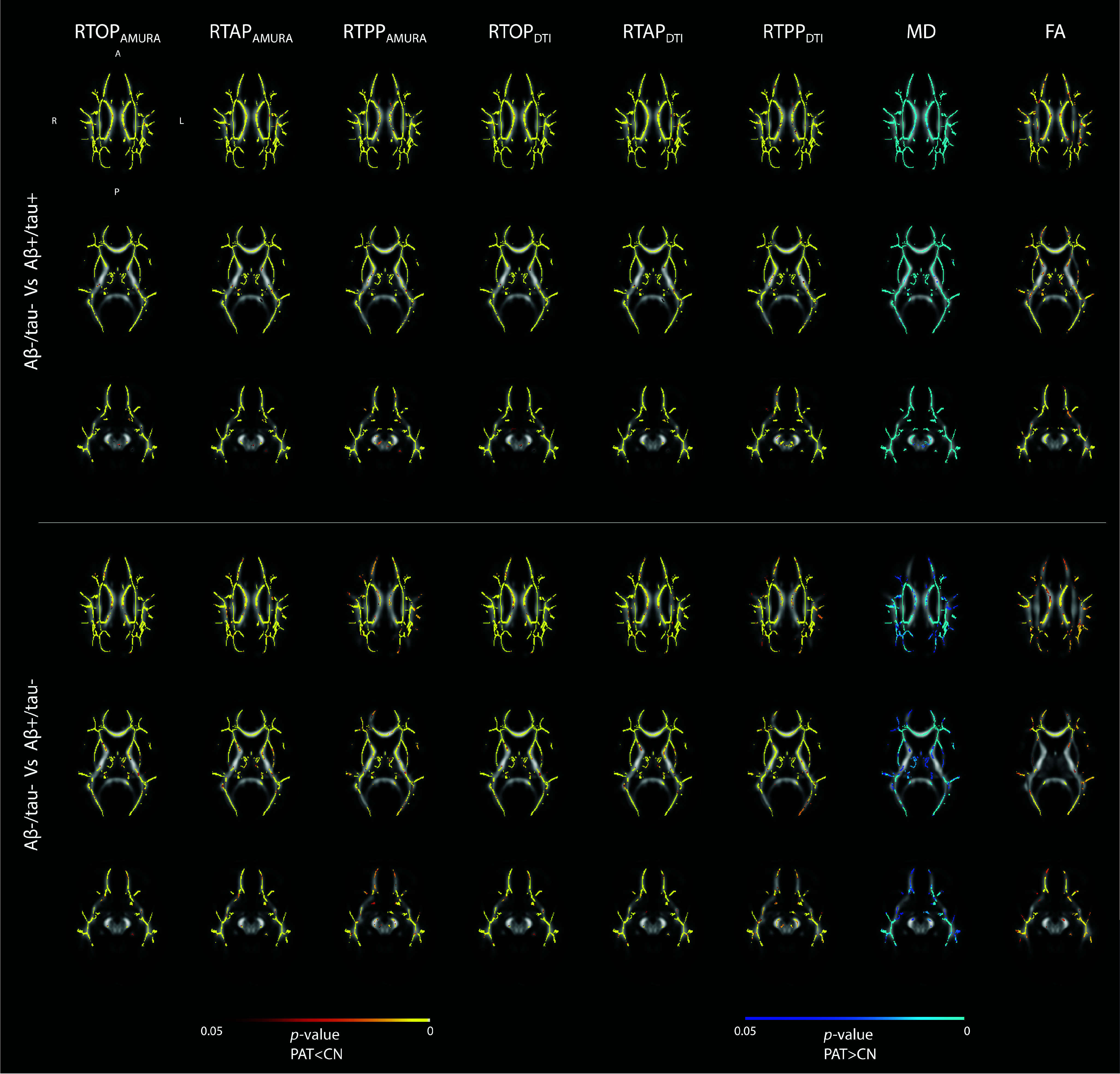
Tract-based Spatial Statistics (TBSS) results. The significant differences for both the contrasts A
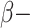 /tau− versus A
/tau− versus A
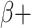 /tau+ and A
/tau+ and A
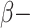 /tau− versus A
/tau− versus A
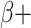 /tau− are shown for the eight diffusion indices (columns). Significant voxels are superimposed on the FA image of the JHU DTI-based WM atlas. Red-yellow and blue-lightblue colormaps indicate statistically significant clusters corrected for multiple comparisons in the CN > patients and CN < patients contrasts, respectively (significant p-value =0.05).
/tau− are shown for the eight diffusion indices (columns). Significant voxels are superimposed on the FA image of the JHU DTI-based WM atlas. Red-yellow and blue-lightblue colormaps indicate statistically significant clusters corrected for multiple comparisons in the CN > patients and CN < patients contrasts, respectively (significant p-value =0.05).
C. SVM Classification Performance
The SVMs performance is illustrated in Fig. 3, showcasing the mean and standard deviation of the measurements across the ten folds. The most challenging task involved discriminating A
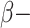 /tau− from A
/tau− from A
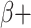 /tau− subjects. MD emerged as the best feature for distinguishing between these two classes, even though with a performance similar to that showed by AMURA and DTI. In particular, it resulted in an accuracy of 0.619. In this classification, the least effective performance was observed in the FA-based classification (accuracy =0.534). As anticipated, superior results were achieved in the A
/tau− subjects. MD emerged as the best feature for distinguishing between these two classes, even though with a performance similar to that showed by AMURA and DTI. In particular, it resulted in an accuracy of 0.619. In this classification, the least effective performance was observed in the FA-based classification (accuracy =0.534). As anticipated, superior results were achieved in the A
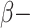 /tau− and A
/tau− and A
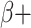 /tau+ condition. Specifically,
/tau+ condition. Specifically,
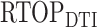 outperformed others, with an accuracy of 0.729, closely followed by
outperformed others, with an accuracy of 0.729, closely followed by
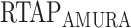 with an accuracy of
with an accuracy of
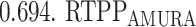 demonstrated the poorest performance (accuracy =0.618). In both cases, the model was affected by a tendency toward imbalanced classification, sometimes labeling A
demonstrated the poorest performance (accuracy =0.618). In both cases, the model was affected by a tendency toward imbalanced classification, sometimes labeling A
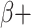 /tau+ or A
/tau+ or A
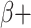 /tau- as A
/tau- as A
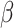 -/tau-. This behavior is emphasized by the high specificity and relatively low sensitivity observed across the ten folds.
-/tau-. This behavior is emphasized by the high specificity and relatively low sensitivity observed across the ten folds.
FIGURE 3.
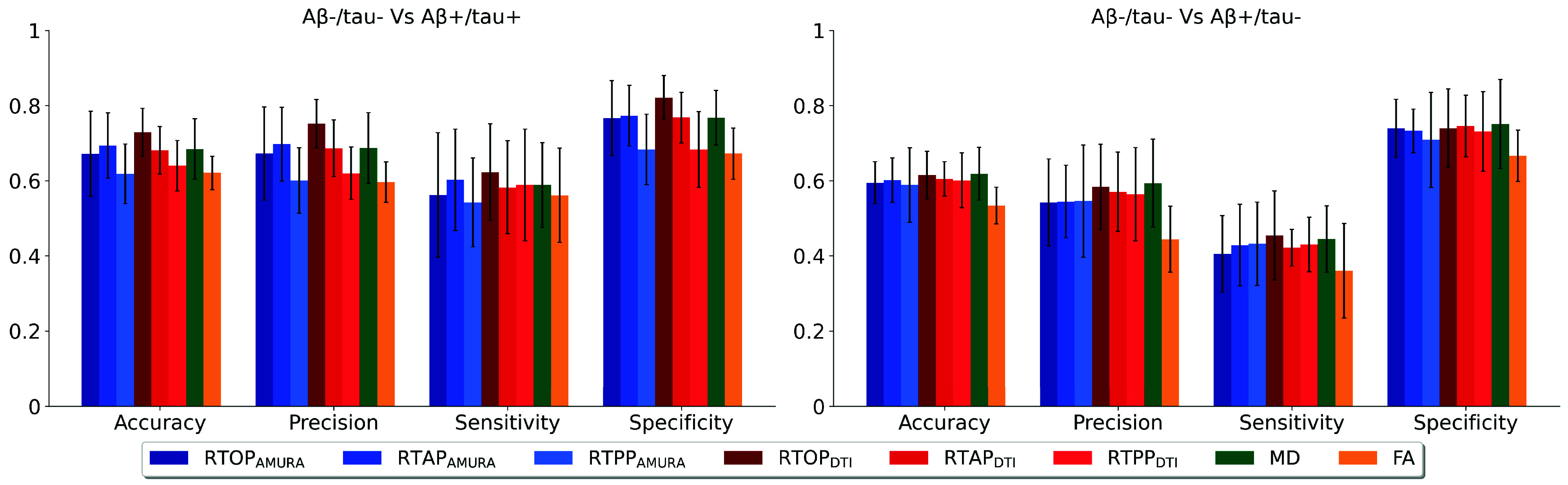
Mean accuracy, precision, sensitivity, and specificity, along with the relative standard deviation, across the 10 folds of the cross-validation step.
D. XAI-Based Post-Hoc Assessment
Fig. 4 represents, for each dMRI index, the top five features found by SHAP and LIME mostly contributing to the classification task that reached the best performance (i.e., A
 /tau− versus A
/tau− versus A
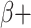 /tau+). As evident, the findings from both the XAI methods are in agreement because at least four among the top five most impactful features are the same across the two approaches for each dMRI index. Only the sorting can vary slightly, and anyway it is preserved for the top two features except for RTPP. Of note, the left cingulum connecting hippocampus appeared as the most important WM tract to consider for distinguishing subjects with amyloid/tau positivity from negative ones. Indeed, in this WM tract, all microstructural indices except RTPP (in both AMURA and DTI versions) showed the highest SHAP value reflecting such a relevance. Instead, the discrepancy with RTPP was in agreement with its derived classification performance, which emerged as the worst. In addition, more generally, respectively RTOP, RTAP, and RTPP, demonstrated a high correspondence between the AMURA and DTI versions, often highlighting the same WM tracts with similar SHAP and LIME values.
/tau+). As evident, the findings from both the XAI methods are in agreement because at least four among the top five most impactful features are the same across the two approaches for each dMRI index. Only the sorting can vary slightly, and anyway it is preserved for the top two features except for RTPP. Of note, the left cingulum connecting hippocampus appeared as the most important WM tract to consider for distinguishing subjects with amyloid/tau positivity from negative ones. Indeed, in this WM tract, all microstructural indices except RTPP (in both AMURA and DTI versions) showed the highest SHAP value reflecting such a relevance. Instead, the discrepancy with RTPP was in agreement with its derived classification performance, which emerged as the worst. In addition, more generally, respectively RTOP, RTAP, and RTPP, demonstrated a high correspondence between the AMURA and DTI versions, often highlighting the same WM tracts with similar SHAP and LIME values.
FIGURE 4.
Mean SHAP (top) and LIME (bottom) value over the validation set for each feature and index-derived SVM classification; Abbreviations: left cingulum connecting the cingulate gyrus (CG (cg) L), left/right cingulum connecting the hippocampus (CG (hi) L/R), right external capsule (EC R), column and body of fornix (FX), genu of corpus callosum (GCC), right inferior cerebellar peduncle (ICP R), left medial lemniscus (ML L), right posterior corona radiata (PCR R), right posterior thalamic radiation including optic radiation (PTR R), left/right retrolenticular part of internal capsule (RIC L/R), splenium of corpus callosum (SCC), right superior cerebellar peduncle (SCP R), left/right superior corona radiata (SCR L/R), left superior fronto-occipital fasciculus that could be a part of anterior internal capsule (SFF (aic) L), right superior longitudinal fasciculus (SLF R), left tapetum (TP L), right anterior corona radiata (ACR R), right anterior limb of internal capsule (AIC R), left inferior cerebellar peduncle (ICP L).
IV. Discussion
In this study, for the first time, we revealed the potential of RTOP/RTAP/RTPP
 as imaging markers for early AD detection by exploiting their WM characterization in subjects with amyloid and possibly tau pathology compared to subject without such a pathology. From a medical point of view, this could represent a further step toward a possible screening at a pre-clinical level without the need of more invasive methods. Pursuing this aim, we took advantage of both classical statistical (i.e., TBSS) and machine learning techniques (i.e., SVMs), using the well-established DTI-based indices as benchmark. Moreover, the SHAP and LIME XAI methods were employed to identify the features most contributing to the SVMs outcomes, enabling the translational value of the present work and identifying the cingulum WM tract as possible target for future clinical research trials. The usage of the two different XAI approaches served for comparing the outcomes and thus testing their reliability. However, the results were additionally critically analysed with respect to the literature to assess their plausibility.
as imaging markers for early AD detection by exploiting their WM characterization in subjects with amyloid and possibly tau pathology compared to subject without such a pathology. From a medical point of view, this could represent a further step toward a possible screening at a pre-clinical level without the need of more invasive methods. Pursuing this aim, we took advantage of both classical statistical (i.e., TBSS) and machine learning techniques (i.e., SVMs), using the well-established DTI-based indices as benchmark. Moreover, the SHAP and LIME XAI methods were employed to identify the features most contributing to the SVMs outcomes, enabling the translational value of the present work and identifying the cingulum WM tract as possible target for future clinical research trials. The usage of the two different XAI approaches served for comparing the outcomes and thus testing their reliability. However, the results were additionally critically analysed with respect to the literature to assess their plausibility.
Classical statistical analysis performed through TBSS further suggested the dMRI derived indices as possible imaging markers of microstructural degeneration from the earliest phases of AD. Indeed, widespread statistically significant differences between A
 -/tau- and A
-/tau- and A
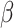 +/tau- or A
+/tau- or A
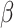 +/tau+ surviving the correction for multiple comparisons were found. In addition, the contrast of such significance evidenced a lower anisotropy and restriction along with a higher diffusivity in A
+/tau+ surviving the correction for multiple comparisons were found. In addition, the contrast of such significance evidenced a lower anisotropy and restriction along with a higher diffusivity in A
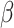 +/tau- or A
+/tau- or A
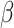 +/tau+ compared to A
+/tau+ compared to A
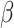 -/tau-, compatible with a clinical picture of neurodegeneration and inline with literature [5].
-/tau-, compatible with a clinical picture of neurodegeneration and inline with literature [5].
In this study, alongside statistical analysis at the population level, we used AI to diagnose the pathology at the subject level. The A
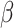 /tau detection task is particularly complex because the mechanisms that lead to the development of AD are still unknown. Despite this, we anyway chose to use a relatively simple machine learning model, focusing instead on the type of information used for classification (i.e., microstructural information). Indeed, the centrality of the role played by the goodness of the chosen features was made evident by the fact that even with a less complex model than the deep generative one used in [16] (i.e., HexaGAN), our SVMs were able to achieve a similar performance to that obtained by Hwang et al. when based on T1w-MRI alone (i.e., macrostructural information). Hwang and colleagues [16] were able to achieve a superior performance only at the cost of more input data than just the images.
/tau detection task is particularly complex because the mechanisms that lead to the development of AD are still unknown. Despite this, we anyway chose to use a relatively simple machine learning model, focusing instead on the type of information used for classification (i.e., microstructural information). Indeed, the centrality of the role played by the goodness of the chosen features was made evident by the fact that even with a less complex model than the deep generative one used in [16] (i.e., HexaGAN), our SVMs were able to achieve a similar performance to that obtained by Hwang et al. when based on T1w-MRI alone (i.e., macrostructural information). Hwang and colleagues [16] were able to achieve a superior performance only at the cost of more input data than just the images.
SVM results interestingly revealed MD as the index leading to the highest accuracy when used to distinguish A
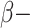 /tau− versus A
/tau− versus A
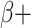 /tau− subjects, suggesting it as possible imaging marker of A
/tau− subjects, suggesting it as possible imaging marker of A
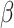 irrespectively from the tau concentration.
irrespectively from the tau concentration.
Of note, a similar finding, although in gray matter (GM), was reported also by Spotorno et al. [4] employing the MAPMRI-derived mean squared displacement (MSD) index in GM. The MSD can be considered as the Ensemle Average Propagator (EAP)-based version of MD since both represent the average amount of diffusion in the unit time and, consequently, holds sensitivity to a lower or higher restriction [4], [44]. In [4], such a measure in GM was found to be correlated with many other markers of amyloid and tau pathology, but in particular the association with A
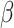 -PET and glial fibrillary acidic protein suggested its relationship to the astrocytic response to A
-PET and glial fibrillary acidic protein suggested its relationship to the astrocytic response to A
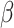 aggregation.
aggregation.
However, in the present study, MD-related mean accuracy was lower compared to that of other indices in the classification of A
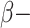 /tau− versus A
/tau− versus A
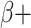 /tau+ subjects (accuracy = 0.685). More specifically,
/tau+ subjects (accuracy = 0.685). More specifically,
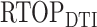 and
and
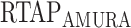 appear to be superior in performance, although not at the level of statistical significance, suggesting their sensitivity to the tau pathology onset (accuracy = 0.729 and 0.694, respectively). Also Chen et al. [3] observed that altered WM, as highlighted by their results using FA and MD, may reflect tau presence. In addition, they found a correlation with tau but not with A
appear to be superior in performance, although not at the level of statistical significance, suggesting their sensitivity to the tau pathology onset (accuracy = 0.729 and 0.694, respectively). Also Chen et al. [3] observed that altered WM, as highlighted by their results using FA and MD, may reflect tau presence. In addition, they found a correlation with tau but not with A
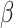 presence enforcing that finding. The present study provides additional evidence to such a hypothesis. All these findings witness in favor of designating indices like
presence enforcing that finding. The present study provides additional evidence to such a hypothesis. All these findings witness in favor of designating indices like
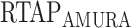 as possible better marker compared to the more standard FA and MD, inline with other results from Moody et al. [7], though these were obtained relying on MAPMRI on a multi-shell acquisition at higher b-values. Hence, according to our results, AMURA allows capturing fine microstructural modulations with a sensitivity comparable to MAPMRI but with data acquisitions requiring a lower number of samples.
as possible better marker compared to the more standard FA and MD, inline with other results from Moody et al. [7], though these were obtained relying on MAPMRI on a multi-shell acquisition at higher b-values. Hence, according to our results, AMURA allows capturing fine microstructural modulations with a sensitivity comparable to MAPMRI but with data acquisitions requiring a lower number of samples.
In this study, XAI aided the decription of the SVM results, demonstrating one time more its important role when artificial intelligence is applied to medicine. Several works already showed the need of this tool, especially in pathology [45]. In the present work, the employment of XAI enabled the discovery of the cingulum WM tract as the most relevant to possibly detect early AD stage subjects. The validation of such a finding consists of its recovery as top feature through both SHAP and LIME, despite the substantial different principles at the basis of the two methods. In particular, by showing that different XAI methods led to the same interpretation of the results, we provided evidence of the stability of the explanations for each microstructural index. On the other hand, the importance of the features also depends on the ML model because of the peculiar assumptions at the basis of the model (e.g., linear relationships rather than others), algorithmic constraints (e.g., presence or absence of regularizations), etc. By studying the explanation that would be obtained by using a ML model different from SVM, it would be defined its so-called consistency [46]. Nevertheless, as also Molnar [46] observed, such an explanation property is controversial. Indeed, even though the algorithmic independence would reflect the robustness of the ranking and should be reached in ideal conditions, a direct comparison of the explanations across models should take into account the model’s complexity with respect to the numerosity of the samples, the impact of the different architectures, the sensitivity to noise and other factors affecting the performance in real conditions when the data is limited and noisy. For this reason, other architectures will be considered in future works while this work aimed at providing a framework ending in the explanation of the most immediate understanding for health and medicine screening applications.
In such a context, we further confirmed the impact of the cingulum by looking for studies in literature which emphasized its role. Microstructural alterations in this tract were found in MCI and individuals at genetic risk or family predisposition for AD [5]. It was also found significantly altered when specifically investigated in subjects with pathological levels of tau presence compared to CN [47]. Very recently, also Chen et al. [3] reported the central role of the cingulum tract when investigating tau pathology. More in detail, in addition to microstructural alterations, they found a significant correlation of these changes with tau burden in the AD continuum. Interestingly, the cingulum is one of the WM tracts most implicated in episodic memory function, that is known to be tipycally impaired in AD. This can be considered as an added form of validation of SHAP and LIME outcomes through literature-based plausibility assessment.
Future works will include other objective assessment methods such as association studies and analyses of the impact of features collinearity. The former are intended to characterize the biological differences in terms of microstructure as depicted by FA, MD, etc. and other terms like functional connectivity; emerging studies in this direction are [32] and [34]. The latter, instead, are aimed at addressing the possible bias on the models’ performance due to the collinearity potentially present in datasets with a high number of features; for example, there exist methods specifically tailored for SHAP [48], [49] or proxies like the modified informative position and the normalized movement ratio formalized by Salih et al. [50], [51] that can be used to quantify the robustness of the feature importance provided by XAI methods with sensitivity with respect to the presence of collinearity.
Concerning the investigation of AMURA model’s potential, the trend’s proximity of its derived indices to their DTI counterparts confirmed the robustness of their characterization also in amyloid/tau pathological tissue. However, additional investigations using data acquired with higher b-value but still clinically feasible number of samples would be required to fully exploit this model. The expectation is to derive indices better approximating those based on the EAP (e.g., MAPMRI-like) with well-known greater sensitivity compared to the ones based on DTI [11]. Moreover, following [17], a future work could include other AMURA indices like the moment-based representations of the diffusion process in brain tissues.
V. Conclusion
This study investigated for the first time AMURA in the characterization of amyloid and tau pathology in AD, revealing their potential as imaging markers for a timely diagnosis relying on SVM classification and XAI-based interpretation of the outcomes. In a translational perspective, findings highly suggest for future clinical works focusing on cingulum WM tract analysed through non-invasive dMRI data acquired with high b-value but still reduced protocol as enabled by AMURA.
Acknowledgment
Dr. Mauro Zucchelli is an employee of Olea-Medical (Research and Development Team). All other authors have no conflicts of interest. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd., and its affiliated company Genentech Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research and Development LLC.; Johnson & Johnson Pharmaceutical Research and Development LLC.; Lumosity; Lundbeck; Merck & Company Inc.; Meso Scale Diagnostics LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education; and the study is coordinated by the Alzheimer’s Therapeutic Research Institute, University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging, University of Southern California.
Open Access provided by 'Università degli Studi di Verona' within the CRUI CARE Agreement
Funding Statement
This work was supported in part by the Fondazione CariVerona (Bando Ricerca Scientifica di Eccellenza 2018, “EDIPO: A computational solution for bringing neuroimaging genetic into translational research” project - reference number 2018.0855.2019), and in part by the Ministero dell’Istruzione e del Merito (MIUR D.M. 737/2021, “AI4Health: empowering neurosciences with eXplainable AI methods” project).
References
- [1].Ballard C., Gauthier S., Corbett A., Brayne C., Aarsland D., and Jones E., “Alzheimer's disease,” Lancet, vol. 377, no. 9770, pp. 1019–1031, Mar. 2011, doi: 10.1016/s0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- [2].Frisoni G. B., et al. , “The probabilistic model of Alzheimer disease: The amyloid hypothesis revised,” Nature Rev. Neurosci., vol. 23, no. 1, pp. 53–66, Jan. 2022, doi: 10.1038/s41583-021-00533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen Q., Abrigo J., Deng M., Shi L., Wang Y.-X., and Chu W. C. W., “Diffusion changes in hippocampal cingulum in early biologically defined Alzheimer's disease,” J. Alzheimer's Disease, vol. 91, no. 3, pp. 1007–1017, Jan. 2023, doi: 10.3233/jad-220671. [DOI] [PubMed] [Google Scholar]
- [4].Spotorno N., Strandberg O., Vis G., Stomrud E., Nilsson M., and Hansson O., “Measures of cortical microstructure are linked to amyloid pathology in Alzheimer's disease,” Brain, vol. 146, no. 4, pp. 1602–1614, Apr. 2023, doi: 10.1093/brain/awac343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alm K. H. and Bakker A., “Relationships between diffusion tensor imaging and cerebrospinal fluid metrics in early stages of the Alzheimer's disease continuum,” J. Alzheimer's Disease, vol. 70, no. 4, pp. 965–981, Aug. 2019, doi: 10.3233/jad-181210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vogt N. M., et al. , “Interaction of amyloid and tau on cortical microstructure in cognitively unimpaired adults,” Alzheimer's Dementia, vol. 18, no. 1, pp. 65–76, Jan. 2022, doi: 10.1002/alz.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Moody J. F., et al. , “Associations between diffusion MRI microstructure and cerebrospinal fluid markers of Alzheimer's disease pathology and neurodegeneration along the Alzheimer's disease continuum,” Alzheimer's Dementia: Diagnosis, Assessment Disease Monitor., vol. 14, no. 1, p. e12381, Jan. 2022, doi: 10.1002/dad2.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Basser P. J. and Pierpaoli C., “Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI,” J. Magn. Reson., Ser. B, vol. 111, no. 3, pp. 209–219, Jun. 1996, doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- [9].Zhang H., Schneider T., Wheeler-Kingshott C. A., and Alexander D. C., “NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain,” NeuroImage, vol. 61, no. 4, pp. 1000–1016, Jul. 2012, doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- [10].Özarslan E., et al. , “Mean apparent propagator (MAP) MRI: A novel diffusion imaging method for mapping tissue microstructure,” NeuroImage, vol. 78, pp. 16–32, Sep. 2013, doi: 10.1016/j.neuroimage.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Aja-Fernández S., de Luis-García R., Afzali M., Molendowska M., Pieciak T., and Tristán-Vega A., “Micro-structure diffusion scalar measures from reduced MRI acquisitions,” PLoS ONE, vol. 15, no. 3, Mar. 2020, Art. no. e0229526, doi: 10.1371/journal.pone.0229526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Loh H. W., Ooi C. P., Seoni S., Barua P. D., Molinari F., and Acharya U. R., “Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022),” Comput. Methods Programs Biomed., vol. 226, Nov. 2022, Art. no. 107161, doi: 10.1016/j.cmpb.2022.107161. [DOI] [PubMed] [Google Scholar]
- [13].van der Velden B. H. M., Kuijf H. J., Gilhuijs K. G. A., and Viergever M. A., “Explainable artificial intelligence (XAI) in deep learning-based medical image analysis,” Med. Image Anal., vol. 79, Jul. 2022, Art. no. 102470, doi: 10.1016/j.media.2022.102470. [DOI] [PubMed] [Google Scholar]
- [14].Shapley L. S., A Value for N-Person Games. Princeton, NJ, USA: Princeton Univ. Press, 1953, pp. 307–318, doi: 10.1515/9781400881970-018. [DOI] [Google Scholar]
- [15].Ribeiro M. T., Singh S., and Guestrin C., “`Why should I trust you?': Explaining the predictions of any classifier,” in Proc. 22nd ACM SIGKDD Int. Conf. Knowl. Discovery Data Mining, New York, NY, USA, 2016, pp. 1135–1144, doi: 10.1145/2939672.2939778. [DOI] [Google Scholar]
- [16].Hwang U., et al. , “Real-world prediction of preclinical Alzheimer's disease with a deep generative model,” Artif. Intell. Med., vol. 144, Oct. 2023, Art. no. 102654, doi: 10.1016/j.artmed.2023.102654. [DOI] [PubMed] [Google Scholar]
- [17].Aja-Fernández S., Pieciak T., Martín-Martín C., Planchuelo-Gómez Á., de Luis-García R., and Tristán-Vega A., “Moment-based representation of the diffusion inside the brain from reduced DMRI acquisitions: Generalized AMURA,” Med. Image Anal., vol. 77, Apr. 2022, Art. no. 102356, doi: 10.1016/j.media.2022.102356. [DOI] [PubMed] [Google Scholar]
- [18].Zavaliangos-Petropulu A., et al. , “Diffusion MRI indices and their relation to cognitive impairment in brain aging: The updated multi-protocol approach in ADNI3,” Frontiers Neuroinform., vol. 13, p. 2, Feb. 2019, doi: 10.3389/fninf.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jenkinson M., Beckmann C. F., Behrens T. E., Woolrich M. W., and Smith S. M., “FSL,” NeuroImage, vol. 62, no. 2, pp. 782–790, 2012, doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- [20].Andersson J. L. R. and Sotiropoulos S. N., “An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging,” NeuroImage, vol. 125, pp. 1063–1078, Jan. 2016, doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Veraart J., Fieremans E., and Novikov D. S., “Diffusion MRI noise mapping using random matrix theory,” Magn. Reson. Med., vol. 76, no. 5, pp. 1582–1593, Nov. 2016, doi: 10.1002/mrm.26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jenkinson M., Bannister P., Brady M., and Smith S., “Improved optimization for the robust and accurate linear registration and motion correction of brain images,” NeuroImage, vol. 17, no. 2, pp. 825–841, Oct. 2002, doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- [23].Avants B. B., Tustison N. J., Stauffer M., Song G., Wu B., and Gee J. C., “The insight ToolKit image registration framework,” Frontiers Neuroinform., vol. 8, p. 44, Apr. 2014, doi: 10.3389/fninf.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nir T. M., et al. , “Fractional anisotropy derived from the diffusion tensor distribution function boosts power to detect Alzheimer's disease deficits,” Magn. Reson. Med., vol. 78, no. 6, pp. 2322–2333, Dec. 2017, doi: 10.1002/mrm.26623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hua K., et al. , “Tract probability maps in stereotaxic spaces: Analyses of white matter anatomy and tract-specific quantification,” NeuroImage, vol. 39, no. 1, pp. 336–347, Jan. 2008, doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Winkler A. M., Ridgway G. R., Webster M. A., Smith S. M., and Nichols T. E., “Permutation inference for the general linear model,” NeuroImage, vol. 92, pp. 381–397, May 2014, doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Géron A., Hands-on Machine Learning With Scikit-Learn, Keras, and TensorFlow. Sebastopol, CA, USA: O'Reilly Media, 2022. [Google Scholar]
- [28].Guido R., Ferrisi S., Lofaro D., and Conforti D., “An overview on the advancements of support vector machine models in healthcare applications: A review,” Information, vol. 15, no. 4, p. 235, Apr. 2024, doi: 10.3390/info15040235. [DOI] [Google Scholar]
- [29].Binson V. A., Thomas S., Subramoniam M., Arun J., Naveen S., and Madhu S., “A review of machine learning algorithms for biomedical applications,” Ann. Biomed. Eng., vol. 52, no. 5, pp. 1159–1183, May 2024, doi: 10.1007/s10439-024-03459-3. [DOI] [PubMed] [Google Scholar]
- [30].Murdoch W. J., Singh C., Kumbier K., Abbasi-Asl R., and Yu B., “Definitions, methods, and applications in interpretable machine learning,” Proc. Nat. Acad. Sci. USA, vol. 116, no. 44, pp. 22071–22080, Oct. 2019, doi: 10.1073/pnas.1900654116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hung S.-C., Wang Y.-T., and Tseng M.-H., “An interpretable three-dimensional artificial intelligence model for computer-aided diagnosis of lung nodules in computed tomography images,” Cancers, vol. 15, no. 18, p. 4655, Sep. 2023, doi: 10.3390/cancers15184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dolci G., et al. , “Diffusion MRI allows capturing the amyloid-α and τ proteins status in Alzheimer's disease continuum,” in Proc. IEEE 21st Int. Symp. Biomed. Imag. (ISBI), May 2024. [Google Scholar]
- [33].Dolci G., et al. , “An interpretable generative multimodal neuroimaging-genomics framework for decoding Alzheimer's disease,” 2024, arXiv:2406.13292.
- [34].Dolci G., et al. , “Multimodal MRI-based detection of amyloid status in Alzheimer's disease continuum,” 2024, arXiv:2406.13305.
- [35].van der Velden B. H. M., Janse M. H. A., Ragusi M. A. A., Loo C. E., and Gilhuijs K. G. A., “Volumetric breast density estimation on MRI using explainable deep learning regression,” Sci. Rep., vol. 10, no. 1, p. 18095, Oct. 2020, doi: 10.1038/s41598-020-75167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu P. and Ogino M., “Guideline-based additive explanation for computer-aided diagnosis of lung nodules,” in Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support, Suzuki K., Reyes M., and Syeda-Mahmood T., Eds., Cham, Switzerland: Springer, 2019, pp. 39–47, doi: 10.1007/978-3-030-33850-3_5. [DOI] [Google Scholar]
- [37].Young K., Booth G., and Simpson B., “Deep neural network or dermatologist?,” in Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support, Suzuki K., Reyes M., and Syeda-Mahmood T., Eds., Cham, Switzerland: Springer, 2019, pp. 48–55, doi: 10.1007/978-3-030-33850-3_6. [DOI] [Google Scholar]
- [38].Zihni E., et al. , “Opening the black box of artificial intelligence for clinical decision support: A study predicting stroke outcome,” PLoS ONE, vol. 15, no. 4, Apr. 2020, Art. no. e0231166, doi: 10.1371/journal.pone.0231166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Agius R., et al. , “Machine learning can identify newly diagnosed patients with CLL at high risk of infection,” Nature Commun., vol. 11, no. 1, p. 363, Jan. 2020, doi: 10.1038/s41467-019-14225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dissanayake T., Fernando T., Denman S., Sridharan S., Ghaemmaghami H., and Fookes C., “A robust interpretable deep learning classifier for heart anomaly detection without segmentation,” IEEE J. Biomed. Health Informat., vol. 25, no. 6, pp. 2162–2171, Jun. 2021, doi: 10.1109/JBHI.2020.3027910. [DOI] [PubMed] [Google Scholar]
- [41].Du Y., Rafferty A. R., McAuliffe F. M., Wei L., and Mooney C., “An explainable machine learning-based clinical decision support system for prediction of gestational diabetes mellitus,” Sci. Rep., vol. 12, no. 1, p. 1170, Jan. 2022, doi: 10.1038/s41598-022-05112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Linardatos P., Papastefanopoulos V., and Kotsiantis S., “Explainable AI: A review of machine learning interpretability methods,” Entropy, vol. 23, no. 1, p. 18, Dec. 2020, doi: 10.3390/e23010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gandolfi M., et al. , “EXplainable AI allows predicting upper limb rehabilitation outcomes in sub-acute stroke patients,” IEEE J. Biomed. Health Informat., vol. 27, no. 1, pp. 263–273, Jan. 2023, doi: 10.1109/JBHI.2022.3220179. [DOI] [PubMed] [Google Scholar]
- [44].Boscolo Galazzo I., Brusini L., Obertino S., Zucchelli M., Granziera C., and Menegaz G., “On the viability of diffusion MRI-based microstructural biomarkers in ischemic stroke,” Frontiers Neurosci., vol. 12, p. 92, Feb. 2018, doi: 10.3389/fnins.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cruciani F., et al. , “Interpretable deep learning as a means for decrypting disease signature in multiple sclerosis,” J. Neural Eng., vol. 18, no. 4, Aug. 2021, Art. no. 0460a6, doi: 10.1088/1741-2552/ac0f4b. [DOI] [PubMed] [Google Scholar]
- [46].Molnar C., Interpretable Machine Learning, 2nd ed., 2022. [Online]. Available: https://christophm.github.io/interpretable-ml-book [Google Scholar]
- [47].Stenset V., et al. , “Cingulum fiber diffusivity and CSF T-tau in patients with subjective and mild cognitive impairment,” Neurobiol. Aging, vol. 32, no. 4, pp. 581–589, Apr. 2011, doi: 10.1016/j.neurobiolaging.2009.04.014. [DOI] [PubMed] [Google Scholar]
- [48].Aas K., Jullum M., and Løland A., “Explaining individual predictions when features are dependent: More accurate approximations to Shapley values,” Artif. Intell., vol. 298, Sep. 2021, Art. no. 103502, doi: 10.1016/j.artint.2021.103502. [DOI] [Google Scholar]
- [49].Mase M., Owen A. B., and Seiler B., “Explaining black box decisions by Shapley cohort refinement,” 2019, arXiv:1911.00467.
- [50].Salih A. M., Galazzo I. B., Raisi-Estabragh Z., Petersen S. E., Menegaz G., and Radeva P., “Characterizing the contribution of dependent features in XAI methods,” IEEE J. Biomed. Health Informat., early access, May 2, 2024, doi: 10.1109/JBHI.2024.3395289. [DOI] [PubMed]
- [51].Salih A., Galazzo I. B., Cruciani F., Brusini L., and Radeva P., “Investigating explainable artificial intelligence for MRI-based classification of dementia: A new stability criterion for explainable methods,” in Proc. IEEE Int. Conf. Image Process. (ICIP), Oct. 2022, pp. 4003–4007, doi: 10.1109/ICIP46576.2022.9897253. [DOI] [Google Scholar]


