Abstract
During the repair of interstrand crosslinks (ICLs) a DNA double-strand break (DSB) is generated. The Fanconi anemia (FA) core complex, which is recruited to ICLs, promotes high-fidelity repair of this DSB by homologous recombination (HR). However, whether the FA core complex also promotes HR at ICL-independent DSBs, for example induced by ionizing irradiation or nucleases, remains controversial. Here, we identified the FA core complex members FANCL and Ube2T as HR-promoting factors in a CRISPR/Cas9-based screen. Using isogenic cell line models, we further demonstrated an HR-promoting function of FANCL and Ube2T, and of their ubiquitination substrate FANCD2. We show that FANCL and Ube2T localize at DSBs in a FANCM-dependent manner, and are required for the DSB accumulation of FANCD2. Mechanistically, we demonstrate that FANCL ubiquitin ligase activity is required for the accumulation of CtIP at DSBs, thereby promoting end resection and Rad51 loading. Together, these data demonstrate a dual genome maintenance function of the FA core complex and FANCD2 in promoting repair of both ICLs and DSBs.
Subject terms: Homologous recombination, Double-strand DNA breaks
Fanconi anemia proteins are required for repair of DNA interstrand crosslinks. Here, the authors show that these proteins are also recruited to DNA double-strand breaks to promote repair by homologous recombination.
Introduction
DNA double-strand breaks (DSBs) are dangerous DNA lesions that separate a chromosome into two fragments. If left unrepaired, DSBs can result in mitotic missegregation of the broken chromosome and subsequent aneuploidy. Hence, efficient repair of DSBs is essential to maintain genome stability. This is ensured by the collective activity of a variety of DSB repair pathways, including homologous recombination (HR)1. DSB repair by HR is initiated by end resection, which involves nuclease-mediated strand removal at the DSB ends to generate 3’ single-strand overhangs2. These overhangs are bound by the recombinase protein Rad51 that drives invasion of the 3’ DNA overhang into a homologous DNA region, most often the sister chromatid3. Subsequently, multiple HR subpathways can be distinguished, all of which involve extension of the DNA overhang on the homologous DNA, followed by untangling of the recombination intermediate and completion of repair.
HR initiation by end resection is a tightly coordinated process2, which starts with the binding of the Mre11-Rad50-Nbs1 (MRN) complex to DSB ends2. Mre11 has endonuclease activity, which is used to nick the DNA adjacent to the DSB, as well as exonuclease activity, which is used to resect the DNA from the nick towards the DSB end2. The endonuclease activity of Mre11 is strongly promoted by its co-factor CtIP4,5. CtIP is a central regulator of end resection that, besides promoting Mre11, can also enhance the activity of the nuclease DNA2, which acts downstream of Mre11 to processively resect the DNA in the 5’ to 3’ direction6. CDK-mediated phosphorylation of CtIP restricts its activity to the S/G2 cell cycle phases, thus synchronizing end resection with the presence of the sister chromatid7,8. CtIP interacts with the Mre11-Rad50-Nbs1 (MRN) complex, as well as with the HR factor BRCA15,9–11. However, neither of these interactions are essential for the recruitment of CtIP to DSBs, indicating that this can be mediated by additional signals at the break site12–14.
In addition to HR, DSBs can be repaired by canonical Non-Homologous End-Joining (c-NHEJ), alternative End-Joining (a-EJ), or Single-Strand Annealing (SSA)1. Whereas c-NHEJ involves very minimal end processing prior to ligation, a-EJ and SSA require end resection to reveal regions of homology that can base pair to join the two opposing DSB ends. Repair by c-NHEJ, a-EJ, and SSA effectively reconnects the broken chromosome fragments, yet mostly at the expense of mutations at the break junction. In contrast, HR is generally considered to faithfully restore the original DNA sequence. Hence, HR forms a barrier against mutagenesis and chromosomal alterations and, as such, is an important tumor suppressor pathway. In agreement, a high frequency of tumors, in particular those derived from breast and ovarian tissue, are HR-deficient due to germline or somatic mutations in HR genes like BRCA1 or BRCA215,16.
Considering the important genome maintenance function of HR, we sought to identify novel genes involved in DSB repair by this pathway. To this end, we performed a targeted CRISPR-based genetic screen in a cell line carrying the DSB repair reporter DSB-Spectrum17. As we show here, the results from this screen suggested that the E2 ubiquitin conjugase Ube2T and the E3 ubiquitin ligase FANCL function as HR-promoting factors. Ube2T and FANCL are both part of the multi-member Fanconi anemia (FA) core complex that plays a well-characterized role during the repair of DNA interstrand crosslinks (ICLs)18. Inactivating mutations in the genes encoding the FA core complex members, or downstream ICL repair factors, are all associated with the hereditary disorder Fanconi anemia19. The FA core complex recognizes crosslinked DNA and subsequently ubiquitinates the FANCD2/FANCI heterodimer18. This is an essential step to recruit nucleases that remove the ICL, but also to promote the downstream repair steps, including HR-mediated repair of the DSB that is generated following ICL removal.
The FA core complex and FANCD2/FANCI have been suggested to also promote HR at DSBs that are generated independently of ICL-repair20–23. However, the relevance of this function has been questioned, as the HR phenotypes observed upon depletion of FA factors were mild or even absent in some studies22,24–28. Moreover, mechanistic insight into the DSB repair function of the FA core complex is lacking. Here, we validate the HR-promoting function of Ube2T and FANCL observed in our CRISPR-based genetic screen using orthogonal approaches in a variety of knock-out cell lines and isogenic control cell lines. Moreover, we show that Ube2T and FANCL activity are required for optimal CtIP-dependent end resection at DSBs, providing a mechanistic explanation for their HR-promoting function. Together, our data indicate that the FA core complex and FANCD2 not only promote ICL repair but are also bona fide DSB repair factors that act during an initial and essential stage of HR.
Results
A DSB-Spectrum reporter screen identifies members of the FA core complex as HR-promoting factors
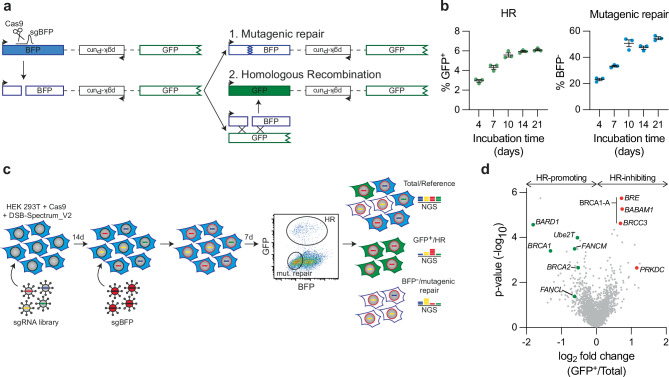
To identify novel genes that drive error-free DSB repair, a genetic screen was performed using DSB-Spectrum_V2, a genomic DSB repair reporter that we had previously created to distinguish mutagenic repair from HR (Fig. 1a)17. The reporter consists of a functional Blue Fluorescent Protein (BFP) gene separated by an ~ 3 kilobase region from a promotorless and truncated Enhanced Green Fluorescent Protein gene (EGFP, hereafter referred to as GFP). In this system, a single DSB is generated by targeting S. pyogenes Cas9 to the BFP gene at a site adjacent to the chromophore-determining amino acids. Mutagenic repair of the DSB, for example by c-NHEJ, will disrupt the BFP gene, resulting in loss of fluorescence (Fig. 1a). Alternatively, repair of the DSB by HR using the downstream, highly homologous truncated GFP gene as a repair template will replace the BFP serine-66 and histidine-67 encoding triplets with threonine and tyrosine-encoding triplets of the truncated GFP gene, causing BFP-to-GFP conversion29. Thus, mutagenic repair or HR-mediated repair of a DSB is detected by total loss of fluorescence or by conversion from BFP to GFP expression, respectively.
Fig. 1. A targeted CRISPR screen in DSB-Spectrum reporter cells identifies FANCM, Ube2T and FANCL as HR-promoting factors.
a Schematic of the DSB-Spectrum_V2 reporter. Adapted from van de Kooij et al.17. BFP=Blue Fluorescent Protein, GFP=Green Fluorescent Protein. b HEK 293T+ Cas9 + DSB-Spectrum_V2 cells were lentivirally infected to express mCherry and the BFP sgRNA targeting the DSB-Spectrum_V2 reporter locus. Next, at indicated time points, BFP and GFP expression were analyzed by flow cytometry. Depicted is the mean ± SEM of a technical triplicate. HR homologous recombination. c Schematic displaying the CRISPR screen layout in HEK 293T + Cas9 + DSB-Spectrum_V2 cells. NGS next-generation sequencing. d Volcano plot showing the gene targets of sgRNAs that were either enriched or depleted from the GFP+ HR population as compared to the reference population. BRCA1/BARD1, BRCA2, and Ube2T/FANCM/FANCL are indicated in green as HR-promoting factors. BRE/BABAM1/BRCC3, all members of the BRCA1-A complex, and PRKDC are indicated in red as HR-inhibiting factors. Enrichment and statistical values were determined by MAGeCK integrated into the BASDaS screening data analysis interface (n = 3 independent biological replicates)81,82. Source data are provided as a Source Data file.
To generate a reporter cell line suitable for genetic screening, HEK 293T cells stably expressing Cas9 were lentivirally transduced to introduce a copy of DSB-Spectrum_V2 into the genome. A single cell clone was expanded to create a homogeneous HEK 293T DSB-Spectrum_V2 cell line. To validate this cell line and establish the kinetics of repair, a BFP-targeting single-guide RNA (sgRNA) or AAVS1-targeting control sgRNA was introduced by lentiviral transduction, and BFP and GFP expression were analyzed by flow cytometry at various time points after transduction. Cells expressing the control sgRNA remained BFP-positive and GFP-negative throughout the experiment (Supplementary Fig. 1a). In contrast, in cells expressing the BFP-targeting sgRNA, both BFP-negative/GFP-negative and BFP-negative/GFP-positive populations could be detected at four days after transduction, indicating mutagenic and HR-mediated DSB repair, respectively (Fig. 1b and Supplementary Fig. 1a). The mutagenic and HR repair populations gradually increased in size until reaching a plateau at 10 days after transduction (Fig. 1b and Supplementary Fig. 1a). As we had previously validated that these distinct fluorescent populations are the consequence of mutagenic DSB repair and HR, respectively17, we concluded that the newly-generated DSB-Spectrum_V2 reporter cell line is functional.
To identify genes that modulate different DNA repair phenotypes, a CRISPR-based genetic screen was performed (Fig. 1c). A custom-generated sgRNA library targeting 2760 genes, with four sgRNAs per gene, was introduced into the DSB-Spectrum_V2 reporter cells by lentiviral transduction (Supplementary Data 1). The targeted genes encoded well-described DNA repair factors, kinases and phosphatases, ubiquitin and SUMO modifiers, and factors that read, write, or remodel chromatin. Cells were cultured for 14 days to allow editing of the target genes and depletion of their protein products, after which the BFP-targeting sgRNA was introduced by lentiviral infection to generate a DSB within the DSB-Spectrum_V2 reporter. At seven days after targeting BFP, the BFP-negative/GFP-positive HR and BFP-negative/GFP-negative mutagenic repair populations were harvested by Fluorescence-Activated Cell Sorting (FACS). In addition, a sample from the total population was collected for use as a reference. The sgRNA counts in all samples were then determined by Illumina sequencing (Supplementary Data 2).
As an initial quality control measure, we determined the level of depletion of sgRNAs targeting essential genes30. The majority of sgRNAs targeting essential genes were strongly depleted from the total surviving population relative to the input library, validating that CRISPR-based gene editing was sufficient to permit a measurable phenotype in our system (Supplementary Fig. 1b). As a second quality control measure, we assessed how sgRNAs targeting known DSB repair factors behaved. A clear depletion of sgRNAs targeting the HR-promoting genes BRCA1, BRCA2, and BARD1 was observed in the GFP-positive HR population compared to the reference population (Fig. 1d and Supplementary Data 2). In contrast, sgRNAs targeting PRKDC or the members of the BRCA1-A complex, which both inhibit HR, were strongly enriched in the HR population (Fig. 1d and Supplementary Data 2). Thus, this screening approach can faithfully detect known factors that promote or impair HR. Theoretically, analysis of sgRNA levels in the GFP-negative/BFP-negative population should allow for the identification of factors involved in mutagenic DSB repair. However, we were not able to validate this branch of the screen, as no enrichment or depletion of known DSB repair factors was observed in this mutagenic repair population (Supplementary Data 2). The selective pressure might be limited on this population given the high frequency of mutagenic repair. This population might furthermore represent a collection of repair outcomes from multiple mutagenic pathways that can compensate for each other’s loss, as we have shown previously17. Considering this result, and based on our primary interest in HR, we therefore decided to focus on the HR branch of this screen.
We generated a list of genes whose sgRNAs were depleted or enriched in the GFP-positive HR population using a False Discovery Rate (FDR) cut-off of ≤0.26 (Supplementary Fig. 1c). The resulting list of hits contained core HR genes like BRCA1, as well as genes that have reported HR functions but are less established HR factors, including VHL, PHF20, USP34, and ASF1A31–34. The screen also identified nine genes with no previously reported function in HR (Supplementary Fig. 1c). To determine which genes to follow up on, we compared our results with those of a recently published CRISPRi screen in K562 cells aimed at identifying regulators of HR using an ectopically provided dsDNA donor23. Although we observed remarkably limited overlap between the two datasets, Ube2T, FANCM, and FANCL were among the candidates identified as HR-promoting factors in both screens (Supplementary Fig. 1d). Ube2T and FANCM were two of the strongest hits identified by our screen, and FANCL-targeting sgRNAs were also clearly depleted from the HR population, even though this did not reach statistical significance (Fig. 1d). These three factors, which function with or within the FA core complex, were the only FA core complex components targeted by the custom-generated sgRNA library used for our screen. Taken together, these data strongly suggest a DSB repair function for the FA core complex that is independent of its role in ICL repair.
Optimal HR requires the expression and ubiquitin ligase activity of FANCL and Ube2T
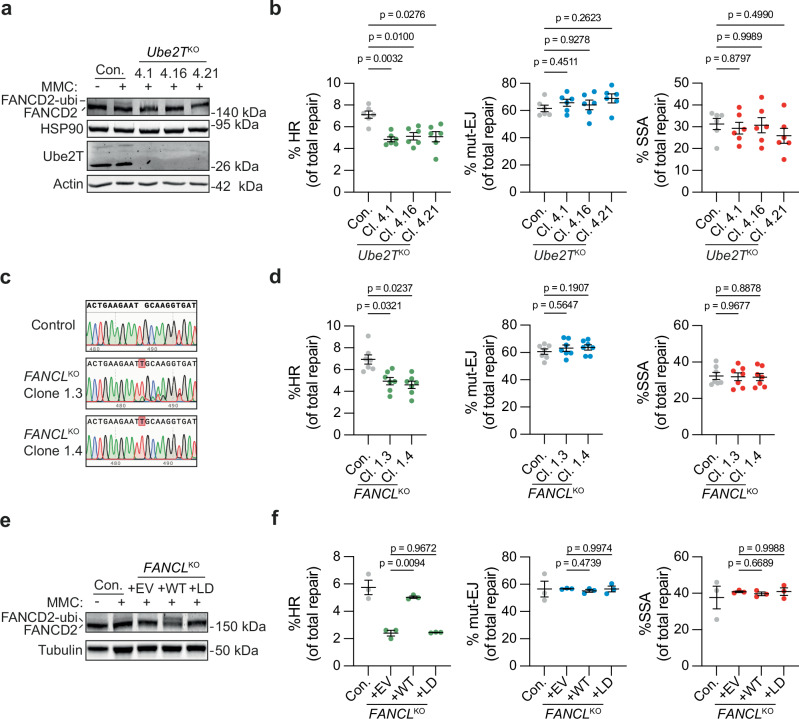
FANCM has been suggested to promote short tract gene conversion during HR by dissolving recombination intermediates like double Holliday junctions35–37. This HR function of FANCM, which is independent of the FA core complex, could explain why FANCM was identified as an HR-promoting factor in our screen. In contrast, the functions of Ube2T and FANCL in HR are largely underexplored. We, therefore, decided to focus on these two FA proteins in more detail. The results from the CRISPR screen were first validated by targeted depletion of Ube2T or FANCL in HR reporter assays using HEK 293T cells carrying the reporter DSB-Spectrum_V3, a next-generation variant of DSB-Spectrum_V2. This V3 reporter contains a mCherry gene between the BFP and truncated GFP genes to allow mutagenic DSB repair resulting from direct end-joining to be distinguished from that resulting from SSA (Supplementary Fig. 2a)17. DSB repair by SSA in this reporter results in the deletion of a mCherry gene, which can be monitored by flow cytometry, in conjunction with repair by HR (GFP-positive) and mutagenic end-joining (mut-EJ; BFP-negative/mCherry-positive population of cells). Monoclonal Ube2T knock-out (Ube2TKO) cell lines were generated using CRISPR technology, and validated by western blot analysis (Fig. 2a). Two Ube2T antibody-responsive bands were detected in the Ube2T wild-type (WT) control cell line (Con.), both of which disappeared in the three individual Ube2TKO clones. To further validate the loss of functional Ube2T, the cells were treated with the DNA cross-linking agent mitomycin C (MMC), and the ubiquitination of the known Ube2T substrate FANCD2 was monitored by western blotting. FANCD2 ubiquitination, indicated by the appearance of a slower migrating FANCD2 species, was observed in the control cells but not in any of the Ube2TKO clones (Fig. 2a), Hence, all three Ube2TKO clones are completely Ube2T-deficient.
Fig. 2. Ube2T and FANCL promote HR-mediated repair of Cas9-induced DSBs.
a HEK 293T + DSB-Spectrum_V3 Ube2TKO clones 4.1, 4.16, and 4.21, as well as the parental control (Con.) were treated with Mitomycin C (MMC, 300 nM) for 24 h. Next, Ube2T expression levels and FANCD2 ubiquitination status were analyzed by western blotting. b Indicated HEK 293T + DSB-Spectrum_V3 cell lines were transfected to express S. pyogenes Cas9 and either a control sgRNA or the sgRNA targeting the BFP gene in the reporter locus. Next, cells were analyzed by flow cytometry to determine the frequency of repair by each of the three indicated pathways (n = 6 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). HR homologous recombination, mut-EJ mutagenic end-joining, SSA single-strand annealing. c DNA sequence alignment of the FANCL sgRNA target site in unedited parental control cells and the HEK 293T + DSB-Spectrum_V3 FANCLKO clones. Depicted are representative sequence chromatograms, red shaded boxes indicate deviations in the DNA sequence of the FANCLKO clone compared to control. d As in panel (b), now analyzing FANCLKO cells (n = 7 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). e HEK 293T + DSB-Spectrum_V3 FANCLKO clones were transduced with an empty vector (EV), FANCL wild-type cDNA (WT), or FANCL Ligase-Dead cDNA (LD), and treated with Mitomycin C (MMC, 1 μM) for 24 h. Next, FANCD2 ubiquitination was analyzed by western blot. f As in panel b, now analyzing the HEK 293T + DSB-Spectrum_V3 FANCLKO cells described in panel (e) (n = 3 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). Source data are provided as a Source Data file.
The Ube2TKO and control cells were transfected to express Cas9, the BFP sgRNA, and the iRFP(670) protein as a marker for transfected cells. After 72–96 h, the frequency of HR, mut-EJ and SSA was determined by flow cytometry. DSB repair by HR was significantly reduced in all Ube2TKO clones as compared to the control cells (Fig. 2b). In contrast, the levels of mut-EJ and SSA were similar between the control and Ube2TKO cell lines. Re-expression of Ube2T rescued the HR defect in the three Ube2TKO clones (Supplementary Fig. 2b, c). We therefore conclude that Ube2T specifically promotes DSB repair by HR, validating the results from the screen.
HEK 293T DSB-Spectrum_V3 cell lines in which FANCL was genetically deleted were generated next. The levels of FANCL protein could not be determined because the commercially available FANCL antibodies that we tried failed to produce reliable FANCL staining on western blot. Therefore, the knock-out status of the selected FANCLKO clones was investigated by PCR amplification of the sgRNA target region in FANCL, followed by Sanger sequencing. Manual inspection of the sequence chromatograms, as well as decomposition of the sequencing results using the TIDE algorithm38, demonstrated that the KO clones contained a + 1 thymine insertion with an additional 8 bp deletion in the case of clone 1.3 (Fig. 2c and Supplementary Fig. 2d). Importantly, no WT sequence trace was identified, indicating that all sequenced FANCL alleles contained out-of-frame mutations. Furthermore, MMC treatment failed to induce FANCD2 ubiquitination in the FANCLKO cells, confirming the absence of functional FANCL in these clones (Supplementary Fig. 2e).
We next investigated DSB repair activity in these FANCLKO cell lines by DSB-Spectrum_V3 reporter assays and found that HR was significantly reduced compared to that in control cells, whereas the levels of mut-EJ and SSA were unaffected (Fig. 2d). To confirm that this phenotype was the specific result of loss of FANCL function, WT FANCL cDNA (+ WT), or an empty vector control (+ EV), was re-introduced into FANCLKO clone 1.4. In addition, we introduced a FANCL mutant carrying a C307A mutation that abrogates its ligase activity (Ligase-Dead, + LD)39. MMC-induced FANCD2 ubiquitination was clearly observed in the FANCLKO cells reconstituted with FANCL WT, but not in the cells reconstituted with either the EV or the FANCL LD mutant (Fig. 2e), indicating that only the + WT cells express functional ligase-competent FANCL. The HR phenotype in the different FANCL cell lines was examined using the DSB-Spectrum_V3 reporter assay. In agreement with the aforementioned data, the FANCLKO cells (+ EV) showed strongly impaired HR compared to the control cells, which was rescued by re-expression of FANCL WT, but not by re-expression of the FANCL LD mutant (Fig. 2f). Taken together, these data indicate that UbeT2 and FANCL expression, as well as FANCL ligase activity, are required for efficient HR.
Loss of Ube2T, FANCL, or FANCD2 sensitizes cells to PARP inhibitor treatment
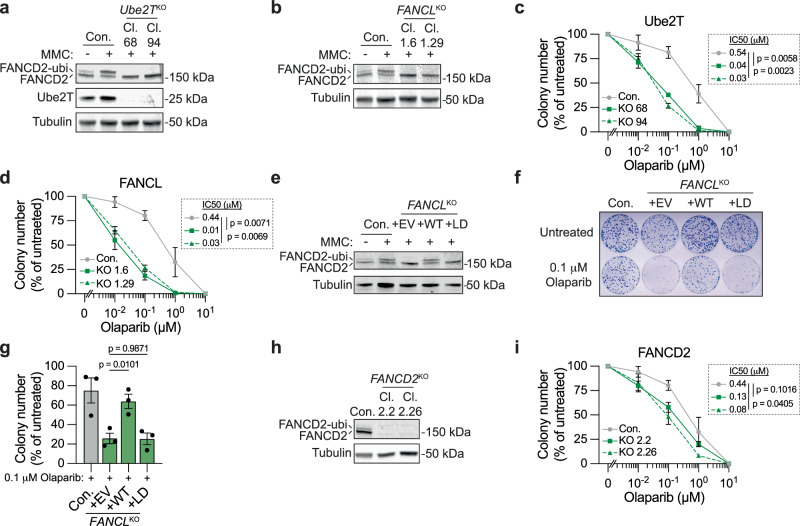
To further validate the HR function of Ube2T and FANCL, we utilized an alternative assay orthogonal to the DSB-Spectrum_V3 reporter assays and explored cells other than HEK 293T34,35. Clonally derived Ube2T and FANCL knock-out U2OS human osteosarcoma cell lines were generated and validated by western blotting and sequence analysis for two Ube2TKO and two FANCLKO clones, respectively (Fig. 3a and Supplementary Fig. 3a). Of note, a single Ube2T species was detected by western blot analysis of the U2OS lysates, in contrast to the doublet that we consistently observed in lysates from HEK 293T cells (Fig. 2a). The knock-out status of the Ube2TKO and FANCLKO cell lines was further validated by the absence of MMC-induced FANCD2 ubiquitination in these cell lines (Fig. 3a, b).
Fig. 3. Loss of Ube2T, FANCL and FANCD2 sensitizes cells to PARP inhibitor-induced toxicity.
a Indicated U2OS Ube2TKO clones (Cl.), as well as the parental control (Con.), were treated with Mitomycin C (MMC, 500 nM) for 24 h. Next, FANCD2 ubiquitination status and protein levels of Ube2T were analyzed by western blot. b As in panel (a), but now for U2OS FANCLKO clones. c, d U2OS cells, either wild-type control (Con.) or knock-out for Ube2T (panel c) or FANCL (panel d), were treated with olaparib for 14 days. Cell viability was assessed by clonogenic survival. Note that the 0 μM value was added manually to the X-axis. Inset shows mean IC50 and p-value (IC50 based on curve fitting of n = 3 independent biological replicates; Ratio paired t test, two-sided. For individual data points mean ± SEM is shown for n = 3 independent biological replicates, expect for the 0.1 μM and 10 μM concentration in panel (c) for which n = 2). e As in panel (b), now for FANCLKO cells that were transduced with an empty vector (EV), FANCL wild-type cDNA (WT), or FANCL ligase-dead cDNA (LD). f, g Indicated cell lines were treated with 0.1 μM olaparib, or left untreated, for 14 days. Cell viability was assessed by clonogenic survival. Panel (f) shows a representative picture of Methylene Blue-stained colonies in 10 cm plates, and panel (g) shows the quantification (n = 3 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). h As in panel (b), but now for untreated FANCD2KO clones. i As in panels (c) and (d), but for FANCD2KO cells (n = 3; mean ± SEM; Ratio paired t test, two-sided). Source data are provided as a Source Data file.
PARP inhibitor (PARPi) treatment is commonly used as a proxy for HR pathway activity because HR defects strongly sensitize cells to the toxic effects of PARP inhibition40,41. The Ube2TKO and FANCLKO clones, as well as control cells, were therefore treated with varying doses of the PARPi olaparib, and clonogenic outgrowth was measured. All Ube2TKO and FANCLKO cell lines were significantly more sensitive to PARPi-induced loss of viability than the control cell line (Fig. 3c, d). Of note, the PARPi sensitivity phenotype of the FANCLKO cells was not as severe as that of BRCA2-depleted cells, which is consistent with the partial HR defect observed in the FANCLKO cells (Supplementary Fig. 3b). To further assess the role of FANCL ligase activity in the olaparib response, FANCLKO clone 1.6 was reconstituted with FANCL WT, the LD mutant, or an EV control. As expected, MMC-induced FANCD2 ubiquitination was observed in the FANCL WT, but not in the EV or FANCL LD cell lines, validating that the re-expressed FANCL WT was functional (Fig. 3e). Moreover, re-expression of FANCL WT, but not of the FANCL LD mutant, rescued the increased PARPi sensitivity of the FANCLKO clone (Fig. 3f, g). These data show that loss of Ube2T or FANCL expression or activity sensitizes cells to PARPi treatment, consistent with an HR-promoting function for both of these factors.
During the repair of ICLs, FANCD2 is the downstream substrate of Ube2T and FANCL. To examine whether FANCD2 functions in DSB repair by HR as well, we generated FANCD2KO cell lines in U2OS cells and measured PARPi sensitivity (Fig. 3h, i). As seen with Ube2T and FANCL, loss of FANCD2 similarly reduced clonogenic outgrowth after olaparib treatment (Fig. 3i). To examine whether FANCD2 mono-ubiquitination was required for the response to PARPi, we introduced GFP-FANCD2 WT or a ubiquitination resistant GFP-FANCD2 K561R mutant into the U2OS FANCD2KO cells (Supplementary Fig. 3c). In addition, a cell line with GFP carrying a Nuclear Localization Signal (NLS) was generated as a negative control. Cell viability after PARPi treatment was assessed using a competition assay. The GFP-NLS, GFP-FANCD2 WT, or GFP-FANCD2 K561R cells were mixed 1:1 with the parental FANCD2KO cells and treated with 1 μM olaparib, or left untreated. Subsequently, the fraction of GFP-positive cells in the mixed population was monitored by flow cytometry. Within the 12-day olaparib treatment period, GFP-FANCD2 WT cells started to outcompete GFP-negative FANCD2KO cells, as indicated by the increased fraction of GFP-positive cells in the mixed population (Supplementary Fig. 3d), suggesting that re-expression of FANCD2 caused resistance towards olaparib. Notably, this increase in GFP-positive cells in the olaparib-treated population was not observed for GFP-NLS cells, and was less prominent for the GFP-FANCD2 K561R mutant cells compared to the GFP-FANCD WT cells (Supplementary Fig. 3d). Hence, FANCD2 expression and mono-ubiquitination promote PARPi resistance. These results suggest a function for FANCD2 in crosslink-independent DSB repair by HR, which we subsequently directly assessed using HEK 293T cells expressing the DSB-Spectrum_V3 reporter. Following the siRNA-mediated depletion of FANCD2, the frequency of HR at Cas9-induced DSBs was significantly reduced, whereas the levels of mutagenic end-joining and SSA remained unaffected (Supplementary Fig. 3e, f). Notably, loss of FANCD2, or of FANCL or Ube2T, did not significantly affect cell-cycle distribution (Supplementary Fig. 3g). Thus, the PARPi sensitivity phenotype of cells following the loss of FANCD2, FANCL, or Ube2T is unlikely to be caused by changes in cell cycle populations. Instead, these results suggest that FANCD2, Ube2T, and FANCL cooperate to directly promote HR repair of DSBs.
The FA core complex is directly recruited to DSBs, where it promotes FANCD2 accumulation
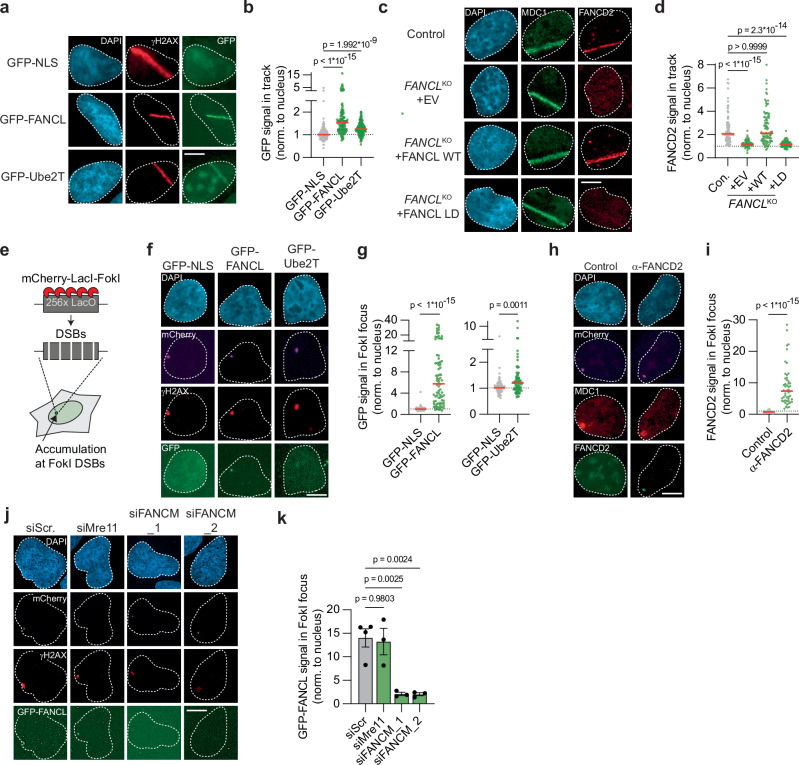
To determine whether Ube2T and FANCL directly promote HR at DSBs, we examined their recruitment to sites of DNA damage using laser micro-irradiation. U2OS cell lines stably expressing GFP-tagged FANCL or Ube2T, as well as a control cell line expressing GFP-NLS, were generated, pre-treated with BrdU, and exposed to UV-A laser micro-irradiation to generate localized stripes of DNA damage, predominantly DSBs42. Subsequently, cells were fixed and analyzed by fluorescence microscopy to study the enrichment of Ube2T or FANCL at the sites of DNA damage. Antibody staining for phosphorylated histone H2AX (γH2AX) was used to identify the DNA damage stripes. Whereas GFP-NLS was equally distributed throughout the nucleus, both FANCL and Ube2T were significantly enriched in the DNA damage stripes (Fig. 4a, b), with the enrichment being somewhat stronger for FANCL than for Ube2T. Thus, both FANCL and Ube2T are recruited to UV-A laser-induced DSBs.
Fig. 4. FANCL, Ube2T and FANCD2 are recruited to DNA double-strand breaks.
a, b U2OS cells expressing either GFP-NLS, GFP-FANCL, or GFP-Ube2T were exposed to UV-A laser micro-irradiation. Next, the GFP signal at γH2AX-positive laser-induced DNA damage tracks was analyzed by fluorescence microscopy. Shown are representative images (a) and quantification (b) of one of two independent biological replicates (scale bar = 10 μM). The dotted line is set at 1 (i.e., no recruitment to the track), red lines indicate median (n = 128, 116, 146 (NLS, FANCL, Ube2T); one-way ANOVA with post-hoc Kruskal-Wallis). c, d As in panels, but now analyzing recruitment of endogenous FANCD2 to MDC1-positive laser-induced DNA damage tracks in U2OS FANCLKO cell lines (EV = empty vector, WT = wild-type, LD = ligase-dead). Shown are representative images (c) and quantification (d) of one of two independent biological replicates (scale bar = 10 μM). Red lines indicate the median (n = 82, 56, 90, 84 (Con., + EV, + WT, + LD); one-way ANOVA with post-hoc Kruskal-Wallis). e Cartoon schematic of a DSB recruitment assay in U2OS 2-6-3 cells. f, g Accumulation of GFP-NLS, GFP-FANCL, or GFP-Ube2T at γH2AX-marked FokI-generated DSBs in U2OS 2-6-3 cells was assessed by fluorescence microscopy. Shown are representative images (f) and quantification (g) of one of two independent biological replicates (scale bar = 10 μM). GFP-FANCL and GFP-Ube2T signals are plotted in individual graphs to optimize the scaling of the Y-axis. Red lines indicate the median (n = 74, 82, 77 (NLS, FANCL, Ube2T); Mann-Whitney test, two-sided). h, i As in panels (f) and (g) but now analyzing endogenous FANCD2 recruitment to MDC1-marked FokI-induced DSBs. Primary α-FANCD2 antibody was omitted from the immuno-staining in the control sample. Shown are representative images (h) and quantification (i) of one of two independent biological replicates. Red lines indicate the median (n = 56, 50 (Control, a-FANCD2); Mann-Whitney test, two-sided). j, k As in panels (f) and (g), but including siRNA transfection. Panel (j) shows representative images (scale bar = 10 μM), and panel (k) shows the quantification of independent biological replicates (n = 4 for siScr, n = 3 for the other conditions; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). Source data are provided as a Source Data file.
FANCD2 has previously been shown to accumulate at DNA damage sites upon DSB-inducing treatments43,44. To examine whether FANCD2 recruitment to DSBs was dependent on FANCL, we assessed FANCD2 localization using laser micro-irradiation assays in the reconstituted U2OS FANCLKO cells described above. Strong enrichment of endogenous FANCD2 was observed in control cells at UV-A laser-induced DNA damage stripes marked by the γH2AX-binding protein MDC1 (Fig. 4c). Importantly, no FANCD2 enrichment was detected in the FANCLKO cells reconstituted with empty vector (Fig. 4c, d). Re-expression of FANCL WT, but not of the FANCL LD mutant, however, rescued the FANCD2 recruitment to sites of DNA damage. Hence, FANCL expression and ligase activity are required for the accumulation of FANCD2 at UV-A laser-induced DSBs. To extend these findings to other members of the FA core complex, we studied the patient-derived head and neck squamous cell carcinoma cell line VU1131, which is deficient for FANCC45. No recruitment of FANCD2 to DNA damage stripes was observed in these cells. However, the re-expression of FANCC completely rescued the FANCD2 recruitment defect (Supplementary Fig. 4a, b). These data indicate that Ube2T and FANCL act as part of the FA core complex to promote FANCD2 recruitment to DSBs. We next tested whether FANCD2 mono-ubiquitination is required for its accumulation at DSBs using the U2OS FANCD2KO cells expressing either GFP-FANCD2 WT or the GFP-FANCD2 K561R mutant (Supplementary Fig. 3c). Although GFP-FANCD2 WT accumulated in UV-A laser stripes, similar to endogenous FANCD2, the GFP-FANCD2 K561R mutant did not (Supplementary Fig. 4c, d). Hence, mono-ubiquitination of FANCD2 is required for its recruitment to UV-A laser-induced DSBs.
UV-A laser micro-irradiation in BrdU-treated cells predominantly causes DSBs, but a minority of other DNA lesions can also be generated that could be at least partially responsible for the accumulation of FA factors at the irradiated sites. To exclude this possibility, the recruitment of FA factors to nuclease-induced DSBs was examined using an alternative approach. GFP-FANCL and GFP-Ube2T expressing cell lines were generated in U2OS 2-6-3 cells that contain a mCherry-tagged, LacI-fused FokI nuclease and a genomically integrated LacO array46. Upon treatment with tamoxifen and Shield-1, the FokI nuclease is stably expressed, translocates to the LacO array, and generates a multitude of DSBs within the LacO array, resulting in the local accumulation of DSB repair factors (Fig. 4e)46. Following treatment of our cell lines with tamoxifen and Shield-1, a single distinct γH2AX focus was observed that co-localized with the mCherry-FokI nuclease (Fig. 4f). In most cells, the GFP-FANCL signal was strongly enriched in these FokI foci (Fig. 4f, g). GFP-Ube2T was also significantly enriched in FokI foci, in contrast to GFP-NLS, which served as a negative control (Fig. 4f, g). Notably, similar to what was observed at UV-A laser-induced DNA damage sites, the accumulation of Ube2T at FokI-induced DSB-sites was substantially less than the accumulation of FANCL (Fig. 4g). Furthermore, strong accumulation of endogenous FANCD2 at FokI-induced DSBs was observed (Fig. 4h, i). Collectively, these results demonstrate that Ube2T, FANCL, and FANCD2 are recruited to bona fide DSBs, suggesting that they act directly at DNA break sites to promote repair by HR.
We next investigated which factors act upstream in the response to DSBs to recruit the FA core complex. We focused on FANCM and the MRN complex, which are the lesion-sensing components during ICL repair and DSB repair by HR, respectively3,18. Expression of FANCM and Mre11 was silenced using siRNAs (Supplementary Fig. 4e, f), and recruitment of GFP-FANCL to FokI-induced DSBs was then assessed. Whereas GFP-FANCL accumulation at the DSB site was unaffected by Mre11 knockdown, it was severely reduced upon knockdown of FANCM (Fig. 4j, k). These results indicate that recruitment of the FA core complex to DSBs is dependent on FANCM, as is its recruitment to ICLs18. These results also suggest that the identification of FANCM as a strong hit in our HR screen is not only due to its function in dissolving HR recombination intermediates (Fig. 1d)35, but also because of its function in recruiting the FA core complex.
FANCL promotes DSB end resection
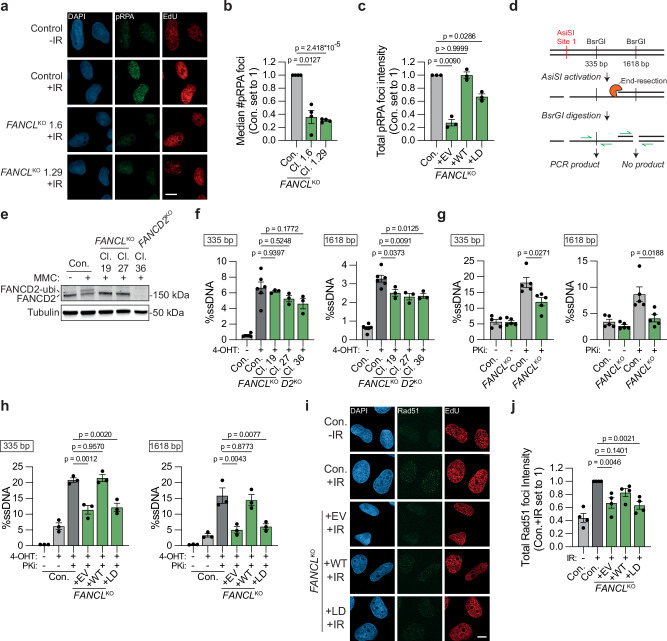
We next explored the mechanism by which FANCL and Ube2T promote DSB repair by HR. Since FANCD2 has previously been suggested to function in DNA end resection47, we hypothesized that this may require the upstream activity of FANCL/Ube2T. To test this, U2OS FANCLKO cells were exposed to ionizing radiation (IR), and the accumulation of serine 4/8 phosphorylated RPA (pRPA) at DNA damage-induced foci was measured as a proxy for the presence of resected ssDNA48. In addition, a short pulse of EdU was administered prior to the IR exposure to allow for the detection of S-phase cells, which are HR-prone. The formation of IR-induced pRPA foci was strongly reduced in the FANCLKO cells as compared to the control cells (Fig. 5a, b and Supplementary Fig. 5a). This reduction in pRPA foci intensity in the FANCLKO cells could be completely rescued by re-expression of FANCL WT, but less so by re-expression of the FANCL LD mutant (Fig. 5c and Supplementary Fig. 5b). A strong reduction in pRPA foci number was also observed in Ube2TKO and FANCD2KO cells (Supplementary Fig. 5c–e). The pRPA foci levels in the FANCD2KO cells were restored by the expression of GFP-FANCD2 WT, but not by the expression of GFP-FANCD2 K561R (Supplementary Fig. 5f).
Fig. 5. FANCL promotes end resection at DNA double-strand breaks.
a, b U2OS FANCLKO clones (Cl.) and wild-type control cells (Con.) were exposed to 10 Gy ionizing radiation (IR), followed by IF microscopy to detect foci containing S4/8 phosphorylated RPA (pRPA) in S-phase (EdU + ) nuclei. Panel (a) shows representative images (scale bar = 10 μM), and panel (b) shows the quantification (n = 4 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). c As in panel (b), but now plotting total pRPA foci intensity per nucleus in FANCLKO cells reconstituted with FANCL WT or LD (n = 3 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). d Schematic of the qPCR-based quantification of end resection in AsiSI cells. e Western blot of MMC-treated (500 nM, 24 h) U2OS AsiSI cells. f Quantification by qPCR of single-strand DNA (ssDNA) at 335 bp or 1618 bp distance from a defined AsiSI-induced DSB (n = 6 for control cells, n = 3 for KO cells, all independent biological replicates; one-way ANOVA with post-hoc Dunnett’s). g As in panel (f), but now including treatment with the DNA-PKcs inhibitor NU7441 (PKi; 2 μM; n = 5 independent biological replicates; mean ± SEM; paired t-test, two-sided). h As in panel (g), now in FANCLKO cells that express either an empty vector (EV), FANCL wild-type (WT), or FANCL ligase-dead (LD; n = 3 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). i, j As in panels (a) and (b), respectively, but now analyzing total Rad51 foci intensity per S-phase nucleus (n = 4 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s; scale bar = 10 μM). Source data are provided as a Source Data file.
To further interrogate an end resection function for FANCL, we quantified resected DNA using U2OS AsiSI cells. In these cells, the site-directed nuclease AsiSI translocates to the nucleus after tamoxifen treatment and induces DSBs at ~ 200 well-defined locations49,50. The extent of end resection can then be quantified as diagrammed in Fig. 5d51. In short, genomic DNA is purified and digested with restriction enzymes that target locations at various distances from a defined AsiSI locus. Unlike dsDNA, resected DNA will remain undigested and can, therefore, be detected by PCR amplification using primers flanking the restriction enzyme target site. U2OS AsiSI FANCLKO and FANCD2KO cells were generated and validated by western blotting for ubiquitinated and total FANCD2 levels (Fig. 5e). The cells were treated with tamoxifen and end resection was quantified at sites located 335 bp or 1618 bp downstream from a defined AsiSI locus (Chr 1: 89231183) by qPCR. Resected DNA was detected specifically in the tamoxifen-treated cells, and at a higher frequency at the 335 bp location than the 1618 bp location, consistent with previous reports (Fig. 5f)51. Compared to control cells, the frequency of resected DNA was reduced in each of the FANCLKO and FANCD2KO clones, significantly so at the 1618 bp distance (Fig. 5f). These results are consistent with an end resection defect in FANCLKO and FANCD2KO cells, although the phenotype in the AsiSI assay was mild compared to the pRPA phenotype. We, therefore, repeated the AsiSI assay in the presence of a DNA-PKcs inhibitor (PKi) to block c-NHEJ and direct DSB repair towards HR. We reasoned that inhibition of canonical end-joining would allow maximum end resection capacity and would, therefore, more potently reveal a potential end resection defect. As anticipated, DNA-PKcs inhibition increased the frequency of resected DNA ~ 3-fold in control cells, both at the 335 bp location as well as at the 1618 bp location (Fig. 5g). In the FANCLKO cells, however, the NU7441-induced increase in end resection was less pronounced, particularly at the 1618 bp location. Consequently, in the presence of DNA-PKcs inhibitor, the frequency of resected DNA was substantially lower in the FANCLKO cells compared to the control cells (Fig. 5g). The end resection defect in FANCLKO cells in presence of DNA-PKcs inhibitor was also observed when resection was quantified at a second AsiSI locus (Chr 1: 109838221; Supplementary Fig. 5g). Importantly, the end resection defect in the U2OS AsiSI FANCLKO cells was completely rescued by re-expression of FANCL WT (Fig. 5h and Supplementary Fig. 5h). Moreover, whereas re-expression of FANCL LD partially rescued the pRPA phenotype of FANCLKO cells (Fig. 5c), it did not rescue end resection at all in the AsiSI assay (Fig. 5h and Supplementary Fig. 5h). Collectively, these results indicate that FANCL ligase activity promotes end resection at DSBs in a manner dependent on FANCD2 ubiquitination.
To assess the downstream effects of the impaired end resection in FANCLKO cells, we examined the loading of the HR recombinase Rad51, which is dependent on ssDNA generation. U2OS FANCLKO cells and their reconstituted controls were exposed to IR, followed by quantification of nuclear Rad51 foci intensity by immunofluorescence microscopy (Fig. 5i, j and Supplementary Fig. 5i). Compared to control cells, the total Rad51 foci intensity per nucleus was significantly reduced in the FANCLKO cells expressing either the empty vector or the FANCL LD mutant, but not in the FANCLKO cells expressing FANCL WT. Finally, Rad51 accumulation at UV-A laser-induced DNA damage sites was examined and found to be significantly lower in the FANCLKO cells than in WT control cells (Supplementary Fig. 5j, k). Taken together, these data indicate that FANCL promotes Rad51 accumulation at DSBs, consistent with a role for FANCL in end resection.
FANCL and Ube2T promote CtIP recruitment to DSBs
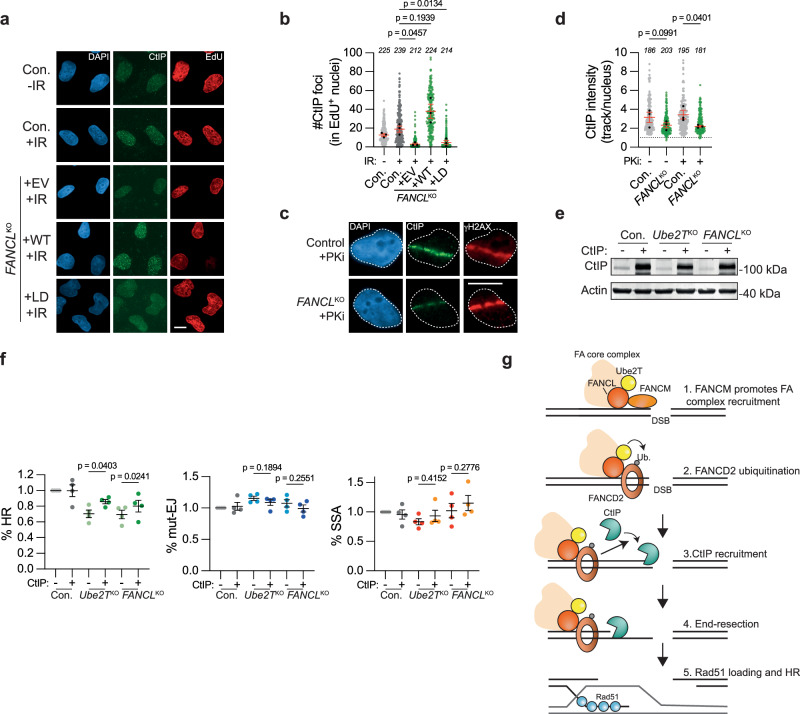
Exactly how the FA core complex promotes end resection at DSBs is unclear. It has previously been shown that FANCD2 directly interacts with the end resection factor CtIP and is required for its recruitment to MMC-induced interstrand crosslinks52,53. We, therefore, hypothesized that FANCL and Ube2T would similarly promote CtIP recruitment to DSBs. To examine this, CtIP accumulation into IR-induced DNA damage foci was quantified in FANCLKO and wild-type control cells. Whereas clear CtIP foci were detected in control cells and in FANCL WT reconstituted FANCLKO cells, a striking absence of CtIP foci was observed in the FANCLKO cells expressing either the empty vector or the FANCL LD mutant (Fig. 6a, b). To further substantiate these findings, CtIP accumulation at UV-A laser-induced DSBs was monitored. CtIP enrichment was reduced at these DSBs in FANCLKO cells compared to their WT counterparts, and this effect was even more pronounced upon inhibition of DNA-PKcs (Fig. 6c, d). Notably, some CtIP accumulation was still observed at UV-A induced DSBs in the FANCLKO cells, indicating that loss of FANCL reduced but did not completely abrogate CtIP recruitment to DSBs. The complete absence of CtIP foci seen following IR exposure is, therefore, most likely explained by a reduction in CtIP accumulation in these foci below the fluorescence imaging threshold. Of note, FANCD2 deficiency has been associated with reduced CtIP expression47. However, we found no difference in CtIP protein levels between control and FANCLKO cells, thus excluding the possibility that FANCL regulates CtIP recruitment indirectly by affecting its expression (Supplementary Fig. 5l). Instead, our results indicate that FANCL ligase activity is required for optimal CtIP recruitment to DSBs.
Fig. 6. FANCL promotes CtIP recruitment to DNA double-strand breaks.
a, b U2OS FANCLKO and wild-type control cells (Con.) were exposed to 10 Gy ionizing radiation (IR), followed by IF microscopy to detect CtIP foci in S-phase (EdU + ) nuclei. Panel (a) shows representative images and panel (b) shows the quantification (scale bar = 10 μM). Plotted are the data from all biological repeats. Each gray or green dot represents an individual nucleus, the total number of nuclei analyzed are indicated in italics, and black dots are the median for each biological repeat (n = 3 independent biological replicates; mean ± SEM; one-way ANOVA with post-hoc Dunnett’s). EV empty vector, WT wild-type, LD ligase-dead. c, d IF-microscopy of UV-A laser micro-irradiated cells. Panel (c) shows representative images, panel (d) shows the quantification (scale bar = 10 μM). Plotted are the data from all biological repeats. Each gray or green dot represents an individual track, the total number of tracks analyzed are indicated in italics, and black dots are the median for each biological repeat (n = 3 independent biological replicates; mean ± SEM; ratio paired t test, two-sided). PKi = DNA-PKcs inhibitor NU7441, 2 μM. e Western blot analysis of CtIP overexpression in the cells described in panel (f). f Indicated HEK 293T + DSB-Spectrum_V3 cell lines were transfected to express either CtIP or an empty vector control, together with Cas9 and the sgRNA targeting the BFP gene in the reporter locus. Next, cells were analyzed by flow cytometry to determine the frequency of repair by each of the three indicated pathways. Data were normalized to the Con.+ EV (n = 4 independent biological replicates; mean ± SEM; One-way ANOVA with post-hoc Dunnett’s). HR homologous recombination, mut-EJ mutagenic end-joining, SSA single-strand annealing. g Model depicting how FANCL/Ube2T promotes the repair of DSBs by homologous recombination. See main text for details. Source data are provided as a Source Data file.
Finally, we sought to determine whether the impaired CtIP recruitment was causal to the HR defect observed in Ube2TKO and FANCLKO cells. As some CtIP recruitment could still take place in the absence of FANCL (Fig. 6c, d), we reasoned that overexpression of CtIP might be sufficient to overcome this recruitment defect. We therefore monitored the frequency of HR repair in control, Ube2TKO or FANCLKO DSB-Spectrum_V3 cells in the absence or presence of CtIP overexpression (Fig. 6e). While CtIP overexpression did not affect the extent of DSB repair by HR in control cells, it significantly enhanced it in Ube2TKO or FANCLKO cells (Fig. 6f). In contrast, CtIP overexpression did not significantly affect the levels of DSB repair by mut-EJ or SSA in any of the FA-deficient cell lines. Hence, CtIP overexpression rescued the HR defect in Ube2TKO and FANCLKO cells. To assess whether CtIP overexpression could also rescue the PARPi sensitivity of these cells, we transfected U2OS FANCLKO and control cells with GFP-NLS or GFP-tagged CtIP and exposed these cells to olaparib. Their survival and proliferative capacity were then determined using clonogenic assays and flow cytometric quantification of live, GFP-positive cells (Supplementary Fig. 6a–c). In both assays, GFP-expressing FANCLKO cells were significantly more sensitive to olaparib than GFP-expressing control cells, consistent with our earlier results (Fig. 3d and Supplementary Fig. 6a, c). In cells overexpressing CtIP, however, FANCL deficiency did not significantly sensitize to Olaparib (Supplementary Fig. 6a, c). Together, these data indicate that impaired CtIP-dependent end resection is causal to the reduced HR capacity and the increased PARPi sensitivity observed in FA core complex-deficient cells.
Discussion
Here, we report that the FA core complex members FANCL and Ube2T, as well as their downstream substrate FANCD2, promote the repair of DSBs through HR. This function is independent of their role in ICL repair, as FANCL and Ube2T are directly recruited to both UV-A laser- and nuclease-induced DSBs. Their recruitment and activity are required for the accumulation of FANCD2 at DSBs. In the absence of FANCL, Ube2T, or FANCD2, DSB end resection is impaired. This can be explained by reduced recruitment of CtIP, whose accumulation at DSBs is highly dependent on the presence of ligase-proficient FANCL. Consequently, FANCL-deficient cells have reduced loading of Rad51 onto IR- or UV-A laser-induced DSBs. Collectively, our data support a model in which Ube2T/FANCL drives the ubiquitination and accumulation of FANCD2 at DSBs to promote CtIP recruitment and end resection during the initial steps of HR (Fig. 6g).
A function for the FA core complex in HR was first suggested approximately two decades ago, based on experiments showing reduced HR in reporter assays in FANCC/FANCG KO chicken DT40 cells20,54. These results were later confirmed in mouse and human cells, and extended to other FA-factors, including FANCL and Ube2T21,22,55–60. Nevertheless, the importance of the FA core complex and FANCD2 for HR has remained controversial for a number of reasons61. Most importantly, the HR phenotype in reporter assays was generally considered mild, when comparing FA-factor deficient cells to those missing canonical HR factors like BRCA121,44,47,61. Similarly, FA-patient derived cell lines were only modestly more sensitive to PARPi treatment than WT control cells26,27. Finally, for FANCA, FANCG, and FANCI, the HR phenotypes were ambiguous22,28,56. Here, FANCL, Ube2T, and FANCM were identified as important HR-promoting factors using an unbiased genetic screening approach targeting 2760 genes. Similarly, a recently published screen that studied HR with an ectopically provided dsDNA donor, also identified FA genes to be strong drivers of this process23. Furthermore, we validated the HR function of FANCL, Ube2T, and FANCD2 using orthogonal assays in a variety of isogenic (non-complemented and complemented) knock-out and control cell lines. These data, therefore, add compelling evidence for an HR-promoting function of the FA core complex and FANCD2.
Although FANCD2KO cells showed an end resection defect, as well as increased PARPi sensitivity, these phenotypes were not as severe as those of the FANCLKO and Ube2TKO cells. This might indicate that FANCL and Ube2T have additional HR-promoting functions that are FANCD2 independent. However, it cannot be excluded that the FANCD2KO clones used in these studies had mild phenotypes due to hyper-activation of alternative, FANCD2-independent genome maintenance mechanisms. Activation of such compensatory pathways might be essential for the outgrowth of FANCD2KO clones, but not of Ube2TKO or FANCLKO clones, if FANCD2’s function in genome maintenance extends beyond that of Ube2T/FANCL60,62,63.
We found that HR is not completely abrogated upon depletion of FANCL or Ube2T, despite the use of validated clonal CRISPR KO cells. The residual HR could be explained by FA factors being required for the repair of a specific subset of DSBs. For example, the FA core complex could be specifically recruited to those DSBs that cause stalling of replication forks, similar to how it is recruited to ICLs upon fork stalling at these lesions18. Alternatively, the FA core complex might primarily be required for the repair of DSBs with blocked ends. This hypothesis could explain why end resection of AsiSI-induced DSBs was more dependent on FANCL when DNA-PKcs was inhibited (Fig. 5g). The latter locks the Ku/DNA-PKcs complex on DSB ends, because auto-phosphorylation of DNA-PKcs is required to loosen its interaction with DNA64.
Throughout our studies, we primarily focused on FANCL, Ube2T, and FANCD2, but we also showed that the accumulation of FANCL and FANCD2 at DSBs is dependent on FANCM and FANCC, respectively. Based on these results, and on published data describing HR functions for other FA factors22,23,65, we consider it likely that FANCL and Ube2T function as subunits of the complete FA core complex in DSB repair. Our finding that FANCM plays an important role in recruiting the complex to a DSB is intriguing because in vitro FANCM binds most strongly to DNA substrates with branched arms and interacts poorly with non-branched dsDNA35. Potentially, accessory factors stabilize FANCM binding to DSBs in vivo. One candidate for such a function would be FANCA, which was shown to bind to blunt dsDNA in vitro28.
Whereas FANCL/Ube2T loss clearly reduced HR, we could not observe any effect on mutagenic end-joining or SSA in our DSB-Spectrum_V3 reporter assays. These end-joining results are in agreement with a study by Howard et al., which reported no effect on total end-joining upon depletion of different FA factors22. However, Howard et al., as well as others, did show a function for the FA core complex and FANCD2 in DSB repair through a-EJ, in studies using reporters specific for this pathway22,57,66. FANCD2 has been shown to promote the accumulation of the a-EJ factor PolΘ to foci induced by UV irradiation or hydroxyurea66. Such an a-EJ function would be consistent with FA factor-dependent recruitment of CtIP, which has been shown to promote a-EJ, but not c-NHEJ67.
We did not detect an a-EJ phenotype in FA factor deficient cells in our studies. This can be explained by the observation that the Cas9 target site in DSB-Spectrum_V3 is predominantly repaired through c-NHEJ, and rarely through a-EJ17. More surprisingly was the absence of an SSA phenotype in our FANCL/Ube2T deficient cells. Although this observation is in agreement with some other studies66, the majority of reporter studies observed a reduction in SSA upon loss of FA-factors21,22,28. Impaired SSA would also be expected, considering its dependency on end resection1. These discrepant results might be explained by the different reporters used in our studies compared to those used in the other studies. We previously observed that knockdown of all established end resection factors reduced SSA-repair of DSB-Spectrum_V3, but this effect was relatively mild for CtIP, despite a strong reduction in HR17. However, when assessed by the SA-GFP reporter, CtIP knockdown strongly reduced SSA22. For both DSB-Spectrum_V3 and SA-GFP, the SSA-repair product that is measured is a repeat-mediated deletion, but the reporters differ with regards to the length of the repeats (517 bp and 280 bp, respectively) and the distance between repeats (3.2 kb and 2.4 kb, respectively). Furthermore, the DSB is generated by the I-SceI nuclease in SA-GFP, which leaves a four nucleotide 3’ overhang, but by Cas9 in DSB-Spectrum_V3, which generally leaves blunt ends and remains associated with the DNA substrate after nucleolysis68. These differences between the reporters could affect the requirements put on the end resection machinery and, hence, the extent to which SSA-mediated DSB repair is dependent on CtIP.
Notwithstanding its function in SSA, CtIP is a core HR factor that interacts with the MRN-complex5, and with BRCA110,11. Perhaps surprisingly, these interactions were demonstrated to be non-essential for CtIP accumulation at DNA lesions13,14. In contrast, our data indicate that CtIP accumulation at DSBs is strongly dependent on the FA core complex and FANCD2. Similarly, it has been shown that FANCD2 is required for the recruitment of CtIP to ICLs, as well as to DNA damage and stalled replication forks induced by hydroxyurea, aphidicolin, or UV irradiation52,53,66,69. Moreover, FANCD2 has been shown to interact directly with CtIP, independent of any DNA damage-inducing treatment52,53,69. Mono-ubiquitination of FANCD2 was reported to promote its interaction with CtIP, but conflicting data were obtained by Yeo et al., who found the interaction to be independent of FANCA or FANCC52,53,69. Nevertheless, the combined studies indicate that promoting the recruitment of CtIP to sites of DNA damage is a key function of the FA core complex and FANCD2.
Our findings emphasize that DSB repair by HR requires the orchestrated activity of a multitude of factors that do not necessarily operate in a linear pathway. These findings furthermore indicate that genome maintenance factors can be involved in multiple pathways, thus causing complex phenotypes when lost or mutated. In line with this notion, inactivating mutations in different multifunctional FA genes cause complex FA disease phenotypes19. FA patients, in general, have defective ICL repair, and an impaired response to replication stress61. In addition, our data adds to a body of evidence suggesting that FA-defective cells have a reduced capacity to repair DSBs by HR. Further investigation into the extent to which improper DSB repair contributes to the FA disease phenotype is therefore warranted.
Methods
Cloning
The cloning design was done using Snapgene 7.02 (Dotmatics). Restriction enzymes and T4 DNA ligase, Phusion Polymerase, and NEBuilder HiFi DNA Assembly Master Mix were all obtained from New England Biolabs and used according to the manufacturer’s instructions. Gel extraction, PCR purification, and DNA mini-, midi- or maxi prepping were done using Qiagen kits according to the manufacturer’s instructions. To generate HEK 293T + Cas9 + DSB-Spectrum_V2 cells, a lentiviral Cas9 plasmid was cloned by NheI/BamHI restriction to isolate 3xFLAG-Cas9 from pCW-3xFLAG-Cas9 (Addgene # 50661)70 followed by ligation into SpeI/BamHI digested pLVX-IRES-Hygromycin. The pLVX-DSB-Spectrum_V2 plasmid was previously described17. To activate the reporter in the genetic screen, pLX-sgRNA (Addgene #50662)70 was modified by replacing the blasticidin resistance cassette with mCherry using BspeI/EcoRI-based restriction ligation. The BspeI site was blunted with Klenow polymerase (New England Biolabs) prior to ligation. Next, the BFP-targeting sgRNA was cloned into pLX-mCherry using the published protocol70. The sgRNA library was cloned into Lentiguide-Blast, which was generated by replacing the EF1a-Puro cassette in Lentiguide-Puro (Addgene #52963)71 with an EF1a-Blasticidin cassette using the Xma/Mlu restriction sites. The CRISPR constructs to make KO cell lines were generated in pSpCas9-2A-iRFP(670), a previously described derivative of pSpCas9-2A-puro17. Cloning of sgRNAs was done by ligating annealed sgRNA primer dimers into BpiI-digested pSpCas9-2A-iRFP(670), as previously described72. The sgRNAs were selected from the Brunello sgRNA library73. To re-express FANCL in the FANCLKO cell lines, a pLVX-hPGK-FANCL-Hygromycin construct was generated. First, the CMV-promoter in the pLVX-Hygromycin lentiviral backbone was replaced with a hPGK promoter by assembling a PCR-derived hPGK fragment into ClaI-digested pLVX-Hygromycin together with FANCL cDNA that was PCR amplified from a cDNA library. For the recruitment assays, GFP-tagged FANCL was cloned by PCR-amplification of eGFP and assembly into XhoI-digested pLVX-hPGK-FANCL-Hygro. GFP-tagged Ube2T was generated in an identical manner, with Ube2T cDNA also derived from a cDNA library. FANCD2 was amplified from a cDNA library and assembled into pLVX-hPGK-GFP-Ube2T digested with XhoI/NotI to replace Ube2T. The pSpCas9-BFP-sgRNA-iRFP(670) plasmids for transient DSB-Spectrum_V3 reporter experiments was previously described17. The pcDNA-CtIP-2xFLAG construct used for CtIP overexpression was a kind gift from Petr Cejka (Institute for Research in Biomedicine, Bellinzona, Switzerland)74. The doxycycline-inducible pCW57.1-GFP-CtIP was obtained from Addgene (#71109)75.
Cell lines
All cell lines were grown at 37 °C with 5% CO2 in DMEM supplemented with 10% fetal calf serum and antibiotics. All cell lines were regularly tested for mycoplasma infection and consistently found mycoplasma-free. U2OS and HEK 293T cells were originally obtained from ATCC (HTB-96 and Crl-3216, respectively) and authenticated by Short Tandem Repeat analysis. The U2OS 2-6-3 cells expressing ER-mCherry-LacI-FokI-DD were a kind gift from Roger Greenberg (University of Pennsylvania, Philadelphia, Pennsylvania, USA)46. The U2OS AsiSI cells were a kind gift from Gaelle Legube (CBI, Toulouse, France)50. The HEK 293T DSB-Spectrum_V3 cell line was previously described17. To generate HEK 293T + Cas9 + DSB-Spectrum_V2 cells, regular HEK 293T cells were lentivirally transduced with pLVX-Cas9-Hygromycin, and selected on Hygromycin B (200 μg/ml; Thermo Fisher Scientific). Next, cells were lentivirally transduced with pLVX-DSB-Spectrum_V2 at low multiplicity-of-infection (MOI) to ensure single integration, and selected on puromycin (1 μg/ml; Invivogen). Subsequently, BFP-positive cells were single-cell sorted by FACS in 96-well plates and expanded. A clone was selected based on the appearance of a sizeable GFP+ and BFP- population, specifically after reporter activation.
Lentiviral transduction
To produce lentivirus, HEK 293T cells were transfected with the lentiviral plasmid and packaging plasmids pCMV-VSVg and pCMV-ΔR8.2 at a 6:1:4 mass ratio. Transfection was done using the CalPhos mammalian transfection kit (Clontech). Alternatively, HEK 293T cells were transfected with the lentiviral plasmid and the packaging plasmids pMDLg/RRE, pRSV-REV, and pMD2.g at a 4:2:1:1 mass ratio using jetPEI transfection reagent (Polyplus). At 48–72 h after transfection, the medium with the virus was harvested and filtered through a 0.45 μM filter. Next, polybrene (4–8 μg/ml) was added and viral supernatant was added to the recipient cells. Infected cells were selected by treatment with Hygromycin B (ThermoFisher Scientific).To obtain similar expression levels of GFP-FANCD2 WT and K561R in the U2OS FANCD2KO cells, these were sorted on equal GFP levels by FACS.
Generation of the custom sgRNA library
To generate the targeted sgRNA library for the genetic screen, the sgRNA sequences of selected targets were copied from the Brunello library73 (Supplementary Data 1). The targeted library was divided into the following four subpools. Subpool 1 targeted the kinome (763 genes). It already existed as a Brunello sub-library and the sequences could be copied directly. Subpool 2 targets chromatin factors (1102 genes). Targets were selected based on association with a chromatin Gene Ontology term or the presence of a bromo-, chromo-, Jumanji, PHD, or TUDOR domain, which was determined using AmiGO, Panther, and SMART76–78. Subpool 3 targeted ubiquitin and SUMO-associated genes (908 genes). This consisted of genes listed in Hutchins et al.79 supplemented with targets missing from the list but having ubiquitin or SUMO-related GO terms, as well as the genes encoding for SUMO or ubiquitin itself. Subpool 4 consisted of phosphatases, and targets were selected using the DEPOD database (144 genes)80. Furthermore, 350 non-targeting control sgRNAs were added, selected from the Brunello library. The total library, therefore, contained 12,018 sgRNAs targeting 2760 non-redundant genes, with four sgRNAs per gene.
Oligos were designed to contain, from 5’ to 3’, a library-specific forward primer sequence (lib FWD), a U6 promoter region for Gibson Assembly (U6 FWD), the sgRNA protospacer sequence, a sgRNA scaffold region for Gibson assembly, and a 10 bp overhang (Supplementary table 1). These 95–96 bp oligo’s were obtained in a pooled format (CustomArray, Inc.), and PCR-amplified using a nested PCR reaction with first the lib FWD primer and an outer reverse primer, followed by a PCR with the U6 FWD and an inner reverse primer. The PCR product was gel-extracted and assembled into BsmBI-digested Lentiguide-Blast using the Quick-Fusion kit (Biotools). The assembled plasmid was transformed by electroporation into Endura competent cells (Biosearch technologies), followed by DNA prepping of the libraries.
Genetic screen in DSB-Spectrum_V2 cells
Throughout the experiment, the representation of the sgRNA library was maintained at 250x. First, HEK 293T + Cas9 + DSB-Spectrum_V2 cells were lentivirally infected with the sgRNA library at an MOI of 0.2 to ensure single integration. Cells were expanded in the presence of Blasticidin S (10 μg/ml; Invitrogen) to high numbers and thereafter stored in liquid nitrogen. Next, three independent replicate experiments were performed. For each replicate of the screen, sgRNA library-infected DSB-Spectrum_V2 cells were taken into the culture and allowed to recover from freeze-thawing for several days. Next, they were lentivirally transduced to introduce pLX-BFP-sgRNA-mCherry at a high MOI, giving 100% infection and strong sgRNA expression. At seven days after BFP sgRNA infection, 4–12*106 cells of the GFP + , BFP- and total population were harvested by FACS on a BD FACSAria II or III (BD Biosciences). Next, genomic DNA was isolated using the Qiagen Blood and Tissue DNeasy kit according to the manufacturer’s protocol. The sgRNA sequences were amplified from the genomic DNA with primers Illumina PCR1 FWD and Ilumina PCR1 REV using Phusion Hot Start Flex polymerase (New England Biolabs), with the annealing temperature set at 58 °C. The resulting product was concentrated by PCR purification using the Qiagen PCR purification kit and subsequently gel-extracted using the Qiagen Gel extraction kit. Next, a short, 12-cycle PCR was performed with Illumina PCR2 FWD and Illumina PCR2 REV to add the Illumina adapters, indices, and sequencing primer binding sites (see Supplementary Data 2 for index-sequences). The resulting PCR product was analyzed by agarose gel electrophoresis and gel-extracted. Next, PCR products were sequenced on a HiSeq 2000 sequencing platform (Illumina). A fraction of the sgRNA plasmid library was also sequenced to determine the sgRNA distribution in the input material. Next, sgRNA counts were extracted from the sequencing fastq files. Prior to analysis, any sgRNA with a read count < 50 in the reference population was excluded from all samples in that particular repeat. Finally, sgRNA fold-change and statistical analysis were determined by uploading the read-count files in the BasDAS analysis system81, a web-based interface to analyze sgRNA depletion and enrichment using the MAGeCK algorithm82. The list of essential genes was obtained from Wang et al.30.
Generation of knock-out cell lines
To generate Ube2T/FANCL and FANCD2 knock-out cell lines, the parental cell lines were transfected using lipofectamine 2000 (Thermo Fisher Scientific) with pSpCas9-iRFP(670) plasmids containing the relevant sgRNAs (Spacer sequences: Ube2T sgRNA TTGCCAACATGTGATGCCTG; FANCL sgRNA ATTTACAACTGAAGAATGCA and FANCD2 sgRNA AGATAATCTAAAATGCCCTG). In all cases, a control cell line transfected with an AAVS1-targeting sgRNA (spacer sequence GGGGCCACTAGGGACAGGAT) was taken along. At 24–72 h after transfection, cells were sorted by FACS for high iRFP(670) expression, followed by culturing and expansion for at least seven days. Next, the AAVS1sg control cells were maintained as a pool, and the Ube2T/FANCL and FANCD2 edited cells were single-cell sorted by FACS in 96-well plates and expanded. Once confluent, growing clones were re-arrayed and split into two 96-well plates, one for maintenance and one for testing. At 24–72 h after replating, the test plate was washed twice with PBS and DirectPCR lysis reagent (Viagen) mixed 1:1 (vol) with ddH2O and supplemented with 0.2 μg/ml proteinase K was added to the wells, followed by overnight incubation at 55 °C. Subsequently, Proteinase K was inactivated by incubation at 85 °C for 1.5 h. Next, a PCR using GoTaq G2 polymerase (Promega) was performed to specifically amplify the WT target locus and not any CRISPR-edited loci. Clones for which no PCR product was obtained were considered candidate knock-out cells and were further expanded. Knock-out status was validated by western blot for Ube2T and FANCD2. For FANCL, the genomic DNA was isolated using the DNAeasy blood and tissue kit (Qiagen), and the target locus was PCR amplified using the FANCL TIDE primers. Next, PCR products were analyzed by Sanger sequencing and by chromatogram deconvolution using TIDE38. FANCL KO status was additionally validated by assessing FANCD2 ubiquitination after MMC treatment on western blot.
DSB-Spectrum reporter assays
For the reporter assays using HEK 293T + Cas9 + DSB-Spectrum_V2 cells, the BFP-targeting sgRNA or AAVS1-targeting control sgRNA was introduced by lentiviral transduction with pLX-sgRNA-mCherry. Next, cells were analyzed by flow cytometry on a BD LSRFortessa (BD Biosciences) running FACS DIVA 5.0.3 or on a Novocyte Quanteon (Agilent) running NovoExpress 1.6.1. Data analysis was done using FlowJo 10.8.0. Gating was done on live cells, single cells, and mCherry-positive events, and the frequency of GFP+ and BFP- cells was determined. The background GFP+ and BFP- frequencies in the AAVS1sg cells were subtracted from those in the BFPsg cells.
For all reporter assays in the Ube2TKO and FANCLKO HEK 293T + DSB-Spectrum_V3 cells, the control cell line (Con.) was the AAVS1sg-transfected and iRFP(670)-sorted pool (see above). DSB-Spectrum_V3 cells were transiently transfected with pSpCas-iRFP(670) containing a BFPsg (spacer sequence ACGGGGTCCAGTGTTTTGCC) or AAVS1sg using lipofectamine 2000 according to manufacturers’ instructions. At 48–96 h after transfection, cells were harvested and analyzed by flow cytometry, gating was done on live cells, single cells, iRFP(670)-positive events. The frequency of GFP+ (HR), BFP-/mCherry+ (mut-EJ), and BFP-/mCherry- (SSA) cells was determined and corrected for background levels in the respective AAVS1sg population. Next, the frequency of each repair population was divided by the sum of frequencies of all three repair populations. For the FANCD2si reporter experiments, HEK 293T + DSB-Spectrum_V3 cells were transfected using lipofectamine RNAiMAX (ThermoFisher Scientific) according to manufacturers’ instructions with either a control siRNA (Control siRNA duplex negative control; Eurogentec SR-CL000-005) or a FANCD2si (Eurogentec; custom sequence 5’-GGUCAGAGCUGUAUUAUUC-3’). This was followed by a second round of siRNA transfection 24 hours later. At 54 h after the first siRNA transfection, the spCas9-iRFP(670) plasmids were transfected and the assay was carried out as described above.
Cell viability assays
For clonogenic survival assays, cells were trypsinized and seeded at low density in a medium with the indicated concentrations of olaparib (Selleckchem, S1060). For the BRCA2 depletion experiment, cells were transfected 24 h prior to plating with luciferase siRNA (Dharmacon; 5’-CGUACGCGGAAUACUUCGA-3’) or BRCA2si (Dharmacon; 5’-GAAGAAUGCAGGUUUAAUA-3’) using lipofectamine RNAiMAX according to manufacturers’ instructions. The medium was replaced with fresh medium containing olaparib 7 days later. At 14 days after plating, cells were washed and colonies were stained with methylene blue (2.5 gr/L in 5% ethanol). Colony number was determined manually, and normalized to the number of colonies in the untreated control plate. Non-linear curve-fitting ([inhibitor] vs. normalized response – variable slopes; no constraints) was performed using GraphPad Prism to determine the IC50.
For the competition assay (Supplementary Fig. 3d), U2OS FANCD2KO cells expressing GFP-NLS, GFP-FANCD2 WT, or GFP-FANCD2 K561R were mixed in a 1:1 ratio with the parental U2OS FANCD2KO cells. Subsequently, mixed cell populations were treated with 1 μM olaparib or left untreated for 12 days. On days 6, 9, and 12, samples were taken from each population, and the fraction of GFP-positive cells was determined by flow cytometry on a Novocyte flow cytometer (Agilent). Subsequently, the fraction of GFP-positive cells in the olaparib-treated population was normalized to the GFP-positive fraction in the untreated population.
To assess the rescue of PARPi sensitivity by CtIP overexpression, U2OS control or FANCLKO cells were transfected to express GFP or GFP-CtIP under the control of a doxycycline-inducible promoter. Next, the GFP-positive population was collected by FACS, and cells were plated for clonogenic survival. Alternatively, cells were reverse-transfected in suspension, and the mixture of cells with transfection reagent was split over multiple wells. Subsequently, the cells were left untreated or treated with olaparib 24 hours after transfection. Four days later, their proliferative capacity was determined by quantifying live, GFP-positive cells on a Novocyte flow cytometer.
Western blotting
Cell pellets were lysed on ice in RIPA buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA, 1% Triton-X100, 0.5% Sodium Deoxycholate, 0.1% SDS, 150 mM NaCl) supplemented with cOmplete EDTA-free Protease Inhibitor Cocktail tablets (Roche), 2 mM MgCl2 and Benzonase Nuclease (100 U/ml; Merck Millipore). Insoluble material was pelleted by centrifugation (21,300 × g, 15 min.), and protein concentration was determined using a BCA assay (Pierce). Next, Laemmli SDS-sample buffer with a reducing agent was added to the lysates. Alternatively, cells were harvested and lysed directly in 2x Laemmli sample buffer. Samples were boiled at 95 °C for 5 min. Next, the protein was separated by SDS-PAGE on a 4–15% Criterion TGX pre-cast midi protein gel (Bio-Rad) and transferred to the nitrocellulose membrane using a standard tank electrotransfer protocol. Membranes were blocked with 5% Blotto non-fat dry milk (Santa-Cruz) (W/V) in Tris-Buffered Saline (TBS). Next, primary as well as secondary antibody staining was performed in a Blocking buffer for fluorescent WB (Rockland), diluted 1:1 in TBS with 0.1% Tween-20 (TBS-T). Membranes were imaged on an Odyssey CLx scanner (LI-COR BioSciences) running ImageStudio V5.2 (LI-COR Biosciences), and image analysis was done using ImageStudio Lite 5.2.5 (LI-COR BioSciences). All antibodies are listed in Supplementary Table 2.
FANCM qPCR
To assess FANCM transcript levels by qPCR, RNA was extracted from siRNA transfected cells using the RNAeasy kit (Qiagen) according to manufacturers’ instructions, followed by cDNA synthesis using the iScript cDNA synthesis kit (BioRad). Next, the qPCR reaction was set up using the PowerUp SYBR green master mix (Applied Biosystems) to amplify FANCM cDNA or Actin cDNA (see Supplementary Table 1 for primer sequences). The reaction was run on a CFX384 C1000 Touch thermal cycler (BioRad). The Ct-values were obtained using CFX Maestro 2.0 software (BioRad), and fold change in FANCM transcript levels was quantified using the ΔΔCt method.
Immunofluorescence microscopy
Cells were plated on coverslips and treated 24–48 hours later. Next, cells were washed with PBS or with ice-cold CSK buffer (10 mM HEPES (pH 7.4), 300 mM Sucrose, 100 mM NaCl, 3 mM MgCl2) in case of Rad51 IF, and pre-extacted with 0.25% Triton-X100 in PBS (or CSK-buffer) on ice for 2 min. Next, cells were fixed with 2% formaldehyde or paraformaldehyde in PBS for 20 min. at room temperature (RT), followed by washing with PBS. Cells were permeabilized again with 0.5% NP-40 in PBS for 5 min. at RT, washed with PBS, and blocked 1% BSA (w/v) in case of Rad51 IF, for 1 h at RT. Next, primary and secondary antibody staining was done in PBS + 1% BSA at RT. In all cases, 0.1 μg/mL 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) was added to the secondary antibody mixture. If needed, incorporated EdU was labeled by a copper-catalyzed click-it reaction. First, cells were fixed again with 2% formaldehyde for 15 min. at RT. Next, the click-it reaction mix (PBS with 100 mM Trs-HCl (pH 8.5), 1 mM CuSO4, 100 mM freshly prepared ascorbic acid) with 1 μl Alexa Fluor™ 647 Azide (Thermo Fisher Scientific) per 5 ml was added to the cells, followed by incubation for 30 min. at RT. Cells were washed with PBS and coverslips were mounted on glass microscopy slides with polymount. Images were acquired on a Zeiss AxioImager M2 wide-field fluorescence microscope with 63x PLAN APO (1.4 NA) oil-immersion objectives, running ZEN 2012 blue edition v1.1.0.0 (Zeiss).
UV-A laser micro-irradiation and FokI nuclease assays
For UV-A laser micro-irradiation, cells were plated on coverslips and first treated with 15 μM 5′-bromo-2-deoxyuridine (BrdU) for 24 h to sensitize for the generation of DSBs. Next, the growth medium was replaced with Leibovitz’s L15 medium supplemented with 10% fetal calf serum (FCS), and cells were placed in a Chamlide TC-A live-cell imaging chamber. UV-A laser tracks were made by a diode-pumped solid-state 355-nm Yttrium Aluminum Garnet laser (average power 14 mW and repetition rate up to 200 Hz). The laser was integrated into a UGA-42-Caliburn/2 L Spot Illumination system (Rapp OptoElectronic). Micro-irradiation was combined with live-cell imaging in an environmental chamber set to 37 °C on an all-quartz wide-field fluorescence Zeiss Axio Observer 7microscope, using a Plan-Neofluar 63 × (1.25 NA) oil-immersion objective (UV-A). One field of view was micro-irradiated per min., for a time period of fifteen minutes. Directly here-after, cells were pre-extracted and fixed as described above. For FokI nuclease assays, U2OS 2-6-3 cells stably expressing ER-mCherry-LacI-FokI-DD were plated on coverslips, and treated with 1 μM 4-hydroxy-tamoxifen (4-OHT; Sigma-Aldrich) and 1 μM Shield1 (Clontech lab, now Fisher Scientific) for 4 h. Next, cells were pre-extracted and fixed as described above. For RNAi-experiments, cells were transfected with a control siRNA (Control siRNA duplex negative control; Eurogentec SR-CL000-005), Mre11 siRNA (ThermoFisher Scientific; Silencer select s8958), or FANCM siRNA (ThermoFisher Scientific; Silencer select s33619 (si1) and s33620 (si2)) using lipofectamine RNAiMAX (ThermoFisher Scientific) according to manufacturers’ instructions. At 48–72 h after transfection, cells were treated with 4-OHT and Shield-1.
All image analysis was performed using ImageJ 2.0.0 software (ImageJ). First, the damaged area was identified in the damage marker channel (γH2AX or MDC1) by thresholding to distinguish the laser stripe or FokI focus, and selection using the wand. Next, the signal intensity of GFP-FANCL/GFP-Ube2T or endogenous FANCD2 in the damaged area was determined (Idamage), as well as their average signal in the nucleoplasm (Inucleoplasm) and the background signal in the imaged area (Ibackground). Subsequently, enrichment was calculated as follows: ((Idamage − Ibackground)/(Inucleoplasm − Ibackground)).
IR-induced foci analysis
Cells on coverslips were treated with ionizing radiation using a Xylon X-ray generator machine (Y.TU225-D02; 200 KV; 12 mA; dose rate 2 Gy/min), and pre-extracted and fixed 4 h later. The pRPA and CtIP foci were quantified using the Image-J macro “Foci-analyzer” (freely available at https://github.com/BioImaging-NKI/Foci-analyzer; created by Bram van den Broek, the Netherlands Cancer Institute, the Netherlands). The Rad51 foci were analyzed with the Olympus ScanR Image Analysis Software (3.3.0). A dynamic background correction was applied, and single-cell nuclei were segmented using an integrated intensity-based object detection module based on the DAPI signal. Rad51 foci segmentation was performed using an integrated spot-detection module to obtain foci counts and foci intensities. Cell cycle staging was performed based on the total DAPI intensity per cell, which scales with DNA content, and the mean EdU intensity per cell, which indicates DNA synthesis. All downstream analyses were performed on properly detected interphase nuclei containing a 2N-4N DNA content as measured by total and mean DAPI intensities. Fluorescence signal intensities are depicted as arbitrary units.
AsiSI end resection assay
The AsiSI end resection assay was done essentially as described in Zhou et al.51. In short, U2OS AsiSI cells were treated with 1 μM 4-OHT, with or without 2μM NU7441 (SelleckChem), and harvested by trypsinization 4 h later. Next, genomic DNA was isolated using the DNeasy blood and tissue kit (Qiagen), and 250 ng DNA was digested with BsrGI (Site 1), BamHI (Site 2), or HindIII (Control; New England Biolabs) overnight at 37 °C. Next, a qPCR was performed using 40 ng digested genomic DNA as a template with the GoTaq qPCR master mix (Promega) according to the manufacturer’s instructions. See the oligo table for primer sequences, which were all identical to the ones described in Zhou et al., except for the DSB2_364 site. The qPCR-reaction was run and quantified on a CFX384 C1000 Touch thermal cycler (BioRad). Data were analyzed to obtain the ΔCt for each reaction using BioRad CFX Manager Software version 3.1, and subsequently, the % of ssDNA% at each location was calculated as follows: ssDNA% = 1/(2^(ΔCt-1) + 0.5)*100.
Cell-cycle distribution
For cell cycle analysis, cells were pulse-labeled with 15 μM BrdU for 1 h, fixed in 70% ethanol, and subsequently denatured in 2 M HCl. Next, cells were stained with α-BrdU antibody and incubated with 0.1 mg/ml propidium iodide, followed by flow cytometric analysis.
Statistics and reproducibility
All statistical testing was done using GraphPad Prism (Dotmatics) version 9.5.1 or 10.2.2 and was two-sided. All indications of “n = ” in the figure captions refer to the number of independent biological replicates, with the exception of Fig. 4b, d, g, i and Supplementary Fig. 4b, for which “n = ” indicates the number of cells imaged and plotted in the depicted experiment. All western blot images shown in the main manuscript are representative of at least two repeats, except for the western blot images shown in Figs. 5e and 6e, which were both done once.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank the LUMC Flow cytometry core facility (LUMC, Leiden, the Netherlands), the UMCG Flow Cytometry Unit (UMCG, Groningen, the Netherlands), and the Robert A. Swanson (1969) Biotechnology Center (Koch Institute, MIT, Cambridge MA, USA) for assistance. We thank Petr Cejka (Institute for Research in Biomedicine, Bellinzona, Switzerland) for sharing the FLAG-CtIP construct. This research was supported by a fellowship from the Dutch Cancer Society (BUIT 2015-7546; B.v.d.K.), by funding from the Leiden University Medical Center regulation for MSCA-IF Seal of Excellence awardees (B.v.d.K.), by Institute Core and Children’s Brain Tumour Center of Excellence grants from Cancer Research UK (C9545/A29580 and C9685/A26398; P.C.), by NWO-VICI grant (09150182110019; M.v.V.), by the Swiss National Science Foundation (310030_197003; M.A), by National Institute of Health grants R01-ES015339 (M.B.Y.), R35-ES028374 (M.B.Y.), R01-CA226898 (M.B.Y.), by the joint Cancer Research UK and Brain Tumor Charity funded Brain Tumor Award C42454/A28596 (M.B.Y.), by the Charles and Marjorie Holloway Foundation (M.B.Y.), by the MIT Center for Precision Cancer Medicine, by the Cancer Center Support Grant P30-CA14051 from the National Cancer Institute, by the Center for Environmental Health Sciences Support Grant P30-ES002109 from the National Institute of Environmental Health Sciences (M.B.Y.), and by ERC Consolidator (ERC-CoG-617485; H.v.A.) and NWO-VICI grants (VI.C.182.052; H.v.A.).
Author contributions
B.v.d.K. and M.B.Y., and H.v.A. conceived the project. B.v.d.K., F.v.d.W., M.v.V., M.B.Y., and H.v.A. designed the experiments. B.v.d.K. and F.v.W. generated the cell lines. B.v.d.K, F.v.d.W., and W.W. validated the KO and rescue status of the cell lines. B.v.d.K. performed the genetic screen and the DBS-Spectrum reporter experiments. P.C. and B.J. assisted with the analysis of the genetic screen results. F.v.d.W. and W.W. performed the clonogenic and proliferation assays. B.v.d.K. and M.B.R. performed the micro-irradiation and FokI recruitment assays. F.v.d.W. and W.W. performed the pRPA foci experiments. F.v.d.W. and M.B.R. performed the AsiSI end-resection assays. B.v.d.K. performed the Rad51 foci experiments, for which data analysis was done by M.S., J.V., and M.A. F.v.d.W. performed the CtIP foci experiments. Unless specified otherwise, data analysis was done by the experimenter, together with B.v.d.K, F.v.d.W., H.v.A., M.v.V., and M.B.Y. B.v.d.K. and H.v.A. wrote the paper. M.B.Y. edited the manuscript. M.A., M.v.V., M.B.Y., and H.v.A. supervised the study. B.v.d.K., M.A., M.v.V., M.B.Y., and H.v.A. obtained funding for the work.
Peer review
Peer review information
Nature Communications thanks Lachaud Christophe and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Sequencing files from the CRISPR/Cas9 screen are available through the Sequence Read Archive of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra), with project number PRJNA1013929, accessions SRX21658367, SRX21658368 and SRX21658369. Note that the sequencing files contain data obtained for two independent HEK 293T DSB-Spectrum_V2 clones (D9 and E5). The representative data presented in this manuscript are from clone D9. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bert van de Kooij, Fenna J. van der Wal.
Contributor Information
Bert van de Kooij, Email: l.w.van.de.kooij@umcg.nl.
Michael B. Yaffe, Email: myaffe@mit.edu
Haico van Attikum, Email: h.van.attikum@lumc.nl.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51090-6.
References
- 1.Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol.20, 698–714 (2019). 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cejka, P. & Symington, L. S. DNA End Resection: Mechanism and control. Annu. Rev. Genet.55, 285–307 (2021). 10.1146/annurev-genet-071719-020312 [DOI] [PubMed] [Google Scholar]
- 3.Jasin, M. & Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol.5, a012740 (2013). [DOI] [PMC free article] [PubMed]
- 4.Anand, R., Ranjha, L., Cannavo, E. & Cejka, P. Phosphorylated CtIP functions as a Co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell64, 940–950 (2016). 10.1016/j.molcel.2016.10.017 [DOI] [PubMed] [Google Scholar]
- 5.Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature450, 509–514 (2007). 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceppi, I. et al. CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection. Proc. Natl. Acad. Sci. USA117, 8859–8869 (2020). 10.1073/pnas.2001165117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huertas, P. & Jackson, S. P. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem284, 9558–9565 (2009). 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huertas, P., Cortés-Ledesma, F., Sartori, A. A., Aguilera, A. & Jackson, S. P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature455, 689–692 (2008). 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, H. et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR–mediated double-strand break repair. PLoS Genet.9, e1003277 (2013). 10.1371/journal.pgen.1003277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu, X., Wu, L. C., Bowcock, A. M., Aronheim, A. & Baer, R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem.273, 25388–25392 (1998). 10.1074/jbc.273.39.25388 [DOI] [PubMed] [Google Scholar]
- 11.Wong, A. K. C. et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene17, 2279–2285 (1998). 10.1038/sj.onc.1202150 [DOI] [PubMed] [Google Scholar]
- 12.You, Z. et al. CtIP Links DNA double-strand break sensing to resection. Mol. Cell36, 954–969 (2009). 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eid, W. et al. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep.11, 962–968 (2010). 10.1038/embor.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polato, F. et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med.211, 1027–1036 (2014). 10.1084/jem.20131939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, H. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med.23, 517–525 (2017). 10.1038/nm.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen, L. & W. M. Martens, J. Van Hoeck, A. & Cuppen, E. Pan-cancer landscape of homologous recombination deficiency. Nat. Commun.11, 5584 (2020). 10.1038/s41467-020-19406-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Kooij, B., Kruswick, A., van Attikum, H. & Yaffe, M. B. Multi-pathway DNA-repair reporters reveal competition between end-joining, single-strand annealing and homologous recombination at Cas9-induced DNA double-strand breaks. Nat. Commun.13, 5295 (2022). 10.1038/s41467-022-32743-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semlow, D. R. & Walter, J. C. Mechanisms of vertebrate DNA interstrand cross-link repair. Annu. Rev. Biochem.90, 107–135 (2021). 10.1146/annurev-biochem-080320-112510 [DOI] [PubMed] [Google Scholar]
- 19.Nalepa, G. & Clapp, D. W. Fanconi anaemia and cancer: an intricate relationship. Nat. Rev. Cancer18, 168–185 (2018). 10.1038/nrc.2017.116 [DOI] [PubMed] [Google Scholar]
- 20.Niedzwiedz, W. et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell15, 607–620 (2004). 10.1016/j.molcel.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi, K. et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA102, 1110–1115 (2005). 10.1073/pnas.0407796102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard, S. M., Yanez, D. A. & Stark, J. M. DNA Damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet.11, e1004943 (2015). 10.1371/journal.pgen.1004943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wienert, B. et al. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat. Commun.11, 2109 (2020). 10.1038/s41467-020-15845-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosedale, G. et al. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol.12, 763–771 (2005). 10.1038/nsmb981 [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi, K. et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat. Struct. Mol. Biol.18, 500–503 (2011). 10.1038/nsmb.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, Y. et al. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood121, 54–63 (2013). 10.1182/blood-2012-07-441212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoepker, C. et al. DNA helicases FANCM and DDX11 are determinants of PARP inhibitor sensitivity. DNA Repair26, 54–64 (2015). 10.1016/j.dnarep.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 28.Benitez, A. et al. FANCA Promotes DNA double-strand break repair by catalyzing single-strand annealing and strand exchange. Mol. Cell71, 621–628 (2018). 10.1016/j.molcel.2018.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaser, A., McColl, B. & Vadolas, J. GFP to BFP Conversion: A versatile assay for the quantification of CRISPR/Cas9-mediated genome editing. Mol. Ther. Nucleic Acids5, e334 (2016). [DOI] [PMC free article] [PubMed]
- 30.Wang, T. et al. Identification and characterization of essential genes in the human genome. Science350, 1096–1101 (2015). 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, K. Y., Im, J.-S., Shibata, E. & Dutta, A. ASF1a Promotes non-homologous end joining repair by facilitating phosphorylation of MDC1 by ATM at double-strand breaks. Mol. Cell68, 61–75 (2017). 10.1016/j.molcel.2017.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Pastor, B. et al. Assessing kinetics and recruitment of DNA repair factors using high content screens. Cell Rep.37, 110176 (2021). 10.1016/j.celrep.2021.110176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sy, S. M. H., Jiang, J. O. W. S., Deng, Y. & Huen, M. S. Y. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res.41, 8572–8580 (2013). 10.1093/nar/gkt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metcalf, J. L. et al. K63-Ubiquitylation of VHL by SOCS1 mediates DNA double-strand break repair. Oncogene33, 1055–1065 (2014). 10.1038/onc.2013.22 [DOI] [PubMed] [Google Scholar]
- 35.Gari, K., Décaillet, C., Stasiak, A. Z., Stasiak, A. & Constantinou, A. The Fanconi anemia protein FANCM can promote branch migration of holliday junctions and replication forks. Mol. Cell29, 141–148 (2008). 10.1016/j.molcel.2007.11.032 [DOI] [PubMed] [Google Scholar]
- 36.Mehta, A., Beach, A. & Haber, J. E. Homology requirements and competition between gene conversion and break-induced replication during double-strand break repair. Mol. Cell65, 515–526 (2017). 10.1016/j.molcel.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deans, A. J. & West, S. C. FANCM Connects the genome instability disorders Bloom’s syndrome and Fanconi anemia. Mol. Cell36, 943–953 (2009). 10.1016/j.molcel.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 38.Brinkman, E. K. et al. Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res.46, e58(2018). 10.1093/nar/gky164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meetei, A. R. et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet.35, 165–170 (2003). 10.1038/ng1241 [DOI] [PubMed] [Google Scholar]
- 40.Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature434, 913–917 (2005). 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- 41.Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature434, 917–921 (2005). 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 42.Limoli, C. L. & Ward, J. F. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res.134, 160–169 (1993). 10.2307/3578455 [DOI] [PubMed] [Google Scholar]
- 43.Nakanishi, K. et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol.4, 913–920 (2002). 10.1038/ncb879 [DOI] [PubMed] [Google Scholar]
- 44.Roques, C. et al. MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J.28, 2400–2413 (2009). 10.1038/emboj.2009.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velimezi, G. et al. Map of synthetic rescue interactions for the Fanconi anemia DNA repair pathway identifies USP48. Nat. Commun.9, 2280 (2018). 10.1038/s41467-018-04649-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, J. et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol.20, 317–325 (2013). 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai, M. Y. et al. Cooperation of the ATM and Fanconi anemia/BRCA pathways in double-strand break End resection. Cell Rep.30, 2402–2415.e5 (2020). 10.1016/j.celrep.2020.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gravel, S., Chapman, J. R., Magill, C. & Jackson, S. P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev.22, 2767–2772 (2008). 10.1101/gad.503108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobbs, F. M. et al. Precision digital mapping of endogenous and induced genomic DNA breaks by INDUCE-seq. Nat. Commun.13, 3989 (2022). 10.1038/s41467-022-31702-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iacovoni, J. S. et al. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J.29, 1446–1457 (2010). 10.1038/emboj.2010.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, Y., Caron, P., Legube, G. & Paull, T. T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res.42, e19 (2014). 10.1093/nar/gkt1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murina, O. et al. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep.7, 1030–1038 (2014). 10.1016/j.celrep.2014.03.069 [DOI] [PubMed] [Google Scholar]
- 53.Unno, J. et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep.7, 1039–1047 (2014). 10.1016/j.celrep.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto, K. et al. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell Biol.23, 5421–5430 (2003). 10.1128/MCB.23.15.5421-5430.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, Y. G. et al. The Fanconi Anemia group A protein modulates homologous repair of DNA double-strand breaks in mammalian cells. Carcinogenesis26, 1731–1740 (2005). 10.1093/carcin/bgi134 [DOI] [PubMed] [Google Scholar]
- 56.Smogorzewska, A. et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell129, 289–301 (2007). 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eccles, L. J., Bell, A. C. & Powell, S. N. Inhibition of non-homologous end joining in Fanconi Anemia cells results in rescue of survival after interstrand crosslinks but sensitization to replication associated double-strand breaks. DNA Repair64, 1–9 (2018). 10.1016/j.dnarep.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis, T. W. et al. Deficiency of the Fanconi anemia E2 ubiqitin conjugase UBE2T only partially abrogates Alu-mediated recombination in a new model of homology dependent recombination. Nucleic Acids Res.47, 3503–3520 (2019). 10.1093/nar/gkz026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alagpulinsa, D. A. et al. Amplification and overexpression of E2 ubiquitin conjugase UBE2T promotes homologous recombination in multiple myeloma. Blood Adv.3, 3968–3972 (2019). 10.1182/bloodadvances.2019000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghunandan, M. et al. Functional cross talk between the Fanconi anemia and ATRX/DAXX histone chaperone pathways promotes replication fork recovery. Hum. Mol. Genet.29, 1083–1095 (2020). 10.1093/hmg/ddz250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ceccaldi, R., Sarangi, P. & D’Andrea, A. D. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol.17, 337–349 (2016). 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- 62.Tian, Y. et al. Constitutive role of the Fanconi anemia D2 gene in the replication stress response. J. Biol. Chem.292, 20184–20195 (2017). 10.1074/jbc.M117.814780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao, X., Joergensen, A. M., Howlett, N. G., Lisby, M. & Oestergaard, V. H. A distinct role for recombination repair factors in an early cellular response to transcription–replication conflicts. Nucleic Acids Res.48, 5467–5484 (2020). 10.1093/nar/gkaa268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu, L. et al. Autophosphorylation transforms DNA-PK from protecting to processing DNA ends. Mol. Cell82, 177–189 (2022). 10.1016/j.molcel.2021.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richardson, C. D. et al. CRISPR–Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet.50, 1132–1139 (2018). 10.1038/s41588-018-0174-0 [DOI] [PubMed] [Google Scholar]
- 66.Kais, Z. et al. FANCD2 Maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep.15, 2488–2499 (2016). 10.1016/j.celrep.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bennardo, N., Cheng, A., Huang, N. & Stark, J. M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet.4, e1000110 (2008). 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang, F. & Doudna, J. A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys.46, 505–529 (2017). 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- 69.Yeo, J. E., Lee, E. H., Hendrickson, E. A. & Sobeck, A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet.23, 3695–3705 (2014). 10.1093/hmg/ddu078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science343, 80–84 (2014). 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods11, 783–784 (2014). 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc.8, 2281–2308 (2013). 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol.34, 184–191 (2016). 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ceppi, I. et al. PLK1 regulates CtIP and DNA2 interplay in long-range DNA end resection. Genes Dev37, 119–135 (2023). 10.1101/gad.349981.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orthwein, A. et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature528, 422–426 (2015). 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas, P. D. et al. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Science31, 8–22 (2022). 10.1002/pro.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carbon, S. et al. AmiGO: online access to ontology and annotation data. Bioinformatics25, 288–289 (2009). 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Letunic, I., Khedkar, S. & Bork, P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res.49, D458–D460 (2021). 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hutchins, A. P., Liu, S., Diez, D. & Miranda-Saavedra, D. The Repertoires of ubiquitinating and deubiquitinating enzymes in eukaryotic genomes. Mol. Biol. Evol.30, 1172–1187 (2013). 10.1093/molbev/mst022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Damle, N. P. & Köhn, M. The human DEPhOsphorylation Database DEPOD: 2019 update. Database2019, baz133 (2019). 10.1093/database/baz133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park, Y.-K., Yoon, B.-H., Park, S.-J., Kim, B. K. & Kim, S.-Y. BaSDAS: a web-based pooled CRISPR-Cas9 knockout screening data analysis system. Genomics Inform.18, e46 (2020). 10.5808/GI.2020.18.4.e46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol.15, 554 (2014). 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Sequencing files from the CRISPR/Cas9 screen are available through the Sequence Read Archive of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra), with project number PRJNA1013929, accessions SRX21658367, SRX21658368 and SRX21658369. Note that the sequencing files contain data obtained for two independent HEK 293T DSB-Spectrum_V2 clones (D9 and E5). The representative data presented in this manuscript are from clone D9. Source data are provided with this paper.