Abstract
Chronic spontaneous urticaria (CSU) is a skin disease caused by mast cells that produce inflammatory mediators. Immune checkpoint receptors such as program death-1 (PD-1) and T-cell immunoglobulin and mucin domain 3 (TIM-3) are essential for the pathophysiology of many autoimmune and allergic diseases. The aim of this study was to investigate the expression of PD-1 and TIM-3 in CSU patients and their relationship to the anti-inflammatory cytokines (TGF-β and IL-10). In the current study, peripheral blood mononuclear cells (PBMCs) from CSU patients and healthy individuals were used and the Urticaria Activity Score 7 (UAS7) was used to assess disease severity. TaqMan-based RT-PCR was used to assess the expression of TIM-3 and PD-1 as well as the anti-inflammatory cytokines transforming growth factor-β (TGF-β) and IL-10. The protein concentrations of TGF-β and IL-10 were also measured by ELISA. The relationship between the expression of TIM-3 and PD-1 as well as TGF- β and IL-10 and the severity of the disease was investigated. The results showed that PD-1 mRNA expression was significantly increased in CSU patients (P<0.0001), while TGF- β and IL-10 levels were higher in CSU patients, but this difference was not significant (p=0.638, p= 0.798). The increase in protein level of IL-10 was significant (P<0.0001). There was also a positive correlation between the expression of PD-1 and TGF- β molecules and disease activity (P=0.0043, P=0.0018). In conclusion, the study found that the immune system expresses inhibitory molecules and anti-inflammatory cytokines to control disease severity. The higher expression of PD-1 molecules and IL-10 is associated with disease severity, suggesting that the immune system is trying to control inflammation and reduce disease severity.
Key Words: TIM-3, PD-1, CSU, IL-10, TGF-β, UAS7
Introduction
Chronic spontaneous urticaria (CSU) is a common skin disorder affecting 0.1% to 3% of the total population in the United States and worldwide (1, 2). CSU is distinguished by the presence of superficial swelling known as wheals and is classified as angioedema with or without deep swelling that lasts longer than six weeks (3). The term "spontaneous" refers to the fact that the disease has arisen for unknown reasons, while there is no clear evidence of the origin of the disease (4) and the exact cause of urticaria is not known, but environmental and genetic factors contribute (5-7). The pathogenesis of CSU is not fully understood, but it is known that skin mast cells play a crucial role in the development of wheals and angioedema (8, 9). The release of histamine from the cutaneous mast cells, together with cytokines and the production of arachidonic metabolites (LTC4, LTD4, LTE4, and PGs), contributes decisively to the consequences of the disease (10, 11). On the other hand, the increase in vascular permeability caused by these mediators leads to infiltration of immune cells such as eosinophils, basophils, neutrophils, monocytes, and CD4+ T cells into the lesion site, causing swelling and itching in response to nerve stimulation (12-14). Quality of life is usually impaired in CSUs, which is directly related to the severity. Antihistamines such as cetirizine commonly alleviate the symptoms of urticaria in children and adults and have few side effects; they are recommended as the first line of therapy in all guidelines (15, 16). If patients do not respond to the first and second lines of treatment with H1 antihistamines at the approved dose or a higher dose than the approved dose, they can be given omalizumab as a third line of treatment. Patients who do not respond to omalizumab are given immunosuppressive medications, such as cyclosporine (17). In this context, autoimmune reactions play an important role in the etiology of CSU (18, 19). The production of autoantibodies against FCεR1 and IgE expressed on mast cells is the cause of peripheral tolerance dysregulation. These autoantibodies can bind to their targets, leading to degranulation of mast cells and basophils (20, 21). The plasma of CSU patients contains IgG autoantibodies directed against IgE or its high-affinity receptor (FcεRI) (α-subunit of high-affinity IgE receptors), leading to degranulation of mast cells and basophils (22). Inadequate suppression of autoreactive responses leads to the development of autoimmune diseases. Recent studies have shown that regulatory molecules play a pivotal role in peripheral tolerance and regulation of autoreactive responses. Among the many regulatory molecules, program death-1 (PD-1) and T-cell immunoglobulin and mucin domain 3 (TIM-3), which have been introduced as immune checkpoints, have received considerable attention and have been extensively studied in the context of autoimmune diseases (23). PD-1 is an inhibitory molecule located on the surface of activated immune cells such as T-CD4+, T-CD8+ and B cells (20). They contribute to the reduction of autoreactive responses, including autoantibodies, as well as the induction of tolerance in autoimmune diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and vitiligo (24). The T-cell immunoglobulin and mucin domain (TIM) family, like PD-1, has an undeniable part in controlling the immune response and is associated with various allergies and autoimmune diseases (25, 26). TIM-3, one of the several members of this family, acts as a negative regulatory molecule by interacting with its receptor GAL-9 (27). As mentioned above, the production of autoantibodies against receptors on the surface of mast cells can lead to chronic urticaria with histamine release. Regarding the self-regulatory functions of immune checkpoints, we hypothesize that these markers have a significant impact in the production of autoantibodies (28). In addition to inhibitory molecules, immunosuppressive cytokines, transforming growth factor-β (TGF-β) and IL-10 also play a role in modulating the immune response and preventing autoimmunological diseases (29-31). TGF-β is a known inhibitory cytokine. TGF-β 1/2/3 isoforms are found in animals. TGF-β1 is a major isoform found mainly in lymphoid tissue and serum. Other isoforms are mainly found in mesenchymal tissues and bones. TGF-β1 is critical for maintaining peripheral tolerance and protecting the host from multiple sclerosis and arthritis (32). IL-10, along with TGF-β, is an important cytokine in the maintenance of immunological tolerance (33). Like TGF-β, this anti-inflammatory cytokine is primarily produced by regulatory T cells (Tregs) and exerts regulatory effects by suppressing mast cell degranulation and releasing inflammatory mediators (34). TGF-β has the potential to inhibit IL-2 production, which may hinder the proliferation and differentiation of autoreactive T cells. Furthermore, studies show that TGF-β can promote the growth and proliferation of Tregs, which has a vital function in the induction of peripheral tolerance (35). IL-10, along with TGF-β, is an important cytokine in the setting of immunologic tolerance (33). These anti-inflammatory cytokines are primarily produced by Tregs and exert regulatory effects by suppressing mast cell degranulation and releasing inflammatory mediators (34). Although inhibitory receptors and inflammatory cytokines are important, the aim of the current study was to clarify the other part of the relationship between inhibitory receptors and inflammatory cytokine expression with immune system dysregulation and impaired immunological tolerance in CSU patients compared to healthy controls. Furthermore, the relationship between the expression of these markers and disease activity (Urticaria Activity Score 7) was also investigated. The initial severity of the disease was assessed using the UAS7 questionnaire. The severity of the itching is indicated from 0 for no itching to 3 for the most severe itching and the number of hives from 0 to 3 (7).
Materials and methods
Patients and control
Thirty CSU patients referred to the second teaching hospital of Buali at Mazadaran University of Medical Sciences in Sari, northern Iran, were recruited for the present study. All subjects gave informed consent and the study methodology was approved by the ethical research council of Mazandaran University of Medical Sciences. All patients were in the early stages of the disease and had not received immunosuppressive drugs. The Urticaria Activity Score 7 (UAS7) was used to assess the disease activity of a CSU patient. Based on clinical and laboratory tests, a specialist physician diagnosed and verified the CSU patients. The exclusion criteria were chronic infections, asthma, other autoimmune diseases and cancer. The control group consisted of healthy volunteers who had no underlying conditions such as allergic diseases, autoimmune disorders, infections, or other chronic problems. Additionally, patients with unhealthy habits such as smoking or an inflammatory diet were excluded from the current study. The protocol of this study was approved by the Ethical Research Committee of Mazandaran University of Medical Sciences (ethical code: IR.MAZUMS.REC.1397.3086)
PBMC isolation and culture
Peripheral blood samples were collected in test tubes containing ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. PBMCs were collected by density gradient centrifugation on Ficoll-Histopaque (Biosera, China) (36). The isolated cells were immediately cultured in four-well flat bottom plates (1×106/well) with RPMI 1640 media supplemented with 1% penicillin/streptomycin and 10% heat-inactivated FBS. After culturing, cells were stimulated with PMA (50 ng/mL) and ionomycin (1 μg/mL) (Sigma) and incubated for 18 hours in 37 °C with 5% CO2.
ELISA and cytokine measurement
PBMCs were cultured and treated with PMA/ionomycin to analyze cytokine production. After 18 hours, culture supernatants were collected, aliquoted and stored at -70 °C. Then the frozen supernatants were removed from the freezer for the first time (avoiding repeated Fr/thw times) and used for the cytokine assay. TGF-β and IL-10 levels were quantified using an ELISA kit from Invitrogen Thermo Fisher Scientific (Waltham, Massachusetts, USA) according to the manufacturer’s instructions.
Real-time PCR
Expression of PD-1, TIM-3, TGF-β, and IL-10 was analyzed by a stem-loop TaqMan real-time PCR assay using a unique sequence index (USI) and a universal probe (37). Total RNA from isolated PBMC was extracted using the RNA isolation kit (Fervorgen, Taiwan) according to the manufacturer’s protocol. The quality and quantity of RNA were determined by agarose gel electrophoresis and absorbance ratio (260nm/280nm), respectively, and stored at -70 oC for further analysis. Extracted RNA was converted to cDNA using mRNA-specific USI RT-PCR primers (Table 1) and a cDNA synthesis kit (GeNet Bio, Korea) according to the manufacturer’s protocol. Gene amplification was performed using the StepOneTM Real-Time PCR instrument and HotStarTaq Plus DNA polymerase (QIAGEN, Germany) (38). The mRNA expression was determined using the Livak method (39). The GAPDH gene is used as an internal control for the quantification of PD-1, TIM-3, TGF-β and IL-10 genes. All primers were designed using Allele ID 6.0 software (Table 1).
Table 1.
Primer sequences used for stem-loop RT-PCR assays.
| Amplicon size (bp) | Gene Name | Primers 5' 3' → |
Accession
Number |
|---|---|---|---|
| 229 | GAPDH | Specific forward primer: TGGAGTCCACTGGCGTCTTCAC USI RT-PCR primer GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACAGGCATTGCTGA |
NM_002046.6 |
| 235 | TIM-3 | Specific forward primer: GAATGTGACTCTAGCAGACAGTGG USI RT-PCR primer GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACCTGGTGGTAAGC |
NM_032782.4 |
| 228 | PD-1 | Specific forward primer: GCCGCACGAGGGACAATAGG USI RT-PCR primer GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACGCATACTCCGTCT |
NM_005018.2 |
| 252 | TGF-B | Specific forward primer GCAAGTGGACATCAACGGGTTC USI RT-PCR primer GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACCGCAGCAGTTCT |
NM_000660.6 |
| 166 | IL-10 | Specific forward primer TTGCTGGAGGACTTTAAGGGTTAC USI RT-PCR primer GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACTGGGTCTTGGTTC |
NM_000572.2 |
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACGGAGGCCATGTAG | USI RT-PCR primer |
Statistical analysis
Qualitative variables were expressed as absolute frequency and relative frequency, and quantitative variables were presented as mean and median. The chi-square test was used to compare qualitative variables between groups. In the case of data with a normal distribution (based on the Kolmogorov-Smirnov test), parametric tests, T-tests, and in the case of abnormally distributed data, non-parametric tests such as Mann-Whitney were used. The data were statistically analyzed using SPSS19. The correlation coefficients were calculated using Pearson correlation tests. The shapes were drawn using PRISM software. A value of p<0.05 was considered significant for all samples.
Results
Demographic characteristics of the patients
Table 2 shows the demographic characteristics of all individuals, including the patients and healthy groups. The age and gender differences between the patient and healthy groups were not statistically significant.
Table 2.
Demographic characteristics of patients.
| Controls | CSU | P- value | |
|---|---|---|---|
| Age | 33.2 ± 7.6 | 34.4 ± 6.4 | 0.535 |
| Gender (male/female) | 9/21 | 8/22 | 0.5 |
| Disease activity | - | 14.7 ± 8.6 | - |
| Severity of pruritus | - | 6.8 ± 4.1 | - |
| Number of urticaria | - | 7.8 ± 5.1 | - |
PD-1 and TIM-3 expression in CSU patient
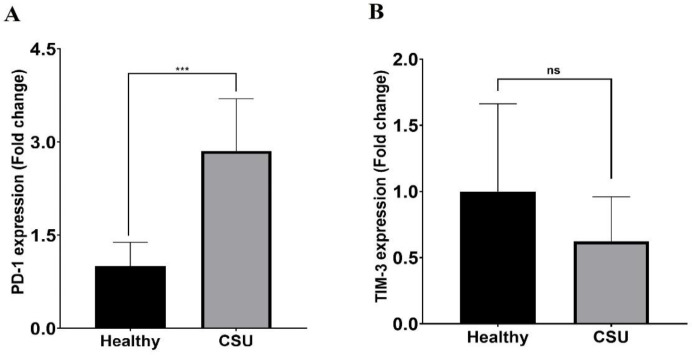
For the analysis of mRNA expression of TIM-3 and PD-1 in CSU patients compared to normal groups, isolated PBMC was cultured and stimulated with PMA/ionomycin and then incubated at 37 C and 5% CO2 for 18 hours. The RNA was extracted and the expression of RNA levels was analyzed. Our results demonstrated significantly higher expression of PD-1 in CSU patients (P<0.0001) (Figure 1A). In addition, the expression of TIM-3 decreased compared to the healthy group, but was not statistically significant (P=0.359) (Figure 1B).
Fig. 1.
PD-1 and TIM-3 mRNA expression in CSU patients. (A) There were significant differences in PD-1 expression in CSU patients compared to the Healthy group (p ≤ 0.0001). (B) The level of TIM-3 expression in CSU patients decreased with no significant differences from the Healthy group (p = 0. 359). (Data are presented as means ± SD, ***p < 0.001, and ns= nonsignificance).
Correlation analysis of TIM-3 and PD-1 expression with disease activity in CSU patients
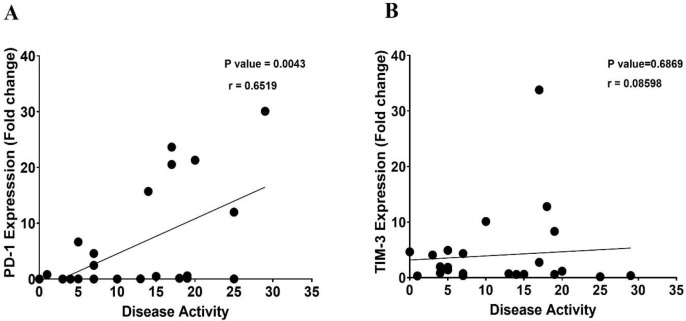
To determine the effect of immune checkpoints on disease severity, the correlation of PD-1 and TIM-3 mRNA expression with disease severity/activity was analyzed. The disease activity of CSU patients was determined using the UAS7. A significant positive correlation was found between disease activity and PD-1 expression (P=0.004, r = 0.6519) (Figure 2A). However, as shown in Figure 2-B, there is no significant correlation between TIM-3 expression and disease activity (p=0.689, r = 0.08598).
Fig. 2.
The linear correlation between TIM-3 and PD-1 expression with disease activity in CSU patients. (A)There was a strong positive correlation (P=0.0043, r=0.6519) between PD-1 mRNA expression and disease activity. However, there was not a significant correlation (P=0.6869, r=0.08598) between TIM-3 mRNA expression and disease activity (B).
Increased expression and production of TGF-β and IL-10 cytokines in PBMC from CSU patients
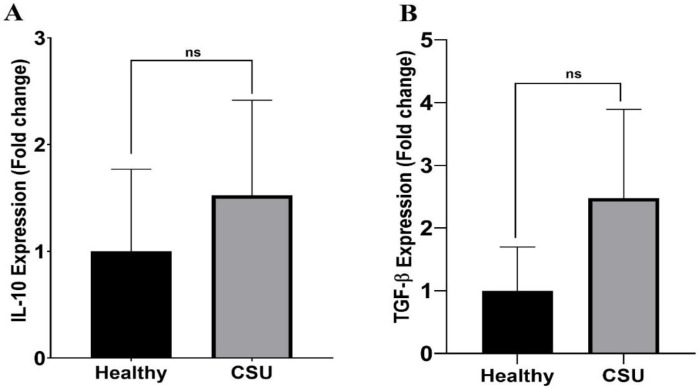
To analyze the mRNA expression of TGF-β and IL-10 in CSU patients compared to normal groups, isolated PBMC was stimulated with PMA/ionomycin and cultured for 18 hours. As illustratedin Figures 3A and B, although the expression of TGF-β and IL-10 is higher in CSU patients thanin healthy controls, these differences are not statistically significant (p=0.638 and p=0.798, respectively).
Fig. 3.
IL-10 and TGF-β mRNA expression in CSU patients and healthy controls. There were no significant differences in (A) IL-10 (p= 0.798) and (B) TGF-β (p=0.638) mRNA expression in the patients compared to healthy controls. Ns. Non-significant. (Data are presented as means ± SD).
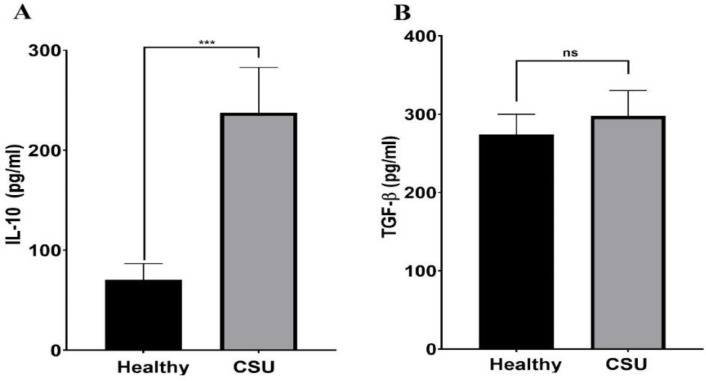
The protein levels of IL-10 and TGF-β in the study groups were also evaluated. As Figure 4 shows, IL-10 levels are significantly higher in CSU patients than in healthy controls (237.6 ± 45.2 vs. 70.45±16.12, p<0.0001). However, no significant difference was observed between TGF-β levels in CSU patients and controls (297.4±32.9 vs. 274.3 ± 25.6 pg/mL, p=0.583).
Fig. 4.
IL-10 and TGF-β protein production in CSU patients and healthy controls. The levels of IL-10 were significantly higher in the patients compared to the controls (p<0.0001) (A). Meanwhile, the levels of TGF-β were not significantly different in the patients than in the controls (p=0.583) (B). (Data are presented as means ± SD, ***: p < 0.001 and ns = non-significant).
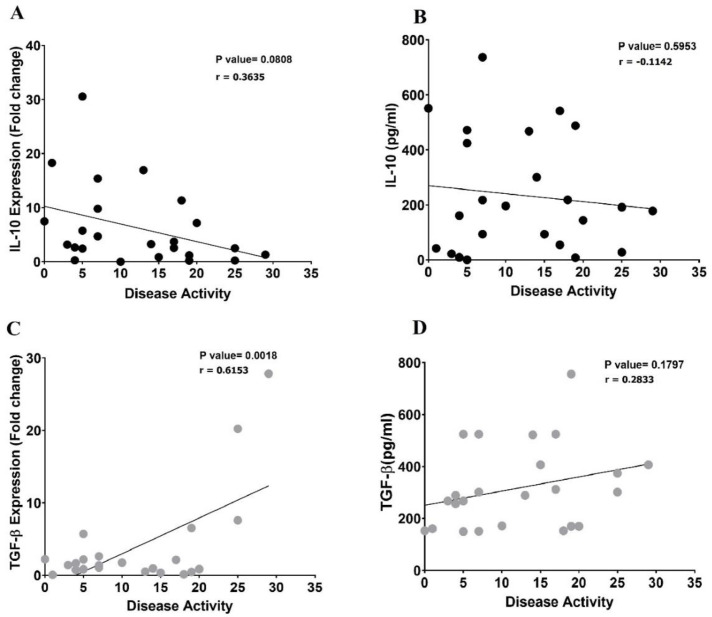
Correlation between cytokine production and disease activity in CSU patients
To evaluate the dysregulation of the immune system in CSU patients, the correlation between the anti-inflammatory cytokines, IL-10 and TGF-β, and disease activity was evaluated. As indicated in Figure 5, there is no significant correlation between either IL-10 mRNA expression (p=0.08, r=-0.3635) (Figure 5A) or its soluble form (p=0.59, r= -0.1142) (Fig 5-B) and disease activity. Nevertheless, a positive significant correlation was seen between TGF-β mRNA expression and disease activity (p=0.0018, r=0.6153) (Figure 5C). Moreover, no significant correlation was observed when soluble TGF-β was the focus of evaluation (p=0.6869, r=0.2833) (Figure 5D).
Fig. 5.
The linear correlation between IL-10 and TGF-β mRNA expression and their soluble levels with the disease activity in CSU patients. There was not a significant correlation between IL-10 mRNA expression (A) nor its soluble form (B) and disease activity in CSU patients. A positive significant correlation was seen between TGF-β mRNA expressions (C) but not in its soluble form and the disease activity (D).
Discussion
The initiation of autoreactive responses is significantly influenced by dysregulations of peripheral tolerance. Immune checkpoints, including CTLA-4, LAG-3, TIM-3 and PD-1, have been extensively studied in this context (40). Recent studies have shown that inflammatory, autoimmune, and allergic diseases such as rheumatoid arthritis, vitiligo and asthma have higher expression of the regulatory molecules PD-1 and TIM-3 (23, 25, 31). In the current study, the relationship between the expression of TIM-3 and PD-1 mRNA and disease activity in the PBMC from CSU patients and normal groups was investigated for the first time. The findings of the ongoing study represented that PBMC from CSU patients had significantly higher PD-1 mRNA expression compared to normal groups. According to studies utilizing mouse models, the PD-1 signaling pathway is crucial for the prevention of autoimmune processes. Collagen-induced arthritis was more frequent and severe in PD-1-deficient mice (25, 41). In the study by A. Bassiouni et al. which is similar to the present study, both PD-1 gene expression and serum sPD-1 levels are significantly increased in individuals with new-onset SLE (42). Measurements of PD-1 and PDL-1 in the peripheral blood of MS patients has revealed that the expression of these inhibitory molecules is significantly lower in MS patients than in healthy individuals (43). In contrast to PD-1, TIM-3 expression was found to be lower than average in the patient groups of the present study, although this was not statistically significant compared to healthy individuals. Recently, studies have reported that downregulation of TIM-3 on the CD4+ T cells is associated with the production of more pro-inflammatory cytokines in SLE (44). Kanai Y et al. investigated the expression of TIM-3 at Th1, T-CD8+ and Th17 levels in psoriasis patients compared to healthy individuals. They discovered that individuals with psoriasis had lower expression of this molecule on cell surfaces (45). Additionally, studies have suggested that TIM-3 plays a protective role in various autoimmune diseases, including multiple sclerosis (MS) and rheumatoid arthritis (RA) (46). In this context, studies have revealed that treatment of experimental autoimmune encephalomyelitis (EAE) with anti-TIM-3 antibodies increases the mortality rate (47). On the other hand, Tembhre et al. examined the expression of TIM-3 on the surface of T-CD4+ cells in the peripheral blood of vitiligo patients and found that vitiligo patients had higher expression of these molecules compared to control groups, suggesting that higher expression of this molecule may be associated with disease complications (48). In addition, TIM-3 may play an inhibitory role as a regulatory molecule on the surface of Th1 cells (49) and in this context redirects immune responses from Th1 to Th2. Treatment with TIM-3 inhibitory antibodies in a mouse model of allergic asthma has been demonstrated to alleviate the symptoms of pneumonia by polarizing Th1 responses (46). According to a study by Tang et al., patients with allergic asthma had higher levels of the regulatory molecule TIM-3 in their TCD4+ lymphocytes, contributing to disease progression due to an imbalance between their Th-1 and Th-2 responses (50). Furthermore, in the present study, the relationship between the disease activity of CSU patients and the expression of PD-1 and TIM-3 was investigated. The results indicated that there was a favorable correlation between the disease activity of CSU patients and the expression of TIM-3 and PD-1. According to the study by Bratke et al. there was a negative correlation between the expression of PD-1/CD4+ T cells and the production of IgE patients with allergic asthma (51). In agreement with the current study, Rahimi et al. discovered that the expression levels of TIM-3 and PD-1 protein and mRNA are positively correlated with disease activity (23). In agreement with other studies, PD-1 inhibits the response to self-antigens and promotes the death of activated T cells. PD-1 expression alleviates disease symptoms, whereas PD-1 deficiency can increase disease severity in murine models of arthritis (52, 53). In contrast to the findings of the current study, Koohini et al. demonstrated negative correlations between the expression of TIM-3 and PD-1 and their co-expression with disease activity in RA patient (25, 41). Earlier studies investigating the association between PD-1 and CTLA-4 gene polymorphism and disease etiology found no statistically significant differences between CSU and healthy groups(12, 54). The autoregulatory and feedback mechanisms cause the increased expression of PD-1 in CSU patients. The researchers discovered that PD-1 on T regulatory cells reduces inflammatory responses in the airways of patients with allergic asthma. They also declared that the therapeutic effect of PD-1 disappears when this receptor is blocked by antibodies (55). Autoimmune diseases and overactive immune responses are prevented by the binding of PD-1 to its receptor, programmed death ligand 1 (PD-L1). However, these interactions inhibit the host antitumor immune response in a tumor environment. In this context, the FDA-approved PD-1 inhibitors pembrolizumab and nivolumab have significantly improved the treatment of several advanced and metastatic tumors. Nivolumab is another PD-1 inhibitor that has been approved by the FDA. It interferes with the ability of PD-1 to negatively control the activation and proliferation of T cells by enabling the binding of multiple PD-1 epitopes to pembrolizumab (17, 56). Considering our results, it would be a therapeutic approach to utilize PD-L1 to alleviate disease severity. As additional immunoregulatory markers, the mRNA expression and levels of the anti-inflammatory cytokines IL-10 and TGF-β were investigated in the ongoing study. In addition to the increased mRNA expression of IL-10, the current study observed the increased IL-10 secretion by PBMC in CSU patients compared to healthy controls. In the context of autoimmune diseases and maintenance of tolerance, IL-10 and TGF-β play an important role (57 , 58). According to studies, these regulatory cytokines can restrict the expression of FCεR1 on mast cell surfaces and thus prevent the release of inflammatory mediators from cutaneous mast cells. Other studies have shown increased levels of IL-10 compared to healthy groups, supporting our study (59-63). However, there are contradictory findings regarding the cytokine IL-10. According to several studies, CSU patient groups produce less IL-10. Papatya et al. have proposed that low IL-10 levels are related to impaired T regulatory function (34). Additionally, although it was not statistically significant, a slight increase was found in TGF- β in the patient group. Moreover, there was a positive correlation between TGF- β production and disease activity in CSU patients. In several other studies, TGF-β levels were lower in the patient groups. Multiple factors, including autoimmune disease type, severity, ethnicity, sample volume, and others, could lead to discrepancies between reports. In this context, IL-10 gene expression showed a trend toward a negative correlation with disease severity, whereas TGF-β was positively correlated with disease severity. It is speculated that in the patient group, due to genetic variations and non-translational modifications, the increase in the expression of IL-10 has led to a decrease in disease severity and a negative correlation, but on the other hand, the increase in the expression of TGF-β is not to the extent that it can reduce the severity of the disease (due to the positive correlation) and it is slow to control the severity and none can overcome the other. The results of the current study have demonstrated that immune checkpoints and anti-inflammatory cytokines play an important role in the development of CSU. In this context, we have explored the concept of feedback and compensatory mechanisms. However, the exact molecular signaling mechanisms that trigger the upregulation of immune checkpoints as a form of compensation remain unclear (28). It can be hypothesized that these processes signal the immune system's struggle to restore immunological tolerance and reduce disease severity. During this process, certain molecules, such as anti-inflammatory cytokines and PD-1, increase to alleviate disease severity, while others, like TIM-3, may exacerbate disease activity. Nevertheless, the specific molecular signaling mechanisms responsible for the upregulation of immune checkpoint as a compensatory response remain uncertain. Unfortunately, due to limitations such as insufficient funding in the present study, it was not possible to analyze the reported markers at the protein level or to distinguish them on distinct T cell subsets. In addition, samples should be taken from the patient's skin at the mast cell level, which is impossible with PBMCs. On the other hand, ethical considerations prevented us from using human samples for the present study. It is expected that these ideas will be incorporated into future studies. Overall, more research is needed to unravel the complexities of the immune system. These discoveries guide us in the right direction to identify effective solutions for therapeutic purposes.
Acknowledgments
The authors would like to thank the patients and their families for their support, cooperation and patience. Our thanks go to the staff in the departments responsible for the care and monitoring of the patients. This study was financially supported by grants (Grant No: 3086 and 3650) from the Research and Technology Council of Mazandaran University of Medical Sciences.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Sánchez-Borges M, Ansotegui IJ, Baiardini I, Bernstein J, Canonica GW, Ebisawa M, et al. The challenges of chronic urticaria part 1: Epidemiology, immunopathogenesis, comorbidities, quality of life, and management. World Allergy Organization Journal. 2021;14(6):100533. doi: 10.1016/j.waojou.2021.100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster L, Rider NL, Archambault ME. Evaluating and managing chronic idiopathic urticaria in adults. JAAPA. 2018;31(7):22–6. doi: 10.1097/01.JAA.0000534976.46365.11. [DOI] [PubMed] [Google Scholar]
- 3.Balp M-M, Halliday AC, Severin T, Leonard SA, Partha G, Kalra M, et al. Clinical remission of chronic spontaneous urticaria (CSU): a targeted literature review. Dermatology and Therapy. 2022;12(1):15–27. doi: 10.1007/s13555-021-00641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maheshwari A, Garg T, Debnath E, Chander R. Chronic spontaneous urticaria: An updated comprehensive review of etiology, pathogenesis and management [Google Scholar]
- 5.Ghaffari J, Farid Hossaini R, Rafatpanah H, Jabbari Azad F, Shahmohammadi S. Chronic urticaria in children: etiologies, clinical manifestations, diagnosis and treatment. Journal of Pediatrics Review. 2013;1(2):55–68. [Google Scholar]
- 6.Hosseini-Farahabadi S, Tavakkol-Afshari J, Ganjali R, Rafatpanah H, Ghaffari J, Farid-Hosseini R. Association between the polymorphism of TGF-β1 gene promoter (-509C> T) and idiopathic chronic urticaria. Iranian Journal of Allergy, Asthma and Immunology. 2006:109–13. [PubMed] [Google Scholar]
- 7.Zuberbier T, Aberer W, Asero R, Abdul Latiff AH, Baker D, Ballmer‐Weber B, et al. The EAACI/GA²LEN/EDF/WAO guideline for the definition, classification, diagnosis and management of urticaria. Allergy. 2018;73(7):1393–414. doi: 10.1111/all.13397. [DOI] [PubMed] [Google Scholar]
- 8.Bracken SJ, Abraham S, MacLeod AS. Autoimmune theories of chronic spontaneous urticaria. Frontiers in immunology. 2019;10:449970. doi: 10.3389/fimmu.2019.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulthanan K, Rujitharanawong C, Munprom K, Trakanwittayarak S, Phumariyapong P, Prasertsook S, et al. Prevalence, clinical manifestations, treatment, and clinical course of chronic urticaria in elderly: A systematic review. Journal of Asthma and Allergy. 2022:1455–90. doi: 10.2147/JAA.S379912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amin K. The role of mast cells in allergic inflammation. Respiratory medicine. 2012;106(1):9–14. doi: 10.1016/j.rmed.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan A, Lebwohl M, Giménez‐Arnau AM, Hide M, Armstrong AW, Maurer M. Chronic spontaneous urticaria: focus on pathophysiology to unlock treatment advances. Allergy. 2023;78(2):389–401. doi: 10.1111/all.15603. [DOI] [PubMed] [Google Scholar]
- 12.Ferrer M. Immunological events in chronic spontaneous urticaria. Clinical and Translational Allergy. 2015;5:1–8. doi: 10.1186/s13601-015-0074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giménez-Arnau AM, de Montjoye L, Asero R, Cugno M, Kulthanan K, Yanase Y, et al. The pathogenesis of chronic spontaneous urticaria: the role of infiltrating cells. The Journal of Allergy and Clinical Immunology: In Practice. 2021;9(6):2195–208. doi: 10.1016/j.jaip.2021.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Ghaffari J, Ghaffari N. Acute and chronic urticaria: prevalence, etiologies, diagnosis, and treatment. Journal of Mazandaran University of Medical Sciences. 2020;30(187):179–95. [Google Scholar]
- 15.Ghaffari J. A review of recent treatment of urticarial in children and adults. 2019. [Google Scholar]
- 16.Ghaffari J. Biologic drugs treatment of chronic urticaria. Journal of Pediatrics Review. 2022;10(4):273–6. [Google Scholar]
- 17.Orzan OA, Popa LG, Mihai MM, Cojocaru A, Giurcăneanu C, Dorobanțu AM. Current and future approaches in management of chronic spontaneous urticaria using anti-ige antibodies. Medicina. 2022;58(6):816. doi: 10.3390/medicina58060816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Qiu L, Wu J, Qi Y, Gao X, He C, et al. GWAS of chronic spontaneous urticaria reveals genetic overlap with autoimmune diseases, not atopic diseases. Journal of Investigative Dermatology. 2023;143(1):67–77. doi: 10.1016/j.jid.2022.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Ghaffari J, Dabbaghzadeh A, Ghaffari N. Comorbidities in patients with chronic urticaria; Clinical and epidemiological review study. Iranian Journal of Health Sciences. 2021 [Google Scholar]
- 20.Brzoza Z, Grzeszczak W, Trautsolt W, Moczulski D. 7 Lack of Association of Programmed Cell Death 1 Gene (PDCD1) Polymorphisms With Susceptibility to Chronic Urticaria in Patients With Positive Autologous Serum Skin Test. Journal of Investigational Allergology and Clinical Immunology. 2012;22(6):432. [PubMed] [Google Scholar]
- 21.Dobrican C-T, Muntean IA, Pintea I, Petricău C, Deleanu D-M, Filip GA. Immunological signature of chronic spontaneous urticaria. Experimental and Therapeutic Medicine. 2022;23(6):1–7. doi: 10.3892/etm.2022.11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulambayar B, Chen Y-H, Ban G-Y, Lee J-H, Jung C-G, Yang E-M, et al. Detection of circulating IgG autoantibody to FcεRIα in sera from chronic spontaneous urticaria patients. Journal of Microbiology, Immunology and Infection. 2020;53(1):141–7. doi: 10.1016/j.jmii.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Rahimi A, Hossein‐Nataj H, Hajheydari Z, Aryanian Z, Shayannia A, Ajami A, et al. Expression analysis of PD‐1 and Tim‐3 immune checkpoint receptors in patients with vitiligo; positive association with disease activity. Experimental dermatology. 2019;28(6):674–81. doi: 10.1111/exd.13952. [DOI] [PubMed] [Google Scholar]
- 24.Singh AK, Stock P, Akbari O. Role of PD‐L1 and PD‐L2 in allergic diseases and asthma. Allergy. 2011;66(2):155–62. doi: 10.1111/j.1398-9995.2010.02458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koohini Z, Hossein-Nataj H, Mobini M, Hosseinian-Amiri A, Rafiei A, Asgarian-Omran H. Analysis of PD-1 and Tim-3 expression on CD4+ T cells of patients with rheumatoid arthritis; negative association with DAS28. Clinical rheumatology. 2018;37:2063–71. doi: 10.1007/s10067-018-4076-4. [DOI] [PubMed] [Google Scholar]
- 26.Dolatkhah K, Alizadeh N, Mohajjel‐Shoja H, Abdoli Shadbad M, Hajiasgharzadeh K, Aghebati‐Maleki L, et al. B7 immune checkpoint family members as putative therapeutics in autoimmune disease: An updated overview. International Journal of Rheumatic Diseases. 2022;25(3):259–71. doi: 10.1111/1756-185X.14273. [DOI] [PubMed] [Google Scholar]
- 27.Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunological reviews. 2010;235(1):172–89. doi: 10.1111/j.0105-2896.2010.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riva A, Chokshi S. Immune checkpoint receptors: homeostatic regulators of immunity. Hepatology international. 2018;12(3):223–36. doi: 10.1007/s12072-018-9867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palomares O, Martin-Fontecha M, Lauener R, Traidl-Hoffmann C, Cavkaytar O, Akdis M, et al. Regulatory T cells and immune regulation of allergic diseases: roles of IL-10 and TGF-β. Genes & Immunity. 2014;15(8):511–20. doi: 10.1038/gene.2014.45. [DOI] [PubMed] [Google Scholar]
- 30.Zheng Z, Huang G, Gao T, Huang T, Zou M, Zou Y, et al. Epigenetic changes associated with interleukin-10. Frontiers in immunology. 2020;11:1105. doi: 10.3389/fimmu.2020.01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlier TDB, Badloe FMS, Ring J, Gutermuth J, Krohn IK. Autoreactive T cells and their role in atopic dermatitis. Journal of autoimmunity. 2021;120:102634. doi: 10.1016/j.jaut.2021.102634. [DOI] [PubMed] [Google Scholar]
- 32.Pyzik M, Piccirillo CA. The TGF-β 1/Foxp3 Regulatory Axis in Immune Self-Tolerance: Implications for Health and Disease. Inflammation & Allergy-Drug Targets (Formerly Current Drug Targets-Inflammation & Allergy)(Discontinued) 2006;5(3):167–77. doi: 10.2174/187152806778256089. [DOI] [PubMed] [Google Scholar]
- 33.Groux H, Cottrez F. The complex role of interleukin-10 in autoimmunity. Journal of autoimmunity. 2003;20(4):281–5. doi: 10.1016/s0896-8411(03)00044-1. [DOI] [PubMed] [Google Scholar]
- 34.Degirmenci PB, Kırmaz C, Vatansever S, Onur E, Nal E, Erdin S, et al. Analysis of the association of chronic spontaneous urticaria with interlekin-4,-10, transforming growth factor-β1, interferon-γ, interleukin-17A and-23 by autologous serum skin test. Advances in Dermatology and Allergology/Postępy Dermatologii i Alergologii. 2017;34(1):70–6. doi: 10.5114/pdia.2016.57679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Yi H, Xia X-P, Zhao Y. Transforming growth factor-beta: an important role in CD4+ CD25+ regulatory T cells and immune tolerance. Autoimmunity. 2006;39(4):269–76. doi: 10.1080/08916930600753903. [DOI] [PubMed] [Google Scholar]
- 36.Kardan M, Rafiei A, Ghaffari J, Valadan R, Morsaljahan Z, Haj-Ghorbani S. Effect of ginger extract on expression of GATA3, T-bet and ROR-γt in peripheral blood mononuclear cells of patients with Allergic Asthma. Allergologia et immunopathologia. 2019;47(4):378–85. doi: 10.1016/j.aller.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Fattahi S, Amirbozorgi G, Lotfi M, Amini Navaei B, Kavoosian S, Asouri M, et al. Development of a universal taqman probe for mRNA gene expression analysis. Iranian Journal of Science and Technology, Transactions A: Science. 2018;42(2):363–70. [Google Scholar]
- 38.Fattahi S, Pilehchian Langroudi M, Samadani AA, Nikbakhsh N, Asouri M, Akhavan-Niaki H. Application of unique sequence index (USI) barcode to gene expression profiling in gastric adenocarcinoma. Journal of cell communication and signaling. 2017;11:97–104. doi: 10.1007/s12079-017-0376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LIVAK KJ. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;43(1):79. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Mohammadi P, Hesari M, Chalabi M, Salari F, Khademi F. An overview of immune checkpoint therapy in autoimmune diseases. International immunopharmacology. 2022;107:108647. doi: 10.1016/j.intimp.2022.108647. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Hu P, Yang J, Shen G, Wu X. The effects of PDL-Ig on collagen-induced arthritis. Rheumatology international. 2011;31:513–9. doi: 10.1007/s00296-009-1249-0. [DOI] [PubMed] [Google Scholar]
- 42.Bassiouni SA, Abdeen HM, Morsi HK, Zaki ME, Abdelsalam M, Gharbia OM. Programmed death 1 (PD-1) serum level and gene expression in recent onset systemic lupus erythematosus patients. The Egyptian Rheumatologist. 2021;43(3):213–8. [Google Scholar]
- 43.Javan MR, Aslani S, Zamani MR, Rostamnejad J, Asadi M, Farhoodi M, et al. Downregulation of immunosuppressive molecules, PD-1 and PD-L1 but not PD-L2, in the patients with multiple sclerosis. Iranian Journal of Allergy, Asthma and Immunology. 2016:296–302. [PubMed] [Google Scholar]
- 44.Cai X, Huang W, Qiao Y, Chen Y, Du S, Chen D, et al. Downregulation of TIM-3 mRNA expression in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Brazilian Journal of Medical and Biological Research. 2014;48(1):77–82. doi: 10.1590/1414-431X20143701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanai Y, Satoh T, Igawa K, Yokozeki H. Impaired expression of Tim-3 on Th17 and Th1 cells in psoriasis. Acta dermato-venereologica. 2012;92(4):367–71. doi: 10.2340/00015555-1285. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Ju Z, Frieri M. The T-cell immunoglobulin and mucin domain (Tim) gene family in asthma, allergy, and autoimmunity. Allergy and asthma proceedings; OceanSide Publications, Inc; 2013. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez‐Manzanet R, DeKruyff R, Kuchroo VK, Umetsu DT. The costimulatory role of TIM molecules. Immunological reviews. 2009;229(1):259–70. doi: 10.1111/j.1600-065X.2009.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tembhre MK, Parihar AS, Sharma A, Gupta S, Chattopadhyay P, Sharma VK. Participation of T cell immunoglobulin and mucin domain-3 (TIM-3) and its ligand (galectin-9) in the pathogenesis of active generalized vitiligo. Immunologic research. 2015;62(1):23–34. doi: 10.1007/s12026-015-8632-6. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Daley D, Akhabir L, Stefanowicz D, Chan-Yeung M, Becker AB, et al. Lack of association of TIM3 polymorphisms and allergic phenotypes. BMC medical genetics. 2009;10(1):1–10. doi: 10.1186/1471-2350-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang F, Wang F, An L, Wang X. Upregulation of Tim-3 on CD4+ T cells is associated with Th1/Th2 imbalance in patients with allergic asthma. International journal of clinical and experimental medicine. 2015;8(3):3809. [PMC free article] [PubMed] [Google Scholar]
- 51.Bratke K, Fritz L, Nokodian F, Geißler K, Garbe K, Lommatzsch M, et al. Differential regulation of PD‐1 and its ligands in allergic asthma. Clinical & Experimental Allergy. 2017;47(11):1417–25. doi: 10.1111/cea.13017. [DOI] [PubMed] [Google Scholar]
- 52.Raptopoulou AP, Bertsias G, Makrygiannakis D, Verginis P, Kritikos I, Tzardi M, et al. The programmed death 1/programmed death ligand 1 inhibitory pathway is up‐regulated in rheumatoid synovium and regulates peripheral T cell responses in human and murine arthritis. Arthritis & Rheumatism. 2010;62(7):1870–80. doi: 10.1002/art.27500. [DOI] [PubMed] [Google Scholar]
- 53.Arnett FC, Edworthy SM, Bloch DA, Mcshane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis & Rheumatism: Official Journal of the American College of Rheumatology. 1988;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 54.Brzoza Z, Grzeszczak W, Rogala B, Trautsolt W, Moczulski D. CTLA-4 polymorphism in the pathogenesis of chronic spontaneous autoreactive urticaria. Allergologia et Immunopathologia. 2014;42(3):241–4. doi: 10.1016/j.aller.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 55.McGee HS, Yagita H, Shao Z, Agrawal DK. Programmed Death-1 antibody blocks therapeutic effects of T-regulatory cells in cockroach antigen-induced allergic asthma. American journal of respiratory cell and molecular biology. 2010;43(4):432–42. doi: 10.1165/rcmb.2009-0258OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandey P, Khan F, Qari HA, Upadhyay TK, Alkhateeb AF, Oves M. Revolutionization in cancer therapeutics via targeting major immune checkpoints PD-1, PD-L1 and CTLA-4. Pharmaceuticals. 2022;15(3):335. doi: 10.3390/ph15030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell. 2008;134(3):392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Commins S, Steinke JW, Borish L. The extended il-10 superfamily: Il-10, il-19, il-20, il-22, il-24, il-26, il-28, and il-29. Journal of Allergy and Clinical Immunology. 2008;121(5):1108–11. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 59.Irinyi B, Aleksza M, Antal-Szalmás P, Sipka S, Hunyadi J, Szegedi A. Cytokine production of CD4+ and CD8+ peripheral T lymphocytes in patients with chronic idiopathic urticaria. Acta dermato-venereologica. 2002;82:4. doi: 10.1080/000155502320323199. [DOI] [PubMed] [Google Scholar]
- 60.Piconi S, Trabattoni D, Iemoli E, Fusi ML, Villa ML, Milazzo F, et al. Immune profiles of patients with chronic idiopathic urticaria. International archives of allergy and immunology. 2002;128(1):59–66. doi: 10.1159/000058004. [DOI] [PubMed] [Google Scholar]
- 61.Caproni M, Cardinali C, Giomi B, Antiga E, D’Agata A, Walter S, et al. Serological detection of eotaxin, IL-4, IL-13, IFN-γ, MIP-1α, TARC and IP-10 in chronic autoimmune urticaria and chronic idiopathic urticaria. Journal of dermatological science. 2004;36(1):57–9. doi: 10.1016/j.jdermsci.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Ying S, Kikuchi Y, Meng Q, Kay AB, Kaplan AP. TH1/TH2 cytokines and inflammatory cells in skin biopsy specimens from patients with chronic idiopathic urticaria: comparison with the allergen-induced late-phase cutaneous reaction. Journal of Allergy and Clinical Immunology. 2002;109(4):694–700. doi: 10.1067/mai.2002.123236. [DOI] [PubMed] [Google Scholar]
- 63.Caproni M, Giomi B, Volpi W, Melani L, Schincaglia E, Macchia D, et al. Chronic idiopathic urticaria: infiltrating cells and related cytokines in autologous serum-induced wheals. Clinical Immunology. 2005;114(3):284–92. doi: 10.1016/j.clim.2004.10.007. [DOI] [PubMed] [Google Scholar]