Correction: BMC Plant Biol 39, 83 (2020)
10.1186/s13046-020-01588-w
Following the publication of the original article [1], the authors identified errors in Fig. 3, specifically:
Fig. 3.
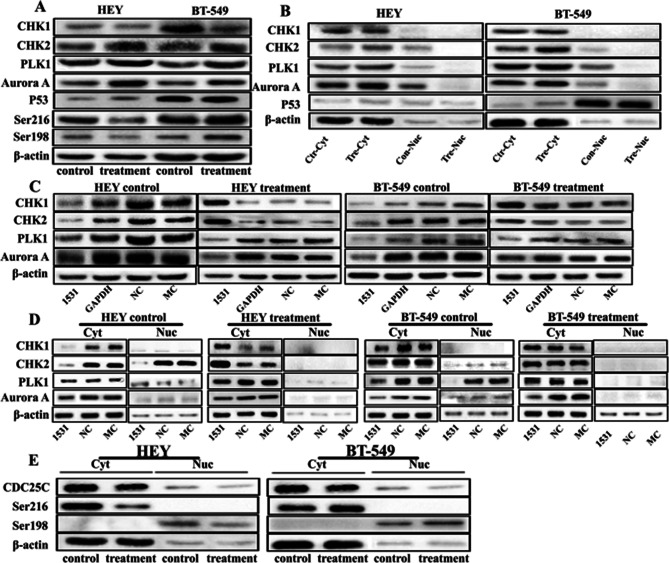
The expression of CHK1, CHK2, PLK1, Aurora A, P53, pCDC25CSer216, and pCDC25CSer198 in HEY and BT-549 control cells and PGCCs with budding daughter cells with and without CDC25C knockdown. a Western blot showed the total protein expression of CHK1, CHK2, PLK1, Aurora A, P53, pCDC25CSer216, and pCDC25CSer198 in HEY and BT-549 control and PGCCs with daughter cells. b The levels of total protein expression of CHK1, CHK2, PLK1, and Aurora A in HEY and BT-549 control and PGCCs with daughter cells, which were transfected with CDC25Ci, siRNA control, and negative control. c Cytoplasmic and nuclear protein expression of CHK1, CHK2, PLK1, Aurora A, P53 in HEY and BT-549 control and PGCCs with daughter cells. d Cytoplasmic and nuclear expression of CHK1, CHK2, PLK1, and Aurora A in HEY and BT-549 control and PGCCs with daughter cells, which were transfected with CDC25Ci, siRNA control, and negative control. e The cytoplasm and nuclear protein expression of pCDC25CSer216 and pCDC25CSer198 in HEY and BT-549 control and PGCCs with daughter cells. Treatment: Cells treated with CoCl2. 1531si: siRNA CDC25C-1531
Figure 3D - the β-actin of HEY and BT-549 control cells were mistakenly reusede.
Figure 3E - the cytoplasm and nuclear protein expression of CDC25C in HEY and BT-549 control and PGCCs with daughter cells the same result was mistakenly placed.
Incorrect Fig. 3
Correct Fig. 3
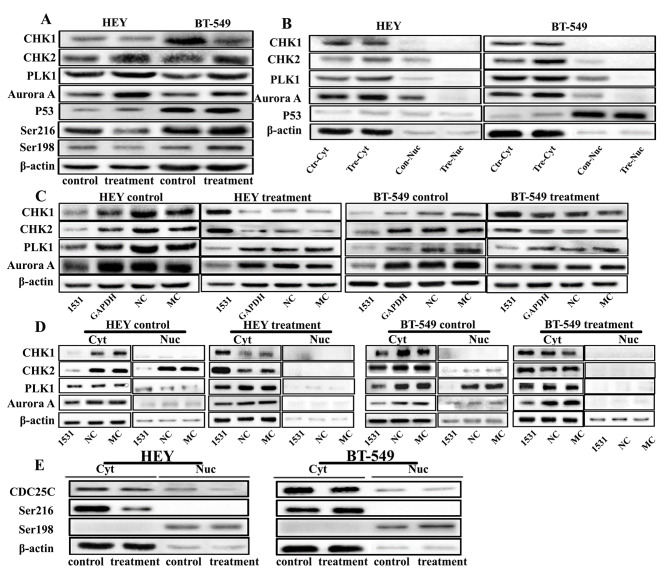
Fig. 3.
The expression of CHK1, CHK2, PLK1, Aurora A, P53, pCDC25CSer216, and pCDC25CSer198 in HEY and BT-549 control cells and PGCCs with budding daughter cells with and without CDC25C knockdown. a Western blot showed the total protein expression of CHK1, CHK2, PLK1, Aurora A, P53, pCDC25CSer216, and pCDC25CSer198 in HEY and BT-549 control and PGCCs with daughter cells. b The levels of total protein expression of CHK1, CHK2, PLK1, and Aurora A in HEY and BT-549 control and PGCCs with daughter cells, which were transfected with CDC25Ci, siRNA control, and negative control. c Cytoplasmic and nuclear protein expression of CHK1, CHK2, PLK1, Aurora A, P53 in HEY and BT-549 control and PGCCs with daughter cells. d Cytoplasmic and nuclear expression of CHK1, CHK2, PLK1, and Aurora A in HEY and BT-549 control and PGCCs with daughter cells, which were transfected with CDC25Ci, siRNA control, and negative control. e The cytoplasm and nuclear protein expression of pCDC25CSer216 and pCDC25CSer198 in HEY and BT-549 control and PGCCs with daughter cells. Treatment: Cells treated with CoCl2. 1531si: siRNA CDC25C-1531
The original article [1] has been corrected.
Footnotes
The online version of the original article can be found at 10.1186/s13046-020-01588-w.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kai Liu, Minying Zheng and Qi Zhao contributed equally to this work.
References
- 1.Liu K, Zheng M, Zhao Q, et al. Different p53 genotypes regulating different phosphorylation sites and subcellular location of CDC25C associated with the formation of polyploid giant cancer cells. J Exp Clin Cancer Res. 2020;39:83. 10.1186/s13046-020-01588-w. 10.1186/s13046-020-01588-w [DOI] [PMC free article] [PubMed] [Google Scholar]