Abstract
Ischemia/reperfusion injury (IRI) is an important risk factor for accelerated cardiac allograft rejection and graft dysfunction1. Utilizing a rat heart isogeneic transplant model, we identified inflammatory pathways involved in IRI in order to identify therapeutic targets involved in disease. Pathway analyses identified several relevant targets, including cytokine signaling by the IL-1 receptor (IL-1R) pathway and inflammasome activation. To investigate the role of IL-1R signaling pathways during I/R injury, we treated syngeneic cardiac transplant recipients at 1-hour post-transplant with Anakinra, an FDA approved IL-1R antagonist, or parthenolide, a Caspase-1 and NF-κB inhibitor that blocks IL-1β maturation. Both Anakinra and parthenolide significantly reduced graft inflammation and cellular recruitment in the treated recipients relative to non-treated controls. Anakinra treatment administered at 1-hour post-transplant to recipients of cardiac allografts from CMV-infected donors significantly increased the time to rejection and reduced viral loads at rejection. Our results indicate that reducing IRI by blocking IL-1R signaling pathways with Anakinra or inflammasome activity with parthenolide provides a promising approach for extending survival of cardiac allografts from CMV-infected donors.
Introduction
Solid organ transplantation (SOT) remains the standard of care for patients with end-stage organ failure. However, chronic allograft rejection (CR) remains a barrier to long-term transplant success. The defining feature of CR in cardiac transplants is development of transplant vascular sclerosis (TVS), affecting approximately 30% of transplant patients. The disease is characterized by a subendothelial low-grade inflammatory process resulting in narrowing of graft coronary vessels and leading to graft loss2. Several risk factors are associated with CR including: donor age, ischemia and reperfusion injury (IRI), acute rejection episodes, hypercholesterolemia and human cytomegalovirus infection2–4. The only effective treatment for CR is re-transplantation. Hence, identifying mechanisms involved in this process is critical to prevent cardiac allograft disease.
IRI is a primary activator of graft inflammation leading to increased risk for both acute and chronic allograft rejection1. During IRI, oxygen and glucose deprivation combined with physical stress result in cell damage and the initiation of apoptotic signaling cascades involving Caspase-1 activation, IL-1β signaling, and NF-κB activation. The resultant pro-inflammatory cascade includes activation and up-regulation of cytokines and chemokines that promote macrophage and neutrophil recruitment1. IRI-induced inflammation promotes acute rejection, leading to tissue fibrosis and increasing the risk of CR. Preservation solutions and limiting graft cold ischemia time (CI) have reduced, but not eliminated, IRI-induced inflammation1. Potential strategies to reduce IRI include pre-conditioning of donor tissue or blocking cytokine/chemokine signaling in the recipient immediately after transplantation1.
Cytomegalovirus (CMV) infection has many deleterious effects on SOT outcomes2,5,6. Importantly, Human CMV (HCMV) infection occurs in 75% of the solid organ donor/recipient population. Both active and latent CMV infections promote allograft rejection2,5,7,8. Currently, prophylactic therapies such as Valganciclovir are used to control CMV infection in transplant recipients. However, this therapy does not prevent late onset CMV disease, and efficacy is attenuated by drug resistance8. A rat transplant model with latent rat CMV (RCMV) donor infection significantly accelerates TVS development and CR7. Latent CMV is reactivated by pro-inflammatory signals that are associated with IRI8,9. CMV infection increases donor graft passenger lymphocyte loads prior to transplantation, which we hypothesize results in a double hit during transplantation10. Despite the similar clinical manifestations of disease associated with IRI and CMV infection, we do not understand the signaling networks that promote IRI-induced graft rejection. Identification of these pathways may identify therapeutic targets to improve graft survival. In this report, we profiled the IRI pro-inflammatory environment that promotes CMV-accelerated rejection. Inflammasome and IL-1R signaling pathways constituted the central node in IRI-induced cardiac allograft injury. Reduction of IRI by treatment of transplant recipients with a single dose of Anakinra improved allograft outcomes and delayed RCMV-infected donor graft CR.
Materials and Methods
Rat Cardiac Transplantation:
OHSU West Campus Small Laboratory Animal Facility is AAALAC accredited and complies with USDA and HHS animal care requirements. Heterotopic syngeneic heart transplantation was performed in Lewis rats in order to characterize IRI without alloimmunity. Heterotopic allogeneic heart transplantation of F344 hearts into Lewis recipients was performed to assess the effect of treating IRI on CR. Rat donor hearts (Lewis syngeneic or F344 allogeneic) were surgically removed, placed in 4°C UW solution for 4 hours, and then transplanted7,11 (Figure 1 and Table 1). Phosphate buffered saline (PBS) served as vehicle for Anakinra (SOBI), whereas 10% Ethanol/90% corn oil served as the parthenolide vehicle (Cayman Chemical). Donor rats for Cohorts 3, 6, and 8–10 were infected at 5 days before transplantation with RCMV Maastricht strain (1×105pfu by intraperitoneal injection). Donor rats for Cohorts 1, 2, 4, 5 and 7 were uninfected. Animals were examined daily for overall health and CR was determined by monitoring graft heartbeat grade11. Blood and tissues from Cohorts 1–8 were harvested at post-operation day (POD) 3; whereas Cohorts 9–10 were harvested at POD14 or at the time of allograft CR. Blood was separated into plasma and peripheral blood mononuclear cell (PBMC) using lymphocyte separation media (Corning). Portions of heart tissues were: fixed for histological evaluation; snap frozen for immunohistochemistry and nucleic acid analysis; and harvested for flow cytometry.
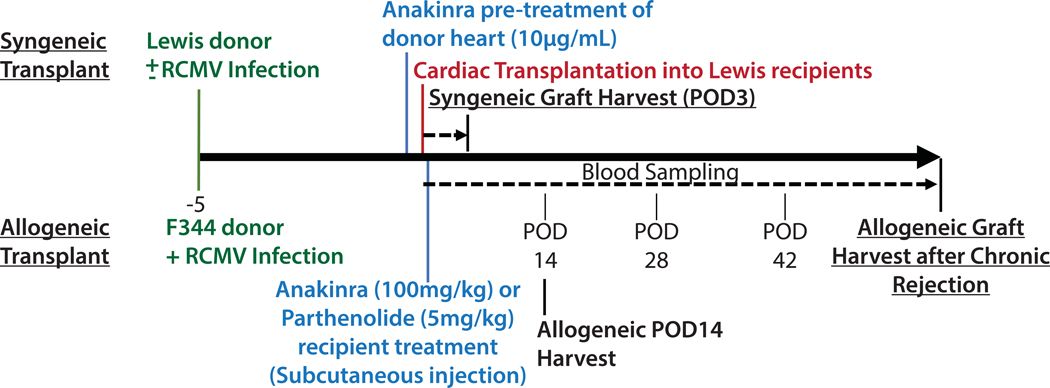
Figure 1.
Study Design. Syngeneic donors (Lewis) were either mock infected or infected with RCMV at 1×105 PFU/animal 5 days prior to transplantation. One cohort of syngeneic cardiac grafts were soaked in UW solution containing 10μg/mL anakinra during the 4-hour cold ischemia time. At 1-hour post-transplantation, groups of recipients were treated by subcutaneous injection with either anakinra (100mg/kg) or parthenolide (5mg/kg) or their respective vehicles. Syngeneic cohorts were harvested at POD3. Allogeneic donors (F344) were infected with RCMV at 1×105 PFU/animal 5 days prior to transplantation. Allogeneic cohorts were treated subcutaneously with anakinra at 100mg/kg or vehicle by subcutaneous injection at 1 hour post-transplantation. Blood samples were taken at POD 14, 28, and 42. Animals were sacrificed at two time-points for analyses: POD14 and at the time of chronic rejection.
Table 1.
Animal Cohorts
| Cohort | n | Graft type | Donor | Recipient | RCMV infection | Drug treatment | Harvest POD |
|---|---|---|---|---|---|---|---|
| 1 | 4–5 | None | Lewis | 3 | |||
| 2 | 4–8 | Syngeneic | Lewis | Lewis | 3 | ||
| 3 | 4 | Syngeneic | Lewis | Lewis | Donor+ | 3 | |
| 4 | 4 | Syngeneic | Lewis | Lewis | Anakinra,Donor Tissue | 3 | |
| 5 | 4–8 | Syngeneic | Lewis | Lewis | Anakinra,Recipient | 3 | |
| 6 | 4 | Syngeneic | Lewis | Lewis | Donor+ | Anakinra,Recipient | 3 |
| 7 | 5 | Syngeneic | Lewis | Lewis | Parthenolide, Recipient | 3 | |
| 8 | 5 | Syngeneic | Lewis | Lewis | Donor+ | Parthenolide, Recipient | 3 |
| 9 | 6,10 | Allogeneic | F344 | Lewis | Donor+ | 14,CR | |
| 10 | 6,10 | Allogeneic | F344 | Lewis | Donor+ | Anakinra,Recipient | 14,CR |
Histological Assessment:
Paraffin embedded heart tissue sections were stained with hematoxylin and eosin (H&E) and evaluated by microscopy. Lymphocytic infiltration and extent of tissue damage were quantified using a graded scale (Supplemental Table 1). Paraffin embedded allograft heart tissue sections were stained with H&E and elastic van Gieson stain. TVS was calculated as the neointimal index (NI=(intima area/lumen + intima area)x100)12–14.
Transcriptomics:
Total RNA was isolated from PBMC and homogenized graft and native hearts using Trizol. RNA deep sequencing analysis (RNASeq) was performed on 1 μg of polyA-fractionated RNA utilizing the TruSeq Stranded mRNA library prep kit (Illumina). Library was validated using Agilent DNA 1000 kit on bioanalyzer according to manufacturer’s protocol. RNA libraries were sequenced by the OHSU Massively Parallel Sequencing Shared Resource Core using their Illumina HiSeq-2500. Bioinformatics analyses and biostatistical comparisons were performed by the Bioinformatics & Biostatistics Core at Oregon National Primate Research Center. The quality of the raw sequencing data was evaluated using FastQC15 combined with MultiQC16 (http://multiqc.info/). The files were imported into ONPRC’s DISCVR-Seq17, LabKey18 Server-based system. PRIMe-Seq Trimmomatic19 removed remaining Illumina adapters. Reads were aligned to the Rattus_norvegicus Rnor_6 genome in Ensembl along with its corresponding annotation, release 90. STAR20 (v020201) aligned reads to the genome using the Two-pass mode with default parameters. STAR calculated the number of reads aligned to each gene. RNA-SeQC21 (v1.1.8.1) ensured alignments were of sufficient quality. Samples had an average of 56M mapped reads, an average exonic rate of 76%, and an average of 15.6K genes detected (>5 raw reads) per sample. Gene-level differential expression analysis was performed in open source software R22. Gene-level raw counts were filtered to remove genes with extremely low counts following the published guidelines23 normalized using the trimmed mean of M-values method (TMM)24 and transformed to log-counts per million with associated sample-wise quality weight and observational precision weights using the Voom method25. Gene-wise linear models comparing the groups were employed for differential expression analyses using Limma with empirical Bayes moderation26 and false discovery rate (FDR) adjustment27. Pathway analysis was performed using Qiagen’s Ingenuity Pathway Analysis (IPA) software.
Pathway Analysis:
IPA was used to evaluate expression data and determine predicted gene interactions. Analysis parameters excluded p-values >0.1, false discovery rates (FDR) >0.01, and log2 fold-change values between −0.9 and 0.9, providing 7,579 (PBMC) and 7, 899 (Heart) analysis ready molecules. P-values were calculated via the Fisher exact test and corrected via the Benjamini-Hochberg procedure. Significantly impacted canonical pathways were explored via IPA core analysis and path explorer to develop a map of predicted down-stream effects from genes regulated following transplantation.
RCMV Quantitative PCR:
Viral genome copies in rat tissues were quantified using real-time PCR with standard cycling parameters with an RCMV DNA polymerase-specific primer and probe set: P1:CCTCACGGGCTACAACATCA; P2:GAGAGTTGACGAAGAACCGACC; Probe:VIC-CGGCTTCGATATCAAGTATCTCCTGCACC-TAMRA. Tissue DNA was extracted using DNAzol (ThermoFisher Scientific 10503027) and diluted to 50ng/μL DNA. qPCR was performed using TaqMan Fast Advanced Master Mix (Invitrogen). RCMV viral DNA served as the quantification standard. Samples were analyzed using an ABI StepOne Real-Time PCR system.
Quantitative RT-PCR:
Gene expression was quantified using real-time RT-PCR using primer and probe sets shown in Table 2. RNA (1μg total RNA for Anakinra cohorts or 5μg for Parthenolide cohorts) was isolated from approximately 20mg of rat heart tissues using Trizol (ThermoFisher Scientific) and DNase treated using TURBO DNase-free kit (Ambion). cDNA was generated using Superscript IV (Invitrogen) and analyzed by real-time PCR. Cloned gene amplicons were used as quantification standards. qRT-PCR was performed using TaqMan Fast Advanced Master Mix. Samples were analyzed using a QuantStudio 7 Flex Real-Time PCR system and data was normalized to the gene encoding ribosomal protein L32.
Table 2.
qRT-PCR primer and probe sets directed against rat inflammatory markers
| Gene | Forward Primer | Reverse Primer | Probe | Dye | Quencher |
|---|---|---|---|---|---|
| CXCL6 | CTGTTCACACTGCCACAGCAT | CAGCGTAGCTCCGTTGCAA | TGCTCCTTTCTCGGCCA | FAM | MGBNFQ |
| CXCL2 | TGAACAAAGGCAAGGCTAACTGA | CTCTTTGATTCTGCCCGTTGA | AAGAACATGGGCTCCTG | FAM | MGBNFQ |
| Cmc2a | GCACTGCACCCAGACAGAAG | CTGGGAGCTTGAGGGTTGAG | TAGCCACTCTCAAGGAT | FAM | MGBNFQ |
| IL-lR | GATTATGAGCCCACGGAATGA | GACGTTGCAGATCAGTTGTATCG | ATGGAAGCTGACCCAGG | FAM | MGBNFQ |
| MMP12 | GAAAGGAGCTGGCACAATGAAG | GCCCCACAGGCAGATACCT | TCCTCGTGCTGGTGCT | FAM | MGBNFQ |
| SPPl | CTCACCTCCCGCATGAAGAG | TCAGACGCTGGGCAACTG | AGTCCGATGAGGCTAT | FAM | MGBNFQ |
| IL-6 | GGAACGAAAGTCAACTCCATCTG | AAGGCAACTGGCTGGAAGTCT | ACAGCTATGAAGTTTCTC | FAM | MGBNFQ |
| L32 | GAAGATTCAAGGGCCAGATCC | GTGGACCAGAAACTTCCGGA | CGGGAGTAACAAGAAAACCAAGCACATGC | VIC | TAMRA |
Multiplex Cytokine Assay:
A 27-Plex rat cytokine assay (Millipore) was performed on POD3 serum samples and heart tissue lysates. Tissues samples weighing 0.1g were homogenized by bead-beating in cold PBS containing 1mM PMSF. Samples were centrifuged at 10,000 RPM in a microcentrifuge at 4°C for 10 minutes. The supernatants were collected for use in the assay.
Flow Cytometry:
Native and graft hearts were harvested from rats at POD3 and digested in PBS containing Collagenase D (200U/mL) (Millipore Sigma) at 37°C for 45 minutes. Digested heart tissues were macerated and strained through 70μm nylon cell strainers (Fisher). Lymphocytes were isolated by centrifugation over lymphocyte separation media (Corning). For each sample, 2×106 cells were stained for 20 minutes at 4°C with fluorescently labeled anti-rat antibodies directed against CD3-APC (BD Biosciences), CD4-APC-Cy7 (BioLegend), CD8-PerCP (BD Biosciences), CD161a-V450 Biotinylated (BD Biosciences), CD68-A700 (Bio-Rad), CD45rA-PE-Cy7 (Invitrogen), His48-FITC (Invitrogen), CD43-PE (BioLegend). Cells were analyzed using an LSRII flow cytometer and data were analyzed using FlowJo software (FlowJo v10.5.3).
Enzyme-linked Immunosorbent Assays (ELISA):
High-binding ELISA plates (Corning 9018) were coated overnight at 4°C with RCMV-infected cellular lysates (10μg/mL) or F344 heart homogenates diluted in PBS. To assess anti-graft antibody levels, hearts from RCMV-naïve F344 rats were homogenized in 2mL/heart of cell lysis buffer (Cell Signaling Technology) containing HALT protease inhibitor (ThermoFisher). A 1/64 dilution of heart homogenate in PBS was used to coat high binding ELISA plates. Plates were blocked with 2% milk in wash buffer (0.05% Tween-PBS) for 2 hours at room temperature. Plates were washed three times with wash buffer and then incubated for 1.5 hours with 2-fold serial dilutions of plasma. After washing, the plates were incubated for 1 hour with HRP-conjugated anti-rat IgG (Invitrogen) or IgM (ThermoScientific) antibodies. Detection and quantification of bound secondary antibodies was performed by adding o-phenylenediamine dihydrochloride substrate (ThermoFisher Scientific). Dilution titers were calculated based on log-log transformations of the linear portion of the dilution curve for each sample.
RCMV Viral Replication Analysis:
NR8383 rat macrophages (ATCC) were maintained in F-12K media (Fisher) with 10%FBS plus PSG. RFL6 fibroblasts (ATCC) were maintained in DMEM (Fisher) with 5%FBS and PSG. Triplicate wells of macrophages or fibroblasts, plated in 24-well plates at 6.5×105 or 1×105 cells/well, respectively, were treated with Anakinra at 10μg/mL or PBS for 2 hours and then mock infected or infected with RCMV-GFP13 at multiplicity of infection (MOI)=0.1 or 0.5 (n=3). Cells were washed and cultured +/−Anakinra (10μg/mL) for 7 days. Supernatant samples from fibroblasts were collected at 24-hour intervals and viral loads were determined by plaque assay. Infected NR8383 macrophages were collected at 7 days post-infection and virus was quantified by qPCR as described above.
Viral plaque Assays:
RFL6 fibroblasts were plated at confluency in 24 well plates. Serial dilutions from 10−1 to 10−6 of viral supernatants were performed in round-bottom 96 well plates. Media was aspirated from RFL6 fibroblasts and 100μL of each viral dilution was added per well. After a 2-hour incubation on a rocker at 37°C, 250μL of carboxymethyl cellulose (CMC) (0.25% low viscosity/0.25% high viscosity (Sigma) in complete DMEM)28 was added to each well and cells were cultured at 37°C. At 7 days, cells were fixed and stained with methylene blue, and plaques were counted to determine viral titers.
Statistical Tests:
A one-way ANOVA with Tukey’s multiple comparisons was used to identify statistically significant differences in histological, qPCR, and POD14 NI data. A one-way ANOVA with Dunnett’s multiple comparisons was used to analyze Luminex assay data. A two-way ANOVA with Tukey’s multiple comparisons was used to analyze flow cytometry results and RCMV RFL6 growth curves. Statistical significance of RCMV NR8383 viral load data and qRT-PCR expression data was determined using two-way ANOVA with Sidak’s multiple comparisons or Dunnett’s multiple comparisons, respectively. Mann-Whitney test was used to analyze CR survival, NI data, and antibody titers. For all tests *(p<0.05), **(p<0.01), ***(p<0.001), ****(p<0.0001). Statistical analysis of animal viral loads was determined as the number of animals with detectable versus non-detectable viral loads by Fisher’s exact test.
Results
IRI Transcriptomic Analysis
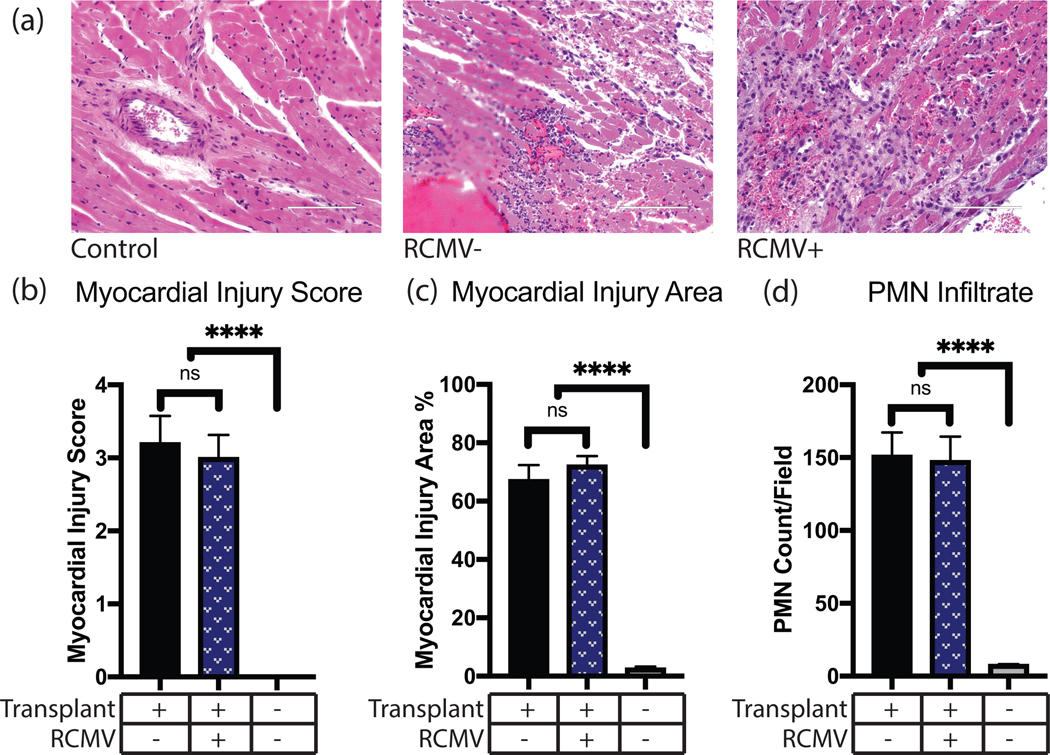
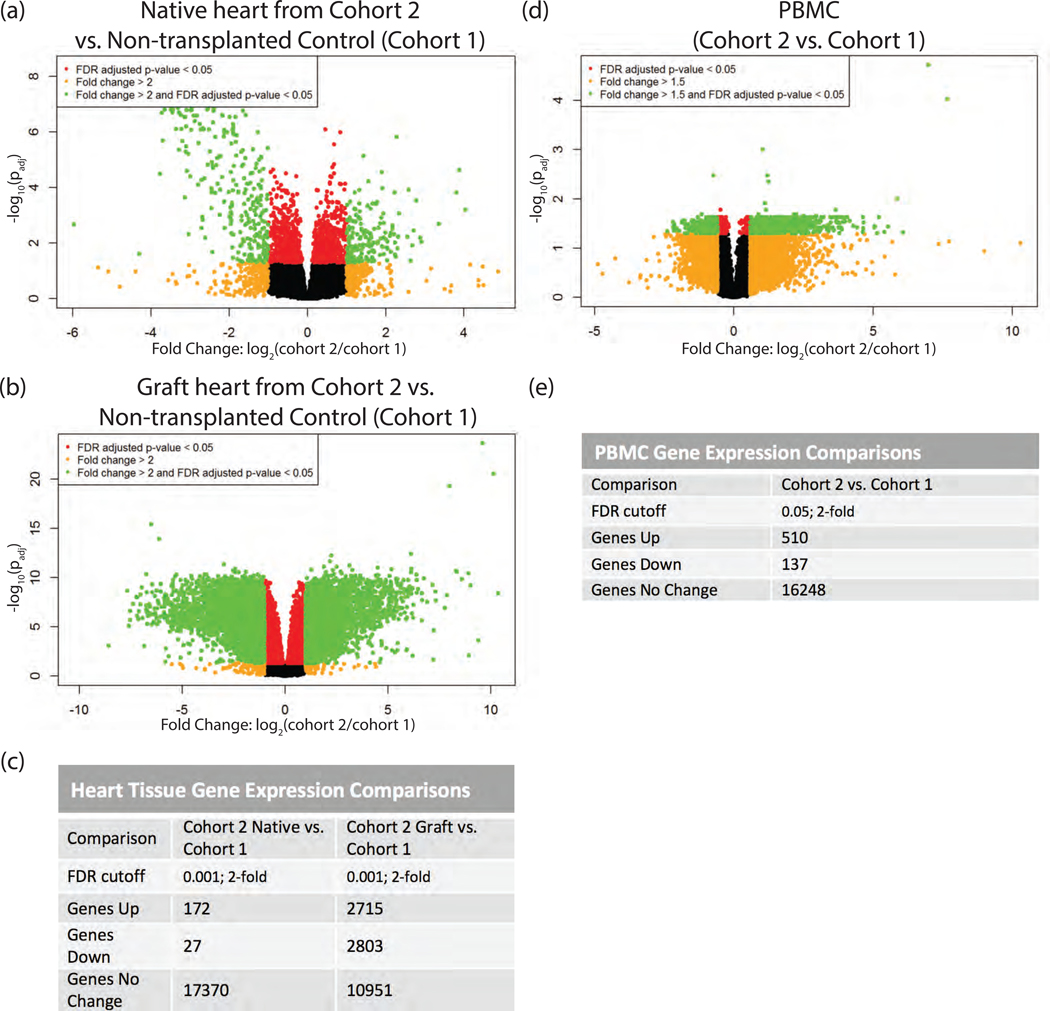
IRI induces cellular responses that promote TVS and accelerate CR. To characterize these responses, a rat model of syngeneic heart transplantation was used to evaluate early graft injury (Figure 1, Table 1). The degree of cardiac graft IRI at POD3 was measured by histological evidence of disease and scored for myocardial injury score, size of myocardial injury area, and level of PMN infiltrate. Significant myocardial injury was observed in transplanted hearts versus non-transplanted controls, with no substantial difference between RCMV-infected and uninfected heart syngeneic grafts (Figure 2). This is consistent with previous findings that CMV acceleration of TVS and CR requires an allogeneic environment11,29. To identify molecular pathways involved in IRI, RNAseq transcriptomics was performed on PBMC and heart tissues from animals in Cohorts 1 and 2 lacking RCMV infection. Differential gene analysis revealed changes in the expression of 5,518 genes in graft heart tissues and 647 genes in PBMC at POD3 (Figure 3). However, fewer genes were transcriptionally altered in syngeneic recipient native hearts (Cohort 2) versus the native hearts of non-transplanted animals (Cohort 1), indicating that the cardiac transplantation surgery causes IRI and its associated transcriptomic changes (Figure 3a,c).
Figure 2.
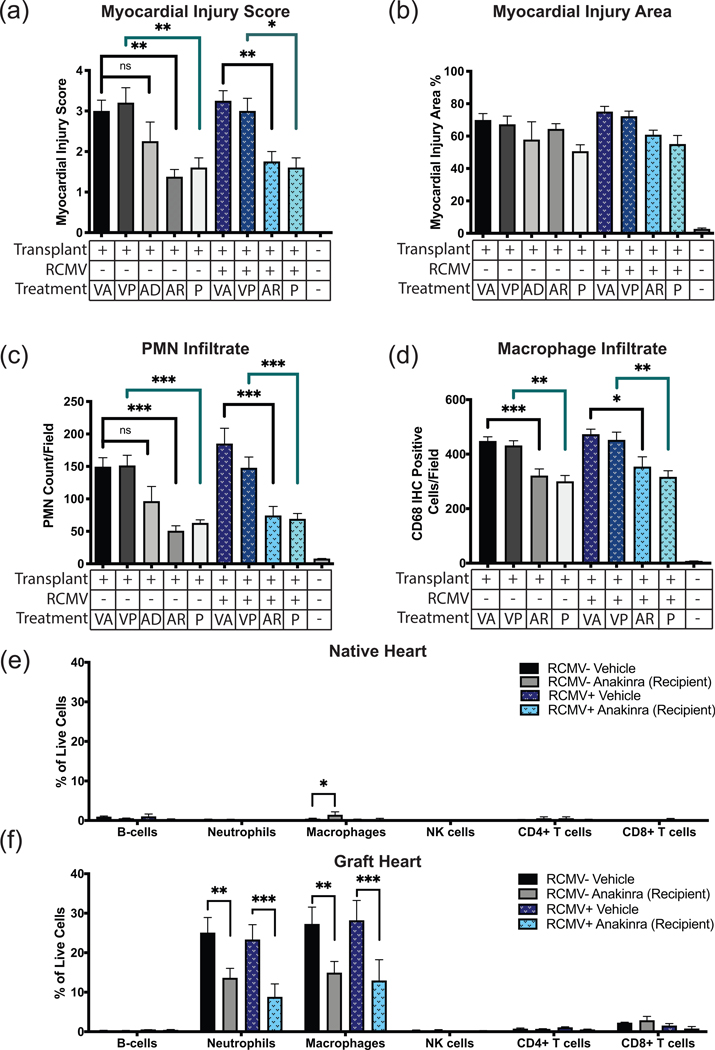
IRI causes myocardial tissue damage and PMN infiltration. Graft or control hearts were harvested at POD3 from isogeneic transplants, fixed in formalin, sectioned, and H&E stained. Tissue sections were then examined and graded for severity of myocardial injury, percent of examined area showing myocardial injury, and PMN infiltrate counts. (a) Representative images of control and I/R injured cardiac tissue at POD3 with or without prior RCMV infection. Control tissue was obtained from a non-transplanted animal. Scale bars represent 100μm. (b) Myocardial injury scores on a scale of 0 (no damage) to 4 (severe damage) as described in Supplemental Table 1, (c) Myocardial injury area determined as percent of examined area showing any degree of myocardial injury, (d) PMN infiltrate as measured by number of PMN cells per field of view at 400x magnification. n=4 for all groups. Error bars represent SEM.
Figure 3.
RNAseq transcriptomic analysis of cardiac tissue and PBMC following IRI. Heart tissues were homogenized in Trizol. PBMC were isolated over lymphocyte separation media and resuspended in Trizol. RNA was extracted from heart and PBMC samples by Trizol preparation with isopropanol precipitation. One μg of total RNA was used for RNAseq. (a) Native heart from RCMV- transplant recipient at POD3 (cohort 2) compared to a non-transplanted control heart (cohort 1), (b) Graft heart from RCMV- transplant recipient at POD3 (cohort 2) compared to a non-transplanted control heart (cohort 1), (c) FDR-cutoffs and regulated gene counts for heart tissue comparisons. (d) PBMC isolated from whole blood of a RCMV- transplant recipient at POD3 (cohort 2) compared to PBMC isolated from whole blood of a non-transplanted control animal (cohort 1), (e) FDR-cutoffs and regulated gene counts for PBMC heart tissues. Genes showing fold-change greater than 2 (Heart) or 1.5 (PBMC) with an FDR adjusted p-value of 0.05 are shown in green in volcano plots (a, b, d) and were selected for further investigation.
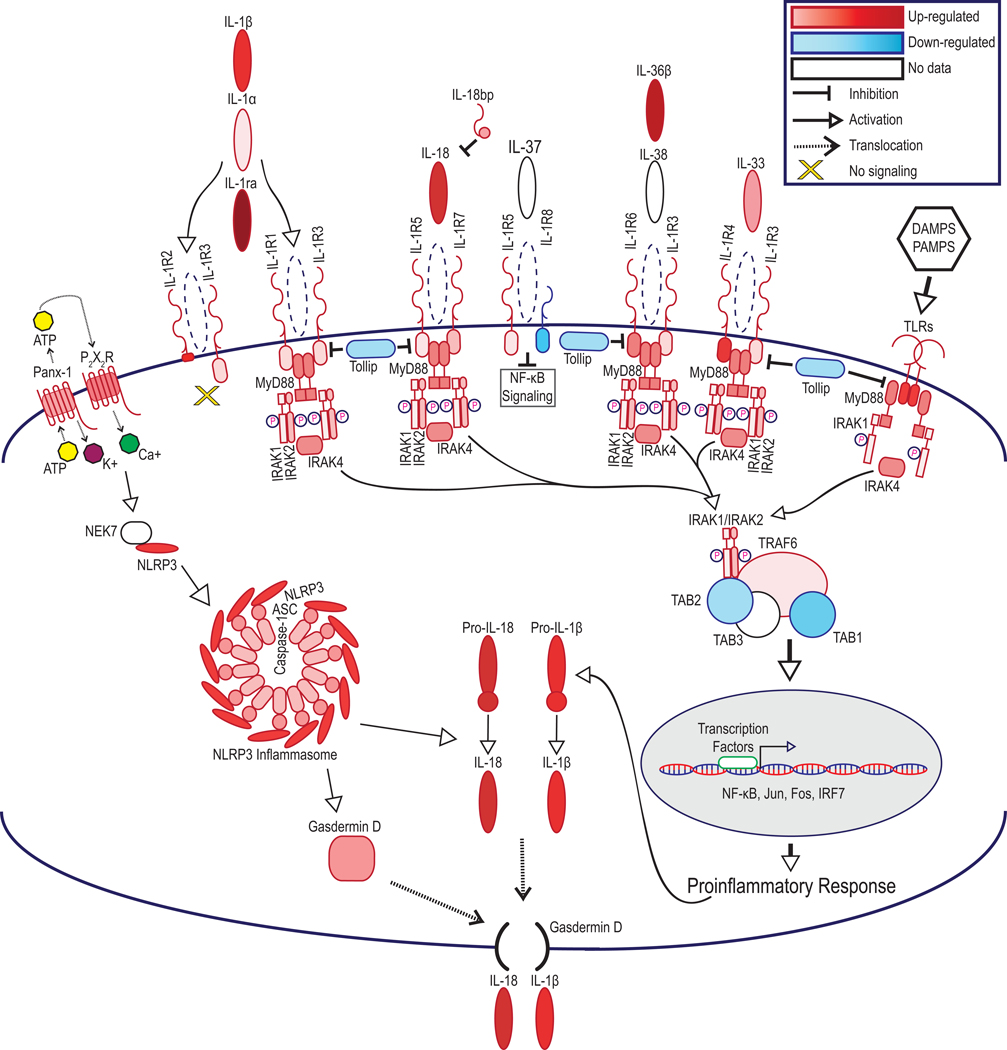
Genes showing differential expression following IRI were organized into functional and disease pathways revealing cellular migration, chemotaxis, and inflammation pathways were altered in Cohort 2 versus Cohort 1 hearts (Supplemental Tables 2.1, 2.2). Top up-regulated hits in graft hearts included acute phase response signaling; NF-κB signaling; and inflammasome pathway. In PBMC samples, up-regulated pathways included NF-κB signaling; acute phase response signaling, and interleukin-signaling (Supplemental Tables 3.1, 3.2). Consideration of upstream regulators and predicted downstream effects revealed that IL-1R up-regulation in recipient PBMC predicted increased expression of interleukins, chemokines, TNF-associated molecules, and IFN factors, matching our RNAseq findings (Supplemental Figure 2). Causal analysis for molecules within these networks predicted increases in the activation of leukocytes, the inflammatory response, leukocyte migration, leuko-/lympho-poiesis, and recruitment of myeloid cells (Supplemental Table 4). Further analysis revealed that members of the IL-1 cytokine family pathway were upregulated in both cardiac grafts and PBMC following IRI (Table 3, Figure 4).
Table 3.
Pro-inflammatory signaling molecules are up-regulated in graft and PBMC data following transplantation. Selected RNAseq data showing graft hearts from transplant recipients (cohort 2) versus non-transplanted native hearts (cohort 1) and PBMC samples from cohort 2 versus cohort 1. Up-regulated genes are color coded in red, down-regulated genes are color coded in blue. Those values that were found to be statistically significant (FDR-p <0.05) are bolded. Functions of each signaling molecule are listed.
| Gene | Heart: Cohort 2 Graft/Cohort 1 | FDR p- value | PBMC: Cohort 2/Cohort 1 | FDR p- value | Function |
|---|---|---|---|---|---|
| IL-1α | 1.66 | 0.4812 | Cytokine, Inflammation | ||
| IL-1β | 19.57 | 0.0000 | 3.63 | 0.4305 | Cytokine, Inflammation |
| IL1rn/IL1ra | 127.41 | 0.0000 | 4.06 | 0.0845 | Cytokine, Inhibitor |
| IL-18 | 29.50 | 0.0000 | 2.11 | 0.6595 | Cytokine, Inflammation |
| IL-18bp | 2.18 | 0.0014 | 1.76 | 0.6741 | Cytokine, Inhibitor |
| IL36β | 95.31 | 0.0000 | 3.48 | 0.0739 | Cytokine, Inflammation |
| IL33 | 3.40 | 0.0000 | 1.17 | 0.9715 | Cytokine, Inflammation |
| IL1R1 | 1.87 | 0.0181 | 12.32 | 0.0503 | Receptor, Inflammation |
| IL1R2 | 41.12 | 0.0000 | 21.46 | 0.1209 | Decoy Receptor, Inhibitor |
| IL1RAP | 1.88 | 0.0028 | 2.36 | 0.1481 | Receptor, Inflammation |
| IL18r1 | 1.69 | 0.0233 | 0.86 | 0.9050 | Receptor, Inflammation |
| IL18Rβ | 2.56 | 0.0000 | 1.95 | 0.4543 | Receptor, Inflammation |
| IL1R8 | 0.56 | 0.0014 | 0.73 | 0.6399 | Receptor, Inhibitor |
| IL1rl1 | 23.25 | 0.0000 | 2.32 | 0.5777 | Receptor, Inflammation |
| IL1rl2 | 4.86 | 0.0001 | 2.49 | 0.4433 | Receptor, Inflammation |
| Myd88 | 5.19 | 0.0000 | 1.84 | 0.0755 | Signaling Adaptor, Inflammation |
| IRAK1 | 1.29 | 0.0000 | 1.20 | 0.5414 | Signaling Adaptor, Inflammation |
| IRAK2 | 2.81 | 0.0000 | 1.24 | 0.7535 | Signaling Adaptor, Inflammation |
| IRAK4 | 4.47 | 0.0000 | 1.64 | 0.0728 | Signaling Adaptor, Inflammation |
| TRAF6 | 1.65 | 0.0006 | 1.26 | 0.8759 | Signaling Adaptor, Inflammation |
| Tollip | 0.80 | 0.0010 | 1.32 | 0.5129 | Signaling Adaptor, Inhibitor |
| Pannexin-1 | 2.32 | 0.0018 | 0.46 | 0.3481 | ATP Release Channel |
| P2X2R | 6.16 | 0.0001 | DAMP, ATP Sensor | ||
| TLR2 | 22.85 | 0.0000 | 3.39 | 0.1182 | PRR, Inflammation |
| TLR3 | 1.87 | 0.0007 | 1.01 | 0.9981 | PRR, Inflammation |
| TLR4 | 4.95 | 0.0000 | 2.00 | 0.5758 | PRR, Inflammation |
| TLR5 | 4.63 | 0.0005 | 7.24 | 0.0170 | PRR, Inflammation |
| TLR6 | 6.51 | 0.0000 | 2.17 | 0.1580 | PRR, Inflammation |
| TLR7 | 22.94 | 0.0000 | 2.25 | 0.3191 | PRR, Inflammation |
| TLR8 | 26.43 | 0.0007 | 2.22 | 0.0443 | PRR, Inflammation |
| TLR9 | 11.48 | 0.0240 | 0.76 | 0.7868 | PRR, Inflammation |
| TLR10 | 20.00 | 0.0000 | 2.24 | 0.2805 | PRR, Inflammation |
| TLR11 | 3.08 | 0.0099 | PRR, Inflammation | ||
| TLR12 | 3.83 | 0.0015 | 0.40 | 0.6166 | PRR, Inflammation |
| TLR13 | 6.92 | 0.0000 | 2.76 | 0.0691 | PRR, Inflammation |
| NLRP3 | 12.35 | 0.0000 | 7.79 | 0.1089 | Sensor, Inflammasome Formation |
| ASC | 3.75 | 0.0000 | 1.15 | 0.8267 | Adaptor, Inflammasome Formation |
| Caspase-1 | 2.75 | 0.0000 | 0.81 | 0.6099 | Enzyme, Inflammasome |
| Gasdermin D | 4.21 | 0.0000 | 0.95 | 0.9080 | Membrane Pore, Cytokine Release |
Cohort 1 = Non-transplanted, non-infected
Cohort 2 = Syngeneic transplant, non-infected
p < 0.05 is significant
Figure 4.
IL-1R signaling molecules are up-regulated in graft and PBMC samples following transplantation leading to a pro-inflammatory response. Statistically significant alterations in gene expression from RNAseq analysis of graft tissues are represented by red (up-regulation) and blue (down-regulation). Molecules which did not meet statistical significance, or for which mapping was unavailable in the rat genome are shown in white. FDR-p<0.05. FDR-corrected p-values and Cohort 2 versus Cohort 1 ratios are listed in Table 3.
Regulated genes from cardiac grafts appearing in multiple pathways were selected for validation by qRT-PCR (Supplemental Figure 1). The up-regulation of MMP12, SPP1, CXCL6, CXCL2, and CXCL3 supported our RNAseq analysis. While IL-1R1 and IL-1R2 were both up-regulated in PBMCs, RNAseq showed only a minor change in POD3 transcriptional expression of IL1R in cardiac grafts. We validated IL-1R expression in cardiac grafts by qRT-PCR and, similarly, found no change between Cohort 2 and Cohort 1 (Supplemental Figure 1). However, IL-1R1 and IL-1R2 were both up-regulated in PBMCs in our RNAseq data (Table 3). These findings indicate that transplant IRI initiates inflammation in the cardiac grafts; and lymphocytes, through their response to IL-1, consequently exacerbate this inflammatory process.
Anakinra and Parthenolide Reduce IRI-Induced Tissue Damage
Transcriptomic analysis suggested a key role for IL-1 signaling in IRI in syngeneic heart transplants. Our data, and prior work by others30–32, implicated NF-κB signaling and the inflammasome as other targets for reducing IRI (Figure 4, Table 3). Two different therapeutics were tested to assess the role of IL-1 signaling in IRI. The first approach used the clinically-approved IL-1R1/2 antagonist Anakinra, which reduces neutrophil and macrophage activation in multiple disease states and in rat models33–36. Additionally, NLRP3 was increased 12.35-fold in cardiac grafts and 7.79-fold in PBMC during transplant-induced IRI (Figure 4, Table 3). The NLRP3 inflammasome activates Caspase-1, which proteolytically activates pro-IL-1β and pro-IL-18. Our second treatment used parthenolide, an inhibitor of Caspase-1/NF-κB, with known anti-inflammatory activity in rats31,37,38.
To determine efficacy of Anakinra two treatment regimens were used: 1) syngeneic donor cardiac grafts during the 4-hour CI period (Cohort 4); or 2) the recipient at 1-hour post-transplant (Cohort 5) (Figure 1, Table 1). Syngeneic graft hearts were harvested at POD 3 and analyzed by IHC and flow cytometry. Pre-transplant donor cardiac graft treatment (Cohort 4) resulted in minimal decreases in myocardial injury (score and area) and PMN infiltration compared to vehicle treated controls (Cohort 2) (Figure 5a–d). In contrast, recipients treated with Anakinra at 1-hour post-transplant (Cohort 5) had a significant decrease in myocardial injury score, PMN and macrophage graft infiltration compared to vehicle treated controls (Cohort 2) (Figure 5a,c,d). Flow cytometric analysis of graft and native heart cellular infiltrates demonstrated that neutrophils and macrophages were the predominant infiltrating immune cell types in the grafts at POD3, which were significantly reduced by Anakinra treatment and absent in native hearts (Figure 5e,f). For parthenolide studies, syngeneic transplant recipients were treated subcutaneously at 1-hour post-transplantation with 5mg/kg (Cohort 7) or vehicle (Cohort 2). Similar to Anakinra, parthenolide significantly reduced POD3 myocardial injury score, as well as PMN and macrophage graft infiltration (Figure 5a–d). RCMV infection did not influence these early events in syngeneic graft hearts (Cohorts 6 and 8), which is consistent with our previous findings that CMV affects allograft rejection starting at POD 739. Our findings demonstrate that syngeneic grafts from RCMV-infected donors respond equally to Anakinra and parthenolide treatments by a reduction in myocardial injury and inflammatory cell infiltration.
Figure 5.
Anakinra treatment of the recipient immediately post-transplantation improves myocardial injury score and PMN infiltrate into cardiac tissue. (a-d) Myocardial injury score (a), myocardial injury area (b), PMN infiltrate (c), and macrophage infiltrate (d). Error bars represent SEM. Statistical significance determined by one-way ANOVA with Tukey’s multiple comparison, *P<0.05, **P<0.01, ***P<0.001 versus vehicle transplant. Representative images are shown in Supplemental Figure 3. Fields counted for (c) and (d) were viewed at magnification equal to 400x. Groups are labeled (Transplant-/RCMV-/Treatment-) Native heart from a non-transplanted, uninfected animal (n=5); (Transplant+/RCMV-/VA) Graft heart from RCMV- donor with anakinra vehicle injection of recipient animals 1-hour post-transplant (n=4); (Transplant+/RCMV-/VP) Graft heart from RCMV- donor with parthenolide vehicle injection of recipient animals 1-hour post-transplant (n=5); (Transplant+/RCMV-/AD) Graft heart from RCMV- donor with anakinra treatment of donor organ pre-transplantation (n=4); (Transplant+/RCMV-/AR) Graft heart from RCMV- donor with anakinra treatment of recipient 1-hour post-transplant (n=4); (Transplant+/RCMV-/P) Graft heart from RCMV- donor with parthenolide treatment of recipient 1-hour post-transplant (n=5); (Transplant+/RCMV+/VA) Donor infected i.p. with 1×105PFU RCMV 5 days prior to transplant, graft heart with anakinra vehicle injection of recipients 1-hour post-transplant (n=4); (Transplant+/RCMV+/VP) Donor infected i.p. with 1×105 PFU RCMV 5 days prior to transplant, graft heart with parthenolide vehicle injection of recipients 1-hour post-transplant (n=4); (Transplant+/RCMV+/AR) Donor infected i.p. with 1×105PFU RCMV 5 days prior to transplant followed by anakinra treatment of recipient 1-hour post-transplant (n=4); (Transplant+/RCMV+/P) Donor infected i.p. with 1×105PFU RCMV 5 days prior to transplant with parthenolide treatment of recipient 1-hour post-transplant (n=5). (e,f) Native (e) and graft (f) heart tissues were harvested at 3 days post-transplant from RCMV- PBS treated (cohort 2), RCMV- Anakinra treated (cohort 4), RCMV+ PBS treated (cohort 5), and RCMV+ Anakinra treated (cohort 6) recipients and processed for flow cytometry staining. Antibodies were directed against cellular markers for T-cells (CD3+, CD4+/CD8+), B-cells (CD45ra+, CD3-, CD161a low), Neutrophils (CD43+, CD3-, CD161a low, CD45rA-), macrophages (CD68+, CD3-), and NK cells (CD161a high, CD3-). Gating strategy as in Supplemental Figure 4. (e) Native heart tissues from transplant recipients at POD3, cell percentages reported as percent of total live cells. (f) Graft heart tissues from transplant recipients at POD3, cell percentages reported as percent of total live cells. Statistical significance determined by two-way ANOVA with Tukey’s multiple comparison, *P<0.05, **P<0.01, ***P<0.001. n=4. Error bars represent SEM.
Anakinra Reduces Inflammatory Cytokine Production Following Transplant IRI
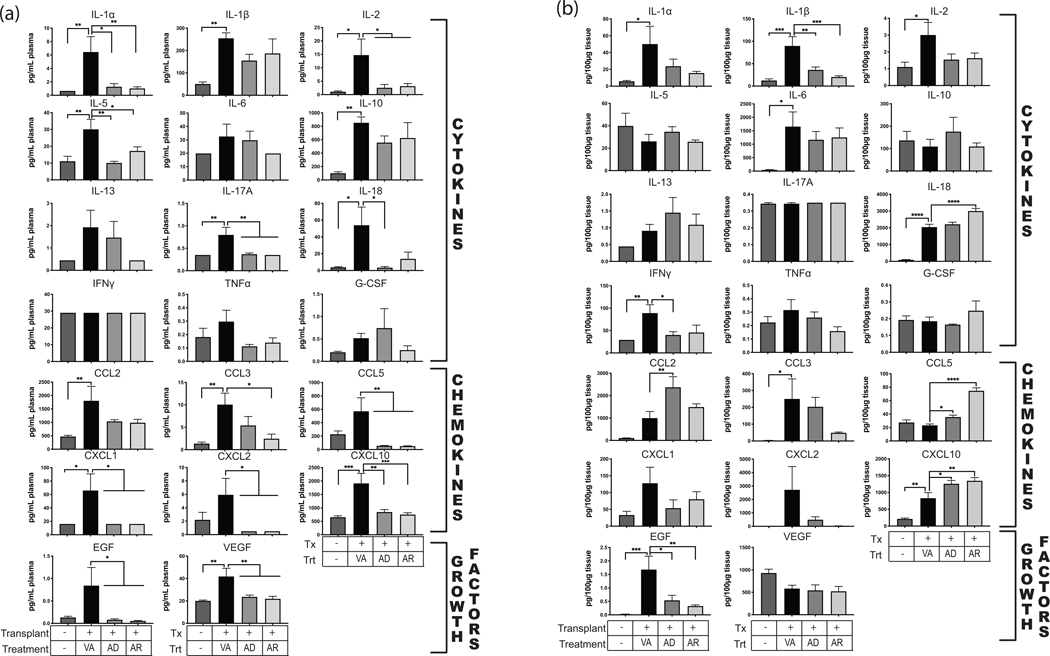
To determine the anti-inflammatory pathways that are activated by IRI and targeted by Anakinra, we performed rat cytokine/chemokine assays on POD3 plasma and graft heart homogenates (Figure 6). Following transplantation, plasma concentrations were elevated above pre-transplant levels for cytokines IL-1β, IL-2, IL-5, IL-10, IL-13, IL-17A, IFNγ, G-CSF, and TNFα; chemokines CCL2, CCL5, CXCL1, CXCL2, and CXCL10; and epidermal growth factor (EGF). Cardiac graft cytokine levels were also increased above those detected in non-transplanted control hearts for IL-1α, IL-1β, IL-2, IL-6, IL-13, IL-18, IFNγ, and TNFα; chemokines CCL2, CCL3, CXCL1, CXCL10; and EGF. Interestingly, while Anakinra treatment reduced the IRI-induced levels of IL-1β, IL-2, TNFα, CXCL1 and EGF, the drug did not have a global effect on all of the upregulated factors and some were differentially affected in plasma versus tissue. For example, IL-18 and CXCL10 were increased in grafts from treated animals compared to transplanted controls whereas plasma levels of CXCL10 were reduced upon treatment. Treatment reduced IL-5, IL-10, IL-13, IL-17A, G-CSF, and CCL2 in plasma. By contrast, graft levels of CCL2 were increased in the treated animals. IL-1α, IL-6, IFNγ, CCL3, and CXCL2 graft levels were reduced as a result of Anakinra treatment. These data indicate that Anakinra reduces plasma and graft cytokine expression associated with IL-1R-driven inflammation.
Figure 6.
Anakinra reduces pro-inflammatory cytokines elevated during transplantation. Luminex cytokine profiling of serum and heart tissue homogenates was analyzed following IL-1R antagonist treatment of recipients compared to non-treated controls and non-transplanted controls. Treatment groups are as indicated with (Transplant-/Treatment-) Native heart from a non-transplanted, uninfected animal (n=5); (Transplant+/VA) Graft heart from RCMV- donor with anakinra vehicle injection of recipient animals 1-hour post-transplant (n=4); (Transplant+/AD) Graft heart from RCMV- donor with anakinra treatment of donor organ pre-transplantation (n=4); (Transplant+/AR) Graft heart from RCMV- donor with anakinra treatment of recipient 1-hour post-transplant (n=4). Error bars represent SEM. Statistical significance determined by one-way ANOVA with Dunnett’s correction for multiple comparisons, *P<0.05, ***P<0.001.
Anakinra Prolongs Survival of RCMV-Infected Cardiac Allografts
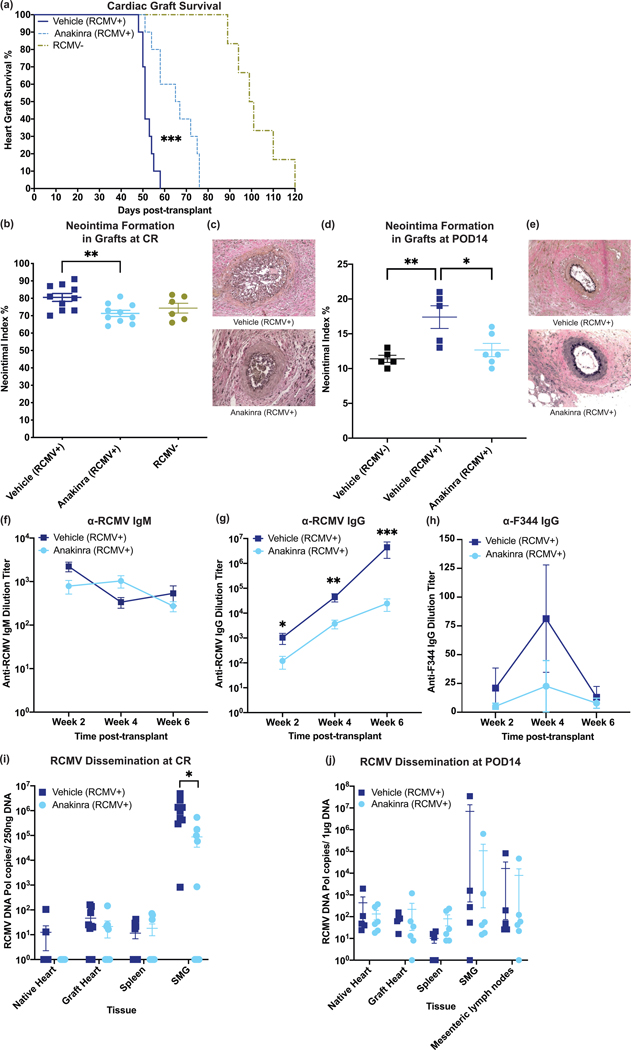
Our findings suggest neutrophil and macrophage infiltration following transplantation is a crucial component of IRI, which may contribute to graft CR. In light of IL-1R signaling blockade to reduce IRI and PMN infiltrate at POD3 from an RCMV+ donor, we determined whether Anakinra immediately following transplantation would improve long-term graft survival in RCMV+ allogeneic cardiac grafts. RCMV-infected F344 donors were transplanted into naïve Lewis rats and the recipients were treated with 100mg/kg Anakinra (Cohort 10) or vehicle (Cohort 9) at 1-hour post-transplantation. Animals were monitored daily for diminishing heart beat as an indicator of graft failure (CR). Time to CR was significantly increased (p<0.001) in Anakinra treated animals (66 days) compared to vehicle controls (51 days) (Figure 7a). Anakinra treatment also reduced the development of TVS (neointimal index (NI) measured at rejection) (Figure 7b,c). To assess kinetics of TVS reduction by Anakinra treatment, we harvested allografts (Cohorts 9 and 10) at POD14. Anakinra treatment reduced neointima formation to near RCMV-naïve transplanted grafts (Figure 7d,e), suggesting that Anakinra abrogates RCMV-induced early graft damaging TVS39.
Figure 7.
One dose of anakinra at 1-hour post-transplant significantly improves survival time and decreases viral loads at rejection. Donor animals were infected with RCMV 5 days prior to transplantation. Recipients were then treated with vehicle or with anakinra at 1-hour post-transplantation. (a) Time to rejection was significantly improved by recipient treatment with anakinra following transplantation. Graft heart rejection was monitored by palpation for heart beat. Animals were sacrificed and tissues were harvested at the time of rejection. Vehicle treated animals had a mean survival time of 52 days and Anakinra treated animals had a mean survival time of 65 days. Statistical significance determined by Mann-Whitney test, ***P<0.001 vs. Vehicle-control, n=10. PBS-treated RCMV- historical controls included for references (light green) (b,c) Neointimal index of coronary arteries in graft hearts at chronic rejection was decreased in Anakinra-treated cohorts. Tissue sections were taken from graft hearts at the time of rejection and fixed in formalin. Tissue sections were stained with H&E and Elastin staining and the neointimal index was determined as an average of 6 coronary artery sections per animal (b). Representative images are shown in (c). Statistical significance was determined by Mann-Whitney test, **P<0.01, n=10. PBS-treated RCMV- historical controls included for references (light green). (d,e) Neointimal index of coronary arteries in graft hearts at POD14 was decreased in Anakinra-treated cohorts. Tissue sections were stained and the neointimal index was scored as in (b/c). Neointimal index scores are summarized in (d). Representative images are shown in (e). Statistical significance was determined by one-way ANOVA with Tukey’s correction for multiple comparisons, *P<0.05, **P<0.01. n=6. (f-h) Anti-RCMV IgG antibody responses were reduced in Anakinra treated animals. Blood was taken from transplant recipients at 2, 4, and 6 weeks post-transplant until graft rejection. Enzyme-linked immunosorbent assays against anti-RCMV IgM (f) and IgG (g) antibodies and anti-F344 cardiac tissue IgG (h) antibodies were performed on serum samples and dilution titers were calculated for each sample. Averages for each cohort at 2, 4, and 6 weeks post-transplant are shown. n=10. Error bars represent SEM. Statistical significance determined by Mann-Whitney test, *P<0.05, **P<0.01, ***P<0.001 vs. Vehicle-control. (i)Viral loads in tissues at rejection showed a significant decrease in the number of animals with detectable viral loads in SMG following Anakinra treatment. Tissues were harvested at time of cardiac graft rejection in RNAlater. Tissues were then homogenized in DNAzol and DNA was extracted. qPCR for RCMV viral DNA polymerase was used to quantitate RCMV genome copies in tissues. Error bars represent SEM. Statistical significance determined by fisher’s exact test, *p=0.0325 (two-tailed). (j) Viral loads in tissues at POD14 did not yet show significant impairment in viral loads in Anakinra-treated cohorts. Tissues were harvested as in (i). n=6.
A possible mechanism behind Anakinra’s reduction in TVS and improved graft survival may be effects on CMV replication because CMV-accelerated TVS/CR are linked to active viral replication40,41. We assessed the effect of Anakinra on RCMV replication and immune response. Serum from RCMV infected graft recipients at 2, 4, and 6 weeks post-transplantation were quantified for RCMV-specific IgG and IgM antibody responses. No significant differences were observed in IgM titers (Figure 7f) at any timepoint, indicating no effect of Anakinra on early antibody responses. However, anti-RCMV IgG antibody levels were reduced with Anakinra versus controls (Figure 7g). Anti-donor IgG antibody responses were very low in all animals, and were not significantly different between treatment groups (Figure 7h). The reduction in antiviral IgG titers may be the result of decreased viral loads due to the impaired recruitment of macrophages into the graft heart or a direct antiviral effect. Accordingly, we first measured RCMV viral DNA loads in native heart, graft heart, spleen, and SMG at the time of rejection. Viral DNA levels in SMG tissues of Anakinra treated animals were reduced compared to controls, with a significantly greater number of animals with undetectable levels in SMG tissues (Figure 7i). Additionally, native heart, graft heart, and spleen tissues from Anakinra-treated animals trended towards decreased detectable levels (Figure 7i). Unexpectedly, Anakinra treatment only modestly reduced viral loads at POD14 in all tissues tested (Figure 7j). To rule out whether Anakinra directly inhibits viral replication, we infected rat macrophages and fibroblasts with RCMV and treated them with Anakinra at 10μg/mL. Treatment did not significantly alter viral titers at any timepoint in fibroblasts (Supplemental Figure 5a), or reduce viral genomes present in macrophages at 7 days post-infection (Supplemental Figure 5b). These results suggest Anakinra treatment in the early post-transplant period improves graft survival from RCMV-infected donors transplanted into naïve recipients by limiting RCMV dissemination, rather than directly impairing viral replication.
Discussion
CR is a major impediment to long-term graft and patient survival in SOT recipients. Alloimmunity, CMV infection, and IRI following transplantation are factors that contribute to acceleration of TVS. Recent work in murine kidney transplants demonstrated that IRI can initiate CMV reactivation, whereas immunosuppression promotes viral dissemination following transplantation29, supporting the hypothesis that IRI combines with CMV-associated pathogenesis to accelerate CR. Reducing transplant-induced IRI has the potential to improve transplant survival rates related to CMV infection; however, IRI lacks specific effective therapies. To develop targeted strategies toward reducing IRI, we identified pathways that contribute to IRI in the absence of alloimmunity in syngeneic cardiac transplants. Importantly, syngeneic grafts, in the setting of CMV infection, do not undergo CR due to the lack of alloimmune responses11. In our current study, we introduced a 4-hour CI time to improve the clinical relevance of our cardiac transplant model. We identified biological changes associated with transplant-induced IRI that included pro-inflammatory cytokines and then utilized targeted therapeutics to enhance graft survival in the context of RCMV infection.
IRI is a multifaceted disease process involving initial injury followed by a cascade of inflammatory steps that mediate further tissue damage. The goal of this study was to identify possible inflammation signaling nodes that, when inhibited, could target the multiple downstream effects of IRI, particularly as they relate to CR. One such node is the IL-1 pathway, which plays an important role in defense against pathogens and immunopathogenic disease32. The IL-1 family includes IL-1α/β, IL-18, IL-33, IL-37, and IL-38, their receptors and downstream activated transcriptional factors that produce other inflammatory molecules. We found upregulation of IL-1 inflammatory pathway members in both PBMC and cardiac grafts that included IL-1α, IL-1β, IL-18, IL-33, and IL-36β; their cognate receptors IL-1R1, IL-1R3–6; and their downstream signaling molecules IRAK1,2,4, MyD88, and TRAF6. Interestingly, IL-1R8, an IL-1R that inhibits NF-κB signaling, was transcriptionally down regulated. Thus, the IL-1 family is highly dysregulated following IRI. The IL-1 pathway has previously been identified as key to development of IRI33,42,43. For example, elevation of IL-1α levels have previously been linked to activation of alloreactive T-cells, suggesting that IL-1α, in addition to our data implicating IL-1β, may be a prominent component of early allograft injury44. Our pathway analysis builds on previous data, but the methods outlined here could be used to identify other targetable pathways for therapy.
Natural mechanisms limit IL-1-induced immunopathogenesis and promote adaptive immunity and wound healing. Soluble IL-1R antagonists and membrane decoys have been exploited for therapeutics that target IL-1 cytokine function during rheumatoid arthritis, autoinflammatory disease, cardiovascular disease, restenosis following angioplasty, systemic sclerosis, IRI, and diabetes42,45,46. In our IRI model, the natural rat IL-1RA was upregulated at POD3, but this is likely too late to prevent immune-mediated damage. However, Anakinra presents a prime candidate for clinical intervention by targeting IRI early inflammatory processes. Anakinra is approved for clinical use and has a short half-life, allowing us to interrogate the impact of a single-treatment on IRI and allograft CR. Parthenolide also targets the IL-1 signaling node, and has previously been tested as an anti-inflammatory drug in a rat myocardial reperfusion injury model31. Our model incorporates a CMV+ donor with active CMV infection of the donor tissue28, as previous studies have shown that CMV infection accelerates CR6, and IL-1Ra therapies have shown promise in other transplant models in the absence of CMV infection47–49. We targeted IRI in the setting of CMV infection in SOT, and demonstrated a reduction in accelerated cardiac graft CR. While the impact of Anakinra on chronic cardiac allograft rejection in the absence of CMV remains to be tested, Anakinra and parthenolide treatments both decreased tissue injury and graft macrophage and PMN recruitment, reducing the downstream events that promote acute and chronic rejection. Given the higher toxicity of parthenolide, Anakinra was a better therapeutic option and it improved graft survival and reduced CMV tissue viral loads at rejection. While Anakinra reduced anti-RCMV IgG responses there was no major impact on anti-donor IgG responses, suggesting that the reduction in inflammation reduced viral load rather than impacting global humoral responses. However, a remaining area to be explored is effect of Anakinra on the role of T-cell responses to viral and/or donor antigens and how this affects TVS and CR. Further work in this area would provide a better understanding of the disease mechanisms at work with and without treatment.
Anakinra solely targets the receptor for IL-1α/β. Thus, our finding that Anakinra did not completely inhibit graft IRI or restore the timing of CR to levels observed for uninfected allografts would suggest that additional IL-1 signaling components may be involved in the tissue injury process. This theory is consistent with our observation that IL-18, IL-33, IL-36, and their receptors are highly expressed during IRI. We hypothesize that combination therapy utilizing Anakinra and newly developed inhibitors of IL-18, IL-33, and IL-36 may have greater impact on reducing graft injury. Furthermore, graft survival may be enhanced with IL-1R antagonists, targetting early events leading to graft dysfunction, in combination with ganciclovir, to prevent CMV-driven, long-term events40,50,51. Future studies will focus on determining the role of Anakinra in CMV-naïve transplants as well as improving our understanding of the effect that Anakinra treatment has on T-cell-mediated immune responses during CR.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health NIAID R01 AI116633. NH and IJ were supported by the OHSU Molecular Microbiology and Immunology Interactions at the Microbe/Host Interface training grant NIH 2T32 AI007472. IJ received additional support from the grant NIH F31 1F311AI145193–01. Short read sequencing assays were performed by the OHSU Massively Parallel Sequencing Shared Resource. The authors acknowledge the support of the Oregon National Primate Research Center Bioinformatics & Biostatistics Core, which is funded in part by NIH grant OD P51 OD011092.
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.Lutz J, Thürmel K, Heemann U. Anti-inflammatory treatment strategies for ischemia/reperfusion injury in transplantation. J Inflamm (Lond). 2010;7(1):27. doi: 10.1186/1476-9255-7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello JP, Mohanakumar T, Nath DS. Mechanisms of chronic cardiac allograft rejection. Texas Hear Inst J. 2013;40(4):395–399. https://pubmed.ncbi.nlm.nih.gov/24082367. [PMC free article] [PubMed] [Google Scholar]
- 3.Colvin-Adams M, Agnihotri A. Cardiac allograft vasculopathy: Current knowledge and future direction. Clin Transplant. 2011;25(2):175–184. doi: 10.1111/j.1399-0012.2010.01307.x [DOI] [PubMed] [Google Scholar]
- 4.Fischer S, Glas KE. A Review of cardiac transplantation. Anesthesiol Clin. 2013;31(2):383–403. doi: 10.1016/j.anclin.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 5.Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. ClinMicrobiolRev. 2009;22(1):76–98. doi: 10.1128/CMR.00034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson I, Andersson R, Friman V, et al. Cytomegalovirus infection and disease reduce 10-year cardiac allograft vasculopathy-free survival in heart transplant recipients. BMC Infect Dis. 2015;15(582):1–9. doi: 10.1186/s12879-015-1321-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orloff SL, Hwee YK, Kreklywich C, et al. Cytomegalovirus latency promotes cardiac lymphoid neogenesis and accelerated allograft rejection in CMV Naive recipients. Am J Transplant. 2011;11(1):45–55. doi: 10.1111/j.1600-6143.2010.03365.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanan P, Razonable RR. Cytomegalovirus infections in solid organ transplantation: a review. Infect Chemother. 2013;45(3):260–271. doi: 10.3947/ic.2013.45.3.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fietze E, Prösch S, Reinke P, et al. Cytomegalovirus infection in transplant recipients: The role of tumor necrosis factor. Transplantation. 1994;58(6):675–680. doi: 10.1097/00007890-199409000-00007 [DOI] [PubMed] [Google Scholar]
- 10.Davis MK, Hunt SA. State of the art: Cardiac transplantation. Trends Cardiovasc Med. 2014;24(8):341–349. doi: 10.1016/j.tcm.2014.08.004 [DOI] [PubMed] [Google Scholar]
- 11.Orloff SL, Streblow DN, Soderberg-Naucler C, et al. Elimination of donor-specific alloreactivity prevents cytomegalovirus-accelerated chronic rejection in rat small bowel and heart transplants. Transplantation. 2002;73(5):679–688. [DOI] [PubMed] [Google Scholar]
- 12.Streblow DN, Kreklywich C, Yin Q, et al. Cytomegalovirus-mediated upregulation of chemokine expression correlates with the acceleration of chronic rejection in rat heart transplants. J Virol. 2003;77(3):2182–2194. doi: 10.1128/jvi.77.3.2182-2194.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vomaske J, Denton M, Kreklywich C, et al. Cytomegalovirus CC chemokine promotes immune cell migration. J Virol. 2012;86(21):11833–11844. doi: 10.1128/JVI.00452-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong AT, Strauch AR, Starling RGC, Sedmak DD, Orosz CG. Morphometric analysis of neointimal formation in murine cardiac allografts. Transplantation. 1997;63(7):941–947. doi: 10.1097/00007890-199704150-00006 [DOI] [PubMed] [Google Scholar]
- 15.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- 16.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bimber B. DISCVR-Seq: LabKey Server Extensions for Management and Analysis of Sequencing Data. 2015. https://github.com/bbimber/discvr-seq/wiki. [Google Scholar]
- 18.Nelson EK, Piehler B, Eckels J, et al. LabKey Server: An open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics. 2011;12(1):71. doi: 10.1186/1471-2105-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Core Team. R: A language and environment for statistical computing. 2017. https://www.r-project.org/. [Google Scholar]
- 23.Chen Y, Lun ATL, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research. 2016;5:1438. doi: 10.12688/f1000research.8987.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3). doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;22(9):561–564. doi: 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47-e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc. 1995. doi: 10.2307/2346101 [DOI] [Google Scholar]
- 28.Kreklywich CN, Smith PP, Jones CB, Cornea A, Orloff SL, Streblow DN. Fluorescence-Based Laser Capture Microscopy Technology Facilitates Identification of Critical In Vivo Cytomegalovirus Transcriptional Programs. In: Yurochko AD, Miller WE, eds. Human Cytomegaloviruses: Methods and Protocols.; 2014:217–237. doi: 10.1007/978-1-62703-788-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Qiu L, Yan S, et al. A clinically relevant murine model unmasks a “two-hit” mechanism for reactivation and dissemination of cytomegalovirus after kidney transplant. Am J Transplant. 2019;19(9):2421–2433. doi: 10.1111/ajt.15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onai Y, Suzuki JI, Kakuta T, et al. Inhibition of IκB phosphorylation in cardiomyocytes attenuates myocardial ischemia/reperfusion injury. Cardiovasc Res. 2004;63(1):51–59. doi: 10.1016/j.cardiores.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 31.Zingarelli B, Hake PW, Denenberg A, Wong HR. Sesquiterpene Lactone Parthenolide, an Inhibitor of IkB Kinase Complex and Nuclear Factor-kB, Exerts Beneficial Effects in Myocardial Reperfusion Injury. SHOCK. 2002;17(2):127–134. [DOI] [PubMed] [Google Scholar]
- 32.Wanderer AA. Rationale and timeliness for IL-1β-targeted therapy to reduce allogeneic organ injury at procurement and to diminish risk of rejection after transplantation. Clin Transplant. 2010;24(3):307–311. doi: 10.1111/j.1399-0012.2010.01256.x [DOI] [PubMed] [Google Scholar]
- 33.Rusai K, Huang H, Sayed N, et al. Administration of interleukin-1 receptor antagonist ameliorates renal ischemia-reperfusion injury. Transpl Int. 2008;21(6):572–580. doi: 10.1111/j.1432-2277.2008.00651.x [DOI] [PubMed] [Google Scholar]
- 34.Vallejo S, Palacios E, Romacho T, Villalobos L, Peiró C, Sánchez-Ferrer CF. The interleukin-1 receptor antagonist anakinra improves endothelial dysfunction in streptozotocin-induced diabetic rats. Cardiovasc Diabetol. 2014;13(1):1–13. doi: 10.1186/s12933-014-0158-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbate A, Salloum FN, Vecile E, et al. Anakinra, a recombinant human interleukin-1 receptor antagonist, inhibits apoptosis in experimental acute myocardial infarction. Circulation. 2008;117(20):2670–2683. doi: 10.1161/CIRCULATIONAHA.107.740233 [DOI] [PubMed] [Google Scholar]
- 36.Dinarello CA, Simon A, Meer JWM van der. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11(8):633–652. doi: 10.1038/jnrd3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong L, Qiao H, Zhang X, et al. Parthenolide is neuroprotective in rat experimental stroke model: Downregulating NF- B, phospho-p38MAPK, and caspase-1 and ameliorating BBB permeability. Mediators Inflamm. 2013;2013. doi: 10.1155/2013/370804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Zhao J, Tan R, et al. Parthenolide inhibits pro-inflammatory cytokine production and exhibits protective effects on progression of collagen-induced arthritis in a rat model. Scand J Rheumatol. 2015;44(3):182–191. doi: 10.3109/03009742.2014.938113 [DOI] [PubMed] [Google Scholar]
- 39.Streblow DN, Kreklywich CN, Andoh T, et al. The Role of Angiogenic and Wound Repair Factors During CMV-Accelerated Transplant Vascular Sclerosis in Rat Cardiac Transplants. Am J Transplant. 2008;8(2):277–287. doi: 10.1111/j.1600-6143.2007.02062.x [DOI] [PubMed] [Google Scholar]
- 40.Valantine HA, Gao S-Z, Menon SG, et al. Impact of Prophylactic Immediate Posttransplant Ganciclovir on Development of Transplant Atherosclerosis: A Post Hoc Analysis of a Randomized, Placebo-Controlled Study. Circ Am Hear Assoc. 1999;100(1):61–66. doi: 10.1161/01.CIR.100.1.61 [DOI] [PubMed] [Google Scholar]
- 41.Merigan TC, Renlund DG, Keay S, et al. A Controlled Trial of Ganciclovir to Prevent Cytomegalovirus Disease After Heart Transplantation. N Engl J Med. 1992;326(18):1182–1186. doi: 10.1056/NEJM199204303261803 [DOI] [PubMed] [Google Scholar]
- 42.Abbate A, Kontos MC, Grizzard JD, et al. Interleukin-1 Blockade With Anakinra to Prevent Adverse Cardiac Remodeling After Acute Myocardial Infarction (Virginia Commonwealth University Anakinra Remodeling Trial [VCU-ART] Pilot Study). Am J Cardiol. 2010;105(10):1371–1377.e1. doi: 10.1016/j.amjcard.2009.12.059 [DOI] [PubMed] [Google Scholar]
- 43.Saxena A, Chen W, Su Y, et al. IL-1 Induces Proinflammatory Leukocyte Infiltration and Regulates Fibroblast Phenotype in the Infarcted Myocardium. J Immunol. 2013. doi: 10.4049/jimmunol.1300725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao DA, Eid RE, Qin L, et al. Interleukin (IL)-1 promotes allogeneic T cell intimal infiltration and IL-17 production in a model of human artery rejection. J Exp Med. 2008;205(13):3145–3158. doi: 10.1084/jem.20081661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westman PC, Lipinski MJ, Luger D, et al. Inflammation as a Driver of Adverse Left Ventricular Remodeling after Acute Myocardial Infarction. J Am Coll Cardiol. 2016;67(17):2050–2060. doi: 10.1016/j.jacc.2016.01.073 [DOI] [PubMed] [Google Scholar]
- 46.Mulders-manders CM, Baas MC, Molenaar FM, Simon A. Peri- and Postoperative Treatment with the Interleukin-1 Receptor Antagonist Anakinra Is Safe in Patients Undergoing Renal Transplantation : Case Series and Review of the Literature. Front Pharmacol. 2017;8(May):1–6. doi: 10.3389/fphar.2017.00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada J, Dana R, Zhu S-N. Interleukin 1 Receptor Antagonist Suppresses Allosensitization in Corneal Transplantation. Arch Ophthalmol. 1998;116(10):1351–1357. doi: 10.1001/archopht.116.10.1351 [DOI] [PubMed] [Google Scholar]
- 48.Dana R. Comparison of topical interleukin-1 vs tumor necrosis factor-alpha blockade with corticosteroid therapy on murine corneal inflammation, neovascularization, and transplant survival (an American Ophthalmological Society thesis). Trans Am Ophthalmol Soc. 2007;105:330–343. [PMC free article] [PubMed] [Google Scholar]
- 49.Dana MR, Yamada J, Streilein JW. Topical Interleukin 1 Receptor Antagonist Promotes Corneal Transplant Survival. Transplantation. 1997;63(10):1501–1507. doi: 10.1097/00007890-199705270-00022 [DOI] [PubMed] [Google Scholar]
- 50.Lemström K, Sihvola R, Bruggeman C, Häyry P, Koskinen P. Cytomegalovirus infection-enhanced cardiac allograft vasculopathy is abolished by DHPG Prophylaxis in the Rat. Circ Am Hear Assoc. 1997;95:2614–2616. doi: 10.1161/01.CIR.95.12.2614 [DOI] [PubMed] [Google Scholar]
- 51.Lemström K, Bruning JH, Bruggeman CA, et al. Cytomegalovirus Infection-Enhanced Allograft Arteriosclerosis Is Prevented by DHPG Prophylaxis in the Rat. Circulation. 1994;90(4):1969–1978. doi: 10.1161/01.CIR.90.4.1969 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.