Abstract
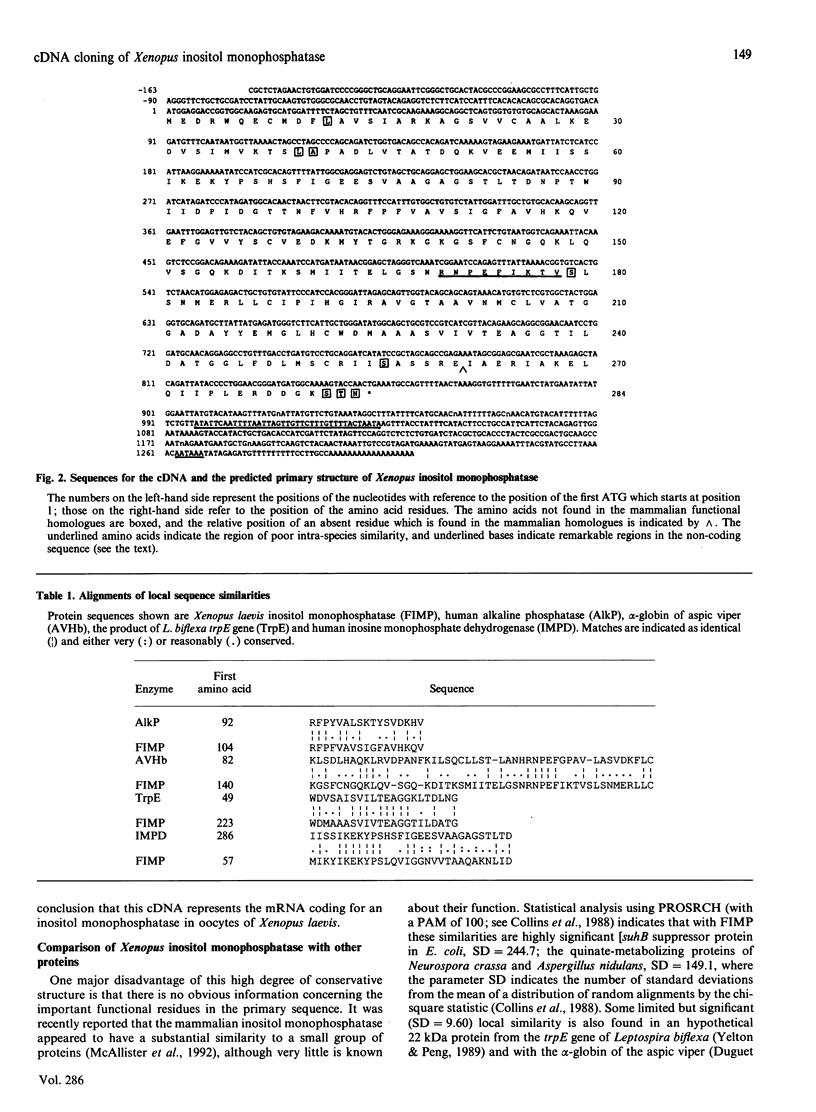
The cDNA coding for an inositol monophosphatase in the oocytes of the African clawed frog, Xenopus laevis, has been isolated and sequenced. The predicted primary structure of this enzyme is markedly conserved when it is compared with its mammalian functional homologues; up to 84% of the amino acid residues are identical, and conservative substitutions increase the similarity to 95%, suggesting that this sequence represents the most parsimonious primary structure for the protein to maintain not only catalytic activity but also perhaps the facility to interact with other macromolecules. Two regions of the protein, each of about 11 residues and separated by about 90 residues, have been identified as a consensus found also in glycerol 3-phosphate dehydrogenase (EC 1.1.1.8). One of these regions is also found to be particularly conserved in the alpha-globin of birds and reptiles; birds and some turtles are known to modulate the oxygen affinity of their haemoglobin with inositol polyphosphate in the same way as with 2,3-bisphosphoglycerate in other species. This region is also conserved in the beta-globin of most species, beginning with lysine-82, which is known to participate in the binding of organic phosphates. These regions of the inositol monophosphatase may represent motifs for the binding of its substrate.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnone A., Perutz M. F. Structure of inositol hexaphosphate--human deoxyhaemoglobin complex. Nature. 1974 May 3;249(452):34–36. doi: 10.1038/249034a0. [DOI] [PubMed] [Google Scholar]
- Arnone A. X-ray diffraction study of binding of 2,3-diphosphoglycerate to human deoxyhaemoglobin. Nature. 1972 May 19;237(5351):146–149. doi: 10.1038/237146a0. [DOI] [PubMed] [Google Scholar]
- Bartlett G. R. Isolation and assay of red-cell inositol polyphosphates. Anal Biochem. 1982 Aug;124(2):425–431. doi: 10.1016/0003-2697(82)90060-4. [DOI] [PubMed] [Google Scholar]
- Berridge M. J., Downes C. P., Hanley M. R. Lithium amplifies agonist-dependent phosphatidylinositol responses in brain and salivary glands. Biochem J. 1982 Sep 15;206(3):587–595. doi: 10.1042/bj2060587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J., Downes C. P., Hanley M. R. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989 Nov 3;59(3):411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- Busa W. B., Gimlich R. L. Lithium-induced teratogenesis in frog embryos prevented by a polyphosphoinositide cycle intermediate or a diacylglycerol analog. Dev Biol. 1989 Apr;132(2):315–324. doi: 10.1016/0012-1606(89)90228-5. [DOI] [PubMed] [Google Scholar]
- Busa W. B. Roles for the phosphatidylinositol cycle in early development. Philos Trans R Soc Lond B Biol Sci. 1988 Jul 26;320(1199):415–426. doi: 10.1098/rstb.1988.0085. [DOI] [PubMed] [Google Scholar]
- Collins J. F., Coulson A. F., Lyall A. The significance of protein sequence similarities. Comput Appl Biosci. 1988 Mar;4(1):67–71. doi: 10.1093/bioinformatics/4.1.67. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl R. E., Whiting P., Potter J., Gee N., Ragan C. I., Linemeyer D., Schoepfer R., Bennett C., Dixon R. A. Cloning and expression of bovine brain inositol monophosphatase. J Biol Chem. 1990 Apr 15;265(11):5946–5949. [PubMed] [Google Scholar]
- Duguet M., Chauvet J. P., Acher R. Phylogeny of hemoglobins: the complete amino acid sequence of an alpha-chain of viper (Vipera aspis) hemoglobin. FEBS Lett. 1974 Oct 15;47(2):333–337. doi: 10.1016/0014-5793(74)81042-2. [DOI] [PubMed] [Google Scholar]
- Fabiny J. M., Jayakumar A., Chinault A. C., Barnes E. M., Jr Ammonium transport in Escherichia coli: localization and nucleotide sequence of the amtA gene. J Gen Microbiol. 1991 Apr;137(4):983–989. doi: 10.1099/00221287-137-4-983. [DOI] [PubMed] [Google Scholar]
- Gee N. S., Ragan C. I., Watling K. J., Aspley S., Jackson R. G., Reid G. G., Gani D., Shute J. K. The purification and properties of myo-inositol monophosphatase from bovine brain. Biochem J. 1988 Feb 1;249(3):883–889. doi: 10.1042/bj2490883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallcher L. M., Sherman W. R. The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain. J Biol Chem. 1980 Nov 25;255(22):10896–10901. [PubMed] [Google Scholar]
- Isaacks R., Harkness D., Sampsell R., Adler J., Roth S., Kim C., Goldman P. Studies on avian erythrocyte metabolism. Inositol tetrakisphosphate: the major phosphate compound in the erythrocytes of the ostrich (Struthio camelus camelus). Eur J Biochem. 1977 Aug 1;77(3):567–574. doi: 10.1111/j.1432-1033.1977.tb11700.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister G., Whiting P., Hammond E. A., Knowles M. R., Atack J. R., Bailey F. J., Maigetter R., Ragan C. I. cDNA cloning of human and rat brain myo-inositol monophosphatase. Expression and characterization of the human recombinant enzyme. Biochem J. 1992 Jun 15;284(Pt 3):749–754. doi: 10.1042/bj2840749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsumeda Y., Ohno S., Kawasaki H., Konno Y., Weber G., Suzuki K. Two distinct cDNAs for human IMP dehydrogenase. J Biol Chem. 1990 Mar 25;265(9):5292–5295. [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders S. E., Burke J. F. Rapid isolation of miniprep DNA for double strand sequencing. Nucleic Acids Res. 1990 Aug 25;18(16):4948–4948. doi: 10.1093/nar/18.16.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Watanabe T., Li W. B., Soong B. W., Chou J. Y. Expression of the germ cell alkaline phosphatase gene in human choriocarcinoma cells. J Biol Chem. 1989 Jul 25;264(21):12611–12619. [PubMed] [Google Scholar]
- Yelton D. B., Peng S. L. Identification and nucleotide sequence of the Leptospira biflexa serovar patoc trpE and trpG genes. J Bacteriol. 1989 Apr;171(4):2083–2089. doi: 10.1128/jb.171.4.2083-2089.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]



