Graphical abstract
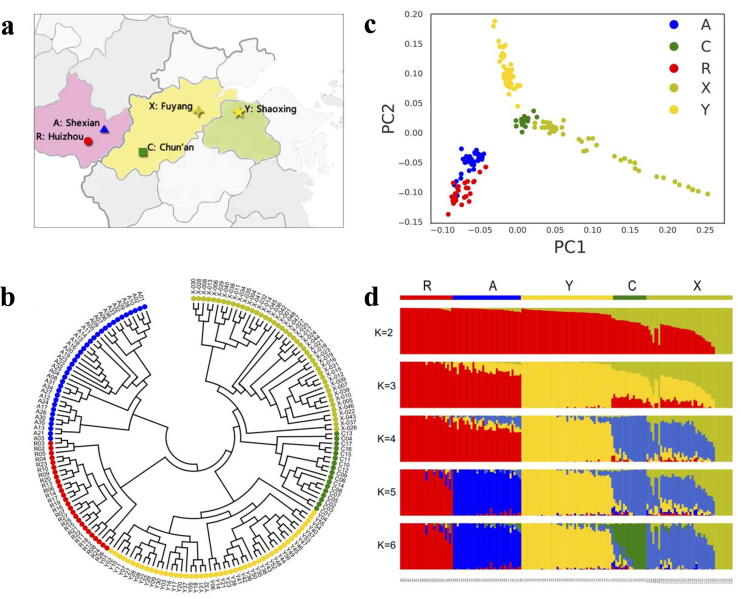
Population genetic analyses of T. grandis. (A) Geographical origin of T. grandis landraces. (B) Neighbor-joining phylogenetic tree constructed using 275,924 SNPs. Colors indicate the following groups: blue, Shexian; green, Chun’an; red, Huizhou; yellow, Shaoxing; yellow-green, Fuyang. (C) PCA plots of the first two components of 170 T. grandis landraces. Each point represents an independent landrace of T. grandis. (D) Population structure analysis with different numbers of clusters.

Keywords: Oil biosynthesis, Torreya grandis, Transcriptome-referenced association study, Single nucleotide polymorphism, RNA sequencing
Highlights
-
•
The genetic architecture of fatty acid and oil content in Torreya grandis was investigated.
-
•
LOB domain-containing protein 40 and surfeit locus protein 1 may involve in the regulation of oil and sciadonic acid biosynthesis.
-
•
Overexpression of TgLBD40 significantly increased seed oil content.
Abstract
Introduction
Torreya grandis is a gymnosperm belonging to Taxodiaceae. As an economically important tree, its kernels are edible and rich in oil with high unsaturated fatty acids, such as sciadonic acid. However, the kernels from different T. grandis landraces exhibit fatty acid and oil content variations.
Objectives
As a gymnosperm, does T. grandis have special regulation mechanisms for oil biosynthesis? The aim of this study was to dissect the genetic architecture of fatty acid and oil content and the underlying mechanism in T. grandis.
Methods
We constructed a high integrity reference sequence of expressed regions of the genome in T. grandis and performed transcriptome-referenced association study (TRAS) for 10 fatty acid and oil traits of kernels in the 170 diverse T. grandis landraces. To confirm the TRAS result, we performed functional validation and molecular biology experiments for oil significantly associated genes.
Results
We identified 41 SNPs from 34 transcripts significantly associated with 7 traits by TRAS (−log10 (P) greater than 6.0). Results showed that LOB domain-containing protein 40 (LBD40) and surfeit locus protein 1 (SURF1) may be indirectly involved in the regulation of oil and sciadonic acid biosynthesis, respectively. Moreover, overexpression of TgLBD40 significantly increased seed oil content. The nonsynonymous variant in the TgLBD40 coding region discovered by TRAS could alter the oil content in plants. Pearson’s correlation analysis and dual-luciferase assay indicated that TgLBD40 positively enhanced oil accumulation by affecting oil biosynthesis pathway genes, such as TgDGAT1.
Conclusion
Our study provides new insights into the genetic basis of oil biosynthesis in T. grandis and demonstrates that integrating RNA sequencing and TRAS is a powerful strategy to perform association study independent of a reference genome for dissecting important traits in T. grandis.
Introduction
The seeds of perennial economic trees have the characteristics of sustained high and stable yield and can synthesize both medicinal and edible nutrients. However, their genetic improvement is seriously hindered because of their complex genetic basis and quantitative traits. As an economically important tree, Torreya grandis (T. grandis) has a long economic life; some trees have lived up to about 1,600 years and still produced a lot of drupe-like fruits with nutty seeds. The seeds have multiple medicinal properties, including antioxidative [1], anti-inflammatory [2], and antiatherosclerotic [3], as a result of their rich nutritive content and bioactive components, especially the polyunsaturated fatty acids. The oil content of T. grandis kernels is more than 50 % of dry mass, of which the contents of total unsaturated fatty acids and sciadonic acid (a newly identified type of non-methylene-interrupted polyunsaturated fatty acid) are more than 80 % and 10 % respectively [4], [5]. Because of its high nutritive content and bioactive components, the kernels have been used as food and traditional medicine for thousands of years in China. However, due to their wide distribution, wind pollination and the breeding of cultivars, T. grandis is morphologically variable in China and exhibits considerable kernel quality variations including fatty acids and oil content [4]. The manipulation of oil content and fatty acid composition has therefore become a key consideration in breeding and biotechnology-assisted improvement of T. grandis.
Oil in plant seeds is generally stored as triacylglycerols (TAGs) and synthesized from fatty acids (FAs). In plastids, the C16:0, C18:0 and C18:1 acyl chain FAs are synthesized by a set of enzymes, including acetyl-CoA carboxylase (ACCase), 3-ketoacyl-ACP synthase (KAS), 3-ketoacyl-ACP reductase (KAR), 3-hydroxyacyl-ACP dehydratase (HAD) and enoyl-ACP reductase (ENR), etc. Then, the nascent FAs are transferred to the endoplasmic reticulum (ER) to form TAGs by a series of enzymes, such as glycerol-3-phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAT), diacylglycerol acyltransferase (DGAT), and the phospholipid: diacylglycerol acyltransferase (PDAT) [6]. Finally, some oil body-associated proteins, such as oleosins (OLE), caleosin (CLO) and steroleosin (SLO), are bond to the resulting TAGs to form oil bodies. The pathway of lipid synthesis is also well understood in a few valuable trees [5]. However, none of the genes involved in natural variation in tree oil have been cloned due to research lags and limitations in research methods.
Association studies between single nucleotide polymorphisms and important economic traits provide a powerful approach for the identification of genes underlying complex traits. Genome-wide association study (GWAS) as an effective association study approach, it has been successfully applied for the study of many plants, such as Arabidopsis [7], rice [8], [9], maize [10], [11], jujube [12], soybean [13] and lettuce [14]. However, to identify candidate genes controlling important economic traits using GWAS, a reference genome or linkage map of the species under study is essential. Moreover, most gymnosperm species have a giant genome, high proportions of repetitive elements and numerous pseudogenes. Therefore, genome sequencing of gymnosperm species is time-consuming and costly. As a result, other approaches are needed to conduct association studies between single nucleotide polymorphisms and economically important traits.
Next-generation sequencing (NGS) technology has revolutionized life science research and been widely applied in various plants, especially plant without a reference genome because it is unconstrained by genomic complexity [15]. Transcriptome analysis by NGS has been used to study the linkage or association between genes and traits for a broad range of species, leading to the identification of many candidate genes responsible for important traits in plants, such as anion homeostasis [16], grain size [17], drought tolerance [18], kernel oil [11], flavonoid biosynthesis [14], erucic acid and tocopherol isoform [19], and clove shape [20]. The ‘transcriptome-referenced association study’ (‘TRAS’) using expressed regions of the genome as a reference sequence that could score population variation at both transcript sequence and expression levels, and identify trait associated transcripts in species for which a reference genome sequence is lacking [20].
To reveal the genetic basis of oil concentration and composition in T. grandis kernels and clarify how oil biosynthesis is regulated, we investigated the oil content and fatty acid variation in 170 diverse T. grandis landraces across five different locations and conducted a TRAS analysis using 275,924 transcriptome-wide single nucleotide polymorphisms (SNP). The reference sequence of expressed regions of the genome was generated from the combination of single-molecule real-time (SMRT) sequencing and Next Generation Sequencing (NGS) technologies. In total, 41 loci from 34 transcripts were identified to be significantly associated with oil and fatty acid content of T. grandis kernels. We also found that an LOB domain-containing protein 40 encoding gene, TgLBD40, from the above 34 transcripts, was highly expressed in developing kernels and that its expression level was significantly correlated with oil content. We subsequently verified that TgLBD40 contributed to oil accumulation in T. grandis kernels by conducting transgenic validation and a series of molecular assays. Our results shed light on the genetic basis of oil biosynthesis in T. grandis and provide useful information for T. grandis breeding programs.
Materials and methods
Plant materials
In mid-August 2017, the seeds with similar maturity were collected from natural populations of T. grandis landraces in five locations which covered the main T. grandis distribution areas in China (Table S1), including Shexian (A), Huizhou (R), Chun’an (C), Shaoxing (Y), and Fuyang (X). The distance between the sampled plants was at least 50 m. In total, 170 diverse landraces were chosen for association analysis. After collection, the arils and seed coats were removed, and the remaining kernels were quickly frozen in liquid nitrogen and then stored at − 80 °C until use. For each landrace, 30 kernels were randomly chosen from a single tree and each of their endosperm was ground into powder in liquid nitrogen for phenotypic measurement. Then, 20 mg powder from each of the 30 endosperms was mixed together for RNA isolation and transcriptome sequencing.
Phenotypic measurement and correlation among 10 traits
For each landrace, their mean value of 30 randomly chosen seeds was calculated to evaluate each landrace. Fatty acid content was determined as described by Wu et al. [4]. Oil content was determined according to Ding et al. [5]. Phenotypic correlation among different traits was calculated as the Pearson’s correlation coefficient of their trait values using SPSS software (version 16.0).
Illumina RNA sequencing
To characterize the allelic variation for this population, such as SNPs, all 170 T. grandis landraces were subjected to Illumina RNA sequencing individually. Total RNA from each sample was isolated by a Plant RNA isolation Kit (DP441, Tiangen Biotech Co. ltd, Beijing, China) and purified with a Dynabeads® Oligo (dT)25 kit (Life Technologies, CA, USA). Then the purified RNA was used to construct cDNA libraries with a NEBNext® UltraTM RNA Library Prep Kit (New England BioLabs, Ipswich, MA, USA) following the manufacturer’s instructions. Library quality was assessed with the Agilent Bioanalyzer 2100 system. Paired-end sequencing was performed for each library using a HiSeq PE Cluster Kit v4 cBot (Illumina, San Diego, CA, USA) in conjunction with the HiSeqTM 4000 Illumina sequencing system. Raw reads in FASTQ format were processed using inhouse Perl scripts to yield clean reads. In this step, reads with adaptor contamination and poly-Ns, as well as low-quality reads with more than 5 % of ambiguous bases (N) were removed.
PacBio SMRTbell library construction and SMRT sequencing
Total RNA was isolated from different organs and tissues of T. grandis landrace “X08” (root, stem, leaf, and flower) using a Plant RNA isolation Kit (DP441, Tiangen Biotech Co. ltd, Beijing, China), following which the RNA samples were combined for PacBio sequencing. Reverse transcription (RT) was conducted using the Clontech SMARTer PCR cDNA Synthesis Kit. The cDNA products of the combined RNA sample were used to construct one SMRTbell library following the manual of the DNA Template Prep Kit 3.0 (Pacific Biosciences, USA). The fragmented cDNA was concentrated by AMPure PB beads and the ends were repaired. Then, blunt hairpin adapters were ligated to the cDNA and exonucleases were added to remove failed ligation products. SMRTbell templates containing cDNA inserts were purified by AMPure PB beads. The sequencing primers and the polymerase were then sequentially annealed to the SMRTbell templates using the DNA/Polymerase Binding Kit P6 v2 (Pacific Biosciences). The MagBead loading Kit (Pacific Biosciences) was used to load the annealed templates onto a Pacific Biosciences RS II sequencer. The sequencing was performed using seven SMRT Cells with the DNA Sequencing Reagent Kit 4.0 v2 (Pacific Biosciences).
Sequence data assembly and annotation
Sequence data were processed using the SMRT analysis software (http://www.pacificbiosciences.com/devnet/). Circular consensus sequence (CCS) reads were generated from the sub-read files using the following parameters: mini Length = 50, read Score = 0.75, artifact = -1000, Min Complete Passes = 2 and Min Predicted Accuracy = 0. After examining for poly(A) signal and 5′ and 3′ adaptors, only the CCS reads with all three signals were considered as an FLNC read [21]. Unmerged subreads were also examined for the three signals, and those with three signals were incorporated into the final FLNC read set. Because PacBio reads have a higher frequency of nucleotide errors than the shorter reads generated by the second generation sequencing technologies, the software proovread [22] was used to correct those errors based on Illumina RNA sequencing data of the T. grandis cultivar X08. Redundant sequences were removed with CD-HIT [23]. The full-length transcripts were subjected to functional annotation by searching against public databases with an E-value ≤ 10−5, including the National Center for Biotechnology Information (NCBI) Non-redundant (Nr) and Nucleotide (Nt) databases, SwissProt protein database, Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/kegg2.html), Cluster of Orthologous Groups of proteins (COG), and TrEMBL. Gene ontologies (GO) were assigned to each full-length transcript using Blast2GO [24].
Reference sequence integration and SNP identification
To obtain a high-quality reference transcriptome, we combined Illumina RNA-sequencing data with single molecule long-read sequencing data together and compared each sequence with Blat. The shorter sequences with over 80 % of matched length were removed. After removing most similar sequences, 140,296 unigenes were collected as reference sequence. The read sequences of Illumina RNA-sequencing were mapped onto the reference transcriptome sequence obtained above and high-quality SNPs were detected using the STAR tool. SNP loci might have missing data for some accessions due to low expression of some genes. For further studies, the missing data were imputed. A total of 275,924 SNPs with missing rates of ≤ 0.5 were filled using fillGenotype.
Population genetics analysis
A neighbor-joining (NJ) phylogenetic tree was constructed by the PHYLIP software using SNPs [25]. A nonparametric bootstrap analysis was performed, with 100 bootstrap replicates. EIGENSOFT software package was used to perform principal component analysis (PCA) of the population [26]. The first two components were plotted for the T. grandis landraces. Population structure was analyzed using the STRUCTURE program [27]. For each K value that ranged from 1 to 20, STRUCTURE was run 20 times with an admixture model and 10,000 burn-in and MCMC replicates. Then the obtained results were imported into CLUMPAK [28]. The final result was presented as a graph which shows the most likely number of populations and the population membership of each landrace. The population-differentiation statistics (FST) were computed as described by Nordborg et al. [29], using a 100-kb window, among the five groups of T. grandis.
TRAS for 10 oil-related traits
To excavate the candidate transcripts for the 10 oil-related traits measured in this study, TRAS was carried out to detect the suggestive loci associated with the 10 traits based on the mapped high-quality 275,924 SNPs for the T. grandis population. The association analysis was conducted using a mixed linear model program and TASSEL tool [30]. The transcriptome-wide significance threshold of the TRAS for all the investigated traits was − log10 (P) greater than 6.0 which was calculated by Bonferroni correction based on the effective number of independent markers [31], [32].
Correlation analysis of expression level and traits
To quantify the expression of each transcript in all 170 diverse T. grandis landraces, the expression level of each transcript in each T. grandis landrace was analyzed by estimating the expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) using RSEM [33]. Pearson’s correlation analysis was used to associate the expression level of transcripts and the trait values.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Expression of TgLBD40 was determined in different T. grandis tissues, including roots, stems, leaves, arils, and developing kernels at different developmental stages (in the middle of May, June, July, August and September of 2018, respectively). The expression level of AtLBD41 was also identified in all used Arabidopsis lines. Total RNA was isolated from the samples by Total RNA Kit (TIANGEN, DP441) with an additional DNase I (TIANGEN), and 1 μg of RNA was used to synthesize the first strand of cDNA using the PrimeScript™RT Master Mix (Takara). Gene expression was determined using the ChamQ SYBR qPCR Master Mix (Vazyme) on a C1000 Touch™ Thermal Cycler (Bio-Rad). The primer pairs are listed in Table S2. The PCR conditions were as follows: 45 cycles of 95 °C for 10 s, 56 °C for 10 s and 72 °C for 20 s. The T. grandis Actin gene was amplified as an internal reference, and the formula 2−△△Cp was uesd to calculate the results. Three biological replicates per sample were used.
Generation of TgLBD40 overexpressing and complemented plants
The full-length coding sequence (CDS) of TgLBD40 was amplified from the cDNA of T. grandis by PCR using gene-specific primers (Table S2) and inserted into the downstream of the 35S promoter of a modified pCAMBIA1300 vector. The resulting recombinant pCAMBIA1300-TgLBD40 construct was transformed into wild type Arabidopsis (Col-0) and atlbd41 mutant (Salk_078678C) via the Agrobacterium tumefaciens-mediated floral dip method. Hygromycin-resistant T1 plants were planted for seed harvesting, and T2 seeds with a hygromycin resistance ratio of 3:1 were selected to collect T3 seeds. T3 seeds with 100 % resistance to hygromycin were used for the following experiments. The expression of TgLBD40 in independent positive transgenic lines was detected by semi-quantitative RT-PCR using the primers listed in Table S2. Three independent lines from each of TgLBD40 overexpressing and complemented plants were selected for further experiment.
Transient expression in tobacco leaves and subcellular localization
The full-length cDNAs of genes were amplified by PCR using gene-specific primers (Table S2) and cloned into the 35S::GFP vector (modified from pCAMBIA1300) to obtain the recombinant constructs under control of the cauliflower mosaic virus (CaMV) 35S promoter. The recombinant vectors were introduced into Agrobacterium tumefaciens strain GV3101 and cultured until the OD600 reached to 0.6. After centrifuging, the agrobacteria were resuspended in the buffer solution containing 10 mM MgCl2, 0.2 mM acetosyringone, and 10 mM MES (pH 5.6) with OD600 at 0.6. For transient expression of the fluorescent proteins, infiltration buffer was injected into leaves of 4-week-old Nicotiana benthamiana. After 3 days of incubation, leaves were harvest for oil determination and subcellular localization analysis. GFP signals were detected using a confocal laser scanning microscope (LSM510, Karl Zeiss).
Total oil content analysis
Following the procedures described by Yeap et al. [34], the total oil content of dried mature seeds and leaves was analyzed by gravimetric lipid assay. Dried samples were weighed carefully on a Semi-Micro analytical balance to ± 0.01 mg. The samples were ground with 2 ml of 6/4 hexane/isopropanol (v/v). The mixture was vortexed for 2 min and sonicated for 15 min at room temperature. Then, 1 ml of aqueous sodium sulphate (2.5 ml of 15 % wt/vol) was added to the mixture, which was vortexed again and centrifuged at 4000 rpm for 5 min, to achieve phase separation. The lower phase was re-extracted following the above method. The upper phases of all extracts were combined and transferred to a clean new glass tube. The lipid extracts were evaporated under oxygen-free nitrogen until a constant weight was obtained. The total oil content was calculated by dividing the weight of the extracted lipid by the initial sample weight.
Dual luciferase assay
To determine the transactivation activity of transcription factors to the promoter of oil biosynthesis genes, a transient dual luciferase assay was performed. The coding regions of transcription factors were cloned into the pGreen II 0029 62-SK vector as an effector. The promoter of oil biosynthesis genes was introduced into the pGreenII 0800-LUC vector, allowing the promoter to be cloned as a transcriptional fusion with the firefly luciferase gene (LUC). All primers are listed in Supplementary Table S2. The constructed effector and reporter plasmids were introduced into Agrobacterium tumefaciens (GV3101) and then cotransformed into N. benthamiana leaves. LUC and REN luciferase activities were measured using a dual luciferase assay kit (Promega). The results were calculated by the ratio of LUC to REN.
Protein extraction and immunoblot analysis
Total proteins were extracted from N. benthamiana leaves using extraction buffer (50 mM Tris-HCl, pH 7.5; 20 mM NaCl; 2 mM PMSF; 20 mM MG132; and protease inhibitor cocktail). Proteins were quantified with the Pierce 660 nm Protein Assay using a BSA standard curve and 20 µg proteins of each sample were loaded for Immunoblot analysis. The blotted gels were incubated with anti-GFP-HRP antibody (Miltenyi Biotec, 130–091-833). Coomassie Brilliant Blue staining of blots was used to control the protein levels after electrotransfer.
Expression and purification of recombinant MBP-TgLBD40 protein.
TgLBD40 coding sequence was cloned in pMAL-c2X vector, using the primers listed in Table S2. The MBP and MBP-TgLBD40 proteins were expressed in Escherichia coli strain BL21 (DE3) and then purified using a MBPtrap HP column (Cytiva) attached to ÄKTA FPLC system (Cytiva) according to the instruction manual of pMAL™ Protein Fusion and Purification System (#E8000S; New England Biolabs, Inc.).
Electrophoretic mobility shift assay (EMSA)
The 5′-Cy5-labeled and unlabeled primers were first synthesized (Table S2). Then the 5′-Cy5-labeled probes and unlabeled competitors were generated by annealing the labeled and unlabeled primers, respectively. The Cy5-labeled probes (100 nM) were incubated with 5 μM of purified MBP-TgLBD40 protein in binding buffer (50 µL) comprising 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 5 % glycerol and 5 mM DTT for 30 min at room temperature. For competition assay, 10-fold, 30-fold and 100-fold molar excess of each competitor was added to the reaction mixture before incubation. Protein-DNA complexes were separated by electrophoresis through 8 % polyacrylamide gel in 0.5X Tris–borate EDTA buffer at 120 V. The Cy5 signals were detected using an Odyssey CLX imaging system (LI-COR).
Results
Phenotypic variation and correlation analyses of oil-related traits
The phenotypic traits we examined included oil content and fatty acid levels (sciadonic acid, eicosadienoic acid, eicosenoic acid, arachidic acid, linolenic acid, linoleic acid, oleic acid, stearic acid and palmitic acid) (Fig. 1a). For all the phenotypes evaluated in this study, we observed almost no global similarities among subpopulations except that the sciadonic acid and oil content were slightly higher in X and Y subpopulations than other subpopulations (Fig. 1a, Fig. 3a and Fig. S1-6a). The populations showed considerable phenotypic variation for most of the traits both in each group and in all groups (Table S3, Fig. 3b and Fig. S1-6b). Pearson correlation analysis demonstrated that eicosadienoic acid and sciadonic acid were strongly and positively correlated with linolenic acid and linoleic acid, and strongly and negatively correlated with palmitic acid and oleic acid (Fig. 1b). In addition, the palmitic acid and oleic acid were also strongly and positively correlated with arachidic acid and eicosenoic acid, and strongly and negatively correlated with linolenic acid and linoleic acid.
Fig. 1.
Phenotypic variation among populations and correlation analysis among traits. (a) Summary of phenotypic distributions among all individuals, with phenotypes grouped by trait category and individuals grouped by population as in Fig. 2. (b) Pairwise correlations of phenotypes across T. grandis landraces, measured as the Pearson correlation. Numbers represent correlation coefficients.
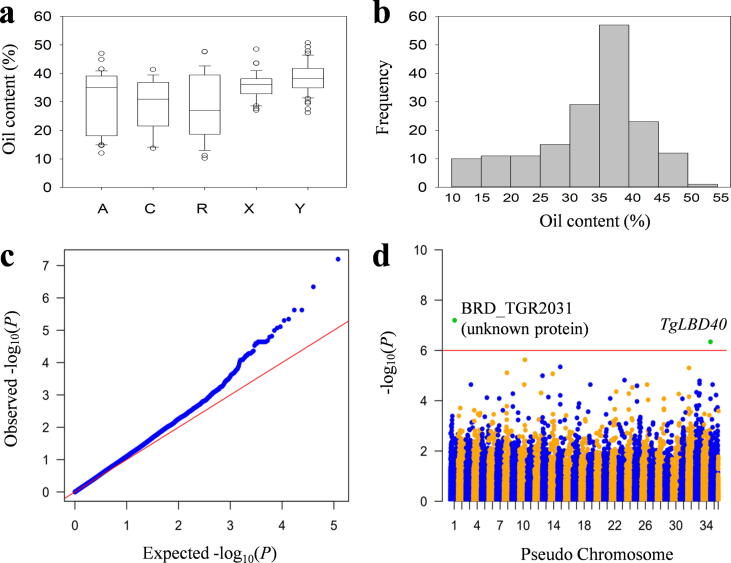
Fig. 3.
Phenotypic distribution and transcriptome-wide association scan for oil content. (a) Boxplots showing the differences in oil content among populations. Box edges represent the upper and lower quantile with the median value shown as a line in the middle of the box. Individuals falling outside the range of the whiskers shown as open dots. (b) Histograms of oil content in all samples. (c) Quantile-Quantile plot for a mixed linear model for oil in all samples. (d) Manhattan plots for oil. The reference transcriptome was generated by integrating Illumina RNA-sequencing data with single molecule long-read sequencing data. The reference sequence was artificially divided into 36 pseudo chromosomes. Candidate genes are shown along the top of the significantly associated signals. The red horizontal line indicates the significance threshold (−log10 (P) = 6.0).
Transcriptome analysis
We sequenced the transcriptome of different organs of T. grandis landrace “X08” using the SMRT sequencing platforms in our previous study [35]. Full-length cDNAs from RNA samples were normalized and subjected to an SMRT sequencing using the PACBIO RS II sequencing platform. A total of 97,211 transcripts with a total length of 235,890,123 bases were obtained as indicated by detection of the poly(A), 5’ and 3’ primers, and sequences.
To characterize the genotypes of the 170 T. grandis landraces in sequence and to perform TRAS analysis, we sequenced their transcriptomes using Illumina technology and obtained approximately 10.64 billion clean reads, with an average number of 62.59 million reads for each landrace (Table S4).
Population structure and differentiation
Neighbor-joining (NJ) phylogenetic tree was constructed based on the SNP genotypes to show the phylogenetic relationships among T. grandis landraces (Fig. 2a). Phylogenetic analysis showed that the 170 T. grandis landraces were resolved into five distinct groups (Fig. 2b), which were consistent with their geographical distribution. This result indicates that T. grandis landraces from different locations have unique genotype variation. Landraces from X are closer to landraces from C than other landraces. The phylogenetic relationships of the different T. grandis groups were also supported by principal component analysis (PCA). We observed clear, deep subpopulation structure in this collection of T. grandis from PCA analysis, which is similar to the NJ phylogenetic tree result (Fig. 2c). The five subpopulations, A, C, R, X and Y, formed clear clusters based on the top two principal components.
Fig. 2.
Population genetic analyses of T. grandis. (a) Geographical origin of T. grandis landraces. (b) Neighbor-joining phylogenetic tree constructed using 275,924 SNPs. Colors indicate the following groups: blue, Shexian; green, Chun’an; red, Huizhou; yellow, Shaoxing; yellow-green, Fuyang. (c) PCA plots of the first two components of 170 T. grandis landraces. Each point represents an independent landrace of T. grandis. (d) Population structure analysis with different numbers of clusters (K = 2–6).
To further investigate the population structure of T. grandis, the Bayesian clustering program STRUCTURE was used through gradually increasing the number of clusters (K). Different numbers of clusters were identified as K was increased, and the ΔK analysis showed that when K < 6, the T. grandis from the five locations could not be separated from the structure completely. At K = 6 and K = 7, T. grandis from C and X were assigned to independent clusters (Fig. 2d). However, R and A, as well as Y, C and X exhibited small admixture proportions, X could be divided into two parts, consistent with the results from our NJ phylogenetic and PCA analyses. Based on transcriptome-wide SNP analysis and phenotypic variation, we speculate that X accessions may have experienced low-frequency gene flow to T. grandis from another place and that the X landraces includes two types of T. grandis.
In order to investigate the population differentiation among A, C, R, X and Y, the population-differentiation statistic (FST) was performed. The transcriptome-wide genetic differentiation between any two groups was weak, with FST index of 0.027 to 0.046 (Fig. S7). The pairwise population differentiation FST between the C and R groups was the highest (FST = 0.046), indicating a relatively higher population differentiation between the C and R groups than between other groups. The lowest FST (0.027) was estimated between the A and R groups. The low population differentiation is advantageous for TRAS in T. grandis.
Transcriptome-referenced association studies for 10 oil-related traits
TRAS analyses on oil-related traits were performed for all the landraces using a mixed linear model (MLM). The method took transcriptome-wide patterns of genetic relatedness into account, greatly reducing false positives, as shown in quantile–quantile plots (Fig. 3c; Fig. S1-6c). In total, we identified 41 SNPs from 34 transcripts that were significantly associated with 10 oil-related traits, with − log10 (P) greater than 6.0 from the compressed MLM (Table S5), including 1 SNP for arachidic acid, 1 SNP for eicosadienoic acid, 23 SNPs from 19 transcripts for eicosenoic acid, 1 SNP for linoleic acid, 2 SNPs from 2 transcripts for oil content, 12 SNPs from 10 transcripts for palmitic acid, and 1 SNP for sciadonic acid. The Manhattan plots for MLM of all the traits are shown in Fig. 3d and Fig. S1-6d.
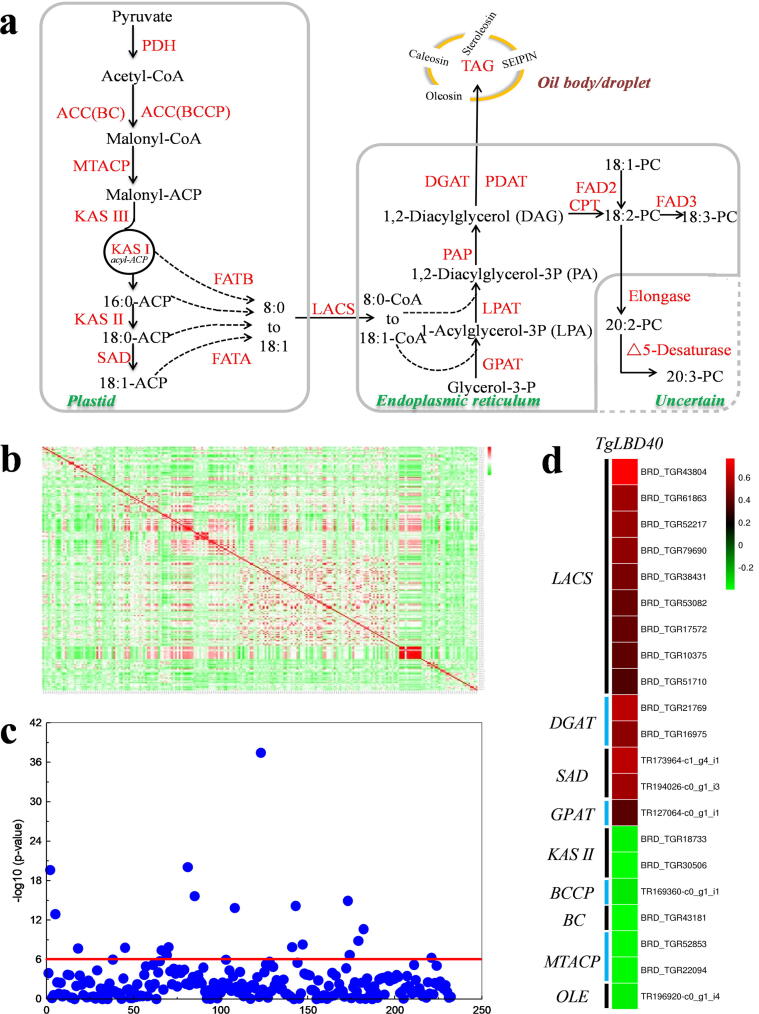
Fig. 6.
Pearson’s correlation analysis between the expression level of TgLBD40 and the expression level of the unigenes for oil biosynthesis pathway enzymes. (a) Schematic diagram of oil biosynthesis pathway. Enzymes are marked in red words. Abbreviations: PDH, pyruvate dehydrogenase; ACC (BC), Biotin carboxylase subunit of heteromeric acetyl-CoA carboxylase (ACCase); ACC(BCCP), biotin carboxyl carrier protein of heteromeric ACCase; KAS, ketoacyl-ACP synthase; SAD, stearoyl-ACP desaturase; FATA, acyl-ACP thioesterase A; FATB, acyl-ACP thioesterase B; LACS, long-chain acyl-CoA sythetase; GPAT, glycerol-3-phosphate acyltransferase; LPAT, 1ysophosphatidic acid acyltransferase; PAP, phosphatidic acid phosphatase; DGAT, diacylglycerol acyhransferase; PDAT, phospholipid: diacylglycerol acyltransferase; CPT, diacylglycerol cholinephosphotransferase; FAD2, v-6 desaturase; FAD3, v-3 desaturase; TAG, triacylglycerol. (b) Heat map of Pearson correlation coefficient at the expression level of TgLBD40 and all unigenes for enzymes in oil biosynthesis pathway. (c) The corresponding P value in (b). The red horizontal line represents P = 1 × 10−6. (d) Unigenes correlated with TgLBD40 at the level of P < 1 × 10−6. The heat map shows the correlation coefficient between unigenes and TgLBD40.
Pearson’s correlation analysis was conducted in order to investigate whether these traits were associated with the expression level of their traits significantly associated transcripts. The results showed that the expression levels of the BRD_TGR26378, BRD_TGR90326 and BRD_TGR17081, BRD_TGR74645, and BRD_TGR57892 and BRD_TGR20025 were positively and significantly associated with contents of sciadonic acid, palmitic acid, arachidic acid, and eicosenoic acid, respectively (Fig. S8). In contrast, the expression levels of BRD_TGR44768 and BRD_TGR88253 were negatively and significantly associated with palmitic acid.
Moreover, we selected the top one to five most significantly associated SNPs for each trait for further analysis. Among them, palmitic acid and linoleic acid shared one SNP (5,167,765 position) which from the transcript was annotated as “unknown protein” (Table 1). Some SNPs are located on the same transcript, such as both of the SNPs in the 5,999,610 bp position and the 5,999,612 bp position were significantly associated with eicosenoic acid and located on the same transcript, BRD_TGR9628, which was annotated as “Shikimate kinase 3”. Likewise, both of the SNPs in the 1,889,693 bp position and the 1,890,393 bp position were significantly associated with palmitic acid and located on the same transcript, BRD_TGR88253, which was annotated as “G patch domain-containing protein TGH homolog” (Table 1). Among the candidate transcripts presented in Table 1, they are all annotated as protein-encoding RNAs that are involved in fatty acid catabolic process (BRD_TGR74645 for arachidic acid), nucleoside metabolic process (BRD_TGR39525 for eicosadienoic acid), shikimate metabolic process (BRD_TGR76587 and BRD_TGR9628 for eicosenoic acid), transcriptional regulation (TR182770-c1_g1_i1 for oil content, and BRD_TGR14127 and BRD_TGR88253 for palmitic acid), cell wall organization (BRD_TGR17081 for palmitic acid), cytochrome c oxidase assembly (BRD_TGR26378 for sciadonic acid), and four encoded uncharacterized proteins.
Table 1.
Transcriptome-wide significant association signals of oil-related traits in T. grandis.
| Traits | SNP ID | Position (bp) | REF/ALT | p value | Sequence ID | Annotation |
|---|---|---|---|---|---|---|
| Arachidic acid | SNP1 | 2,997,193 | C/T | 1.42 × 10−9 | BRD_TGR74645 | Acyl-CoA thioesterase |
| Eicosadienoic acid | SNP2 | 8,620,054 | C/G | 5.92 × 10−8 | BRD_TGR39525 | UKL1: Uridine kinase-like protein 1 |
| Eicosenoic acid | SNP3 | 3,813,471 | C/T | 9.23 × 10−10 | BRD_TGR20025 | unknown |
| SNP4 | 7,129,377 | C/T | 9.23 × 10−10 | BRD_TGR57892 | unknown | |
| SNP5 | 15,989 | A/G | 1.46 × 10−9 | BRD_TGR76587 | SK3: Shikimate kinase 3 | |
| SNP6 | 5,999,610 | C/T | 9.82 × 10−10 | BRD_TGR9628 | SK3: Shikimate kinase 3 | |
| SNP7 | 5,999,612 | T/C | 1.08 × 10−9 | BRD_TGR9628 | SK3: Shikimate kinase 3 | |
| Linoleic acid | SNP8 | 5,167,765 | T/A | 1.21 × 10-7 | BRD_TGR94687 | unknown |
| Oil content | SNP9 | 220,350 | T/G | 4.54 × 10-7 | TR182770-c1_g1_i1 | LBD40: LOB domain-containing protein 40 |
| SNP10 | 5,546,666 | G/C | 6.33 × 10-8− | BRD_TGR2031 | unknown | |
| Palmitic acid | SNP11 | 8,112,400 | G/T | 1.05 × 10−8 | BRD_TGR14127 | UBA2C: UBP1-associated protein 2C |
| SNP12 | 6,023,941 | A/C | 1.42 × 10−7 | BRD_TGR17081 | ARAD1: Probable arabinosyltransferase | |
| SNP13 | 1,889,693 | T/C | 1.64 × 10−8 | BRD_TGR88253 | TGHH: G patch domain-containing protein TGH homolog | |
| SNP14 | 1,890,393 | G/A | 2.34 × 10−8 | BRD_TGR88253 | TGHH: G patch domain-containing protein TGH homolog | |
| SNP8 | 5,167,765 | T/A | 1.75 × 10−7 | BRD_TGR94687 | unknown | |
| Sciadonic acid | SNP15 | 853,862 | A/T | 5.62 × 10−7 | BRD_TGR26378 | SURF1: Surfeit locus protein 1 |
Potential regulatory networks for traits
The correlations between the expression levels of the transcripts and the traits were also used to excavate potential candidate regulators. Table S6 showed that there were 11 transcripts significantly correlated with three traits with correlation coefficient ≥ 0.7. Most of the traits were significantly correlated with the expression level of at least two transcripts, suggesting that the different transcripts correlated with the same trait interacted to control the trait.
TgLBD40 is localized to the nucleus and is highly expressed in oil accumulating tissues
Given that TgLBD40 was a strong candidate gene related to kernel oil content, we further characterized the expression pattern, amino acid sequence and phylogeny of TgLBD40. The expression levels of TgLBD40 were examined in different tissues and different developmental stages of kernels. The results of qRT-PCR and oil content determination showed that TgLBD40 was highly expressed in tissues with high oil content, including the leaves, arils and the kernels before reaching full maturity (Fig. 4a). Both the expression level of TgLBD40 and oil content increased with the extension of kernel development time, and reached the highest level in September, which was just before the full maturity of kernels (Fig. 4a). We also detected the expression levels of TgLBD40 in nine T. grandis plants with different oil content using qRT-PCR, and the results showed that there was a significant correlation between the expression of TgLBD40 and oil content (Fig. S9). These results indicated that the transcript abundance of TgLBD40 showed a similar trend with oil content, and TgLBD40 might be involved in oil body formation or oil accumulation in T. grandis. Phylogenetic analysis revealed that LBD40-like proteins from gymnosperms, mosses, Gramineae, Brassicaceae, Fabidae, and woody angiosperm clades clustered separately, suggesting functional conservation within the clade and possible functional diversity between clades (Fig. 4b). Furthermore, the alignment analysis of amino acid sequence was also conducted between TgLBD40 and its homologs in Arabidopsis and Glycine max. The result showed that TgLBD40 was highly homologous with the LBD40-like proteins from Arabidopsis and Glycine max, and possesses the conserved domain known as LOB domain (Fig. 4c). However, it has not been reported that LBD40-like proteins are involved in oil accumulation and oil body formation. In order to investigate the subcellular localization of TgLBD40, we expressed a 35S::TgLBD40-GFP construct in N. benthamiana leaf epidermal cells by agro-infiltration. The TgLBD40-GFP fusion protein was found only in the nucleus. When TgLBD40-GFP protein was co-expressed with nuclear-localized transcription factor, OsART1-RFP [36], they were colocalized (Fig. 4d). Taken together, these results indicate that TgLBD40 may be a transcriptional activator involved in the regulation of oil biosynthesis in T. grandis.
Fig. 4.
Expression and sequence analysis of TgLBD40. (a) The relative expression level of TgLBD40 and the oil content in different tissues and different developmental stages of kernels. (b) Phylogenetic relationship between TgLBD40 and its orthologs. All amino acid sequences were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/). (c) Domain structure and full-length amino acid alignment of representative LBD protein orthologs, including TgLBD40, XP_003541285.1 (Glycine max) and NP_566175.1 (Arabidopsis thaliana). The conserved LOB domain is highlighted by red line. (d) Subcellular localization analysis of TgLBD40 in N. benthamiana leaves. Both 35S::TgLBD40-GFP and 35S::OsART1-RFP (nuclear marker) constructs were transiently coexpressed in N. benthamiana leaves. GFP green fluorescence, bright field, RFP red fluorescence, and merged images are shown. Fluorescence signals were analyzed by using confocal microscopy. Scale bar = 50 μm.
Overexpression of TgLBD40 promotes oil accumulation
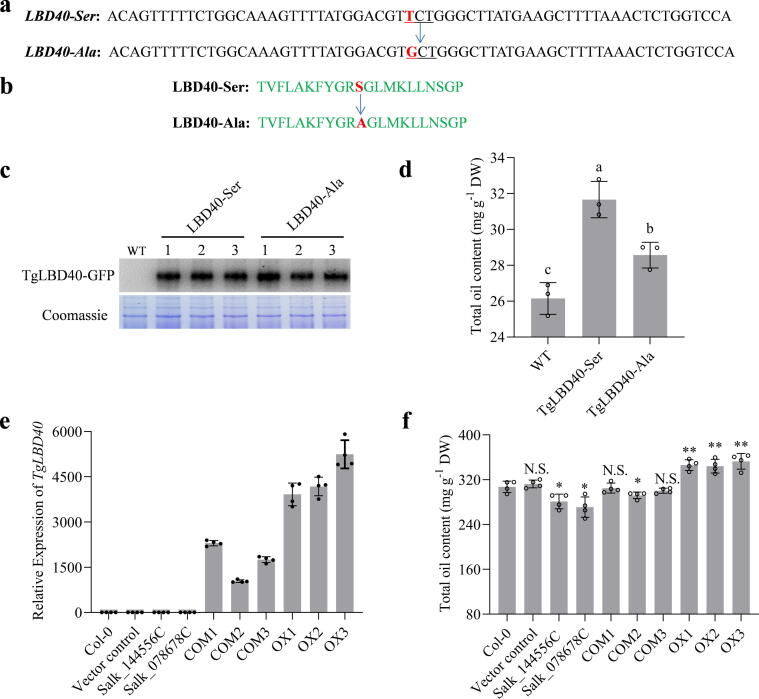
Sequence analysis suggested that the 220,350 position SNP significantly associated with oil content is a nonsynonymous polymorphism in TgLBD40 (T > G variant with amino acid change from Ser to Ala). To demonstrate whether TgLBD40 is functionally involved in oil accumulation, and whether the nonsynonymous variant in the TgLBD40 coding region alters gene function, the two cDNAs of TgLBD40 with only T > G variant at 220,350 position SNP (here the cDNAs of TgLBD40 with T and TgLBD40 with G are designated as TgLBD40-Ser and TgLBD40-Ala, respectively) were transiently expressed in N. benthamiana leaves individually (Fig. 5a, b). TgLBD40-Ser-GFP and TgLBD40-Ala-GFP fusion protein expression in the tobacco leaves was identified by immunoblot analysis and the oil content of samples with relatively consistent protein expression was determined (Fig. 5c). Results showed that both TgLBD40-Ser and TgLBD40-Ala elevated the oil content of tobacco leaves, but TgLBD40-Ser was the most effective one (Fig. 5d). We also found that the proportions of TgLBD40-ser and TgLBD40-Ala natural variant in the A, C, R, X and Y were 20:16, 17:0, 21:6, 36:1 and 53:0, respectively (Fig. S10). Results showed that in the five T. grandis populations except for C, the higher the proportion of TgLBD40-ser and TgLBD40-Ala, the higher the oil content (Fig. S10 and Fig. 3a). The higher proportion of TgLBD40-ser and TgLBD40-Ala in C is probably because it has only 17 samples. These results suggest that the 220,350 position SNP may be one of the functional polymorphisms responsible for the variation in oil levels. However, the influence of unidentified polymorphisms cannot be ruled out. To further demonstrate the role of TgLBD40 in oil accumulation, we generated TgLBD40-Ser overexpressing Arabidopsis lines. Three independent transgenic lines were analyzed in the T3 generation. qRT-PCR analyses indicated that TgLBD40 was strongly expressed in transgenic lines but not in Col-0 and vector control plants (Fig. 5e). We subsequently compared oil content of the overexpressing, Col-0 and vector control Arabidopsis seeds. As expected, the oil content of the overexpressing plants was increased with a statistical significance in comparison with Col-0 and vector control plants (Fig. 5f). To check whether TgLBD40 is a functional homologue of AtLBD41 which has the highest homology with TgLBD40 in Arabidopsis, we investigated the seed oil contents of two atlbd41 mutants (SALK_144556C and Salk_078678C). The mutants were identified by two paired reactions and homozygous mutant lines were selected for oil content determination (Fig. S11). Results showed that both mutants have significantly lower oil content than Col-0 and vector control plants (Fig. 5f). Moreover, we also performed transgenic complementation of Salk_078678C with TgLBD40-Ser and found two complemented lines with oil contents to the level of the wild type. The expression levels of AtLBD41 in all used lines were detected by qRT-PCR (Fig. S12a). Results showed that there was almost no AtLBD41 expression in the mutants and the transgenic lines had no significant effect on AtLBD41 expression (Fig. S12b). These results suggest that there is functional conservation in oil biosynthesis regulation of TgLBD40 and its homolog in Arabidopsis, although they clustered in different clades (Fig. 4b).
Fig. 5.
TgLBD40 promotes oil accumulation. (a) Partial nucleotide sequences of TgLBD40, red letters represent the SNP mined by TRAS. (b) The protein sequences corresponding to the nucleotide sequences in (a). (c) Immunoblot analysis of TgLBD40-Ser-GFP and TgLBD40-Ala-GFP fusion protein expression in tobacco leaves. The total proteins were isolated for immunoblot assays with anti-GFP antibody. Coomassie brilliant blue staining indicates that similar amounts of proteins were loaded. (d) Oil content of wild type (WT), LBD40-Ala and LBD40-Ser transient expressed tobacco leaves. Values are averages and SD of three individual experiments. Different letters indicate significant difference at P < 0.05. (e) The relative expression level of TgLBD40-Ser was identified by qRT-PCR in all used Arabidopsis lines. (f) Oil percentage of seed dry weight. Values are averages and SD of four individual experiments. Statistical significance was determined by Student’s two-tailed t test (*P < 0.05, ***P < 0.01). N.S. means no significant difference compared to Col-0.
The expression of TgLBD40 is significantly correlated with the expression of oil synthesis pathway genes
To understand the potential molecular mechanism by which TgLBD40 increased oil accumulation in T. grandis, the correlation between the expression level of TgLBD40 and the expression level of the unigenes for oil biosynthesis pathway enzymes was investigated by Pearson’s correlation analysis. The results showed that in the unigenes with a correlation level of P < 1 × 10−6, most of the unigenes positively correlated with TgLBD40 were annotated as long-chain acyl-CoA sythetase (LACS) (9 unigenes), DGAT (2 unigenes), stearoyl-ACP desaturase (SAD) (2 unigenes) and GPAT (1 unigene), respectively; there were 2, 1, 1, 2, and 1 unigenes negatively correlated with TgLBD40 that were annotated as KASⅡ, biotin carboxyl carrier protein (BCCP), biotin carboxylase subunit of heteromeric acetyl-CoA carboxylase (BC), MTACP and OLE, respectively (Fig. 6). These results indicated that TgLBD40 may act as a transcription factor to affect the expression of TAG biosynthesis-related genes, thus promoting oil biosynthesis.
TgLBD40 enhanced transcription of TgDGAT1 by combining with TgWRI1
Dual-luciferase assay was used to investigate the regulation mechanism of TgLBD40 on oil biosynthesis in T. grandis. The results showed that significantly enhanced transcription from the promoter of the oil biosynthetic gene TgDGAT1 could be detected when TgWRI1, a key transcription factor in the regulation of plant oil biosynthesis, co-transformed with TgLBD40-Ser or TgLBD40-Ala, although TgLBD40-Ser, TgLBD40-Ala and TgWRI1 alone had no effect on the activation of TgDGAT1 promoter (Fig. 7a, b). Interestingly, the positive regulatory effect of TgLBD40-Ala combined with TgWRI1 on TgDGAT1 promoter was significantly weaker than that of TgLBD40-Ser combined with TgWRI1 (Fig. 7b), indicating that the 220,350 position SNP identified from TRAS could alter the transcriptional activation activity of TgLBD40. Phylogenetic analysis results showed that TgDGAT1 was closely clustered with NP_001237684.2 (Glycine max), NP_001302732.1 (Brassica napus) and NP_179535.1 (Arabidopsis thaliana) (Fig. S13a), and TgWRI1 was closely clustered with XP_013647955.1 (Brassica napus) and NP_001035857.1 (Arabidopsis thaliana) (Fig. S14a). A comparison of protein sequences indicates that TgDGAT1 contains a conserved MOBAT domain and shares high amino acid identity to AtDGAT1 (NP_179535.1) and ZmDGAT1 (EU039830.1) (Fig. S13b). The TgWRI1 protein sequence contains two highly conserved AP2 domains and a highly conserved 14–3-3 binding motif, which were similar to AtWRI1 (NP_001035857.1) and BnWRI1 (XP_013647955.1) (Fig. S14b). These results suggest that TgDGAT1 and TgWRI1 may have functions similar to their homologs in other plants.
Fig. 7.
TgLBD40 directly binds to promoter of TgDGAT1 and its enhancement of transcription of TgDGAT1 requires combination with TgWRI1. (a) Diagrams of the reporter and effector vectors used in the dual-luciferase assay. (b) Analysis of the regulatory effects of TgLBD40-Ser, TgLBD40-Ala and TgWRI1 alone or TgWRI1 combined with TgLBD40-Ser or TgLBD40-Ala on the TgDGAT1 promoter by dual luciferase assay. Each value represents the mean ± SD of four independent experiments. Statistical significance was determined by Student’s two-tailed t test (**P < 0.01, ***P < 0.001). (c) The promoter fragments used to generate probes. Red letters represent the LBD binding site like DNA motifs. (d) Results of EMSAs confirming TgLBD40 bound to the promoter fragments of TgDGAT1. The unlabeled probe was taken as competitor. MBP alone was used as negative control of the binding. Red arrowhead indicates the DNA-protein complex.
Previous studies showed that LBDs can recognize HCGGCG/GCGGCW sites to regulate the expression of genes involved in plant growth, development and metabolic processes [37], [38]. As shown in Fig. 7c, two LBD binding site-like DNA motifs in the TgDGAT1 promoter were found. To test whether TgLBD40 could directly bind to the TgDGAT1 promoter, EMSA assays were performed. Results showed that the purified MBP-tagged TgLBD40 protein but not the MBP alone could directly bind to the TgDGAT1 promoter fragments containing LBD binding site-like DNA motifs in vitro (Fig. 7d). Furthermore, the unlabeled probes could effectively compete with the binding (Fig. 7d). These results indicating that TgLBD40 can directly bind to the TgDGAT1 promoter, but its enhancement of transcription of TgDGAT1 requires combination with TgWRI1.
Discussion
In this study, a TRAS approach that is independent of a reference genome was used to identify traits associated with SNPs and transcripts based on association mapping and regulatory networks. TRAS has its advantages although it cannot identify SNPs in an intron or regulatory regions of genes. TRAS can directly associate the trait with candidate transcripts and their expression. In comparison, the loci controlling the trait identified by GWAS is only a genome region [20]. Moreover, TRAS can detect potential interaction of trait associated-transcripts by co-expression analysis. To perform the TRAS approach, a reference transcriptome of T. grandis was generated by integrating Illumina RNA-sequencing data with single molecule long-read sequencing data in this study. Based on this reference transcriptome and the RNA sequencing of 170 diverse T. grandis landraces, a total of 275,924 high-quality SNPs and a large gene expression profile library were generated. Our results showed that the traits we focused on exhibit a broad variation within sub-populations and this makes it possible for us to identify loci controlling traits in T. grandis populations successfully.
At present, research on fatty acid and oil synthesis in plants has focused almost entirely on oil seeds in angiosperm species. Several transcription factors like LEAFY COTYLEDON1 (LEC1), LEC2, WRINKLED1 (WRI1), FUSCA3 (FUS3) and ABSCISIC ACID3 (ABI3) are reported to act as positive regulators of oil biosynthesis, while Transparent Testa 2 (TT2), TT8, GLABRA2 (GL2), Arabidopsis six-b-interacting protein 1-like 1 (ASIL1), and APETALA2 (AP2) act as negative regulators of oil biosynthesis in angiosperm [39]. Target genes of these transcription factors in oil biosynthesis include FATB (acyl-ACP thioesterase B), PKb1 (pyruvate kinase), BCCP2 (BIOTIN CARBOXYL CARRIER PROTEIN2), ACP1 (acyl-carrier protein1), OLE1 (oleosin 1), KASI, and KASII [39]. Oil biosynthesis in gymnosperm species is difficult to study because their effective population sizes are very large and their genomes are highly heterozygous [40]. To date, there are few studies on oil synthesis in gymnosperms. In our previous study, we found that three genes, TgOLEO1, TgCLO1 and TgSLO1, encoding oil body-associated proteins in Torreya grandis can affect the oil content [5]. The difference between gymnosperms and angiosperms in oil composition is that the oil of gymnosperms contains sciadonic acid, while angiosperm oil does not. This is because almost all angiosperm species have lost the capability to introduce supplementary Δ5-desaturation into unsaturated C20 fatty acids [41], [42]. The candidate genes encoding the last two enzymes of sciadonic acid biosynthesis have been identified using transcriptome sequencing [4].
In this study, we identified 41 SNPs from 34 transcripts that were significantly associated with seven oil-related traits using TRAS. None of the reported homologous genes that have been identified through forward genetics to control related traits were identified, suggesting that it is the first findings, to our knowledge, that the natural variation of the genes identified. Some of the genes identified by TRAS in this study may indirectly influence oil accumulation and composition. For example, enhancement of gibberellin (GA) signaling or exogenous gibberellic acid can affect the total seed oil content [43]. In Arabidopsis, LOB domain-containing protein 40 (LBD40) was identified as an early gibberellin-responsive gene [44]. In this study, we identified an oil content associated transcript (TR182770-c1_g1_i1) encoding a protein which homology with Arabidopsis LBD41, suggesting that TgLBD40 may mediate the regulation of gibberellin on oil biosynthesis. Sciadonic acid biosynthesis needs a desaturase AL10 or AL21 which has a cytochrome b5 domain and may have cytochrome b5 activity [45]. Cytochrome b5 could interact with cytochrome C to form a complex which is believed to involve the formation of salt linkages between specific carboxylic acid residues of cytochrome b5 with lysine residues on cytochrome C [46]. It was reported that surfeit locus protein 1 (SURF1) can be involved in the biogenesis of cytochrome c oxidase [47]. Here, we identified a sciadonic acid associated transcript (BRD_TGR26378) encoding a protein which homology with human SURF1, suggesting that TgSURF1 may be involved in the regulation of sciadonic acid biosynthesis.
The Pearson’s correlation analysis showed that the gene expression levels of eight of the 34 transcripts were significantly correlated with their corresponding traits (Fig. S8), suggesting that the association of these 8 transcripts were authentic. However, the remaining 26 transcripts were associated with their corresponding trait only in terms of the SNPs, but not in terms of gene expression, which may be caused by the linkage disequilibrium [20]. Alternatively, these transcripts may regulate the trait through changing the protein function but not changing the transcriptional expression level.
In addition to the identification of trait-associated SNPs and the transcripts where the SNPs are located, we also revealed the association networks across different traits. For example, we identified the 5,167,765 position SNP that functioned as a key node for connecting palmitic acid and linoleic acid. It is noted that palmitic acid is strongly and negatively correlated with linoleic acid (Fig. 1b), suggesting that the gene at this position may mediate the metabolic process that converts palmitic acid to linoleic acid.
To confirm the TRAS result, we performed functional validation and molecular biology experiments for TgLBD40. The plant-specific LBD gene family plays an important role in the regulation of lateral organ development and participates in the regulation of anthocyanin and nitrogen metabolism [48]. However, a role for the LBD gene family in the regulation of plant oil biosynthesis has not been reported so far. Transient expression assay demonstrated that polymorphisms of the 220,350 position SNP in the TgLBD40 coding region could alter the oil content in plants. When TgLBD40 was stably overexpressed in Arabidopsis, seeds accumulated more oil content than Col-0 seeds (Fig. 5). Pearson’s correlation analysis showed that the expression level of TgLBD40 was positively and significantly correlated with the expression level of LACS, DGAT and GPAT encoding unigenes, while negatively and significantly correlated with the expression level of OLE encoding unigenes. It has been shown in other plants that overexpression of LCS, DGAT and GPAT and mutations of some OLE members can increase oil content [49], [50], [51], [52], [53]. LOB domain proteins are suggested to act as transcription factors based on their nuclear localization [54], [55] and their ability to bind DNA motif HCGGCG/GCGGCW [37], [38]. We have demonstrated that TgLBD40 was localized in the nucleus and could directly bind to the TgDGAT1 promoter, but its enhancement of transcription of TgDGAT1 requires combination with TgWRI1. However, whether TgLBD40 can directly or indirectly activate the expression of other oil synthesis pathway related enzyme coding genes needs to be further studied.
Conclusions
In conclusion, our TRAS analysis yielded dozens of genes that are potentially associated with traits of interest in T. grandis, a species without a reference genome. While some of the identified genes have unknown functions, almost all of the others are not directly related to oil biosynthesis. The result of functional validation of TgLBD40 demonstrates that TRAS combined with transgenic technology provides a powerful tool for rapid identification of new genes related to oilseed quality.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (U20A2049, 32271923, 31971699), the Key Research and Development Program of Zhejiang Province (2021C02001, 2020C02019), the Fundamental Research Funds for the Provincial Universities of Zhejiang (2020YQ003), the Breeding of New Varieties of Torreya grandis Program (2021C02066-11), the Scientific Research Startup Fund Project of Zhejiang A&F University (2018FR028), the Special Fund for Forest Scientific Research in the Public Welfare (201504708), the Young Elite Scientists Sponsorship Program by China Academy of Space Technology (CAST) (2018QNRC001).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.01.007.
Contributor Information
Jianbo Xiao, Email: jianboxiao@yahoo.com.
Jiasheng Wu, Email: wujs@zafu.edu.cn.
Lili Song, Email: lilisong@zafu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Dong D.D., Wang H.F., Xu F., Xu C., Shao X.F., Li H.S. Supercritical carbon dioxide extraction, fatty acid composition, oxidative stability, and antioxidant effect of Torreya grandis seed oil. J Am Oil Chem Soc. 2014;91:817–825. [Google Scholar]
- 2.Chen B.Q., Cui X.Y., Zhao X., Zhang Y.H., Piao H.S., Kim H., et al. Antioxidative and acute anti-inflammatory effects of Torreya grandis. Fitoterapia. 2006;27:262–267. doi: 10.1016/j.fitote.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y.J., Wang J.F., Li G.L., Zheng Z.H., Su W.J. Antitumor and antifungal activities in endophytic fungi isolated from pharmaceutical plants, Taxus mairei, Cephalataxus fortunei and Torreya grandis. Fems Immunol Med Mic. 2001;31:163–167. doi: 10.1111/j.1574-695X.2001.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 4.Wu J., Huang J., Hong Y., Zhang H., Ding M., Lou H., et al. De novo transcriptome sequencing of Torreya grandis reveals gene regulation in sciadonic acid biosynthesis pathway. Ind Crop Prod. 2018;120:47–60. [Google Scholar]
- 5.Ding M., Lou H., Chen W., Zhou Y., Zhang Z., Xiao M., et al. Comparative transcriptome analysis of the genes involved in lipid biosynthesis pathway and regulation of oil body formation in Torreya grandis kernels. Ind Crop Prod. 2020;145 [Google Scholar]
- 6.Chapman K.D., Ohlrogge J.B. Compartmentation of triacylglycerol accumulation in plants. J Biol Chem. 2012;287:2288–2294. doi: 10.1074/jbc.R111.290072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atwell S., Huang Y.S., Vilhjalmsson B.J., Willems G., Horton M., Li Y., et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W., Gao Y., Xie W., Gong L., Lu K., Wang W., et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46:714–721. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- 9.Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 10.Buckler E.S., Holland J.B., Bradbury P.J., Acharya C.B., Brown P.J., Browne C., et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Peng Z., Yang X., Wang W., Fu J., Wang J., et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013;45:43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 12.Guo M., Zhang Z., Li S., Lian Q., Fu P., He Y., et al. Genomic analyses of diverse wild and cultivated accessions provide insights into the evolutionary history of jujube. Plant Biotechnol J. 2021;19:517–531. doi: 10.1111/pbi.13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang C., Ma Y., Wu S., Liu Z., Wang Z., Yang R., et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017;18:161. doi: 10.1186/s13059-017-1289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Su W., Tao R., Zhang W., Chen J., Wu P., et al. RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat Commun. 2017;8:2264. doi: 10.1038/s41467-017-02445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutz K., Heilkenbrinker A., Lonne M., Walter J., Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotech. 2013;24:22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Koprivova A., Harper A.L., Trick M., Bancroft I., Kopriva S. Dissection of the control of anion homeostasis by associative transcriptomics in Brassica napus. Plant Physiol. 2014;166:442–450. doi: 10.1104/pp.114.239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Si L., Chen J., Huang X., Gong H., Luo J., Hou Q., et al. OsSPL13 controls grain size in cultivated rice. Nat Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 18.Mao H., Wang H., Liu S., Li Z., Yang X., Yan J., et al. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun. 2015;6:8326. doi: 10.1038/ncomms9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havlickova L., He Z., Wang L., Langer S., Harper A.L., Kaur H., et al. Validation of an updated Associative Transcriptomics platform for the polyploid crop species Brassica napus by dissection of the genetic architecture of erucic acid and tocopherol isoform variation in seeds. Plant J. 2018;93:181–192. doi: 10.1111/tpj.13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Liu X., Zhu S., Tang S., Mei S., Chen J., et al. Transcriptome-referenced association study of clove shape traits in garlic. DNA Res. 2018;25:587–596. doi: 10.1093/dnares/dsy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minoche A.E., Dohm J.C., Schneider J., Holtgrawe D., Viehover P., Montfort M., et al. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 2015;16:184. doi: 10.1186/s13059-015-0729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackl T., Hedrich R., Schultz J., Forster F. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014;30:3004–3011. doi: 10.1093/bioinformatics/btu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Götz S., García-Gómez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felsentein J., Phylip ð. Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 26.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratifification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopelman N.M., Mayzel J., Jakobsson M., Rosenberg N.A., Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordborg, M. Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 2005; 3: e196. [DOI] [PMC free article] [PubMed]
- 30.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 31.Duggal P., Gillanders E.M., Holmes T.N., Bailey-Wilson J.E. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;9:516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Guo Z., Huang C., Duan L., Chen G., Jiang N., et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun. 2014;5:5087. doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B., Dewey C. RSEM: accurate transcript quantifification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeap W., Lee F., Shan D.K., Musa H., Appleton D.R., Kulaveerasingam H. WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 2017;91:97–113. doi: 10.1111/tpj.13549. [DOI] [PubMed] [Google Scholar]
- 35.Lou H., Ding M., Wu J., Zhang F., Chen W., Yang Y., et al. Full-Length Transcriptome Analysis of the Genes Involved in Tocopherol Biosynthesis in Torreya grandis. J Agr Food Chem. 2019;67:1877–1888. doi: 10.1021/acs.jafc.8b06138. [DOI] [PubMed] [Google Scholar]
- 36.Yamaji N., Huang C.F., Nagao S., Yano M., Sato Y., Nagamura Y., et al. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell. 2009;21:3339–3349. doi: 10.1105/tpc.109.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chalfun-Junior A., Franken J., Mes J.J., Marsch-Martinez N., Pereira A., Angenent G.C. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximaldistal patterning in Arabidopsis petals. Plant Mol Biol. 2005;57:559–575. doi: 10.1007/s11103-005-0698-4. [DOI] [PubMed] [Google Scholar]
- 39.Manan S., Chen B., She G., Wan X., Zhao J. Transport and transcriptional regulation of oil production in plants. Crit Rev Biotechnol. 2017;37:641–655. doi: 10.1080/07388551.2016.1212185. [DOI] [PubMed] [Google Scholar]
- 40.Nystedt B., Street N.R., Wetterbom A., Zuccolo A., Lin Y.C., Scofield D.G., et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- 41.Wolff R.L. The phylogenetic significance of sciadonic (all-cis 5, 11, 14–20: 3) acid in gymnosperms and its quantitative significance in land plants. J Am Oil Chem Soc. 1999;76:1515–1516. [Google Scholar]
- 42.Aitzetmuller K. Fatty acid patterns of Ranunculaceae seed oils: phylogenetic relationships. Plant Syst Evol. 1995;9:229–240. [Google Scholar]
- 43.Chen M., Du X., Zhu Y., Wang Z., Hua S., Li Z., et al. Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 2012;35:2155–2169. doi: 10.1111/j.1365-3040.2012.02546.x. [DOI] [PubMed] [Google Scholar]
- 44.Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., et al. Global Analysis of DELLA Direct Targets in Early Gibberellin Signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayanova O., Haslam R., Caleron M.V., Napier J.A. Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: identification of an acyl-coenzyme A C20 Δ5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol. 2007;144:455–467. doi: 10.1104/pp.107.098202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodgers K.K., Pochapsky T.C., Sligar S.G. Probing the mechanisms of macromolecular recognition: the cytochrome b5-cytochrome c complex. Science. 1988;240:1657–1659. doi: 10.1126/science.2837825. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 48.Majer C., Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Ding L., Gu S., Zhu F., Ma Z., Li J., Li M., et al. Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus. BMC Plant Biol. 2020;20:21. doi: 10.1186/s12870-020-2240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shockey J.M., Gidda S.K., Chapital D.C., Kuan J.C., Dhanoa P.K., Bland J.M., et al. Tung tree DGATl and DGAT2 have nonredundant functions in triaeylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cakes J., Brackenridge D., Colletti R., Daley M., Hawkins D.J., Xiong H., et al. Expression of fungal diacylglycerol acyhransferase2 genes to increase kernel oil in maize. Plant Physiol. 2011;155:1146–1157. doi: 10.1104/pp.110.167676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao J., Li J.L., Li D., Tobin J.F., Gimeno R.E. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. P Natl Acad Sci USA. 2006;103:19695–19700. doi: 10.1073/pnas.0609140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miquel M., Trigui G., Dandrea S., Kelemen Z., Baud S., Berger A., et al. Specialization of Oleosins in Oil Body Dynamics during Seed Development in Arabidopsis Seeds. Plant Physiol. 2014;164:1866–1878. doi: 10.1104/pp.113.233262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H.W., Kim N.Y., Lee D.J., Kim J. LBD18/ASL20 Regulates Lateral Root Formation in Combination with LBD16/ASL18 Downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009;151:1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. ARF7 and ARF19 Regulate Lateral Root Formation via Direct Activation of LBD/ASL Genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.