Abstract
The pathogenesis of atopic dermatitis (AD) is multifactorial, involving a dynamic interplay between genetic susceptibility, skin-barrier dysfunction, microbiome alterations, and immune dysregulation, whereas food allergy (FA) arises from the interplay of transcutaneous sensitization to food allergens and failure in the induction of oral tolerance. Skin epicutaneous sensitization is commonly involved in the development of AD and FA. Although clinical trials have been conducted to prevent AD or FA by applications of emollients on the skin after birth, the results are not consistent. For more effective preventive strategies, reliable biomarkers are required to identify high-risk individuals. Skin tape stripping (STS) is a non-invasive technique for identifying these biomarkers in the skin. By analyzing the stratum corneum collected via STS, researchers can gain molecular or cellular insights into the early pathogenesis and potential progression of AD and FA. This review aims to elucidate the critical aspects of AD and FA, underlying their pathogenesis, early manifestations, and STS's potential as a tool for identifying predictive non-invasive biomarkers in infants prior to onset of clinical disease.
Keywords: Biomarker, atopic dermatitis, food allergy
INTRODUCTION
Atopic dermatitis (AD) and food allergy (FA) are common diseases in children, affecting up to 20% and 10% globally, respectively.1,2,3 The rising incidence of AD and FA in infants and young children has profound implications for their long-term health and quality of life, as well as for their caregivers.3,4,5 These conditions pose significant public health challenges, particularly when they manifest early in life.
AD and FA are closely associated, with both conditions commonly manifesting within the first year of life.6 Food-specific immunoglobulin E (IgE) responses can be detected within the first few months of life and reach a prevalence of around 10% by 1 year of age.7 AD usually precedes the development of FA, a progression known as the atopic march.6 This progression may be attributed to the unique immune phenotype of early-onset AD, which differs from that seen in older children and adults.8
Animal studies provide evidence that the application of ovalbumin or peanut protein on disrupted skin facilitates a type 2 immune response through antigen-presenting cells, resulting in FA and anaphylaxis.9,10 This sequence of events underscores the need for early and accurate diagnosis and highlights the potential for interventions that may modify disease trajectory and improve outcomes.
PATHOGENETIC MECHANISM AND RISK FACTORS OF AD AND FA
The pathogenesis of AD is multifactorial, involving a dynamic interplay between genetic susceptibility, skin-barrier dysfunction, microbiome alterations, and immune dysregulation, whereas FA arises from the interplay of transcutaneous sensitization and failure in the induction of oral tolerance.11,12 These mechanistic factors can interact and exacerbate each condition.13 Genetically, loss-of-function mutations in the filaggrin (FLG) gene are frequently associated with AD, leading to skin barrier disruption that facilitates allergen penetration and sensitization.1 T helper 2 (Th2) immune activation, which is predominant in both AD and FA, is known to downregulate the expression of epidermal proteins including filaggrin, alter epidermal lipid metabolism, reduce antimicrobial peptide production by keratinocytes, and inhibit keratinocyte differentiation.14
Epidermal barrier dysfunction is commonly found in both lesional and non-lesional skin of patients with AD, as evidenced by increased transepidermal water loss (TEWL) and pH disturbance, elevated permeability, reduced water retention, and altered lipid profiles.15 The barrier dysfunction is further exacerbated by microbial dysbiosis, including colonization with Staphylococcus aureus.16,17 Research indicates that 60%–100% of AD patients have skin colonized by S. aureus, and up to 10%–30% of these isolates are methicillin-resistant S. aureus (MRSA).16,17 S. aureus colonization worsens AD and may lead to microbial dysbiosis, increased allergen sensitization, polarization towards Th2/Th17 responses, progression of the atopic march, and the development of FA in patients with AD.18 A recent study showed S. aureus induces cytokines such as interleukin (IL)-1β, tumor necrosis factor (TNF)-α, IL-6, and IL-33, leading to changes in epidermal lipid composition and subsequent skin barrier dysfunction.17 This occurs through the inhibition of fatty acid elongase enzymes, such as elongation of very long chain fatty acids protein (ELOVL) 3 and ELOVL4, with these effects being more pronounced after exposure to MRSA compared to methicillin-sensitive S. aureus (MSSA).17 Additionally, toxins produced by Staphylococcus on the skin enhance transcutaneous sensitization to food allergens, thereby promoting the development of FA.19,20
Environmental factors significantly contribute to the development of AD and FA. External factors such as scratching, low environmental humidity, allergens, and topical irritants further damage the skin barrier in individuals with AD.11,14 Importantly, early-life exposure of the skin to high levels of environmental food allergens, known as epicutaneous exposure, increases the risk of food sensitization, particularly in children with damaged skin and low filaggrin levels.13,21,22 Therefore, key strategies to prevent food sensitization and allergies include skin barrier restoration, aggressive treatment of AD, and the early introduction of foods through the gut.23,24,25
A recent study utilizing a murine model showed that transcutaneous sensitization triggers the recruitment of activated dendritic cells to the draining lymph nodes, resulting in the formation of allergen-specific IgG1+ germinal center B cells and the production of serum IgG1.9 The allergen-specific IgG1+ memory compartment predominantly displayed an immature, pro-germinal center phenotype, and subsequent exposures to the food allergen triggered the development of IgE+ germinal center B cells, increased serum IgE levels, and activated the classical anaphylaxis pathway.9 This indicates a positive correlation between food sensitization and increased AD severity and duration.2 However, clinical allergy can only be confirmed through actual food consumption, and since the timing of food antigen intake varies among individuals, it is difficult to accurately assess the age of FA onset.
WHY SKIN PREDICTIVE BIOMARKERS?
The potential of the skin as a site for predictive biomarkers in AD and FA is significant due to its accessibility and central role in immune interactions.26 Early changes in the integrity and function of the skin barrier often signal the onset of both AD and FA.6 The immune environment of the skin, rich in dendritic cells, T cells, and mast cells, reflects and possibly anticipates the complex immunological shifts during the development of these conditions.27 Therefore, the expression of specific molecules such as cytokines, chemokines, and other mediators from genomics, transcriptomics, proteomics, and lipidomics in the skin can serve as early biomarkers to predict the risk of developing AD and FA.17,28,29,30,31 Various techniques can be employed to analyze skin biomarkers, including invasive methods like biopsies, less invasive methods like epidermal curettage or microneedle patches, and non-invasive methods like skin tape stripping (STS).30 Among these, STS stands out as an ideal non-invasive method for early and repeated assessments, particularly important for managing children effectively and sensitively.26,32
To effectively prevent AD and FA from an early stage, it is crucial to establish strong biomarkers associated with the early detection of molecular signatures in the skin.28,29,33 This proactive strategy can enhance patient outcomes through personalized treatment and prevention plans targeting specific immunological pathways involved in each patient’s disease process.26 Thus, identifying predictive skin biomarkers using non-invasive sampling and various omics approaches, where AD initially manifests, holds promises for improving the management of allergic diseases. Furthermore, understanding these skin biomarkers elucidates the pathogenetic mechanisms underlying AD and FA.
STS: A SUITABLE METHOD FOR SKIN SAMPLING
There are several non-invasive methods for collecting skin samples, including skin surface washing, the thin film headspace sampling method, and STS.34,35 Skin surface washing is effective for evaluating skin cytokines, but is inconvenient and difficult to apply to children due to the relatively long time required (about 30 minutes). Additionally, it has limitations in obtaining RNAs and lipids. The thin film headspace sampling method is suitable for longitudinal studies to measure volatile organic compounds in the skin, but cannot be used to analyze proteins, RNAs, or lipids. STS method offers several advantages over these methods.
A specialized skin tape disc, such as D-Squame® tape disc (22 mm diameter; CuDerm, Dallas, TX, USA), is used for STS method. Up to 20 consecutive D-Squame tape discs are applied to the same site on the skin. The D-Squame pressure instrument D500 (CuDerm) ensures consistent application of equivalent pressure (e.g., 225 g/cm2) across all tape strips, as previously described.36 The number of STS applications can be adjusted based on the specific goals of the procedure. Kim et al.32 conducted side-by-side studies comparing skin biopsies and STS to determine the depth reached by the STS procedure in both healthy subjects and patients with AD. They found that 20 STS applications can reach the upper granular layer of epidermis, where RNA and cytokines are present. Thus, 15–20 applications of STS are needed to evaluate RNA expression from the upper granular layer of the epidermis. To obtain epidermal lipids and proteins, only 8 STS applications are sufficient. These comparative studies were conducted only in healthy subjects and those with AD. Therefore, future research may be needed to evaluate the efficacy of the STS method in other skin conditions, such as psoriasis, urticaria, and contact dermatitis. Twenty consecutive STS applications do not cause bleeding or scarring in non-lesional skin and allows analysis of gene transcription. It is not painful and only minor skin irritation is observed during STS.
Various samples including RNA, proteins, and lipids can be harvested from STS. Gene expression of epidermal barrier proteins and cytokines can be analyzed by polymerase chain reaction. Protein levels of epidermal cytokines can be detected by ELISA or a Meso Scale Discovery immunoassay. Lipids can be dissolved in methanol for liquid chromatography electrospray ionization tandem mass spectrometry analyses. The STS method offers several advantages over conventional skin biopsy. It is a useful strategy for collecting skin samples multiple times from the same subjects. Due to its non-invasive nature and low cost, the STS method enables the collection of a large number of samples. Additionally, it facilitates the collection of skin samples from subjects of diverse ages, including young infants.14,17,29,33
The STS method is an acceptable strategy for examining the molecular profiles of the skin across all age groups, from young infants to senior adults. Traditionally, the primary method for studying the pathophysiology of inflammatory skin diseases, such as AD and psoriasis, has been skin biopsy.32,37,38 However, the invasive and painful nature of skin biopsy poses significant limitations for its use in all subjects with skin diseases. Ethical concerns and the discomfort associated with the procedure make it particularly challenging to obtain skin samples from children, even though AD is most prevalent in this age group. Consequently, this can lead to bias and errors in the analysis of data from children with AD. Major characteristic findings of AD include abnormal keratinocyte differentiation and aberrant epidermal lipid profiles in the upper granular layer and cornified layer of the skin.28,38,39 Therefore, a non-invasive and convenient STS method could be useful to investigate the expression of specific genes, lipids, and proteins in the epidermis.28,32,33,40,41,42,43 In summary, STS is an excellent strategy for evaluating the genes, proteins, and lipids of epidermis across all age groups.
PREDICTIVE BIOMARKERS FOR AD
AD in children is a major public health concern due to its high prevalence, significant impact on quality of life, substantial socioeconomic burden, and frequent progression to respiratory allergies. Consequently, identifying predictive biomarkers for preclinical asymptomatic target organ damage is crucial for preventing AD. These biomarkers represent an intermediate step between exposure to risk factors and subsequent development of clinical events. Given that the primary target organ of AD is the skin, a biomarker that detects skin changes in a preclinical stage could serve as a useful predictor.
Skin barrier function: TEWL
TEWL is a noninvasive method to measure skin barrier function. Multiple commercially available instruments can measure TEWL.44,45 The accuracy of these measurements is influenced by several factors, including temperature, humidity, ventilation, and intrinsic factors.44,46,47 To ensure reliable results, measurements should be performed under standard conditions: maintaining a relative humidity of 40%−60% and acclimating the subject at an ambient temperature of 20°C−22°C for at least 20–30 minutes before measuring TEWL.44,46,47 Additionally, subjects should not apply topical lotions or cosmetics to the skin areas being measured and should avoid exercise and caffeine intake for at least three hours prior to measurement.44,46,47
In a Japanese cohort study, TEWL was measured on the forehead under ambient environmental conditions (24°C−27°C, 11%−58% humidity) within the first week of life in 116 infants, who were followed until 32 weeks after birth. When the probability of AD incidence was stratified by TEWL, the high TEWL group (TEWL ≥ 6.50 g/m2/h) showed a higher AD incidence than the low TEWL group (TEWL < 6.50 g/m2/h) (P < 0.05).48 In a study using a cohort of infants living in south-east Norway, TEWL was measured on the lateral part of one upper arm using an open chamber DermaLab USB (Cortex, Hadsund, Denmark) system under reasonable environmental conditions (20°C−25°C, 20%−50% humidity).49 This study found that high TEWL (> 9.33 g/m2/h) at less than 3 months was significantly associated with atopic eczema at a mean 24 months (17.5–35.2 months) (odds ratio [OR], 7.67; 95% confidence interval [CI], 1.04–56.77) although the cohort size was small (n = 32).49 These studies suggest that TEWL at 2 or 3 months of age can be used to predict the future development of AD before the age of 12 months.
In contrast, a Korean birth cohort study of 87 infants measured TEWL on the volar surface of the forearm using a Tewameter TM300 (Courage & Khazaka) at 2 months of age and found no statistical significance between infants who later developed AD and those who did not.29 Another study nested within the prospective Copenhagen Baby Skin birth cohort measured TEWL on the central part of the flexor forearm using a portable device (AquaFlux model AF200; Biox Systems Ltd, London, UK).31 In this study, the median TEWL at 2 months of age did not differ between 44 infants diagnosed with AD in their first year and 44 healthy controls matched by sex and season of birth (14.0 vs. 13.3 g/m2/h, P = 0.9).31 In the Preventing Atopic Dermatitis and Allergies in Children prospective birth cohort study involving 1,150 mother-child pairs, TEWL was measured on both affected and unaffected skin of AD using an open chamber DermaLab USB (Cortex) at room temperature (20°C−25°C) with variable humidity (6%−73%).50 A multivariate analysis demonstrated that high TEWL (> 11.3 g/m2/h) at 3 months of age was not predictive for AD at 6 months.50 Additionally, a study measuring TEWL on the central volar forearm with a portable device (AquaFlux model AF200; Biox Systems Ltd) reported that an elevated mean TEWL at 2 months of age did not increase the risk of AD among term (adjusted OR [aOR], 1.29; 95% CI, 0.81–2.05; P = 0.3) or preterm children (aOR, 0.53; 95% CI, 0.17–1.64; P = 0.3).51
TEWL measurement is technically demanding and varies significantly across individuals and anatomical locations. It is influenced by environmental factors such as temperature, humidity and pollution. This variability may explain the inconsistent results from previous studies regarding the efficacy of TEWL as a predictive biomarker of AD. Additionally, because functional changes in the skin barrier often follow biochemical or molecular alterations, assessing TEWL at 2 months of age may be too early to reliably predict the future development of AD, particularly in mild cases.
Structural proteins of skin barrier: FLG
Filaggrin plays a major role in skin barrier function. Filaggrin, synthesized from the precursor profilaggrin encoded by the FLG gene on chromosome 1q21, binds with cytoskeletal keratins to maintain corneocyte structural integrity. Upon degradation, filaggrin yields breakdown products such as free amino acids, pyrrolidone carboxylic acid (PCA), and urocanic acid (UCA), which are key components of the epidermal natural moisturizing factor.52
FLG mutations were strongly associated with AD, and meta-analyses have confirmed these associations with an overall OR ranging from 3.12 to 4.78.53 However, AD develops in only 42% of all FLG heterozygotes,54 and approximately 54% of FLG null mutation carriers in the United States remain asymptomatic for AD.55 Therefore, targeted screening for FLG mutation would be beneficial only in a limited number of patients. Among AD patients, cases without FLG mutations are much more than those with FLG mutations. Indeed, filaggrin expression is downregulated by type 2 cytokines including IL-4 and IL-13, and most patients with AD have an acquired defect in filaggrin expression.56
In a Danish birth cohort, STS were collected at 2 months of age and revealed that low levels of UCA increased the risk of developing AD until the age of 2 years (adjusted hazard ratio [aHR], 1.68; 95% CI, 1.07–2.64).51 In contrast, a Korean birth cohort study demonstrated that filaggrin levels measured in STS at 2 months did not predict future development of AD in a logistic regression model.29 Another Korean birth cohort also showed that no difference in the levels of UCA and PCA of stratum corneum (SC) was observed between healthy and future AD subjects.28 These findings indicate that filaggrin expression is not reduced at 2 months of age in AD patients without FLG mutations, but may be decreased before this age in those with FLG mutations.
Immune responses
Thymic stromal lymphopoietin (TSLP) and IL-33, collectively known as alarmin cytokines, facilitate communication between keratinocytes and immune cells, thereby regulating immune activity in the skin. TSLP is produced mainly in epithelial cells and epidermal keratinocytes in response to both environmental and endogenous triggers.57 TSLP induces Th2 differentiation,58 downregulates filaggrin expression,59 and contributes to allergic skin inflammation in AD.60 In a Korean birth cohort study, skin samples were obtained from 87 infants using STS at the age of 2 months on their forearms where clinically apparent AD did not appear and followed them for 2 years. This study revealed that high TSLP expression at 2 months was associated with higher risk of AD with the aOR of 5.3 (95% CI, 1.3–21.4). In addition, aOR reached 20.2 (95% CI, 1.5–272.3) when high TSLP expression was combined with positive family history.29 This observation was replicated in another Korean birth cohort by showing that TSLP level in the skin increased the aOR of 4.1 (95% CI, 1.7–10.1). These results indicated that epidermal TSLP protein expression level can be used as an early biomarker to predict AD development.28
Thymus and activation regulated chemokine (TARC)/chemokine (C-C motif) ligand 17 (CCL17) is constitutively expressed in the thymus and is produced by dendritic cells, endothelial cells, keratinocytes and fibroblasts.61 TARC/CCL17 plays a crucial role in the migration of chemokine (C-C motif) receptor 4 (CCR4)-expressing T cells to the skin.62 There is a significant correlation between serum TARC levels and disease severity in patients with AD.63 Indeed, higher levels of TARC/CCL17 were observed in umbilical cord serum of neonates who subsequently developed AD during infancy compared to those who did not develop AD, with median values of 1,586.9 vs. 819.6 pg/mL, respectively (P < 0.001).64 This finding was replicated in a separate Japanese birth cohort study.65 Recent research suggests the possibility that TARC/CCL17 levels in the skin of newborn babies may predict the development of AD during infancy. A Danish prospective birth cohort demonstrated that elevated TARC/CCL17 levels in the skin at 2 months of age increased the risk of overall AD (aHR, 1.85; 95% CI, 1.18–2.89) and moderate-to-severe AD (aHR, 4.65; 95% CI, 1.91–11.31) during the first 2 years of life.51 A nested case-control study within the Copenhagen Baby Skin birth cohort revealed that, at 2 months, skin TARC/CCL17 levels were slightly but significantly higher in children who developed AD by 12 months compared to those who did not (0.02 vs. 0.01 pg/μg, P = 0.01).31
S100A8/A9, heterodimeric members of the calcium-binding S100 protein family, are released from keratinocytes. These proteins function as proinflammatory alarmins or damage-associated molecular pattern molecules.66,67 An in vitro experiment showed that S100A8/A9 elevated levels of IL-6, IL-8, and monocyte chemoattractant protein-1 in keratinocytes and concurrently diminished the expression of the skin barrier proteins filaggrin and loricrin.68 Additionally, elevated levels of S100A8/A9 in both lesional skin and serum were correlated with disease severity in patients with AD.69,70 A study of an Irish birth cohort (n = 86) showed that higher levels of S100A8/A9 in the antecubital fossa at 2 months of age, not at birth, in infants with the normal FLG genotype predicted the development of AD in the first year of life (P = 0.033). This association was not observed in infants with filaggrin loss-of-function mutations.71
In STS from Korean infants at two months, cytokines including IL-13, TNF-α, macrophage-derived chemokine (MDC), and IL-6 were detected before AD onset.33 Levels of IL-1Ra, TNF-β, IL-8, IL-18, IL-22, CCL2, and vascular endothelial growth factor A (VEGF-A) were elevated in the skin of German infants who later developed AD,72 while IL-1Ra, IL-8, IL-18, IL-1α, IL-1β, CCL27, and chemokine (C-X-C motif) ligand (CXCL) 2 were not associated with later AD development in Irish birth cohort study.71 Various cytokines and chemokines may serve as potential predictive biomarkers, but further studies are required.
Skin microbiome
It is well known that skin microbial dysbiosis is linked to AD. Notably, S. aureus is colonized on the lesional skin of 70%–100% of patients with AD, and S. aureus colonization is correlated with AD severity.73 A few observational studies have delineated changes in skin microbiome signatures that may influence AD onset during infancy. In a cohort of 50 children from BASELINE longitudinal birth cohort, colonization of the antecubital fossa with commensal staphylococci at 2 months was inversely associated with the incidence of AD at 12 months.74 A Swiss birth cohort study involving 149 infants revealed that culture-proven S. aureus colonization at 3 months was significantly more prevalent in infants who later developed AD by the age of 2 years. This study also noted that the prevalence of S. aureus was higher on the skin of infants at the onset of AD and even 2 months prior, compared with age-matched, unaffected infants.75 Similarly, in a Japanese birth cohort, whole-genome sequencing of S. aureus strains isolated from the cheek skin of 268 infants was performed at 1 and 6 months after birth. The findings indicated that colonization by S. aureus at 6 months significantly elevated the risk of developing AD over the subsequent 2 years.76 Recently, Fonfara et al. 72 examined skin swab samples from a German birth cohort study (n = 50) using 16S amplicon sequencing and found that before AD onset, levels of various Staphylococcus epidermidis amplicon sequence variants (ASVs) were higher, while those of Streptococcus oralis, Streptococcus vestibularis, and several Streptococcus mitis ASVs were lower compared to children with no subsequent AD. Of note, even at AD onset, the skin microbiome predominantly featured S. epidermidis, with virtually no S. aureus sequences detected. These longitudinal studies suggest that S. aureus colonization in the skin may contribute to AD onset in infants and serve as early indicators of AD onset. Nonetheless, it is plausible that S. aureus colonization is secondary to subclinical skin changes and may not represent the initial pathophysiological event preceding the clinical manifestation of AD.
Skin lipid profiles
Lipid is a critical component of the epidermal barrier. The intercellular multilayered lipid lamellae are located within the SC and are primarily composed of 50% ceramides (CERs), 25% cholesterol, and 15% free fatty acids. Epidermal CERs, in particular, consisting of a long-chain sphingoid bases linked to fatty acids, are critical for skin barrier function.77 Human CERs are categorized into 20 classes, distinguished by their specific sphingoid bases (SB) and fatty acids. Aside from these CER classes in lipid lamellae, SC includes 5 classes of protein-bound CERs that are covalently linked to the cornified envelope proteins of corneocytes, which are terminally differentiated keratinocytes.78
In a case-control nested study in the prospective Copenhagen Baby Skin birth cohort (n = 300), 44 random children with onset of AD in the first year of life were compared with 44 children who did not develop AD. STS samples were obtained from the dorsal side of the hands at 2 months of age. They found that phytosphingosine ([P]) levels were much lower in children who developed AD compared with children who did not (238 vs. 535 pmol/mg, P < 0.001). Most lipid ratios in children who developed AD showed significantly increased concentrations of shorter SBs as compared to longer SBs. The ratio of CER[DS]-(d18:1)/CER[S]-(d20:1) was lower in children who developed AD and the ratio of CER[S]-(d18:1)/CER[S]-(d20:1) was the single best predicting lipid biomarker. The combination of 5 lipid ratios ([DS]-(d17:0)/[DS]-(d18:0), [DS]-(d18:0)/[S]-(d18:1), [P]-(t18:0)/[S]-(d18:1), CER[DS]-(d17:0)/CER[DS]-(d18:0), CER[S]-(d17:1)/CER[S]-(d18:1)) gave an prediction accuracy of 89.4%.31
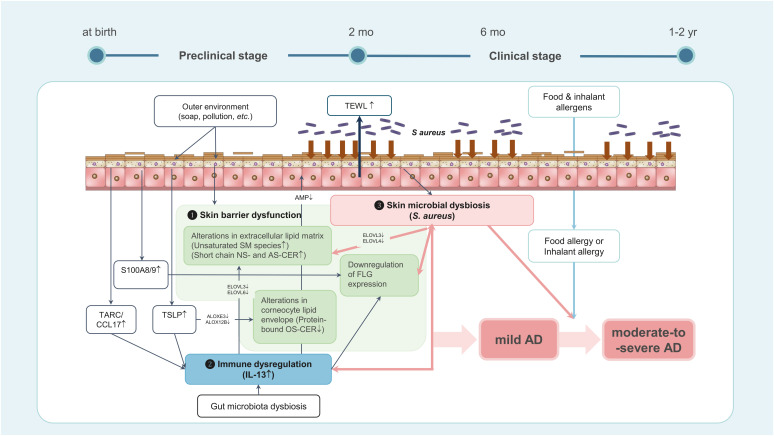
In a Korean birth cohort study, STS samples were collected from the forearms of newborns (n=111) at 2 months of age, before any clinical signs of AD, with and without a family history of atopic diseases. These infants were monitored until age 2 years to confirm the presence or absence of AD. In this study, 22 out of 74 (29.7%) in the risk group and 5 out of 37 (13.5%) in the control group developed AD. In future AD cases, protein-bound CERs decreased (P < 0.001), while unsaturated sphingomyelin species increased (P < 0.0001), along with elevated short-chain nonhydroxy fatty acid sphingosine CERs and alpha-hydroxy fatty acid sphingosine CERs (P < 0.01 and P < 0.05, respectively). Of note, multivariable logistic regression analysis demonstrated that AD was strongly predicted from the combination of family history, type 2 cytokines, and dysregulated lipids, with the OR of 54.0 (95% CI, 9.2–317.5).28 Therefore, integrating data from STS, skin swab, and TEWL, following standard protocols, may offer more accurate predictions of AD development. Table summarizes the skin biomarkers that predict the development of AD during infancy. These findings suggest the early pathogenetic mechanisms of AD development in infants, as shown in Fig. 1.
Table. Potential biomarkers to predict the development of atopic dermatitis at age 12–24 months.
| Population | Follow-up | Samples | Biomarkers | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Barrier proteins | Barrier lipids | Cytokines | Barrier function | Skin microbiome | ||||
| Japan (n = 116) | 32 wk | TEWL (forehead) at 7 days | TEWL* | Horimukai et al. 48 (2016) | ||||
| South Korea (n = 75) | 2 yr | STS and TEWL (forearm) at 2 months | Filaggrin (MS)† | TSLP↑ (MS)* | TEWL† | Kim et al. 29 (2016) | ||
| Norway (n = 32) | 1 yr | TEWL (lateral part of upper arm) at less than 3 months | TEWL* | Berents et al. 49 (2017) | ||||
| Ireland (n = 50) | 1 yr | Skin swab for bacterial 16S ribosomal RNA sequencing in antecutibal fossa at 2 months | Colonization with commensal staphylococci: negatively significant | Kennedy et al. 74 (2017) | ||||
| Colonization with S. aureus† | ||||||||
| Swiss (n = 149) | 2 yr | Skin swab for bacterial culture in axillary fossa at 3 months | Colonization with S. aureus* | Meylan et al. 75 (2017) | ||||
| Japan (n = 268) | 2 yr | Skin swab for whole-genome sequencing in cheek skin at 6 months | Colonization with S. aureus* | Nakamura et al. 76 (2020) | ||||
| Norway/Sweden (n = 1,150) | 6 mon | TEWL (lateral part of left upper arm) at 3 months | High TEWL (> 90th percentile)† | Rehbinder et al. 50 (2020) | ||||
| South Korea (n = 111) | 2 yr | STS (forearm) at 2 months | Filaggrin breakdown products (UCA, PCA)† | Protein-bound OS-CER↓, unsaturated sphingomyelin↑, short-chain NS-CER↑, short-chain AS-CER↑* | TSLP↑, IL-13↑* | Berdyshev et al. 28 (2023) | ||
| IL-4† | ||||||||
| Denmark (n = 88) | 2 yr | STS (dorsal side of hands) and TEWL (central part of the flexor forearm) at 2 months | Phytosphingosine ↓, shorter sphingoid base↑, the ratio of CER[S]-(d18:1)/CER[S]-(d20:1)↓* | TARC/CCL17↑* | TEWL† | Rinnov et al. 31 (2023) | ||
| Denmark (n = 426) | 2 yr | STS (dorsal side of hands) and TEWL (central part of the flexor forearm) at 2 months | Filaggrin breakdown products (UCA)↓ (LC)* | TARC/CCL17↑* | TEWL† | Halling et al. 51 (2023) | ||
| IL-8↑, IL-18↑* for moderate-to-severe AD | ||||||||
| Ireland (n = 86) | 1 yr | Skin swab (antecubital fossa) at 2 months | S100A8/9↑* | Stamatas et al. 71 (2024) | ||||
| IL-1Ra, IL-8, IL-18, IL-1a, IL-1b, CCL27, CXCL2† | ||||||||
| Germany (n = 50) | 2 yr | STS and skin swab (antecubital fossa) at 2–21 days, 6 months, 12 months, and 24 months | IL-1Ra↑, TNF-β↑, IL-8↑, IL-18↑, IL-22↑, CCL2↑, TARC↑, TSLP↑, VEGF-A↑* | S. epidermidis ASVs↑, S. oralis, S. vestibularis, and S. mitis ASVs ↓* | Fonfara et al. 72 (2024) | |||
| Colonization with S. aureus† | ||||||||
TEWL, transepidermal water loss; STS, skin tape stripping; MS, mass spectrometry; TSLP, thymic stromal lymphopoietin; UCA, urocanic acid; PCA, pyrrolidone carboxylic acid; OS-CER, ω-hydroxy fatty acid sphingosine ceramide; NS-CER, nonhydroxy fatty acid sphingosine ceramide; AS-CER, alpha-hydroxy fatty acid sphingosine ceramide; IL, interleukin; TARC, thymus and activation regulated chemokine; CCL17, chemokine (C-C motif) ligand 17; LC, liquid chromatography; AD, atopic dermatitis; CXCL, chemokine (C-X-C motif) ligand; TNF, tumor necrosis factor; VEGF-A, vascular endothelial growth factor A; ASV, amplicon sequence variant.
*Significant; †Not significant.
Fig. 1. A hypothesis concerning the early events in the skin leading to AD development during early infancy. Levels of TSLP, TARC/CCL17 and type 2 cytokines such as IL-13 are elevated in the skin within the first 2 months of life. This initiates immune dysregulation and alterations in the protein and lipid composition of the skin barrier. Subsequently, there is a decrease in filaggrin expression accompanied by changes in the skin microbiome, culminating in the onset of AD during infancy. Conversely, in infants with filaggrin mutation, diminished filaggrin expression may be evident before 2 months of age, potentially leading to the early onset of severe AD.
AD, atopic dermatitis; TSLP, thymic stromal lymphopoietin; TARC, thymus and activation regulated chemokine; CCL17, chemokine (C-C motif) ligand 17; IL, interleukin; TEWL, transepidermal water loss; AMP, antimicrobial peptide; SM, sphingomyelin; NS-CER, nonhydroxy fatty acid sphingosine ceramide; AS-CER, alpha-hydroxy fatty acid sphingosine ceramide; FLG, filaggrin; OS-CER, ω-hydroxy fatty acid sphingosine ceramide.
PREDICTIVE BIOMARKERS FOR FA
It is well known that AD is a major risk factor for FA.79 In the same context, filaggrin loss-of-function mutations demonstrated a significant association with peanut allergy (OR, 5.3; 95% CI, 2.8–10.2). This finding remained significant (P = 0.0008) after adjusting for coexistent AD.80 In the Learning Early About Peanut Allergy (LEAP) study, peanut allergy was reduced among high-risk infants with AD who were introduced to peanuts early in life, whereas about 40% of the placebo group of children with AD developed FA by age 5.12
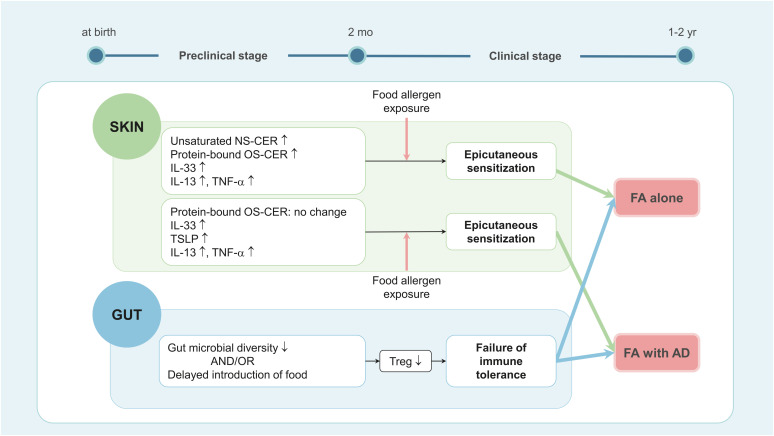
Although AD often precedes the development of FA, not all FA patients have AD. In a Korean birth cohort study, STS samples were obtained from their forearms at 2 months of age, prior to any clinical signs of FA or AD, and analyzed to find biomarkers predictive of FA, either alone (FA+AD−) or with AD (FA+AD+).33 In this study, 18 out of 129 newborns developed FA by the age of 2, with 9 having FA+AD− and 9 having FA+AD+. Levels of unsaturated (N24:1)(C18-sphingosine)CER and (N26:1)(C18-sphingosine)CER were elevated, both in absolute amounts and as a proportion of their molecular group, in the SC of children who later developed FA (both FA+AD− and FA+AD+) compared to the healthy controls (all P < 0.05), but not in the SC of those who later developed AD without FA (FA−AD+). In contrast to infants with future AD, who exhibited decreased levels of protein-bound ω-hydroxy fatty acid sphingosine CERs (OS-CERs), the SC of infants predisposed to FA+AD− demonstrated a significant increase in OS-CERs levels compared to that of healthy children. Levels of unsaturated sphingomyelin species were not increased in the SC of infants who later developed FA, whereas they were significantly elevated in the SC of infants who later developed FA−AD+. While levels of TSLP, IL-13, and TNF-α were increased in infants who later developed either FA or AD, IL-33 levels were elevated in infants who later developed FA but not in those who developed AD alone. Logistic regression analysis indicated that a combination of dysregulated lipids and cytokines strongly predicts FA, evidenced by the OR of 101.4 (95% CI, 5.4–1,910.6). These results suggest that changes in cytokines and lipid profiles in the skin at 2 months of age can serve as biomarkers to predict the development of FA during infancy (Fig. 2).
Fig. 2. A hypothesis concerning the early events in the skin leading to FA development during early infancy. FA development is linked to concurrent epicutaneous sensitization to food allergens and failure to induce oral tolerance. AD is a strong risk factor for FA development and FA is often accompanied by AD. Thus, early AD-associated skin changes—elevated TSLP, IL-13 and TNF-α levels—within the first 2 months of life, increase the risk of developing FA. However, in the case of FA occurring independent of AD, increased levels of IL-33, IL-13, TNF-α, unsaturated NS-CER, and protein-bound OS-CER are observed within 2 months of age.
FA, food allergy; AD, atopic dermatitis; TSLP, thymic stromal lymphopoietin; IL, interleukin; TNF, tumor necrosis factor; NS-CER, nonhydroxy fatty acid sphingosine ceramide; OS-CER, ω-hydroxy fatty acid sphingosine ceramide; Treg, regulatory T cell.
So far, there are few studies to identify predictive biomarkers for FA development. Recently, omics technology has been applied to uncover the molecular mechanisms of FA.81 This approach will not only validate existing findings but also contribute to the discovery of new biomarkers for FA development.
CONCLUSION
Non-invasive methods such as skin swabs, STS, and TEWL are effective strategies for analyzing the skin microbiome, filaggrin degradation products, skin lipid profiles, skin cytokines, and skin barrier function, all of which are related to the development of AD and FA. Studies utilizing multi-omics approaches and STS have demonstrated the ability to detect early alterations in infant immune responses and epidermal barrier function, which precede the development of AD and FA during infancy. In other words, as early as 2 months of age, certain epidermal proinflammatory cytokines, type 2 cytokines, and lipid profiles may be used as effective biomarkers for identifying infants at elevated risk of developing AD and FA later in life.
However, there are not many studies related to predictive biomarkers yet. Expanding research to include diverse cohorts of children from various ethnic backgrounds in different regions is crucial to corroborate these findings and to determine whether the skin biomarkers identified are specific to Asian children living in Asia-Pacific regions or applicable universally across different ethnicities. Finally, early identification of children at high risk allows for the implementation of targeted preventive strategies before the age of 2 months, potentially preventing the onset of clinical AD and FA during infancy.
ACKNOWLEDGMENTS
This research was supported and funded by Seoul National University Hospital Lee Kun-hee Child Cancer & Rare Disease Project, Republic of Korea (grant number: 22B-005-0100) and the Edelstein Family Chair of Pediatric Allergy-Immunology. Also, the authors would like to thank Sang-eun Lee from Multimedia Services Part, Samsung Medical Center (Seoul, Korea) for drafting the graphical abstract.
Footnotes
Disclosure: There are no financial or other issues that might lead to conflict of interest.
References
- 1.Tokura Y, Hayano S. Subtypes of atopic dermatitis: from phenotype to endotype. Allergol Int. 2022;71:14–24. doi: 10.1016/j.alit.2021.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Tsakok T, Marrs T, Mohsin M, Baron S, du Toit G, Till S, et al. Does atopic dermatitis cause food allergy? A systematic review. J Allergy Clin Immunol. 2016;137:1071–1078. doi: 10.1016/j.jaci.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, Kim M, Kim J, Park B, Min N, Jung M, et al. Quality of life in food allergy: validation of the Korean Version of the Food Allergy Quality of Life Questionnaire Parent Form (K-FAQLQ-PF) and risk factor analysis. Allergy Asthma Immunol Res. 2023;15:43–54. doi: 10.4168/aair.2023.15.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fishbein AB, Silverberg JI, Wilson EJ, Ong PY. Update on atopic dermatitis: diagnosis, severity assessment, and treatment selection. J Allergy Clin Immunol Pract. 2020;8:91–101. doi: 10.1016/j.jaip.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung M, Kim S, Yoo HW, Kim HY, Kim M, Lee JY, et al. Validity and reliability of the Korean versions of the Food Allergy Quality of Life Questionnaire-child form and teenager form. Allergy Asthma Immunol Res. 2024;16:202–210. doi: 10.4168/aair.2024.16.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tham EH, Rajakulendran M, Lee BW, Van Bever HP. Epicutaneous sensitization to food allergens in atopic dermatitis: what do we know? Pediatr Allergy Immunol. 2020;31:7–18. doi: 10.1111/pai.13127. [DOI] [PubMed] [Google Scholar]
- 7.Kulig M, Bergmann R, Klettke U, Wahn V, Tacke U, Wahn U. Natural course of sensitization to food and inhalant allergens during the first 6 years of life. J Allergy Clin Immunol. 1999;103:1173–1179. doi: 10.1016/s0091-6749(99)70195-8. [DOI] [PubMed] [Google Scholar]
- 8.Czarnowicki T, He H, Canter T, Han J, Lefferdink R, Erickson T, et al. Evolution of pathologic T-cell subsets in patients with atopic dermatitis from infancy to adulthood. J Allergy Clin Immunol. 2020;145:215–228. doi: 10.1016/j.jaci.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiménez-Saiz R, Ellenbogen Y, Koenig JF, Gordon ME, Walker TD, Rosace D, et al. IgG1+ B-cell immunity predates IgE responses in epicutaneous sensitization to foods. Allergy. 2019;74:165–175. doi: 10.1111/all.13481. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Liu B, Huang J, He H, Zhou L, He Y, et al. Succinate and mitochondrial DNA trigger atopic march from atopic dermatitis to intestinal inflammation. J Allergy Clin Immunol. 2023;151:1050–1066.e7. doi: 10.1016/j.jaci.2022.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Kim BE, Leung DY. Pathophysiology of atopic dermatitis: clinical implications. Allergy Asthma Proc. 2019;40:84–92. doi: 10.2500/aap.2019.40.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Toit G, Sampson HA, Plaut M, Burks AW, Akdis CA, Lack G. Food allergy: update on prevention and tolerance. J Allergy Clin Immunol. 2018;141:30–40. doi: 10.1016/j.jaci.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Hui-Beckman JW, Goleva E, Berdyshev E, Leung DY. Endotypes of atopic dermatitis and food allergy. J Allergy Clin Immunol. 2023;151:26–28. doi: 10.1016/j.jaci.2022.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Ahn K, Kim BE, Kim J, Leung DY. Recent advances in atopic dermatitis. Curr Opin Immunol. 2020;66:14–21. doi: 10.1016/j.coi.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langan SM, Irvine AD, Weidinger S. Atopic dermatitis. Lancet. 2020;396:345–360. doi: 10.1016/S0140-6736(20)31286-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Kim BE, Ahn K, Leung DY. Interactions between atopic dermatitis and Staphylococcus aureus infection: clinical implications. Allergy Asthma Immunol Res. 2019;11:593–603. doi: 10.4168/aair.2019.11.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Kim BE, Berdyshev E, Bronova I, Bin L, Bae J, et al. Staphylococcus aureus causes aberrant epidermal lipid composition and skin barrier dysfunction. Allergy. 2023;78:1292–1306. doi: 10.1111/all.15640. [DOI] [PubMed] [Google Scholar]
- 18.Shi B, Leung DY, Taylor PA, Li H. Methicillin-resistant Staphylococcus aureus colonization is associated with decreased skin commensal bacteria in atopic dermatitis. J Invest Dermatol. 2018;138:1668–1671. doi: 10.1016/j.jid.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada H, Kaitani A, Izawa K, Ando T, Kamei A, Uchida S, et al. Staphylococcus aureus δ-toxin present on skin promotes the development of food allergy in a murine model. Front Immunol. 2023;14:1173069. doi: 10.3389/fimmu.2023.1173069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J, Tong P, Wang Z, Xu X, Zhao X, Meng X, et al. Staphylococcus aureus enterotoxin B is a cofactor of food allergy beyond a superantigen. J Immunol. 2023;211:1287–1297. doi: 10.4049/jimmunol.2200549. [DOI] [PubMed] [Google Scholar]
- 21.Brough HA, Simpson A, Makinson K, Hankinson J, Brown S, Douiri A, et al. Peanut allergy: effect of environmental peanut exposure in children with filaggrin loss-of-function mutations. J Allergy Clin Immunol. 2014;134:867–875.e1. doi: 10.1016/j.jaci.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenhawt M. Environmental exposure to peanut and the risk of an allergic reaction. Ann Allergy Asthma Immunol. 2018;120:476–481.e3. doi: 10.1016/j.anai.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Chalmers JR, Haines RH, Bradshaw LE, Montgomery AA, Thomas KS, Brown SJ, et al. Daily emollient during infancy for prevention of eczema: the BEEP randomised controlled trial. Lancet. 2020;395:962–972. doi: 10.1016/S0140-6736(19)32984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe A, Su J, Tang M, Lodge CJ, Matheson M, Allen KJ, et al. PEBBLES study protocol: a randomised controlled trial to prevent atopic dermatitis, food allergy and sensitisation in infants with a family history of allergic disease using a skin barrier improvement strategy. BMJ Open. 2019;9:e024594. doi: 10.1136/bmjopen-2018-024594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto-Hanada K, Kobayashi T, Mikami M, Williams HC, Saito H, Saito-Abe M, et al. Enhanced early skin treatment for atopic dermatitis in infants reduces food allergy. J Allergy Clin Immunol. 2023;152:126–135. doi: 10.1016/j.jaci.2023.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Bakker D, de Bruin-Weller M, Drylewicz J, van Wijk F, Thijs J. Biomarkers in atopic dermatitis. J Allergy Clin Immunol. 2023;151:1163–1168. doi: 10.1016/j.jaci.2023.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nat Rev Immunol. 2004;4:211–222. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berdyshev E, Kim J, Kim BE, Goleva E, Lyubchenko T, Bronova I, et al. Stratum corneum lipid and cytokine biomarkers at age 2 months predict the future onset of atopic dermatitis. J Allergy Clin Immunol. 2023;151:1307–1316. doi: 10.1016/j.jaci.2023.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Kim BE, Lee J, Han Y, Jun HY, Kim H, et al. Epidermal thymic stromal lymphopoietin predicts the development of atopic dermatitis during infancy. J Allergy Clin Immunol. 2016;137:1282–1285.e4. doi: 10.1016/j.jaci.2015.12.1306. [DOI] [PubMed] [Google Scholar]
- 30.Mortlock RD, Ma EC, Cohen JM, Damsky W. Assessment of treatment-relevant immune biomarkers in psoriasis and atopic dermatitis: toward personalized medicine in dermatology. J Invest Dermatol. 2023;143:1412–1422. doi: 10.1016/j.jid.2023.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinnov MR, Halling AS, Gerner T, Ravn NH, Knudgaard MH, Trautner S, et al. Skin biomarkers predict development of atopic dermatitis in infancy. Allergy. 2023;78:791–802. doi: 10.1111/all.15518. [DOI] [PubMed] [Google Scholar]
- 32.Kim BE, Goleva E, Kim PS, Norquest K, Bronchick C, Taylor P, et al. Side-by-side comparison of skin biopsies and skin tape stripping highlights abnormal stratum corneum in atopic dermatitis. J Invest Dermatol. 2019;139:2387–2389.e1. doi: 10.1016/j.jid.2019.03.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berdyshev E, Kim J, Kim BE, Goleva E, Lyubchenko T, Bronova I, et al. Skin biomarkers predict the development of food allergy in early life. J Allergy Clin Immunol. 2024;153:1456–1463.e4. doi: 10.1016/j.jaci.2024.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portugal-Cohen M, Kohen R. Non-invasive evaluation of skin cytokines secretion: an innovative complementary method for monitoring skin disorders. Methods. 2013;61:63–68. doi: 10.1016/j.ymeth.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Jiang R, Cudjoe E, Bojko B, Abaffy T, Pawliszyn J. A non-invasive method for in vivo skin volatile compounds sampling. Anal Chim Acta. 2013;804:111–119. doi: 10.1016/j.aca.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 36.Leung DY, Calatroni A, Zaramela LS, LeBeau PK, Dyjack N, Brar K, et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med. 2019;11:eaav2685. doi: 10.1126/scitranslmed.aav2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim BE, Howell MD, Guttman-Yassky E, Gilleaudeau PM, Cardinale IR, Boguniewicz M, et al. TNF-α downregulates filaggrin and loricrin through c-Jun N-terminal kinase: role for TNF-α antagonists to improve skin barrier. J Invest Dermatol. 2011;131:1272–1279. doi: 10.1038/jid.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, DeBenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2009;124:R7–12. doi: 10.1016/j.jaci.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Leung DY, Berdyshev E, Goleva E. Cutaneous barrier dysfunction in allergic diseases. J Allergy Clin Immunol. 2020;145:1485–1497. doi: 10.1016/j.jaci.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berdyshev E, Goleva E, Bronova I, Dyjack N, Rios C, Jung J, et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight. 2018;3:e98006. doi: 10.1172/jci.insight.98006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broccardo CJ, Mahaffey SB, Strand M, Reisdorph NA, Leung DY. Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. J Allergy Clin Immunol. 2009;124:1113–1115.e1. doi: 10.1016/j.jaci.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyjack N, Goleva E, Rios C, Kim BE, Bin L, Taylor P, et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol. 2018;141:1298–1309. doi: 10.1016/j.jaci.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman-Yassky E, Diaz A, Pavel AB, Fernandes M, Lefferdink R, Erickson T, et al. Use of tape strips to detect immune and barrier abnormalities in the skin of children with early-onset atopic dermatitis. JAMA Dermatol. 2019;155:1358–1370. doi: 10.1001/jamadermatol.2019.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwab H, Flora J, Mayrovitz HN. Impacts of skin eccrine glands on the measured values of transepidermal water loss. Cureus. 2022;14:e32266. doi: 10.7759/cureus.32266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fluhr JW, Feingold KR, Elias PM. Transepidermal water loss reflects permeability barrier status: validation in human and rodent in vivo and ex vivo models. Exp Dermatol. 2006;15:483–492. doi: 10.1111/j.1600-0625.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 46.Rogiers V EEMCO Group. EEMCO guidance for the assessment of transepidermal water loss in cosmetic sciences. Skin Pharmacol Appl Skin Physiol. 2001;14:117–128. doi: 10.1159/000056341. [DOI] [PubMed] [Google Scholar]
- 47.Pinnagoda J, Tupker RA, Agner T, Serup J. Guidelines for transepidermal water loss (TEWL) measurement. A report from the Standardization Group of the European Society of Contact Dermatitis. Contact Dermat. 1990;22:164–178. doi: 10.1111/j.1600-0536.1990.tb01553.x. [DOI] [PubMed] [Google Scholar]
- 48.Horimukai K, Morita K, Narita M, Kondo M, Kabashima S, Inoue E, et al. Transepidermal water loss measurement during infancy can predict the subsequent development of atopic dermatitis regardless of filaggrin mutations. Allergol Int. 2016;65:103–108. doi: 10.1016/j.alit.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Berents TL, Lødrup Carlsen KC, Mowinckel P, Skjerven HO, Rolfsjord LB, Bradley M, et al. Transepidermal water loss in infancy associated with atopic eczema at 2 years of age: a population-based cohort study. Br J Dermatol. 2017;177:e35–e37. doi: 10.1111/bjd.15157. [DOI] [PubMed] [Google Scholar]
- 50.Rehbinder EM, Advocaat Endre KM, Lødrup Carlsen KC, Asarnoj A, Stensby Bains KE, Berents TL, et al. Predicting skin barrier dysfunction and atopic dermatitis in early infancy. J Allergy Clin Immunol Pract. 2020;8:664–673.e5. doi: 10.1016/j.jaip.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 51.Halling AS, Rinnov MR, Ruge IF, Gerner T, Ravn NH, Knudgaard MH, et al. Skin TARC/CCL17 increase precedes the development of childhood atopic dermatitis. J Allergy Clin Immunol. 2023;151:1550–1557.e6. doi: 10.1016/j.jaci.2022.11.023. [DOI] [PubMed] [Google Scholar]
- 52.Stefanovic N, Irvine AD. Filaggrin and beyond: new insights into the skin barrier in atopic dermatitis and allergic diseases, from genetics to therapeutic perspectives. Ann Allergy Asthma Immunol. 2024;132:187–195. doi: 10.1016/j.anai.2023.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med. 2011;365:1315–1327. doi: 10.1056/NEJMra1011040. [DOI] [PubMed] [Google Scholar]
- 54.Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–513. 513.e1–507. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margolis DJ, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, Campbell LE, et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J Allergy Clin Immunol. 2012;130:912–917. doi: 10.1016/j.jaci.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, Debenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2007;120:150–155. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takai T. TSLP expression: cellular sources, triggers, and regulatory mechanisms. Allergol Int. 2012;61:3–17. doi: 10.2332/allergolint.11-RAI-0395. [DOI] [PubMed] [Google Scholar]
- 58.Liu YJ, Soumelis V, Watanabe N, Ito T, Wang YH, Malefyt RW, et al. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol. 2007;25:193–219. doi: 10.1146/annurev.immunol.25.022106.141718. [DOI] [PubMed] [Google Scholar]
- 59.Kim JH, Bae HC, Ko NY, Lee SH, Jeong SH, Lee H, et al. Thymic stromal lymphopoietin downregulates filaggrin expression by signal transducer and activator of transcription 3 (STAT3) and extracellular signal-regulated kinase (ERK) phosphorylation in keratinocytes. J Allergy Clin Immunol. 2015;136:205–208.e9. doi: 10.1016/j.jaci.2015.04.026. [DOI] [PubMed] [Google Scholar]
- 60.Margolis DJ, Kim B, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, et al. Thymic stromal lymphopoietin variation, filaggrin loss of function, and the persistence of atopic dermatitis. JAMA Dermatol. 2014;150:254–259. doi: 10.1001/jamadermatol.2013.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saeki H, Tamaki K. Thymus and activation regulated chemokine (TARC)/CCL17 and skin diseases. J Dermatol Sci. 2006;43:75–84. doi: 10.1016/j.jdermsci.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 62.Imai T, Nagira M, Takagi S, Kakizaki M, Nishimura M, Wang J, et al. Selective recruitment of CCR4-bearing Th2 cells toward antigen-presenting cells by the CC chemokines thymus and activation-regulated chemokine and macrophage-derived chemokine. Int Immunol. 1999;11:81–88. doi: 10.1093/intimm/11.1.81. [DOI] [PubMed] [Google Scholar]
- 63.Thijs J, Krastev T, Weidinger S, Buckens CF, de Bruin-Weller M, Bruijnzeel-Koomen C, et al. Biomarkers for atopic dermatitis: a systematic review and meta-analysis. Curr Opin Allergy Clin Immunol. 2015;15:453–460. doi: 10.1097/ACI.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 64.Miyahara H, Okazaki N, Nagakura T, Korematsu S, Izumi T. Elevated umbilical cord serum TARC/CCL17 levels predict the development of atopic dermatitis in infancy. Clin Exp Allergy. 2011;41:186–191. doi: 10.1111/j.1365-2222.2010.03634.x. [DOI] [PubMed] [Google Scholar]
- 65.Sato N, Yamaide F, Nakano T, Yonekura S, Okamoto Y, Shimojo N. Association of umbilical cord serum TARC/CCL17 with childhood allergies: a birth cohort study. Allergol Int. 2023;72:551–556. doi: 10.1016/j.alit.2023.04.002. [DOI] [PubMed] [Google Scholar]
- 66.Kerkhoff C, Voss A, Scholzen TE, Averill MM, Zänker KS, Bornfeldt KE. Novel insights into the role of S100A8/A9 in skin biology. Exp Dermatol. 2012;21:822–826. doi: 10.1111/j.1600-0625.2012.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saito-Sasaki N, Sawada Y. S100 proteins in the pathogenesis of psoriasis and atopic dermatitis. Diagnostics (Basel) 2023;13:3167. doi: 10.3390/diagnostics13203167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim MJ, Im MA, Lee JS, Mun JY, Kim DH, Gu A, et al. Effect of S100A8 and S100A9 on expressions of cytokine and skin barrier protein in human keratinocytes. Mol Med Rep. 2019;20:2476–2483. doi: 10.3892/mmr.2019.10454. [DOI] [PubMed] [Google Scholar]
- 69.Jin S, Park CO, Shin JU, Noh JY, Lee YS, Lee NR, et al. DAMP molecules S100A9 and S100A8 activated by IL-17A and house-dust mites are increased in atopic dermatitis. Exp Dermatol. 2014;23:938–941. doi: 10.1111/exd.12563. [DOI] [PubMed] [Google Scholar]
- 70.Biagini Myers JM, Sherenian MG, Baatyrbek Kyzy A, Alarcon R, An A, Flege Z, et al. Events in normal skin promote early-life atopic dermatitis-the MPAACH cohort. J Allergy Clin Immunol Pract. 2020;8:2285–2293.e6. doi: 10.1016/j.jaip.2020.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stamatas GN, Sato T, Chaoimh CN, Oddos T, Insel R, Hourihane JO, et al. Early skin inflammatory biomarker is predictive of development and persistence of atopic dermatitis in infants. J Allergy Clin Immunol. 2024;153:1597–1603.e4. doi: 10.1016/j.jaci.2024.02.018. [DOI] [PubMed] [Google Scholar]
- 72.Fonfara M, Hartmann J, Stölzl D, Sander N, Harder I, Rodriguez E, et al. Stratum corneum and microbial biomarkers precede and characterize childhood atopic dermatitis. J Eur Acad Dermatol Venereol. 2024 doi: 10.1111/jdv.19932. [DOI] [PubMed] [Google Scholar]
- 73.Tham EH, Chia M, Riggioni C, Nagarajan N, Common JE, Kong HH. The skin microbiome in pediatric atopic dermatitis and food allergy. Allergy. 2024;79:1470–1484. doi: 10.1111/all.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WH, Murray D, et al. Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017;139:166–172. doi: 10.1016/j.jaci.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J Invest Dermatol. 2017;137:2497–2504. doi: 10.1016/j.jid.2017.07.834. [DOI] [PubMed] [Google Scholar]
- 76.Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med. 2020;12:eaay4068. doi: 10.1126/scitranslmed.aay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goleva E, Berdyshev E, Leung DY. Epithelial barrier repair and prevention of allergy. J Clin Invest. 2019;129:1463–1474. doi: 10.1172/JCI124608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suzuki M, Ohno Y, Kihara A. Whole picture of human stratum corneum ceramides, including the chain-length diversity of long-chain bases. J Lipid Res. 2022;63:100235. doi: 10.1016/j.jlr.2022.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paller AS, Spergel JM, Mina-Osorio P, Irvine AD. The atopic march and atopic multimorbidity: many trajectories, many pathways. J Allergy Clin Immunol. 2019;143:46–55. doi: 10.1016/j.jaci.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 80.Brown SJ, Asai Y, Cordell HJ, Campbell LE, Zhao Y, Liao H, et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J Allergy Clin Immunol. 2011;127:661–667. doi: 10.1016/j.jaci.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sindher SB, Chin AR, Aghaeepour N, Prince L, Maecker H, Shaw GM, et al. Advances and potential of omics studies for understanding the development of food allergy. Front Allergy. 2023;4:1149008. doi: 10.3389/falgy.2023.1149008. [DOI] [PMC free article] [PubMed] [Google Scholar]