Abstract
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors of the digestive system. RNA methylation plays an important role in tumorigenesis and metastasis, which could alter gene expression and even function at multiple levels, such as RNA splicing, stability, translocation, and translation. In this study, we aimed to conduct a comprehensive analysis of RNA methylation-related genes (RMGs) in HCC and their relationship with survival and clinical features.
Methods
A retrospective analysis was performed using publicly available HCC-related datasets. The differentially expressed genes (DEGs) between HCC and controls were identified from TCGA-LlHC and intersected with RMGs to obtain differentially expressed RNA methylation-related genes (DERMGs). Regression analysis was used to screen for prognostic genes and construct risk models. Simultaneously, clinical, immune infiltration and therapeutic efficacy analyses were performed. Finally, multivariate cox regression was used to identify independent risk factors, and quantitative real-time polymerase chain reaction (qRT-PCR) was used to validate the expression levels of the core genes of the model.
Results
A 21-gene risk model for HCC was established with excellent performance based on ROC curves and survival analysis. Risk scores correlated with tumor grade, pathologic T, and TNM stage. Immune infiltration analysis showed correlations with immune scores, 11 immune cells, and 30 immune checkpoints. Low-risk patients showed a higher susceptibility to immunotherapy. The risk score and TNM stage were independent prognostic factors. qRT-PCR confirmed higher expression of PRDM9, ALPP, and GAD1 in HCC.
Conclusions
This study identified RNA methylation-related signature genes in HCC and constructed a risk model that predicts patient outcomes and reflects the immune microenvironment. Prognostic genes are involved in complex regulatory mechanisms, which may be useful for cancer diagnosis, prognosis, and therapy.
Keywords: hepatocellular carcinoma, RNA methylation-related genes, risk score, prognosis, immune infiltration
Introduction
Primary hepatocellular carcinoma (HCC) is the sixth most common malignancy worldwide and also the third leading cause of cancer-related deaths, which poses a significant threat to human health. 1 A multitude of factors contribute to the development of HCC, including hepatitis virus infection, alcohol abuse, non-alcoholic fatty liver disease, and aflatoxin B1. 2 Although surgery-based comprehensive treatment is currently the main therapeutic modality for HCC, patient outcomes remain unsatisfactory, which poses a greater challenge to clinical caregivers. 3 Immune intervention represents a novel breakthrough in the treatment paradigm of HCC in recent years, mainly by reducing T cell-mediated tumor immune escape and improving local immune cell response. However, the effective immune targets for HCC are limited.3–5 The type and abundance of tumor-infiltrating immune cells have been reported to influence patient disease prognosis.6–8 Consequently, it is of significant and imperative importance to investigate tumor-related immune genes and develop novel HCC markers and models in order to provide suitable therapeutic strategies and enhance the prognosis of patients.
RNA methylation encompasses a diverse array of chemical modifications, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), N7-methylguanosine (m7G), 5-methylcytosine (m5C), and 5-hydroxymethylcytosine (hm5C). These modifications occur at distinct positions on RNA molecules, collectively referred to as the methylation phenomenon. One of the most prevalent modifications observed in mRNA of higher organisms is the methylation of the sixth nitrogen atom of adenine (m6A). The evidence is mounting that this process of RNA methylation is intimately linked to cancer cell proliferation, cellular stress, metastasis, and the immune response. Moreover, RNA methylation-related proteins have emerged as a promising target for cancer therapy. In HCC, YTHDF2, a protein that recognizes methylation, was demonstrated to stimulate the liver cancer stem cell phenotype and metastasis by regulating OCT4 expression via m6A RNA methylation. 9 The m1A-related signals are markedly elevated in HCC patient tumor tissues and liver cancer stem cells (CSCs), and TRMT6/TRMT61A enhances m1A methylation in a subset of tRNA, thereby activating Hedgehog signaling and driving the self-renewal of liver CSCs and tumorigenesis. 10 The m7G methyltransferase WD repeat domain 4 (WDR4) has been demonstrated to promote HCC cell proliferation by inducing the G2/M cell cycle transition and inhibiting apoptosis, in addition to enhancing metastasis and sorafenib resistance through epithelial-mesenchymal transition (EMT). 11 However, the impact of RNA methylation-related genes (RMGs) on the prognosis and treatment of HCC patients remains to be fully elucidated.
The objective of this study is to investigate RNA methylation-related markers that affect the prognosis and treatment of HCC patients and to explore the potential mechanisms regulating HCC.
Materials and Methods
Study Design
In this study, we constructed prognostic features associated with RNA methylation in HCC based on the TCGA-LIHC dataset and RMGs. It also explored the impact of prognostic genes on immune cell infiltration, immunotherapy, etc in HCC. Then the expression of core genes was detected by quantitative real-time polymerase chain reaction (qRT-PCR). The detailed study is shown in the Supplementary Figure 1. The reporting of this study conforms to REMARK guidelines. 12
Data Source
The clinical data, transcription profiles of 365 HCC patients, and 50 controls were downloaded from the TCGA database and utilized as the training set (TCGA-LIHC dataset). The GSE14520 dataset, comprising 212 samples of HCC, was employed as an external validation set. A total of 2007 RMGs were identified from the GeneCards database (https://www.genecards.org/).
Acquirement of Differentially Expressed Genes (DEGs)
The sequencing data of expression were analysed by R software after normalization. The R package “DEseq2” was used to identify DEGs between controls and HCC samples. This was achieved by setting a threshold of |log2FC| > 1.5 and a P.adj.val < 0.05. 13
Functional Enrichment Analysis and Protein-Protein Interaction (PPI) Network Construction
To screen for genes associated with RNA methylation in DEGs, differentially expressed RMGs were obtained by overlapping the DEGs with the RMGs. Furthermore, to elucidate the functions and pathways that involved in DERMGs, GO annotation and KEGG enrichment analysis were conducted using the “clusterProfiler” package.14,15 The GO terms were comprised of the following three categories: biological process (BP), cellular component (CC), and molecular function (MF). While the KEGG pathway enrichment analysis was prone to describe gene function at the genomic and molecular levels and show the correlated genes. The adjusted P-value (Adj.P.Val) was set at 0.05, which was regarded as the threshold for statistical significance. Subsequently, we conducted a pathway interconnection analysis using ClueGO.
To investigate the interactions between the aforementioned DERMGs at the protein level, we employed the Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org/) to construct a PPI network. 16 Then, Cytoscape plugin-MCODE was used to screen the significant modules in the PPI network, with the degree cut off = 2, node score cut off = 0.2, k-core cut off = 2, and max. depth cut off = 100.
Establishment and Validation of the Prognostic Model
A total of 365 HCC samples with survival information from TCGA-LIHC were utilized as the training set to construct the risk model. A univariate Cox regression analysis was employed to identify prognosis-related genes with a p-value of less than 0.05. These genes were then subjected to least absolute shrinkage and selection operator (LASSO) analysis using glmnet16 to further refine the prognostic genes and construct prognostic models. 17 The patients were divided into distinct risk groups based on the median risk score. Subsequently, Kaplan-Meier (K-M) survival curves were plotted using the survival package in R to assess differences in patient survival between risk groups. 18 The predictive power of the model was then assessed by plotting receiver operating characteristic (ROC) curves using the survival ROC software. 19 The external validation set was employed to test the efficacy of the risk model. The relationships between the risk score and the clinical characteristics were analysed with the Wilcoxon test and Kruskal-Wallis test in the TCGA-LIHC dataset.
Hierarchical Analysis and Independent Prognostic Analyses
To assess the prognostic value of clinical features (tumor grade, pathologic T, cancer status, TNM stage, gender, age, inflammation, and BMI), we evaluated the level of difference in risk scores between subtypes of clinical features. Subsequently, we conducted a multivariate cox regression analysis to identify independent prognostic factors for HCC patients. A nomogram for predicting 1-, 3-, and 5-year survival of HCC patients was established using rms, based on independent prognostic factors. 20 The performance of the nomogram was evaluated by a calibration curve.
Gene Set Enrichment Analysis (GSEA)
In order to further elucidate the functions and pathways involved in differential genes between high- and low-risk groups, DEseq2 was initially employed to analyze the discrepancy between LIHC samples in the training set belonging to the high- and low-risk groups. The discrepancy was then calculated and sorted from the largest to the smallest according to log2FC. Then, the c2.cp.kegg.v2023.1.Hs.symbols.gmt in Molecular Signatures Database (MSigDB, https://www.gsea-msigdb.org) was used as the background gene set to perform GSEA. The signal pathway with p.adjust < 0.05 was significantly enriched. Subsequently, enrichplot was employed to show the top five signaling pathways with the largest and smallest normalized enrichment scores (NES). 21
Evaluation of the Immune Microenvironment (IME) Landscape
The immune score, stromal score, and estimate score have been demonstrated to be effective in predicting the content of immune and stromal components in tumors or diseases. The immune and stromal scores of HCC samples were calculated using the ESTIMATE algorithm, 22 which was provided in the ESTIMATE. The estimate score was obtained by adding the immune score and stromal score and was also used to assess tumor purity. Subsequently, the CIBERSORT algorithm was employed to assess the extent of immune infiltration in patients and to identify differential immune cells between high- and low-risk groups. Additionally, the correlation between differential immune cells was calculated. Furthermore, we conducted a comparative analysis of the differential expression of immune checkpoint molecules between the high- and low-risk groups.
Drug Sensitivity Analysis
The sensitivity of each patient to chemotherapeutic agents was predicted by pRRophetic, which included eight chemotherapy drugs. 23 The estimated half-maximal inhibitory concentration (IC50) value for each patient treated with a specific chemotherapy drug was obtained through the function “pRRopheticPredict”. Subsequently, the correlation between risk scores and chemotherapy drugs was calculated.
Evaluation of the Therapeutic Efficacy
To assess treatment sensitivity between two risk groups of patients, we evaluated the effect of immunotherapy between the different groups. The current ImmunoScale Score (IPS) has been shown to predict patient response to immune checkpoint inhibitor (ICI) therapy, downloaded from The Cancer Immunome Atlas (TCIA, https://tcia.at). Higher IPS scores indicate a better response to immunotherapy. Tumor Immune Dysfunction and Exclusion (TIDE, http://tide.dfci.harvard.edu/), which is employed to assess mechanisms of immune evasion, is another powerful biomarker designed to predict response to immunotherapy. Higher TIDE scores imply that tumor cells are more likely to evade immune surveillance, suggesting a lower rate of response to immunotherapy. TIDE scores were obtained after offloading input data as described in the instructions. Furthermore, an immunotherapy cohort (IMvigor210 cohort) was included to validate tumor response to PD-L1 blockade. 24 The survival impact of patients in risk group in IMvigor210 was assessed by K-M survival curves and the validity of risk score in this cohort was by ROC curves.
Clinical Samples Collection
In this retrospective study, we consecutively collected 10 patients with HCC tumor and adjacent tissues who underwent partial hepatectomy from September 14, 2023, to December 31, 2023 (after ethical approval). Samples were stored at −80 ° C with liquid nitrogen. The inclusion criteria were: (1) pathologically confirmed HCC diagnosis; (2) no neoadjuvant interventional therapy, radiotherapy, immunotherapy or targeted therapy before surgery. We excluded patients who had: (1) merged with other systemic malignant tumors; (2) incomplete resection. All patient details have been de-identified. This study was approved by the Ethics Committee of Hainan General Hospital, Hainan Medical University. All participants were required to sign a written informed consent form, and the relevant information was used for medical research purposes. All methods were carried out in accordance with relevant guidelines and regulations.
Expression Validation of Core Genes
Tissue samples were collected from 10 patients with HCC tumor and adjacent tissues. The LASSO regression model was ranked according to the absolute value of the coefficients, and the top five genes were selected as core genes for qRT-PCR experiments to verify their expression. All samples were lysed with TRIzol Reagent, and total RNA was isolated following the manufacturer's instructions. The extracted RNA was reverse-transcribed to cDNA using the SureScript-First-strand-cDNA-synthesis-kit before qRT-PCR. The qRT-PCR reaction consisted of 3 µl of reverse transcription product, 5 µL of 2xUniversal Blue SYBR Green qPCR Master Mix, and 1 µl each of forward and reverse primer. The PCR was performed in a BIO-RAD CFX96 Touch TM PCR detection system (Bio-Rad Laboratories, Inc., USA) under the following conditions: initial denaturation at 95°Cfor 1 min, followed by 40 cycles that each involved incubation at 95 °C for 20 s, 55 °C for 20 s, and 72 °C for 30 s. All primers were synthesized by Servicebio (Servicebio, Wuhan, China). The detailed forward and reverse primers are presented in Table 1. The GAPDH gene served as an internal control, and the relative expression of five biomarkers was determined using the 2−ΔΔCt method. The experiment was repeated in triplicate on independent occasions.
Table 1.
the Primer Sequences of Genes in qRT-PCR
| Primer | Sequence |
|---|---|
| PRDM9 F | AGGTCAAGCCTCTGCTATCAC |
| PRDM9 R | GTCCCCCGAACACTTACAGA |
| SLC22A6 F | CATGACTGCCGAGCTCTACC |
| SLC22A6 R | ATATCCCTGCTTCTTTCTGAGTGG |
| ALPP F | AACGGTCCAGGCTATGTGCT |
| ALPP R | CATGACGTGCGCTATGAAGG |
| GAD1 F | GCGGACCCCAATACCACTAAC |
| GAD1 R | CACAAGGCGACTCTTCTCTTC |
| MYCN F | GTCGCAGAAACCACAACATCC |
| MYCN R | GGCGGCCTTCTCATTCTTTAC |
| GAPDH F | CGAAGGTGGAGTCAACGGATTT |
| GAPDH R | ATGGGTGGAATCATATTGGAAC |
Statistical Analysis
All of this study was analysed using R language. A differential analysis was conducted between the HCC and control samples using the DESeq2. The LASSO regression analysis was performed using the glmnet. An enrichment analysis was conducted using the clusterProfiler. The ROC curves and K-M survival curves were plotted using the survivalROC and survminer, respectively. The IC50 values of the drugs were assessed using the pRRophetic. The statistical differences in the core genes between HCC samples and adjacent samples were detected by paired t-test using GraphPad Prism 5, and the level of statistical significance was tested and expressed * as P < 0.05, ** means P < 0.01, *** means P < 0.001.
Results
Identification and Functional Enrichment of 185 DERMGs
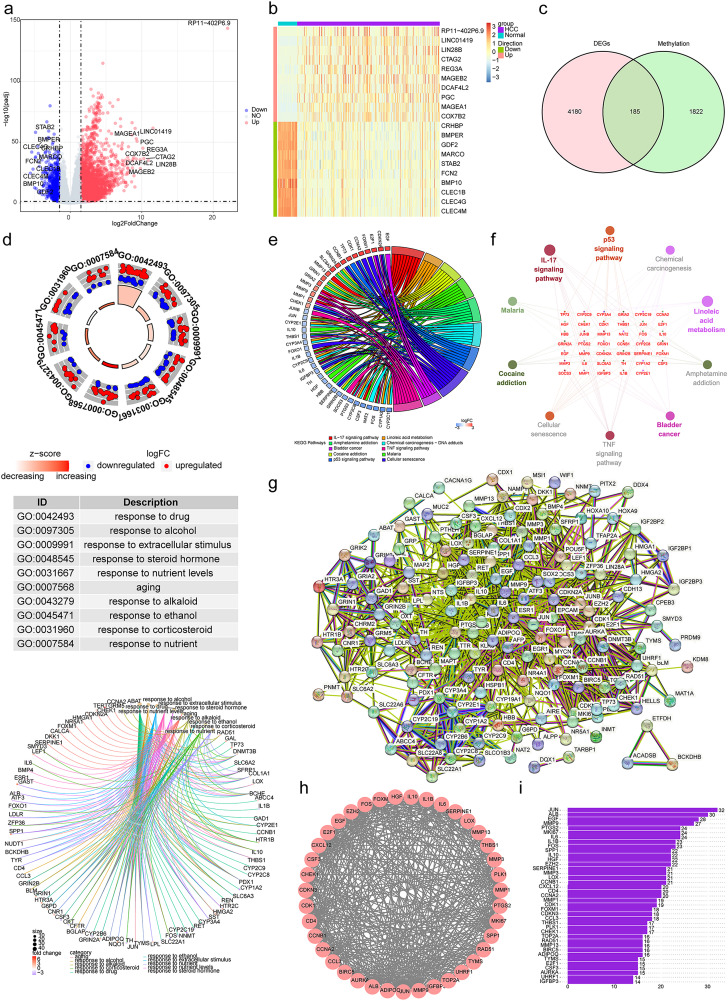
By comparing tumor and normal tissue samples, 4365 DEGs including 3615 upregulated genes and 750 downregulated genes were detected as shown in Figure 1a, and b showed the expression of the top 10 up-regulated and down-regulated DEGs by heatmap. After overlapping DEGs with 2007 RMGs, we obtained 185 DERMGs (Figure 1c). Further, GO annotation and KEGG enrichment analysis were conducted. GO function analysis of these 185 genes showed that they were involved in aging, response to alcohol, and response to alkaloid (Figure 1d). KEGG pathway analysis showed that they were involved in IL-17 signaling pathway, linoleic acid metabolism, and cocaine addiction (Figure 1e). Then we analyzed the distribution of the functions and pathways among the core genes. The enrichment analysis indicated that these genes were significantly enriched in IL-17 signaling pathway and linoleic acid metabolism (Figure 1f).
Figure 1.
Differential, enrichment analysis and construction of protein-protein interaction (PPI) network. (a) The volcano map of differentially expressed genes (DEGs) between hepatocellular carcinoma (HCC) and control groups, including 3615 upregulated genes and 750 downregulated genes. Red dots represent up-regulated genes, blue dots represent down-regulated genes, and grey indicates non-significant genes. (b) The heat map of top10 up- and down-regulated DEGs in HCC and control groups. (c) The venn diagram of 185 RNA methylation-related DEGs (DERMGs) via overlap of DEGs and RNA methylation-related genes (RMGs). (d) The circular graph and chord diagram of top 10 most significant biological functions enriched by Gene Ontology (GO) annotation. (e, f) The string diagram (e) and cluego network diagram (f) of the top 10 most significant pathways enriched by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. (g) The PPI network of 185 DERMGs. (h) By using MCODE plugin, 39 potential hub genes were screened as key modules. (i) The bar chart showed the number of protein interactions for each of the 39 genes.
Obtainment of 39 Potential hub Genes via PPI
We constructed PPI networks to specify the interactions between DERMGs (Figure 1). Further, 39 genes were selected as potential hub genes via MCODE plugin (Figure 1 h). Then we analyzed the interactions among 39 genes, and JUN had the most interactions with others (Figure 1i).
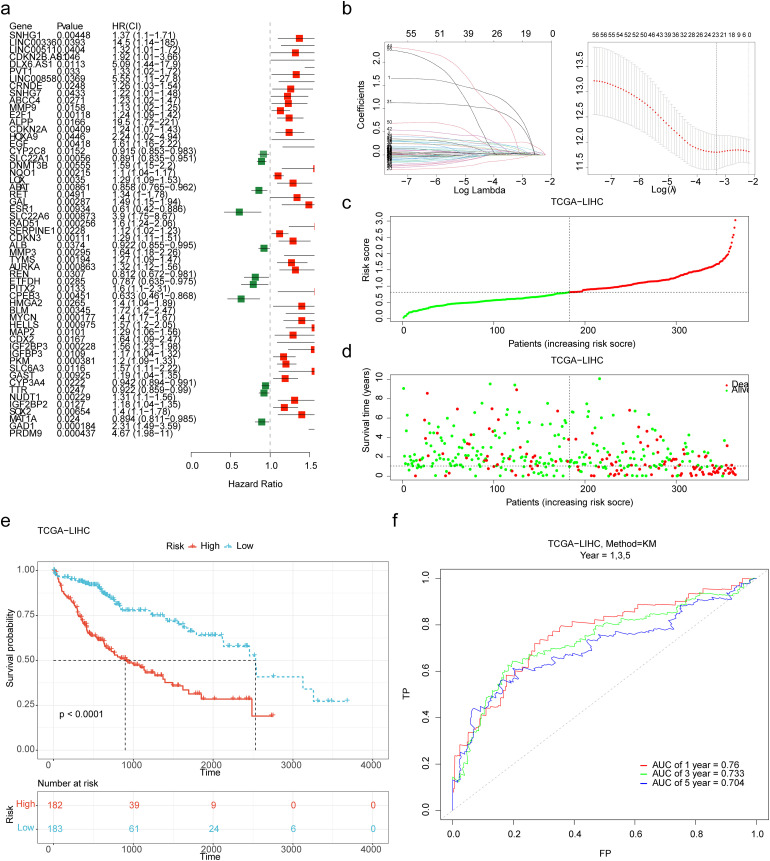
Establishment and Validation of the Effective HCC Prognostic Model
The results of univariate cox regression analysis showed that 56 DERMGs were correlated with overall survival (OS) of HCC patients (p < 0.05) (Figure 2a). Next, 21 prognostic genes screened by LASSO analysis (Figure 2b), PRDM9, GAD1, SOX2, SLC6A3, IGF2BP3, MAP2, MYCN, CPEB3, AURKA, MMP3, SERPINE1, SLC22A6, GAL, RET, LOX, NQO1, SLC22A1, EGF, CDKN2A, ALPP and E2F1 were identified and were selected to develop a prognostic model. The risk score = (0.986 × expression of PRDM9) + (0.273 × expression of GAD1) + (0.054 × expression of SOX2) + (0.103 ×expression of SLC6A3) + (0.028 × expression of IGFBP3) + (0.071 × expression of MAP2) + (0.190 × expression of MYCN) +(− 0.084 × expression of CPEB3) + (0.039 × expression of AURKA) + (0.060 × expression of MMP3) + (0.030 × expression of SERPINE1) + (0.769 × expression of SLC22A6) +(0.148 × expression of GAL) + (0.180 × expression of RET) + (0.066 ×expression of LOX) + (0.033 × expression of NQO1) + (− 0.015 ×expression of SLC22A1) + (0.024× expression of EGF) + (0.023× expression of CDKN2A) + (0.440 × expression of ALPP) + (0.043 × expression of E2F1).
Figure 2.
Establishment of the prognostic model based on 21 prognostic genes. (a) The 56 DERMGs associated with overall survival (OS) of HCC patients were obtained by univariate cox regression analysis. (b) The plot of gene coefficients and error plots for 10-fold cross-validation in least absolute shrinkage and selection operator (LASSO) analysis. (c) The distribution of the risk score in the high- and low-risk groups. (d) The survival status distribution of samples in the high- and low-risk groups. (e) Kaplan-Meier survival curve demonstrated that the high-risk group showed poor overall survival compared to the low-risk group. (f) The receiver operating characteristic (ROC) curves of the prognostic model with 1, 3, 5 years as survival time points. AUC, area under the curve.
The distribution of the risk score and the survival status in the high- and low-risk groups are presented in Figure 2c-d. The K-M survival curve demonstrated that the high-risk group showed poor OS (Figure 2e). The ROC curve showed that the risk score had good performance in predicting patient's survival status, the AUC of 1-, 3-, 5-year were 0.760, 0.733, and 0.704 (Figure 2f). In the validation set, the distribution of the risk score and the survival status in the high- and low-risk groups were presented in Supplementary Figure 2a-b. K-M analysis also showed a significant difference in survival status of patients between the two risk groups (Supplementary Figure 2c). The ROC curve displayed that the risk score had good performance in predicting patient's survival status of 1-, 3-, and 5- year, the AUC values were 0.630, 0.657, and 0.660, respectively (Supplementary Figure 2d).
In addition, we analyzed the expression of model genes between HCC and controls in the training and validation sets, and the expression trends of model genes in the training and the validation sets were consistent (Supplementary Figure 3a-b).
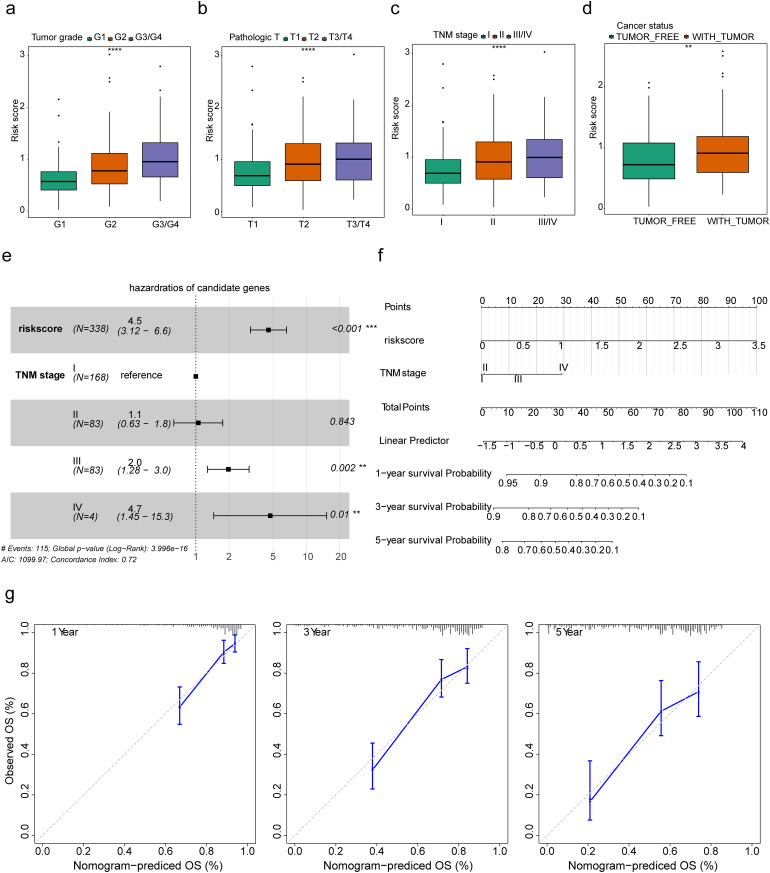
Construction of a Nomogram That Effectively Predicts Survival in HCC Patients
We assessed the relevance between the risk score and clinicopathological traits. The risk score was significantly increased in tumor grade, pathologic T, and TNM stage (Figure 3a-c), the risk score was significantly different between tumor free and with tumor group in cancer status. (Figure 3d). Further, we performed multivariate analysis to detect independent prognostic factors. Risk score, tumor grade, TNM stage, and age were included in multivariate analysis, the result indicated that risk score and stage were independent prognostic factors in HCC (Figure 3e). Thereafter, we constructed a nomogram to predict the 1-, 3-, and 5-year survival of HCC patients by using risk score and TNM stage (Figure 3f). The calibration curves for 1-, 3-, and 5-year (Figure 3 g) showed that the nomogram-predicted probability of survival was close to the actual survival.
Figure 3.
Clinical features and independent prognostic analyses. (a-d) Comparison of the risk score among different subtypes for tumor grade, pathologic T, TNM stage, and cancer status. **p < 0.01; ****p < 0.0001. (e) The result of multivariate analysis indicated that risk score and stage were independent prognostic factors in HCC. ***p < 0.001. (f) The nomogram was constructed based on risk score and TNM stage to predict the 1-, 3-, and 5-year survival of HCC patients. (g) The calibration curves of the nomogram show that the model has validity and accuracy.
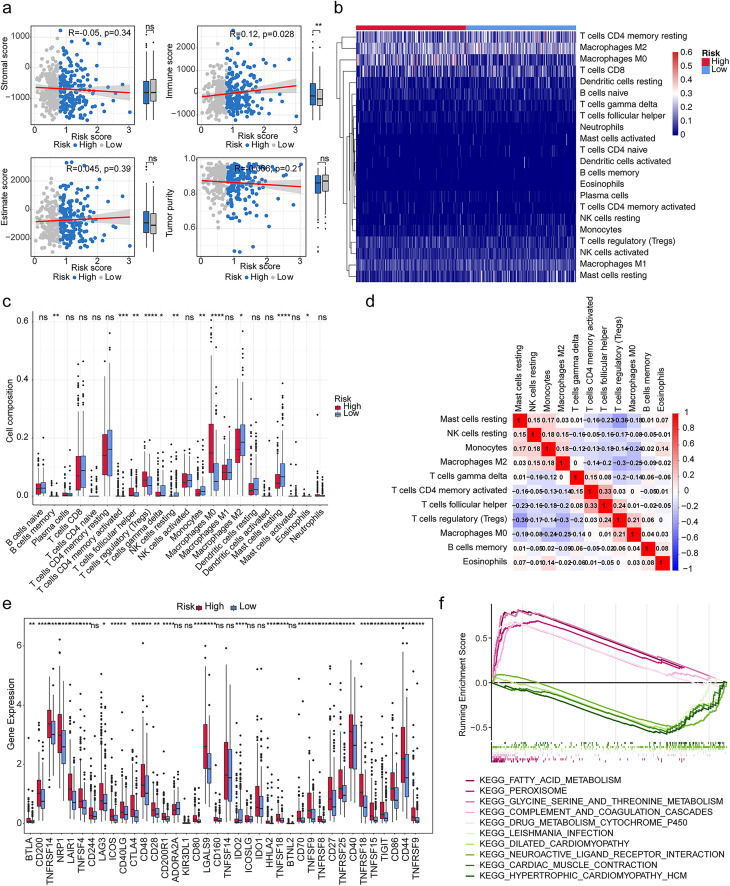
Certain Correlation of Risk Module with IME Landscape
We calculated the immune score, stromal score, estimate score, tumor purity, and their correlation with risk scores. The results showed that the immune score (cor = 0.12) was remarkably positively correlated with risk score (p < 0.05) (Figure 4a). Then, we used CIBERSORT databases to assess the percentage of immune infiltrating cells in HCC patients (Figure 4b). We obtained 11 differential immune cells. The difference of immune cells in high and low-risk groups mainly included B cells memory, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), T cells gamma delta, NK cells resting, Monocytes, Macrophages M0, Macrophages M2, Mast cells resting and Eosinophils (Figure 4c). We calculated the correlations between the differential immune infiltrating cells. The results indicated that T cells regulatory (Tregs) and Mast cells resting (cor = −0.36) had significantly negative correlations (Figure 4d). Furthermore, we compared the expression of 48 immune checkpoints between high-risk and low-risk groups, and the results showed that 40 immune checkpoint molecules were significantly different between high and low-risk groups, except CD244, ADORA2A, KIR3DL1, CD160, TNFSF14, ICOSLG, IDO1 and BTNL2 (Figure 4e).
Figure 4.
Immune infiltration analysis and enrichment analysis. (a) Comparison of stromal score, immune score, estimate score, and tumor purity between high- and low-risk groups. ns, not significant; *p < 0.05; **p < 0.01. (b) The heat map of immune infiltrating cells in HCC patients. (c) The discrepancies of the cell composition of immune cells in high and low-risk groups. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (d) The correlation analysis among differential immune cells. (e) The expression of 48 immune checkpoints was compared between high-risk and low-risk groups. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (f) Enrichment of multiple signaling pathways involved in differential genes between high and low risk groups.
Enrichment of multiple signaling pathways involved in differential genes between different risk groups
We further probed the signaling pathway of differential genes between high and low risk groups. These genes were found to be markedly enriched in fatty acid metabolism, peroxisome, glycine-serine and threonine metabolism, complement and coagulation cascade, drug metabolism cytochrome P450, and neuroactive ligand receptor interactions (Figure 4f).
Response and Outcome to Chemotherapeutic Agents and Treatment Efficacy Varies among Patients in Different Risk Groups
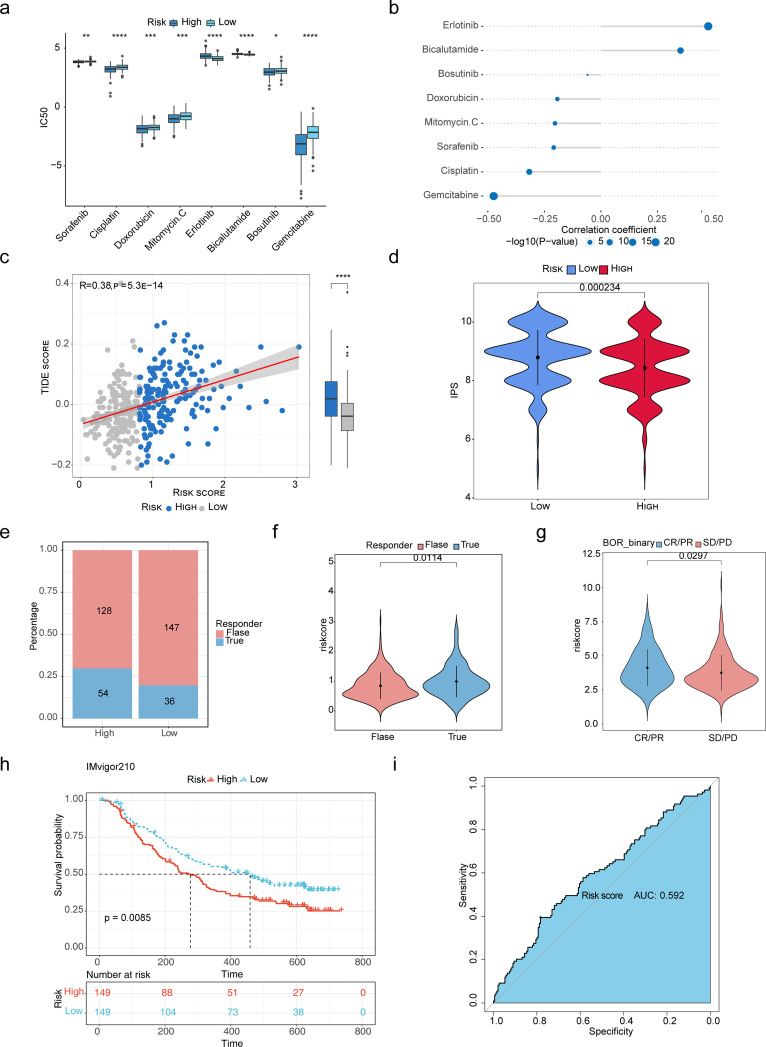
To explore the relationship between risk score and chemoresistance, the IC50 was used to predict the treatment response to 8 drugs in the TCGA-LIHC cohort. Among them, low-risk score samples were more sensitive to Sorafenib, Cisplatin, Doxorubicin, Mitomycin.C, Bosutinib, and Gemcitabine (Figure 5a). The result of the correlation between IC50 and risk score showed that Erlotinib had the strongest positive correlation to risk score, and Gemcitabine had the strongest negative correlation to risk score (Figure 5b).
Figure 5.
Assessment of immunotherapy response and chemoresistance in high- and low-risk groups. (a) The low-risk score samples were more sensitive to Sorafenib, Cisplatin, Doxorubicin, Mitomycin, Bosutinib, and Gemcitabine. *p < 0.05; ** p < 0.01; ***p < 0.001; ****p < 0.0001. (b) The result of the correlation between IC50 and risk score showed that Erlotinib had the strongest positive correlation to risk score, and Gemcitabine had the strongest negative correlation to risk score. (c) The risk score was significantly positively correlated to TIDE scores. ****p < 0.0001. (d) Patients in the high-risk group tended to achieve lower IPS scores in the TCGA cohort. (e) The proportion of people responding to PD-L1 therapy in the high-risk score group was greater than that of the low-risk score group. (f, g) The discrepancies of risk score between samples in the “TURE” and “FALSE” groups (f), and between the complete response (CR)/partial response (PR) and stable disease (SD)/progressive disease (PD) group (g). (h) Kaplan-Meier survival analysis of high- and low-risk groups in the IMvigor210 database. (i) The ROC curve of risk score in the IMvigor210 database.
We further explored the predictive potential of the risk model for immunotherapy response by analyzing the correlation between risk model and recognized immunotherapy predictors. We discovered that risk scores were significantly positively correlated to TIDE scores (Figure 5c). Moreover, patients in the high-risk group tended to achieve lower IPS scores in the TCGA cohort (Figure 5d).
The potential PD-LI response was predicted using the TIDE algorithm. The results suggested that the proportion of people who responded to PD-L1 therapy in the high-risk score group was greater than that of the low-risk score group (Figure 5e), and the “TURE” group had a significantly higher risk score than the “FALSE” group (Figure 5f). In addition, we verified the reliability of the risk score in predicting the benefit of immunotherapy in HCC patients receiving anti-PD-L1 immunotherapy in IMvigor210 cohorts. The complete response (CR)/partial response (PR) group had a high-risk score than the stable disease (SD)/progressive disease (PD) group (Figure 5), and the low-risk score group had a longer survival time after anti-PD-L1 therapy than the high-risk score group (Figure 5 h). The ROC curve suggested that the risk score could moderately predict the benefit of anti-PD-L1 therapy (AUC = 0.592) Figure 5i.
Validation of the Core Genes by qRT-PCR
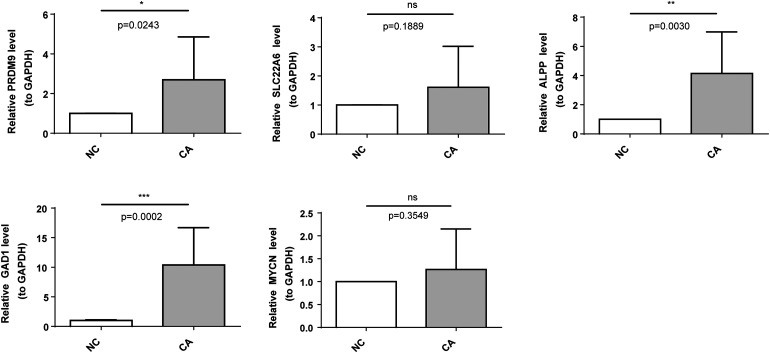
To further validate the differences of model genes in HCC and control samples, we selected five core genes (PRDM9, SLC22A6, ALPP, GAD1, and MYCN) to probe their expression by qRT-PCR. Figure 6 showed that the expression of five core genes was higher in HCC than in the control group, in which there were significant differences in PRDM9, ALPP and GAD1 (p < 0.05). The expression trends of these five genes in qRT-PCR remained consistent with those in the TCGA-LIHC dataset. As for the two genes that were not statistically significant between HCC and control groups, we believed that this might be due to the effects of sample size and individual variation.
Figure 6.
Validation of the expression levels of signature core genes by quantitative real-time polymerase chain reaction (qrt-PCR). ns, not significant; *p < 0.05; ** p < 0.01; ***p < 0.001.
Discussion
Methylation modifications of RNA play an integral role in post-transcriptional regulation. In recent years, an increasing number of studies have demonstrated the significance of RNA methylation modifications in tumor invasion and metastasis, as well as in the IME. These modifications alter molecular functions through interactions with different methylation regulators, exerting tumor-promoting or suppressive effects. Nevertheless, the functions and mechanisms of RMGs in HCC remain to be fully elucidated. In this study, we constructed an RNA methylation risk model and analyzed the correlation between HCC and tumor immunity, with the intention of providing new ideas for the development of effective modern immunotherapeutic approaches.
Functional Enrichment and Pathway Analysis of 185 DERMGs
A total of 4365 genes were identified as differentially expressed in cancer samples and control samples in this study. We crossed the 2004 RMGs identified with the differential genes to obtain 185 DERMGs. The biological processes and signaling pathways in which they were involved were explored, and it was found that they were mainly involved in the IL-17 signaling pathway, the p53 signaling pathway, and cellular senescence, indicating that these DERMGs play an important role in HCC. As reported in the relevant literature, IL-17A acts as an important regulator of the inflammatory response of macrophages (blast cells and bone marrow-derived monocytes) and cholesterol synthesis in steatosis hepatocytes, as well as a tumor-promoting cytokine in alcohol-induced HCC. 25 The tumor suppressor protein p53 maintains cholesterol ester homeostasis by inhibiting the ubiquitin-specific peptidase 19 (USP19) and sterol O-acyltransferase (SOAT) 1. Dysregulation of P53 activates the P53-USP19-SOAT1 signaling axis, which increases cholesterol esterification and is involved in the development of HCC. 26 In addition, cellular senescence affects energy metabolism, immune infiltration, chemokines, expression of immune checkpoint-related genes, and immunotherapeutic response in HCC. 27 The above evidence suggests that these DERMGs may promote HCC through the above signaling pathways.
Establishment of Risk Models Based on 21 Prognostic Genes and Related Research Reports
Subsequently, by one-way cox and LASSO regression analysis, we obtained 21 RNA methylation-related markers associated with the outcome of HCC patients. These markers were utilized to construct a risk model and column plot, which enabled more accurate prediction of the prognosis of HCC. In part of them, GAD1, SOX2, IGF2BP3, MYCN, AURKA, MMP3, LOX, NQO1, EGF, CDKN2A, E2F1 were up-regulated and CPEB3, SLC22A1 were down-regulated in HCC samples, which is consistent with previous studies.28–42 In RNA methylation-related research, the N6-methyladenosine (m6A) modification is activated by AURKA through the inhibition of METTL14 ubiquitination and degradation, while AURKA binding to the DROSHA transcript further enhances the attachment of the m6A reader IGF2BP2, which serves to stabilize the m6A modification of DROSHA. 43 The mRNA decay of CDKN2A is mediated by the recruitment of the m6A reader YTHDF2 and its direct interaction via circMET. 44 IGF2BP3 functions as a recognition protein for RNA N6-methyladenosine and promotes the stability and translation of target mRNA. 45 In addition, the NQO1/p53/SREBP1 axis, which is mediated by NQO1, promotes the progression and metastasis of HCC by regulating snail stability. 38 The E2F1-mediated upregulation of AUF1 has been demonstrated to promote the development of HCC and enhance drug resistance via the stabilization of AKR1B10. 46 The SOX2 expression predicts poor survival of HCC or post-hepatectomy patients and it promotes HCC cell invasion by activating Slug.29,47,48 The above indicates that these RNA methylation-related markers may exert a potential influence on the development, invasion, metastasis, and prognosis of HCC patients.
Comparison of Immune Cell Infiltration Characteristics in Different Risk Modules
Subsequently, the patients were divided into two risk groups based on risk scores and the immunological characteristics of the high- and low-risk groups were explored. The results of the immune analysis suggest that the immune cells that exhibited the most significant differences between the high- and low-risk groups were mainly B cells memory, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), T cells gamma delta, NK cells resting, Monocytes, Macrophages M0, Macrophages M2, Mast cells resting and Eosinophils. Among these, T cells regulatory (Tregs) and Mast cells resting showed significantly negative correlations. These differential immune cells may exert a potential influence in TME of HCC. Studies have demonstrated that regulatory T cells (Tregs) represent a subset of T cells that regulate autoimmune responsiveness within the body. These cells were previously known as suppressor T cells, and they are involved in a multitude of immune regulatory processes. Peripheral Tregs upregulate checkpoint inhibitors and contribute to systemic immune dysfunction and antitumor activity by several inhibitory pathways including PDL1/CTLA-4. This process is presumed to facilitate HCC development at young age. 49 Macrophages are derived from monocytes. Macrophages M2 represent a significant proportion of tumor mesenchymal cells and are an essential component of the tumor microenvironment (TME). They interact with tumor cells and secrete a range of cytokines including EGF, VEGF, PDGF, MMP family, and CCL family proteins. 50 Tumor-associated macrophages (TAMs) can be stimulated to transform M2-like polarization via canonical Wnt/β-catenin signaling by tumor cells-derived Wnt ligands, which results in tumor growth, migration, metastasis, and immunosuppression in HCC. 51 Additionally, eosinophils can readily respond to diverse stimuli and are capable of synthesizing and secreting numerous molecules, including unique granule proteins that may kill tumor cells. Alternatively, they can secrete soluble mediators of pro-angiogenesis and matrix remodeling that facilitate tumor growth. 52 Furthermore, tumor-associated neutrophils have been reported that they can recruit macrophages and Tregs to promote the progression of HCC and resistance to sorafenib. 53 The above findings further suggest that these differences in immune cells may influence the prognosis of patients with HCC.
Prediction Analysis of Response to Chemotherapy and Immunotherapy and Clinical Outcomes in Patients with Different Risk Groups
Drug sensitivity analysis revealed that the low-risk group exhibited greater sensitivity to Sorafenib, Cisplatin, Doxorubicin, Mitomycin C, Bosutinib, and Gemcitabine than the high-risk group. Conversely, Erlotinib and Bicalutamide showed higher sensitivity in the high-risk group. These agents are commonly used in the clinical treatment of hepatocellular carcinoma (HCC), with evidence supporting their efficacy in this indication.54–56 Our results also demonstrate their value in the application of HCC. To assess the reliability of the risk score in predicting immunotherapeutic efficacy, the results of TIDE and IPS scores indicated that patients in the high-risk group were more likely to develop immune escape and exhibit lower immunogenicity. The proportion of patients who responded to PD-L1 therapy in the high-risk group was greater than that of the low-risk group. The low-risk group exhibited a longer survival time following anti-PD-L1 therapy than the high-risk group. Furthermore, the ROC curve indicated that the risk score could predict the benefit of anti-PD-L1 therapy to a moderate extent (AUC = 0.592). Upon review of the literature, SOX2 inhibited SOCS3 and PTPN1 transcription and induced sustained activation of the JAK-STAT pathway, thereby leading to overexpression of interferon-stimulated genes resistance signature. SOX2 has been demonstrated to induce immune evasion of CD8+ T-cell killing, and it has been identified as an independent prognostic factor for poor survival and resistance to anti-PD-1 therapy in melanoma with PD-L1 high expression. 57 Nuclear AURKA elevated PD-L1 expression, decreased CD8+ T cell proliferation and activity via an MYC-dependent pathway, and contributed to immune evasion in triple-negative breast cancer. 58 This suggests that, due to varying degrees of immune escape, the prognosis of patients with different responses to immunotherapy is also distinct in the two risk groups. These findings also provide new insights into the relationship between chemotherapy and cancer immunotherapy and increase our capacity to select clinical drug-therapy strategies. Furthermore, multivariate analysis indicated that risk score and stage were independent prognostic factors in HCC. And a nomogram was constructed to predict precisely the 1-, 3-, and 5-year survival of HCC patients by using risk score and TNM stage. Previous studies have shown that SOX2, IGF2BP3, MYCN, CPEB3, AURKA, GAL, RET, LOX, NQO1, SLC22A1, CDKN2A, and E2F1 are associated with the prognosis of HCC patients.29–33,35–39,41,42 Furthermore, Li et al demonstrated that the RNA m6A/m5C/m1A-regulated genes could be utilized to construct a risk signature for the assessment of HCC prognosis and immune status. 59 Our study also revealed that the RNA methylation-based risk score can accurately predict survival outcomes and evaluate the immune infiltration level.
Review and Reflection of the Literatures Related to RNA Methylation
In contrast to previous studies, Nie et al demonstrated that the m6A-modified binding protein YTHDF1 could promote prostate cancer progression by regulating androgen function-related gene TRIM68. 60 Maimaiti et al constructed a m7G score signature index based on the m7G regulators-mediated methylation modification patterns and analyzed the characterization of the immune microenvironment in lower-grade glioma, 61 which were not entirely different from our findings. Our study identified 21 RNA methylation-related signature genes (including m6A, m7G, and regulators) and developed a risk model that moderately predicted the prognosis of patients and reflected the immune microenvironment of HCC. Zhang et al investigated the potential functions of RNA methylation writers and established an RNA methylation pattern-based risk model for HCC patients, 62 which was similar to our study. The preceding sections have highlighted the pivotal role of RMGs in TME infiltration, targeted therapy, and immunotherapy, suggesting that they may serve as potential targets for tumors.
The Limitations of our Study
The present study had certain limitations. Primarily, the sample size of the tissue specimens utilized for the expression validation was insufficient, and the biological behavior of the core genes in HCC was not fully elucidated. Secondly, our findings indicate that these DERMGs play distinct roles in the multiple BP, such as IME landscape, drug sensitivity, and immunotherapeutic efficacy. However, the potential molecular mechanisms were not evaluated, and further investigation is warranted to elucidate the detailed mechanisms of DERMGs in HCC. Thirdly, the RMGs clusters and risk model may improve the prognosis of HCC patients in a higher-risk group for immunotherapy. Therefore, further research is necessary to explore whether these DERMGs could be used as diagnostic markers or therapeutic targets in HCC and guide more effective immunotherapy strategies.
Conclusion
Our research identified 21 RNA methylation-related signature genes and developed a risk model that moderately predicted the prognosis of HCC patients and screened for independent prognostic factors. Our findings are expected to be useful for cancer diagnosis, prognosis, and therapy.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338241276895 for A Novel RNA Methylation-Related Prognostic Signature and its Tumor Microenvironment Characterization in Hepatocellular Carcinoma by Luzheng Liu, Jiacheng Chen, Fei Ye, Yanggang Yan, Yong Wang and Jincai Wu in Technology in Cancer Research & Treatment
Acknowledgments
Not applicable.
Footnotes
Authors’ Note: All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by CJC, YF, YYG, WY and WJC. The first draft of the manuscript was written by LLZ and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data Availability Statement: The TCGA-LIHC dataset analyzed in this study was downloaded from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) and GSE14520 dataset was downloaded from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval and Consent Statement: Study protocols were approved by the Medical Ethics Committee of Hainan Affiliated Hospital of Hainan Medical University (address: No.19 Xiuhua Road, Haikou, China; approval number: Med-Eth-Re[2023] 319; date: September 13, 2023), based on the ethical principles for medical research involving human subjects of the Helsinki Declaration. Written informed consent was obtained from all participating patients prior to enrollment into the study.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Hainan Province Science and Technology Special Fund, the National Natural Science Foundation of China, the special research fund of The Innovation Platform for Academicians of Hannan Province (grant number ZDYF2020134, ZDYF2022SHFZ283, 81960331, 81660489, YSPTZX202005).
ORCID iD: Luzheng Liu https://orcid.org/0000-0003-1746-4517
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2.Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151‐172. [DOI] [PubMed] [Google Scholar]
- 5.Hao X, Sun G, Zhang Y, et al. Targeting immune cells in the tumor microenvironment of HCC: New opportunities and challenges. Front Cell Dev Biol. 2021;9:775462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Quan F, Xu J, Xiao Y, Li X, Li Y. Combination of multiple tumor-infiltrating immune cells predicts clinical outcome in colon cancer. Clin Immunol. 2020;215:108412. [DOI] [PubMed] [Google Scholar]
- 7.Erdag G, Schaefer JT, Smolkin ME, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012;72(5):1070‐1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obeid JM, Erdag G, Smolkin ME, et al. PD-L1, PD-L2 and PD-1 expression in metastatic melanoma: Correlation with tumor-infiltrating immune cells and clinical outcome. Oncoimmunology. 2016;5(11):e1235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Huang S, Zhuang H, et al. YTHDF2 Promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020;39(23):4507‐4518. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wang J, Li X, et al. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. 2021;12(1):6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia P, Zhang H, Xu K, et al. MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. 2021;12(7):691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387‐391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331‐D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Franceschini A, Wyder S, et al. STRING V10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447‐D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Lu W, Xu C, et al. PTGIS May be a predictive marker for ovarian cancer by regulating fatty acid metabolism. Comput Math Methods Med. 2023;2023:2397728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou RS, Zhang EX, Sun QF, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19(1):779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu TT, Li R, Huo C, et al. Identification of CDK2-related immune forecast model and ceRNA in lung adenocarcinoma, a pan-cancer analysis. Front Cell Dev Biol. 2021;9:682002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu Q, She H, Chen X, Zhong L, Zhu J, Fang L. Identification and experimental validation of mitochondria-related genes biomarkers associated with immune infiltration for sepsis. Front Immunol. 2023;14:1184126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Wang D, Yang L, et al. Cuproptosis related genes associated with Jab1 shapes tumor microenvironment and pharmacological profile in nasopharyngeal carcinoma. Front Immunol. 2022;13:989286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geeleher P, Cox N, Huang RS. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma HY, Yamamoto G, Xu J, et al. IL-17 signaling in steatotic hepatocytes and macrophages promotes hepatocellular carcinoma in alcohol-related liver disease. J Hepatol. 2020;72(5):946‐959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, Gu L, Lin X, et al. P53 deficiency affects cholesterol esterification to exacerbate hepatocarcinogenesis. Hepatology. 2023;77(5):1499‐1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao B, Wang Y, Lu S. Cellular senescence affects energy metabolism, immune infiltration and immunotherapeutic response in hepatocellular carcinoma. Sci Rep. 2023;13(1):1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan H, Tang G, Wang H, et al. DNA Methylation reactivates GAD1 expression in cancer by preventing CTCF-mediated polycomb repressive complex 2 recruitment. Oncogene. 2016;35(30):3995‐4008. [DOI] [PubMed] [Google Scholar]
- 29.Hosseini-Khah Z, Babaei MR, Tehrani M, et al. SOX2 And bcl-2 as a novel prognostic value in hepatocellular carcinoma progression. Curr Oncol. 2021;28(4):3015‐3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeng YM, Chang CC, Hu FC, et al. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology. 2008;48(4):1118‐1127. [DOI] [PubMed] [Google Scholar]
- 31.Qin XY, Suzuki H, Honda M, et al. Prevention of hepatocellular carcinoma by targeting MYCN-positive liver cancer stem cells with acyclic retinoid. Proc Natl Acad Sci U S A. 2018;115(19):4969‐4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Zou C, Qiu Z, et al. CPEB3-mediated MTDH mRNA translational suppression restrains hepatocellular carcinoma progression. Cell Death Dis. 2020;11(9):792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Z, Yin L, Zhou H, et al. Combined inhibition of AURKA and HSF1 suppresses proliferation and promotes apoptosis in hepatocellular carcinoma by activating endoplasmic reticulum stress. Cell Oncol (Dordr). 2021;44(5):1035‐1049. [DOI] [PubMed] [Google Scholar]
- 34.Zhang B, Zhou J. CircSEC24A (hsa_circ_0003528) interference suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells via miR-421/MMP3 axis. Bioengineered. 2022;13(4):9049‐9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai YT, Li CY, Huang YH, et al. Galectin-1 orchestrates an inflammatory tumor-stroma crosstalk in hepatoma by enhancing TNFR1 protein stability and signaling in carcinoma-associated fibroblasts. Oncogene. 2022;41(21):3011‐3023. [DOI] [PubMed] [Google Scholar]
- 36.Ye S, Zhao XY, Hu XG, et al. TP53 And RET may serve as biomarkers of prognostic evaluation and targeted therapy in hepatocellular carcinoma. Oncol Rep. 2017;37(4):2215‐2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Huang S, Wu G, et al. Lysyl oxidase is predictive of unfavorable outcomes and essential for regulation of vascular endothelial growth factor in hepatocellular carcinoma. Dig Dis Sci. 2015;60(10):3019‐3031. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Liu Y, Han A, et al. The NQO1/p53/SREBP1 axis promotes hepatocellular carcinoma progression and metastasis by regulating snail stability. Oncogene. 2022;41(47):5107‐5120. [DOI] [PubMed] [Google Scholar]
- 39.Heise M, Lautem A, Knapstein J, et al. Downregulation of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) in human hepatocellular carcinoma and their prognostic significance. BMC Cancer. 2012;12:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Chen D, Ning F, Du J, Wang H. EGF Is highly expressed in hepatocellular carcinoma (HCC) and promotes motility of HCC cells via fibronectin. J Cell Biochem. 2018;119(5):4170‐4183. [DOI] [PubMed] [Google Scholar]
- 41.Luo JP, Wang J, Huang JH. CDKN2A Is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Biosci Rep. 2021;41(10):BSR20211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Y, Liang ZY, Zhou WX, Zhou L. Plasminogen activator inhibitor 2 (PAI2) inhibits invasive potential of hepatocellular carcinoma cells in vitro via uPA- and RB/E2F1-related mechanisms. Hepatol Int. 2019;13(2):180‐189. [DOI] [PubMed] [Google Scholar]
- 43.Peng F, Xu J, Cui B, et al. Oncogenic AURKA-enhanced N6-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells. Cell Res. 2021;31(3):345‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Chen Y, Liu N, et al. CircMET promotes tumor proliferation by enhancing CDKN2A mRNA decay and upregulating SMAD3. Mol Cancer. 2022;21(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T, Guan G, Zhang J, et al. E2F1-mediated AUF1 upregulation promotes HCC development and enhances drug resistance via stabilization of AKR1B10. Cancer Sci. 2022;113(4):1154‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang P, Qiu J, Li B, et al. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin Biochem. 2011;44(8–9):582‐589. [DOI] [PubMed] [Google Scholar]
- 48.Sun C, Sun L, Li Y, Kang X, Zhang S, Liu Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating slug. Med Oncol. 2013;30(2):503. [DOI] [PubMed] [Google Scholar]
- 49.Langhans B, Nischalke HD, Krämer B, et al. Role of regulatory T cells and checkpoint inhibition in hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68(12):2055‐2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caux C, Ramos RN, Prendergast GC, Bendriss-Vermare N, Ménétrier-Caux C. A milestone review on how macrophages affect tumor growth. Cancer Res. 2016;76(22):6439‐6442. [DOI] [PubMed] [Google Scholar]
- 51.Yang Y, Ye YC, Chen Y, et al. Crosstalk between hepatic tumor cells and macrophages via Wnt/β-catenin signaling promotes M2-like macrophage polarization and reinforces tumor malignant behaviors. Cell Death Dis. 2018;9(8):793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grisaru-Tal S, Itan M, Klion AD, Munitz A. A new Dawn for eosinophils in the tumour microenvironment. Nat Rev Cancer. 2020;20(10):594‐607. [DOI] [PubMed] [Google Scholar]
- 53.Zhou SL, Zhou ZJ, Hu ZQ, et al. Tumor-Associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology. 2016;150(7):1646‐1658.e17. [DOI] [PubMed] [Google Scholar]
- 54.Hou Z, Liu J, Jin Z, et al. Use of chemotherapy to treat hepatocellular carcinoma. Biosci Trends. 2022;16(1):31‐45. [DOI] [PubMed] [Google Scholar]
- 55.Saviano A, Habersetzer F, Lupberger J, et al. Safety and antiviral activity of EGFR inhibition by erlotinib in chronic hepatitis C patients: A phase ib randomized controlled trial. Clin Transl Gastroenterol. 2022;13(6):e00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuchs BC, Hoshida Y, Fujii T, et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology. 2014;59(4):1577‐1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu R, Wang C, Li Z, et al. SOX2 Promotes resistance of melanoma with PD-L1 high expression to T-cell-mediated cytotoxicity that can be reversed by SAHA. J Immunother Cancer. 2020;8(2):e001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun S, Zhou W, Li X, et al. Nuclear Aurora kinase A triggers programmed death-ligand 1-mediated immune suppression by activating MYC transcription in triple-negative breast cancer. Cancer Commun (Lond). 2021;41(9):851‐866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D, Li K, Zhang W, et al. The m6A/m5C/m1A regulated gene signature predicts the prognosis and correlates with the immune Status of hepatocellular carcinoma. Front Immunol. 2022;13:918140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nie Q, Wu X, Huang Y, Guo T, Kuang J, Du C. RNA N6-methyladenosine-modified-binding protein YTHDF1 promotes prostate cancer progression by regulating androgen function-related gene TRIM68. Eur J Med Res. 2023;28(1):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maimaiti A, Feng Z, Liu Y, et al. N7-methylguanosin regulators-mediated methylation modification patterns and characterization of the immune microenvironment in lower-grade glioma. Eur J Med Res. 2023;28(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Gao J, Hu M, et al. Integrated investigation of the clinical implications and targeted landscape for RNA methylation modifications in hepatocellular carcinoma. Eur J Med Res. 2023;28(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338241276895 for A Novel RNA Methylation-Related Prognostic Signature and its Tumor Microenvironment Characterization in Hepatocellular Carcinoma by Luzheng Liu, Jiacheng Chen, Fei Ye, Yanggang Yan, Yong Wang and Jincai Wu in Technology in Cancer Research & Treatment