Summary
Although thousands of genes have been identified or cloned in rice (Oryza sativa) in the last two decades, the majority of them have only been separately characterized in specific varieties or single‐gene modified backgrounds, thus limiting their practical application. We developed an optimized multiplex genome editing (MGE) toolbox that can efficiently assemble and stably express up to twelve sgRNA targets in a single plant expression vector. In this study, we established the MGE‐based Rapid Directional Improvement (MRDI) strategy for directional improvement of complex agronomic traits in one small‐scale rice transformation. This approach provides a rapid and practical procedure, encompassing sgRNA assembly, transgene‐free screening and the creation of promising germplasm, by combining the precision of gene editing with phenotype‐based field breeding. The MRDI strategy was used to generate the full diversity of twelve main agronomic genes in rice cultivar FXZ for the directional improvement of its growth duration and plant architecture. After applying the MRDI to FXZ, ideal plants with the desired traits of early heading date reduced plant height, and more effective panicles were generated without compromising yield, blast resistance and grain quality. Furthermore, the results of whole‐genome sequencing (WGS), including the analysis of structural variations (SVs) and single nucleotide variations (SNVs) in the MGE plants, confirmed the high specificity and low frequency of unwanted mutations associated with this strategy. The MRDI breeding strategy would be a robust approach for exploring and applying crucial agronomic genes, as well as for generating novel elite germplasm in the future.
Keywords: multiplex genome editing, rapid directional improvement, heading date, plant architecture, rice
CRISPR/Cas9‐based multiplex gene editing (MGE) has the potential to simultaneously incorporate several beneficial agronomic genes in order to breed ideal varieties (Zhu et al., 2020). However, many significant agronomic traits, including yield, growth period, and quality, are often influenced by the combined effects of multiple loci with additive effects and genetic interactions. Regarding heading date, a crucial trait for rice (Oryza sativa L.) breeding, hundreds of heading date QTLs had been identified in recent decades (Vicentini et al., 2023). The core regulator of Hd1/Ehd1, in cooperation cooperated with Ghd7/DTH8/DTH7, have been unveiled and well‐illustrated in controlling photoperiod sensitivity and geographic adaption of rice varieties. However, many minor effect genes that control heading, either dependent or independent of the Hd1/Ehd1 regulatory pathway, have been identified, including DTH2, Se14, and Ef7. The establishment of mutation pools comprising these relevant genes is crucial for their potential utilization in germplasm improvement and creation, which was difficultly achieved by traditional approach.
In this regard, we try to develop a strategy called MGE‐based Rapid Directional Improvement (MRDI) for mutation pool and germplasm creation through a combination of MGE and crossbreeding, and expedite its prospective practical application in crop breeding (Figure 1a). FXZ is a newly developed varieties (CERTIFICATE NO. 20200011) that possesses exceptional quality and high resistance to blast disease. However, its prolonged growth period (>140 days) and the slender leaf shape limit its adaptability and practical application. We are aiming to generate new FXZ germplasm with moderately early‐maturation trait and better plant architecture through the MRDI strategy with multiplex edited genes that control heading date and plant architecture.
Figure 1.
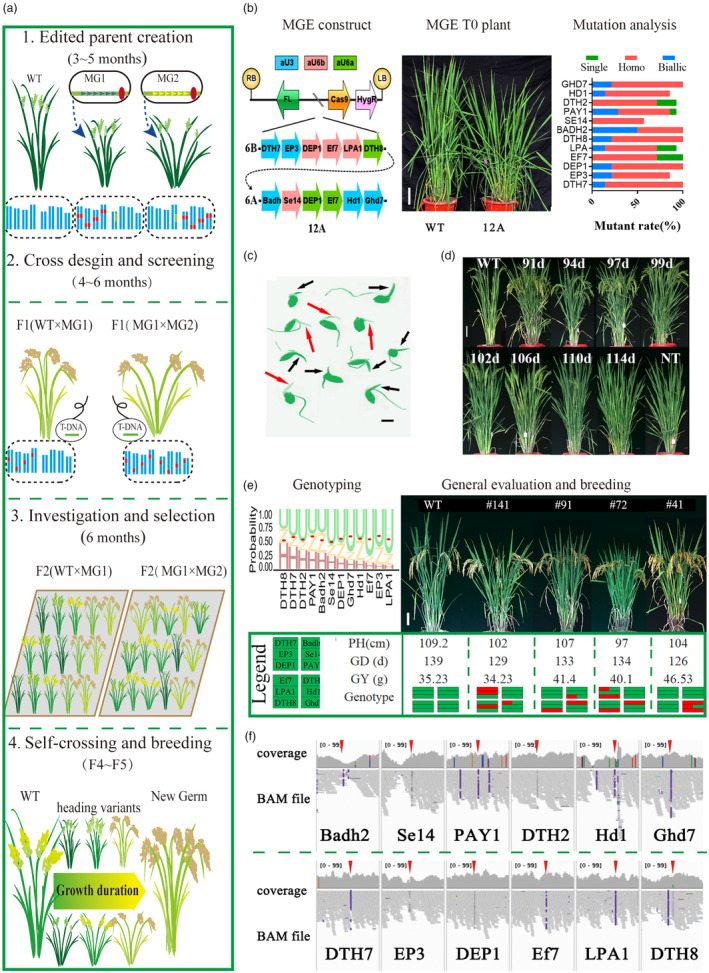
MGE‐based Rapid Directional Improvement (MRDI) in the study. (a) Flow diagram of MRDI, which consists of four fundamental steps, as indicated in the timeline below the title. MG1 and MG2 represent different MGE plants with distinct plant type and heading date. Triangle and oval indicate sgRNAs cassette and Cas9 cassette, respectively; red and green box indicate edited allele and T‐DNA insert, respectively. (b ~ e) The steps from 1st to 4th of MRDI in our study. (b) The left panel: MGE vectors 6A, 6B and 12A. The vector 12A contains 12 main agronomic trait genes, while 6A and 6B each contain a set of six different genes. LB/RB: left/right border of T‐DNA; Cas9: spCas9 cassette; Ubi: ubiquitin promoter; Hyg, Hygromycin ‐resistance cassette; FL, fluorescent cassette; aU3/aU6a/aU6b, sgRNA cassette driven by artificial snRNA promoter. The middle panel: the rice plant carrying 12 edited genes. Scale bar, 15 cm. The right panel: the overall mutagenesis efficiency recorded from 41 T0 lines of 6A/6B and 12A transgenic plants. (c) GFP screening of transgene‐free F1 hybrid seed. Red/black arrow indicate shoot of fluorescent/non‐fluorescent hybrid seed. Scale bar, 5 mm. (d) Heading phenotypes of F2 plant from 12A/WT under normal long‐day (NLD) conditions. Plants were grown for 114 days. Scale bar, 15 cm. Representative plants were selected and arranged according to heading dates, with the heading dates annotated above the plants. (e) The left panel: seqlogo of 12 genes illustrating the percentage of homozygous alleles in 88 lines of 12A F2:3 population with desired agronomic traits, and the red ovals indicate the average distribution of edited genes based on pooled sequencing results from the 12A population (~50 individual plants). The letters U/H/Z represent unedited/ homozygous‐edited/heterozygous‐edited site. The right panel: representatives of F2:3 progeny plants selected for early heading date, moderate plant architecture, and good yield potential. Scale bar, 15 cm. PH, plant height; GD, growth duration; GYP, grain yield per plant; Genotype, detailed allele type; red block indicates edited alleles, while green block indicates unedited alleles. (f) The IGV view of adjacent regions containing twelve targets in 12A T1 plant. The red triangle indicates the mutation of target sites directly detected by NGS analysis. The complete IGV view has been included in Figure S9.
The MGE structure is critical for the production of stable and homozygous mutants of multiplex target sites. We chose to use a multiplex stand‐alone sgRNA unit, with each sgRNA driven by an individual snRNA promoter (Hao et al., 2020; Ma et al., 2015; Xing et al., 2014). To simplify the construct process, we have introduced unique non‐palindromic stick‐ends by shifting the cleavage sites of the three artificial promoters along the 3′–5′ direction at the termini (Figure S1a). This modification allows for the assembly of over 24 potential units in a single round of PCR (Figure S1b; Table S1). We optimize the sgRNA cassette assembly combination for high ligation efficiency, enabling the assembly of up to 16 sgRNAs using golden‐gate ligation (Figure S1c–f). Overall, this vector structure provides consistent expression levels and comparable mutant efficiency, although some sites had positon effect in transient system (Figure S2a–c).
To implement the MRDI strategy for improving heading date and comprehensive traits, we selected 12 main agronomic trait genes, including seven heading date related genes, four plant architecture genes and one quality gene (Figure 1b). These genes were physically distant from each other (Figure S2d), to facilitate complete segregation and minimize the potential hazard of chromosome structural variation caused by simultaneous editing of tandemly arrayed genes (Zhou et al., 2023).The sgRNAs of these genes were designed and assembled to the single vector “12A” for simultaneously knocking out 12 genes in rice. To prevent the instability of the duodecuplex structure, we divided the targets from the same chromosome into two paired sextuple vectors 6A and 6B, by reusing and rearranging the cassettes (Figure 1b; Figure S2d).
Upon transformation with FXZ, a total of 41 independent T0 plants were acquired. Among these, 17 plants harboured vector 6A, 10 plants carried vector 6B, and 14 plants contained vector 12A (Figure 1b; Figure S2e). In summary, except for Se14, which exhibited lower mutant efficiency in protoplasts, the remaining targets of 12A displayed similar mutant rates to those observed in sextuplex‐edited plants, ranging from 85.2% to 100% (Figure 1b; Figures S2f and S3; Table S2). The top‐two off‐target sites for each gene were selected and verified (Table S3), and there were no discernible mutated peaks in all T0 plants.
Considering the high mutagenesis efficiency in T0 plants, we have designed a cross plan for the second step of the MRDI. Initially, we screened T0 lines with all target sites edited and low T‐DNA insert copy number. These lines were either crossed as 6A × WT, 12A × WT and 6A × 6B to generate a segregation population of multiplex genes. Through the utilization of a GFP‐based non‐invasive sorting method, we successfully identified transgene‐free seeds from the hybrid seeds within 2 days after germination (Figure 1c). Subsequently, we harvested and selected the F1 plants with fully heterozygous alleles of 12A and 6A for population construction and germplasm creation.
The seven heading date related genes could generate 128 (27) combinations for homozygous segregation and 2187 (37) combinations for intermediate heterozygous progeny due to combinations of seven gene interactions. We initially investigated the T2 plants from 6A T1 parents carrying heterozygous alleles of the minor effect gene of DTH2 and Se14. The results showed that the knockout DTH2 and Se14 could individually and coordinately affect heading date in the background of the Hd1/Gdh7 mutant (Figure S4a–c). Therefore, a pool of plants with rich diversity in heading time would be generated from F2 population when the gene collections of heading date were segregated.
We planted and investigated two F2 population of 6A/FXZ and 12A/FXZ under NLD (natural long day) conditions, respectively. Compared with the wild type FXZ (107.2 days of heading date), two populations exhibited large variations in heading date (Figure 1d; Figure S5a–d), ranging from 91 to 121 days in the 12A population and from 94 to 122 days in the 6A population. In two population, early heading plants (a week earlier than WT) accounted for 30% (84 of 280 plants) in the 12A population and 13% (39 of 293 plants) in the 6A; while late heading plants (a week later than WT) accounted for 7.5% (21 of 280 plants) in 12 A and 19% (43 of 293 plants) in 6A, respectively (Figure S5b–d).
With directional improvement via specific gene pool, in practice we initially selected approximately 25% of the plants (142 plants from two populations) that exhibited early maturity, reduced plant height, wide short blades, and a moderate plant type in the primary population, which differed from traditional cross breeding with selection ratios typically lower than 5% (Jennings, 1979). To gain insights into the distribution of edited genes within the selected candidate breeding lines, we analysed the genotypes of 88 lines from the 12A population. The wild type alleles were predominant for most of the genes, indicating that the unedited alleles comprised the majority of the 12A MGE plants and a low percentage of edited genes would be favourable for the desired traits of heading date and grain yield. Interestingly, the dth8, dth7 and dth2 was the highest percentage of homozygote edited allele, i.e. potential “hot spot” genes for promoting heading date in the selected population (Figure 1e). From the F2:3 populations, we screened ideal progeny plants with the desired trait of early heading date and moderate plant architecture, specifically reduced plant height and more effective panicles (Figure 1e). In these lines, #41, #72 and #91 harboured homozygous alleles for dth2 and displayed early head date and short flag leaf, and #72 and #91 also harboured dth8 alleles. Importantly, most of MGE progeny maintained blast resistance and high grain quality, indicating that the MRDI of FXZ could accurately and efficiently promote heading date without comprising general agronomic traits (Tables S4 and S5).
We continue to track the traits of the progeny plants from the F3:4 sub‐population to further confirm the combined effects of these genes in a homozygous background. dth2 has a moderately positive effect on yield in rice with delayed heading date, and could alleviate the yield loss caused by the deletion of DTH7, DTH8 and Ghd7 (Figure S6). By contrast, the knockout of Ef7 has a stronger photosensitive effect compared to that of DTH2. When combined with the double mutant DTH7/DTH8, ef7 mutant exhibits a moderate yield trait and plant architecture in long‐day conditions, making it a minor allele that helps balance both yield and heading date in the selected population (Figure S6; Table S6). In addition, we observed that that LPA1, EP3 and DEP1 have interactive effects on plant type, yield trait and heading date. EP3 and LPA1, two minor selected genes, exhibited a strong interaction in plant type and provided more tiller numbers and moderately loose architecture in double mutants. In line #141 with EP3/LPA1 double mutant, the plant had a less compact plant shape, and the single mutants of EP3 or LPA1 lead to overly compact or loose plant type, respectively (Figure S7; Table S6). Overall, lines #72‐1, #41‐1, #41‐2, #91‐1 and #139‐1 show great potential as germplasm resources for early heading and improved plant architecture through the MRDI breeding strategy.
Recently, the occurrence of chromosome rearrangements and mutations resulting from concurrent DSBs induced by CRISPR‐Cas9 has raised concerns about the potential adverse effects on CRISPR‐based precision breeding (Zhou et al., 2023). Although the targets selected for our MRDI strategy have been optimized and re‐organized, concerns regarding simultaneous dozen of DSBs remained. To address this, we performed whole‐genome sequencing (WGS) on T1 and F4 plants to investigate structural variants. Our analysis revealed no significant structural variations (SVs) across the entire genome, and the T1 samples with sextuple or duodecuplex editing shared a similar distribution of SVs with the wild‐type FXZ and single mutant plants (Figure S8). Additionally, the mutation patterns within regions containing the target sites further validated the specificity of our MRDI design in three types of MGE plants (Figure 1f; Figure S9a,b). We also examined single nucleotide variations (SNVs) and small Indels in F4 plants and newly generated T0 plants of double, sextuple, and nonuple mutants, and overall mutations do not accumulate significantly in MGE plants compared to the control (empty vector) (Figure S10), consistent with previous reports on the high specificity of CRISPR in rice (Tang et al., 2018; Zhang et al., 2023). These results support the potential of utilizing MGE strategy with optimized targets for breeding purposes.
Notably, this strategy offers the advantages of almost identical genetic background and distinct allele genotypes, making it also a powerful tool for analysing gene interactions at the phenotype level. We believe that the MRDI breeding strategy has the potential to be a powerful approach for exploring and applying important agronomic genes in the future.
Funding
The work was supported by the Natural Science Foundation of Fujian Province (Grant No. 2020J011354), the National Rice Industry Technology System of Modern Agriculture for China (Grant No. CARS‐01‐20), Key Science and Technology Projects of Fujian Academy of Agricultural Sciences (Grant No. KJZD202401), the “5511” Collaborative Innovation project for High‐quality Development and Surpasses of Agriculture between Government of Fujian and Chinese Academy of Agricultural Sciences (Grant No. XTCXGC2021001), Key program of Science and Technology in Fujian province, China (Grant No. 2020NZ08016), Open Project Fund of Key Laboratory of Ministry of Education for Genetics, Breeding, and Multiple Utilization of Crops of Fujian Agricultrue and Forestry University (GBMUC‐2019‐002).
Author contributions
JFZ and WYD planned and designed the experiment; WYD and JFZ wrote the manuscript; WYD and ZHM design and construct the vector for MRDI, WYD and FJX construct and CQH, ZZX, WJL, ZM, YFT, JJH, XHG, LX and JFZ performed experiments and analysed the data; WYD, WLY, LYL, HW, QMY, ZXX, ZYS, XHA and JFZ were in charge of collection and interpretation some materials.
Conflict of interest
The authors declare no competing interests.
Supporting information
Data S1 Experimental procedures.
Figure S1 Optimization of multiplex sgRNA expression cassette assembly.
Figure S2 High mutagenesis efficiency and uniformity expression of MGE toolbox.
Figure S3 Genotype of the T0 plant of three vectors with completed mutation in all targets.
Figure S4 Effect of DTH2/Se14 on heading phenotype in 6A background.
Figure S5 Variation of heading dates of F2 population.
Figure S6 Plant morphology of F3:4 plant harbouring dth2 or ef7 homozygote alleles.
Figure S7 Plant morphology of F3:4 plant harbouring ep3 and lpa1 homozygote alleles.
Figure S8 Circos representation of structural genome variations in multiplex editing plants.
Figure S9 Validation of the specific editing pattern of MGE plants.
Figure S10 Genome‐wide analysis of SNVs and InDels caused by multiplex genome editing.
Table S1 List of unique non‐palindromic sticky ends.
Table S2 List of edited site patterns of MGE T0 plants.
Table S3 Potential Off target sites of 12 genes.
Table S4 Evaluation of blast resistance for MGE T2 plants and F2 population.
Table S5 Quality traits for MGE T2 plants.
Table S6 Agronomic traits of F3:4 plants.
Table S7 Primers for mutagenesis and transgene detection.
Table S8 Sticky‐end sets for optimization of 12/16 cassette assembly.
Table S9 Optimal primer sets for 16 sgRNAs accessibly.
Table S10 Cassettes and primers for targeting 12 genes of agronomic traits in rice.
Acknowledgements
We thank Prof. Qijun Chen at College of Biological Sciences, China Agricultural University for kindly donation of PHUE411 and PBUE411 vector and instructions. We thank Prof. Caixia Gao at Institute of Genetics and Developmental Biology, CAS for technical support of mutagenesis efficiency by protoplast system. We thank Prof. Yaoguang Liu at College of Life Sciences, South China Agricultural University for kindly donation of artificial short promoter vector system. We thank Prof. Zhiqiang WU and Prof. Kai Chen at Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences for providing valuable guidance on NGS analysis.
Data availability statement
The Data will be Available in the manuscript.
References
- Hao, Y. , Zong, W. , Zeng, D. , Han, J. , Chen, S. , Tang, J. , Zhao, Z. et al. (2020) Shortened snRNA promoters for efficient CRISPR/Cas‐based multiplex genome editing in monocot plants. Sci. China Life Sci. 63, 933–935. [DOI] [PubMed] [Google Scholar]
- Jennings, P.R. (1979) Rice improvement. Los Baños: International Rice Research Institute. [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A Robust CRISPR/Cas9 System for Convenient, High‐Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Liu, G. , Zhou, J. , Ren, Q. , You, Q. , Tian, L. , Xin, X. et al. (2018) A large‐scale whole‐genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini, G. , Biancucci, M. , Mineri, L. , Chirivì, D. , Giaume, F. , Miao, Y. , Kyozuka, J. et al. (2023) Environmental control of rice flowering time. Plant Commun. 4, 100610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wu, Y. , Li, G. , Qi, A. , Zhang, Y. , Zhang, T. and Qi, Y. (2023) Genome‐wide investigation of multiplexed CRISPR‐Cas12a‐mediated editing in rice. Plant Genome 16, e20266. [DOI] [PubMed] [Google Scholar]
- Zhou, S. , Cai, L. , Wu, H. , Wang, B. , Gu, B. , Cui, S. , Huang, X. et al. (2023) Fine‐tuning rice heading date through multiplex editing of the regulatory regions of key genes by CRISPR‐Cas9. Plant Biotechnol. J. 22, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Li, C. and Gao, C. (2020) Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Experimental procedures.
Figure S1 Optimization of multiplex sgRNA expression cassette assembly.
Figure S2 High mutagenesis efficiency and uniformity expression of MGE toolbox.
Figure S3 Genotype of the T0 plant of three vectors with completed mutation in all targets.
Figure S4 Effect of DTH2/Se14 on heading phenotype in 6A background.
Figure S5 Variation of heading dates of F2 population.
Figure S6 Plant morphology of F3:4 plant harbouring dth2 or ef7 homozygote alleles.
Figure S7 Plant morphology of F3:4 plant harbouring ep3 and lpa1 homozygote alleles.
Figure S8 Circos representation of structural genome variations in multiplex editing plants.
Figure S9 Validation of the specific editing pattern of MGE plants.
Figure S10 Genome‐wide analysis of SNVs and InDels caused by multiplex genome editing.
Table S1 List of unique non‐palindromic sticky ends.
Table S2 List of edited site patterns of MGE T0 plants.
Table S3 Potential Off target sites of 12 genes.
Table S4 Evaluation of blast resistance for MGE T2 plants and F2 population.
Table S5 Quality traits for MGE T2 plants.
Table S6 Agronomic traits of F3:4 plants.
Table S7 Primers for mutagenesis and transgene detection.
Table S8 Sticky‐end sets for optimization of 12/16 cassette assembly.
Table S9 Optimal primer sets for 16 sgRNAs accessibly.
Table S10 Cassettes and primers for targeting 12 genes of agronomic traits in rice.
Data Availability Statement
The Data will be Available in the manuscript.


