Abstract
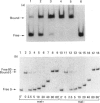
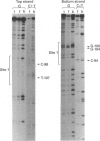
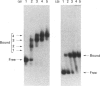
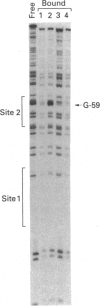
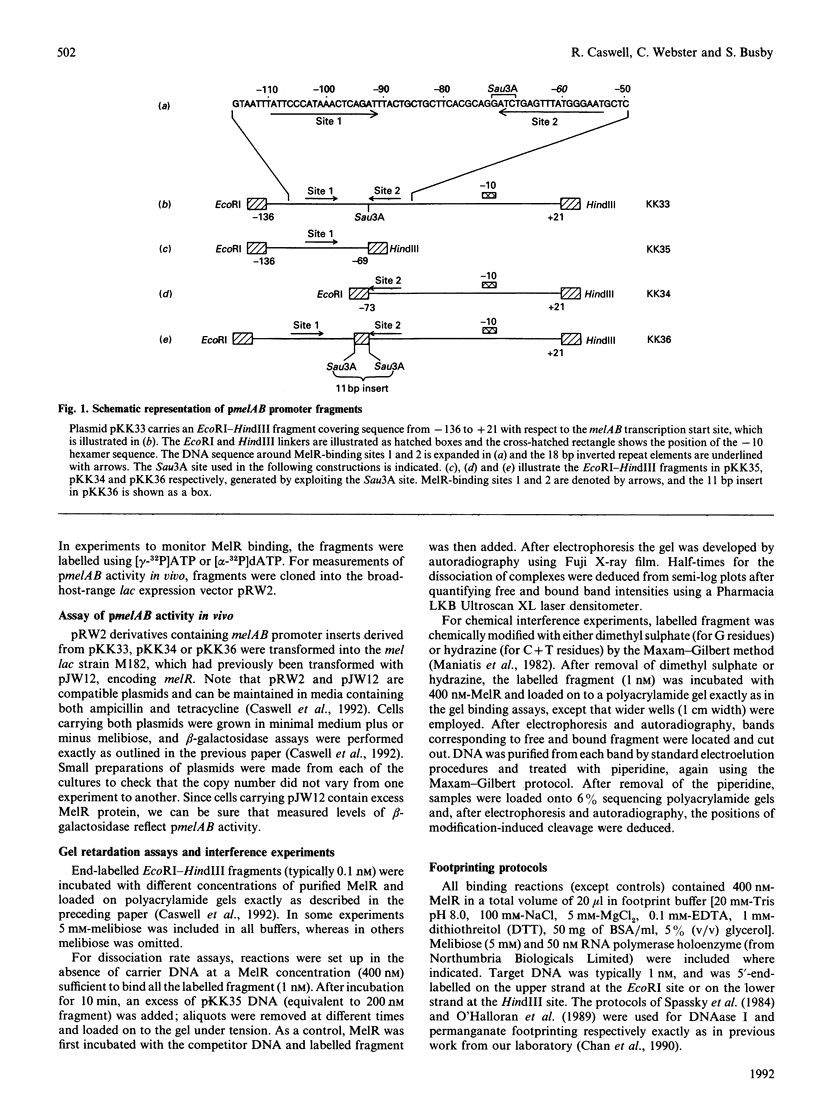
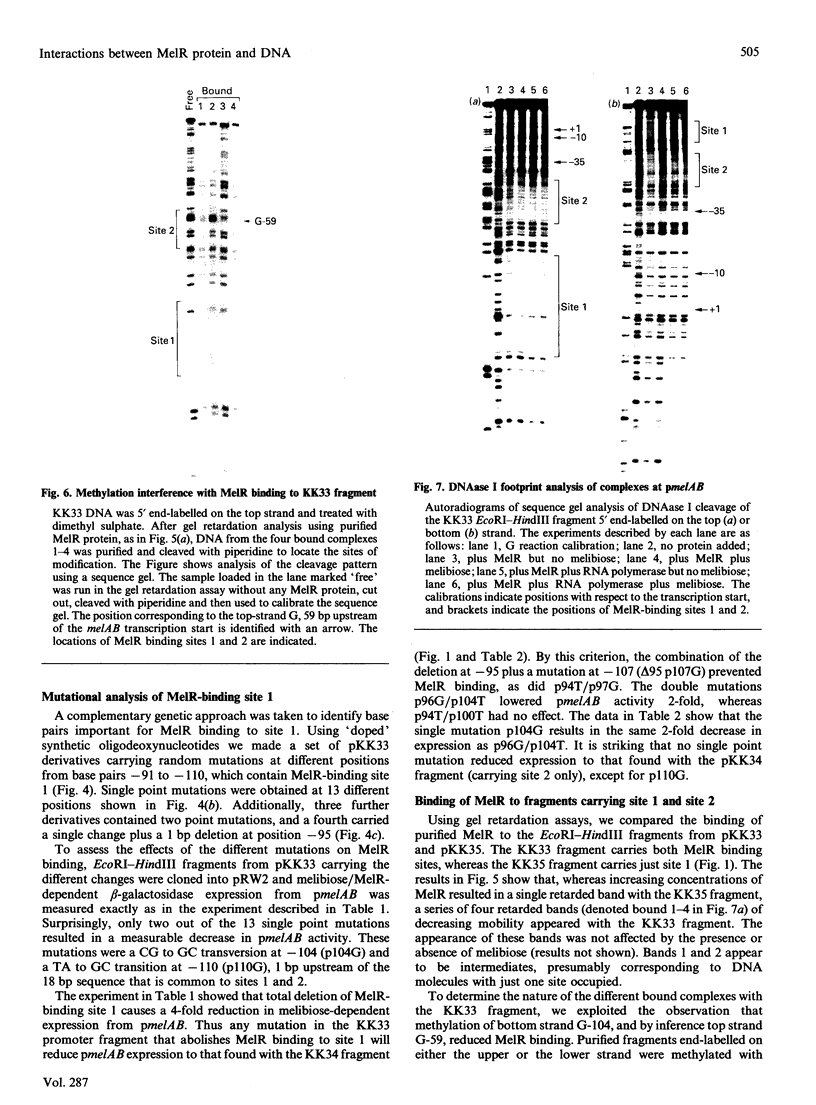
Escherichia coli MelR protein binds to two sites located upstream of the melAB transcription start site. Although both sites are required for optimal melibiose-dependent expression from the melAB promoter, some MelR-dependent expression is found if the upstream site is deleted or if the spacing between the two sites is altered. Gel retardation assays have been exploited to study MelR binding to a DNA fragment carrying just the upstream site. Methylation interference analysis was used to identify one guanine (at -104) which is important for MelR binding. Mutational analysis confirmed the importance of this base and revealed a second position (at -110) where mutations interfere with melAB promoter activity. Experiments using potassium permanganate as a probe suggested that the DNA sequence around -110 adopts a distorted conformation. We propose that the mutation at -104 alters MelR binding by interfering with a direct contact, whereas the mutation at -110 primarily affects DNA conformation. The binding of purified MelR protein to a melAB promoter fragment carrying both binding sites has also been studied: binding results in four retarded bands in gel assays. Methylation interference experiments have been exploited to identify the binding sites occupied in each complex. Although both binding sites share a common 18 bp sequence, MelR binding to the more upstream site is stronger. We could find no evidence for co-operative interactions between MelR and RNA polymerase and no major effects of melibiose. Some evidence for melibiose-dependent distortion in complexes between MelR and the melAB promoter is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansari A. Z., Chael M. L., O'Halloran T. V. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992 Jan 2;355(6355):87–89. doi: 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Busby S., Buc H. Positive regulation of gene expression by cyclic AMP and its receptor protein in Escherichia coli. Microbiol Sci. 1987 Dec;4(12):371–375. [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Caswell R., Williams J., Lyddiatt A., Busby S. Overexpression, purification and characterization of the Escherichia coli MelR transcription activator protein. Biochem J. 1992 Oct 15;287(Pt 2):493–499. doi: 10.1042/bj2870493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B., Spassky A., Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus -35 region sequences. Biochem J. 1990 Aug 15;270(1):141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., O'Halloran T. V. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990 May 22;29(20):4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- Gaston K., Kolb A., Busby S. Binding of the Escherichia coli cyclic AMP receptor protein to DNA fragments containing consensus nucleotide sequences. Biochem J. 1989 Jul 15;261(2):649–653. doi: 10.1042/bj2610649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton E. P., Lee N. Three binding sites for AraC protein are required for autoregulation of araC in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1749–1753. doi: 10.1073/pnas.85.6.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanatani M., Yazyu H., Shiota-Niiya S., Moriyama Y., Kanazawa H., Futai M., Tsuchiya T. Physical and genetic characterization of the melibiose operon and identification of the gene products in Escherichia coli. J Biol Chem. 1984 Feb 10;259(3):1807–1812. [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci U S A. 1985 May;82(10):3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman P. S., Cole J. A., Busby S. J. Mutational analysis of the nucleotide sequence at the FNR-dependent nirB promoter in Escherichia coli. Nucleic Acids Res. 1989 Jan 11;17(1):135–145. doi: 10.1093/nar/17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Spassky A., Chapon C., Blazy B., Buc H. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 1983 Nov 25;11(22):7833–7852. doi: 10.1093/nar/11.22.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Brown N. L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990 Sep 11;18(17):5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O. Nucleoprotein structures at positively regulated bacterial promoters: homology with replication origins and some hypotheses on the quaternary structure of the activator proteins in these complexes. Mol Microbiol. 1989 Mar;3(3):455–458. doi: 10.1111/j.1365-2958.1989.tb00191.x. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Spassky A., Busby S., Buc H. On the action of the cyclic AMP-cyclic AMP receptor protein complex at the Escherichia coli lactose and galactose promoter regions. EMBO J. 1984 Jan;3(1):43–50. doi: 10.1002/j.1460-2075.1984.tb01759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., de Crombrugghe B. Interactions of RNA polymerase and the cyclic AMP receptor protein on DNA of the E. coli galactose operon. Nucleic Acids Res. 1983 Aug 11;11(15):5165–5180. doi: 10.1093/nar/11.15.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin J. F., Schleif R. F. Purification and properties of RhaR, the positive regulator of the L-rhamnose operons of Escherichia coli. J Mol Biol. 1990 Jan 5;211(1):75–89. doi: 10.1016/0022-2836(90)90012-B. [DOI] [PubMed] [Google Scholar]
- Webster C., Gardner L., Busby S. The Escherichia coli melR gene encodes a DNA-binding protein with affinity for specific sequences located in the melibiose-operon regulatory region. Gene. 1989 Nov 30;83(2):207–213. doi: 10.1016/0378-1119(89)90106-6. [DOI] [PubMed] [Google Scholar]
- Webster C., Gaston K., Busby S. Transcription from the Escherichia coli melR promoter is dependent on the cyclic AMP receptor protein. Gene. 1988 Sep 7;68(2):297–305. doi: 10.1016/0378-1119(88)90032-7. [DOI] [PubMed] [Google Scholar]
- Webster C., Kempsell K., Booth I., Busby S. Organisation of the regulatory region of the Escherichia coli melibiose operon. Gene. 1987;59(2-3):253–263. doi: 10.1016/0378-1119(87)90333-7. [DOI] [PubMed] [Google Scholar]