Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common pathological type of non-Hodgkin lymphoma, and is closely associated with hepatitis B virus (HBV) infection status and hepatitis B X (HBx) gene integration. This project investigated the cellular biological effects and molecular mechanisms responsible for lymphomagenesis and the progression of HBx integration in DLBCL. The data showed that clinical DLBCL cells demonstrated HBx integration, and the sequencing analysis of integrated sites validated HBx integration in the constructed HBx-transfected cells. Compared with control cells, HBx-transfected cells had a significantly reduced proportion of mitochondrial membrane potential, signals of chromosomal DNA breaks, and proportion of apoptotic cells. Further studies found that this decreased apoptosis level was associated with a significant reduction of cleaved Caspase-3 and downstream poly ADP-ribose polymerase (PARP) proteins, revealing the molecular mechanisms of HBx-associated apoptosis in DLBCL. Animal experiments also demonstrated that the protein expression of cleaved Caspase-3 and PARP was prominently reduced in HBx-transfected cells from subcutaneous tumors in mice. Furthermore, the HBx-integrated cells in clinical tissues had significantly lower cleaved PARP levels than the HBx-negative samples. Therefore, HBx integration inhibits cell apoptosis through the Caspase-3-PARP pathway in DLBCL indicating a potential biomarker and therapeutic target in HBV related DLBCL.
Keywords: Diffuse large B cell lymphoma, Hepatitis B virus, HBx integration, Apoptosis
1. Introduction
Non-Hodgkin lymphoma (NHL) is one of the most common cancers, accounting for 4 % of all cancers [1,2]. Diffuse large B-cell lymphoma (DLBCL) is the primary pathological type accounting for 30–50 % of NHL [3,4]. The cause of this disease is strongly correlated with host infection status, with HIV infection having a 650-fold increased risk of developing DLBCL [5]. Some clinical studies have shown that a higher prevalence of hepatitis B surface antigen (HBsAg) is associated with lymphomagenesis in DLBCL patients [6,7]. However, regardless of HBsAg clearance, the infected population has a higher risk of B-cell NHL [8]. Unlike the hepatitis C virus, which is associated with extrahepatic manifestations of autoimmune-related lymphoproliferative disorders [9], hepatitis B virus (HBV) infection is correlated with DLBCL in part from HBV directly entering B lymphocytes [10,11].
HBV infection is a worldwide health problem, with an estimated 296 million carriers of chronic hepatitis B infection (WHO, 2019). HBV straddles the line between DNA and RNA viruses, with a DNA genome replicated by reverse transcription. Double-stranded linear DNA molecules can also integrate into the host cell genome at random sites by non-homologous end joining [12]. Therefore, it is speculated that intracellular HBV DNA may be integrated in DLBCL.
Hepatitis B also results in an estimated 820 thousand deaths yearly (WHO, 2019), mostly from cirrhosis and hepatocellular carcinoma (HCC). The mechanisms of HBV-driven carcinoma include the expression of viral proteins from viral gene X (hepatitis B X, HBx) to modulate cell viability, integration of HBV DNA into the host genome to alter gene function, and accumulation of genetic damage due to inflammation [13,14]. HBx integration is observed more frequently in HCC and is associated with cancer-related gene expression and patient survival [15,16]. However, the effect of HBx integration on DLBCL remains unclear.
To assess the cellular biological effects and molecular mechanism of HBx gene integration in DLBCL, integration of the HBx gene in clinical DLBCL tissues was studied. HBx integrated cell lines were constructed to evaluate the biological effects and related molecular mechanisms by ex vivo experiments, verifying the results in experimental animals and clinical cases. This study aimed to investigate the etiology of a novel interpretation of HBV-related DLBCL and to facilitate the development of new strategies to prevent and treat this disease.
2. Materials and methods
2.1. Cell culture
Originally from the American Type Culture Collection, Pfeiffer cells were cultured in RPMI 1640 (Gibco, USA) containing 10 % FBS and 1 % penicillin/streptomycin, and maintained at 37 °C in a humidified 5 % CO2 incubator. Cells were passaged every three days and maintained at a density of 3 × 105 and 3 × 106 cells/ml.
2.2. Electroporation of HBx gene
HBx plasmid (Addgene 24931) was transfected into the DLBCL cells of the Pfeiffer strain by electroporation [17]. Pfeiffer cells were resuspended and maintained in RPMI 1640 medium supplemented with 10 % FBS at room temperature. The cell density was 2 × 107 cells per ml, and the volume used per electroporation was 0.4 ml in the 0.4 cm cuvettes. A 20 μl dilution of 15 μg HBx or control plasmid (pEGFP-C2) was used for electroporation. A capacitance of 960 μF was used, with a voltage of 250 V. Cells were placed into growth media for culture, and 800 μg/ml G418 was added into the media at 48 h post-electroporation to screen positive cells, for two weeks.
2.3. HBx gene detection in cells
DNA from transfected cultured cells and DLBCL tissues was extracted using a supporting magnetic kit (EmerTher Company, China). PCR was performed via a protocol previously described, with some modifications [18]. In particular, primer pairs for detecting the HBx gene (1689F: CGACCGACCTTGAGGCATAC and 1826R: AAAAGTTGCATGGTGCTGGT, reference site from NC_003977.2) were validated and used for PCR, which was performed using an Ex Taq kit (Takara Bio) on an ABI ProFlex instrument (Applied Biosystems). Amplification of the 138 bp products was performed using routine denaturation, annealing (60 °C), and elongation. The products were separated using electrophoresis and sequenced for further verification.
The Genome Walking Kit (Takara Bio) was used to detect HBx integration. With an HBx gene primer at one end and the human genome random primer at the other, specific HBx-integrated fragments in the human genome were amplified after three nested PCR cycles, according to the manufacturer's instructions [19]. Based on the known HBx sequence, three specific interval primers were designed in the same direction (F1: CTGGTGCGCAGACCAATTTATGCC, F2: TAATCTCCTCCCCCAACTCCTCCC, and F3: GGTCGGTCGTTGACATTGCTGAGA). The reverse primer was AP1 for all PCR reactions, which was supplied in the kit. The PCR products were separated and visualized by electrophoresis. Due to the complexity of the integration event, there may be limitations of false negative cases existence.
2.4. Next-generation sequencing for HBx transfected cells
A total amount of 0.5 μg DNA per sample was used as input material for whole-genome sequencing (WGS). DNA libraries were sequenced on the Illumina HiSeq platform following cluster generation. Genomic DNA was purified by paired-end sequencing with at least 30-fold coverage. The paired-end reads were mapped to the human reference genome (UCSC hg38) and HBx plasmid genome (Addgene 24931). If a cluster of multiple (≥2) read pairs was identified with close mapping positions linking an end of hg38 to an end of the HBx plasmid, it was considered as a candidate integration breakpoint.
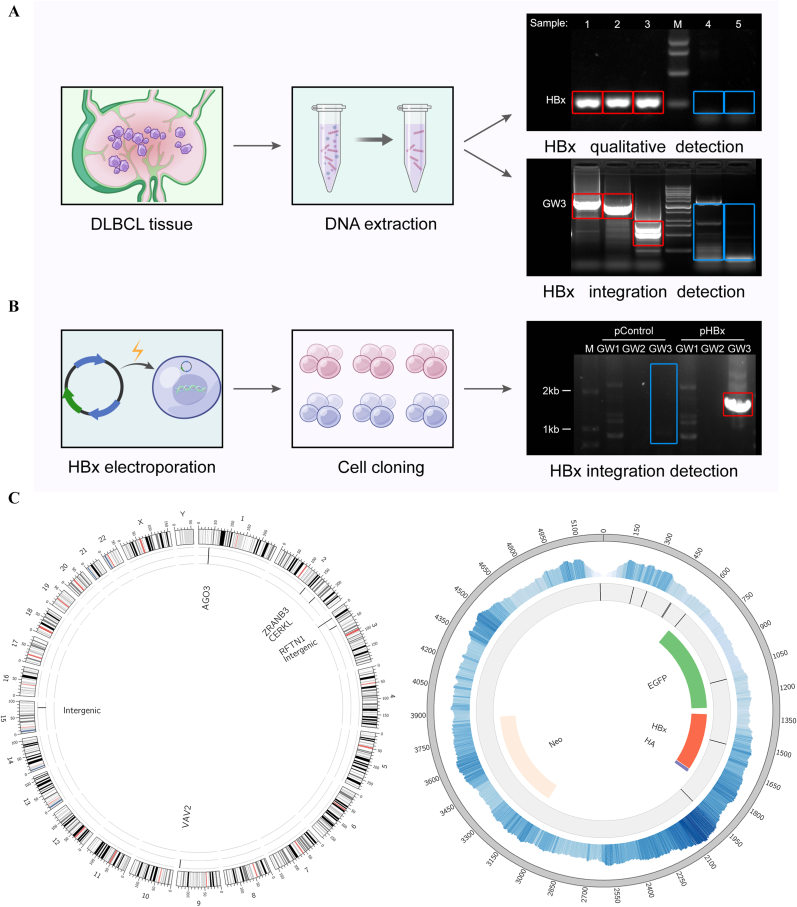
The Circos graph shows the frequency of HBx plasmid integration breakpoints at a particular locus in the human genome. Chromosome numbers and scale bars indicate the number of reads discovered. The frequency of the integration breakpoints at different loci in the HBx plasmid genome is depicted using a blue histogram. Genomic positions are numbered, and the locations of breakpoints and EGFP, HBx, HA, and neo genes are shown.
2.5. Cell proliferation by CCK8
Cell proliferation was analyzed using the CCK8 assay (Beyotime, China). Triplicate cells were cultured at a density of 2.5 × 103/well in 100 μl of medium in 96-well microplates. Sufficient wells for monitoring cell growth over six days were reserved, with the necessary blank wells. The absorbance was analyzed at 450 nm with a microplate reader (Spectramax M5, Molecular Devices, USA) after a 2-h-treatment of 10 μl CCK8 reagent.
2.6. Phenotype detection of apoptosis
Pfeiffer cells and transfected cells were cultured and harvested at a density of 1 × 106 cells per test. The cells were washed twice with PBS. Mitochondrial membrane potential was assessed using flow cytometry. Cells were suspended in warm RPMI 1640 and stained with 200 nM mitochondrial dye MitoTracker Red CMXRos (Invitrogen, USA) at 37 °C for 20 min. After centrifuging and discarding the staining solution, the pellets were gently resuspended in warm culture medium and analyzed using CytoFlex S (Beckman Coulter, USA).
Apoptosis levels were determined using the Annexin V/7-AAD staining method and detected using flow cytometry. Cells were stained with PE labelled Annexin V and nucleic acid dye 7-AAD on ice, according to the instructions for the PE Annexin V Apoptosis Detection Kit I (BD Biosciences, USA). The stained cells were immediately analyzed using CytoFlex S (Beckman Coulter, USA).
The Tunnel assay was used to assess the level of apoptosis using a laser confocal microscope and a Tunnel apoptosis detection kit (Yeasen, China). The cells were fixed with 4 % paraformaldehyde on ice for 20 min and permeabilized with 0.2 % Triton X-100 for 5 min at room temperature. Cells were incubated with Alexa Fluor 640-12-dUTP labelling mix and recombinant TdT enzyme in the dark for 1 h, according to the manufacturer's instructions. EDTA was added to terminate the reaction, followed by washing with 0.1 % Triton X-100 containing 5 mg/ml BSA. Finally, the images were visualized and acquired using DMI6000B/DFC365FX (Leica, Germany).
2.7. Apoptosis array
Proteins in the human apoptosis signaling pathway were detected using a human apoptosis antibody array (R&D Systems, USA) following the manufacturer's instructions. Briefly, total proteins of wild-type Pfeiffer cells (Pfeiffer), HBx-transfected (pHBx), and control plasmid-transfected (pControl) Pfeiffer cells were extracted, and the concentrations were measured using a BCA protein quantification kit (Yeasen, China) adjusted to 300 μg in 250 μl lysis buffer per array. The membrane array was blocked and incubated with the protein samples overnight at 4 °C. The membranes were washed and incubated with an antibody cocktail and streptavidin–HRP solution. Chemi Reagent Mix was used for visualization, and the intensity was measured using an Amersham Imager 600 (General Electric Company, USA).
2.8. Western blot
Whole-cell proteins were extracted with Western IP lysis buffer (Beyotime, China) supplemented with protease inhibitors, and the protein concentrations were measured using a BCA protein quantification kit (Yeasen, China). Protein samples were separated by 10 % polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes (Merck Millipore, USA), and blocked with 5 % non-fat milk (Sangon Biotech, China). The primary antibodies used were β-actin (catalogue 66009-1-Ig, Proteintech, USA), Bax (5023, Cell Signaling Technology (CST), USA), poly ADP-ribose polymerase (PARP) (9532, CST), cleaved PARP (5626, CST), Caspase-3 (SC-7272, Santa Cruz Biotechnology, USA), and cleaved Caspase-3 (9664, CST). The secondary antibodies were HRP-conjugated AffiniPure goat anti-mouse IgG (SA00001-1, Proteintech) and HRP-conjugated AffiniPure goat anti-rabbit IgG (SA00001-2, Proteintech). Signals were detected using an Amersham ECL chemiluminescence system. The relative expression levels of apoptosis-related proteins were represented by signal intensity of each band, which was normalized by the respective control signal intensities, for example, Bax/β-actin, cleaved PARP/PARP, and cleaved Caspase-3/Caspase-3.
2.9. Animal experiment
Four-week-old SPF male BALB/c nude mice (GemPharmatech Co., Ltd.) were randomly divided into three groups with four mice in each group. Mouse xenograft models were established by subcutaneous injection of 100 μl (1 × 107/mouse) of cell suspensions of Pfeiffer, pControl, and pHBx. The weight, tumor length, and width of mice were measured periodically, and the tumor volume was calculated. Four weeks post-injection, the mice were euthanized by carbon dioxide asphyxiation, and the tumors were removed and fixed with paraformaldehyde. HE staining was performed on tumor tissues, and tumor cells were selected to create a tissue chip for subsequent experiments. All animal experiments (FUSCC-IACUC-2022010) performed in the current study were approved by the Ethics Committee of the Fudan University Shanghai Cancer Center.
2.10. Fluorescence in situ hybridization (FISH)
The experiment was performed using FISH on paraffin sections of tissue chips using a specific fluorescence probe of the HBx gene (GTGAAAAAGTTGCATGGTGCTGGT and ACCAATTTATGCCTACAGCCTCCTA) labelled with Cy3 (red). After dewaxing, dehydration, and digestion, hybridization solution was added to the sections with a probe concentration of 600 nM and incubated in a humidity chamber overnight at 40 °C. The solution was discarded, and the sample was washed with an SCC solution. The probe2 hybridization solution was added and incubated in a humidity chamber at 40 °C for 45 min. The same procedure was performed for signal probe hybridization. The cell nuclei were stained with DAPI in the dark. FISH slides were examined using a fluorescence microscope (NIKON ECLIPSE CI, Japan) and scanned using a whole-section in situ imaging system (PANNORAMIC MIDI, 3DHISTECH, Hungary) to analyze fluorescent signals using the matching software (CaseViewer version 2.4, 3DHISTECH). Experiments using clinical DLBCL tissue chips (HLymB085PT01, Shanghai Outdo Biotech, China) were approved by the Ethics Committee of Shanghai Outdo Biotech (YB M-05-01).
2.11. Immunohistochemistry
Primary antibodies against HBx antigens, Bax, cleaved Caspase-3, and cleaved PARP were used for immunohistochemistry (IHC) following the standard recommended protocols. Antigen expression was categorized by determining a previously described immunoreactive score (IRS) [20]. Each spot was assigned an intensity score from 0 to 3, and the proportion of tumor staining for that intensity was recorded in 25 % increments in the range of 0–100 (P0, P1–4), with less than 5 % recorded as zero. A final IRS (range, 0–12) was obtained by adding the products of scores obtained for each intensity multiplied by the proportion of the area stained.
2.12. Statistical analysis
Statistical analysis was performed using SPSS version 20.0 (IBM Corporation, USA), and figures were generated using the GraphPad Prism software version 5.0. The data are expressed as mean ± standard deviation. The mean values obtained from the experiments were compared using one-way analysis of variance with a post-hoc test. Two-way ANOVA with Sidak's multiple comparisons test was used to compare the growth curves. A P-value of less significance was set at P < 0.05.
3. Results
3.1. HBx can be integrated in DLBCL
DLBCL is an HBV-defining condition characterized by strong HBx expression [11]. To explore stable intracellular existence of HBx, we performed PCR experiments on DNA extracted from clinical DLBCL tissues. Three HBx-positive tissues derived from previous study [11] were detected owing to HBx integration from serum HBV-positive patients (numbers 1 to 3, marked with a red box). In contrast, two HBV-negative patients (numbers 4–5, marked with a blue box) were negative for PCR detection and genome working experiments (Fig. 1A).
Fig. 1.
HBx integration in DLBCL.
(A) Five clinical tissue samples of DLBCL from serum HBsAg positive patients (Number 1 to 3) and negative patients (Number 4 and 5) were produced following nucleic acid extraction. HBx gene (HBx) was detected using the PCR method, and a Genome Walking analysis of integration (GW3) was performed. (B) HBx plasmid was electrotransferred into DLBCL cells, Pfeiffer strain. Genome walking was used to verify the integration of the HBx gene into the genome of clonal cells. (C) WGS analysis of HBx transfected DLBCLs performed using the next-generation sequencing platform (Illumina HiSeq). The left panel shows the breakpoint location of HBx integration found in the human genome. The right panel shows the distribution of cytoplasmic reads during sequencing and the breakpoint location of the plasmid during HBx integration.
For further studies, an HBx-integrated model in vitro was developed. HBx plasmid (pHBx, Addgene 24931) and control plasmid (pControl) were transfected into DLBCL Pfeiffer cells by electroporation. Pfeiffer pHBx cells were found with specific HBx integrated fragments (marked in the red box) through Genome Walking method, confirming the integration of the HBx gene into the constructed cells (Fig. 1B). WGS was performed using Illumina HiSeq and seven breakpoints of plasmid fragment integration were found in the human genome (Fig. 1C, left). After analyzing the distribution of plasmid fragments, the middle and downstream fragments of HBx were mainly enriched, and breakpoints on the plasmid during integration were obtained (Fig. 1C, right). This complex genetic profile confirmed the integration of HBx into transfected cells.
3.2. HBx integration induces anti-apoptosis effects
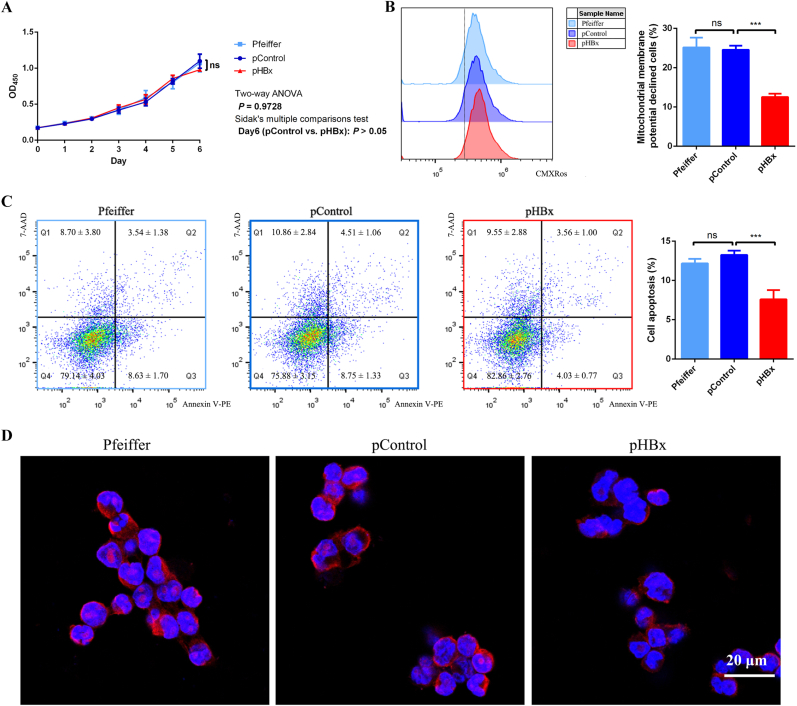
To evaluate the function of the HBx gene in DLBCL cells, variation in biological phenotypes of HBx-transfected (pHBx) Pfeiffer cells compared to wild-type DLBCL Pfeiffer cells (Pfeiffer) and control plasmid-transfected (pControl) Pfeiffer cells were monitored. The proliferative ability of pHBx cells was no significant changes in comparison against Pfeiffer and pControl cells using the CCK8 method (Fig. 2A). Regarding the level of cell apoptosis, mitochondrial membrane potentials were investigated using flow cytometry. The proportion of pHBx cells with decreased mitochondrial membrane potential was significantly reduced (Fig. 2B). Flow cytometric analysis of Annexin V/7-AAD also showed a significant reduction in the proportion of apoptotic cells in pHBx cells (Fig. 2C). Data using the Tunnel method (confocal microscopy) showed that the chromosomal DNA break signal was significantly reduced in pHBx cells (Fig. 2D). These findings indicated that HBx integration significantly reduced apoptosis in DLBCL cells.
Fig. 2.
HBx inhibited apoptosis of DLBCL cells strain Pfeiffer.
Wild-type DLBCL cells Pfeiffer strain (Pfeiffer), HBx-transfected (pHBx), and control plasmid-transfected (pControl) Pfeiffer cells were tested for proliferative and apoptotic phenotypes. (A) Cell proliferation was detected using CCK8 method and the proliferation curve was plotted according to the absorbance. (B) Mitochondrial membrane potential was detected using flow cytometry method and the proportion of cells exhibiting decreased mitochondrial membrane potential was determined based on the threshold line in histogram. (C) The level of apoptosis was detected using the Annexin V/7-AAD method by flow cytometry and the proportion of apoptotic cells was calculated according to the Q2 and Q3 cells in scatter plot. (D) The level of apoptosis was detected by laser confocal microscopy using the TUNEL method. ***P < 0.001, ns for no statistical difference.
3.3. Anti-apoptosis effects are related to the Caspase-3-PARP pathway
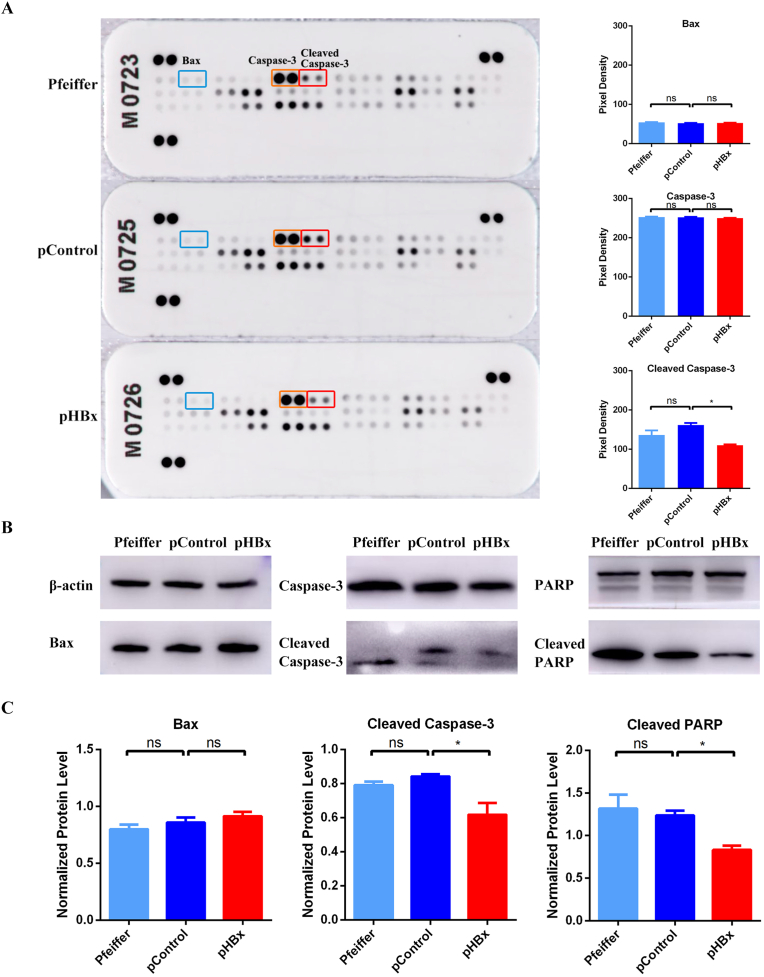
Since HBx can influence the apoptotic phenotype, we further analyzed the molecular mechanisms that cause phenotypic changes. Apoptosis-related proteins were detected in the Pfeffer-transfected cells. Changes in the differential proteins were screened using antibody hybridization of the human apoptosis array (Fig. 3A). Cleaved Caspase-3 levels were lower in pHBx cells than in Pfeiffer and pControl cells, which was consistent with the phenotype of reduced apoptosis levels. No significant differences were detected in the molecular levels of other upstream proteins, such as Bax (marked in the blue box). Western blots of apoptosis-related proteins, such as Bax, Caspase-3 and PARP, were performed (Fig. 3B). Variance analyses and post-hoc tests on the Western blot data revealed that downstream proteins of cleaved Caspase-3 and cleaved PARP, rather than Bax protein, were significantly reduced in pHBx cells, differentiating the Caspase-3-PARP pathway of apoptosis as the most likely molecular mechanism induced by HBx (Fig. 3C).
Fig. 3.
Related molecules of HBx inhibiting apoptosis in Pfeiffer cells.
Proteins from wild-type Pfeiffer cells (Pfeiffer), HBx-transfected (pHBx), and control plasmid-transfected (pControl) Pfeiffer cells were extracted to detect apoptosis-related proteins. (A) Differentially expressed molecules screened by an apoptosis array. (B) Western blot analysis was used to verify the apoptosis-related proteins. (C) Western blot results were analyzed using one-way ANOVA and post-hoc statistics. *P < 0.05, ns indicates no statistically significant difference.
3.4. HBx integrated DLBCLs inhibit apoptosis in vivo
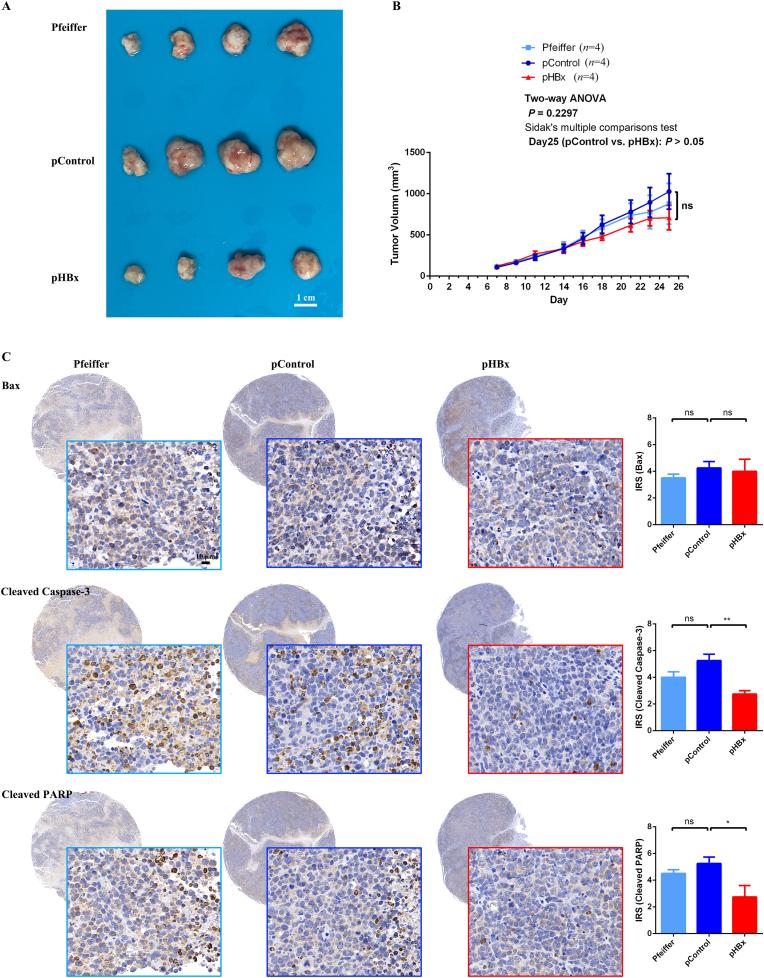
Experiments on the constructed cells in vivo were performed to validate the data obtained in this study. Following subcutaneous injection of Pfeiffer, pControl, and pHBx cells, tumor growth in the mice was monitored. Compared with control cells, pHBx cells showed no significant difference in proliferative ability measured by tumor size (Fig. 4A–B). Regarding the apoptotic molecular pathways, cleaved Caspase-3 and cleaved PARP expression significantly decreased in HBx-transfected cells compared with control cells, whereas Bax expression did not change significantly according to the IRS (Fig. 4C). These results were consistent with the in vitro cell experiments, in which HBx-integrated DLBCLs inhibited apoptosis related to the Caspase-3-PARP pathway.
Fig. 4.
HBx-integrated DLBCLs inhibited apoptosis in vivo.
Suspensions of wild-type Pfeiffer cells (Pfeiffer), HBx-transfected (pHBx), and control plasmid-transfected (pControl) Pfeiffer cells were subcutaneously injected into the backs of BALB/c nude mice (1 × 107/animal, with four mice in each group). (A) Tumor tissue was taken for observation of tumor size; (B) Length and width of the tumor in mice were measured three times per week, and the volume change was calculated; (C) Tumor tissue was fixed, and the expression of Bax, cleaved Caspase-3 and cleaved PARP was detected using the IHC method.
3.5. HBx integration is clinically related to DLBCL apoptosis
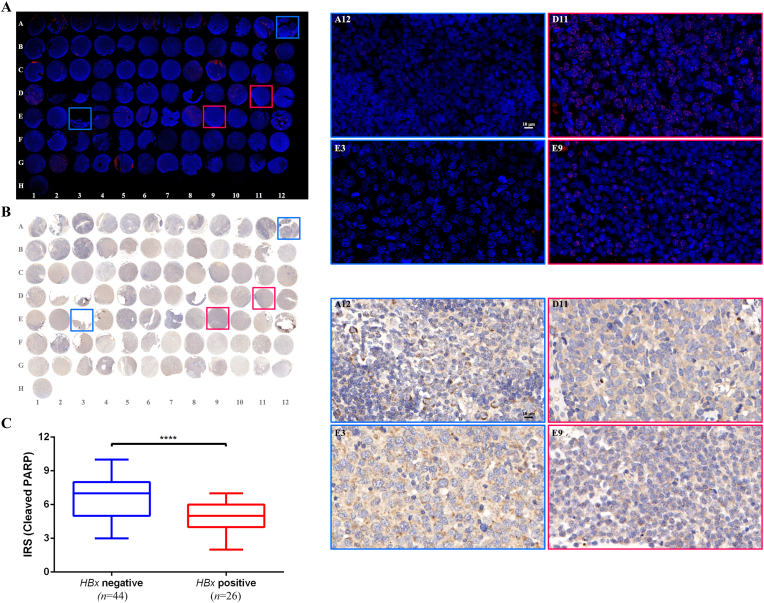
Finally, the results of previous experiments were validated in clinical DLBCL patients. The HBx gene in situ was detected using FISH in a tissue chip (HLymB085PT01) containing 15 tonsillitis lymph node tissues (A1-B3) and 70 DLBCL tissues (B4–H1). HBx gene integration, illustrated in red, was observed in 26 DLBCL cases (37.1 %, 26/70) (Fig. 5A). The cleaved PARP protein was assessed using IHC method and visualized under the corresponding field of view (Fig. 5B). The apoptotic molecular levels in HBx-integrated and HBx-negative DLBCL samples were compared. The HBx-integrated cells had lower cleaved PARP levels than the HBx-negative samples, according to the IRS (P < 0.0001) (Fig. 5C). Notably, the decreased apoptosis induced by HBx observed in the experimental samples was validated in the clinical samples.
Fig. 5.
HBx integrated into clinical DLBCL tissues and was associated with apoptosis marker.
(A) HBx molecules were detected using FISH method in the DLBCL tissue chip (HLymB085PT01), and the integration of HBx genes in situ was demonstrated using specific Cy3 probe-red. The HBx positive (n = 26, 37.1 %) and negative (n = 44, 62.9 %) groups were stratified based on the results of FISH test. (B) The tumor tissue chip was fixed, and the expression of cleaved PARP was detected using IHC method. The IRS between HBx integration and negative group was compared in (C). And the framed spots labelled with coordinates in (A) and (B) were enlarged and displayed in (D) and (E) respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Therefore, HBx gene integration inhibits apoptosis and is correlated with the Caspase-3-PARP pathway in DLBCL. These data validate the critical role of HBx in DLBCL, which requires close clinical monitoring and active intervention.
4. Discussion
This study demonstrated the existence of HBx integration in clinical DLBCL tissues (Fig. 1, Fig. 5), and the constructed HBx-integrated cells exhibited a significantly reduced apoptosis cytological phenotype (Fig. 2). The critical molecules of HBx-related apoptosis were associated with cleaved Caspase-3 and PARP proteins (Fig. 3) and were validated in vivo using a mouse model (Fig. 4) and in clinical cases (Fig. 5). Notably, the wild-type DLBCL Pfeiffer strain and control plasmid transfected cells simultaneously used in this study contribute to assessing the effects of long-term drug screening and cell homeostasis maintenance to avoid controversy from potential integration of plasmid drug resistance gene as compared between wide-type Pfeiffer and pControl cells, and to ensure the effectiveness of integrated HBx function as compared between pHBx and pControl cells.
Integrated HBV DNA has been investigated as a driver of hepatocarcinogenesis [21]. The integration of viral DNA into the host genome may induce genome instability and influence gene expression profile changes near the integration site [22]. However, regardless of whether HBV integration was at a promoter, intron, or exon, some genes were recurrently affected in tumor samples. Sung et al. reported that samples with HBV integration had significantly higher expression levels of TERT, MLL4, and CCNE1 than tumors without HBV DNA integration [15]. Regardless, the integration sites leading to the same phenotypic outcomes could be caused by the integrated viral fragment itself. Therefore, although the constructed HBx integration DLBCL cells showed various integration sites in the current study, data for cells with no differentiation of integration origin were involved, reflecting the changes in the overall level brought about by HBx itself. It is essential to elucidate the viral integration segment of HBx and the integration site within the cell genome in order to further investigate the functional mechanism underlying the interaction between HBV and DLBCL in the future study.
The role of HBx in modulating cell proliferation and programing cell death is replete with controversies regarding different molecular pathways. Decreased apoptosis level may not necessarily be correlated with the level of cell proliferation [23]. Our experiment demonstrated a simultaneous decrease in both apoptotic cells and proliferating mitotic cells of pHBx (raw data available upon request), potentially explaining the lack of difference in total cell proliferation as measured by CCK8 methods. Further exploration of the intermediate regulatory mechanisms is warranted. Regarding the molecular pathway of apoptosis, HBx-induced hepatic apoptosis was determined through the activation of the TNFR1-NFκB pathway [24], although cell malignancy caused by HBx integration was related to reduced apoptosis levels in PARP-related pathways in HCC [25]. The expression of cleaved PARP is efficiently suppressed by HBx [26]. Ren et al. showed a distinctive set of mutated genes in HBsAg-positive DLBCLs, affecting multiple key pathways involving lymphomagenesis [27]. The current study demonstrated that HBx in DLBCL decreased apoptosis through the PARP pathway, which is consistent with the findings in HCC pathogenesis. Declining cleaved PAPR indicated an increase in active PARP, which is essential for maintaining cell stability, and suggests a potential therapeutic target for PARP in DLBCL.
Due to the lack of a lymphoma protein expression database and prognostic information on experimental tissues, no prognostic analysis was performed in this study. However, a considerable number of DLBCL patients are infected with HBV, which is correlated with poor clinical outcomes [28]. The prognostic potential of HBx integration and HBV-related DLBCL remains unknown. HBV infection confers resistance to chemotherapeutic agents that induce S-phase arrest by explicitly inhibiting the activation of CHK2 signaling in DLBCL [29]. Further investigation into apoptotic mechanisms is warranted to elucidate the role of HBx's function in chemotherapy and its impact on prognosis of DLBCL. Given that cleaved PARP levels were reduced in HCC and that the decrease in apoptosis level is beneficial to the survival of tumor cells, a PARP inhibitor was used to induce cell apoptosis in HCC treatment, particularly for HBV-associated tumors [30]. In DLBCL, prophylaxis using antiviral agents in HBsAg-positive patients improves the survival of patients with HBV infection [31]. Therefore, it is reasonable to speculate that treatment targeting PARP in HBV-related DLBCL may have favorable outcomes and warrants further investigation. Investigating whether the integration of HBx impact the treatment efficacy for DLBCL will also be a focus of future research.
In summary, HBx integration inhibits apoptosis through the Caspase-3-PARP pathway in DLBCL. However, the value of PARP as a potential biomarker and therapeutic target in DLBCL requires further study. Thorough characterization of the HBx integration pathway that induces the development of DLBCL remains the focus of improved treatment strategies.
Funding
This work was supported by the National Natural Science Foundation of China (grant Nos. 81800190, 82072876, and 82002618) and the Project of Science and Technology Commission of Shanghai Municipality (grant Nos. 22YF1407900 and 21JM0010404).
Ethical approval
All animal experiments performed in the current study were approved by the Ethics Committee of Fudan University Shanghai Cancer Center (FUSCC-IACUC-2022010). The experiments using clinical DLBCL tissue chips in this study were approved by the Ethics Committee of Shanghai Outdo Biotech (YB M-05-01).
CRediT authorship contribution statement
Yanchun Wang: Writing – review & editing, Writing – original draft, Funding acquisition, Formal analysis, Data curation, Conceptualization. Xiaolin Guan: Writing – original draft, Formal analysis, Data curation. Fangfang Lv: Supervision, Resources. Yi Rong: Writing – review & editing, Formal analysis. Xin Meng: Methodology. Ying Tong: Methodology, Formal analysis. Xiaolu Ma: Funding acquisition, Formal analysis. Hui Zheng: Methodology. Cuncun Chen: Methodology. Suhong Xie: Methodology. Heng Zhang: Methodology, Funding acquisition. Feng Dong: Funding acquisition, Data curation. Lin Guo: Validation, Supervision. Renquan Lu: Writing – review & editing, Validation, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge the Department of Precision Medicine, Fudan University Shanghai Cancer Center for sequencing analysis support.
Contributor Information
Lin Guo, Email: guolin500@hotmail.com.
Renquan Lu, Email: renquanlu@fudan.edu.cn.
Data availability
Data will be made available on request.
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023 Jan;73(1):17–48. doi: 10.3322/caac.21763. Epub 2023/01/13. Cited in: Pubmed; PMID 36633525. [DOI] [PubMed] [Google Scholar]
- 2.Xia C., Dong X., Li H., Cao M., Sun D., He S., Yang F., Yan X., Zhang S., Li N., Chen W. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin. Med. J. 2022 Feb 9;135(5):584–590. doi: 10.1097/CM9.0000000000002108. Epub 2022/02/11. Cited in: Pubmed; PMID 35143424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thandra K.C., Barsouk A., Saginala K., Padala S.A., Barsouk A., Rawla P. Epidemiology of non-hodgkin's lymphoma. Med. Sci. 2021 Jan 30;9(1) doi: 10.3390/medsci9010005. Epub 2021/02/13. Cited in: Pubmed; PMID 33573146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry A.M., Diebold J., Nathwani B.N., MacLennan K.A., Muller-Hermelink H.K., Bast M., Boilesen E., Armitage J.O., Weisenburger D.D. Non-Hodgkin lymphoma in the Far East: review of 730 cases from the international non-Hodgkin lymphoma classification project. Ann. Hematol. 2016 Jan;95(2):245–251. doi: 10.1007/s00277-015-2543-4. Epub 2015/11/06. Cited in: Pubmed; PMID 26537613. [DOI] [PubMed] [Google Scholar]
- 5.Coté T.R., Biggar R.J., Rosenberg P.S., Devesa S.S., Percy C., Yellin F.J., Lemp G., Hardy C., Geodert J.J., Blattner W.A. Non-Hodgkin's lymphoma among people with AIDS: incidence, presentation and public health burden. Int. J. Cancer. 1997;73(5):645–650. doi: 10.1002/(SICI)1097-0215(19971127)73:5<645::AID-IJC6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 6.Huang X., Young K.H., Guo W., Wang Y., Wang X., Xi Y., Wang L., Bai O. Identification of hepatitis B virus aetiologic antigens, HBx and Pre-S2, in diffuse large B-cell lymphoma. J. Viral Hepat. 2020 Sep;27(9):948–950. doi: 10.1111/jvh.13301. Epub 2020/04/14. Cited in: Pubmed; PMID 32281709. [DOI] [PubMed] [Google Scholar]
- 7.Deng L., Song Y., Young K.H., Hu S., Ding N., Song W., Li X., Shi Y., Huang H., Liu W., Zheng W., Wang X., Xie Y., Lin N., Tu M., Ping L., Ying Z., Zhang C., Sun Y., Zhu J. Hepatitis B virus-associated diffuse large B-cell lymphoma: unique clinical features, poor outcome, and hepatitis B surface antigen-driven origin. Oncotarget. 2015 Sep 22;6(28):25061–25073. doi: 10.18632/oncotarget.4677. Epub 2015/09/01. Cited in: Pubmed; PMID 26314957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X., Pan H., Yang P., Ye P., Cao H., Zhou H. Both chronic HBV infection and naturally acquired HBV immunity confer increased risks of B-cell non-Hodgkin lymphoma. BMC Cancer. 2019 May 22;19(1):477. doi: 10.1186/s12885-019-5718-x. Epub 2019/05/23. Cited in: Pubmed; PMID 31113483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zignego A.L., Giannini C., Gragnani L. HCV and lymphoproliferation. Clin. Dev. Immunol. 2012;2012 doi: 10.1155/2012/980942. Epub 2012/08/02. Cited in: Pubmed; PMID 22852020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabrerizo M., Bartolome J., Caramelo C., Barril G., Carreno V. Molecular analysis of hepatitis B virus DNA in serum and peripheral blood mononuclear cells from hepatitis B surface antigen-negative cases. Hepatology. 2000 Jul;32(1):116–123. doi: 10.1053/jhep.2000.8541. Epub 2000/06/28. Cited in: Pubmed; PMID 10869298. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Wang H., Pan S., Hu T., Shen J., Zheng H., Xie S., Xie Y., Lu R., Guo L. Capable infection of hepatitis B virus in diffuse large B-cell lymphoma. J. Cancer. 2018;9(9):1575–1581. doi: 10.7150/jca.24384. Epub 2018/05/16. Cited in: Pubmed; PMID 29760795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bill C.A., Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc. Natl. Acad. Sci. U. S. A. 2004 Jul 27;101(30):11135–11140. doi: 10.1073/pnas.0403925101. Epub 2004/07/20. Cited in: Pubmed; PMID 15258290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brechot C., Gozuacik D., Murakami Y., Paterlini-Brechot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) Semin. Cancer Biol. 2000 Jun;10(3):211–231. doi: 10.1006/scbi.2000.0321. Epub 2000/08/11. Cited in: Pubmed; PMID 10936070. [DOI] [PubMed] [Google Scholar]
- 14.Brechot C., Kremsdorf D., Soussan P., Pineau P., Dejean A., Paterlini-Brechot P., Tiollais P. Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC): molecular mechanisms and novel paradigms. Pathol. Biol. 2010 Aug;58(4):278–287. doi: 10.1016/j.patbio.2010.05.001. Epub 2010/07/30. Cited in: Pubmed; PMID 20667665. [DOI] [PubMed] [Google Scholar]
- 15.Sung W.K., Zheng H., Li S., Chen R., Liu X., Li Y., Lee N.P., Lee W.H., Ariyaratne P.N., Tennakoon C., Mulawadi F.H., Wong K.F., Liu A.M., Poon R.T., Fan S.T., Chan K.L., Gong Z., Hu Y., Lin Z., Wang G., Zhang Q., Barber T.D., Chou W.C., Aggarwal A., Hao K., Zhou W., Zhang C., Hardwick J., Buser C., Xu J., Kan Z., Dai H., Mao M., Reinhard C., Wang J., Luk J.M. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012 May 27;44(7):765–769. doi: 10.1038/ng.2295. Epub 2012/05/29. Cited in: Pubmed; PMID 22634754. [DOI] [PubMed] [Google Scholar]
- 16.Zheng B., Liu X.L., Fan R., Bai J., Wen H., Du L.T., Jiang G.Q., Wang C.Y., Fan X.T., Ye Y.N., Qian Y.S., Wang Y.C., Liu G.J., Deng G.H., Shen F., Hu H.P., Wang H., Zhang Q.Z., Ru L.L., Zhang J., Gao Y.H., Xia J., Yan H.D., Liang M.F., Yu Y.L., Sun F.M., Gao Y.J., Sun J., Zhong C.X., Wang Y., Kong F., Chen J.M., Zheng D., Yang Y., Wang C.X., Wu L., Hou J.L., Liu J.F., Wang H.Y., Chen L. The landscape of cell-free HBV integrations and mutations in cirrhosis and hepatocellular carcinoma patients. Clin. Cancer Res. 2021 Jul 1;27(13):3772–3783. doi: 10.1158/1078-0432.CCR-21-0002. Epub 2021/05/06. Cited in: Pubmed; PMID 33947693. [DOI] [PubMed] [Google Scholar]
- 17.Kumar P., Nagarajan A., Uchil P.D. Electroporation. Cold Spring Harb. Protoc. 2019 Jul 1;2019(7) doi: 10.1101/pdb.top096271. Epub 2019/07/03. Cited in: Pubmed; PMID 31262965. [DOI] [PubMed] [Google Scholar]
- 18.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., Fu L., Song M., Chen P., Gao W., Ren B., Sun Y., Cai T., Feng X., Sui J., Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012 Nov 13;1 doi: 10.7554/eLife.00049. Epub 2012/11/15. Cited in: Pubmed; PMID 23150796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapter F.M., Waters D.L. Genome walking. Methods Mol. Biol. 2014;1099:133–146. doi: 10.1007/978-1-62703-715-0_12. Epub 2013/11/19. Cited in: Pubmed; PMID 24243201. [DOI] [PubMed] [Google Scholar]
- 20.Halon A., Donizy P., Surowiak P., Matkowski R. ERM/Rho protein expression in ductal breast cancer: a 15 year follow-up. Cell. Oncol. 2013 Jun;36(3):181–190. doi: 10.1007/s13402-013-0125-9. Epub 2013/02/20. Cited in: Pubmed; PMID 23420497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu T., Zhang H., Urban S. Hepatitis B virus DNA integration: in vitro models for investigating viral pathogenesis and persistence. Viruses. 2021 Jan 26;13(2) doi: 10.3390/v13020180. Epub 2021/02/04. Cited in: Pubmed; PMID 33530322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuveut C., Wei Y., Buendia M.A. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 2010 Apr;52(4):594–604. doi: 10.1016/j.jhep.2009.10.033. Epub 2010/02/27. Cited in: Pubmed; PMID 20185200. [DOI] [PubMed] [Google Scholar]
- 23.Pich K., Respekta N., Dawid M., Mlyczynska E., Kurowska P., Rak A. New insights into cell apoptosis and proliferation: the potential role of vaspin. J. Physiol. Pharmacol. : Off. j. Polish Phy. Soc. 2021 Dec;72(6) doi: 10.26402/jpp.2021.6.02. eng. Epub 2022/04/05. Cited in: Pubmed; PMID 35377336. [DOI] [PubMed] [Google Scholar]
- 24.Kim J.Y., Song E.H., Lee H.J., Oh Y.K., Choi K.H., Yu D.Y., Park S.I., Seong J.K., Kim W.H. HBx-induced hepatic steatosis and apoptosis are regulated by TNFR1- and NF-kappaB-dependent pathways. J. Mol. Biol. 2010 Apr 9;397(4):917–931. doi: 10.1016/j.jmb.2010.02.016. Epub 2010/02/17. Cited in: Pubmed; PMID 20156456. [DOI] [PubMed] [Google Scholar]
- 25.Al-Anazi M.R., Nazir N., Colak D., Al-Ahdal M.N., Al-Qahtani A.A. Deletion and functional analysis of hepatitis B virus X protein: evidence for an effect on cell cycle regulators. Cell. Physiol. Biochem. 2018;49(5):1987–1998. doi: 10.1159/000493670. Epub 2018/09/21. Cited in: Pubmed; PMID 30235448. [DOI] [PubMed] [Google Scholar]
- 26.Yang C.H., Cho M. Hepatitis B virus X gene differentially modulates cell cycle progression and apoptotic protein expression in hepatocyte versus hepatoma cell lines. J. Viral Hepat. 2013 Jan;20(1):50–58. doi: 10.1111/j.1365-2893.2012.01625.x. Epub 2012/12/13. Cited in: Pubmed; PMID 23231084. [DOI] [PubMed] [Google Scholar]
- 27.Ren W., Ye X., Su H., Li W., Liu D., Pirmoradian M., Wang X., Zhang B., Zhang Q., Chen L., Nie M., Liu Y., Meng B., Huang H., Jiang W., Zeng Y., Li W., Wu K., Hou Y., Wiman K.G., Li Z., Zhang H., Peng R., Zhu S., Pan-Hammarstrom Q. Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood. 2018 Jun 14;131(24):2670–2681. doi: 10.1182/blood-2017-11-817601. Epub 2018/03/17. Cited in: Pubmed; PMID 29545328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D.G., Chen G., Wang C., Ke L.F., Wu H., He H.M., Yang Y., Chen Y.P. Clinicopathological and prognostic features of hepatitis B virus-associated diffuse large B-cell lymphoma: a single-center retrospective study in China. Infect. Agents Cancer. 2021 Aug 17;16(1):57. doi: 10.1186/s13027-021-00396-x. Epub 2021/08/19. Cited in: Pubmed; PMID 34404436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X., Guo X., Xing L., Yue W., Yin H., He M., Wang J., Yang J., Chen J. HBV infection potentiates resistance to S-phase arrest-inducing chemotherapeutics by inhibiting CHK2 pathway in diffuse large B-cell lymphoma. Cell Death Dis. 2018 Jan 19;9(2):61. doi: 10.1038/s41419-017-0097-1. Epub 2018/01/21. Cited in: Pubmed; PMID 29352124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerossier L., Dubois A., Paturel A., Fares N., Cohen D., Merle P., Lachuer J., Wierinckx A., Saintigny P., Bancel B., Selves J., Schnitzler A., Ouine B., Cartier A., de Koning L., Puard V., Bieche I., Hernandez-Vargas H., Hall J., Chemin I. PARP inhibitors and radiation potentiate liver cell death in vitro. Do hepatocellular carcinomas have an achilles' heel? Clin Res Hepatol Gastroenterol. 2021 Sep;45(5) doi: 10.1016/j.clinre.2020.09.014. Epub 2020/11/14. Cited in: Pubmed; PMID 33183998. [DOI] [PubMed] [Google Scholar]
- 31.Huang H.H., Hsiao F.Y., Chen H.M., Wang C.Y., Ko B.S. Antiviral prophylaxis for hepatitis B carriers improves the prognosis of diffuse large B-cell lymphoma in Taiwan - a population-based study. Br. J. Haematol. 2021 Jan;192(1):110–118. doi: 10.1111/bjh.17142. Epub 2020/11/02. Cited in: Pubmed; PMID 33131074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.