Abstract
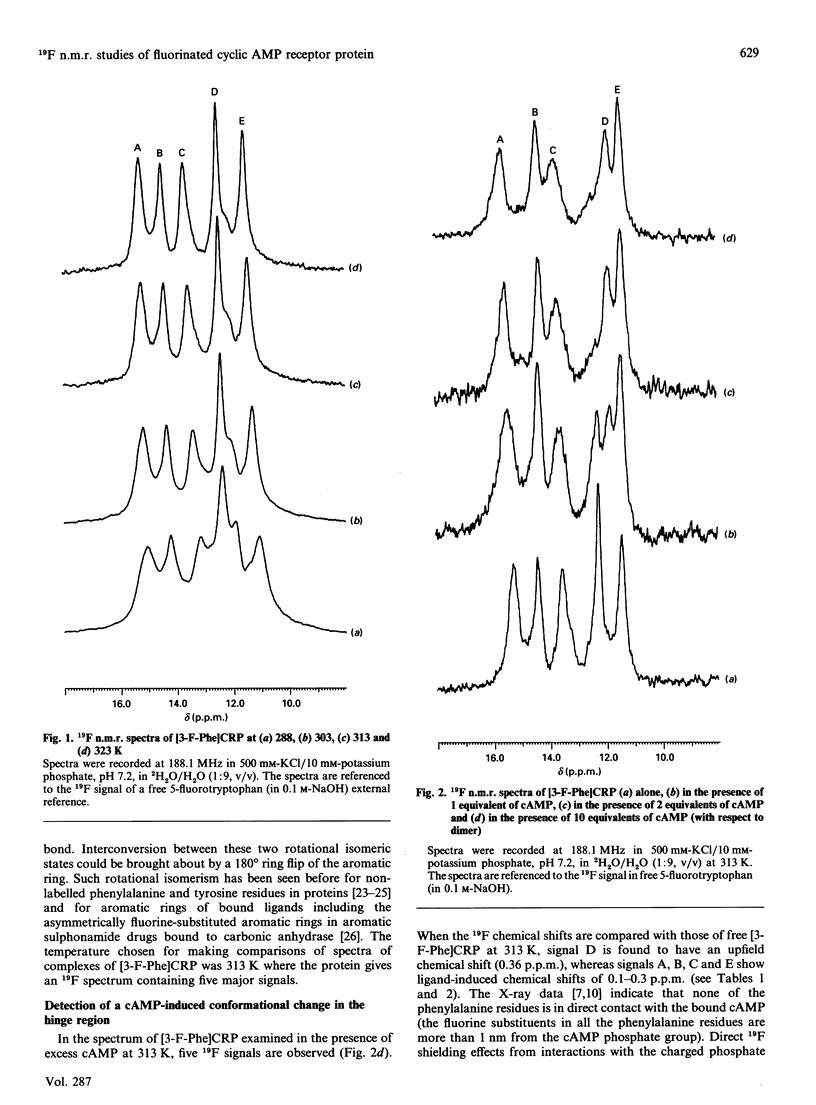
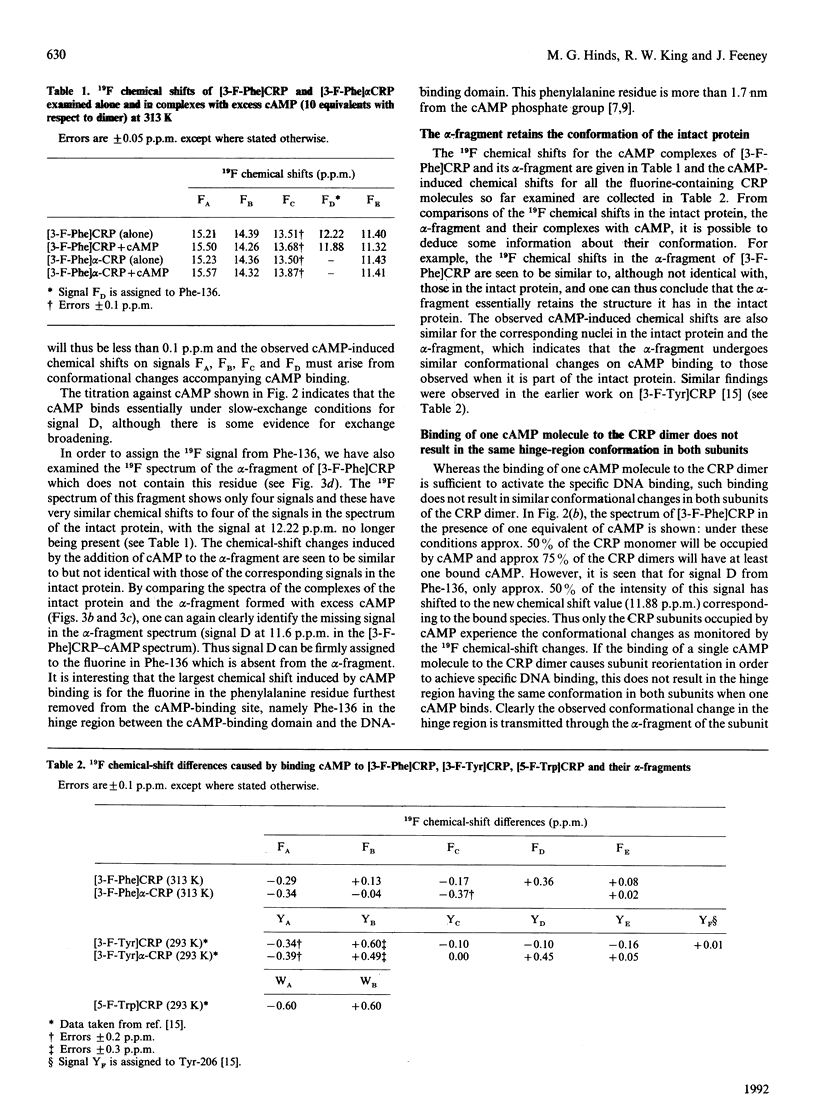
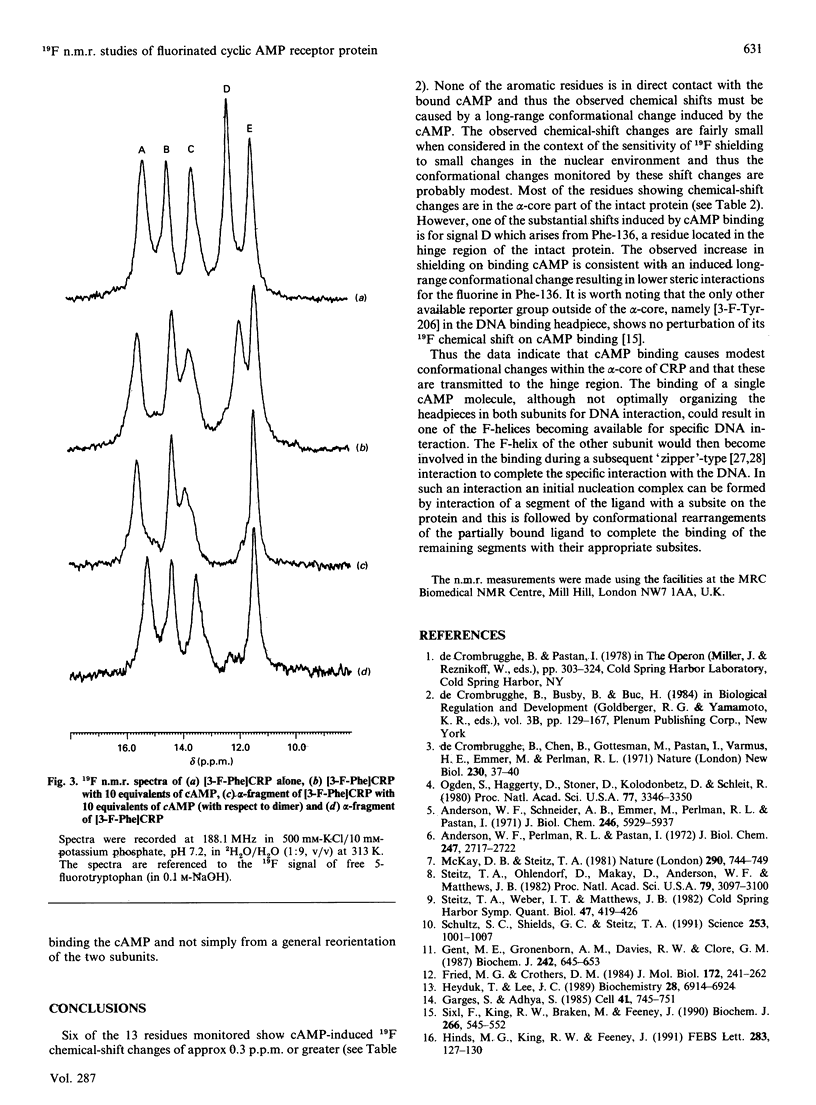
A fluorine-containing analogue of the cyclic AMP (cAMP) receptor protein (CRP) from Escherichia coli was prepared by biosynthetic incorporation of 3-fluorophenylalanine (3-F-Phe). 19F n.m.r. studies on this protein have provided direct evidence for cAMP-induced conformational changes not only within the cAMP-binding domain but also within the hinge region connecting the cAMP-binding domain to the DNA-binding headpiece. At 313 K, the 19F n.m.r. spectrum of [3-F-Phe]CRP showed five signals corresponding to the five phenylalanine residues as expected for a symmetrical dimer. Proteolysis of [3-F-Phe]CRP with subtilisin produced a fragment (the alpha-fragment) containing the cAMP-binding domain. The alpha-fragment contains all the phenylalanines except for Phe-136, a residue located in the hinge region. By comparing the 19F spectra of [3-F-Phe]CRP and its alpha-fragment, the signal for Phe-136 was assigned. The chemical shifts of the corresponding signals in the two spectra are similar, indicating that the alpha-fragment retains the structure it has in the intact protein. The largest cAMP-induced shift was observed for the signal from Phe-136 providing direct evidence for a conformational change in the hinge region. However, whereas binding of a single cAMP molecule to a CRP dimer is known to be sufficient to activate the DNA binding, the n.m.r. data indicate that the hinge region does not have the same conformation in both subunits when only one cAMP molecule is bound.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. B., Perlman R. L., Pastan I. Effect of adenosine 3',5'-monophosphate analogues on the activity of the cyclic adenosine 3',5'-monophosphate receptor in Escherichia coli. J Biol Chem. 1972 May 10;247(9):2717–2722. [PubMed] [Google Scholar]
- Burgen A. S., Roberts G. C., Feeney J. Binding of flexible ligands to macromolecules. Nature. 1975 Feb 27;253(5494):753–755. doi: 10.1038/253753a0. [DOI] [PubMed] [Google Scholar]
- Campbell I. D., Dobson C. M., Williams R. J. Proton magnetic resonance studies of the tyrosine residues of hen lysozyme-assignment and detection of conformational mobility. Proc R Soc Lond B Biol Sci. 1975 Jun 17;189(1097):503–509. doi: 10.1098/rspb.1975.0070. [DOI] [PubMed] [Google Scholar]
- De Crombrugghe B., Chen B., Gottesman M., Pastan I., Varmus H. E., Emmer M., Perlman R. L. Regulation of lac mRNA synthesis in a soluble cell-free system. Nat New Biol. 1971 Mar 10;230(10):37–40. doi: 10.1038/newbio230037a0. [DOI] [PubMed] [Google Scholar]
- Donoso-Pardo J. L., Turner P. C., King R. W. Cyclic nucleotide binding to cAMP receptor protein from Escherichia coli. Optical and ligand-binding studies. Eur J Biochem. 1987 Nov 2;168(3):687–694. doi: 10.1111/j.1432-1033.1987.tb13470.x. [DOI] [PubMed] [Google Scholar]
- Dugad L. B., Cooley C. R., Gerig J. T. NMR studies of carbonic anhydrase-fluorinated benzenesulfonamide complexes. Biochemistry. 1989 May 2;28(9):3955–3960. doi: 10.1021/bi00435a049. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Garges S., Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985 Jul;41(3):745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Gent M. E., Gronenborn A. M., Davies R. W., Clore G. M. Probing the sequence-specific interaction of the cyclic AMP receptor protein with DNA by site-directed mutagenesis. Biochem J. 1987 Mar 15;242(3):645–653. doi: 10.1042/bj2420645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk T., Lee J. C. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry. 1989 Aug 22;28(17):6914–6924. doi: 10.1021/bi00443a021. [DOI] [PubMed] [Google Scholar]
- Hinds M. G., King R. W., Feeney J. 19F NMR evidence for interactions between the c-AMP binding sites on the c-AMP receptor protein from E. coli. FEBS Lett. 1991 May 20;283(1):127–130. doi: 10.1016/0014-5793(91)80569-o. [DOI] [PubMed] [Google Scholar]
- Hull W. E., Sykes B. D. Fluorine-19 nuclear magnetic resonance study of fluorotyrosine alkaline phosphatase: the influence of zinc on protein structure and a conformational change induced by phosphate binding. Biochemistry. 1976 Apr 6;15(7):1535–1546. doi: 10.1021/bi00652a027. [DOI] [PubMed] [Google Scholar]
- Laiken N., Némethy G. A model for the binding of flexible ligands to the surfaces of proteins and other macromolecules. I. Statistical-mechanical treatment. J Phys Chem. 1970 Dec 10;74(25):4421–4431. doi: 10.1021/j100719a022. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Sixl F., King R. W., Bracken M., Feeney J. 19F-n.m.r. studies of ligand binding to 5-fluorotryptophan- and 3-fluorotyrosine-containing cyclic AMP receptor protein from Escherichia coli. Biochem J. 1990 Mar 1;266(2):545–552. doi: 10.1042/bj2660545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder G. H., Rowan R., 3rd, Karplus S., Sykes B. D. Complete tyrosine assignments in the high field 1H nuclear magnetic resonance spectrum of the bovine pancreatic trypsin inhibitor. Biochemistry. 1975 Aug 26;14(17):3765–3777. doi: 10.1021/bi00688a008. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Weber I. T., Matthew J. B. Catabolite gene activator protein: structure, homology with other proteins, and cyclic AMP and DNA binding. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):419–426. doi: 10.1101/sqb.1983.047.01.049. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Wagner G. NMR investigations of the dynamics of the aromatic amino acid residues in the basic pancreatic trypsin inhibitor. FEBS Lett. 1975 Feb 1;50(2):265–268. doi: 10.1016/0014-5793(75)80504-7. [DOI] [PubMed] [Google Scholar]


