ABSTRACT
The major human pathogen Streptococcus pneumoniae has been the subject of intensive clinical and basic scientific study for over 140 years. In multiple instances, these efforts have resulted in major breakthroughs in our understanding of basic biological principles as well as fundamental tenets of bacterial pathogenesis, immunology, vaccinology, and genetics. Discoveries made with S. pneumoniae have led to multiple major public health victories that have saved the lives of millions. Studies on S. pneumoniae continue today, where this bacterium is being used to dissect the impact of the host on disease processes, as a powerful cell biology model, and to better understand the consequence of human actions on commensal bacteria at the population level. Herein we review the major findings, i.e., puzzle pieces, made with S. pneumoniae and how, over the years, they have come together to shape our understanding of this bacterium’s biology and the practice of medicine and modern molecular biology.
KEYWORDS: Streptococcus pneumoniae, discovery, pathogenesis, vaccinology, molecular biology, genetics, model organism
INTRODUCTION
Sir William Osler described pneumonia as “the captain of the men of death” (1). If so, then Streptococcus pneumoniae (the pneumococcus) has long been and remains a stalwart soldier in this legion of death. Most likely first described by Klebs in 1875 (2), it was not until 1881 that both George M. Sternberg and Louis Pasteur independently described the ability of these lancet-shaped, ovoid diplococci to kill their host (3, 4). Pasteur injected saliva from a child who had died of rabies into rabbits, whereas Sternberg did the same with his own saliva. In both instances, the rabbits died, and diplococci were isolated from the bloodstream. Sternberg named the isolated bacteria Micrococcus pasteuri, while Pasteur named this new pathogen microbe septicemique de la salive. By 1886, it was referred to in publications by Fraenkel as “pneumococcus” due to its frequent isolation from individuals with pneumonia (5). In 1920, it was renamed Diplococcus pneumoniae (6). Only in 1974 was this bacterium given the moniker Streptococcus pneumoniae due to its growth as short chains when grown in media (7).
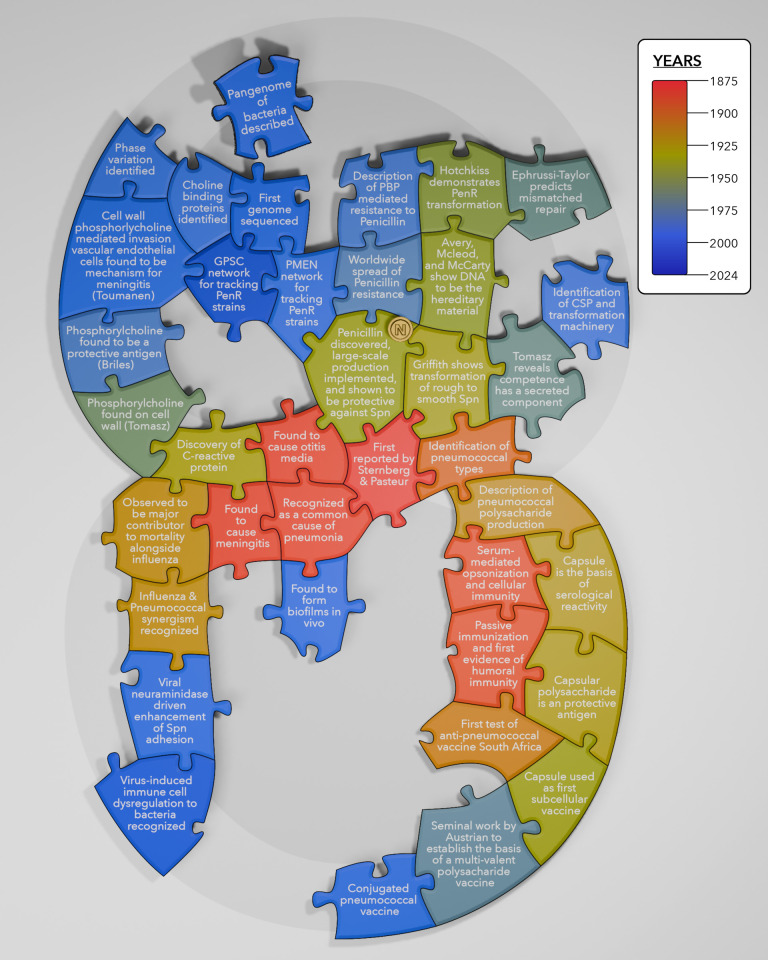
Soon following its discovery, S. pneumoniae was recognized to be a common cause not only of pneumonia but also of otitis media and meningitis (8, 9). Importantly, during the late 1800s and early 1900s, and with a notable spike as a result of the 1918 influenza pandemic, pneumonia was the third leading cause of death overall with S. pneumoniae as a primary culprit (10, 11). Accordingly, S. pneumoniae was extensively studied, and during this golden era of discovery, it was in many instances the microbe used to first describe and subsequently characterize fundamental biological, clinical, and immunological phenomena. These lines of study ultimately resulted in the development of polysaccharide-based vaccines, which have saved the lives of millions (12–14). Additionally, studies revealed DNA as the unit of inheritance (15) marking the start of modern molecular genetics. Subsequent work with S. pneumoniae has pushed the boundaries of our understanding of genetic plasticity, species diversity, and the evolution of bacteria in response to human action. These and other major milestones associated with our understanding pneumococcal pathogenesis are detailed below (Fig. 1).
Fig 1.
The pneumococcus-shaped puzzle highlights major discoveries associated with this bacterium and captures the following key areas of discovery: identification of the species and its association with pathogenesis (central), its central role in the discovery and validation of polysaccharide vaccines (lower right), discoveries related to the transforming principle and gene transfer (upper right), genome sequencing, strain tracking and pangenome studies (top), viral-bacterial interactions (lower left), and characterization of cell wall components (upper left). Puzzle pieces are color coded according to time of the discovery, with empty areas representing yet undiscovered facets of pneumococcal biology or pathogenesis. Created by Emily Krueger, reproduced with permission.
SEROTYPING AND POLYSACCHARIDE-BASED VACCINES
In 1891, Klemperer and Klemperer reported that serum from rabbits injected with heat-killed pneumococci or in vitro broth culture filtrates contained a factor that when infused into another animal conferred protection against lethal challenge with the same strain but not necessarily other clinical isolates (16). In turn, Metchnikoff and Issaeff separately showed that this generated serum could aggregate the bacteria and enhanced the uptake of pneumococci by phagocytes, respectively (17, 18). These were among the first demonstrations of humoral and cellular immunity. Subsequent work by Neufeld showed that S. pneumoniae consisted of distinct “types” to which serum-based immunity could be categorized; i.e., serum to bacteria in one type would not react with bacteria belonging to another type and, when bacteria were mixed with specific antiserum against the same type, would cause agglutination visible to the human eye, and that under a microscope, “quellung” or swelling of the bacterium’s capsule occurred (19). Differential reactivity to antiserum became the basis of pneumococcal serotyping, which remains the most common form of classification for S. pneumoniae. It was not until 1917 that Dochez and Avery first determined that the pneumococcus produced copious levels of polysaccharide during infection (20). Subsequently in 1923, Avery and Heidelberger showed that this polysaccharide was the antigenic basis of serological reactivity (21). This discovery revealed that factors other than proteins could be antigens and expanded the potential repertoire of future vaccine formulations. Today, more than 100 distinct serotypes of S. pneumoniae have been identified. What is more, the biochemical composition of most capsule types has been defined and is now known to be responsible for the antibody-based differences in reactivity (22–24).
Even before capsule was identified as a protective antigen, efforts were under way to determine whether immunization was a viable means to block pneumococcal disease. In 1911, Sir Almroth Wright tested if whole killed pneumococci protected African gold miners against pneumococcal infection (25). His failure to demonstrate efficacy was likely the result of a low dose of antigen administered to the miners as well as the mixture of pneumococcal serotypes causing disease, versus the single strain used for immunization (26). In 1937, immunization with purified capsular polysaccharide was used to stop an outbreak of pneumonia in a state hospital (27). This was the first demonstration of a subcellular vaccine having efficacy against disease. Others who published findings supporting the efficacy of immunization with S. pneumoniae products during this time include Macleod, Heidelberger, and Kaufmann and their respective colleagues (26, 28). In later years, Robert Austrian published seminal work on the topic showing that a polysaccharide vaccine was efficacious for at-risk adults (29, 30). The work was soon extended to include not only capsule vaccine against pneumococcal disease but also capsule vaccine targeting Haemophilus influenzae and Neisseria meningitidis. Ultimately, this body of work led to the creation and licensing of a purified polysaccharide vaccine containing capsules corresponding to 14 different serotypes, which at the time accounted for 80% of pneumococcal disease in the United States. This was expanded to 23-valent in 1983 (31), and this formulation remains licensed for use in adults today.
Following the introduction and success of the protein conjugated Haemophilus influenza type b vaccine developed by Robbins et al. (32, 33), a new seven-valent conjugate vaccine against S. pneumoniae was tested in the 1990s. This version was composed of purified capsular polysaccharide conjugated to the diphtheria toxoid CRM197, conferring reactivity as a T cell-dependent antigen and thus protection of children under the age of 2 years. This new pneumococcal vaccine, subsequently licensed in 2000, was found not only to be efficacious against pneumonia and invasive disease caused by the included serotypes (34, 35) but also to prevent colonization of the nasopharynx, thereby blocking transmission and conferring herd immunity to unvaccinated individuals (36, 37). Based upon this success, but also to thwart the escalation of non-vaccine serotypes, which moved into this unoccupied niche (38, 39), two new conjugate vaccines containing 10 and 13 capsule types were introduced in 2010 (40). The newest versions of these vaccines, 15- and 20-valent, were approved for use in the United States in 2022. Notably, in other countries, S. pneumoniae conjugate vaccines are designed to address their local distribution of serotypes and thus contain a different set of capsular polysaccharides. The preponderance of data indicates that the pneumococcal conjugate vaccine is a major public health triumph, with rates of disease caused by almost all serotypes included within these vaccines having plummeted since their introduction.
Importantly, the licensing of these newer conjugate vaccine formulations was not dependent on large clinical trials testing efficacy against disease, but instead on demonstration that the new vaccine generated a comparable immune response to the serotypes covered by the older conjugate vaccine, that antibody titers against any new capsule types reached a predetermined titer considered to be protective (41), and that the newly generated antibody demonstrated efficacy in an in vitro opsonophagocytosis killing assay (42). This change in licensing requirement and its acceptance by the World Health Organization (WHO) were made possible due to the development of reliable assays for measuring antibody levels and their function (43, 44). In the wake of the success of these vaccines, the approaches used for the pneumococcal vaccines have become the model for developing polysaccharide-based vaccines against many other pathogens.
PENICILLIN AND THE TREATMENT OF PNEUMOCOCCUS
In 1926, Felton and Bailey were able to purify capsular polysaccharide and show that this was the subcellular fraction in heat-killed bacteria responsible for conferring immunity (45). With the understanding that serum containing antibody against capsule was protective, serotherapy, the use of animal-generated antiserum, was in use to treat S. pneumoniae infections by 1913. This treatment reduced mortality from 25.0% to 7.5% (46). Serum therapy relied on first obtaining sputum, followed by serotyping to select the matching antiserum. Thus, this process is perhaps the first example of what is now referred to as personalized medicine (47). Serum therapy was subsequently abandoned due to efficacy of a powerful new form of treatment: antimicrobials (28). In 1931, sulfapyridine was introduced and even used in 1942 to treat Winston Churchill’s pneumonia (48). However, the use of sulfapyridine was soon replaced with penicillin. Penicillin was discovered in 1929, when Fleming characterized a mold contaminant that induced lysis of staphylococcus colonies (49). Twelve years after Fleming’s discovery, a team of British scientists led by Florey and Chain published a groundbreaking and comprehensive study on this cell wall-acting antibiotic, which set the stage for penicillin to revolutionize human medicine. The study described methods for purification of penicillin, its bacterial targets, its efficacy in killing bacteria on cells, animal models, and five patients with pneumonia (50). Fleming and Florey and Chain were awarded the 1945 Nobel Prize in Physiology and Medicine for these contributions. The potential of this new medicine for treatment of wounded World War II soldiers was immediately recognized, with Florey leading a trial in North African military hospitals in 1942 (51). In 1943, penicillin was administered to 500 patients for the treatment of streptococcal infections in the United States and was found to be highly effective against pneumococcal pneumonia (28, 52). Subsequent widespread implementation of penicillin resulted in a 40%–50% decrease in S. pneumoniae-associated mortality among those 12 years of age to the elderly (28, 53). In fact, vaccine development came to a complete halt because of the effectiveness of penicillin. However, in 1964, Austrian and Gold reported that penicillin had no effect on the outcome of bacteremic pneumococcal pneumonia over the first 5 days of infection (54). This meant that the arsenal against the “captain of the men of death” needed to be expanded.
While penicillin analogs today remain a common treatment for pneumococcal infections, the second half of the 20th century was marked by global waves of penicillin resistant (PenR) S. pneumoniae. The first clinical cases of resistance for S. pneumoniae were seen in Boston in 1965, albeit they were not recognized as such (55). Subsequently, PenR S. pneumoniae was reported in Australia in 1967, and by 1974, resistant strains were reported worldwide, with high incidence in South Africa, Hungary, and Spain (55–58). Levels of resistance increased dramatically over the next decades. For example, in a Spanish hospital, the incidence of PenR S. pneumoniae went from 4% in 1979 to 40% in 1990 (57). While rates of PenR invasive infections in the United States remain highly variable across time and geographic regions (59), it is clear that the conjugate vaccine has impacted antimicrobial resistance rates by reducing the prevalence of S. pneumoniae serotypes whose genome encodes antibiotic resistance markers. In this manner, the inclusion of select serotypes in the vaccine is also a means to block the spread of antibiotic-resistant strains. Highlighting the potential risks associated with the spread of this pathogen, today, penicillin non-susceptible pneumococci are categorized as priority 3 by the WHO and drug-resistant S. pneumoniae as a serious threat by the Centers for Disease Control and Prevention (60, 61).
In contrast to most PenR bacteria, the pneumococcus does not produce a β-lactamase that destroys the antimicrobial. Instead, as described in the 1980s in clinical isolates of pneumococci and Neisseria gonorrhoeae, PenR in the pneumococcus arises from modifications in wall transpeptidases [the penicillin binding proteins (PBPs)] that decrease their affinity to this antibiotic (62–64). This alternative mechanism of resistance is common in multiple species of enterococci, Neisseria, and other streptococci, as well as Staphylococcus aureus (65, 66). Sequence comparisons and genetic studies comparing PenR and sensitive strains in the 1980s and 1990s provided global and fine-tuned understanding of the structure-function relationships between PBPs and penicillin and revealed recombination of the DNA encoding PBPs between S. pneumoniae and related Viridans streptococci that resulted in this trait (57, 67–74). These studies lead to the prevailing model that intra- and interspecies recombination events are a prominent mechanism for development of PenR in pneumococci (70, 75). This model continues to be widely accepted and is further supported by in vitro evolution studies (76). Together, the high fitness costs of de novo generation of PenR and the high rates of gene transfer in S. pneumoniae have led to enormous diversity in the array of PBPs found in PenR pneumococci.
Given that the capsule gene locus is frequently a site for exchange by genetic recombination, genetic lineages of S. pneumoniae often encompass multiple different serotypes. Along such lines, the need to track antibiotic resistance for S. pneumoniae beyond their serotype was a major impetus for developing tools to categorize the genetic distribution of specific genetic lineages. One of the first comprehensive ways to determine differences was via pulse-field electrophoresis of genomic DNA (77). Subsequently, and as PCR and DNA sequencing became readily available, multilocus sequence typing became the dominant methodology (78). In 1997, the Pneumococcal Molecular Epidemiology Network was established to better characterize and standardize the identification of antimicrobial-resistant pneumococcal clones (79). More recently, with the emergence of what is perhaps the most extensive collection of sequenced genomes among clinical bacterial isolates as well as the widespread feasibility to sequence new isolates, the Global Pneumococcal Sequencing (GPS) Project (GPS Database) has established a worldwide surveillance network to collect genomic sequence and epidemiological data on strains, including antimicrobial susceptibility (80) (https://www.pneumogen.net/gps/index.html). With over 21,000 whole genomes of S. pneumoniae now publicly available, our understanding of pneumococcal variability and the extent of horizontal gene transfer can be studied at an unprecedented level of resolution. All in all, the discovery and industrial-scale implementation of antimicrobials to treat bacterial infection were revolutionary in human medicine. The subsequent spread of PenR S. pneumoniae highlights the stark ability of this pathogen to change population structure in response to human-based intervention. Finally, the human need to track the epidemiology of resistant strains has in turn served as a window to understand intraspecies genomic variability and plasticity (81).
ANTIBIOTIC TOLERANCE, HETERORESISTANCE, AND AUTOLYSINS
Not all bacteria that survive exposure to antibiotics are resistant, i.e., able to grow in the presence of antimicrobials. Instead, some are tolerant, capable of resuming growth after removal of the antimicrobial. In 1970, Tomasz et al. observed that suppression of the autolytic enzyme LytA led to absence of lysis and killing by cell wall active antibiotics (82). His group went further to describe that escaping the killing activity of antibiotics could be by virtue of genetic mutation (irreversible) or phenotypic growth conditions (reversible), that non-growing bacteria do not necessarily die, and that the rate of growth was directly proportional to the rate of death (83, 84). These findings underpin the field of antibiotic tolerance.
The pneumococcus also exhibits heteroresistance (85). In this case, subpopulations within a monoclonal culture exhibit the ability to grow at antibiotic concentrations above the minimal inhibitory concentration. This resistance is reversible, consistent with a non-heritable response. Yet, the property of heteroresistance is also strain specific, suggesting heteroresistance is influenced by factors encoded in the genome. Along such lines, this property has been associated with the number of altered PBPs, specific low-affinity alleles of pbp2x, and induction of phosphate ABC transporter genes (85, 86). It is tempting to speculate that heteroresistance results from hedge betting behaviors associated with survival in the presence of penicillin. At this time, the genetic determinants or molecular networks that determine the probability of heteroresistance within a population and their clinical consequences remain poorly understood. While the spread of PenR is an undeniable threat to global human health, the clinical consequences of tolerance and heteroresistance remain less clear, yet they likely contribute to relapsing infections and may serve as an intermediate step toward the evolution of resistance.
THE TRANSFORMING PRINCIPLE AND GENETIC PLASTICITY OF S. PNEUMONIAE
Transformation was first discovered in the pneumococcus, and today this bacterium still stands as a paradigm for genomic plasticity. Whereas the clinical importance of horizontal gene transfer is highlighted in the evolution of PenR, our understanding of it started in 1928 when Griffith showed that co-injection of live, attenuated, rough (unencapsulated) pneumococci, formerly type II, with heat-killed encapsulated virulent pneumococci belonging to type III, resulted in death of challenged mice and that only encapsulated bacteria carrying type III capsule were recovered. Thus, rough pneumococci were able to incorporate an element, a “transforming principle,” from the dead bacteria that allowed them to acquire a distinct capsule type and the virulent phenotype (87). Subsequently, in 1944, the nature of this transforming principle was determined in experiments by Avery et al. Using pneumococcal extracts which had carbohydrates and lipids removed, they found that it was the samples treated with DNAse and not proteases that lost their capacity to transform rough pneumococci (15). In 1951, these results were validated by Hotchkiss, who demonstrated that DNA was also responsible for the transformation of PenR (in a capsule-independent manner) (88). The discovery and characterization of DNA as the transforming principle are considered by most to be the start of molecular genetics.
The transfer of DNA across S. pneumoniae strains set the stage for subsequent discoveries. Ephrussi-Taylor and Gray puzzled over the different efficiencies in transformation among strains and in 1966 proposed the existence of the cellular mismatch repair system, which they referred to as the “destruction choice” due to strain-dependent bias in the frequency of incorporated DNA sequences in the recombination site (89). Similarly, and also in the 1960s, Tomasz and colleagues established that a secreted protein was required for the ability of S. pneumoniae to take up DNA and undergo homologous recombination, providing the first evidence for the mechanism underlying transformation and the first example of a pheromone establishing communication between bacteria (90–92). Two decades later, the nature and processing of the competence-inducing peptide were established by Morrison et al. and Havarstein et al. (93, 94). This secreted peptide, a quorum signal, directs population-level behavior when in sufficient concentration, triggering the assembly of a channel for DNA capture and import, as well as transcriptional changes in over 5% of the genome (95, 96). Notably, and in mixed strain populations, activation of competence can activate a second quorum signal that triggers bacteriocin release and the death of neighboring subpopulations (fratricide), which serve as a source of DNA for genetic exchange (97–99).
It is now clear that S. pneumoniae population-level behaviors rely on a multitude of cell-cell communication peptides. These secreted peptides function not only to regulate DNA uptake and competition but also for physiological processes, including nutritional responses, biofilm development, and capsule levels (100–109). Accordingly, many of these peptides are required for colonization and virulence (110). Thus, throughout the 20th century, studies on the transfer of genetic material in the pneumococcus were pivotal for the identification of DNA as the hereditary molecule, shed light on the molecular mechanisms underlying capsule switching (which remained relevant in the context of conjugate vaccine design), and contributed to our understanding of penicillin resistance, cellular mismatch systems, as well as components of quorum sensing and fratricide in bacterial communities.
STUDIES ON CELL MORPHOLOGY
The diversity of bacterial cell shapes in nature results from varied mechanisms of cell growth and division; the pneumococcus being a prototype for ovoid bacteria (111). This shape is formed by two modes of peptidoglycan synthesis: peripheral and septal. These processes are carried out by the elongasome and divisome, respectively (111). Recent studies in S. pneumoniae have shown that the movement of the septal peptidoglycan synthase occurs along a single track at the midcell, propelled by cell wall synthesis (112). Additionally, studies using high-resolution fluorescence microscopy have uncovered the spatial and temporal coordination of both types of peptidoglycan synthesis (113, 114). Highlighting its role as a model organism, studies are uncovering similar patterns of cell wall in other Gram-positive species (115, 116). Finally, while some aspects of S. pneumoniae cell division and growth are widespread, this bacterium also displays atypical features regarding the peptidoglycan, such as minimal turnover, differences in metabolic control of the synthesis precursor pathway, and specialization of its synthesis proteins. Current models suggest ovoid shaped bacteria evolved from a rod-shaped ancestor by gene reduction (117).
THE PNEUMOCOCCAL PANGENOME
In the 1670s, van Leeuwenhoek focused his microscope and observed the bacterial world. Similarly, the development of high-throughput DNA sequencing has revealed another layer of the unseen, the bacterial genome. The first gap-free annotated S. pneumoniae genomes were released in 2001, placing them among the first 30 complete genomes released for a human pathogen (118, 119). By 2005, two groups working in parallel concluded that the pangenome of a species extended well beyond the genes of a single strain, leading to the concept of the core genome, accessory genome, and pangenome (also referred to as supragenome in earlier studies) (120–122). The pneumococcal pangenome was one of the early ones to be described (123, 124). Today, as one of the bacterial species with the greatest number of sequenced genomes, it serves as a valuable model for studies on the characterization and evolution of pangenomes.
The pangenome of S. pneumoniae is both large and highly diverse (125). It encompassed the entire set of genes in a population, and the genes are classified based on their distributions, where core genes are encoded in all strains and accessory genes are encoded in a subset of strains. In an individual strain, approximately one-fifth of the genome consists of accessory genes, and together with single nucleotide polymorphisms (SNPs), these underlie much of strain diversity. The percentage of core and accessory genes in the pangenome is database dependent. Early estimates suggested 50% of the genes in the pangenome were core. Yet, in some data sets, the pangenome is over 10,000 genes, making the numbers of core genes as low as 5% (123, 124, 126). Diversity in the pangenome is likely restricted to gene variants that allow for colonization of the human upper respiratory tract, as this niche is the primary reservoir for S. pneumoniae and the source for its transmission. Our personal observation is that often regions of the genome encode functionally related genes that are not homologs, for instance, restriction enzymes or bacteriocins, highlighting a high degree of interchangeability and specialization among accessory genes (97, 127). Notably, analyses of genetic lineages generally do not reveal a robust correlation between gene content and the ability to successfully colonize a specific niche in the human host. Exceptions exist, such as the classic non-typable strains which lack capsule, i.e., non-encapsulated S. pneumoniae: these form a distinct phyletic group and are associated with eye infections (128–131). The plasticity of the S. pneumoniae genome extends beyond gene diversity and also includes duplication events, as illustrated by a recent study where suppressor mutations have arisen via dosage effects incurred from chromosome duplications (132). Studies of the pneumococcal pangenome have not only shed light on the diversity and plasticity of this species but have also served as an important and mathematical framework for the study of evolution. Mathematical models suggest that the frequencies of accessory genes are shaped by negative frequency dependency, where rare genes are selected for until they become common, at which point they incur a cost, for example, as result of antibody recognition due to prior colonization by another strain carrying the same gene product (133). This suggests a very robust distribution of genes within the pangenome with an equilibrium population composition. Modeling this state provides a framework within which to assess the threat of emerging lineages and predict the impact of interventions, i.e., antimicrobial and vaccine-based, on pneumococcal populations (133). Overall, the plasticity of single strains, incurred by horizontal gene transfer and supported by a diverse pangenome, may allow strains to adapt to host niches and therapeutics. However, the structure of the pangenome, including the frequency at which individual genes are observed within the population, may be constrained. Thus, the S. pneumoniae population is shaped by factors that promote both flexibility and constraint.
Classically, a molecule that when blocked (by gene deletion or inhibition of function) leads to a decrease in virulence or virulence-associated phenotypes is termed a virulence factor. However, it is noteworthy that an estimated 80% of S. pneumoniae virulence determinants, including the genes encoding capsular polysaccharide production and the pore-forming toxin pneumolysin, have orthologs encoded in closely related commensal species or are part of the core genome and found in non-virulent versions of S. pneumoniae, suggesting that their presence alone does not confer pathogenicity (123). Along such lines, transposon-based mutagenesis studies performed in the 1990s and 2000s supplemented by comparative genomic studies and targeted mutagenesis of genes encoded on regions of diversity or pathogenicity islands further support the idea that disease can be potentiated by different bacterial factors and their interplay with diverse host factors (134–136).
Differences between infected individuals also determine what a virulence determinant is. A recent genome-wide association study on a large number of samples of patients and pathogens of pneumococcal meningitis revealed that the genetic diversity of the host may explain almost 50% of the variation in disease severity, shedding light on the extent to which host and bacterial components contribute to invasive disease (137). Moreover, there is also evidence that the fitness landscape within the host drives the genomic composition of the pneumococcus. This is elegantly illustrated by a study of strains isolated from patients with sickle cell disease over a 20-year period (these patients display a 600-fold increase in pneumococcal mortality) (138). The analyses revealed a set of virulence determinants that is distinct from the set in the general population and likely selected for by the high-iron environment, the chronic activated endothelium, and the long-term penicillin pressure in individuals with sickle cell disease. In this context, virulence can be conceptualized as an emergent property driven by both host and pathogen determinants, where the molecular determinants of the pathogen can be shaped by host conditions.
In summation, studies focused on determining the basis of antimicrobial resistance made it clear that S. pneumoniae should not be sorted based on serotype alone. Pangenome studies emphasized this perspective by revealing unexpected diversity across genetic lineages and even within strains belonging to the same lineage. Further, characterization of the pangenome has offered a conceptual framework to consider S. pneumoniae as a dynamic community that exchanges genetic material across strains and related species, resulting in genomic variability and the ability to overcome new and unpredicted selective pressures imposed by the host. Not surprisingly, it is noteworthy that S. pneumoniae is a prototype for the development of CRISPR-based gene expression knockdown systems to assess the contribution of gene products to bacterial fitness without the limitation imposed by gene deletion-mediated bottlenecks (139, 140).
BIOFILM FORMATION DURING COLONIZATION AND GROWTH WITHIN TISSUES
In vivo studies with S. pneumoniae have affirmed that the biofilm state is of major consequence during nasopharyngeal colonization, not necessarily associated with a disease state, and a major contributor to the antimicrobial recalcitrance seen during otitis media. Host factor-triggered dispersal of S. pneumoniae in a biofilm is also a mechanism as to how pneumococci transition from asymptomatic colonizers to potentially deadly opportunistic pathogens. The concept and term “biofilm” was coined and crystalized by J. Willian “Bill” Costerton during his studies of environmental bacteria (141, 142). Work in the 1980s introduced the importance of this mode of bacterial growth in chronic infections including cystic fibrosis and infected internal prosthetic devices (143). In a series of four studies, Ehrlich and colleagues demonstrated the role of biofilms in pneumococcal disease when they demonstrated that bacterial biofilms contribute to the pathogenesis of chronic otitis media (144–147). Over the next decade, biofilms were characterized on abiotic surfaces, on cultured cells, and in vivo (148–151). Further, the genomic determinants of biofilms were explored, demonstrating, for example, that the capsule is highly inhibitory to biofilm formation (152). For S. pneumoniae, the phenotypic implications of growing as biofilms include heightened transformation efficiency, increased tolerance to antimicrobial agents, resistance to desiccation, facilitation of survival on inanimate objects (fomites), and a diminished capacity for invasiveness and immunoreactivity (153–158). Today, biofilm formation is thought to occur as a metabolic response to the bacterium’s environment and to be a key part of the complex interplay between bacterial and host factors. Correspondingly, changes in the host, such as viral infections, microbiota switches, or inflammatory responses, are thought to trigger biofilm dispersal (157). Bacteria dispersed from biofilms exhibit gene expression signatures that differ from both planktonic and biofilm modes of growth and display an enhanced ability to cause infection in murine models of disease (159). Interestingly, there is evidence that S. pneumoniae that are attached to tissue are behaving like biofilms, perhaps due to depletion of nutrients at the level of the infected microenvironment (160). Importantly, capsule production is one of the virulence determinants most strongly affected by carbon availability, and since its presence inhibits attachment and biofilm formation, there is a direct link between metabolism, physiology, and virulence that is increasingly reinforced by emerging metabolism-related studies.
VIRAL BACTERIAL SYNERGY
Respiratory viruses have long been recognized to potentiate bacterial infection with the most severe forms of pneumococcal pneumonia superimposed or closely following a preceding viral infection (161–164). The latter is especially the case for influenza A virus (165), and S. pneumoniae superinfection contributed substantially to the mortality associated with the 1918–1919 influenza pandemic (10, 11). As a result and over the years, scores of studies having explored the molecular mechanisms underlying this synergism, and in many ways, influenza and S. pneumoniae have together served as the prototypes to understand how respiratory viruses exacerbate airway bacterial disease. For example, influenza A virus is known to bind to the surface of S. pneumoniae and other bacteria, enhancing their capacity to adhere to host cells (166). Viral-induced inflammation and influenza neuraminidase activity frees nutrients and exposes receptors for aspirated bacteria to co-opt (167, 168). Influenza disrupts the function of the mucociliary escalator (169) and also causes macrophages to lose efficiency in the uptake of bacteria via scavenger receptors such as MARCO (170). Influenza-induced ion channel dysregulation also causes an increase in the pH of mucosal secretions that reduces the activity of antimicrobial components (171). Pneumococcal gene expression is altered following interactions with influenza and influenza-infected cells. This exposure triggers dispersal from biofilms as well as altered expression of virulence genes including increased expression of pneumococcal surface protein A (PspA), which mediates pneumococcal attachment to dying host cells (157, 172, 173). The latter is important for transmission, as bacteria attached to host cells are shed during colonization, and this enhances their resistance to desiccation (174, 175). Further, influenza has been shown to prime mucosal epithelial cells for pneumolysin-mediated necroptosis and NLRP3-inflammasome-driven pyroptosis during co-infection (158, 176), enhancing disease severity in the airway by altering localization of the bacteria and further releasing factors S. pneumoniae uses for its metabolic benefit (177). Notably, and alongside other work showing that other viruses such as respiratory syncytial virus, rhinovirus, and metapneumovirus potentiate pneumococcal disease, recent work suggests there is also a lethal synergy between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and S. pneumoniae (178–181). Moreover, while superinfections are relatively rare, they drive a meaningful fraction of SARS-CoV-2-associated mortality (182). One explanation is that S. pneumoniae colonization dampens the antiviral response, in turn negatively impacting the generation of antibodies against SARS-Cov-2 (183). Thus, pneumococcus is again at the forefront of attempts to understand the basis of severe respiratory disease.
COLONIZATION FACTORS, PHASE VARIATION, AND INADVERTENT DISEASE
The study of S. pneumonia has also contributed to our general and molecular understanding of how asymptomatic colonizers of the upper airway adapt to the host as disease develops. Sequencing of the S. pneumoniae genome revealed that a major portion of open reading frames was dedicated to acquisition and utilization of diverse carbon sources (119), a feature that most likely reflects the paucity of glucose in the human nasopharynx (184). Importantly, the purpose of S. pneumoniae virulence factors is to promote colonization and transmission to the next host (185, 186). The fact that these determinants are also responsible for disease is incidental to their role in colonization and transmission. With this mindset, investigators have performed genetic screens to identify the factors that are important for transmission and not surprisingly have identified several non-canonical virulence determinants. These included the dlt locus, a determinant of antimicrobial peptide resistance, which enhances pneumococcal shedding by adding d-alanine onto lipoteichoic acid and thereby increasing toll-like receptor recognition and localized inflammation, which promotes mucous secretion and with it bacterial expulsion (187, 188). Likewise, genes involved in DNA repair pathways were found to be required for desiccation resistance (189). Genes involved in fatty acid metabolism, oligopeptide transport, biosynthesis of amino acids, and iron transport have also been identified as being critical. Altogether, these studies suggest that resilience and metabolic aspects are key for the transmission process and that bacteria must be able to accommodate the harsh transitionary environment on a fomite as well as the naïve host from a metabolic perspective (190, 191).
In 1994, Weiser et al. described pneumococcal phase variation for the first time. In brief, the bacterium was found to spontaneously, and at low incidence, change between a transparent and opaque colony phenotype when grown as single colonies on clear agar plates and viewed with oblique light (192). Since then, phase variants have been shown to be different with regard to their expression of multiple surface proteins, levels of capsular polysaccharide and cell wall teichoic acid (or C-polysaccharide), and hydrogen peroxide production, which is a by-product of pyruvate oxidase activity, among other aspects (193–195). Further work led to the understanding that the transparent phenotype, which is more adhesive and carried less capsule, is the predominant phenotype present in the nasopharynx during colonization. In contrast, the opaque phenotype is the version found in the bloodstream (196). Subsequent work showed the opaque phase variants are better protected from immune cells that are present under inflammatory conditions such as otitis media and following an exacerbating event, such as during viral co-infection (197). Importantly, the mechanism for phase variation has been identified as a reversible shift in the methylation pattern caused by DNA inversions in three homologous DNA methyltransferases that are part of a restriction modification system (198, 199). These modifications impact the transcription level of genes across the genome and illustrate how pneumococcal epigenetics serve as an additional layer of single-cell diversity.
In summary, and despite being a major cause of human mortality, S. pneumoniae should be considered in the context of its adaptation to its obligate human host, as a pathobiont—typically asymptomatic colonizer that can cause disease under proper circumstances—which must persist in the upper respiratory tract long enough for transmission to a new host via aerosols and fomites. In turn, we must also consider the epigenomic, transcriptional, and post-transcriptional adaptation that occur once the pneumococcus disseminates away from the nasopharynx to other tissues, causing opportunistic disease (200). The nature and regulation of these virulence factors are most likely selected by fitness advantages associated with robust chronic colonization and greater transmission rates, and not for tissue dissemination or pathogenesis.
CHOLINE-BINDING PROTEINS AND AN INTRACELLULAR ROLE OF THE PNEUMOCOCCUS
C-reactive protein (CRP) was discovered in 1930 in sera of patients with acute pneumococcal pneumonia and was so named as it bound to the C-polysaccharide, i.e., wall teichoic acid, a component of S. pneumoniae cell wall. CRP is now known to be produced by the liver in response to interleukin-6, and the binding of CRP to its ligand, phosphorylcholine residues on wall teichoic and lipoteichoic acid, serves to activate the complement system and opsonize the bacterium (201). In 1967, Tomasz first showed that choline was incorporated on the pneumococcal cell wall (202). Clark and Weiser extended the impact of this finding to show that all respiratory pathogens harbor phosphorylcholine somewhere on their surfaces, not only to bind CRP but also to adhere to the platelet-activating factor receptor on the pulmonary epithelium (203).
Subsequent work by multiple groups, spearheaded by work done on the bacterium’s autolysin LytA and PspA, identified a family of proteins known as choline-binding proteins (204–206). These molecules play critical roles in pneumococcal cell wall remodeling, autolysis, immune evasion, host cell adhesion, and the invasion of host cells and, for this reason, have been considered to be potential vaccine antigens (207). For the most severe forms of disease, S. pneumoniae binds and translocates across vascular endothelial cells. Crossing the blood-brain barrier results in meningitis, and crossing into the bloodstream provides a doorway to other organ systems (208–210). Key virulence determinants for pneumococcal binding and translocation across vascular endothelial cells include the aforementioned phosphorylcholine residues present on wall teichoic and lipoteichoic acid, which bind to platelet-activating factor receptor on host cells (210, 211); choline-binding protein A (alternatively PspC), which binds to host laminin receptor and polymeric immunoglobulin receptor (212, 213); and the pneumococcal pilus, when present, which binds to platelet endothelial cell adhesion molecule 1 (214, 215). Critically, these interactions also take place with mucosal epithelial cells and, as such, are important for colonization. Notably, capsule, which is a requisite for pneumococcal survival in the bloodstream, is generally inhibitory of host cell invasion (210). Along such lines, autolysin-mediated capsule shedding was recently discovered as a way for the pneumococcus not only to evade killing by host cationic antimicrobial peptides but also to facilitate its adhesion to lung cells (216). Yet, still, the production of capsular polysaccharide by S. pneumoniae and other pathogens has also been shown to be important for translocation across vascular endothelial cells and to delay bacterial killing that results from the maturation of endolysosomes (217). Thus, work with S. pneumoniae has shown that the role of capsular polysaccharide on bacterial pathogenesis is multifaceted, its benefits context dependent, and surprisingly includes intracellular survival.
While S. pneumoniae is thought of as an extracellular pathogen and paracellular translocation of tissues is known to occur, a transitionary intracellular role is also a key aspect of its pathogenesis (218, 219). Within vascular endothelial cells, S. pneumoniae spp. exist within clathrin-coated endosomes which evade degradation by acidic lysosomes in a pneumolysin-dependent manner (220, 221). Current work suggests that the pneumococcus is co-opting aspects of LC3-mediated endocytosis to enter the cell and persist (222). Notably, work with S. pneumoniae led to an improved understanding of how bacteria translocate across the blood-brain barrier and even as to why other respiratory tract pathogens are neurotropic. Along such lines, S. pneumoniae, Haemophilus influenzae, and Neisseria meningitidis were all found to bind to platelet-activating factor receptor via phosphorylcholine residues and to laminin receptor (211, 212, 223, 224), albeit adhesion to the latter is through distinct proteins. These interactions initiate the uptake and translocation of bacteria across vascular endothelial cells including the blood-brain barrier (208, 210). Notably, while the majority of these bacteria are killed as a result of this process, some survive and are shuttled to the parenchyma of the organ or released into the central nervous system (210, 217). Intracellular growth within immune cells may also serve as a reservoir for S. pneumoniae during invasive disease. Oggioni and colleagues suggest that intracellular survival in CD169+ splenic macrophages drive pneumococcal bacteremia following an initial clearance event (37). Intracellular pneumococci have also been implicated in the death of cardiomyocytes and heart failure during severe infections (225). Thus, a better understanding of the often-overlooked intracellular aspect of disease is warranted.
CHARACTERIZATION OF CHOLINE-BINDING PROTEINS AND THE POTENTIAL FOR A PROTEIN-BASED VACCINE CANDIDATE
Until the 1980s, when Briles et al. showed that antibody against phosphorylcholine was protective against S. pneumoniae, capsule was generally thought as the only truly protective antigen (226). In 1984, McDaniel, Briles, and colleagues (227) identified a monoclonal antibody against a protease-sensitive pneumococcal antigen that was protective against challenge (16). The knowledge of this molecule, subsequently called pneumococcal surface protein A (18) and shown to be a choline-binding protein, along with the earlier newfound ability to purify the bacterium’s previously discovered pore-forming toxin pneumolysin (228), opened the possibility of creating a protein-based vaccine against the pneumococcus. Subsequent studies, aided in their identification of targets by newly available access to the bacterial genome, and methodologies to screen transposon mutant libraries ultimately resulted in the identification and characterization of virulence associated factors including additional choline-binding proteins, pathogenicity islands, metal acquisition factors, pili, a serine-rich repeat proteins, and other virulence determinants (134, 135, 205, 229–232). Many of these proteins demonstrated protection as antigens in preclinical animal models of pneumonia and sepsis, particularly when used in combination (233–236). However, enthusiasm for a protein-based vaccines ultimately became tempered by the discovery that considerable variation existed for many of these proteins, and individual versions were not always cross-protective (237). Moreover, evidence emerged that many of these proteins were not uniformly present in all pneumococci (238) . What is more, transcriptomic analyses of pneumococcal gene expression in vivo showed considerable variability in gene expression across distinct anatomical sites (160, 239), indicating that the targeted antigen may not be present during some facets of disease. This enhanced understanding of the complexity of any protein-based vaccine occurred in the backdrop of simultaneous continued success of the conjugate vaccine against S. pneumoniae, and, in turn, its steady expansion to include the majority of virulent serotypes, further reduced industry support for a protein-based vaccine. Nonetheless, this work resulted in considerable advancement of our understanding of the molecular mechanisms underlying S. pneumoniae pathogenesis and how the bacterium interacts with host cells and evades the host defense. Work on protein-based antigens continues today, and most of the effort is focused on identification of antigen(s) to be administered alongside the current conjugate vaccine and thereby provides protection against pneumococci whose serotypes are not included in the current conjugate vaccine formulation. There are also studies on the use of synthetic CPS serotypes coupled to immunogenic proteins (240), as well as the possibility of using pneumococcal extracellular vesicles, as these simultaneously display an array of proteins (241). Potential antigens include well-characterized and established molecules such as PspA and pneumolysin, as well as recombinant hybrid proteins that combine multiple antigens (242).
SUMMARY
For over a century, studies focused on pneumococcus have sought to reduce this pathogen’s morbidity and mortality. Yet the impact of this work has extended well beyond pneumococcus as associated findings have yielded major breakthroughs in our understanding of genetics, immunology, antibiotic resistance, and evolution among other areas. The ability to induce competence in the laboratory dramatically facilitated the genetic manipulation of S. pneumoniae, thereby transforming it into a model organism for multiple areas of study. Current vaccines against S. pneumoniae have had remarkable success in reducing the burden of disease and should be considered major public health victories. Yet, despite this, pneumococcus remains a major human pathogen, highlighting the need to improve vaccines and pursue novel antimicrobial targets. Ongoing studies incorporate molecular mechanisms, account for the pneumococcal mode of growth, and consider the impact of the host in the process of pathogenesis. This global perspective on pneumococcal virulence takes into consideration the contribution of bacterial and host nutritional status, immune responses, and surrounding microbes to the pathogenic state. The hope is that these studies will not only address the existing gaps in protection but also continue to uncover fundamental biological tenets.
ACKNOWLEDGMENTS
We thank David E. Briles, Moon Nahm, Hervé Tettelin, Elaine Tuomanen, Jeffery N. Weiser, and Malcolm Winkler for their critical reading of the manuscript and their insights on the biology and history of the pneumococcus. We thank Emily Krueger for the design and implementation of Fig. 1.
Contributor Information
Carlos J. Orihuela, Email: corihuel@uab.edu.
George O'Toole, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire, USA.
REFERENCES
- 1. Osler W. 1901. The principles and practice of medicine. D. Appleton and Co, USA. [Google Scholar]
- 2. Klebs E. 1875. Beitrage zur kenntniss der Schistomyceten. Die Monadinen Archiv fur Exp Path Pharm (Leizpag) 4:409–488. doi: 10.1007/BF01929859 [DOI] [Google Scholar]
- 3. Pasteur L. 1881. Note sur la maladie nouvelle provoquee de la salive d’un enfant mort de la rage. Bulletin de l’Acad Med (Paris) 58:120–129. doi: 10.3406/rmv.1881.2052 [DOI] [Google Scholar]
- 4. Sternberg GM. 1881. A fatal form of septicaemia in the rabbit, produced by the subcutaneous injection of human saliva. Annl Rep Natl Board Health 3:87–108. [Google Scholar]
- 5. Fraenkel A. 1886. Beitrage zu lehre von den mikrococcen der genuinen fibrinosen pneumoniae. Zeitschrift fur Klinische Medicin 11:437–458. [Google Scholar]
- 6. Winslow C-E, Broadhurst J, Buchanan RE, Krumwiede C, Rogers LA, Smith GH. 1920. The families and genera of the bacteria final report of the committee of the society of american bacteriologists on characterization and classification of bacterial types. J Bacteriol 5:191–229. doi: 10.1128/jb.5.3.191-229.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deibel RH, Seeley HWJ. 1974. Bergey’s manual of determinative bacteriology (Eds R. E. Buchanan & N. E. Gibbons). 8th ed, p 490–517. Williams & Wilkens. [Google Scholar]
- 8. Netter . 1887. De la meningite due au pneumocoque (avec ou sans pneumoniae). Archiv General Med Series 7:423–255. [Google Scholar]
- 9. Zaufal E. 1887. Mikrooganismen im secrete der Otitis media acuta. Prager Medicinische Wochenschrif 12 [Google Scholar]
- 10. Chien Y-W, Klugman KP, Morens DM. 2009. Bacterial pathogens and death during the 1918 influenza pandemic. N Engl J Med 361:2582–2583. doi: 10.1056/NEJMc0908216 [DOI] [PubMed] [Google Scholar]
- 11. GBD 2019 Antimicrobial Resistance Collaborators . 2022. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400:2221–2248. doi: 10.1016/S0140-6736(22)02185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Izurieta P, Scherbakov M, Nieto Guevara J, Vetter V, Soumahoro L. 2022. Systematic review of the efficacy, effectiveness and impact of high-valency pneumococcal conjugate vaccines on Otitis media. Hum Vaccin Immunother 18:2013693. doi: 10.1080/21645515.2021.2013693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavia M, Bianco A, Nobile CGA, Marinelli P, Angelillo IF. 2009. Efficacy of pneumococcal vaccination in children younger than 24 months: a meta-analysis. Pediatrics 123:e1103–10. doi: 10.1542/peds.2008-3422 [DOI] [PubMed] [Google Scholar]
- 14. Shinefield HR, Black S. 2000. Efficacy of pneumococcal conjugate vaccines in large scale field trials. Pediatr Infect Dis J 19:394–397. doi: 10.1097/00006454-200004000-00036 [DOI] [PubMed] [Google Scholar]
- 15. Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klemperer G, Klemperer F. 1891. Versuche uber immunisirung und heilung bei der pneumokokkeninection. Berliner Klinische Wochenschrift 28:869–875. [Google Scholar]
- 17. Issaeff B. 1893. Contribution a l’etude de l’immunite acquise contre la pneumocoque. Annales de l’Institut Pasteur 7:260–279. [Google Scholar]
- 18. McDaniel LS, Yother J, Vijayakumar M, McGarry L, Guild WR, Briles DE. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J Exp Med 165:381–394. doi: 10.1084/jem.165.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neufeld F. 1902. Ueber die agglutination der pneumokokken und uber die theorien der aglutination. Zeitschrift fur Hygiene und Infecktionfrnkheiten (Leizpig) 40:54–72. doi: 10.1007/BF02140530 [DOI] [Google Scholar]
- 20. Dochez AR, Avery OT. 1917. The elaboration of specific soluble substance by pneumococcus during growth. J Exp Med 26:477–493. doi: 10.1084/jem.26.4.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Avery OT, Heidelberger M. 1923. Immunological relationships of cell constituents of pneumococcus. J Exp Med 38:81–85. doi: 10.1084/jem.38.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ganaie FA, Saad JS, Lo SW, McGee L, van Tonder AJ, Hawkins PA, Calix JJ, Bentley SD, Nahm MH. 2023. Novel pneumococcal capsule type 33E results from the inactivation of glycosyltransferase WciE in vaccine type 33F. J Biol Chem 299:105085. doi: 10.1016/j.jbc.2023.105085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ganaie F, Saad JS, McGee L, van Tonder AJ, Bentley SD, Lo SW, Gladstone RA, Turner P, Keenan JD, Breiman RF, Nahm MH. 2020. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral Streptococcus. mBio 11:e00937-20. doi: 10.1128/mBio.00937-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright Almroth E, Parry Morgan W, Colebrook L, Dodgson R. 1914. Observations on prophylactic inoculation against pneumococcus infections and on the results which have been achieved by it. The Lancet 183:87–95. doi: 10.1016/S0140-6736(01)56449-1 [DOI] [Google Scholar]
- 26. Austrian R. 1981. Some observations on the pneumococcus and on the current status of pneumococcal disease and its prevention. Clin Infect Dis 3:S1–S17. doi: 10.1093/clinids/3.Supplement_1.S1 [DOI] [PubMed] [Google Scholar]
- 27. Smillie WG, Warnock GH, White HJ. 1938. A study of a type I pneumococcus epidemic at the state hospital at Worcester, mass. Am J Public Health Nations Health 28:293–302. doi: 10.2105/ajph.28.3.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watson DA, Musher DM, Jacobson JW, Verhoef J. 1993. A brief history of the pneumococcus in biomedical research: a panoply of scientific discovery. Clin Infect Dis 17:913–924. doi: 10.1093/clinids/17.5.913 [DOI] [PubMed] [Google Scholar]
- 29. Austrian R. 1977. Prevention of pneumococcal infection by immunization with capsular polysaccharides of Streptococcus pneumoniae: current status of polyvalent vaccines. J Infect Dis 136 Suppl:S38–42. doi: 10.1093/infdis/136.supplement.s38 [DOI] [PubMed] [Google Scholar]
- 30. Austrian R. 1977. Pneumococcal infection and pneumococcal vaccine. N Engl J Med 297:938–939. doi: 10.1056/NEJM197710272971712 [DOI] [PubMed] [Google Scholar]
- 31. Robbins JB, Austrian R, Lee CJ, Rastogi SC, Schiffman G, Henrichsen J, Mäkelä PH, Broome CV, Facklam RR, Tiesjema RH. 1983. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis 148:1136–1159. doi: 10.1093/infdis/148.6.1136 [DOI] [PubMed] [Google Scholar]
- 32. Robbins JB, Schneerson R, Anderson P, Smith DH. 1996. The 1996 Albert Lasker medical research awards prevention of systemic infections, especially meningitis, caused by Haemophilus influenzae type B. impact on public health and implications for other polysaccharide-based vaccines. JAMA 276:1181–1185. doi: 10.1001/jama.276.14.1181 [DOI] [PubMed] [Google Scholar]
- 33. Morris SK, Moss WJ, Halsey N. 2008. Haemophilus influenzae type B conjugate vaccine use and effectiveness. Lancet Infect Dis 8:435–443. doi: 10.1016/S1473-3099(08)70152-X [DOI] [PubMed] [Google Scholar]
- 34. Koshy E, Murray J, Bottle A, Sharland M, Saxena S. 2010. Impact of the seven-valent pneumococcal conjugate vaccination (PCV7) programme on childhood hospital admissions for bacterial pneumonia and empyema in England: national time-trends study, 1997-2008. Thorax 65:770–774. doi: 10.1136/thx.2010.137802 [DOI] [PubMed] [Google Scholar]
- 35. Whitney CG, Pilishvili T, Farley MM, Schaffner W, Craig AS, Lynfield R, Nyquist A-C, Gershman KA, Vazquez M, Bennett NM, Reingold A, Thomas A, Glode MP, Zell ER, Jorgensen JH, Beall B, Schuchat A. 2006. Effectiveness of seven-valent pneumococcal conjugate vaccine against invasive pneumococcal disease: a matched case-control study. The Lancet 368:1495–1502. doi: 10.1016/S0140-6736(06)69637-2 [DOI] [PubMed] [Google Scholar]
- 36. Centers for Disease Control and Prevention (CDC) . 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease--United States, 1998-2003. MMWR Morb Mortal Wkly Rep 54:893–897. [PubMed] [Google Scholar]
- 37. Millar EV, Watt JP, Bronsdon MA, Dallas J, Reid R, Santosham M, O’Brien KL. 2008. Indirect effect of 7‐valent pneumococcal conjugate vaccine on pneumococcal colonization among unvaccinated household members. Clin Infect Dis 47:989–996. doi: 10.1086/591966 [DOI] [PubMed] [Google Scholar]
- 38. Ghaffar F, Barton T, Lozano J, Muniz LS, Hicks P, Gan V, Ahmad N, McCracken GH. 2004. Effect of the 7‐valent pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae in the first 2 years of life. Clin Infect Dis 39:930–938. doi: 10.1086/423379 [DOI] [PubMed] [Google Scholar]
- 39. Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784–1792. doi: 10.1001/jama.297.16.1784 [DOI] [PubMed] [Google Scholar]
- 40. Berman-Rosa M, O’Donnell S, Barker M, Quach C. 2020. Efficacy and effectiveness of the PCV-10 and PCV-13 vaccines against invasive pneumococcal disease. Pediatrics 145:e20190377. doi: 10.1542/peds.2019-0377 [DOI] [PubMed] [Google Scholar]
- 41. Jódar L, Butler J, Carlone G, Dagan R, Goldblatt D, Käyhty H, Klugman K, Plikaytis B, Siber G, Kohberger R, Chang I, Cherian T. 2003. Serological criteria for evaluation and licensure of new pneumococcal conjugate vaccine formulations for use in infants. Vaccine 21:3265–3272. doi: 10.1016/s0264-410x(03)00230-5 [DOI] [PubMed] [Google Scholar]
- 42. Romero-Steiner S, Frasch CE, Carlone G, Fleck RA, Goldblatt D, Nahm MH. 2006. Use of opsonophagocytosis for serological evaluation of pneumococcal vaccines. Clin Vaccine Immunol 13:165–169. doi: 10.1128/CVI.13.2.165-169.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burton RL, Nahm MH. 2006. Development and validation of a fourfold multiplexed opsonization assay (MOPA4) for pneumococcal antibodies. Clin Vaccine Immunol 13:1004–1009. doi: 10.1128/CVI.00112-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wernette CM, Frasch CE, Madore D, Carlone G, Goldblatt D, Plikaytis B, Benjamin W, Quataert SA, Hildreth S, Sikkema DJ, Käyhty H, Jonsdottir I, Nahm MH. 2003. Enzyme-linked immunosorbent assay for quantitation of human antibodies to pneumococcal polysaccharides. Clin Diagn Lab Immunol 10:514–519. doi: 10.1128/cdli.10.4.514-519.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Felton LD, Bailey GH. 1926. Biological significance of the soluble specific substances of pneumococci. J Infect Dis 38:131–144. doi: 10.1093/infdis/38.2.131 [DOI] [Google Scholar]
- 46. Avery OT, Chickering HT, Cole R, Dochez AR. 1917. Acute Lobar pneumonia. Rockefeller Institute for Medical Research. [Google Scholar]
- 47. Scott HP. 2006. Pneumonia before antibiotics therapeutic evolution and evaluation in twentieth-century America. J Clin Invest 116:2311. doi: 10.1172/JCI29920 [DOI] [Google Scholar]
- 48. Christensen SB. 2021. Drugs that changed society: history and current status of the early antibiotics: salvarsan, sulfonamides, and β-lactams. Molecules 26:6057. doi: 10.3390/molecules26196057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fleming A. 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. Influenzæ. Br J Exp Pathol 10:226–236. [Google Scholar]
- 50. Abraham EP, Chain E, Fletcher CM, Gardner AD, Heatley NG, Jennings MA, Florey HW. 1941. Further observations on penicillin. Lancet 238:177–189. doi: 10.1016/S0140-6736(00)72122-2 [DOI] [PubMed] [Google Scholar]
- 51. Lobanovska M, Pilla G. 2017. Penicillin’s discovery and antibiotic resistance: lessons for the future? Yale J Biol Med 90:135–145. [PMC free article] [PubMed] [Google Scholar]
- 52. Keefer CS. 1943. Penicillin in the treatment of infections: a report of 500 cases. JAMA 122:1217. doi: 10.1001/jama.1943.02840350001001 [DOI] [Google Scholar]
- 53. Zumla A. 2010. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Lancet Infect Dis 10:303–304. doi: 10.1016/S1473-3099(10)70089-X [DOI] [Google Scholar]
- 54. Austrian R, Gold J. 1964. Pneumococcal bacteremia with especial reference to bacteremic pneumococcal pneumonia. Ann Intern Med 60:759–776. doi: 10.7326/0003-4819-60-5-759 [DOI] [PubMed] [Google Scholar]
- 55. Appelbaum PC. 1992. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis 15:77–83. doi: 10.1093/clinids/15.1.77 [DOI] [PubMed] [Google Scholar]
- 56. Hansman D, Devitt L, Miles H, Riley I. 1974. Pneumococci relatively insensitive to penicillin in Australia and New Guinea. Med J Aust 2:353–356. doi: 10.5694/j.1326-5377.1974.tb70836.x [DOI] [PubMed] [Google Scholar]
- 57. Liñares J, Pallares R, Alonso T, Perez JL, Ayats J, Gudiol F, Viladrich PF, Martin R. 1992. Trends in antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Bellvitge hospital Barcelona, Spain (1979-1990). Clin Infect Dis 15:99–105. doi: 10.1093/clinids/15.1.99 [DOI] [PubMed] [Google Scholar]
- 58. Marton A. 1992. Pneumococcal antimicrobial resistance: the problem in hungary. Clin Infect Dis 15:106–111. doi: 10.1093/clinids/15.1.106 [DOI] [PubMed] [Google Scholar]
- 59. Reyburn R, Maher J, von Mollendorf C, Gwee A, Mulholland K, Russell F, ARI Review group . 2023. The impact of the introduction of ten- or thirteen-valent pneumococcal conjugate vaccines on antimicrobial-resistant pneumococcal disease and carriage: a systematic literature review. J Glob Health 13:05001. doi: 10.7189/jogh.13.05001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. CDC . 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 61. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 62. Dougherty TJ, Koller AE, Tomasz A. 1980. Penicillin-binding proteins of penicillin-susceptible and intrinsically resistant Neisseria gonorrhoeae. Antimicrob Agents Chemother 18:730–737. doi: 10.1128/AAC.18.5.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hakenbeck R, Tarpay M, Tomasz A. 1980. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 17:364–371. doi: 10.1128/AAC.17.3.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zighelboim S, Tomasz A. 1980. Penicillin-binding proteins of multiply antibiotic-resistant South African strains of Streptococcus pneumoniae. Antimicrob Agents Chemother 17:434–442. doi: 10.1128/AAC.17.3.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Spratt BG. 2012. The 2011 Garrod lecture: from penicillin-binding proteins to molecular epidemiology. J Antimicrob Chemother 67:1578–1588. doi: 10.1093/jac/dks109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zapun A, Contreras-Martel C, Vernet T. 2008. Penicillin-binding proteins and β--lactam resistance. FEMS Microbiol Rev 32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x [DOI] [PubMed] [Google Scholar]
- 67. Coffey TJ, Dowson CG, Daniels M, Zhou J, Martin C, Spratt BG, Musser JM. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol 5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x [DOI] [PubMed] [Google Scholar]
- 68. Dowson CG, Hutchison A, Brannigan JA, George RC, Hansman D, Liñares J, Tomasz A, Smith JM, Spratt BG. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA 86:8842–8846. doi: 10.1073/pnas.86.22.8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Laible G, Spratt BG, Hakenbeck R. 1991. Interspecies recombinational events during the evolution of altered PBP 2X genes in penicillin‐resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol 5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x [DOI] [PubMed] [Google Scholar]
- 70. Reichmann P, König A, Liñares J, Alcaide F, Tenover FC, McDougal L, Swidsinski S, Hakenbeck R. 1997. A global gene pool for high‐level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J Infect Dis 176:1001–1012. doi: 10.1086/516532 [DOI] [PubMed] [Google Scholar]
- 71. Severin A, Figueiredo AM, Tomasz A. 1996. Separation of abnormal cell wall composition from penicillin resistance through genetic transformation of Streptococcus pneumoniae. J Bacteriol 178:1788–1792. doi: 10.1128/jb.178.7.1788-1792.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sibold C, Henrichsen J, König A, Martin C, Chalkley L, Hakenbeck R. 1994. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol 12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x [DOI] [PubMed] [Google Scholar]
- 73. Smith AM, Klugman KP. 1998. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 42:1329–1333. doi: 10.1128/AAC.42.6.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smith AM, Klugman KP. 2005. Amino acid mutations essential to production of an altered PBP 2X conferring high-level β-lactam resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob Agents Chemother 49:4622–4627. doi: 10.1128/AAC.49.11.4622-4627.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hakenbeck R, Chhatwal S. 2007. Molecular biology of Streptococci. Horizon Scientific Press. [Google Scholar]
- 76. Nishimoto AT, Dao TH, Jia Q, Ortiz-Marquez JC, Echlin H, Vogel P, van Opijnen T, Rosch JW. 2022. Interspecies recombination, not de novo mutation, maintains virulence after β-lactam resistance acquisition in Streptococcus pneumoniae. Cell Rep 41:111835. doi: 10.1016/j.celrep.2022.111835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lefevre JC, Faucon G, Sicard AM, Gasc AM. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J Clin Microbiol 31:2724–2728. doi: 10.1128/jcm.31.10.2724-2728.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shi ZY, Enright MC, Wilkinson P, Griffiths D, Spratt BG. 1998. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J Clin Microbiol 36:3514–3519. doi: 10.1128/JCM.36.12.3514-3519.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, Hakenbeck R, Hryniewicz W, Lefévre JC, Tomasz A, Klugman KP. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol 39:2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gladstone RA, Lo SW, Lees JA, Croucher NJ, van Tonder AJ, Corander J, Page AJ, Marttinen P, Bentley LJ, Ochoa TJ, et al. 2019. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine 43:338–346. doi: 10.1016/j.ebiom.2019.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Andam CP, Hanage WP. 2015. Mechanisms of genome evolution of Streptococcus. Infect Genet Evol 33:334–342. doi: 10.1016/j.meegid.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tomasz A, Albino A, Zanati E. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138–140. doi: 10.1038/227138a0 [DOI] [PubMed] [Google Scholar]
- 83. Charpentier E, Tuomanen E. 2000. Mechanisms of antibiotic resistance and tolerance. Microbes Infect 2:1855–1864. doi: 10.1016/S1286-4579(00)01345-9 [DOI] [PubMed] [Google Scholar]
- 84. Tuomanen E. 1986. Phenotypic tolerance: the search for -lactam antibiotics that kill nongrowing bacteria. Clin Infect Dis 8:S279–S291. doi: 10.1093/clinids/8.Supplement_3.S279 [DOI] [PubMed] [Google Scholar]
- 85. Morand B, Mühlemann K. 2007. Heteroresistance to penicillin in Streptococcus pneumoniae. Proc Natl Acad Sci USA 104:14098–14103. doi: 10.1073/pnas.0702377104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Engel H, Mika M, Denapaite D, Hakenbeck R, Mühlemann K, Heller M, Hathaway LJ, Hilty M. 2014. A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 58:3934–3941. doi: 10.1128/AAC.02547-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159. doi: 10.1017/s0022172400031879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hotchkiss RD. 1951. Transfer of penicillin resistance in pneumococci by the desoxyribonucleate derived from resistant cultures. Cold Spring Harb Symp Quant Biol 16:457–461. doi: 10.1101/sqb.1951.016.01.032 [DOI] [PubMed] [Google Scholar]
- 89. Ephrussi-Taylor H, Gray TC. 1966. Genetic studies of recombining DNA in pneumococcal transformation. J Gen Physiol 49:211–231. doi: 10.1085/jgp.49.6.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tomasz A, Hotchkiss RD. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA 51:480–487. doi: 10.1073/pnas.51.3.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tomasz A. 1965. Control of the competent state in pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208:155–159. doi: 10.1038/208155a0 [DOI] [PubMed] [Google Scholar]
- 92. Tomasz A. 1970. Cellular metabolism in genetic transformation of pneumococci: requirement for protein synthesis during induction of competence. J Bacteriol 101:860–871. doi: 10.1128/jb.101.3.860-871.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA 92:11140–11144. doi: 10.1073/pnas.92.24.11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Håvarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x [DOI] [PubMed] [Google Scholar]
- 95. Claverys J-P, Martin B, Polard P. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x [DOI] [PubMed] [Google Scholar]
- 96. Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x [DOI] [PubMed] [Google Scholar]
- 97. Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun 75:443–451. doi: 10.1128/IAI.01775-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Steinmoen H, Teigen A, Håvarstein LS. 2003. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J Bacteriol 185:7176–7183. doi: 10.1128/JB.185.24.7176-7183.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Steinmoen H, Knutsen E, Håvarstein LS. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci USA 99:7681–7686. doi: 10.1073/pnas.112464599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Abdullah IT, Ulijasz AT, Girija UV, Tam S, Andrew P, Hiller NL, Wallis R, Yesilkaya H. 2022. Structure‐function analysis for the development of peptide inhibitors for a gram‐positive quorum sensing system. Mol Microbiol 117:1464–1478. doi: 10.1111/mmi.14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Aggarwal SD, Eutsey R, West-Roberts J, Domenech A, Xu W, Abdullah IT, Mitchell AP, Veening J-W, Yesilkaya H, Hiller NL. 2018. Function of BriC peptide in the pneumococcal competence and virulence portfolio. PLoS Pathog 14:e1007328. doi: 10.1371/journal.ppat.1007328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cuevas RA, Eutsey R, Kadam A, West-Roberts JA, Woolford CA, Mitchell AP, Mason KM, Hiller NL. 2017. A novel streptococcal cell-cell communication peptide promotes pneumococcal virulence and biofilm formation. Mol Microbiol 105:554–571. doi: 10.1111/mmi.13721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hu D, Laczkovich I, Federle MJ, Morrison DA. 2023. Identification and characterization of negative regulators of Rgg1518 quorum sensing in Streptococcus pneumoniae. J Bacteriol 205:e0008723. doi: 10.1128/jb.00087-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kadam A, Eutsey RA, Rosch J, Miao X, Longwell M, Xu W, Woolford CA, Hillman T, Motib AS, Yesilkaya H, Mitchell AP, Hiller NL. 2017. Promiscuous signaling by a regulatory system unique to the pandemic PMEN1 pneumococcal lineage. PLoS Pathog 13:e1006339. doi: 10.1371/journal.ppat.1006339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Motib AS, Al-Bayati FAY, Manzoor I, Shafeeq S, Kadam A, Kuipers OP, Hiller NL, Andrew PW, Yesilkaya H. 2019. TprA/PhrA quorum sensing system has a major effect on pneumococcal survival in respiratory tract and blood, and its activity is controlled by CcpA and GlnR. Front Cell Infect Microbiol 9:326. doi: 10.3389/fcimb.2019.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Motib A, Guerreiro A, Al-Bayati F, Piletska E, Manzoor I, Shafeeq S, Kadam A, Kuipers O, Hiller L, Cowen T, Piletsky S, Andrew PW, Yesilkaya H. 2017. Modulation of quorum sensing in a gram‐positive pathogen by linear molecularly imprinted polymers with anti‐Infective properties. Angew Chem Int Ed Engl 56:16555–16558. doi: 10.1002/anie.201709313 [DOI] [PubMed] [Google Scholar]
- 107. Shlla B, Gazioglu O, Shafeeq S, Manzoor I, Kuipers OP, Ulijasz A, Hiller NL, Andrew PW, Yesilkaya H. 2021. The Rgg1518 transcriptional regulator is a necessary facet of sugar metabolism and virulence in Streptococcus pneumoniae. Mol Microbiol 116:996–1008. doi: 10.1111/mmi.14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhi X, Abdullah IT, Gazioglu O, Manzoor I, Shafeeq S, Kuipers OP, Hiller NL, Andrew PW, Yesilkaya H. 2018. Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis. Sci Rep 8:6369. doi: 10.1038/s41598-018-24910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Laczkovich I, Mangano K, Shao X, Hockenberry AJ, Gao Y, Mankin A, Vázquez-Laslop N, Federle MJ. 2022. Discovery of unannotated small open reading frames in Streptococcus pneumoniae D39 involved in quorum sensing and virulence using ribosome profiling. mBio 13:e0124722. doi: 10.1128/mbio.01247-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Aggarwal SD, Yesilkaya H, Dawid S, Hiller NL. 2020. The pneumococcal social network. PLoS Pathog 16:e1008931. doi: 10.1371/journal.ppat.1008931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pinho MG, Kjos M, Veening J-W. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. doi: 10.1038/nrmicro3088 [DOI] [PubMed] [Google Scholar]
- 112. Perez AJ, Cesbron Y, Shaw SL, Bazan Villicana J, Tsui H-CT, Boersma MJ, Ye ZA, Tovpeko Y, Dekker C, Holden S, Winkler ME. 2019. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc Natl Acad Sci USA 116:3211–3220. doi: 10.1073/pnas.1816018116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Perez AJ, Boersma MJ, Bruce KE, Lamanna MM, Shaw SL, Tsui H-C, Taguchi A, Carlson EE, VanNieuwenhze MS, Winkler ME. 2021. Organization of peptidoglycan synthesis in nodes and separate rings at different stages of cell division of Streptococcus pneumoniae. Mol Microbiol 115:1152–1169. doi: 10.1111/mmi.14659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Trouve J, Zapun A, Arthaud C, Durmort C, Di Guilmi AM, Söderström B, Pelletier A, Grangeasse C, Bourgeois D, Wong Y-S, Morlot C. 2021. Nanoscale dynamics of peptidoglycan assembly during the cell cycle of Streptococcus pneumoniae. Curr Biol 31:2844–2856. doi: 10.1016/j.cub.2021.04.041 [DOI] [PubMed] [Google Scholar]
- 115. Schäper S, Brito AD, Saraiva BM, Squyres GR, Holmes MJ, Garner EC, Hensel Z, Henriques R. 2023. Processive movement of Staphylococcus aureus essential septal peptidoglycan synthases is independent of FtsZ treadmilling and drives cell constriction. doi: 10.1101/2023.06.29.547026 [DOI] [PMC free article] [PubMed]
- 116. Whitley KD, Grimshaw J, Roberts DM, Karinou E, Stansfeld PJ, Holden S. 2023. A one-track model for spatiotemporal coordination of Bacillus subtilis septal cell wall synthesis. doi: 10.1101/2023.06.29.547024 [DOI]
- 117. Massidda O, Nováková L, Vollmer W. 2013. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division?. Environ Microbiol 15:3133–3157. doi: 10.1111/1462-2920.12189 [DOI] [PubMed] [Google Scholar]
- 118. Hoskins J, Alborn WE, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, et al. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
- 120. Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. 2005. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin Orthop Relat Res 437:20–24. doi: 10.1097/00003086-200508000-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ehrlich GD. 2001. “The biofilm and distributed genome paradigms provide a new theoretical structure for understanding chronic bacterial infections” Interscience Conference on Antimicrobials Agents and Chemotherapy (ICAAC); Chicago, Illinois [Google Scholar]
- 122. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome. Proc Natl Acad Sci USA 102:13950–13955. doi: 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol 11:R107. doi: 10.1186/gb-2010-11-10-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol 189:8186–8195. doi: 10.1128/JB.00690-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hiller NL, Sá-Leão R. 2018. Puzzling over the pneumococcal pangenome. Front Microbiol 9:2580. doi: 10.3389/fmicb.2018.02580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. van Tonder AJ, Bray JE, Jolley KA, Jansen van Rensburg M, Quirk SJ, Haraldsson G, Maiden MCJ, Bentley SD, Haraldsson Á, Erlendsdóttir H, Kristinsson KG, Brueggemann AB. 2019. Genomic analyses of >3,100 nasopharyngeal pneumococci revealed significant differences between pneumococci recovered in four different geographical regions. Front Microbiol 10:317. doi: 10.3389/fmicb.2019.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Eutsey RA, Powell E, Dordel J, Salter SJ, Clark TA, Korlach J, Ehrlich GD, Hiller NL. 2015. Genetic stabilization of the drug-resistant PMEN1 pneumococcus lineage by its distinctive DpnIII restriction-modification system. mBio 6:e00173. doi: 10.1128/mBio.00173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Antic I, Brothers KM, Stolzer M, Lai H, Powell E, Eutsey R, Cuevas RA, Miao X, Kowalski RP, Shanks RMQ, Durand D, Hiller NL. 2017. Gene acquisition by a distinct phyletic group within Streptococcus pneumoniae promotes adhesion to the ocular epithelium. mSphere 2:e00213-17. doi: 10.1128/mSphere.00213-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hilty M, Wüthrich D, Salter SJ, Engel H, Campbell S, Sá-Leão R, de Lencastre H, Hermans P, Sadowy E, Turner P, et al. 2014. Global phylogenomic analysis of nonencapsulated Streptococcus pneumoniae reveals a deep-branching classic lineage that is distinct from multiple sporadic lineages. Genome Biol Evol 6:3281–3294. doi: 10.1093/gbe/evu263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Keller LE, Robinson DA, McDaniel LS. 2016. Nonencapsulated Streptococcus pneumoniae: emergence and pathogenesis. mBio 7:e01792. doi: 10.1128/mBio.01792-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Valentino MD, McGuire AM, Rosch JW, Bispo PJM, Burnham C, Sanfilippo CM, Carter RA, Zegans ME, Beall B, Earl AM, Tuomanen EI, Morris TW, Haas W, Gilmore MS. 2014. Unencapsulated Streptococcus pneumoniae from conjunctivitis encode variant traits and belong to a distinct phylogenetic cluster. Nat Commun 5:5411. doi: 10.1038/ncomms6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tsui H-CT, Joseph M, Zheng JJ, Perez AJ, Manzoor I, Rued BE, Richardson JD, Branny P, Doubravová L, Massidda O, Winkler ME. 2023. Negative regulation of MurZ and MurA underlies the essentiality of GpsB‐And StkP ‐mediated protein phosphorylation in Streptococcus pneumoniae. Mol Microbiol 120:351–383. doi: 10.1111/mmi.15122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Azarian T, Grant LR, Arnold BJ, Hammitt LL, Reid R, Santosham M, Weatherholtz R, Goklish N, Thompson CM, Bentley SD, O’Brien KL, Hanage WP, Lipsitch M. 2018. The impact of serotype-specific vaccination on phylodynamic parameters of Streptococcus pneumoniae and the pneumococcal pan-genome. PLoS Pathog 14:e1006966. doi: 10.1371/journal.ppat.1006966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.t01-1-03106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. 2006. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun 74:4766–4777. doi: 10.1128/IAI.00316-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. van Opijnen T, Camilli A. 2012. A fine scale phenotype–genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lees JA, Ferwerda B, Kremer PHC, Wheeler NE, Serón MV, Croucher NJ, Gladstone RA, Bootsma HJ, Rots NY, Wijmega-Monsuur AJ, et al. 2019. Joint sequencing of human and pathogen genomes reveals the genetics of Pneumococcal meningitis. Nat Commun 10:2176. doi: 10.1038/s41467-019-09976-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Carter R, Wolf J, van Opijnen T, Muller M, Obert C, Burnham C, Mann B, Li Y, Hayden RT, Pestina T, Persons D, Camilli A, Flynn PM, Tuomanen EI, Rosch JW. 2014. Genomic analyses of pneumococci from children with sickle cell disease expose host-specific bacterial adaptations and deficits in current interventions. Cell Host Microbe 15:587–599. doi: 10.1016/j.chom.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. de Bakker V, Liu X, Bravo AM, Veening J-W. 2022. CRISPRi-Seq for genome-wide fitness quantification in bacteria. Nat Protoc 17:252–281. doi: 10.1038/s41596-021-00639-6 [DOI] [PubMed] [Google Scholar]
- 140. Liu X, Kimmey JM, Matarazzo L, de Bakker V, Van Maele L, Sirard J-C, Nizet V, Veening J-W. 2021. Exploration of bacterial bottlenecks and Streptococcus pneumoniae pathogenesis by CRISPRI-Seq. Cell Host Microbe 29:107–120. doi: 10.1016/j.chom.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Costerton JW, Geesey GG, Cheng K-J. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86 [DOI] [PubMed] [Google Scholar]
- 142. McCoy WF, Bryers JD, Robbins J, Costerton JW. 1981. Observations of fouling biofilm formation. Can J Microbiol 27:910–917. doi: 10.1139/m81-143 [DOI] [PubMed] [Google Scholar]
- 143. Costerton JW. 1984. The etiology and persistence of cryptic bacterial infections: a hypothesis. Clin Infect Dis 6:S608–S616. doi: 10.1093/clinids/6.Supplement_3.S608 [DOI] [PubMed] [Google Scholar]
- 144. Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ, Daigle BJ, Ehrlich MD, Post JC. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of Otitis media. JAMA 287:1710–1715. doi: 10.1001/jama.287.13.1710 [DOI] [PubMed] [Google Scholar]
- 145. Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic Otitis media. JAMA 296:202–211. doi: 10.1001/jama.296.2.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Post JC, Preston RA, Aul JJ, Larkins-Pettigrew M, Rydquist-White J, Anderson KW, Wadowsky RM, Reagan DR, Walker ES, Kingsley LA, Magit AE, Ehrlich GD. 1995. Molecular analysis of bacterial pathogens in Otitis media with effusion. JAMA 273:1598–1604. [PubMed] [Google Scholar]
- 147. Rayner MG, Zhang Y, Gorry MC, Chen Y, Post JC, Ehrlich GD. 1998. Evidence of bacterial metabolic activity in culture-negative Otitis media with effusion. JAMA 279:296–299. doi: 10.1001/jama.279.4.296 [DOI] [PubMed] [Google Scholar]
- 148. Blanchette-Cain K, Hinojosa CA, Akula Suresh Babu R, Lizcano A, Gonzalez-Juarbe N, Munoz-Almagro C, Sanchez CJ, Bergman MA, Orihuela CJ. 2013. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio 4:e00745–13. doi: 10.1128/mBio.00745-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, Johnson C, Hu FZ, Stoodley P, Ehrlich GD, Post JC. 2008. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol 8:173. doi: 10.1186/1471-2180-8-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Moscoso M, García E, López R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol 188:7785–7795. doi: 10.1128/JB.00673-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew PW, Pozzi G. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol 61:1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Muñoz-Elías EJ, Marcano J, Camilli A. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun 76:5049–5061. doi: 10.1128/IAI.00425-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gilley RP, Orihuela CJ. 2014. Pneumococci in biofilms are non-invasive: implications on nasopharyngeal colonization. Front Cell Infect Microbiol 4:163. doi: 10.3389/fcimb.2014.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Marks LR, Parameswaran GI, Hakansson AP. 2012. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect Immun 80:2744–2760. doi: 10.1128/IAI.00488-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Marks LR, Reddinger RM, Hakansson AP. 2012. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. mBio 3:e00200-12. doi: 10.1128/mBio.00200-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Marks LR, Reddinger RM, Hakansson AP. 2014. Biofilm formation enhances fomite survival of Streptococcus pneumoniae and Streptococcus pyogenes. Infect Immun 82:1141–1146. doi: 10.1128/IAI.01310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Pettigrew MM, Marks LR, Kong Y, Gent JF, Roche-Hakansson H, Hakansson AP. 2014. Dynamic changes in the Streptococcus pneumoniae transcriptome during transition from biofilm formation to invasive disease upon influenza A virus infection. Infect Immun 82:4607–4619. doi: 10.1128/IAI.02225-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Trappetti C, Oggioni MR. 2015. Biofilm formation under in vitro conditions, p 245–255. In Streptococcus pneumoniae. Elsevier. [Google Scholar]
- 159. Marks LR, Davidson BA, Knight PR, Hakansson AP. 2013. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4:e00438-13. doi: 10.1128/mBio.00438-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. D’Mello A, Riegler AN, Martínez E, Beno SM, Ricketts TD, Foxman EF, Orihuela CJ, Tettelin H. 2020. An in vivo atlas of host–pathogen transcriptomes during Streptococcus pneumoniae colonization and disease. Proc Natl Acad Sci USA 117:33507–33518. doi: 10.1073/pnas.2010428117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Hament J-M, Aerts PC, Fleer A, van Dijk H, Harmsen T, Kimpen JLL, Wolfs TFW. 2005. Direct binding of respiratory syncytial virus to pneumococci: a phenomenon that enhances both pneumococcal adherence to human epithelial cells and pneumococcal invasiveness in a murine model. Pediatr Res 58:1198–1203. doi: 10.1203/01.pdr.0000188699.55279.1b [DOI] [PubMed] [Google Scholar]
- 162. Gwaltney JM, Sande MA, Austrian R, Hendley JO. 1975. Spread of Streptococcus pneumoniae in families. II. relation of transfer of S. pneumoniae to incidence of colds and serum antibody. J Infect Dis 132:62–68. doi: 10.1093/infdis/132.1.62 [DOI] [PubMed] [Google Scholar]
- 163. Avadhanula V, Rodriguez CA, Devincenzo JP, Wang Y, Webby RJ, Ulett GC, Adderson EE. 2006. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol 80:1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. McCullers JA. 2014. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12:252–262. doi: 10.1038/nrmicro3231 [DOI] [PubMed] [Google Scholar]
- 165. Klugman KP, Chien Y-W, Madhi SA. 2009. Pneumococcal pneumonia and influenza: a deadly combination. Vaccine 27 Suppl 3:C9–C14. doi: 10.1016/j.vaccine.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 166. Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. 2019. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol 4:1328–1336. doi: 10.1038/s41564-019-0447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. McCullers JA, Bartmess KC. 2003. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis 187:1000–1009. doi: 10.1086/368163 [DOI] [PubMed] [Google Scholar]
- 168. Sender V, Hentrich K, Henriques-Normark B. 2021. Virus-induced changes of the respiratory tract environment promote secondary infections with Streptococcus pneumoniae. Front Cell Infect Microbiol 11:643326. doi: 10.3389/fcimb.2021.643326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Pittet LA, Hall-Stoodley L, Rutkowski MR, Harmsen AG. 2010. Influenza virus infection decreases tracheal mucociliary velocity and clearance of Streptococcus pneumoniae. Am J Respir Cell Mol Biol 42:450–460. doi: 10.1165/rcmb.2007-0417OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sun K, Metzger DW. 2008. Inhibition of pulmonary antibacterial defense by interferon--γ during recovery from influenza infection. Nat Med 14:558–564. doi: 10.1038/nm1765 [DOI] [PubMed] [Google Scholar]
- 171. Earnhardt EY, Tipper JL, D’Mello A, Jian M-Y, Conway ES, Mobley JA, Orihuela CJ, Tettelin H, Harrod KS. 2023. Influenza A–induced cystic fibrosis transmembrane conductance regulator dysfunction increases susceptibility to Streptococcus pneumoniae. JCI Insight 8:e170022. doi: 10.1172/jci.insight.170022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Park S-S, Gonzalez-Juarbe N, Riegler AN, Im H, Hale Y, Platt MP, Croney C, Briles DE, Orihuela CJ. 2021. Streptococcus pneumoniae binds to host GAPDH on dying lung epithelial cells worsening secondary infection following influenza. Cell Rep 35:109267. doi: 10.1016/j.celrep.2021.109267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Platt MP, Lin Y-H, Penix T, Wiscovitch-Russo R, Vashee I, Mares CA, Rosch JW, Yu Y, Gonzalez-Juarbe N. 2022. A multiomics analysis of direct interkingdom dynamics between influenza a virus and Streptococcus pneumoniae uncovers host-independent changes to bacterial virulence fitness. PLoS Pathog 18:e1011020. doi: 10.1371/journal.ppat.1011020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Lane JR, Tata M, Yasmin R, Im H, Briles DE, Orihuela CJ. 2023. PspA-mediated aggregation protects Streptococcus pneumoniae against desiccation on fomites. mBio 14. doi: 10.1128/mbio.02634-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. French AJ, Rockey NC, Sage VL, Brown KM, Shephard MJ, Frizzell S, Myerburg MM, Hiller NL, Lakdawala SS. 2023. Detection of influenza virus and Streptococcus pneumoniae in air sampled from co-infected ferrets and analysis of their influence on pathogen stability. mSphere 8:e00039-23. doi: 10.1101/2023.02.24.529988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Gonzalez-Juarbe N, Riegler AN, Jureka AS, Gilley RP, Brand JD, Trombley JE, Scott NR, Platt MP, Dube PH, Petit CM, Harrod KS, Orihuela CJ. 2020. Influenza-induced oxidative stress sensitizes lung cells to bacterial-toxin-mediated necroptosis. Cell Rep 32:108062. doi: 10.1016/j.celrep.2020.108062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Park S-S, Gonzalez-Juarbe N, Martínez E, Hale JY, Lin Y-H, Huffines JT, Kruckow KL, Briles DE, Orihuela CJ. 2021. Streptococcus pneumoniae binds to host lactate dehydrogenase via PspA and PspC to enhance virulence. mBio 12:e00673-21. doi: 10.1128/mBio.00673-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Barman TK, Singh AK, Bonin JL, Nafiz TN, Salmon SL, Metzger DW. 2022. Lethal synergy between SARS-Cov-2 and Streptococcus pneumoniae in hACE2 mice and protective efficacy of vaccination. JCI Insight 7:e159422. doi: 10.1172/jci.insight.159422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Kukavica-Ibrulj I, Hamelin M-E, Prince GA, Gagnon C, Bergeron Y, Bergeron MG, Boivin G. 2009. Infection with human metapneumovirus predisposes mice to severe pneumococcal pneumonia. J Virol 83:1341–1349. doi: 10.1128/JVI.01123-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Karppinen S, Teräsjärvi J, Auranen K, Schuez-Havupalo L, Siira L, He Q, Waris M, Peltola V. 2017. Acquisition and transmission of Streptococcus pneumoniae are facilitated during rhinovirus infection in families with children. Am J Respir Crit Care Med 196:1172–1180. doi: 10.1164/rccm.201702-0357OC [DOI] [PubMed] [Google Scholar]
- 181. Dagan R, van der Beek BA, Ben-Shimol S, Greenberg D, Shemer-Avni Y, Weinberger DM, Danino D. 2023. The COVID-19 pandemic as an opportunity for unravelling the causative association between respiratory viruses and pneumococcus-associated disease in young children: a prospective study. EBioMedicine 90:104493. doi: 10.1016/j.ebiom.2023.104493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Patton MJ, Orihuela CJ, Harrod KS, Bhuiyan MAN, Dominic P, Kevil CG, Fort D, Liu VX, Farhat M, Koff JL, Lal CV, Gaggar A, Richter RP, Erdmann N, Might M, Gaggar A. 2023. COVID-19 bacteremic co-infection is a major risk factor for mortality, ICU admission, and mechanical ventilation. Crit Care 27:34. doi: 10.1186/s13054-023-04312-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Mitsi E, Reiné J, Urban BC, Solórzano C, Nikolaou E, Hyder-Wright AD, Pojar S, Howard A, Hitchins L, Glynn S, et al. 2022. Streptococcus pneumoniae colonization associates with impaired adaptive immune responses against SARS-CoV-2. J Clin Invest 132:e157124. doi: 10.1172/JCI157124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Blanchette KA, Shenoy AT, Milner J, Gilley RP, McClure E, Hinojosa CA, Kumar N, Daugherty SC, Tallon LJ, Ott S, King SJ, Ferreira DM, Gordon SB, Tettelin H, Orihuela CJ. 2016. Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect Immun 84:2922–2932. doi: 10.1128/IAI.00277-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Briles DE, Novak L, Hotomi M, van Ginkel FW, King J. 2005. Nasal colonization with Streptococcus pneumoniae includes subpopulations of surface and invasive pneumococci. Infect Immun 73:6945–6951. doi: 10.1128/IAI.73.10.6945-6951.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Weiser JN, Ferreira DM, Paton JC. 2018. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 16:355–367. doi: 10.1038/s41579-018-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Cooper VS, Honsa E, Rowe H, Deitrick C, Iverson AR, Whittall JJ, Neville SL, McDevitt CA, Kietzman C, Rosch JW. 2020. Experimental evolution in vivo to identify selective pressures during pneumococcal colonization. mSystems 5:e00352-20. doi: 10.1128/mSystems.00352-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zafar MA, Hammond AJ, Hamaguchi S, Wu W, Kono M, Zhao L, Weiser JN. 2019. Identification of pneumococcal factors affecting pneumococcal shedding shows that the dlt locus promotes inflammation and transmission. mBio 10:e01032-19. doi: 10.1128/mBio.01032-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Matthews AJ, Rowe HM, Rosch JW, Camilli A. 2021. A Tn-seq screen of Streptococcus pneumoniae uncovers DNA repair as the major pathway for desiccation tolerance and transmission. Infect Immun 89:e0071320. doi: 10.1128/IAI.00713-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Rowe HM, Karlsson E, Echlin H, Chang T-C, Wang L, van Opijnen T, Pounds SB, Schultz-Cherry S, Rosch JW. 2019. Bacterial factors required for transmission of Streptococcus pneumoniae in mammalian hosts. Cell Host Microbe 25:884–891. doi: 10.1016/j.chom.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Verhagen LM, de Jonge MI, Burghout P, Schraa K, Spagnuolo L, Mennens S, Eleveld MJ, van der Gaast-de Jongh CE, Zomer A, Hermans PWM, Bootsma HJ. 2014. Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLoS ONE 9:e89541. doi: 10.1371/journal.pone.0089541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62:2582–2589. doi: 10.1128/iai.62.6.2582-2589.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Overweg K, Pericone CD, Verhoef GGC, Weiser JN, Meiring HD, De Jong APJM, De Groot R, Hermans PWM. 2000. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect Immun 68:4604–4610. doi: 10.1128/IAI.68.8.4604-4610.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idänpään‐Heikkilä I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x [DOI] [PubMed] [Google Scholar]
- 195. Weiser JN, Markiewicz Z, Tuomanen EI, Wani JH. 1996. Relationship between phase variation in colony morphology, Intrastrain variation in cell wall physiology, and nasopharyngeal colonization by Streptococcus pneumoniae. Infect Immun 64:2240–2245. doi: 10.1128/iai.64.6.2240-2245.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis 177:368–377. doi: 10.1086/514205 [DOI] [PubMed] [Google Scholar]
- 197. Tong HH, Weiser JN, James MA, DeMaria TF. 2001. Effect of influenza A virus infection on nasopharyngeal colonization and Otitis media induced by transparent or opaque phenotype variants of Streptococcus pneumoniae in the Chinchilla model. Infect Immun 69:602–606. doi: 10.1128/IAI.69.1.602-606.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Li J, Li J-W, Feng Z, Wang J, An H, Liu Y, Wang Y, Wang K, Zhang X, Miao Z, Liang W, Sebra R, Wang G, Wang W-C, Zhang J-R. 2016. Epigenetic switch driven by DNA Inversions dictates phase variation in Streptococcus pneumoniae. PLoS Pathog 12:e1005762. doi: 10.1371/journal.ppat.1005762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055. doi: 10.1038/ncomms6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Chaguza C, Senghore M, Bojang E, Gladstone RA, Lo SW, Tientcheu P-E, Bancroft RE, Worwui A, Foster-Nyarko E, Ceesay F, Okoi C, McGee L, Klugman KP, Breiman RF, Barer MR, Adegbola RA, Antonio M, Bentley SD, Kwambana-Adams BA. 2020. Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation. Nat Commun 11:3442. doi: 10.1038/s41467-020-17327-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Kushner I. 2023. C-reactive protein – my perspective on its first half century, 1930-1982. Front Immunol 14:1150103. doi: 10.3389/fimmu.2023.1150103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Tomasz A. 1967. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in pneumococcus. Science 157:694–697. doi: 10.1126/science.157.3789.694 [DOI] [PubMed] [Google Scholar]
- 203. Clark SE, Weiser JN. 2013. Microbial modulation of host immunity with the small molecule phosphorylcholine. Infect Immun 81:392–401. doi: 10.1128/IAI.01168-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Briese T, Hakenbeck R. 1985. Interaction of the pneumococcal amidase with lipoteichoic acid and choline. Eur J Biochem 146:417–427. doi: 10.1111/j.1432-1033.1985.tb08668.x [DOI] [PubMed] [Google Scholar]
- 205. Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun 68:5690–5695. doi: 10.1128/IAI.68.10.5690-5695.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Yother J, White JM. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J Bacteriol 176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Sempere J, Llamosí M, Del Río Menéndez I, López Ruiz B, Domenech M, González-Camacho F. 2021. Pneumococcal choline-binding proteins involved in virulence as vaccine candidates. Vaccines (Basel) 9:181. doi: 10.3390/vaccines9020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Brown AO, Mann B, Gao G, Hankins JS, Humann J, Giardina J, Faverio P, Restrepo MI, Halade GV, Mortensen EM, Lindsey ML, Hanes M, Happel KI, Nelson S, Bagby GJ, Lorent JA, Cardinal P, Granados R, Esteban A, LeSaux CJ, Tuomanen EI, Orihuela CJ. 2014. Streptococcus pneumoniae translocates into the myocardium and forms unique Microlesions that disrupt cardiac function. PLOS Pathog. 10:e1004383. doi: 10.1371/journal.ppat.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis 190:1661–1669. doi: 10.1086/424596 [DOI] [PubMed] [Google Scholar]
- 210. Ring A, Weiser JN, Tuomanen EI. 1998. Pneumococcal trafficking across the blood-brain barrier. molecular analysis of a novel bidirectional pathway. J Clin Invest 102:347–360. doi: 10.1172/JCI2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435–438. doi: 10.1038/377435a0 [DOI] [PubMed] [Google Scholar]
- 212. Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala’Aldeen DAA, Tuomanen EI. 2009. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest 119:1638–1646. doi: 10.1172/JCI36759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Zhang J-R, Mostov KE, Lamm ME, Nanno M, Shimida S, Ohwaki M, Tuomanen E. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827–837. doi: 10.1016/s0092-8674(00)00071-4 [DOI] [PubMed] [Google Scholar]
- 214. Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA 103:2857–2862. doi: 10.1073/pnas.0511017103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Iovino F, Nannapaneni P, Henriques-Normark B, Normark S. 2020. The impact of the ancillary pilus‐1 protein RrgA of Streptococcus pneumoniae on colonization and disease. Mol Microbiol 113:650–658. doi: 10.1111/mmi.14451 [DOI] [PubMed] [Google Scholar]
- 216. Kietzman CC, Gao G, Mann B, Myers L, Tuomanen EI. 2016. Dynamic capsule restructuring by the main pneumococcal autolysin LytA in response to the epithelium. Nat Commun 7:10859. doi: 10.1038/ncomms10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Brissac T, Martínez E, Kruckow KL, Riegler AN, Ganaie F, Im H, Bakshi S, Arroyo-Diaz NM, Spencer BL, Saad JS, Nahm MH, Orihuela CJ. 2021. Capsule promotes intracellular survival and vascular endothelial cell translocation during invasive pneumococcal disease. mBio 12:e0251621. doi: 10.1128/mBio.02516-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Xu S, Mo D, Rizvi FZ, Rosa JP, Ruiz J, Tan S, Tweten RK, Leong JM, Adams W. 2023. Pore-forming activity of S. pneumoniae pneumolysin disrupts the paracellular localization of the epithelial adherens junction protein E-cadherin. Infect Immun 91:e0021323. doi: 10.1128/iai.00213-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Devraj G, Guérit S, Seele J, Spitzer D, Macas J, Khel MI, Heidemann R, Braczynski AK, Ballhorn W, Günther S, Ogunshola OO, Mittelbronn M, Ködel U, Monoranu CM, Plate KH, Hammerschmidt S, Nau R, Devraj K, Kempf VAJ. 2020. HIF-1α is involved in blood–brain barrier dysfunction and paracellular migration of bacteria in pneumococcal meningitis. Acta Neuropathol 140:183–208. doi: 10.1007/s00401-020-02174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Radin JN, Orihuela CJ, Murti G, Guglielmo C, Murray PJ, Tuomanen EI. 2005. β-arrestin 1 participates in platelet-activating factor receptor-mediated endocytosis of Streptococcus pneumoniae. Infect Immun 73:7827–7835. doi: 10.1128/IAI.73.12.7827-7835.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Surve MV, Bhutda S, Datey A, Anil A, Rawat S, Pushpakaran A, Singh D, Kim KS, Chakravortty D, Banerjee A. 2018. Heterogeneity in pneumolysin expression governs the fate of Streptococcus pneumoniae during blood-brain barrier trafficking. PLoS Pathog 14:e1007168. doi: 10.1371/journal.ppat.1007168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Shizukuishi S, Ogawa M, Ryo A, Ohnishi M. 2020. The multi-step mechanism and biological role of noncanonical autophagy targeting Streptococcus pneumoniae during the early stages of infection. Autophagy 16:1152–1153. doi: 10.1080/15548627.2020.1743937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Jen FEC, Warren MJ, Schulz BL, Power PM, Swords WE, Weiser JN, Apicella MA, Edwards JL, Jennings MP. 2013. Dual pili post-translational modifications synergize to mediate meningococcal adherence to platelet activating factor receptor on human airway cells. PLoS Pathog 9:e1003377. doi: 10.1371/journal.ppat.1003377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, Gibson BW, Apicella MA. 2000. Non‐typeable haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol 37:13–27. doi: 10.1046/j.1365-2958.2000.01952.x [DOI] [PubMed] [Google Scholar]
- 225. Brissac T, Shenoy AT, Patterson LA, Orihuela CJ. 2018. Cell invasion and pyruvate oxidase-derived H2O2 are critical for Streptococcus pneumoniae-mediated cardiomyocyte killing. Infect Immun:e00569. doi: 10.1128/IAI.00569-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Briles DE, Forman C, Hudak S, Claflin JL. 1982. Anti-phosphorylcholine antibodies of the T15 Idiotype are optimally protective against Streptococcus pneumoniae. J Exp Med 156:1177–1185. doi: 10.1084/jem.156.4.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. McDaniel LS, Scott G, Kearney JF, Briles DE. 1984. Monoclonal antibodies against protease-sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J Exp Med 160:386–397. doi: 10.1084/jem.160.2.386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Mitchell TJ, Walker JA, Saunders FK, Andrew PW, Boulnois GJ. 1989. Expression of the pneumolysin gene in Escherichia coli: rapid purification and biological properties. Biochim Biophys Acta 1007:67–72. doi: 10.1016/0167-4781(89)90131-0 [DOI] [PubMed] [Google Scholar]
- 229. Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, Bryant AP, McDevitt D, Morrison DA, Holden DW. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol 40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x [DOI] [PubMed] [Google Scholar]
- 230. Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun 66:5620–5629. doi: 10.1128/IAI.66.12.5620-5629.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Seiberling M, Bologa M, Brookes R, Ochs M, Go K, Neveu D, Kamtchoua T, Lashley P, Yuan T, Gurunathan S. 2012. Safety and immunogenicity of a pneumococcal histidine triad protein D vaccine candidate in adults. Vaccine 30:7455–7460. doi: 10.1016/j.vaccine.2012.10.080 [DOI] [PubMed] [Google Scholar]
- 232. Corsini B, Aguinagalde L, Ruiz S, Domenech M, Antequera ML, Fenoll A, García P, García E, Yuste J. 2016. Immunization with LytB protein of Streptococcus pneumoniae activates complement-mediated phagocytosis and induces protection against pneumonia and sepsis. Vaccine 34:6148–6157. doi: 10.1016/j.vaccine.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 233. Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis 188:339–348. doi: 10.1086/376571 [DOI] [PubMed] [Google Scholar]
- 234. Briles DE, Ades E, Paton JC, Sampson JS, Carlone GM, Huebner RC, Virolainen A, Swiatlo E, Hollingshead SK. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun 68:796–800. doi: 10.1128/IAI.68.2.796-800.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Miyaji EN, Oliveira MLS, Carvalho E, Ho PL. 2013. Serotype-independent pneumococcal vaccines. Cell Mol Life Sci 70:3303–3326. doi: 10.1007/s00018-012-1234-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Ogunniyi AD, Folland RL, Briles DE, Hollingshead SK, Paton JC. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect Immun 68:3028–3033. doi: 10.1128/IAI.68.5.3028-3033.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Crain MJ, Waltman WD, Turner JS, Yother J, Talkington DF, McDaniel LS, Gray BM, Briles DE. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun 58:3293–3299. doi: 10.1128/iai.58.10.3293-3299.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Selva L, Ciruela P, Blanchette K, Del Amo E, Pallares R, Orihuela CJ, Muñoz-Almagro C. 2012. Prevalence and clonal distribution of pcpA, psrP and Pilus-1 among pediatric isolates of Streptococcus pneumoniae. PLoS ONE 7:e41587. doi: 10.1371/journal.pone.0041587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun 72:5582–5596. doi: 10.1128/IAI.72.10.5582-5596.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Mann B, Thornton J, Heath R, Wade KR, Tweten RK, Gao G, El Kasmi K, Jordan JB, Mitrea DM, Kriwacki R, Maisonneuve J, Alderson M, Tuomanen EI. 2014. Broadly protective protein-based pneumococcal vaccine composed of pneumolysin toxoid-CbpA peptide recombinant fusion protein. J Infect Dis 209:1116–1125. doi: 10.1093/infdis/jit502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Narciso AR, Iovino F, Thorsdottir S, Mellroth P, Codemo M, Spoerry C, Righetti F, Muschiol S, Normark S, Nannapaneni P, Henriques-Normark B. 2022. Membrane particles evoke a serotype-independent cross-protection against pneumococcal infection that is dependent on the conserved lipoproteins MalX and PrsA. Proc Natl Acad Sci USA 119:e2122386119. doi: 10.1073/pnas.2122386119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Zhang F, Lu Y-J, Malley R. 2013. Multiple antigen-presenting system (MAPS) to induce comprehensive B- and T-cell immunity. Proc Natl Acad Sci U S A 110:13564–13569. doi: 10.1073/pnas.1307228110 [DOI] [PMC free article] [PubMed] [Google Scholar]