Abstract
Immunostaining with p57KIP2 is a widely used diagnostic technique to differentiate complete hydatidiform moles (CHMs) from partial hydatidiform moles (PHM) and non-molar hydropic abortion. However, distinguishing between PHMs and non-molar hydropic abortions using histopathology alone is often challenging. This study aimed to evaluate the technical validity and additional benefits of using fluorescence in situ hybridization (FISH) in combination with p57KIP2 immunostaining to diagnose molar and non-molar conceptuses. The study involved 80 specimens, which underwent genetic diagnosis using short tandem repeat analysis, including 44 androgenetic CHMs, 20 diandric monogynic PHMs, 14 biparental non-molar hydropic abortions, 1 monoandric digynic triploid abortion, and 1 vaginal specimen of gestational trophoblastic neoplasia. Two pathologists independently diagnosed the cases based on morphology and p57KIP2 immunostaining while the clinical information was masked. FISH analysis was performed using 3 probes (CEP17, CEPX, and CEPY), which revealed that all androgenetic CHM and biparental diploid non-molar hydropic abortion specimens were diploid. Among the 20 diandric monogynic PHM cases examined by analyzing short tandem repeat polymorphisms, 18 were triploid, and the remaining 2 were diploid. These two specimens were possibly androgenetic/biparental mosaics based on FISH analysis, where the three-signal ratios counting 50 cells were clearly within the diploid ranges. Eight of the 20 genetic PHMs and 2 of the 14 genetically confirmed non-molar hydropic abortions that were falsely diagnosed based on morphology and immunohistochemistry by at least 1 pathologist were correctly diagnosed as PHM and non-molar hydropic abortion, respectively, by FISH analysis. However, 1 monoandric digynic villus was classified as triploid by FISH analysis, leading to a false PHM diagnosis. In conclusion, the combination of FISH analysis with p57KIP2 immunostaining helps in diagnosing molar and non-molar conceptuses in numerous cases; nevertheless, exceptional cases should be considered.
Key Words: Fluorescence in situ hybridization, Hydatidiform moles, p57KIP2, Immunohistochemistry, Short tandem repeat polymorphism
Hydatidiform mole (HM) is a trophoblastic disease characterized by the proliferation of trophoblasts and the presence of hydropic villous structures. These moles are classified into 2 categories based on their chromosomal composition: complete hydatidiform moles (CHMs) and partial hydatidiform moles (PHMs). While normal conceptions are biparental and diploid, CHMs and PHMs are mostly androgenetic diploids and diandric monogynic triploids, respectively1. Distinguishing between CHMs and PHMs is crucial, as 15% to 20% and 1% to 4% of cases of these conditions, respectively, can develop into gestational trophoblastic neoplasia (GTN). In contrast, GTN rarely develops after non-molar conceptions, making the differential diagnosis between HMs and non-molar conceptions necessary, particularly for older women with a likelihood of diminished fertility who desire to conceive2,3. Morphologic assessments of villous specimens are commonly used for diagnosis; however, the accuracy of hematoxylin-eosin (HE) staining-based pathologic diagnosis can be insufficient in practice4–6. Therefore, auxiliary techniques are necessary to improve and refine pathologic diagnoses6–8.
Molecular genotyping, which employs the analysis of short tandem repeat (STR) polymorphisms, is considered the most reliable method for diagnosing molar pregnancies and can accurately distinguish between molar and non-molar pregnancies1,9–13. However, STR analysis has some limitations such as its high cost compared with that of immunohistochemistry. The specialized techniques and instruments required to extract genomic DNA, amplify STR fragments, and separate polymerase chain reaction (PCR) products by capillary electrophoresis contribute to the high cost of the analysis14,15. Consequently, in Japan, STR analysis for the molecular diagnosis of HM using formalin-fixed paraffin-embedded (FFPE) specimens is not commonly used in routine clinical practice in the pathologic department and is instead reserved for challenging cases or research purposes.
Immunostaining for p57KIP2, a cyclin-dependent kinase inhibitor 1C gene-encoded imprinting product transcribed exclusively from the maternal chromosome, has emerged as a precise method for identifying CHM10,11,16–19. Unlike biparental products of conception (POC), such as PHMs and non-molar specimens, androgenetic CHMs contain only paternal chromosomes, resulting in the loss of expression of p57KIP2 in villous cytotrophoblasts and stromal cells. This method is almost as effective as STR analysis in distinguishing CHMs from PHMs and non-molar specimens10,14. However, differentiating PHM from hydropic abortion based on p57KIP2 immunohistochemistry is impractical as both entities exhibit preserved p57KIP2 expression. This distinction can be extremely difficult in numerous cases, for which STR analysis remains the gold standard5,11,20.
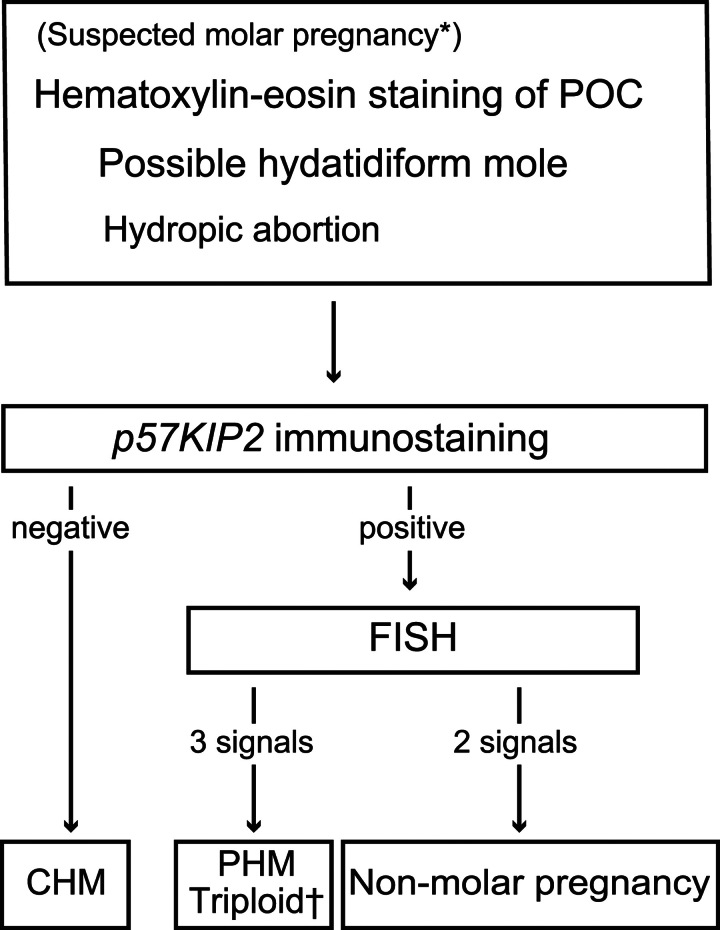
Determining the ploidy of a POC can aid in differentiating between CHM and PHM or between PHM and non-molar conceptuses. While PHM is triploid, CHM and non-molar conceptuses are diploid, but rare exceptions exist, such as tetraploid CHM21, monoandric digynic triploid conceptions, or non-molar trisomic abortus. Various techniques, including karyotyping, flow cytometry, and fluorescence in situ hybridization (FISH), have traditionally been used for this purpose22–25. The diagnostic workflow, combined with p57KIP2 immunohistochemistry and FISH analysis, can theoretically distinguish between CHM, PHM, and non-molar abortus in almost all cases except for digynic non-molar triploid or trisomy cases. The algorithm involves p57KIP2-negative diploid cells, p57KIP2-positive triploid cells, including trisomies with congruent chromosomal probes, and p57KIP2-positive diploid cells (Fig. 1)26,27. Although some studies have reported using p57KIP2 immunohistochemistry and FISH analysis for trophoblastic pathology, some cases analyzed using p57KIP2 staining and FISH analysis were not confirmed by genetic diagnosis28–31. Therefore, this study aimed to evaluate the technical validity and efficacy of the use of additional FISH analysis using masked samples with a predetermined molecular diagnosis by STR analysis.
FIG. 1.
Diagnostic workflow of the study *Based on sonographic or macroscopic findings. †Monoandric digynic triploidy would be theoretically categorized here. POC indicates product of conception; FISH, fluorescent in situ hybridization; CHM, complete hydatidiform mole; PHM, partial hydatidiform mole.
MATERIALS AND METHODS
Ethical Statement
This study was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the institutional ethical committee of the Graduate School of Medicine, Chiba University, Japan (approval numbers 1198 and 2113). All participants provided written informed consent.
Participants and Samples
In this study, 153 patients who had undergone evacuation of a POC due to suspected molar pregnancy at Chiba University Hospital between 2009 and 2015 were included. The POCs were fixed in formalin neutral buffer solution for 24 to 48 h and then embedded in paraffin for histologic diagnosis, as per the routine procedure. The specimens were diagnosed by certified pathologists at the Department of Pathology of Chiba University Hospital.
Genetic Diagnosis of POCs
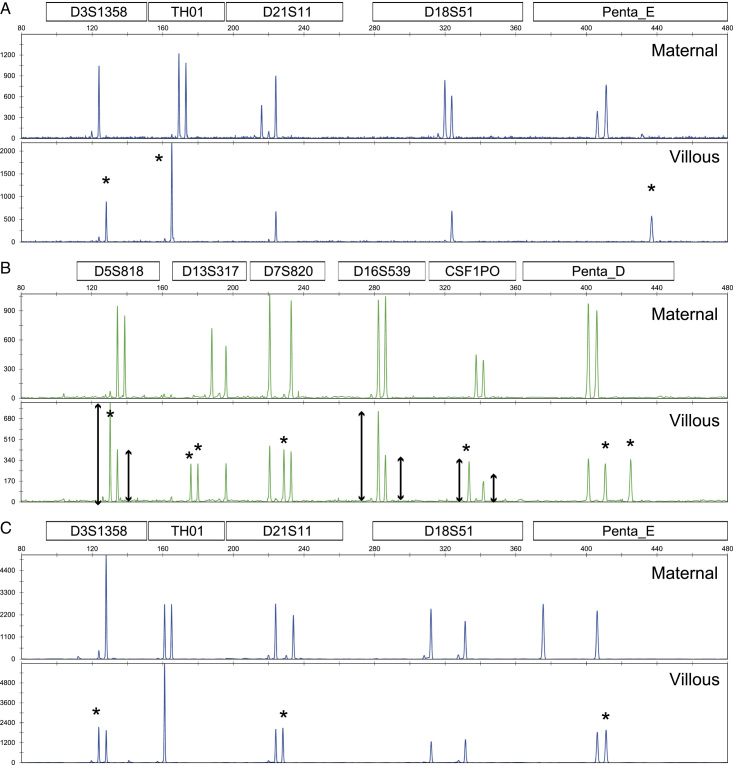
Molecular diagnostic analysis was performed on blood and fresh POC samples from 152 patients. Villous tissues were isolated from blood clots and decidua tissues under a stereomicroscope when villous tissues were scarce. Genomic DNA was extracted from both blood and tissue samples, and STR polymorphic alleles were amplified with the PowerPlex 16 HS Kit (Promega, Madison). The amplified fragments were electrophoresed on the ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City) and analyzed through the GeneMapper 4.0 software (Applied Biosystems). The classification of HM and non-molar villi was based on previous research and definitions from other institutions9,10. Cases with one or more villous loci without maternal alleles were considered androgenetic and categorized as genetic CHM; otherwise, they were identified as biparental conceptuses. The triploid or diploid classification of biparental concepts was based on parental contribution. The peak heights of 2 allelic loci were evaluated in all loci. A biparental disomic chromosome was indicated by an even peak height, whereas a trisomic chromosome was indicated by a peak height ratio of 2:1 (Fig. 2). Cases estimated to be trisomic in almost all loci with 2 paternal and 1 maternal parental contribution were assigned as diandric monogynic PHM (genetic PHM). Cases estimated to be disomic in almost all loci were assigned as genetically confirmed non-molar diploid conceptus.
FIG. 2.
Electropherograms of short tandem repeat PCR fragments. (A) Complete hydatidiform mole, (B) partial hydatidiform mole, and (C) abortion. Upper and lower lanes indicate maternal and villous electropherograms, respectively. Asterisks are the alleles of paternal contribution. PCR indicates polymerase chain reaction.
Study Flow
Of the 153 POCs, genetic assessment through STR analysis was performed on 150 villous tissues, as 3 samples could not be successfully analyzed. The analysis of the 150 POCs revealed that 111 were androgenetic CHMs, 20 were diandric monogynic PHMs, 18 were biparental diploid abortions, and 1 was a monoandric digynic triploid abortion. Because the number of androgenetic CHMs was significantly higher compared with the other groups, we selected 78 samples of intrauterine conceptuses comprising 44 CHMs, 20 PHMs, and 14 non-molar abortions for further analysis. In addition, 2 villous samples, including an aborted monoandric digynic triploid villous specimen and a vaginal specimen of low-risk GTN after CHM, were included as validation controls. Consequently, the analysis included a total of 80 samples.
Transfer of Formalin-fixed Paraffin-embedded Samples and Pathologic Diagnosis
The 80 FFPE blocks selected for the study (1 block per sample) were anonymized and sent to the Pathologic Laboratory of Kotobiken Medical Laboratories, Inc., without revealing any clinical information or diagnosis. The pathologists were only informed that the specimens were villous tissues, including CHM, PHM, and non-molar villi. All pathologic evaluations and FISH analyses were performed independently of Chiba University Hospital at the Pathological Laboratory of Kotobiken Medical Laboratories.
FFPE sections (3 μm thick) were deparaffinized with xylene, rehydrated with a graded ethanol series, and stained with HE for histologic analyses. Immunostaining was performed using an Autostainer Link 48 (Dako, Glostrup, Denmark). Immunohistochemistry of p57KIP2 was conducted using a rabbit polyclonal antibody (p57Kip2 Ab-7 #RB-1637-R7, 1:10; Thermo Fisher Scientific, Waltham, MA, USA). Antigen retrieval was performed using Target Retrieval Solution (pH 9) on PT Link 200 (Dako). Endogenous peroxidase was quenched with 3% H2O2 in distilled water for 5 min, and then the slides were incubated with the primary antibody for 30 min at room temperature. The sections were stained using EnVision FLEX (Dako), according to the manufacturer’s protocol, and counterstained with hematoxylin.
Positive immunohistochemical staining of p57KIP2 was indicated by diffuse and distinct nuclear staining of villous cytotrophoblasts and stromal cells. Lack of staining or <10% nuclear staining was considered negative. Stained extravillous trophoblasts and maternal decidua were used as positive internal controls. The 2 pathologists independently evaluated the specimens using the combination of HE and p57KIP2 staining without access to clinical information or primary pathologic diagnosis from Chiba University Hospital. Subsequently, the specimens were diagnosed as CHM, PHM, or non-molar abortus, including hydropic abortion.
Fluorescent in situ Hybridization
The ploidy pattern of specimens from all 80 patients was analyzed. To minimize confounding factors, we selected the centromeric FISH probe for chromosome 17, as trisomy of this chromosome is exceedingly rare12,32. The FFPE tissue specimens were cut into 4 µm-thick sections and placed on coated slides. The slides were treated with hydrochloric acid and a pre-treatment solution, followed by proteolytic digestion with pepsin. FISH was then performed for chromosomes 17 and XY using Vysis CEP17 (D17Z1) and CEPX Spectrum Orange/Y Spectrum Green DNA Probe Kits (Abbott Molecular, Chicago), respectively. After hybridization, the tissue specimens on the slides were counterstained with 4′,6-diamidino-2-phenylindole, and examined under a fluorescence microscope (Olympus, Tokyo).
To ensure objectivity, we counted at least 50 non-overlapping nuclei in interphase and metaphase chromosomes to determine the number of signals. The three-signal rate of CEP17 was calculated by dividing the number of cells presenting 3 signals by the total number of counted cells. For CEPX/Y probes, cells presenting 3 signals were defined as those with 3 signals on CEPX and none on CEPY, 2 signals on CEPX and 1 on CEPY, 1 signal on CEPX and 2 on CEPY, or no signal on CEPX and 3 on CEPY.
Statistical Analysis
Statistical analyses and data visualization were conducted using the R software (version 4.2.2: https://www.R-project.org).
RESULTS
Short Tandem Repeats Polymorphism Analysis
Fig. 2 shows electropherograms of representative multiplex STR-PCR fragments. Fig. 2A–C show genetic CHM, PHM, and non-molar conceptuses, respectively. As shown in Fig. 2A, only paternal alleles were detected at 3 loci, leading to the classification of the case as a genetic CHM. The triploid diagnosis was based on the almost equal peak heights of 3 allelic loci (D13S317, D7S820, and Penta D in Fig. 2B). Moreover, the longer peak height of the 2 allelic loci (D5S818, D16S539, and CSF1PO in Fig. 2B) was almost twice the height of the shorter one, consistent with diandric monogynic triploid as genetic PHM10,33. In Fig. 2C, the peak heights of 2 allelic loci (D13S317, D21S11, D18S51, and Penta E) were almost equal, indicating non-molar conceptus. Finally, 78 enrolled samples were classified as 44 genetic CHMs, 20 genetic PHMs, and 14 genetically confirmed non-molar conceptuses. In addition, 1 invasive HM (genetic CHM) of a vaginal specimen and 1 monoandric digynic triploid villous specimen were further analyzed.
Pathologic Diagnosis Combined with Hematoxylin-eosin Staining and p57KIP2 Immunostaining
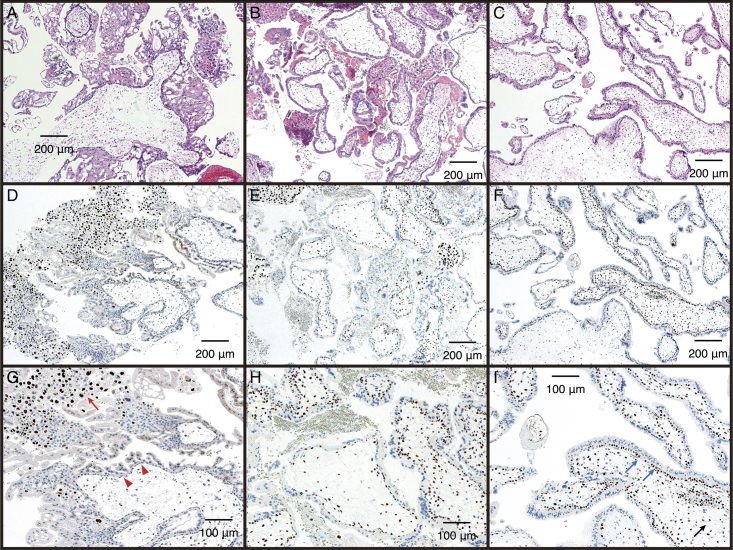
Table 1 summarizes the results of the independent diagnosis of specimens by 2 pathologists using HE staining and p57KIP2 immunostaining. Representative images of HE staining and p57KIP2 immunostaining are presented in Fig. 3. The villous cytotrophoblasts and stromal cells in androgenetic CHM specimens were negative for p57KIP2 (Fig. 3D and G). Conversely, villous cytotrophoblasts and stromal cells were positive for p57KIP2 in diandric monogynic PHM and non-molar villous specimens (Fig. 3E, F, H, and I), which contained the maternal chromosomes. For CHM cases, extravillous trophoblasts were typically stained as an internal positive control (Fig. 3G). The villous samples were classified as CHM, PHM, or non-molar conceptus based on their HE staining and p57KIP2 immunostaining patterns (Table 1). Among the 18 diandric genetic PHM cases (cases #45–62), at least 1 pathologist determined 7 cases to be non-molar hydropic abortions but not PHMs. Two cases of genetically confirmed abortus (cases #77 and #78) were identified as PHM.
TABLE 1.
Summary of genetic, pathologic, and FISH ploidy analyses
| Pathologic diagnosis involving p57KIP2 immunostaining | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic diagnosis by STR analysis | Pathologist 1 | Pathologist 2 | ||||||||
| Case | Genetic diagnosis | Parental origin | HE+p57 | p57 | HE+p57 | p57 | Discordant cases between pathologists | FISH ploidy | HE +p57KIP2 +FISH | Final diagnosis (1) |
| 1–38 | CHM | P1P1 | CHM | - | CHM | - | — | 2 | CHM | CHM |
| 39–42 | CHM | P1P2 | CHM | - | CHM | - | — | 2 | CHM | CHM |
| 43 | CHM | P1P1 | CHM | - | PHM | + | CHM/PHM | 2 | CHM | CHM (2) |
| 44 | CHM | P1P2 | PHM | + | PHM | + | — | 2 | Abortion* | CHM (2) |
| 45–53 | PHM | MP1P2 | PHM | + | PHM | + | — | 3 | PHM | PHM |
| 54–57 | PHM | MP1P2 | PHM | + | HA | + | PHM/HA | 3 | PHM† | PHM |
| 58, 59 | PHM | MP1P2 | HA | + | PHM | + | PHM/HA | 3 | PHM† | PHM |
| 60 | PHM | MP1P2 | AB | + | AB | + | — | 3 | PHM† | PHM |
| 61 | PHM | MP1P2 | HA | + | HA | + | — | 3 | PHM† | PHM |
| 62 | PHM | MP1P2 | AB | + | HA | + | HA/ AB | 3 | PHM† | PHM |
| 63 | PHM | MP1P1 | PHM | +/-‡ | HA | + | PHM/HA | 2 | Not specified | Mosaic (3) |
| 64 | PHM | MP1P1 | PHM | +/-‡ | PHM | + | — | 2 | Not specified | Mosaic (3) |
| 65 | Abortion | MP | AB | + | AB | + | — | 2 | Abortion | Abortion |
| 66–71 | Abortion | MP | AB | + | HA | + | — | 2 | Abortion | Abortion |
| 72–76 | Abortion | MP | HA | HA | — | 2 | Abortion | Abortion | ||
| 77 | Abortion | MP | HA | + | PHM | + | PHM/HA | 2 | Abortion† | Abortion |
| 78 | Abortion | MP | PHM | + | PHM | + | — | 2 | Abortion† | Abortion |
| 79 | CHM | P1P1 | CHM | - | CHM | - | — | 2 | CHM | CHM (4) |
| 80 | Abortion | M1M2P | AB | + | AB | + | — | 3 | PHM* | Abortion (5) |
Falsely revised cases based on FISH results.
Correctly revised cases based on FISH results.
Cytotrophoblasts (+), stroma cells (-).
(1) Final diagnosis was determined based on all genetic, histologic, immunohistochemical, and FISH information.
(2) p57KIP2-positive androgenetic CHM cases previously reported (ref.33; Am J Clin Pathol. 2020;154(6):776-83).
(3) Estimated biparental diploid and androgenetic diploid mosaic cases.
(4) Vaginal specimen of low-risk GTN after evacuation of CHM.
(5) Monoandric digynic triploid case.
AB indicates abortion; CHM, complete hydatidiform mole; FISH, fluorescence in situ hybridization; HE, hematoxylin-eosin; M, maternal; P, paternal; p57, p57KIP2; PHM, partial hydatidiform mole; STR, short tandem repeat.
FIG. 3.
Representative pathological photographs. Hematoxylin-eosin staining (A–C, ×50) and p57KIP2 immunostaining (D–F, ×50, G–I, ×100). Complete hydatidiform mole (A, D, and G), partial hydatidiform mole (B, E, and H), and non-molar villous (C, F, and I). Red arrows indicate positive staining of extravillous trophoblasts, red triangles indicate negative staining of villous cytotrophoblasts, blue arrows indicate positive staining of villous cytotrophoblasts, and black arrows indicate positive staining of villous stromal cells.
Fluorescent in situ Hybridization Analysis
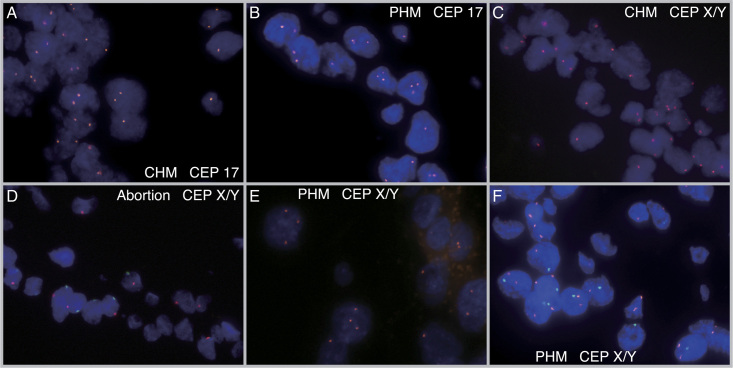
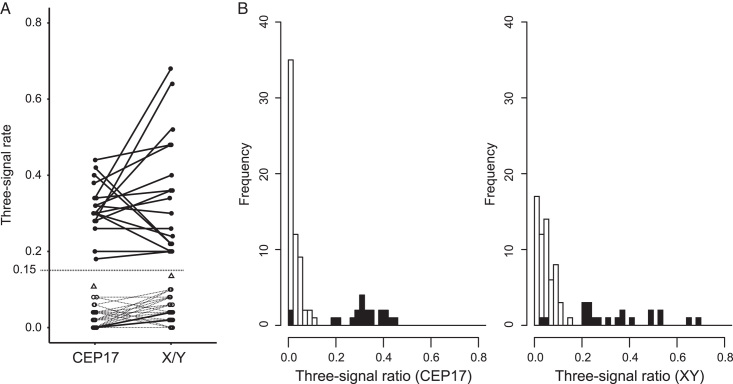
All cases underwent successful FISH analysis using CEP17 and CEPX/Y probes, including the oldest FFPE blocks that were up to 5 years old. Fig. 4 shows representative FISH results where CHM and PHM cases displayed 2 and 3 FISH signals of CEP17, respectively (Fig. 4A and B). Similarly, CHM, non-molar hydropic abortion, PHM, and PHM cases showed 2, 2, 3, and 3 FISH signals of CEPX/Y, respectively (Fig. 4C-F). Supplementary Table S1, Supplemental Digital Content 1, http://links.lww.com/IJGP/A157 details the signal counts of 50 nuclei, and Fig. 5 illustrates the distribution of the three-signal ratios in the diploid and triploid samples, which showed a bimodal pattern with a recognizable boundary zone (Fig. 5A). The threshold for the three-signal ratio was determined as 0.15, although 2 triploid cases (cases #63 and #64) diagnosed by STR analysis were in the diploid region with 2 FISH signals, where the three-signal ratios counting 50 cells were 0.00 and 0.00 on CEP17 and 0.04 and 0.02 on CEPX/Y probes, respectively (Fig. 5B and Supplementary Table S1, Supplemental Digital Content 1, http://links.lww.com/IJGP/A157). The median and range of the three-signal ratio of CEP17 in diploid and triploid samples were 0.00 [0.00–0.08] and 0.31 [0.18–0.44], respectively, whereas the median and range of the three-signal ratio of CEPX/Y in diploid and triploid samples were 0.04 [0.00–0.10] and 0.32 [0.20–0.68], respectively. In addition the vaginal specimen of low-risk GTN after CHM showed slightly higher three-signal ratios of 0.10 on CEP17 and 0.14 on CEPX/Y, as depicted in Fig. 5A, although the ratios were within the diploid range.
FIG. 4.
Images of fluorescence in situ hybridization with the CEP17 and CEPX/Y probes. Red signals indicated CEP17 or CEPX probes, whereas blue signals indicated CEP Y probes. (A) CEP17 probe on CHM shows 2 signals. (B) CEP17 probe on PHM shows 3 signals. (C) CEPX/Y probe on CHM shows 2 signals. The sex karyotype of this case, determined by STR analysis, was estimated to be XX. (D) CEPX/Y probe on abortion shows 2 signals. The sex karyotype of this case was estimated to be XY by STR analysis. (E) The CEPX/Y probe in PHM showed 3 signals. The sex karyotype of this case was estimated to be XXX by STR analysis. (F) The CEPX/Y probe on the PHM shows three signals. The sex karyotype of this case was estimated to be XXY by STR analysis. CHM indicates complete hydatidiform mole; PHM, partial hydatidiform mole.
FIG. 5.
Distribution of the three-signal rates. Distribution of the three-signal rates. (A) Comparison of the three-signal rates between CEP17 and X/Y probes. Filled circles and solid lines show the triploid cases. Open circles and dotted lines show the diploid cases. Open triangles indicate the three-signal rates of a vaginal GTN lesion (case #79). The discrimination borderline (0.15) is shown as a dotted line. (B) The distributions of three-signal rates using the CEP17 and CEPX/Y probes. The filled and unfilled bars indicate the triploid and diploid cases determined using short tandem repeat polymorphism analysis.
Evaluation of the Diagnostic Workflow Involving p57KIP2 Immunostaining and FISH
We evaluated the results of the diagnostic workflow along with those of p57KIP2 immunostaining and FISH analysis, which are presented in Table 1. We identified a single androgenetic heterozygous CHM case (case #44) as a non-molar hydropic abortion due to the diploid signal in the FISH analysis and retained expression of p57KIP2 (Fig. 6A and B). We observed discordant p57KIP2 immunostaining patterns, with positive staining in the villous cytotrophoblasts and negative staining in villous stromal cells. Although single nucleotide polymorphism array data, in addition to STR analysis, indicated no possibility of genetic mosaicism, we noted a mosaic pattern of p57KIP2 staining, as previously detailed33.
FIG. 6.
Representative images of misdiagnosed cases. p57KIP2-positive androgenetic CHM (case #44) is shown. Hematoxylin-eosin staining (A, ×100) and p57KIP2 immunostaining (B, ×100). Short tandem repeat polymorphism analysis confirmed this case as heterozygous androgenetic CHM. Villous cytotrophoblasts and extravillous trophoblasts are positively stained, as reported in reference33 (Am J Clin Pathol 2020;154:776-83). (C) Diandric monogynic triploid case (#60, ×50). It was misdiagnosed as abortion based on morphology at primary diagnosis. (D) Biparental diploid case (#78, ×50). It was misdiagnosed as PHM at primary diagnosis. CHM indicates complete hydatidiform mole; PHM, partial hydatidiform mole.
Initially, 8 genetically confirmed PHM cases were diagnosed as non-molar abortions (cases #54–62) based on morphology and immunohistochemistry by at least 1 pathologist but were later identified as PHM based on the FISH ploidy information (Fig. 6C). In addition, 2 genetically confirmed non-molar abortion cases (cases #77 and #78) were initially misdiagnosed as PHM but later corrected to abortion (Fig. 6D). We classified 1 exceptional case of monoandric digynic triploidy as PHM because of the triploid signal in the FISH analysis and positive staining of p57KIP2 (case #80).
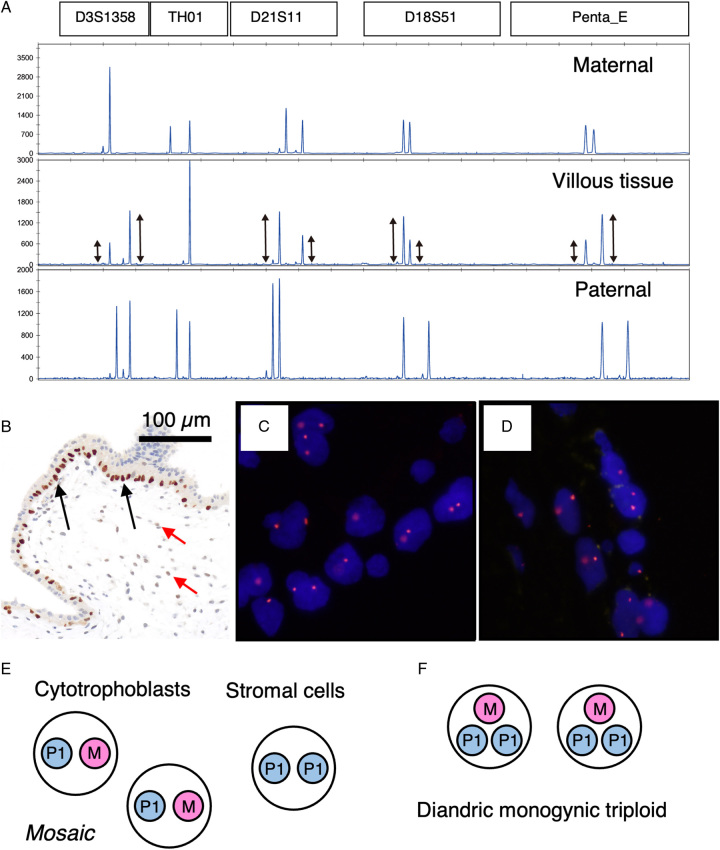
Two cases of diandric monogynic triploidy (cases #63 and #64) classified by STR analysis (Fig. 7A) were indeterminate. All loci showed 1 or 2, but not 3, alleles. Both cases showed a swollen villus and a discordant p57KIP2 staining pattern, with positive staining observed in the villous cytotrophoblasts and negative staining in the stromal cells (Fig. 7B). The peak heights on the STR electropherogram were not uniform in 2 allelic loci but were roughly in the ratio of 2:1 or 1:2 (Fig. 7A, middle lane), which could be consistent with triploid PHM. However, we observed 2 clear FISH signals for CEP17 and CEPX (Fig. 7C and D). Therefore, we concluded that these 2 cases were androgenetic/biparental mosaic (MP1/P1P1) (Fig. 7E) but not diandric monogyny triploid PHM (MP1P1, paternally homozygous) (Fig. 7F).
FIG. 7.
Discordance between molecular genotyping using STR analysis and FISH. (A) Electropherograms of STR-PCR fragments for case #63. All villous loci (middle lane) show imbalanced 2 alleles. Villous peak height ratio is almost 2 or 0.5. (B) p57KIP2 immunostaining of sample #63 (×100). Cytotrophoblasts exhibited distinct staining (indicated by black arrows). One pathologist concluded that villous stromal cells were negative for staining, while another pathologist observed minimal staining intensity (highlighted by red arrows). (C) FISH image with CEP17 probe. Two signals in 1 nucleus were observed. (D) FISH image with CEPX/Y probes. Two signals of CEPX (orange) in 1 nucleus were observed. (E) Possible explanation for discordant cases (#63 and #64). Villous cytotrophoblasts would be biparental diploid, and stromal cells would have identical androgenetic chromosomes of the biparental cytotrophoblasts. (F) Cytogenetic constitution of homozygous diandric monogyny triploid PHM. Paternal chromosomes were from a single sperm. FISH indicates fluorescence in situ hybridization; PHM, partial hydatidiform mole; STR, short tandem repeat.
DISCUSSION
The diagnostic methodology proposed in this study, which incorporates p57KIP2 immunostaining with FISH analysis, can be a useful tool for distinguishing diploid and triploid conceptuses. Our results indicated that although p57KIP2 immunostaining correctly identified almost all androgenetic CHM cases, almost half of the diandric monogynic PHMs were incorrectly diagnosed as abortion based on morphology and p57KIP2 staining information by at least 1 pathologist. This study demonstrates that FISH analysis can overcome the limitations of p57KIP2 immunostaining; in cases where the differential diagnosis between PHM and non-molar conceptuses presents difficulty, FISH ploidy information can aid in assigning cases to PHM, except for exceptional cases, such as monoandric digynic triploid cases. The incidence of digynic triploidy accounts for one-third of all triploid cases observed during the first trimester1. Therefore, it is important to acknowledge the occurrence of monoandric digynic triploid cases.
The availability and reproducibility of diagnostic procedures are essential factors to consider when selecting a clinically applicable diagnostic method. Numerous institutions subject genomic DNA, extracted from chorionic villous tissues and decidua (maternal) tissues using FFPE blocks, to STR analysis12,15,34. However, in many pathologic departments, FISH analysis can be performed more easily than STR analysis, as it is technically available. Although FISH probes are expensive, FISH analysis has been widely used in pathologic departments and laboratories to diagnose breast cancer subtypes and some types of sarcoma or leukemia using FISH probes for human epidermal growth factor receptor 2 and fusion genes, respectively35–37. In this study, we used only 2 FISH probes, chromosomes 17 and XY, to determine the ploidies of POCs. The result obtained from 1 FISH probe does not strictly represent the ploidy but represents the disomy or trisomy of the selected probe chromosome. Moreover, aneuploidy is the main factor contributing to early pregnancy loss. Therefore, caution is required when using single-probe FISH assays to determine ploidy in the villous tissues of POCs, which may include numerous trisomy cases. In this study, we selected the CEP17 probe because it rarely harbors trisomy of chromosome 17, which has a reported frequency below 1%32,38. Thus, a single CEP17 probe is one of the best choices for evaluating ploidy. Furthermore, the three-signal ratio of FISH was found to be a critical parameter for determining the ploidy (diploid vs. triploid) of the samples. When histologic sections are prepared for examination by FISH analysis, some parts of the cells are inevitably lost during sectioning, leading to artificial deletions39. The frequency of occurrence of this artificial effect, known as “truncation artifacts,” may be high. We overcame this problem by using the three-signal ratio as an objective indicator. When the boundary zones are sufficiently large, suspected molar conceptuses can be directly categorized as diploid or triploid (Fig. 5). However, the conditions for performing FISH, including section thickness at each laboratory, must be adjusted to achieve reproducible FISH results. Owing to its availability, ease, and reproducibility, FISH may be a feasible diagnostic tool in clinical settings and pathologic departments.
In the present study, we have uncovered a caveat regarding STR analysis; although STR analysis offers a significant advantage in distinguishing between PHM and non-molar abortus, it has limitations. The significance of STR analysis was introduced in the book “World Health Organization Classification of Tumours, Female Genital Tumours,” which presented the STR electropherogram of genetic PHM13. However, in our study, the 2 cases (case #63 and #64) previously diagnosed as genetic PHM without three allelic loci (diandric monogynic triploid) using STR analysis were found to be androgenetic/biparental mosaic diploid cases using FISH. The STR patterns of our androgenetic/biparental mosaic diploid cases were similar to those of previously reported diandric monogynic triploids without three allelic loci26,34,40,41. Therefore, based on the STR analysis, biparental and androgenetic diploid cells in the cases of biparental/androgenetic conceptus could include one paternal haploid in both cell populations. The discrepancy in the p57KIP2 immunohistochemistry pattern and the absence of three allelic loci in the villi by STR analysis suggest the presence of androgenetic/biparental mosaic diploid, as mentioned by Xing et al12. In cases where biparental/androgenetic mosaic gestation is suspected, accurate diagnosis can be facilitated through STR analysis, employing laser microdissection, which separately collects villous trophoblasts and stromal cells34. Nevertheless, FISH can still be helpful for precise diagnosis.
The limitation of the diagnostic workflow in this study is its propensity to generate perplexing outcomes in exceptional cases involving paradoxical p57KIP2 immunostaining. The most notable limitation of this workflow is its inability to distinguish between monoandric digynic triploid (non-molar) (case #80) and diandric monogynic triploid (PHM). STR analysis is deemed more specific in diagnosing PHM than p57KIP2 and FISH analysis, as up to 1/3 of triploid abortions were digynic, non-molar conceptuses42,43. However, three signals in FISH analysis strongly support the decision to diagnose PHM. In addition, androgenetic diploid CHMs that retain maternal chromosome 11 have been reported to exhibit positive p57KIP2 immunostaining. Previous studies have indicated that 1% to 2% of androgenetic CHM cases could manifest sustained expression of p57KIP210,18,44,45. In addition to p57KIP2-positive outcomes, 2 FISH signals could be interpreted as non-molar abortus. Moreover, a diandric monogynic triploid PHM or uniparental disomy case with loss of maternal chromosome 11 was reported to be negative in p57KIP2 immunostaining46,47. In the present study, vaginal villous specimens of invasive moles displayed a slightly elevated three-signal ratio (Fig. 5A), attributable to their high proliferation potential. However, our diagnostic workflow is not applicable if the initial histologic (HE) examination did not suspect HM. For example, in this study, 2 independent pathologists evaluated case #60 (diandric monogynic triploid PHM) as a non-molar hydropic abortion. However, this case would not have been diagnosed as an HM if it had not been included for analysis using the diagnostic workflow; case #60 was referred to our hospital based on sonographic findings suggesting a molar pregnancy. Therefore, clinical information is a critical determinant for applying the diagnostic workflow.
In conclusion, this study demonstrates that a comprehensive analysis using p57KIP2 immunostaining and FISH is helpful in the pathologic diagnosis of POC. Further validation in multiple laboratories and with multiple pathologists is required to confirm our findings. Reliable differentiation between molar and non-molar pregnancies is crucial for accurate patient management, early detection of GTN, and ensuring the safety of subsequent pregnancies.
Supplementary Material
Acknowledgments
The authors thank the staff of the Department of Obstetrics and Gynecology and the Department of Pathology at Chiba University Hospital for treating the patients and performing the pathologic diagnosis. They also thank Editage for the English language editing.
Footnotes
This study received research grants from JSPS KAKENHI grant numbers JP17K16831 (A.S.), JP18K09281 (H.U.), JP21K19551 (H.U.), and JP22H03220 (H.U.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Current Address: Y.N. Department of Pathology, Yokosuka Kyosai Hospital, Yokosuka, Kanagawa, Japan.
Current Address: M.S. Department of Evolution and Reproductive Medicine, Medical Mycology Research Center, Chiba University, Chiba, Chiba, Japan.
K.H., M.K., and T.F. are employees of Kotobiken Medical Laboratories Inc. The remaining authors declare no conflict of interest.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.intjgynpathology.com.
Contributor Information
Hirokazu Usui, Email: hirokazu-usui@faculty.chiba-u.jp.
Kazufusa Hoshimoto, Email: hoshimoto.fam@gmail.com.
Asuka Sato, Email: aschiba@chiba-u.jp.
Motofumi Kano, Email: m.kano@koutou-biken.co.jp.
Toshio Fukusato, Email: fukusato@koutou-biken.co.jp.
Yukio Nakatani, Email: ynak2012@gmail.com.
Makio Shozu, Email: shozumakio@chiba-u.jp.
REFERENCES
- 1.Hui P, Buza N, Murphy KM, et al. Hydatidiform moles: Genetic basis and precision diagnosis. Annu Rev Pathol 2017;12:449–85. [DOI] [PubMed] [Google Scholar]
- 2.Savage PM, Sita-Lumsden A, Dickson S, et al. The relationship of maternal age to molar pregnancy incidence, risks for chemotherapy and subsequent pregnancy outcome. J Obstet Gynaecol 2013;33:406–11. [DOI] [PubMed] [Google Scholar]
- 3.Albright BB, Shorter JM, Mastroyannis SA, et al. Gestational trophoblastic neoplasia after human chorionic gonadotropin normalization following molar pregnancy: a systematic review and meta-analysis. Obstet Gynecol 2020;135:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukunaga M, Katabuchi H, Nagasaka T, et al. Interobserver and intraobserver variability in the diagnosis of hydatidiform mole. Am J Surg Pathol 2005;29:942–7. [DOI] [PubMed] [Google Scholar]
- 5.Usui H, Kiyokawa T, Qu J, et al. Comparison between pathological diagnosis and cytogenetic diagnosis by short tandem repeat polymorphism analysis of suspected molar pregnancies. J Reprod Med 2016;61:219–23. [PubMed] [Google Scholar]
- 6.Ronnett BM, DeScipio C, Murphy KM. Hydatidiform moles: Ancillary techniques to refine diagnosis. Int J Gynecol Pathol 2011;30:101–16. [DOI] [PubMed] [Google Scholar]
- 7.Usui H. Auxiliary and experimental diagnostic techniques for hydatidiform moles. J Obstet Gynaecol Res 2022;48:3077–86. [DOI] [PubMed] [Google Scholar]
- 8.Laviola GM, Fortini AS, Salles D, et al. Complementary tool in diagnosis of hydatidiform mole: Review. Pathol Res Pract 2022;237:154041. [DOI] [PubMed] [Google Scholar]
- 9.Usui H, Qu J, Sato A, et al. Gestational trophoblastic neoplasia from genetically confirmed hydatidiform moles: Prospective observational cohort study. Int J Gynecol Cancer 2018;28:1772–80. [DOI] [PubMed] [Google Scholar]
- 10.Banet N, DeScipio C, Murphy KM, et al. Characteristics of hydatidiform moles: Analysis of a prospective series with p57 immunohistochemistry and molecular genotyping. Mod Pathol 2014;27:238–54. [DOI] [PubMed] [Google Scholar]
- 11.Ronnett BM. Hydatidiform moles: Ancillary techniques to refine diagnosis. Arch Pathol Lab Med 2018;142:1485–502. [DOI] [PubMed] [Google Scholar]
- 12.Xing D, Adams E, Huang J, et al. Refined diagnosis of hydatidiform moles with p57 immunohistochemistry and molecular genotyping: updated analysis of a prospective series of 2217 cases. Mod Pathol 2021;34:961–82. [DOI] [PubMed] [Google Scholar]
- 13.Buza N, Colgan TJ, Sebire NJ, et al. Partial hydatidiform moles In: WHO Classification of Tumours Editorial Board, ed World Health Organization Classification of Tumours, Female Genital Tumours, 5th edition. Lyon: IARC publication; 2020:317–8. [Google Scholar]
- 14.Kaur B, Sebire NJ. p57(KIP2) immunostaining for diagnosis of hydatidiform mole. BJOG 2018;125:1234. [DOI] [PubMed] [Google Scholar]
- 15.Fisher RA, Tommasi A, Short D, et al. Clinical utility of selective molecular genotyping for diagnosis of partial hydatidiform mole; a retrospective study from a regional trophoblastic disease unit. J Clin Pathol 2014;67:980–4. [DOI] [PubMed] [Google Scholar]
- 16.Kihara M, Matsui H, Seki K, et al. Genetic origin and imprinting in hydatidiform moles. Comparison between DNA polymorphism analysis and immunoreactivity of p57KIP2. J Reprod Med 2005;50:307–12. [PubMed] [Google Scholar]
- 17.Landolsi H, Missaoui N, Brahem S, et al. The usefulness of p57(KIP2) immunohistochemical staining and genotyping test in the diagnosis of the hydatidiform mole. Pathol Res Pract 2011;207:498–504. [DOI] [PubMed] [Google Scholar]
- 18.McConnell TG, Murphy KM, Hafez M, et al. Diagnosis and subclassification of hydatidiform moles using p57 immunohistochemistry and molecular genotyping: Validation and prospective analysis in routine and consultation practice settings with development of an algorithmic approach. Am J Surg Pathol 2009;33:805–17. [DOI] [PubMed] [Google Scholar]
- 19.Castrillon DH, Sun D, Weremowicz S, et al. Discrimination of complete hydatidiform mole from its mimics by immunohistochemistry of the paternally imprinted gene product p57KIP2. Am J Surg Pathol 2001;25:1225–30. [DOI] [PubMed] [Google Scholar]
- 20.Vang R, Gupta M, Wu LS, et al. Diagnostic reproducibility of hydatidiform moles: Ancillary techniques (p57 immunohistochemistry and molecular genotyping) improve morphologic diagnosis. Am J Surg Pathol 2012;36:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajii T, Ohama K. Androgenetic origin of hydatidiform mole. Nature 1977;268:633–4. [DOI] [PubMed] [Google Scholar]
- 22.Lage JM, Mark SD, Roberts DJ, et al. A flow cytometric study of 137 fresh hydropic placentas: Correlation between types of hydatidiform moles and nuclear DNA ploidy. Obstet Gynecol 1992;79:403–10. [DOI] [PubMed] [Google Scholar]
- 23.Jeffers MD, Michie BA, Oakes SJ, et al. Comparison of ploidy analysis by flow cytometry and image analysis in hydatidiform mole and non-molar abortion. Histopathology 1995;27:415–21. [DOI] [PubMed] [Google Scholar]
- 24.Cheville JC, Greiner T, Robinson RA, et al. Ploidy analysis by flow cytometry and fluorescence in situ hybridization in hydropic placentas and gestational trophoblastic disease. Hum Pathol 1995;26:753–7. [DOI] [PubMed] [Google Scholar]
- 25.Hoffner L, Parks WT, Swerdlow SH, et al. Simultaneous detection of imprinted gene expression (p57(KIP2)) and molecular cytogenetics (FICTION) in the evaluation of molar pregnancies. J Reprod Med 2010;55:219–28. [PubMed] [Google Scholar]
- 26.LeGallo RD, Stelow EB, Ramirez NC, et al. Diagnosis of hydatidiform moles using p57 immunohistochemistry and HER2 fluorescent in situ hybridization. Am J Clin Pathol 2008;129:749–55. [DOI] [PubMed] [Google Scholar]
- 27.Maggiori MS, Peres LC. Morphological, immunohistochemical and chromosome in situ hybridization in the differential diagnosis of Hydatidiform Mole and Hydropic Abortion. Eur J Obstet Gynecol Reprod Biol 2007;135:170–6. [DOI] [PubMed] [Google Scholar]
- 28.Bifulco C, Johnson C, Hao L, et al. Genotypic analysis of hydatidiform mole: An accurate and practical method of diagnosis. Am J Surg Pathol 2008;32:445–51. [DOI] [PubMed] [Google Scholar]
- 29.Kipp BR, Ketterling RP, Oberg TN, et al. Comparison of fluorescence in situ hybridization, p57 immunostaining, flow cytometry, and digital image analysis for diagnosing molar and nonmolar products of conception. Am J Clin Pathol 2010;133:196–204. [DOI] [PubMed] [Google Scholar]
- 30.Chen KH, Hsu SC, Chen HY, et al. Utility of fluorescence in situ hybridization for ploidy and p57 immunostaining in discriminating hydatidiform moles. Biochem Biophys Res Commun 2014;446:555–60. [DOI] [PubMed] [Google Scholar]
- 31.Segawa T, Kuroda T, Kato K, et al. Cytogenetic analysis of the retained products of conception after missed abortion following blastocyst transfer: a retrospective, large-scale, single-centre study. Reprod Biomed Online 2017;34:203–10. [DOI] [PubMed] [Google Scholar]
- 32.Murphy KM, McConnell TG, Hafez MJ, et al. Molecular genotyping of hydatidiform moles: Analytic validation of a multiplex short tandem repeat assay. J Mol Diagn 2009;11:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Usui H, Sato A, Ota M, et al. Androgenetic complete hydatidiform moles with p57KIP2-positive immunostaining. Am J Clin Pathol 2020;154:776–83. [DOI] [PubMed] [Google Scholar]
- 34.Hodgson A, Dube V, Strickland S, et al. Androgenetic/biparental mosaic/chimeric conceptions with a molar component: A diagnostic and clinical challenge. Int J Gynecol Pathol 2021;40:510–7. [DOI] [PubMed] [Google Scholar]
- 35.Ragazzi M, Bisagni A, Gasparini E, et al. Impact of 2013 ASCO/CAP guidelines on HER2 determination of invasive breast cancer: A single institution experience using frontline dual-color FISH. Breast 2017;34:65–72. [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Perez JA, Nielsen GP, Antonescu C, et al. EWSR1/FUS-NFATc2 rearranged round cell sarcoma: Clinicopathological series of 4 cases and literature review. Hum Pathol 2019;90:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boer JM, den Boer ML. BCR-ABL1-like acute lymphoblastic leukaemia: From bench to bedside. Eur J Cancer 2017;82:203–18. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Cheng Q, Meng L, et al. Clinical application of SNP array analysis in first-trimester pregnancy loss: A prospective study. Clin Genet 2017;91:849–58. [DOI] [PubMed] [Google Scholar]
- 39.Ventura RA, Martin-Subero JI, Jones M, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn 2006;8:141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis GH, DeScipio C, Murphy KM, et al. Characterization of androgenetic/biparental mosaic/chimeric conceptions, including those with a molar component: morphology, p57 immnohistochemistry, molecular genotyping, and risk of persistent gestational trophoblastic disease. Int J Gynecol Pathol 2013;32:199–214. [DOI] [PubMed] [Google Scholar]
- 41.Sunde L, Niemann I, Hansen ES, et al. Mosaics and moles. Eur J Hum Genet 2011;19:1026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redline RW, Hassold T, Zaragoza MV. Prevalence of the partial molar phenotype in triploidy of maternal and paternal origin. Hum Pathol 1998;29:505–11. [DOI] [PubMed] [Google Scholar]
- 43.Zaragoza MV, Surti U, Redline RW, et al. Parental origin and phenotype of triploidy in spontaneous abortions: Predominance of diandry and association with the partial hydatidiform mole. Am J Hum Genet 2000;66:1807–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher RA, Hodges MD, Rees HC, et al. The maternally transcribed gene p57(KIP2) (CDNK1C) is abnormally expressed in both androgenetic and biparental complete hydatidiform moles. Hum Mol Genet 2002;11:3267–72. [DOI] [PubMed] [Google Scholar]
- 45.DeScipio C, Haley L, Beierl K, et al. Diandric triploid hydatidiform mole with loss of maternal chromosome 11. Am J Surg Pathol 2011;35:1586–91. [DOI] [PubMed] [Google Scholar]
- 46.Xing D, Miller K, Beierl K, et al. Loss of p57 expression in conceptions other than complete hydatidiform mole: a case series with emphasis on the etiology, genetics, and clinical significance. Am J Surg Pathol 2022;46:18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebire NJ, May PC, Kaur B, et al. Abnormal villous morphology mimicking a hydatidiform mole associated with paternal trisomy of chromosomes 3,7,8 and unipaternal disomy of chromosome 11. Diagn Pathol 2016;11:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.