Abstract
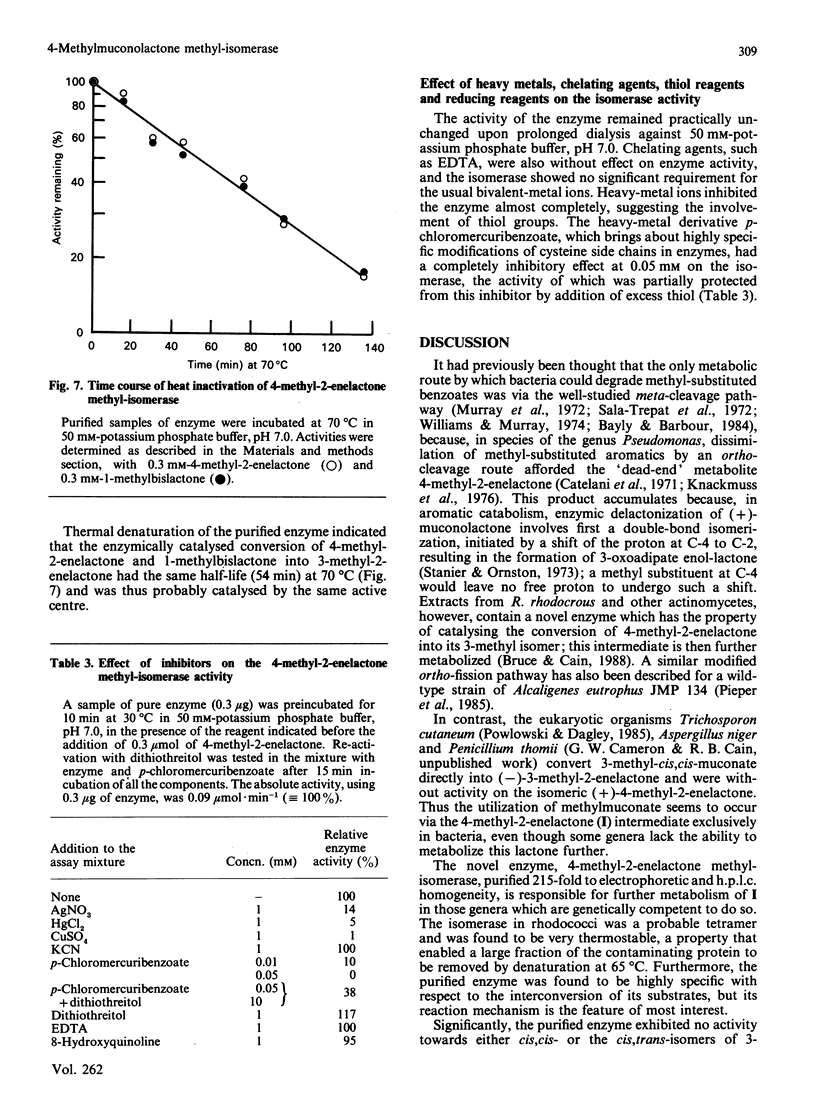
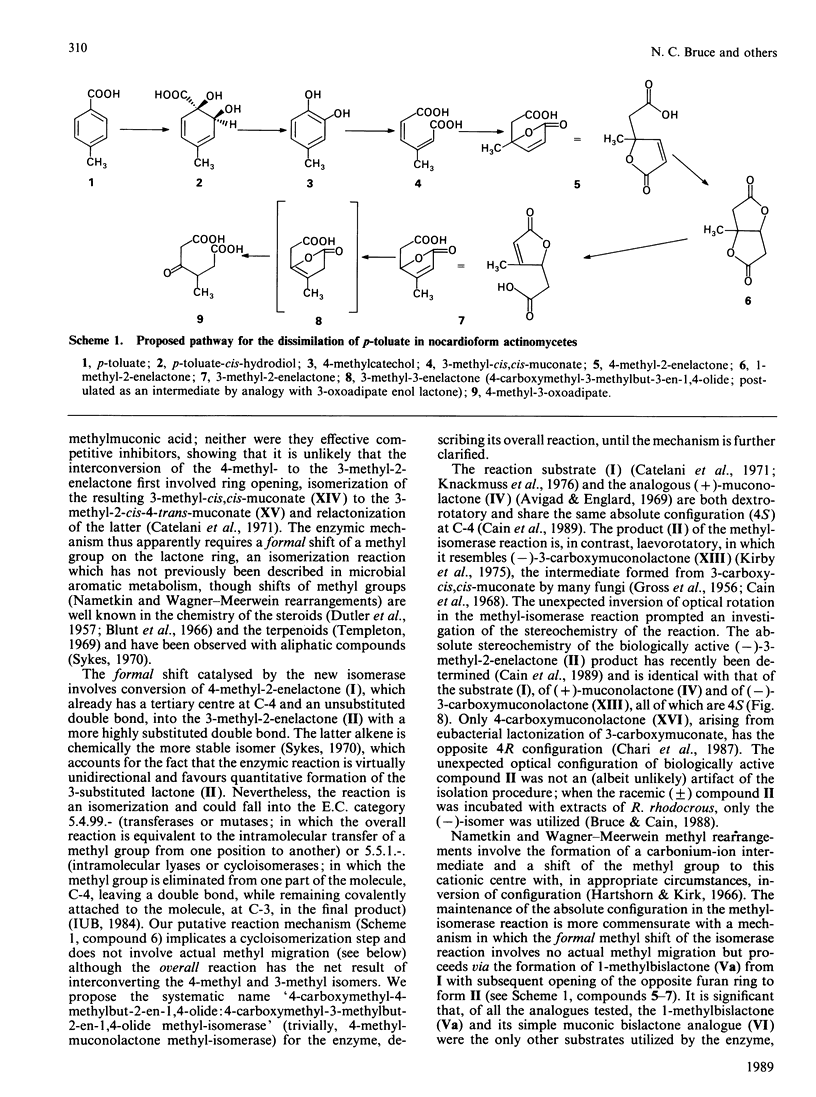
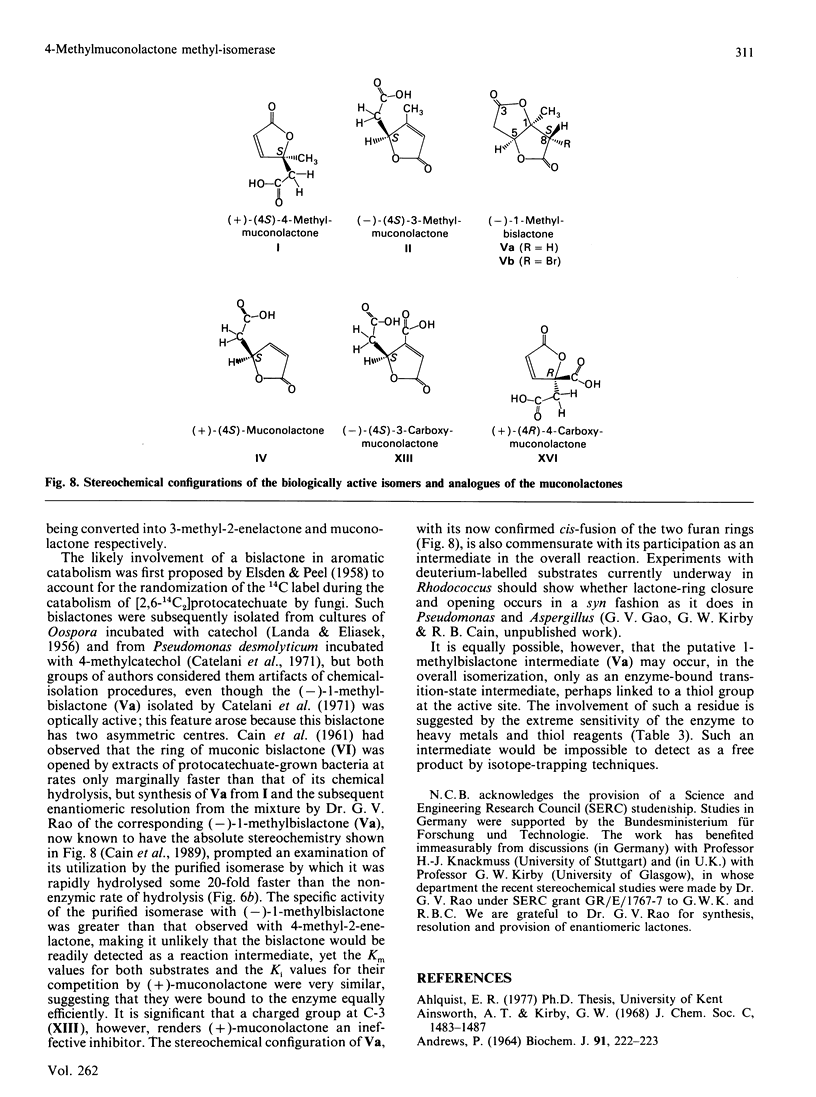
The novel enzyme 4-methyl-2-enelactone methyl-isomerase was detected in, and purified to electrophoretic homogeneity from, p-toluate-grown cells of Rhodococcus rhodocrous N75, a nocardioform actinomycete. The enzyme was very thermostable and had a native Mr of 75,500; as the monomer had an Mr of 17,000, the enzyme is probably tetrameric. The new isomerase is highly specific with respect to its lactone substrate, only accepting (+)-(4S)-4-methylmuconolactone (4-carboxymethyl-4-methylbut-2-en-1,4-olide), and the putative isomerization reaction intermediate 1-methylbislactone ((-)-1-methyl-3,7-dioxo-2,6-dioxabicyclo-[3.3.0]octane) as substrates, and yielding (-)-(4S)-3-methylmuconolactone (4-carboxymethyl-3-methylbut-2-en-1,4-olide) as product. Some other lactone analogues acted as competitive inhibitors. Our data suggest that the isomerization does not involve actual methyl migration, but proceeds via the 1-methybislactone.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R. B., Bilton R. F., Darrah J. A. The metabolism of aromatic acids by micro-organisms. Metabolic pathways in the fungi. Biochem J. 1968 Aug;108(5):797–828. doi: 10.1042/bj1080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R. B. The metabolism of protocatechuic acid by certain micro-organisms. A reassessment of the evidence for the participation of 2:6-dioxa-3:7-dioxobicyclo[3:3:0]octane as an intermediate. Biochem J. 1961 May;79(2):312–316. doi: 10.1042/bj0790312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catelani D., Fiecchi A., Galli E. Dextro-gamma-carboxymethyl-gamma-methyl-delta-alpha-butenolide. A 1,2-ring-fission product of 4-methylcatechol by Pseudomonas desmolyticum. Biochem J. 1971 Jan;121(1):89–92. doi: 10.1042/bj1210089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELSDEN S. R., PEEL J. L. Metabolism of carbohydrates and related compounds. Annu Rev Microbiol. 1958;12:145–202. doi: 10.1146/annurev.mi.12.100158.001045. [DOI] [PubMed] [Google Scholar]
- GROSS S. R., GAFFORD R. D., TATUM E. L. The metabolism of protocatechuic acid by Neurospora. J Biol Chem. 1956 Apr;219(2):781–796. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Murray K., Duggleby C. J., Sala-Trepat J. M., Williams P. A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972 Jul 24;28(3):301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- Powlowski J. B., Dagley S. beta-Ketoadipate pathway in Trichosporon cutaneum modified for methyl-substituted metabolites. J Bacteriol. 1985 Sep;163(3):1126–1135. doi: 10.1128/jb.163.3.1126-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Murray K., Williams P. A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972 Jul 24;28(3):347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Ornston L. N. The beta-ketoadipate pathway. Adv Microb Physiol. 1973;9(0):89–151. [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


