Abstract
Background
Certain paediatric nervous system malignancies have dismal prognoses. Retinoic acid (RA) is used in neuroblastoma treatment, and preclinical data indicate potential benefit in selected paediatric brain tumour entities. However, limited single-agent efficacy necessitates combination treatment approaches.
Methods
We performed drug sensitivity profiling of 76 clinically relevant drugs in combination with RA in 16 models (including patient-derived tumouroids) of the most common paediatric nervous system tumours. Drug responses were assessed by viability assays, high-content imaging, and apoptosis assays and RA relevant pathways by RNAseq from treated models and patient samples obtained through the precision oncology programme INFORM (n = 2288). Immunoprecipitation detected BCL-2 family interactions, and zebrafish embryo xenografts were used for in vivo efficacy testing.
Results
Group 3 medulloblastoma (MBG3) and neuroblastoma models were highly sensitive to RA treatment. RA induced differentiation and regulated apoptotic genes. RNAseq analysis revealed high expression of BCL2L1 in MBG3 and BCL2 in neuroblastomas. Co-treatments with RA and BCL-2/XL inhibitor navitoclax synergistically decreased viability at clinically achievable concentrations. The combination of RA with navitoclax disrupted the binding of BIM to BCL-XL in MBG3 and to BCL-2 in neuroblastoma, inducing apoptosis in vitro and in vivo.
Conclusions
RA treatment primes MBG3 and NB cells for apoptosis, triggered by navitoclax cotreatment.
Subject terms: Embryonal neoplasms, Targeted therapies
Introduction
Although past decades have seen a remarkable increase in childhood cancer survival to above 80%, it remains the leading cause of death from disease in children beyond infancy [1]. Particularly relapsed tumours of the peripheral and central nervous system, still carry dismal prognoses and significantly contribute to childhood cancer mortality [1, 2]. In the case of tumour recurrence, few therapeutic options remain with precision oncology approaches, such as the INFORM (INdividualized Therapy FOr Relapsed Malignancies in Childhood) programme, identifying survival-prolonging actionable targets in just up to 10% of cases [3]. Importantly, disease- and treatment-related morbidities, such as chronic cardiovascular disease, secondary malignancy, and neurological impairment, also burden the lives of many childhood cancer survivors, including patients treated for CNS malignancies [4].
The most common diagnoses of CNS malignancies in children are medulloblastoma (MB), high-grade glioma (HGG) and ependymoma (EPN). To accurately and reliably differentiate between the numerous heterogeneous subtypes of childhood brain tumours, molecular analyses of the genome and methylome are now routinely incorporated into diagnostics [5]. In MB, four distinct molecular subgroups have been identified, with Group 3 MB (MBG3) patients having the worst prognosis [6, 7]. Despite surgical resection, craniospinal irradiation, and intensive chemotherapy, the 5-year overall survival is less than 50% in these high-risk MB cases [7, 8]. Exceptionally poor survival can be observed in the diverse spectrum of paediatric HGG, where molecularly informed classification is based on histone 3 (H3) mutation status [9, 10]. In EPN, the two particularly aggressive subgroups, supratentorial ZFTA fusion-positive and posterior fossa group A (PFA) EPN, mostly occur in childhood [10, 11].
The most common extracranial tumour in childhood, neuroblastoma (NB), originates during embryogenesis from blocked differentiation of neural crest-derived precursor cells of the developing sympathetic nervous system [12]. It has a large spectrum of disease courses, ranging from spontaneous regression and maturation to aggressively progressing disease despite intensive multimodal treatment [13]. Risk-adapted treatment may only consist of observation or surgical resection and chemotherapy in low- to intermediate-risk cases [14]. In high-risk NB, which is often driven by amplification of the MYCN oncogene (NBMYCN), however, intensive chemotherapy alongside surgery, myeloablation and autologous stem-cell transplantation, radiation, and maintenance therapy with anti-GD2 immunotherapy and retinoic acid (RA) only achieves a 5-year overall survival of approximately 60% [13, 15]. As first-line therapeutic in high-risk NB care, RA is a component of maintenance therapy in current treatment recommendations from the North American Children’s Oncology Group and the International Society of Paediatric Oncology Europe Neuroblastoma Group [15–17]. The standard paediatric treatment regimen for high-risk NB with 13-cis-RA leads to serum concentrations in the low micromolar range (trough to peak: 4.1–7.2 µM) and is well tolerated by patients [18]. Furthermore, 13-cis-RA permeates the blood-brain barrier (BBB) well, with peak white matter concentrations reaching approximately half of the peak serum concentrations in a preclinical rat model [19]. Additionally, it has demonstrated antiproliferative, differentiating, and proapoptotic effects against preclinical models of multiple other paediatric nervous system tumour entities [20–24].
To identify novel treatment approaches, mechanisms by which relapsed paediatric nervous system tumours evade current treatment approaches, are of particular interest. The B-cell lymphoma 2 (BCL-2) family of proteins plays an important role in maintaining homoeostasis in apoptosis by regulating mitochondrial outer membrane permeabilization (MOMP) through the interaction of its pro- and anti-apoptotic members [25]. Importantly, the main anti-apoptotic proteins BCL-2, B-cell lymphoma-extra large (BCL-XL) and myeloid cell leukaemia 1 (MCL-1) are often upregulated across many adult and paediatric cancer types [26]. To address this mechanism, small-molecule BCL-2 homology 3 (BH3) mimetics, such as navitoclax and venetoclax, bind to the hydrophobic groove of antiapoptotic and pore-forming BCL-2 proteins, thus antagonising BCL-2 and BCL-XL and inducing MOMP [25, 27]. These therapeutics display compelling strategies to target cancer cells and have been under intensive clinical investigation in a variety of cancers as single agents and combined with other therapies [25].
As many subtypes of the diverse spectrum of paediatric nervous system cancers have high rates of aggressive progression or relapse and current standard-of-care approaches result in severe chronic health conditions, novel targeted treatment strategies need to be explored to potentially improve efficacy and reduce toxicity. Here, we investigated a molecularly diverse panel of high-risk paediatric nervous system tumours for their RA response as a single drug and in combination with a library of 76 clinically relevant compounds, with the aim of identifying improved combination therapies for this unmet clinical need.
Materials and methods
Cell culture
Cell culture was performed as described previously [28]. The N = 16 culture models applied for the drug screen included, besides established cell lines, the patient-derived cultures NB-S-124 (NB), SU-DIPG-13 (HGG) and SU-DIPG-25 (HGG) (both kindly provided by Michelle Monje, Stanford, CA, USA), and INF_R_1073_relapse1_LTC (HGG) and INF_R_859_primary_LTC (HGG) (both derived from primary INFORM tumour samples; LTC: longterm culture). Further validation experiments were performed using the patient-derived tumouroid cultures INF_R_1632_relapse1_PDX_LTC (NB; from mouse PDX), INF_R_1887_relapse1_LTC (EPN), Med_2112fh_PDX_FTC (MBG3; from mouse PDX; FTC: fresh tissue culture), and RCMB28 (MBG3). INFORM long-term cultures (LTC) were established from primary tumour samples from the INFORM programme as previously described [3, 28, 29]. All patient-derived models were cultured organoid-like as free-floating and semiadherent spheroids under serum-free conditions in stem-cell medium. The Supplementary Information gives a detailed description about methods and models.
Drugs and reagents
13-cis-RA (MedChem Express, Monmouth Junction, NJ, USA), A-1155463 (Selleckchem, Houston, TX, USA), all-trans-RA (Selleckchem), entinostat (Biomol GmbH, Hamburg, Germany), I-BET151 (Selleckchem), navitoclax (TargetMol, Wellesley Hills, MA, USA), staurosporine (Selleckchem), and venetoclax (Selleckchem) were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich) to a 10 mM stock concentration and stored as aliquots at −80 °C protected from light.
Metabolic activity assays and medium-throughput drug combination screening
The assays were performed as described previously [28, 30, 31]. All culture models were seeded as three-dimensional tumouroids on round-bottom 384-well ultralow attachment plates (Corning Inc., Corning, NY, USA). The Supplementary Information gives a detailed description.
Bulk RNA sequencing
For bulk RNA sequencing, two medulloblastoma (MB) (D425, HD-MB03) and two neuroblastoma (NB) (NB-S-124, SK-N-BE(2)-C) cultures were chosen. Cells were seeded on 10 cm dishes, incubated for 24 h and subsequently treated for 144 h with 0.5/1 µM (MB/NB) all-trans-RA or DMSO solvent control in two independent replicate experiments. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA integrity and quantity were assessed with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). After passing initial quality control at the in-house DKFZ Genomics & Proteomics Core Facility, RNAseq was performed on Illumina HiSeq 4000 Single-read 50 bp (Illumina, San Diego, CA, USA) using the TruSeq stranded Protocol. Read quality control was passed with Q30 > 97.5% in all samples. Read mapping and counting were carried out by the RSEM pipeline using the Bowtie2 alignment programme and Expectation Maximization (EM) algorithm for gene-level read estimates [32, 33]. The data is available on the online platform R2 (http://r2.amc.nl): Exp Medulloblastoma / Neuroblastoma RA - Oehme - 20 - rsem - gencode19. Subsequent differential expression analysis was performed using the DESeq2 package [34] in R. Gene Ontology (GO) analysis was performed using clusterProfiler package [35].
Synergy analysis
For synergy analysis, a 5 × 5 matrix and seven ray design of RA combined with entinostat, I-BET151 or navitoclax was used. Cells were seeded as three-dimensional tumouroids on round-bottom 384-well plates and treated for 144 h, and readout was performed as described in section “Metabolic activity assays and medium-throughput drug combination screening”. To assess combination effects, the experimentally measured antitumour effect was compared to the expected response determined using the Loewe reference model, as independent drug effects cannot be assumed in these combinations. Synergy scores were calculated and visualised with SynergyFinder web-application 2.0 (https://synergyfinder.fimm.fi) [36] for analysis of the matrix and a custom R pipeline for ray designs based on the DRC R-package [37] (sample R script included in supplementary information). Loewe Combination Indices were calculated according to Lee et al [38].
Colony formation assay
To assess clonogenic growth and sustained treatment effect after drug removal, cells were seeded on six-well plates at 2000 cells per well and treated after 24 h for 96 h. Then, drugs were removed by medium change and cells were cultured for another 7 d before undergoing fixation and staining with 1% crystal violet solution. Colonies per well were quantified using the ITCN plugin in ImageJ Fiji version 2.5.0.
Morphology assays
To observe morphological effects upon prolonged treatment, cells were seeded on six-well plates (100,000 cells/well HD-MB03; 20,000 cells/well SK-N-BE(2)-C; 10,000 cells/well SJ-GBM2) and treated the next day for additional 8 d. Fixation and staining was performed with 1% crystal violet solution. Three random sites per replicate were captured with light microscopy (Nikon Eclipse TS2 10x magnification).
High-content fluorescence microscopy (HCM)
For HCM, cells were seeded as a monolayer on a black high-content imaging ½ area 96-well microplate with a flat glass bottom (Corning Inc.). To achieve monolayer attachment for NB-S-124 cells, corresponding wells underwent surface coating with 1:100 diluted Matrigel (Corning Inc.) before seeding. Treatment was carried out the day after seeding using the TECAN D300e drug dispenser. After 72 h of treatment, cells were stained with fluorescence dyes for 30 min as indicated in Supplementary Table 1. Live cell imaging was performed with an ImageXpress Micro Confocal High-content microscope (Molecular Devices, San José, CA, USA) at 10x magnification. CellProfiler Version 4.2.5 [39] and CellProfiler Analyst Version 3.0.4 [40] was used for automated image analysis. To quantify the number of Caspase positive cells and fragmented nuclei, a custom CellProfiler pipeline and the CellProfiler Analyst machine-learning tool Classifier was used as described in Supplementary Table 2. To quantify the length of neurite-like protrusions, a custom CellProfiler pipeline was used as described in Supplementary Table 3. The high-content imaging of patient-derived tumouroids for validation was performed as described in the Supplementary Information.
Caspase-3/7 activity assay
To measure caspase-3/7 activity, the Caspase-3/7 Fluorometric Assay (BioVision, Abcam, Cambridge, UK) was performed following the manufacturer’s instructions and as described previously [41]. Details are listed in the Supplementary Information.
Trypan blue assays
To detect cell viability and cell death, trypan blue assays were performed as described previously [41]. Details are listed int the Supplementary Information.
Western blot analysis and immunoprecipitation
Protein expression analysis was performed as reported previously [41]. The immunoprecipitation method was described previously [42]. Details are listed in Supplementary Information.
Zebrafish lines, toxicity assays, embryo xenotransplantation, and treatment
In vivo zebrafish (Danio rerio) embryo xenograft was performed as previously described [43]. Details are listed in the Supplementary Information.
Patient-derived tumour xenotransplantation
Mouse PDX were performed as previously described [44]. Details are listed in the Supplementary Information.
Statistical analysis
If not indicated otherwise, experiments were carried out as independent triplicates. GraphPad Prism 5 software (Version 5.01, GraphPad Software, San Diego, CA, USA) was used to calculate dose-response curves and corresponding absolute half-maximal inhibiting concentrations (IC50). RStudio (Version 2022.07.1 + 554, RStudio PBC, Boston, MA, USA) was used for statistical analysis and plotting with R (Version 4.2.1, R Foundation for Statistical Computing, Vienna, Austria) and R packages tidyr, dplyr and ggplot2. To compare data, a two-tailed unpaired t-test or, where appropriate, one-way analysis of variance (ANOVA) with Tukey’s post hoc corrections were applied. p < 0.05 were considered significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Gene expression analysis R2
Web-based gene expression analysis R2 (R2: microarray analysis and visualisation platform; http://r2.amc.nl) was used to investigate the expression of regulated genes in the MB and NB cell models after RA treatment (Medulloblastoma / Neuroblastoma RA - Oehme - 20 - rsem - gencode19; 2021-12-01). Further R2 based gene expression analyses were performed using the Brain Tumor (2023-01-23) - Kool - 123 - MAS5.0 - u133p2 data set produced by Marcel Kool and Neuroblastoma - Westermann - 39 - TMM - ensh37e75 data set produced by Frank Westermann and in the INFORM 2288 patient cohort (2023-07-05), produced and kindly provided by the INFORM programme [3, 29, 31, 45].
Results
High-risk neuroblastoma and Group 3 medulloblastoma models share RA sensitivity
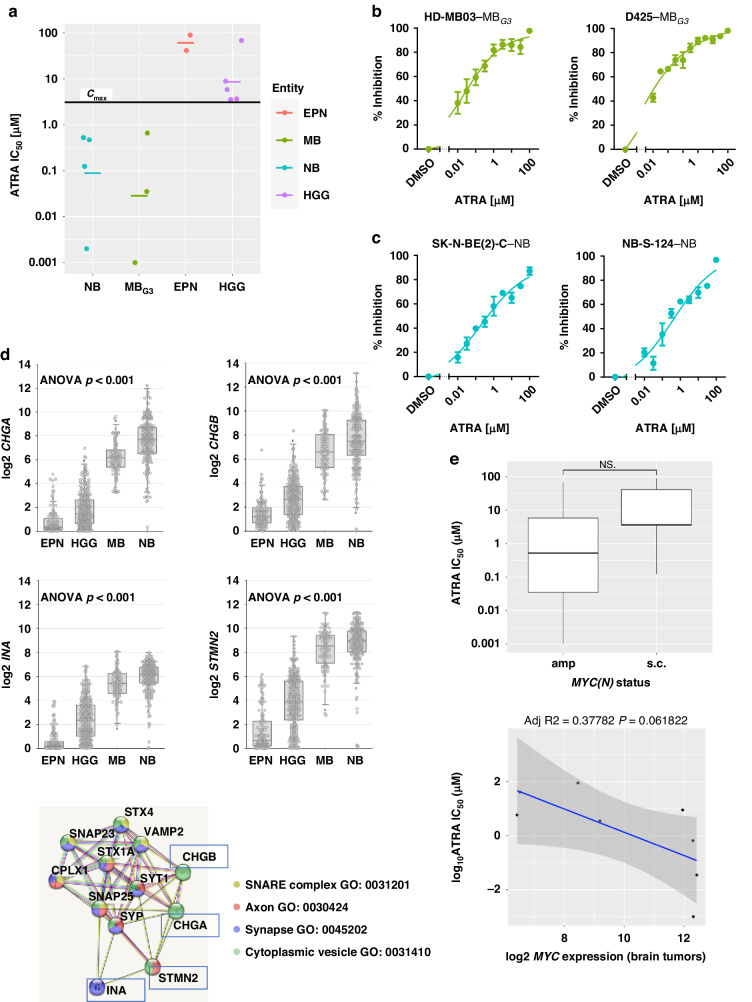
To assess the retinoic acid (RA) sensitivity of a diverse range of hard-to-treat paediatric nervous system tumours, a panel of 16 models (cell lines and ex vivo patient-derived models) was selected for investigation. The panel comprised four neuroblastoma (NB), four medulloblastoma (MB), two ependymoma (EPN) and six paediatric high-grade glioma (HGG) models reflecting different high-risk subgroups, molecular characteristics and growth kinetics (Fig. 1a, Supplementary Table S4). Testing for all models was performed uniformly on three-dimensional spheroids in round-bottom plates to better reflect the in vivo conditions with growth patterns, metabolism, cell-cell contacts, and treatment resistance [31, 46]. Upon 144 h of treatment with the single agent all-trans-RA (ATRA), tumour cells responded differently depending on the (sub)entity. High-risk NB and MYC-amplified MBG3 models demonstrate good RA sensitivity with absolute half-maximal inhibitory concentrations (IC50 values) well below the peak serum concentration (Cmax) for the standard dosing regimen in paediatric oncology [18] (Fig. 1a–c, Supplementary Table S4). The most pronounced antitumoural activity was observed in CHP134 (NBMYCN, IC50 = 2 nM), D425 (MBG3, IC50 = 1 nM), and HD-MB03 (MBG3, IC50 = 35 nM) (Fig. 1b, c). The one model for Sonic hedgehog MB (MBSHH), and the models for EPN subgroups (ZFTA and PFA), and HGG subgroups (K27M, G34V/R and wildtype), however, showed little to no RA response at clinically relevant concentrations (Supplementary Table S4). Thus, we defined RA responsive models as those achieving at least 50% inhibition at concentrations below the Cmax (7.2 µM for NB, 3.1 µM for CNS tumours as levels are half as high beyond the blood-brain barrier [19]), which includes all NB and all MBG3 models, while the remaining nine models were classified as RA less-responsive. To capture common features of the responsive entities MB and NB versus differences to the less-responsive entities HGG and EPN, we used the INFORM patients gene expression data set of mainly relapsed tumours on the online platform R2 (Genomics Analysis and Visualisation Platform (http://r2.amc.nl)) and identified the top 50 differentially regulated genes (TOP50) between two groups for the comparison MB versus HGG, MB versus EPN, NB versus HGG and NB versus EPN (Supplementary Tables 5–8). From all four TOP50 gene lists, only four genes overlapped, which were significantly upregulated in NB and MB (especially in G3/G4) for all comparisons, namely CHGA, CHGB, INA and STMN2 (Fig. 1d; Supplementary Fig. S1). The protein interaction tool STRING (string-db.org) [47] identified GO terms relevant to synaptic secretory vesicles as a connecting theme (Fig. 1d). While the six MYC(N)-amplified MB and NB cell lines plus high-MYC-expressing SH-SY5Y cells showed the highest RA sensitivity of all models, it is important to note that MYC(N) amplification is not a suitable biomarker of RA sensitivity, as this feature was also found in less-responsive models (Fig. 1e; Supplementary Fig. S2). Hence, ATRA responsiveness appears not to be restricted to MYC(N) amplification nor growth kinetics in a given tumour model but rather favours embryonal tumours of the nervous system with synaptic vesicle formation capacity.
Fig. 1. Neuroblastoma and Group 3 medulloblastoma cells share antitumoural response patterns to retinoic acid treatment.
a Retinoic acid sensitivity as measured after 144 h of ATRA treatment through the metabolic activity CellTiter Glo assay. Absolute IC50 values of N = 14 culture models (including the primary patient-derived cultures NB-S-124, SU-DIPG-13, SU-DIPG-25, INF_R_1073_relapse1_LTC, and INF_R_859_primary_LTC) depicted separated by entity and compared to the estimated ATRA white matter Cmax (3.1 µM). All cells were seeded as three-dimensional tumouroids. The bar indicates the corresponding mean. In two out of N = 16 tested culture models RA IC50 values could not be calculated (>100 µM) due to no dose-dependent response (ONS-76, SU-DIPG-13). b, c ATRA dose-response curves of selected MBG3 and NB cultures after 144 h. The % Inhibition was calculated using the raw values of the metabolic activity at a respective drug concentration compared to the positive and negative controls. Error bars indicate standard deviation. N = 3 d Dot box plots of genes that are significantly upregulated in both relapsed neuroblastoma and medulloblastoma cases compared to ependymoma and high-grade glioma from the INFORM programme as measured by RNA sequencing (log2-fold-change expression). EPN, N = 128; HGG, N = 298; MB, N = 103; NB, N = 223. Connecting GO terms identified from the STRING protein interaction analysis of the four shared significantly upregulated genes. e Box plot of absolute RA IC50 value of N = 14 culture models compared to known genomic amplification status of either MYC/MYCN or single copy variant, and dot plot depicting absolute RA IC50 values and microarray-derived log2 MYC expression of 8 brain tumour culture models with linear regression analysis. Out of the N = 10 brain tumour culture models with measurable RA IC50 values two were excluded due to missing expression data (SJ-GBM-2, KNS-42). Amp amplified, ANOVA analysis of variance, ATRA all-trans-retinoic acid, Cmax peak concentration, EPN ependymoma, GO gene ontology, HGG high-grade glioma, IC50 half-maximal inhibitory concentration, MB medulloblastoma, MBG3 Group 3 medulloblastoma, NB neuroblastoma, s.c. single copy.
Combination drug screening identifies BCL-XL inhibitors (A-1155463, navitoclax) as top hits across RA-sensitive models
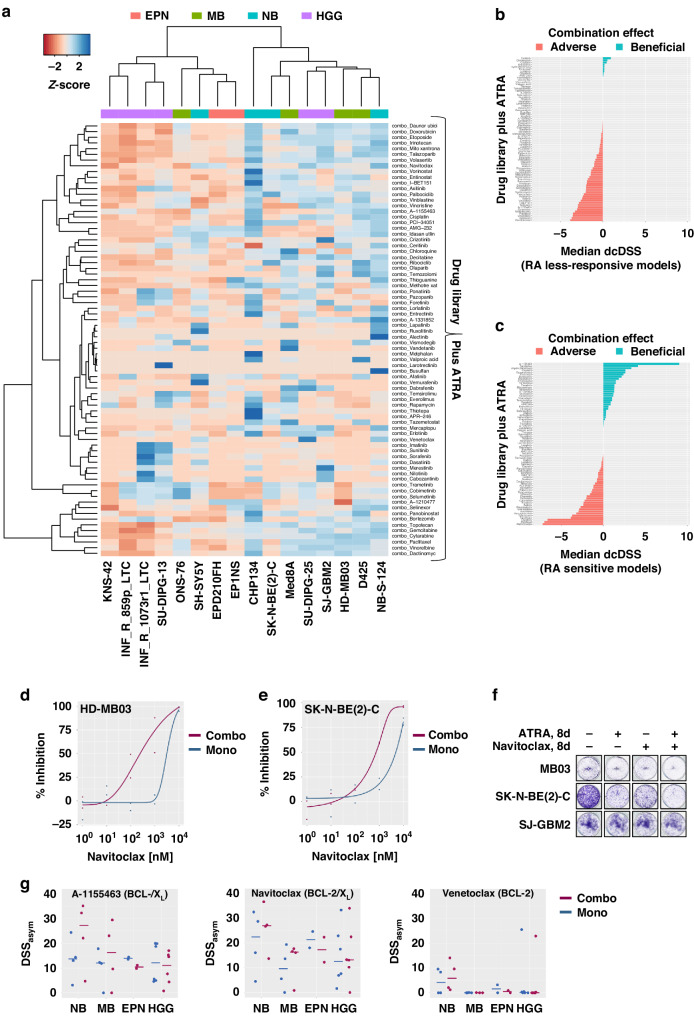
To identify beneficial combination treatments to either significantly enhance RA treatment in sensitive tumours or overcome RA resistance, a medium-throughput drug combination screen was performed on the above-described panel of 16 models. To this end, we combined the KiTZ drug library of 76 clinically relevant compounds, most of which are already approved or in late-stage clinical trials [31], with a fixed ATRA concentration (1 µM in NB, 0.5 µM in CNS tumours) or solvent control and treated cells for 144 h. The modified drug sensitivity score (DSSasym) was used to assess the respective treatment efficacy, and the differential combination DSS (dcDSS) was used to detect possible beneficial or adverse effects of drug combinations [30]. Hierarchical cluster analysis of DSSasym (Combo) values revealed two response pattern groups of eight models each, with the six MYC(N)-amplified MB and NB models clustering together (Fig. 2a). Selecting effective combinations by dcDSS (>2) and DSSasym (>10) for treatment efficacy, no overall beneficial combination was identified in RA less-responsive models (highest median dcDSSCeritinib = 0.95) with mostly negative dcDSS values (Fig. 2b, Supplementary Fig. S3). In RA-responsive models, the BCL-2 family inhibitor navitoclax was identified as top hit (median dcDSS 4.1) (Fig. 2c–g, Supplementary Fig. S4) and confirmed by longterm growth assay (Fig. 2f). The selective BCL-XL inhibitor A-1155463 also classified as top hit (median dcDSS 9.1) (Fig. 2g, Supplementary Fig. S4). Other apoptotic modulators, such as the selective BCL-2 inhibitor venetoclax (median dcDSS 1.2) and MCL-1 inhibitor A-1210477 (median dcDSS 0.0), showed no consistent combination benefit across different models (Fig. 2g, Supplementary Fig. S4). When entities were analysed separately, entinostat (class I HDAC inhibitor) and I-BET151 (BET bromodomain inhibitor) displayed entity-specific combination benefits with RA in MB and NB, respectively (Supplementary Figs. S5–6). In the two EPN (highest median dcDSSRapamycin = 2.9, DSSasym < 5) and six HGG (highest median dcDSSSelumetinib = 1.9) models, no entity-specific combination partner for RA treatment was identified. In these RA less-responsive models, the combination with RA had a rather adverse effect on the treatment efficacy of most drugs (Fig. 2b, Supplementary Fig. S7–8). In RA-sensitive models, the least favourable combination partners for RA treatment were the antimetabolite methotrexate and the conventional chemotherapeutics paclitaxel, topotecan, mercaptopurine, vincristine, and gemcitabine (Fig. 2c).
Fig. 2. Retinoic acid combination drug library screening reveals beneficial combination partners.
a Hierarchical clustering analysis of modified drug sensitivity scores for 76 clinically relevant drugs in combination with retinoic acid in N = 16 culture models. Treatment on three-dimensional tumouroids was performed for 144 h with the addition of a fixed RA concentration or solvent control (DMSO). b, c Median dcDSS values of all 76 library drugs for N = 9 RA less-responsive models and N = 7 RA sensitive models, respectively. Positive dcDSS values indicate beneficial (blue) and negative values indicate adverse (blue) combination effects. d, e Navitoclax dose-response curves navitoclax with (combo) or without (mono) the fixed RA concentration in cell lines HD-MB03 and SK-N-BE(2)-C. f Representative longterm growth assay wells of HD-MB03, SK-N-BE(2)-C and SJ-GBM2 cells treated as indicated for 8 days. g Dotplots of DSSasym values for BH3 mimetics A-1155463 (selective BCL-XL-inhibitor), navitoclax (BCL-2/XL-inhibitor), and venetoclax (selective BCL-2-inhibitor) with (combo) or without (mono) the fixed RA concentration in N = 16 culture models separated by entity (NB, N = 4; MB, N = 4; EPN, N = 2; HGG, N = 6). The bar indicates the corresponding median DSSasym. DSSasym modified drug sensitivity score, dcDSS differential combination DSS, RA retinoic acid.
RA/navitoclax cotreatment synergistically inhibits MYC(N)-amplified MB and NB proliferation
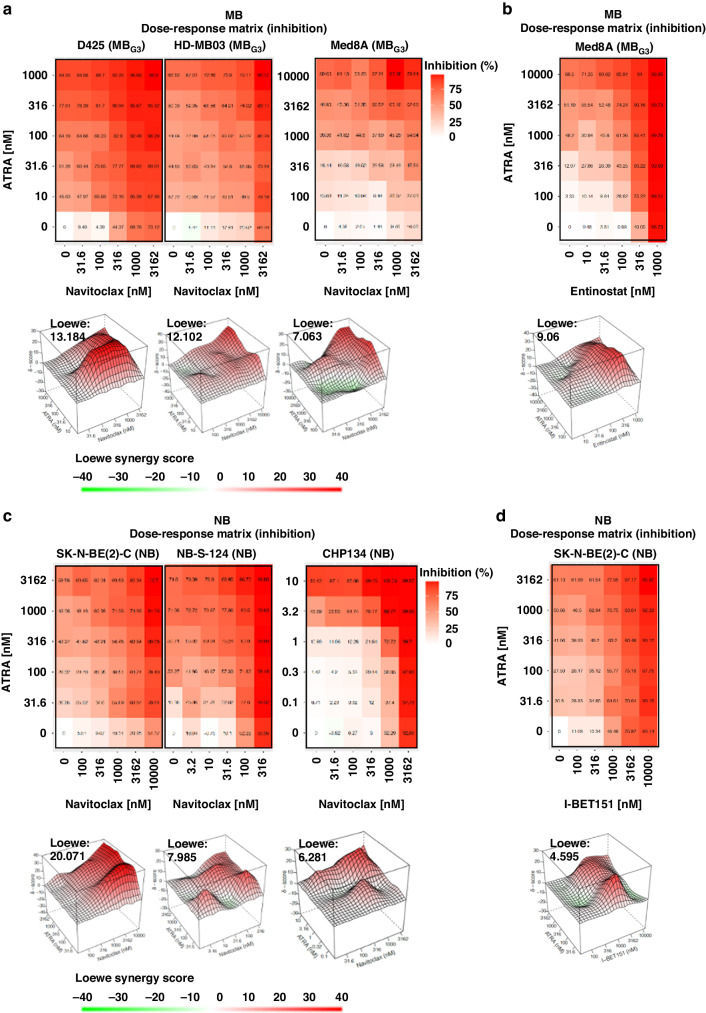
Due to its experimental setup with the use of one single RA concentration, the results of this drug combination screen with the most promising compounds were confirmed in a setup suitable for synergism analysis. We analysed drug combinations through a 5 × 5 matrix design and a self-generated seven-ray design based on the respective IC50 concentrations of the combination partners for antiproliferative synergy in the 6 MYC(N)-amplified MB and NB cell lines. As top hits for RA-responsive models, we chose navitoclax, which is currently under investigation in clinical trials (e.g., NCT05192889 in a paediatric cohort), entinostat (MB specific) and I-BET151 (NB specific). A Loewe synergy score >10 indicated synergy for ATRA/navitoclax (Fig. 3a) and additivity for ATRA/entinostat (Fig. 3b) in MBs. For NBs, additivity to synergy was indicated for ATRA/navitoclax (Fig. 3c), and additivity was indicated for ATRA/I-BET151 (Fig. 3d). The highest values were obtained for the ATRA/navitoclax combination in the MBG3 models D425 (13.18) and HD-MB03 (12.10) and the NBMYCN model SK-N-BE(2)-C (20.07). The ray design analysis confirmed these results (Supplementary Fig. S9).
Fig. 3. Retinoic acid/navitoclax cotreatment synergistically inhibits viability in culture models of both MBG3 and NB.
a %Inhibition heatmaps of 5 × 5 ATRA/navitoclax dose-response matrices and 3D synergy maps annotated with overall Loewe synergy scores in N = 3 MBG3 tumouroid cell cultures after 144 h of treatment. Red depicts positive and green negative Loewe synergy scores. Values larger than 10 are interpreted as synergistic, −10 to 10 as additive, and below −10 as antagonistic. b 5 x 5 ATRA/entinostat dose-response matrix and 3D synergy map of Med8A. c 5 x 5 ATRA/navitoclax dose-response matrix and 3D synergy maps in N = 3 NB tumouroid cell cultures. d 5 × 5 ATRA/I-BET151 dose-response matrix and 3D synergy map of SK-N-BE(2)-C.
RA-induced gene expression patterns in MYC(N)-amplified MB and NB involve differentiation and regulation of proliferation and apoptosis
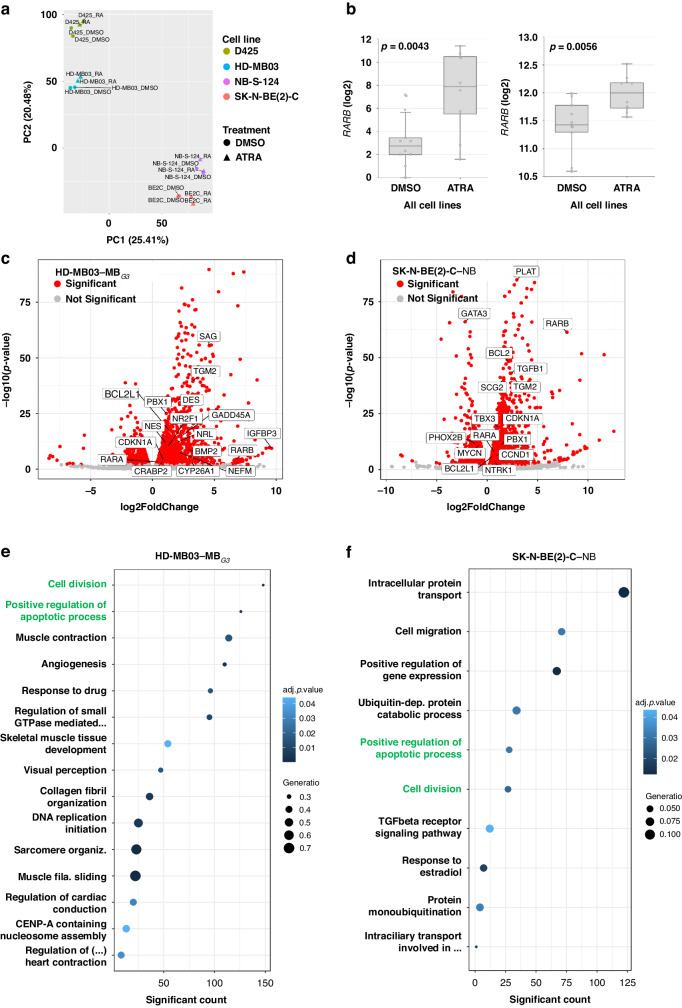
To assess the impact of RA treatment on RA-responsive tumour cells on a transcriptional level, two MYC-amplified MBG3 and two MYCN-amplified NB models were chosen for bulk RNA sequencing. Cells were sequenced after 144 h of treatment with ATRA (1 µM in NB, 0.5 µM in MB) or solvent control. While the principal component analysis (PCA) dimension reduction indicated that the largest expression differences depend on the entity, followed by the cell line and last, the treatment condition (Fig. 4a), there are also shared response patterns to RA treatment. On a single gene level, RA induced the expression of genes involved in RA signalling (e.g., RARB and RARA, Fig. 4b) and metabolism, cell cycle regulation, and apoptotic regulation. In both NB and MB cell lines, the induction of genes involved in differentiation was observed. While RA induced signs of neuronal differentiation in NB, biochemical markers of both neuronal and myogenic differentiation were observed in MB cell lines upon RA treatment (Fig. 4c, d).
Fig. 4. Retinoic acid induces transcriptomic changes in apoptotic regulation and differentiation.
a Principal component analysis of expression patterns from RNA sequencing of N = 2 MBG3 and N = 2 NB culture models treated with ATRA (MB 0.5 µM/ NB 1 µM) or DMSO solvent for 144 h. Two replicates were performed for each condition. b Boxplots of RARB/RARA expression (log2) in all five cell lines comparing DMSO with 144 h ATRA treatment. c, d Volcano plots of differentially expressed genes upon 144 h of ATRA treatment depicting log2-(fold change) and -log10(p-value) in HD-MB03 and SK-N-BE(2)-C cells, respectively. Significantly up- or downregulated genes are depicted in red, not significant in grey. Selected genes of interest are annotated. e, f Significantly enriched GO terms upon 144 h of ATRA treatment in HD-MB03 and SK-N-BE(2)-C cells, respectively. GO terms are ordered by count of significantly differentially expressed genes, dot size represents gene ratio (proportion of significant genes in the respective GO term), and colour indicates the adjusted p-values. Overlapping GO terms are highlighted green.
To identify the biological processes underlying the transcriptional changes, we performed gene ontology (GO) analysis, which revealed significant deregulation of the biological processes cell division, gene expression, positive regulation of apoptotic process, and drug response in both MB and NB models. In line with the induction of differentiation marker genes, MB cell lines were also found to be enriched in GO terms linked to myogenic differentiation (Fig. 4e, f). Overall, RA treatment led to gene expression changes implying hampered cell division, reduced proliferation, signs of differentiation and positive regulation of apoptotic processes.
Combining RA treatment with BCL-XL inhibition leads to a shift from differentiating effects to increased apoptosis
To visualise and quantify changes in cell morphology and cell death of two MBG3 (D425, HD-MB03) and NBMYCN (SK-N-BE(2)-C, NB-S-124) models, high-content confocal fluorescence microscopy was used with fluorescent dyes for cell membrane (CellMask, red), nuclei (Hoechst, blue) and nuclei of apoptotic (caspase-3/7 active) cells (green). In line with previous findings of the effect of retinoids on NB morphology [21], a single RA treatment induced the formation of larger cell bodies and neurite-like cellular protrusions, a morphologic sign of differentiation. Longterm morphology assays confirmed these morphological changes for MB and NB cells, but not HGG nor EPN cells (Fig. 5a, Supplementary Figs. 10–13). In combination with navitoclax, the cellular morphology shifted from this differentiated phenotype to programmed cell death, as evidenced by decreased formation of neurite-like protrusions (Fig. 5b, Supplementary Figs. 10–11) and an increase in the fraction of apoptotic cells and fragmented nuclei (Supplementary Figs. S13, S14E, S15A) compared to RA single treatment. Moreover, the combinational treatment resulted in significantly decreased colony formation ability (Supplementary Fig. S15B). The fluorometric quantification of caspase-3/7 activity in the abovementioned MYC(N)-driven models, confirmed that treatment with navitoclax increased the number of apoptotic cells, which was enhanced when combined with RA in cell lines of both entities (Fig. 5c, Supplementary Fig. S14E). Consistently, navitoclax alone and especially RA/navitoclax cotreatment resulted in substantial apoptotic PARP cleavage (Fig. 5d). To further characterise the combination treatment with organoid-like patient-derived models, another round of high-content imaging was performed. The models (one MYCamp-MBG3, one NB, one EPN) were seeded as three-dimensional tumouroids. Treatment started three days after seeding and consisted of two cycles of four days presence followed by three days absence of drug for 14 days in total. The combination treatment substantially decreased spheroid sizes in MBG3 and NB models, but not in the EPN model (Supplementary Fig. S16). Notably, a RA non-responsive MYCamp-MBG3 organoid model did not reveal resistance-breaking combination benefits with navitoclax in the metabolic activity assay, supporting the conclusions that only RA responsive cells profit from RA/navitoclax cotreatment and that MYC(N) amplification alone does not predict RA sensitivity reliably (Supplementary Fig. S17A).
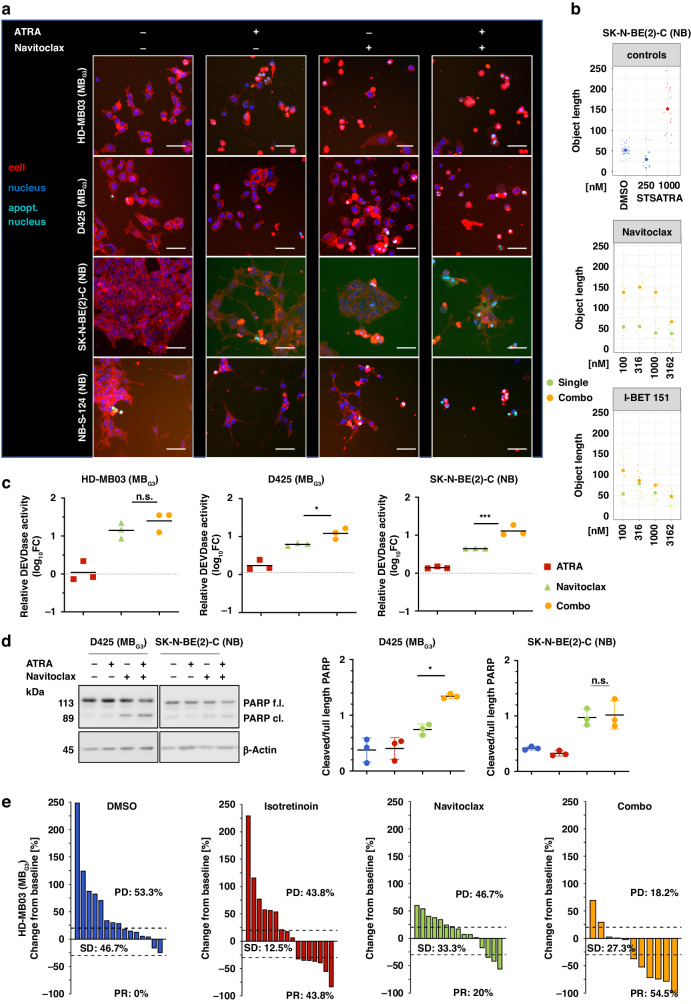
Fig. 5. ATRA differentiating effects shift to increased induction of apoptosis in combination with navitoclax.
a Representative images from high-content fluorescence microscopy of HD-MB03, D425, SK-N-BE(2)-C and NB-S-124 after 72 h of treatment with DMSO control, 500/1000 nM ATRA (MB/NB), 1000/3162 nM navitoclax (MB/NB), and a combination of both. Blue fluorescence shows nuclei, red shows cell bodies, and green (in overlay with blue: turquoise) shows apoptotic nuclei. Scale bar: 100 µm. b Graphs depicting the median object length of the cell including the cellular neurite-like protrusions after 72 h of DMSO, death control STS (staurosporine), ATRA, navitoclax, and I-BET151 treatment in SK-N-BE(2)-C cells. Green indicates single treatment, yellow indicates combination treatment. N = 3. c Log10-fold change of relative DEVDase activity compared to DMSO control in HD-MB03, D425, SK-N-BE(2)-C. Cells were treated with 500/1000 nM ATRA (MB/NB, red), 1000/3000 nM navitoclax (green), or combination (yellow) for 48 h. The bar indicates the corresponding mean. N = 3. d One representative Western blot of SK-N-BE(2)-C and D425 cells treated with 500/1000 nM ATRA (D425/SK-N-BE(2)-C), 1000/3000 nM navitoclax (green), or combination (yellow) for 72 h. PARP protein can be seen in its full-length (f.l.) and cleaved (cl.) version. Actin was used as a loading control. Dot plots on the right depict the ratio of cleaved to full-length PARP. The bar indicates the mean. N = 3. e Waterfall plots representing the change from baseline growth of individual HD-MB03 zebrafish embryo medulloblastoma xenografts treated with DMSO (blue), 500 nM isotretinoin (red), 10 µM navitoclax (green), and combination (yellow) for 48 h. Each bar represents a single xenograft tumour. DMSO, N = 15; isotretinoin, N = 16; navitoclax, N = 15; combo, N = 11. PD progressive disease, SD stable disease, PR partial response.
The combination treatment substantially impaired tumour growth in vivo. We transplanted either fluorescently labelled HD-MB03 or NB-S-124 cells into the yolk sac of zebrafish embryos and started treatment 24 h after injection for 48 h with solvent control, RA, navitoclax and combination. We applied the RECIST (Response Evaluation Criteria in Solid Tumours) criteria adapted to zebrafish for the definition of partial response (PR, tumour size ≥ 30% below basal value) and progressive disease (PD, tumour size ≥ 20% above basal value) [48]. While most tumours grew progressively in the control group (53.3% PD MB, 44.0% PD NB), less growth occurred in the single treatments with retinoic acid (43.8% PD MB, 21.4% PD NB) and navitoclax (46.7% PD MB, 32.1% PD NB). For the MBG3 model with the combination treatment more than half of the tumours decreased significantly in size (54.5% PR, partial response) and the number of tumours with progressive growth decreased significantly (18.2% PD). Combination treatment, thus, successfully led to a disease control rate (DCR: SD + PR) of over 80% of cases and achieved tumour regression (PR) in over 50% of cases (Fig. 5e). Similar results were obtained for the NB-S-124 NB model, where the combination treatment led to a DCR of 92% (Supplementary Fig. S17B).
Medulloblastomas - in contrast to neuroblastomas - mainly depend on BCL-XL
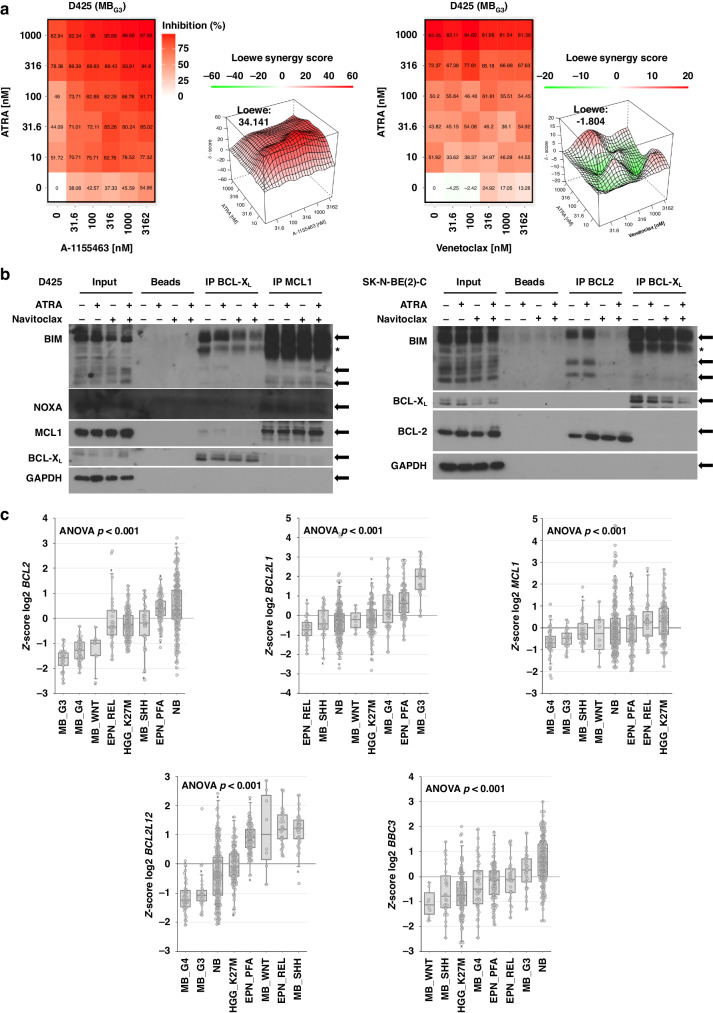
To further narrow down the role of the navitoclax target proteins BCL-2 and BCL-XL in combination with RA treatment, we performed a synergy analysis of the selective BCL-2 inhibitor venetoclax and selective BCL-XL inhibitor A-1155463 combined with RA in MBG3 D425 cells. Here, venetoclax demonstrated neither single-agent efficacy nor synergy in the RA combination (Loewe synergy score −1.80). A-1155463, on the other hand, was both active and highly synergistic with RA (Loewe synergy score 34.14) (Fig. 6a). Thus, the combined benefit of RA/navitoclax in MBG3 is likely to be caused by targeting BCL-XL and not BCL-2.
Fig. 6. BCL-XL is an important targetable vulnerability in MBG3.
a %Inhibition heatmaps of 5 × 5 ATRA/A-1155463 (selective BCL-XL inhibitor) and ATRA/venetoclax (selective BCL-2 inhibitor) dose-response matrices and 3D synergy maps annotated with overall Loewe synergy scores of D425 cells after 144 h of treatment. Red depicts positive and green negative Loewe synergy scores. Values larger than 10 are interpreted as synergistic, −10 to 10 as additive, and below −10 as antagonistic. b Representative immunoprecipitation plots of BCL-XL and MCL-1 in D425 and BCL-2 and BCL-XL in SK-N-BE(2)-C cells and detection of selected protein interaction partners. Cells were treated for 48 h with DMSO, 500/1000 nM ATRA (MBG3/NB), 1000/3000 nM navitoclax, and a combination thereof. GAPDH served as loading control in the input lysate. N = 3. c Boxplots of log2 z-score expression of anti-apoptotic genes BCL2, BCL2L1, and MCL1 and putative BH3 mimetic response biomarkers BCL2L12, and BBC3 as measured by RNA sequencing from N = 2288 relapsed paediatric tumours from the INFORM study. Boxplots depict selected entities of paediatric nervous system tumours. EPN_PFA posterior fossa ependymoma A, EPN-REL ZFTA-fusion positive ependymoma, HGG_K27M K27M-mutated high-grade glioma, MB_G3 Group 3 medulloblastoma, MB_G4 Group 4 medulloblastoma, MB_SHH sonic hedgehog medulloblastoma, MB_WNT wingless medulloblastoma, NB neuroblastoma.
To assess interactions between pro- and anti-apoptotic proteins in single- and cotreatment conditions, immunoprecipitation (IP) was performed in D425 (MBG3) and SK-N-BE(2)-C (NBMYCN) cells after 48 h of treatment (Fig. 6b). As D425 cells depend on BCL-XL and hardly express BCL-2, IPs were performed for the anti-apoptotic proteins BCL-XL and MCL-1. The pro-apoptotic BIM, which is mainly expressed in the BIM-EL form and less as BIM-L/S in D425, was associated with BCL-XL. Upon treatment with navitoclax, some displacement of BIM from BCL-XL was observed. In SK-N-BE(2)-C cells, which strongly express BCL-2, IPs were performed for BCL-2 and BCL-XL. Here, BIM was mainly bound to BCL-2 and hardly any to BCL-XL. Treatment with navitoclax displaced BIM-L and BIM-S from BCL-2. Thus, the on-target activity of navitoclax is evident in both MB and NB models, leading to substantial apoptosis when combined with RA treatment.
To explore the role of BCL-2, BCL-XL and MCL-1 in a large paediatric patient pancancer cohort, we investigated their expression in RNA sequencing of n = 2288 patient samples from the INFORM registry. The international precision oncology programme INFORM enrols relapsed/refractory paediatric cancer patients for comprehensive molecular analysis. It revealed that NB was the solid tumour with the highest expression of BCL2, while MBG3 demonstrated among the lowest BCL2 expression. BCL2L1 (BCL-XL), on the other hand, was most highly expressed by MBG3, while NB tumours showed average expression. The below average expression of MCL1 suggests that this anti-apoptotic protein may play only a minor role in relapse of the embryonal tumours MBG3 and NB (Fig. 6c). As mere expression of the anti-apoptotic target proteins has not been clearly demonstrated to predict response to BH3-mimetic treatment, we investigated the expression of the recently suggested predictive biomarkers BCL2L12 and BBC3 for paediatric tumours [31]. Consistently, MBG3 and NB tumours of the INFORM cohort expressed BCL2L12 at comparatively low levels and BBC3 at above-average levels (Fig. 6c; Supplementary Fig. S18). Comparing the four canonical MB subgroups in detail, Group 3 tumours both expressed BCL2L1 (BCL-XL) and BBC3 at significantly higher levels than the other subgroups and showed significantly lower BCL2L12 expression than the MBSHH and MBWNT subgroups (Fig. 6c).
Overall, these results suggest that RA and navitoclax cotreatment not only elicits strong antitumoural effects in the MBG3 and NB preclinical models studied here, but also may provide a novel treatment option for relapsed patients with these high-risk cancers.
Discussion
MBG3 and NB are embryonal tumours that result from impaired differentiation of neuronal lineage progenitors during fetal development of the cerebellum and sympathetic nervous system, respectively [12, 49]. Despite highly intensive treatment approaches, the current standard of care for these diseases does not yield acceptable outcomes in high-risk and relapsed cases [7, 13]. Therefore, we investigated the potential of the differentiation-inducing drug RA and sought to identify a combinatorial compound that might efficiently enhance its effects.
In accordance with other preclinical investigations we demonstrated RA sensitivity in NB and MBG3, while MBSHH models show a very limited response [20, 22, 50, 51]. Conversely, paediatric HGGs and PFA- and ZFTA fusion-positive EPN models resisted antitumoural RA effects, contrary to one previous study of two PFA cultures [24]. This might be explained by the three-dimensional tumouroid model used in our study, which mimics drug-resistant phenotypes similar to the in vivo situation. Our data indicate that RA is especially effective in tumour subtypes capable of neuronal differentiation and that RA treatment supports neuronal differentiation in MBG3 and NB as demonstrated by translational regulation of marker genes, and morphological changes in the neuroblastoma models, independent of the growth rate. In the transcriptomic data of NB cells, we observed changes consistent with the recently described RA-induced reprogramming of the adrenergic core regulatory circuit [52]. Our results also point toward RA-induced upregulation of anti-apoptotic members of the BCL-2 family, such as BCL2 and BCL2L1, which are druggable with BH3 mimetics. Thus, RA treatment also induces novel targetable vulnerabilities, such as the BCL-2 family as evidenced by cotreatment with the BCL-2/BCL-XL inhibitor navitoclax, which shifted the differentiated phenotype to cell death. RA/navitoclax effectively inhibited tumour growth in an in vivo zebrafish embryo xenograft model. This model is characterised by high evolutionary conservation of human genetic pathways, an immature immune system preventing rejection, and transparency allowing live visualisation of in vivo growth and migration [53]. The low amount of cells used per injection and cost- and time-effectiveness make it an attractive model for high-throughput precision oncology approaches [43, 48].
BCL2L1 and markers of the BH3-mimetic response are highly expressed in relapsed MBG3 patients, suggesting clinical potential for BCL-XL inhibition in these patients. Targeting the BCL-2 protein family provides a promising area of investigation for the treatment of paediatric nervous system tumours [27], as the anti-apoptotic BCL-2 family members BCL-2, BCL-XL, and MCL-1 are frequently upregulated in cancer cells, including MB and NB [26]. Of note, across various cancers, MYC amplification is often associated with upregulation of BCL-XL and MCL-1, rescuing cells from MYC-induced apoptosis [26]. In NB, BCL-2 plays an important role in the RA response, with low BCL-2 expressing cells undergoing RA-mediated apoptosis, while differentiating cells increase BCL-2 expression and consequently evade cell death [54]. RA-induced differentiation in NB has also been linked to decreased chemosensitivity to antineoplastic agents, such as topotecan, doxorubicin and cisplatin, which is mediated through BCL-2 and BCL-XL upregulation [55].
BH3 mimetics have been investigated in many preclinical studies regarding their efficacy in paediatric nervous system tumours as single and combinatorial agents [27]. The combination of the investigational synthetic retinoid fenretinide with selective BCL-2 inhibition was shown to synergistically induce cell death and increase survival in an in vivo model through NOXA upregulation in NB cells that express high levels of BCL-2 [56]. The sensitivity of NB cells to selective BH3 mimetics targeting BCL-2, BCL-XL, and MCL-1, however, is heterogeneous and all three antiapoptotic proteins are equally relevant targets for apoptotic modulation [42]. Notably, selective BCL-XL inhibition was most effective against primary patient-derived NB cells [42]. In addition, in a study of selective BH3 mimetics across 18 paediatric solid tumour cell lines, the selective BCL-2 inhibitor venetoclax was the least efficacious, and concomitant antagonism of BCL-XL and MCL-1 produced the most pronounced induction of cell death [57]. In MB, BCL-XL is a direct transcriptional target of the MBG3 master transcription factor NRL and has been described as a potential subgroup-specific vulnerability [58]. Indeed, targeting BCL-XL can induce apoptosis in MBG3 cell culture and impede growth in orthotopic xenografts [58]. Furthermore, subsequent studies of BH3 mimetics in MBG3 demonstrated that navitoclax and selective inhibition of BCL-XL, but not BCL-2, while having modest single-agent antitumoural activity, render cells more vulnerable to the effects of cytotoxic stimuli, chemotherapy, and targeted therapeutics by increasing the induction of intrinsic apoptosis [59–61]. Consistent with these findings, our study provides evidence that effective targeting of apoptotic regulation in MBG3 is primarily achieved by antagonising BCL-XL and should be combined with other treatment approaches, such as RA, that synergise with BH3 mimetics in inducing cell death. In NB, RA treatment may benefit most from combination with a BCL-2-/BCL-XL-inhibitor such as navitoclax or APG-1252, as both proteins seem to be involved in the RA response and provide relevant targets.
To evade apoptotic effects of oncogenic MYC(N), tumour cells may upregulate pro-survival BCL-2 family members or suppress TP53 signalling. In colorectal cancer cells it has been shown that p53, which undergoes entinostat induced post-transcriptional modifications, can directly interact with BCL-2-family proteins to trigger apoptosis [62]. In this study, we did not investigate the treatment-induced protein interactions of p53, however, TP53 status did not seem to correlate with distinct response patterns to the investigated apoptotic induction.
Besides BH3 mimetics we identified therapeutic potential in combining the HDAC inhibitor entinostat with RA in MBG3 models. Noteworthy, the class I inhibitor entinostat was far more beneficial for the combination with ATRA as the pan-HDAC inhibitor vorinostat, but no clear class-specific combination effect was shown. Furthermore, class I HDAC inhibition can also result in adverse combination effects with RA through antagonism between differentiation and cytotoxicity in other entities [63–66]. In acute promyelocytic leukaemia cells, RA-induced BCL-XL activity has been described as cytoprotective against HDAC class I inhibition but less so against pan-HDAC inhibition [63]. Notably, a recent MBG3 cell culture study demonstrated synergistic activity between BCL-XL-inhibition and entinostat [61]. Thus, further studies would be needed to provide clear evidence and predictive biomarkers for clinical translation in selected cancer (sub-)entities.
From a clinical perspective, it is important to note that the RA/navitoclax cotreatment concentrations that synergistically decreased MB and NB viability to relevant levels (<50%) are achievable in patients [18, 67]. While there are already decades of experience with isotretinoin/13-cis-RA therapy and its safety and efficacy in high-risk NB patients [15, 18], its use has been limited to experimental approaches in MB. However, recently, Leary et al. demonstrated that oral isotretinoin treatment analogous to NB protocols could be safely incorporated into an intensive cytotoxic chemotherapy regimen for infant embryonal brain tumours, including MBG3 alongside the HDAC inhibitor vorinostat, with acceptable toxicity [68]. Moreover, a randomised-controlled Children’s Oncology Group trial tested carboplatin and isotretinoin in high-risk MB and supratentorial primitive neuroectodermal tumours [69]. However, the isotretinoin treatment arm was closed before molecular subgroup analyses were introduced into diagnostic routine and no subgroup-specific analyses were reported [69]. It is conceivable that RA might only provide benefit for a certain subgroup of MB patients, which according to the evidence provided here would most likely be found within MBG3. In such a group, RA would likely exert its strongest potential when given with a suitable targeted therapeutic, rather than conventional chemotherapy.
Navitoclax is known to induce dose-limiting thrombocytopenia, which has been attributed to its targeting of BCL-XL [67], yet intermittend or lower navitoclax dosing [70] and novel dual BCL-2-/BCL-XL [71] inhibitors show promising clinical potential, as these strategies may provide better tolerance. Nevertheless, navitoclax was chosen for our investigations as it is the only BCL-2-/BCL-XL-inhibitor with described pharmocokinetics in the paediatric oncology population [70], where it is also currently investigated in ongoing clinical trials (e.g., NCT05192889 or NCT05740449). Finally, the penetration of BH3 mimetics through the BBB into the CNS is still in question. A disruption of the BBB, as is observed in paediatric brain tumours clinically, however, might provide a way for BH3 mimetics to treat these cancer cells despite high molecular weights [61, 72].
Unlike with current first line therapies in paediatric oncology, there is an unmet clinical need for innovative treatment strategies in recurring childhood cancer, which shows poor survivaI [3, 29, 31, 45]. Thus, many of the culture models in our study are derived from relapsed disease, which is often characterised by accumulating genetic mutations confering treatment resistance e.g. through p-glykoprotein activity [41] and changed BCL-2 family dependencies [73]. As translation of newly identified treatment approaches to early phase clinical trials in paediatric oncology mainly focuses on treating recurrent or refractory disease, we investigated potential predictive biomarkers in the comprehensive INFORM cohort. Nevertheless, our study does not allow for a clear conclusion regarding treatment potential in primary as compared to relapsed disease, as primary in contrast to relapsed tumours are characterised mainly by amplifications and deletions.
In conclusion, we demonstrate that RA treatment modulates apoptotic regulation in MBG3 cells and subsequently increases vulnerability to selective BCL-XL inhibition. While both compounds show limited single-agent activity, RA/BCL-XL-inhibition cotreatment synergistically induces apoptotic cell death and improves in vivo antitumoural efficacy. Together with molecular data from relapsed patients, our results suggest that BCL-XL is a particularly important target in MBG3, and the combination treatment investigated here may provide clinical benefit in these patients.
Supplementary information
Acknowledgements
We would like to thank Marko Weidmann and Aileen Friedenauer for excellent technical assistance. We thank the High Throughput Sequencing unit of the Genomics & Proteomics Core Facility, German Cancer Research Center (DKFZ) for providing excellent RNA sequencing services. We thank Laura Turunen and Jani Saarela from the High Throughput Biomedicine Unit and Vilja Pietiäinen (Institute for Molecular Medicine Finland, Helsinki Institute of Life Science, University of Helsinki, Helsinki, Finland and Biocenter Finland) for the support with drug-plate printing and the provision of ready-to-go drug plates. We would like to convey our heartfelt thanks to the INFORM (bio-)informatics: Carsten Maus, Erjia Wang (Genomics and Proteomics Core Facility, DKFZ), Lena Weiser, Gregor Warsow (Omics IT and Data Management Core Facility, DKFZ), Robert Autry, Gnanaprakash Balasubramanian, Christopher Previti and Rolf Kabbe (Division of Pediatric Neurooncology, DKFZ). We thank the patients and their families for their consent to collect and use tumour material. We thank all participating INFORM sites for providing tumour tissue, molecular, and clinical data, and we thank Karen Frese and Benjamin Meder for zebrafish husbandry and overall support in zebrafish studies.
Author contributions
Conceptualisation: OW and IO; methodology: TS, CZ, SH, SN, AS-D, RS-A, CM, NM, MK, TM, MV and HP; software: CZ, SC, SH, DN, TH-L and MS; validation: TS, CZ, DN, SC, SH, AS-D; formal analysis: TS, CZ, SC, SH, DN; investigation: TS, CZ, SC, SH, SN, RS-A, and DN; resources: FW, MS, TM, MK, OW and IO; data curation: HP and IO; writing—original draught preparation: TS and IO; writing—review and editing: TS, CZ, DN, SC, SH, SN, AS-D, RS-A, FW, CM, NM, MS, TM, MK, TH-L, MV, HP, OW and IO; visualisation: TS, CZ, DN, SC, SH, CM, TH-L, MV and IO; supervision: HP, OW and IO; project administration: OW and IO; funding acquisition: IO. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Monika Kutzner Foundation and a doctoral scholarship by the Kind-Philipp-Foundation. The work was also supported by SFB 1389-UNITE Glioblastoma, Work Package A02 (O. Witt) and D02 (M. Schlesner). The INFORM programme is financially supported by the German Cancer Research Center (DKFZ), several German health insurance companies, the German Cancer Consortium (DKTK), the German Federal Ministry of Education and Research (BMBF), the German Federal Ministry of Health (BMG), the Ministry of Science, Research and the Arts of the State of Baden-Württemberg (MWK BW); the German Cancer Aid (DKH), the German Childhood Cancer Foundation (DKS), RTL television, the aid organisation BILD hilft e.V. (Ein Herz für Kinder) and the generous private donation of the Scheu family. Open Access funding enabled and organized by Projekt DEAL.
Data availability
Gene expression data are available through the web-based gene expression analysis R2 (R2: microarray analysis and visualisation platform; http://r2.amc.nl) platform: Medulloblastoma / Neuroblastoma RA - Oehme - 20 - rsem - gencode19; 2021-12-01.
Competing interests
Olaf Witt participated in advisory boards of Novartis, BMS, Janssen and receives research grants from BVD, Day One Therapeutics. Ina Oehme receives research grants from PreComb, BVD and Day One Therapeutics.
Ethics approval
Institutional Review Board Statement Zebrafish husbandry (permit number 35-9185.64/BH Hassel) and experiments (permit number 35-9185.81/G-126/15) were performed according to local animal welfare standards (Tierschutzgesetz §11, Abs. 1, No. 1) and in accordance with European Union animal welfare guidelines (EU Directive 2010/63/EU). All applicable national and institutional guidelines for the care and use of zebrafish were followed. Xenograft of tumour cell suspension was carried out in immune-compromised mice (Nod-scid IL2Rgammanull) immediately following processing (permit number G91/20). NSG mice were obtained from the laboratory breeding colony (German Cancer Research Centre). All mice were maintained in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals with approval from the Fred Hutchinson Cancer Research Centre Institutional Animal Care and Use Committee (IR no. 1457). All procedures involving animals were performed in accordance with the ethical standards of the institution.
Informed consent
The study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. All patients, their legally acceptable representatives, or both (if possible) provided written informed consent. Approval for the study protocol (and any modifications thereof) was obtained from independent ethics committees and the institutional review board at each participating centre. INFORM was registered with the German Clinical Trial Register, number DRKS00007623.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-024-02740-5.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.Kaatsch P, Grabow D, Spix C. German Childhood Cancer Registry-Anual Report 2018 (1980–2017). Institute of Medical Biostatistics, Epidemiology and Informatics (IMBEI) at the University Medical Center of the Johannes Gutenberg University Mainz. 2019.
- 3.van Tilburg CM, Pfaff E, Pajtler KW, Langenberg KPS, Fiesel P, Jones BC, et al. The pediatric precision oncology INFORM registry: clinical outcome and benefit for patients with very high-evidence targets. Cancer Discov. 2021;11:2764–79. 10.1158/2159-8290.CD-21-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oeffinger KC, Mertens AC, Sklar CA, Kawashima T, Hudson MM, Meadows AT, et al. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355:1572–82. 10.1056/NEJMsa060185 [DOI] [PubMed] [Google Scholar]
- 5.Sturm D, Capper D, Andreiuolo F, Gessi M, Kolsche C, Reinhardt A, et al. Multiomic neuropathology improves diagnostic accuracy in pediatric neuro-oncology. Nat Med. 2023. 10.1038/s41591-023-02255-1. [DOI] [PMC free article] [PubMed]
- 6.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72. 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajjar A, Robinson GW, Smith KS, Lin T, Merchant TE, Chintagumpala M, et al. Outcomes by Clinical and Molecular Features in Children With Medulloblastoma Treated With Risk-Adapted Therapy: Results of an International Phase III Trial (SJMB03). J Clin Oncol. 2021:Jco2001372. 10.1200/jco.20.01372. [DOI] [PMC free article] [PubMed]
- 8.Ramaswamy V, Remke M, Bouffet E, Bailey S, Clifford SC, Doz F, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131:821–31. 10.1007/s00401-016-1569-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32:520–537.e525. 10.1016/j.ccell.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23:1231–51. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017;133:5–12. 10.1007/s00401-016-1643-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansky S, Sharma AK, Korber V, Quintero A, Toprak UH, Wecht EM, et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat Genet. 2021;53:683–93. 10.1038/s41588-021-00806-1 [DOI] [PubMed] [Google Scholar]
- 13.Irwin MS, Naranjo A, Zhang FF, Cohn SL, London WB, Gastier-Foster JM, et al. Revised neuroblastoma risk classification system: a report from the children’s oncology group. J Clin Oncol. 2021;39:3229–41. 10.1200/JCO.21.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon T, Hero B, Schulte JH, Deubzer H, Hundsdoerfer P, von Schweinitz D, et al. 2017 GPOH Guidelines for diagnosis and treatment of patients with neuroblastic tumors. Klin Padiatr. 2017;229:147–67. 10.1055/s-0043-103086 [DOI] [PubMed] [Google Scholar]
- 15.Ladenstein R, Pötschger U, Valteau-Couanet D, Luksch R, Castel V, Yaniv I, et al. Interleukin 2 with anti-GD2 antibody ch14.18/CHO (dinutuximab beta) in patients with high-risk neuroblastoma (HR-NBL1/SIOPEN): a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19:1617–29. 10.1016/S1470-2045(18)30578-3 [DOI] [PubMed] [Google Scholar]
- 16.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363:1324–34. 10.1056/NEJMoa0911123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DuBois SG, Macy ME, Henderson TO. High-risk and relapsed neuroblastoma: toward more cures and better outcomes. Am Soc Clin Oncol Educ Book. 2022;42:1–13. [DOI] [PubMed] [Google Scholar]
- 18.Khan AA, Villablanca JG, Reynolds CP, Avramis VI. Pharmacokinetic studies of 13-cis-retinoic acid in pediatric patients with neuroblastoma following bone marrow transplantation. Cancer Chemother Pharmacol. 1996;39:34–41. 10.1007/s002800050535 [DOI] [PubMed] [Google Scholar]
- 19.Le Doze F, Debruyne D, Albessard F, Barre L, Defer GL. Pharmacokinetics of all-trans retinoic acid, 13-cis retinoic acid, and fenretinide in plasma and brain of Rat. Drug Metab Dispos. 2000;28:205–8. [PubMed] [Google Scholar]
- 20.Hallahan AR, Pritchard JI, Chandraratna RA, Ellenbogen RG, Geyer JR, Overland RP, et al. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med. 2003;9:1033–8. 10.1038/nm904 [DOI] [PubMed] [Google Scholar]
- 21.Sidell N, Altman A, Haussler MR, Seeger RC. Effects of retinoic acid (RA) on the growth and phenotypic expression of several human neuroblastoma cell lines. Exp Cell Res. 1983;148:21–30. 10.1016/0014-4827(83)90184-2 [DOI] [PubMed] [Google Scholar]
- 22.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–24. 10.1158/0008-5472.919.65.3 [DOI] [PubMed] [Google Scholar]
- 23.Karmakar S, Banik NL, Patel SJ, Ray SK. Combination of all-trans retinoic acid and taxol regressed glioblastoma T98G xenografts in nude mice. Apoptosis. 2007;12:2077–87. 10.1007/s10495-007-0116-2 [DOI] [PubMed] [Google Scholar]
- 24.Donson AM, Amani V, Warner EA, Griesinger AM, Witt DA, Levy JMM, et al. Identification of FDA-approved oncology drugs with selective potency in high-risk childhood ependymoma. Mol Cancer Ther. 2018;17:1984–94. 10.1158/1535-7163.MCT-17-1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. 10.1038/cdd.2017.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgerald MC, O’Halloran PJ, Connolly NMC, Murphy BM. Targeting the apoptosis pathway to treat tumours of the paediatric nervous system. Cell Death Dis. 2022;13:460. 10.1038/s41419-022-04900-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller M, Rosch L, Najafi S, Gatzweiler C, Ridinger J, Gerloff XF, et al. Combining APR-246 and HDAC-inhibitors: a novel targeted treatment option for neuroblastoma. Cancers (Basel). 2021;13:4476. [DOI] [PMC free article] [PubMed]
- 29.Worst BC, van Tilburg CM, Balasubramanian GP, Fiesel P, Witt R, Freitag A, et al. Next-generation personalised medicine for high-risk paediatric cancer patients—The INFORM pilot study. Eur J cancer. 2016;65:91–101. 10.1016/j.ejca.2016.06.009 [DOI] [PubMed] [Google Scholar]
- 30.ElHarouni D, Berker Y, Peterziel H, Gopisetty A, Turunen L, Kreth S, et al. iTReX: interactive exploration of mono- and combination therapy dose response profiling data. Pharm Res. 2022;175:105996. 10.1016/j.phrs.2021.105996 [DOI] [PubMed] [Google Scholar]
- 31.Peterziel H, Jamaladdin N, ElHarouni D, Gerloff XF, Herter S, Fiesel P, et al. Drug sensitivity profiling of 3D tumor tissue cultures in the pediatric precision oncology program INFORM. NPJ Precis Oncol. 2022;6:94. 10.1038/s41698-022-00335-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–93. 10.1093/nar/gkaa216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-response analysis using R. PLoS One. 2016;10:e0146021. 10.1371/journal.pone.0146021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JJ, Kong M, Ayers GD, Lotan R. Interaction index and different methods for determining drug interaction in combination therapy. J Biopharm Stat. 2007;17:461–80. 10.1080/10543400701199593 [DOI] [PubMed] [Google Scholar]
- 39.Stirling DR, Swain-Bowden MJ, Lucas AM, Carpenter AE, Cimini BA, Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinforma. 2021;22:433. 10.1186/s12859-021-04344-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dao D, Fraser AN, Hung J, Ljosa V, Singh S, Carpenter AE. CellProfiler Analyst: interactive data exploration, analysis and classification of large biological image sets. Bioinformatics. 2016;32:3210–2. 10.1093/bioinformatics/btw390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosch L, Herter S, Najafi S, Ridinger J, Peterziel H, Cinatl J, et al. ERBB and P-glycoprotein inhibitors break resistance in relapsed neuroblastoma models through P-glycoprotein. Mol Oncol. 2023;17:37–58. 10.1002/1878-0261.13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bierbrauer A, Jacob M, Vogler M, Fulda S. A direct comparison of selective BH3-mimetics reveals BCL-XL, BCL-2 and MCL-1 as promising therapeutic targets in neuroblastoma. Br J Cancer. 2020. 10.1038/s41416-020-0795-9. [DOI] [PMC free article] [PubMed]
- 43.Gatzweiler C, Ridinger J, Herter S, Gerloff XF, ElHarouni D, Berker Y, et al. Functional therapeutic target validation using pediatric zebrafish Xenograft models. Cancers (Basel). 2022;14:849. [DOI] [PMC free article] [PubMed]
- 44.Brabetz S, Leary SES, Grobner SN, Nakamoto MW, Seker-Cin H, Girard EJ, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med. 2018;24:1752–61. 10.1038/s41591-018-0207-3 [DOI] [PubMed] [Google Scholar]
- 45.Heipertz AE, Pajtler KW, Pfaff E, Schramm K, Blattner-Johnson M, Milde T, et al. Outcome of children and adolescents with relapsed/refractory/progressive malignancies treated with molecularly informed targeted drugs in the pediatric precision oncology registry INFORM. JCO Precis Oncol. 2023;7:e2300015. 10.1200/PO.23.00015 [DOI] [PubMed] [Google Scholar]
- 46.Bingel C, Koeneke E, Ridinger J, Bittmann A, Sill M, Peterziel H, et al. Three-dimensional tumor cell growth stimulates autophagic flux and recapitulates chemotherapy resistance. Cell Death Dis. 2017;8:e3013. 10.1038/cddis.2017.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–d646. 10.1093/nar/gkac1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrobel JK, Najafi S, Ayhan S, Gatzweiler C, Krunic D, Ridinger J, et al. Rapid in vivo validation of HDAC inhibitor-based treatments in neuroblastoma zebrafish xenografts. Pharmaceuticals (Basel). 2020;13:345. [DOI] [PMC free article] [PubMed]
- 49.Smith KS, Bihannic L, Gudenas BL, Haldipur P, Tao R, Gao Q, et al. Unified rhombic lip origins of group 3 and group 4 medulloblastoma. Nature. 2022;609:1012–20. 10.1038/s41586-022-05208-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai R, Siu IM, Tyler BM, Staedtke V, Gallia GL, Riggins GJ. Evaluation of retinoic acid therapy for OTX2-positive medulloblastomas. Neuro Oncol. 2010;12:655–63. 10.1093/neuonc/nop062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garner EF, Stafman LL, Williams AP, Aye JM, Goolsby C, Atigadda VR, et al. UAB30, a novel RXR agonist, decreases tumorigenesis and leptomeningeal disease in group 3 medulloblastoma patient-derived xenografts. J Neurooncol. 2018;140:209–24. 10.1007/s11060-018-2950-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmerman MW, Durbin AD, He S, Oppel F, Shi H, Tao T, et al. Retinoic acid rewires the adrenergic core regulatory circuitry of childhood neuroblastoma. Sci Adv. 2021;7:eabe0834. 10.1126/sciadv.abe0834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miserocchi G, Mercatali L, Liverani C, De Vita A, Spadazzi C, Pieri F, et al. Management and potentialities of primary cancer cultures in preclinical and translational studies. J Transl Med. 2017;15:229. 10.1186/s12967-017-1328-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niizuma H, Nakamura Y, Ozaki T, Nakanishi H, Ohira M, Isogai E, et al. Bcl-2 is a key regulator for the retinoic acid-induced apoptotic cell death in neuroblastoma. Oncogene. 2006;25:5046–55. 10.1038/sj.onc.1209515 [DOI] [PubMed] [Google Scholar]
- 55.Hadjidaniel MD, Reynolds CP. Antagonism of cytotoxic chemotherapy in neuroblastoma cell lines by 13-cis-retinoic acid is mediated by the antiapoptotic Bcl-2 family proteins. Mol Cancer Ther. 2010;9:3164–74. 10.1158/1535-7163.MCT-10-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen TH, Koneru B, Wei SJ, Chen WH, Makena MR, Urias E, et al. Fenretinide via NOXA induction, enhanced activity of the BCL-2 inhibitor venetoclax in high BCL-2-expressing neuroblastoma preclinical models. Mol Cancer Ther. 2019;18:2270–82. 10.1158/1535-7163.MCT-19-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kehr S, Haydn T, Bierbrauer A, Irmer B, Vogler M, Fulda S. Targeting BCL-2 proteins in pediatric cancer: dual inhibition of BCL-XL and MCL-1 leads to rapid induction of intrinsic apoptosis. Cancer Lett. 2020;482:19–32. 10.1016/j.canlet.2020.02.041 [DOI] [PubMed] [Google Scholar]
- 58.Garancher A, Lin CY, Morabito M, Richer W, Rocques N, Larcher M, et al. NRL and CRX define photoreceptor identity and reveal subgroup-specific dependencies in medulloblastoma. Cancer Cell. 2018;33:435–449.e436. 10.1016/j.ccell.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westhoff MA, Schuler-Ortoli M, Zerrinius D, Hadzalic A, Schuster A, Strobel H, et al. Bcl-XL but not Bcl-2 is a potential target in medulloblastoma therapy. Pharmaceuticals (Basel). 2022;15:91. [DOI] [PMC free article] [PubMed]
- 60.Levesley J, Steele L, Bruning-Richardson A, Davison A, Zhou J, Ding C, et al. Selective BCL-XL inhibition promotes apoptosis in combination with MLN8237 in medulloblastoma and pediatric glioblastoma cells. Neuro Oncol. 2018;20:203–14. 10.1093/neuonc/nox134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeuner S, Vollmer J, Sigaud R, Oppermann S, Peterziel H, ElHarouni D, et al. Combination drug screen identifies synergistic drug interaction of BCL-XL and class I histone deacetylase inhibitors in MYC-amplified medulloblastoma cells. J Neurooncol. 2024;166:99–112. 10.1007/s11060-023-04526-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marx C, Sonnemann J, Beyer M, Maddocks ODK, Lilla S, Hauzenberger I, et al. Mechanistic insights into p53-regulated cytotoxicity of combined entinostat and irinotecan against colorectal cancer cells. Mol Oncol. 2021;15:3404–29. 10.1002/1878-0261.13060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hennig D, Müller S, Wichmann C, Drube S, Pietschmann K, Pelzl L, et al. Antagonism between granulocytic maturation and deacetylase inhibitor-induced apoptosis in acute promyelocytic leukaemia cells. Br J Cancer. 2015;112:329–37. 10.1038/bjc.2014.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noack K, Mahendrarajah N, Hennig D, Schmidt L, Grebien F, Hildebrand D, et al. Analysis of the interplay between all-trans retinoic acid and histone deacetylase inhibitors in leukemic cells. Arch Toxicol. 2017;91:2191–208. 10.1007/s00204-016-1878-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bug G, Ritter M, Wassmann B, Schoch C, Heinzel T, Schwarz K, et al. Clinical trial of valproic acid and all-trans retinoic acid in patients with poor-risk acute myeloid leukemia. Cancer. 2005;104:2717–25. 10.1002/cncr.21589 [DOI] [PubMed] [Google Scholar]
- 66.Leiva M, Moretti S, Soilihi H, Pallavicini I, Peres L, Mercurio C, et al. Valproic acid induces differentiation and transient tumor regression, but spares leukemia-initiating activity in mouse models of APL. Leukemia. 2012;26:1630–7. 10.1038/leu.2012.39 [DOI] [PubMed] [Google Scholar]
- 67.Wilson WH, O’Connor OA, Czuczman MS, LaCasce AS, Gerecitano JF, Leonard JP, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–59. 10.1016/S1470-2045(10)70261-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leary SES, Kilburn L, Geyer JR, Kocak M, Huang J, Smith KS, et al. Vorinostat and isotretinoin with chemotherapy in young children with embryonal brain tumors: a report from the Pediatric Brain Tumor Consortium (PBTC-026). Neuro Oncol. 2022;24:1178–90. 10.1093/neuonc/noab293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leary SES, Packer RJ, Li Y, Billups CA, Smith KS, Jaju A, et al. Efficacy of carboplatin and isotretinoin in children with high-risk medulloblastoma: a randomized clinical trial from the children’s oncology group. JAMA Oncol. 2021;7:1313–21. 10.1001/jamaoncol.2021.2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pullarkat VA, Lacayo NJ, Jabbour E, Rubnitz JE, Bajel A, Laetsch TW, et al. Venetoclax and navitoclax in combination with chemotherapy in patients with relapsed or refractory acute lymphoblastic leukemia and lymphoblastic lymphoma. Cancer Discov. 2021;11:1440–53. 10.1158/2159-8290.CD-20-1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balachander SB, Criscione SW, Byth KF, Cidado J, Adam A, Lewis P, et al. AZD4320, a dual inhibitor of Bcl-2 and Bcl-x(L), induces tumor regression in hematologic cancer models without dose-limiting thrombocytopenia. Clin Cancer Res. 2020;26:6535–49. 10.1158/1078-0432.CCR-20-0863 [DOI] [PubMed] [Google Scholar]
- 72.Selt F, Sigaud R, Valinciute G, Sievers P, Zaman J, Alcon C, et al. BH3 mimetics targeting BCL-XL impact the senescent compartment of pilocytic astrocytoma. Neuro Oncol. 2023;25:735–47. 10.1093/neuonc/noac199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacob M, Wiedemann S, Brucher D, Pieper NM, Birkhold M, Sarchen V, et al. Increased MCL1 dependency leads to new applications of BH3-mimetics in drug-resistant neuroblastoma. Br J Cancer. 2023;129:1667–78. 10.1038/s41416-023-02430-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data are available through the web-based gene expression analysis R2 (R2: microarray analysis and visualisation platform; http://r2.amc.nl) platform: Medulloblastoma / Neuroblastoma RA - Oehme - 20 - rsem - gencode19; 2021-12-01.