Abstract
The aim of this study was to develop a medical imaging and comprehensive stacked learning-based method for predicting high- and low-risk thymoma. A total of 126 patients with thymomas and 5 patients with thymic carcinoma treated at our institution, including 65 low-risk patients and 66 high-risk patients, were retrospectively recruited. Among them, 78 patients composed the training cohort, while the remaining 53 patients formed the validation cohort. We extracted 1702 features each from the patients’ arterial-, venous-, and plain-phase images. Pairwise subtraction of these features yielded 1702 arterial-venous, arterial-plain, and venous-plain difference features each. The Mann‒Whitney U test and least absolute shrinkage and selection operator (LASSO) and SelectKBest methods were employed to select the best features from the training set. Six models were built with a stacked learning algorithm. By applying stacked ensemble learning, three machine learning algorithms (XGBoost, multilayer perceptron (MLP), and random forest) were combined by XGBoost to produce the the six basic imaging models. Then, the XGBoost algorithm was applied to the six basic imaging models to construct a combined radiomic model. Finally, the radiomic model was combined with clinical information to create a nomogram that could easily be used in clinical practice to predict the thymoma risk category. The areas under the curve (AUCs) of the combined radiomic model in the training and validation cohorts were 0.999 (95% CI 0.988–1.000) and 0.967 (95% CI 0.916–1.000), respectively, while those of the nomogram were 0.999 (95% CI 0.996–1.000) and 0.983 (95% CI 0.990–1.000). This study describes the application of CT-based radiomics in thymoma patients and proposes a nomogram for predicting the risk category for this disease, which could be advantageous for clinical decision-making for affected patients.
Keywords: Machine learning, CT, Thymoma
Subject terms: Cancer, Oncology
Introduction
Thymoma, a rare neoplasm of thymic epithelial origin, is the predominant malignancy of the anterior mediastinum1, accounting for approximately 47% of all neoplasms in this region2,3; in Asia, its prevalence is approximately 0.49 per 100,000 person-year4,5. Notably, thymoma is associated with paraneoplastic syndromes6, particularly myasthenia gravis. In 2015, the World Health Organization (WHO) introduced a new classification system for thymic epithelial tumours, which includes six categories: types A, AB, B1, B2 and B3 and thymic carcinoma7. Based on the biological behaviour of the tumour, the categories can be simplified into low-risk thymoma (types A, AB, and B1) and high-risk thymoma (types B2 and B3)8.
Surgery is the primary treatment for thymoma, with complete resection resulting in the best survival rates9,10. Patients with low-risk thymoma typically do not require adjuvant therapy, whereas those in the high-risk group may require multimodal therapy11. Early and accurate diagnosis and differentiation between the risk groups are therefore crucial. However, tissue biopsy is limited by the spatiotemporal heterogeneity of the tumour and the risks associated with deep and transpleural biopsy. Computed tomography (CT) is a noninvasive imaging modality with wide applicability12. Radiomics enables the noninvasive quantification of tumour heterogeneity and identification of malignant characteristics13.
In recent years, numerous studies have focused on the use of radiomics for predicting the risk category of thymomas14. These studies include that by Tian et al.15, who investigated the performance of radiomic-based CT phenomics in predicting the pathological stage and survival outcomes of thymic epithelial tumour patients, achieving integrated areas under the curve (AUCs) of 0.935 and 0.811. Xiao et al.16 developed a comprehensive radiomic diagnostic model using multivariate logistic regression analysis that incorporates clinical and conventional MR imaging characteristics, apparent diffusion coefficient (ADC) values, and radiomic features and demonstrated excellent performance in distinguishing low- from high-risk thymoma patients. Feng et al.17 utilized 14 machine learning models with different feature selection strategies to establish a three-class model based on radiomic features, predicting simplified risk categories of thymic epithelial tumours (TETs). MM et al.14 trained a support vector machine (SVM)-based classification model to differentiate between thymomas and thymic carcinomas. Integration of traditional and radiomic features in the model achieved the highest diagnostic performance18.
However, few studies have been conducted to extract and analyse characteristic differences between the features in plain scan and arterial and venous phase CT images. The objective of this study was to propose an imaging-based radiomic and machine learning approach to predict high- and low-risk thymoma19. To achieve this aim, we extracted imaging features and their paired differences among plain-, arterial-, and venous-phase CT images and input these data into machine learning algorithms to establish robust predictive models20. By combining radiomic features with clinical characteristics, we sought to provide clinicians with more refined diagnostic and prognostic insights into thymoma, thereby enabling them to make more precise personalized treatment decisions21.
Materials and methods
Patient cohort and pathological evaluation
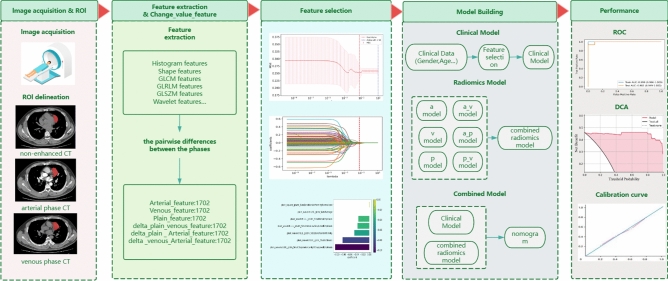
This retrospective study was approved by the Ethical Review Committee of the Affiliated Hospital of Guangdong Medical University. Because of the retrospective nature of the study, the Ethics Committee waived the need for written informed consent. The study design and pipeline are illustrated in Fig. 1. We acquired data from a cohort of 126 patients diagnosed with thymoma and 5 patients diagnosed with thymic carcinoma (Fig. 2) obtained exclusively from the hospital’s picture archiving and communication system (PACS). The data were collected from patients who were seen at the hospital from 2015 to 2023, including 74 male and 57 female patients, with ages ranging from 16 to 80 years. The inclusion criteria were as follows: (1) archival data indicating that the patient was postoperatively pathologically diagnosed with thymoma between January 2015 and October 2023; and (2) complete CT images and clinicopathological data. The following exclusion criteria were applied: (1) CT imaging artefacts; (2) no relevant treatment prior to the preoperative CT scan; and (3) incomplete clinical data.
Figure 1.
Study design and pipeline.
Figure 2.
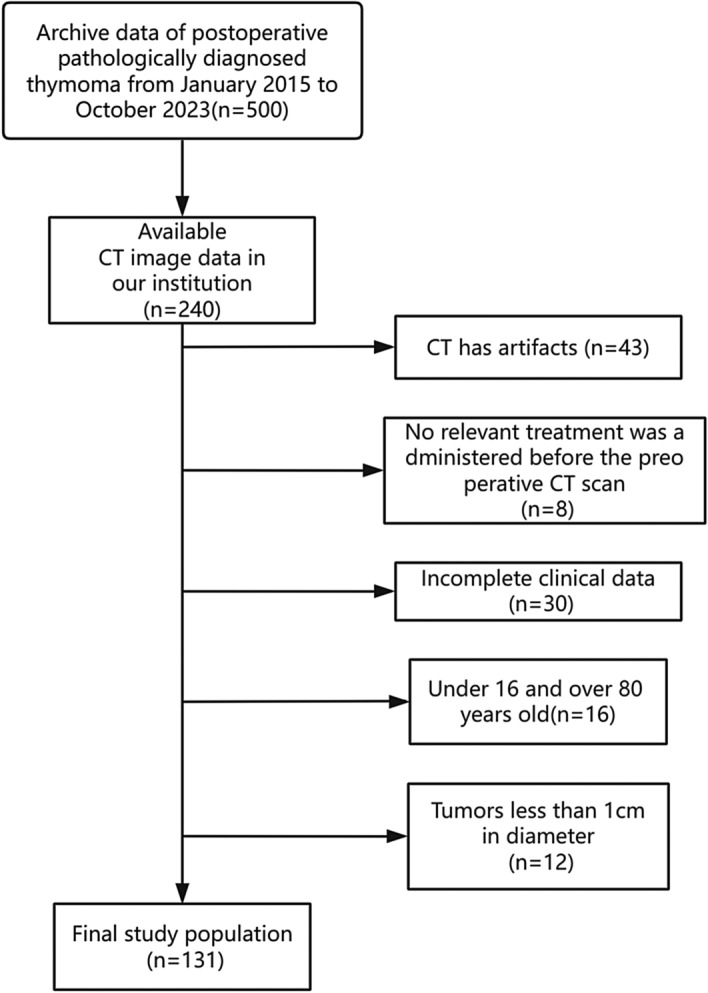
Flowchart of patient selection. CT, computed tomography.
CT imaging protocol
CT scans were performed with a GE Medical Systems Optima CT680 series scanner at the Affiliated Hospital of Guangdong Medical University. The imaging protocol followed standardized procedures to ensure consistent image acquisition across all patients. For each patient, a series of axial images were acquired with the following settings: slice thickness: 0.625 mm, tube voltage: 120 kV, tube current: 261 mA, reconstruction diameter: 380.00. For enhanced scanning, iodohexanol was injected into the median cubital vein at a flow rate of 4 ml/s and a dose of 0.9–1.0 ml/kg. Phase triggering was performed with aortic tracking: when the CT value reached and exceeded 100 HU, the arterial phase was initiated, and the venous phase followed after a 15-s delay.
Image segmentation and feature extraction
The original images from the plain phase (PP), arterial phase (AP) and venous phase (VP) were stored in corresponding folders in DICOM format. A radiologist with 5 years of experience used ITK-SNAP 3.8 software (https://www.itksnap.org) to manually delineate the lesions layer by layer; these delineations were then verified by a radiologist with more than 20 years of experience, and if the findings were disputed, a third radiologist with more than 30 years of experience made the final decision. The window width and window level were set to 35 and 450, respectively. Features were extracted from the segmented images with PyRadiomics using the following settings: Partition width: 25; Resampling pixel spacing: [1, 1, 1] (in millimetres); interpolator: nearest neighbour; normalization: enabled. The RadiomicsFeatureExtractor class was used to extract features from each phase with all features and image types enabled. The extracted features mainly included first-order histogram features, morphological features, texture features, and Gaussian wavelet transform filter features.
By integrating the ITK-SNAP and PyRadiomics libraries, thymoma feature information can be accurately extracted from CT images . We used PyRadiomics to extract features from the PP, VP and AP images separately, obtaining 1702 features from each phase for a total of 5106 features. The pairwise differences between the features from each phase were then calculated to generate an additional 5106 features, resulting in a dataset containing a total of 10,212 features.
Feature selection
The comprehensive set of 10,212 radiomic features consisted of the features extracted from the plain-, arterial-, and venous-phase images and the between-phase differences in those features. A total of 1702 features each were extracted from the arterial-phase, venous-phase, and plain scan-phase images. These features were then pairwise subtracted, resulting in 1702 arterial-venous difference features, 1702 arterial-plain difference features, and 1702 venous-plain difference features. These features are hereafter referred to as plain, arterial, venous, delta_arterial_venous, delta_plain_arterial, and delta_plain_venous features, encompassing a diverse spectrum of quantitative imaging characteristics.
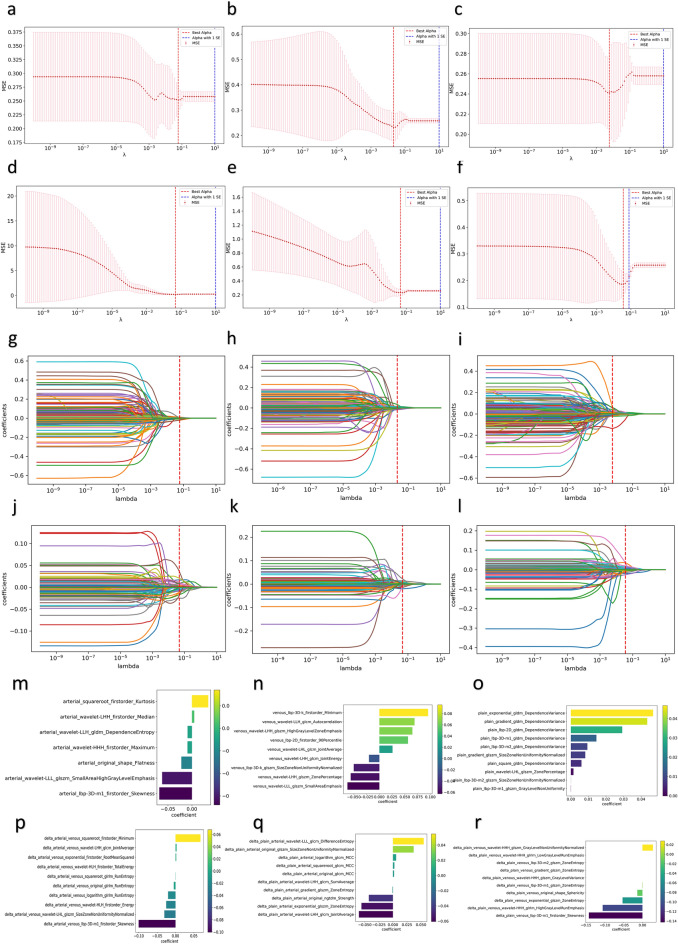
To select the most informative features, a multistep approach was employed. First, the Mann‒Whitney U test was applied to compare features between the risk group; features with p values less than 0.05 were retained22. The least absolute shrinkage and selection operator (LASSO) method was subsequently utilized to further streamline the feature set based on their coefficients; however, the size of the resulting feature set remained substantial. To achieve optimal performance and interpretability, the SelectKBest method, which selects the top ten features within a feature set with the highest discriminatory potential, was employed (Suppl Appendix A1) The six sets of features described above underwent the aforementioned feature selection processes, and the relevant results are shown in Fig. 3.
Figure 3.
Radiomic feature selection. (a–f) Cross-validation curves of the LASSO regression model. (a) Arterial-phase features, (b) venous-phase features, (c) plain scan-phase features, (d) differences between the arterial-phase features and the venous-phase features, (e) differences between the arterial-phase features and the plain scan-phase features, (f) differences between the plain scan-phase features and the venous-phase features. (g–l) Coefficient curves for the radiomic features. (g) Arterial-phase features, (h) venous-phase features, (i) plain scan-phase features, (j) differences between the arterial-phase features and the venous-phase features, (k) differences between the arterial-phase features and the plain scan-phase features, (l) differences between the plain scan-phase features and the venous-phase features. (m–r) Coefficients in the LASSO model. (m) Arterial-phase features, (n) venous phase features, (o) plain scan-phase features, (p) differences between the arterial-phase features and the venous-phase features, (q) differences between the arterial-phase features and the plain scan-phase features, (r) differences between the plain scan-phase features and the venous-phase features.
Model building based on stacked ensemble learning
To verify the stability of the model, Bootstrapping method was used for model evaluation given the small size of the sample. Bootstrapping is an effective resampling technique that can generate multiple sample sets through repeated sampling from the original dataset with repositions. These sample sets are used to train and validate the models, thus providing estimates of the performance of multiple models23. To scale up the predictive power of multiple machine learning algorithms, we used a stacked ensemble learning approach to build a robust and accurate model for predicting high-risk thymoma. In the first layer, three different machine learning algorithms were selected to develop the six radiomic models, including XGBoost, random forest, and multilayer perceptron (MLP). The results were fed into the second layer, which was then trained on the inputs with XGBoost24, yielding the final model. We chose XGBoost as the meta-learner to summarize the prediction results of the base model (Suppl Appendix A3).
During the construction of the between-phase features, data from the plain, arterial, and venous phases were systematically analysed. Pairwise subtraction was performed to derive the corresponding difference features between these phases. These difference features serve as pivotal metrics for comprehending the characteristic changes in thymoma across distinct stages.
The above basic model was trained with the features from the plain, arterial and venous phases and the three sets of difference features. The base models were integrated using XGBoost to generate the final six independent models25. The radiomic signature is the output of the integrated image model, which is constructed as follows. We use stacked learning methods to learn the arterial, venous, plain, and difference features. The first layer of stacked learning consists of three basic learners: XGBoost, random forest, and MLP, and the second layer is XGBoost. The final output was used to construct six independent models with the arterial phase, venous phase, plain phase, and three sets of difference features. XGBoost was subsequently used to integrate the six imaging-based models to output the third layer model, the combined radiomics model (the seventh model). Through multiple logistic regression, a nomogram (the eighth model) was constructed based on the combined radiomic model as well as age and sex. The model building process is shown in Fig. S2.
Statistical analysis
The Mann‒Whitney U test was used to compare continuous data (the radiomic features) between groups in Python via the SciPy library, and a unilateral p value < 0.05 was considered to indicate statistical significance. The Chi-square test and t test were conducted in Excel to compare sex and age, respectively, and two-tailed p values < 0.05 were considered to indicate statistical significance. Python (3.9.12) was used to implement the LASSO and SelectKBest algorithms for filtering imaging omics features and the MLP, random forest, and XGBoost algorithms to develop imaging omics models. The nomogram was constructed via the code at the following address: "https://github.com/Hhy096/nomogram". The AUCs of the models were compared with the DeLong test in Python (3.9.12), whose results are shown in Table 1. Decision curve analysis (DCA) was performed in Python (3.9.12) to evaluate the clinical utility of the models, and calibration curves were drawn to describe the calibration ability of the models in the training and validation sets.
Table 1.
DeLong test results for each model.
| Model (AUC Value) |
a (0.820) |
a_p (0.795) |
a_v (0.853) |
p (0.738) |
p_v (0.800) |
v (0.783) |
nomogram (0.983) |
radiomics (0.967) |
|---|---|---|---|---|---|---|---|---|
|
a (0.820) |
1 | – | – | – | – | – | – | – |
|
a_p (0.795) |
< 0.01 | 1 | – | – | – | – | – | – |
|
a_v (0.853) |
0.525 | 0.037 | 1 | – | – | – | – | – |
|
p (0.738) |
0.273 | < 0.01 | 0.485 | 1 | – | – | – | – |
|
p_v (0.800) |
0.108 | < 0.01 | 0.897 | 0.101 | 1 | – | – | – |
|
v (0.783) |
0.593 | 4.913 | 0.162 | 0.887 | < 0.01 | 1 | – | – |
|
nomogram (0.983) |
0.014 | 0.651 | 0.142 | 0.027 | 0.137 | 0.012 | 1 | – |
|
radiomics (0.967) |
0.026 | 0.542 | 0.446 | 0.074 | 0.580 | 0.084 | 0.368 | 1 |
Ethics approval and consent to participate
This retrospective clinical study was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical University, and was carried out in accordance with the Declaration of Helsinki. The requirement for the informed consent was waived.
Results
Patient characteristics
This study included 131 patients with thymomas who received treatment at our hospital, of whom 65 were low risk and 66 were high risk. Among these patients, 78 were assigned to the training cohort, while the remaining 53 formed the validation cohort. Table 2 presents the baseline characteristics of the thymoma patients at the onset of the study. The clinical and pathological characteristics did not differ significantly between the training and validation cohorts.
Table 2.
Baseline patient characteristics.
| Characteristic | Types | N | Mean age (± standard deviation) |
P-value | Sex | P-value |
|---|---|---|---|---|---|---|
| WHO classification | ||||||
| Low risk thymoma | A | 19 | 55.5 (± 14.653) | 0.034 | Male 29; female 36 | 0.472 |
| AB | 32 | |||||
| B1 | 14 | |||||
| High risk thymoma | B2 | 39 | 51.0 (± 13.214) | Male 45; female 21 | ||
| B3 | 14 | |||||
| B2B3 | 5 | |||||
| Thymic carcinoma | 8 | |||||
| Total | 131 | 53.3 (± 14.073) | Male 74; female 57 | |||
Feature selection
After a comprehensive feature set was extracted with the PyRadiomics library, a multistage feature selection process was implemented to identify the most informative features26. First, we normalized the features of the arterial-phase, venous-phase, and plain scan phase images and the pairwise differences between the phases; a total of 1702 features were extracted from each of the six sets of images. We subsequently used the Mann‒Whitney U test to exclude features with p values greater than 0.0527 and retained 19, 27 and 45 features in the arterial phase, venous phase and plain scan phase, respectively. Additionally, 231 features were retained from the arterial-venous phase set, 190 from the arterial-plain phase set, and 41 from the venous-plain phase set. Then, we implemented LASSO with tenfold cross-validation for further feature screening, retaining 7, 9 and 45 features from the arterial-phase, venous-phase and plain scan-phase features, respectively. Furthermore, 19, 17, and 14 features were retained from the arterial-venous phase, arterial-plain phase, and venous-plain phase sets, respectively. Finally, we selected the most relevant features with the SelectKBest method. A total of 7, 9 and 10 features were retained form the arterial-phase, venous-phase, and plain scan-phase sets, respectively, while 10 features each were retained from the three difference sets.
Radiomic model development
By exploring the key stages of radiomic model development, the features selected above were used to create a robust predictive model for predicting high-risk thymoma.
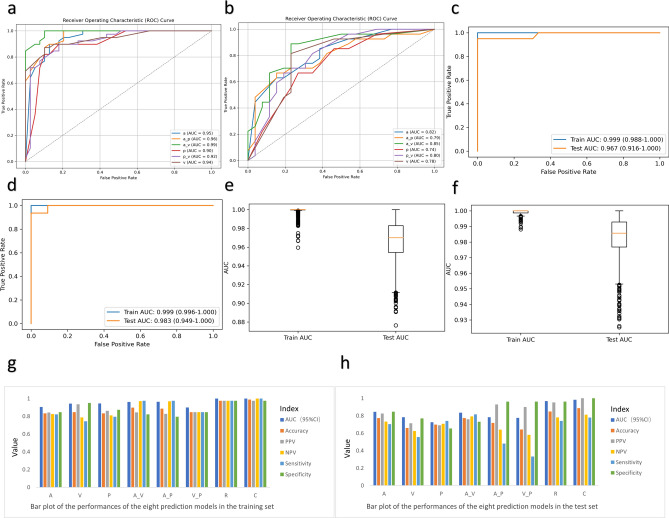
The six feature sets were used to construct a feature-enhanced dataset that encapsulates the essence of the radiomic attributes for each patient. To build the radiomic model, a series of machine learning algorithms (random forest, XGBoost, and MLP) were employed, each customized to exploit the potential of the curated features. Stacked integrated learning methods were used to integrate the outputs of individual machine learning models to create powerful metamodels. The performance of the radiomic models was rigorously evaluated through a variety of metrics, including accuracy, positive predictive value, negative predictive value, sensitivity, specificity, and AUC. The performance of the models is shown in Fig. 4, and the detailed values are shown in Table 3.
Figure 4.
Model performance. (a–d) ROC curves of the models. (a,b) Models based on the plain scan-phase, venous-phase, and arterial-phase features and the corresponding pairwise differences in the features between imaging phases in the training (a) and test sets (b). (c,d) Combined radiomic model (c) and nomogram (d) in the training and test sets. (e,f) Comparison of the AUCs in the training and test sets for the radiomic model (e) and the combined model (f). (g,h) Bar plot of the performance of the eight prediction models in the training set and test set.
Table 3.
Performance of the prediction models.
| Training dataset | Test dataset | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Accuracy | PPV | NPV | Sensitivity | Specificity | AUC (95% CI) | Accuracy | PPV | NPV | Sensitivity | Specificity | |
| Arterial phase | 0.945 | 0.833 | 0.842 | 0.825 | 0.821 | 0.846 | 0.822 | 0.774 | 0.826 | 0.733 | 0.704 | 0.846 |
| Venous phase | 0.943 | 0.846 | 0.935 | 0.787 | 0.744 | 0.949 | 0.782 | 0.660 | 0.714 | 0.625 | 0.556 | 0.769 |
| Unenhanced phase | 0.901 | 0.833 | 0.861 | 0.810 | 0.795 | 0.872 | 0.743 | 0.698 | 0.690 | 0.708 | 0.741 | 0.654 |
| The change between the arterial phase and the venous phase | 0.991 | 0.897 | 0.844 | 0.970 | 0.974 | 0.821 | 0.845 | 0.774 | 0.759 | 0.792 | 0.815 | 0.731 |
| The change between the arterial phase and the unenhanced phase | 0.963 | 0.885 | 0.826 | 0.969 | 0.974 | 0.795 | 0.785 | 0.717 | 0.929 | 0.641 | 0.481 | 0.962 |
| The change between the unenhanced phase and the venous phase | 0.922 | 0.846 | 0.846 | 0.846 | 0.846 | 0.846 | 0.775 | 0.642 | 0.900 | 0.581 | 0.333 | 0.962 |
Ensemble model development and validation
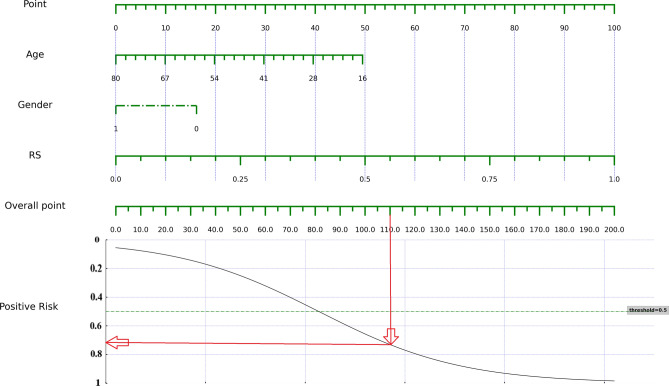
The ensemble model is a combination of the plain, arterial and venous phase models, exploiting the collective strengths of the predictive capabilities of each imaging phase28. XGBoost was chosen as the meta-learner to aggregate the predictions of the base models. The nomogram combining the age, sex, and the radiomics model output is shown in Fig. 5. The performance of the integrated model was thoroughly evaluated in the training and independent validation datasets. The results of DCA are shown in Fig. 6. To determine the stability and generalizability of the model in different datasets, cross-validation and external validation were performed. Feature importance analysis was performed on the ensemble model to reveal the impact of individual radiomic attributes on the ensemble prediction and to facilitate model interpretation29.
Figure 5.
Combined radiomic nomogram for predicting the risk category of thymoma. After calculating the total score, the probability of high-risk thymoma can be derived from the point on the curve corresponding to the total score on the x-axis.
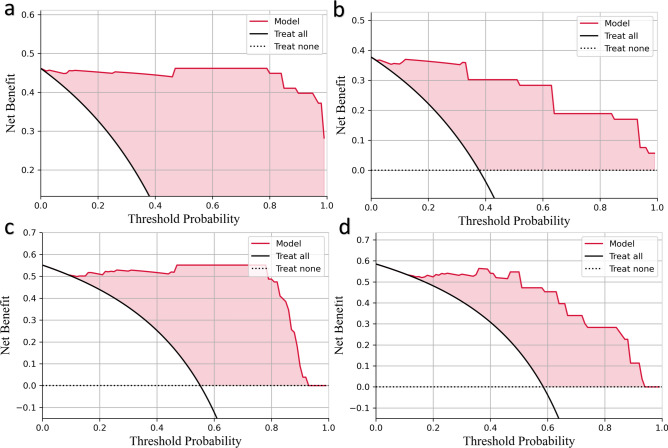
Figure 6.
Decision curve analysis. The net benefit of each model is plotted on the y-axis, and the x-axis indicates the threshold values. The black and dashed lines indicate the assumptions that all or no patients have thymoma, respectively. (a,b) Combined radiomic model in the training (a) and test sets (b). (c,d) Nomogram in the training (c) and test set (d).
Discussion
In the field of medical imaging research, differential analysis of CT imaging across different phases has become a valuable diagnostic tool. Previous studies have shown that differences in scan and enhancement CT values can form the basis for optimizing contrast agent injection plans and increasing the quality and accuracy of diagnostic imaging30. Tang et al. reported that the difference in CT values between the arterial phase and portal venous phase (PVPMAP) was an independent factor in stratifying the risk of stomach gastrointestinal stromal tumours (GISTs)31. With this background, this study is the first to propose the use of an ensemble learning method to evaluate the risk category of thymoma by combining the CT radiomics features from three different phases (plain phase, arterial phase and venous phase) with those from the pairwise differences in the phases to construct models with good predictive performance that could serve as an innovative and more accurate method for assessing the risk of thymoma.
In this study, the AUCs of the models constructed with the differential features tended to be greater than those of the three-phase models. The reason may be that the three-phase CT images provided information about the morphology and density of the tumours in different blood flow states, whereas the differential values provided information about dynamic changes. By using the differential values, subtle differences in tumour growth and angiogenesis could be better captured. Second, the differential images may allow the early detection of lesions or abnormalities more easily, as small changes can be masked in the static phases. Finally, the differential signature better reflects the enhancement pattern produced by the contrast agent within the tumour, which is very valuable for evaluating the tumour blood supply and aggressiveness. Our model effectively demonstrated the changes in the heterogeneity within the area of the tumour.
Additionally, in this study, we utilized stacked ensemble learning in the prediction of high- and low-risk thymomas. Previously, Liu et al.32 used transfer learning with clinical, radiomic, and deep features to establish an SVM classifier-based model for predicting high- and low-risk thymomas, achieving AUCs of 0.99 and 0.95, respectively. The nomograms we exported via stacked learning achieved AUC values of 0.99 and 0.98, respectively, exceeding the AUCs reported in previous studies. The reason may be that stacked learning integrates predictions from multiple base models, effectively improving the accuracy and robustness over individual radiomic models in predicting thymoma risk. This approach not only handles complex imaging data well but also produces models with good generalization abilities, providing clinicians with a more reliable tool for assessing thymoma risk. Compared with biopsy, machine learning models have significant value in diagnosing benign and malignant tumours due to their noninvasive nature, providing efficient clinical assessment tools for patient comfort and diagnostic effectiveness (Suppl Appendix A2). We used the interpretable machine learning algorithm Random Forest to build a model based on features selected during the arterial phase. The model achieved an AUC of 0.78 on the test set, which is lower than the AUC of 0.84 achieved by the model constructed using the stacked learning algorithm.
Through the analysis of the selected features, we noted that various types of image features, including texture features (such as arterial_original_gldm_SmallDependenceEmphasis), morphological features (such as shape-based (3D)), and first-order statistical features (such as first-order statistics) were screened. Features extracted from the arterial phase, such as arterial_original_gldm_SmallDependenceEmphasis and arterial_wavelet-LLH_gldm_DependenceEntropy (abbreviated feature names), depict specific morphological and textural characteristics of the lesion during arterial perfusion that are closely related to vascular perfusion. Features extracted from the venous phase, such as venous_wavelet-LHH_glszm_SizeZoneNonUniformity and venous_wavelet-HHH_gldm_Dependence Variance, highlight the specificity in identifying the lesion during venous perfusion. Plain scan phase features such as plain_wavelet-LHL_gldm_SmallDependence_HighGrayLevelEmphasis and delta_plain_venous_exponential_glszm_LargeArea_HighGrayLevelEmphasis provide baseline image information independent of the use of contrast agent. Differential features, such as delta_plain_arterial_original_glcm_MCC and delta_plain_arterial_original_glrlm_RunEntropy, may reflect significant changes occurring between the two corresponding phases that may be associated with malignant transformation or other lesion features, providing strong clues for diagnosing benign and malignant thymomas33. These features appear to prioritize the basic morphological characteristics of the lesion, helping identify the inherent properties of lesions without the influence of contrast agent. Furthermore, these features reveal significant changes that occur within thymomas under different blood supply states. This multilevel feature extraction aids in comprehensively describing the complex characteristics of thymomas, providing clinicians with more information about the lesions and demonstrating potential for influencing clinical decision-making and in the formulation of personalized treatment strategies.
Although we did not assess pathological molecular markers such as Ki-67 and TdT in the construction of the predictive models in this study, their importance should not be ignored. Instead, in future studies, we can combine machine learning with medical imaging features not only to predict thymoma risk but also to determine the pathological type of the tumour. This combined approach would be conducive to providing personalized therapies, optimizing treatment regimens, and improving patient survival. Our ultimate goal is to develop a universal predictive model for all types of cancer, opening new horizons for cancer research and treatment. Achieving this goal, however, requires in-depth research into the role of pathological molecular markers in cancer and their application in predictive models. This is a challenging but promising mission that could provide new perspectives for understanding and fighting cancer.
In this study, we encountered challenges related to the small sample size and complexity of the model design, both of which can lead to overfitting and consequently degrade model performance on new data. To increase the robustness and predictive accuracy of the model, we employed the bootstrap method, mitigating the dependence on a single dataset through repeated sampling and providing confidence intervals for various performance metrics. By effectively utilizing limited data, this strategy improved model robustness.
However, this study has several other limitations that should be noted. First, its single-centre nature limits the generalizability of the results, since the clinical and demographic characteristics of patients may differ across regions. Second, the retrospective design and limited sample size resulted in a relatively small dataset, which may have affected model training and performance evaluation, increased the risk of overfitting, and reduced external validity. Furthermore, the lack of inclusion of genomic data is an important limitation, as it may provide key insights into the biological mechanisms of thymomas.
To overcome these limitations, future research should adopt a multicentre and prospective design to collect more comprehensive and consistent data, increase the sample size to improve the statistical strength of the data, and integrate genomic data to complement the radiological and clinical features. Such an approach would facilitate a more complete understanding of thymoma, identifying molecular biomarkers associated with disease prognosis and treatment response. With these improvements, the accuracy and reliability of the thymoma risk prediction models could be improved, leading to improvements in clinical decision-making and patient outcomes.
In conclusion, our study revealed that radiomics can effectively predict the risk level of thymic tumour patients, with clinical differential radiomic signatures demonstrating stronger predictive power than the single-phase radiomic signatures. This knowledge can aid clinicians in guiding the selection of personalized treatment plans for early-stage thymoma patients. The proposed approach provides robust support for personalized therapy, with important implications for future clinical practice.
Supplementary Information
Abbreviations
- LASSO
Least absolute shrinkage and selection operator
- AUC
Area under the curve
- DCA
Decision curve analysis
- ROC
Receiver operating characteristic
- PAS
Picture archiving and communication system
- ADC
Apparent diffusion coefficient
- TETs
Thymic epithelial tumors
- SVM
Support vector machine
- MLP
Multilayer perceptron
Author contributions
Zhu Liang , Jiamin Li, YihanTang, Shuyan He: Conceptualization, Methodology, Software, Writing-Original draft preparation. Yaxuan Zhang, Chunyuan Chen, Siyuan Li, Xuefeng Wang, Ziye Zhuang, Xinyan Xu: Data curation, Writing-Original draft preparation. Shuyan He, Biao Deng: Supervision, Software, Validation. All authors reviewed the manuscript.
Data availability
The other original contributions presented in the study were included in the article/Supplementary Material. For more inquiries can contact the corresponding authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhu Liang and Jiamin Li.
Contributor Information
Shuyan He, Email: 1012027045@qq.com.
Biao Deng, Email: 15760562638@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-69735-3.
References
- 1.Roden, A. C. et al. Distribution of mediastinal lesions across multi-institutional, international, radiology databases. J. Thorac. Oncol.15, 568–579. 10.1016/j.jtho.2019.12.108 (2020). 10.1016/j.jtho.2019.12.108 [DOI] [PubMed] [Google Scholar]
- 2.Du, X. et al. Expression and diagnostic value of NPTX1 in thymoma patients. Zhongguo Fei Ai Za Zhi24, 1–6. 10.3779/j.issn.1009-3419.2021.102.03 (2021). 10.3779/j.issn.1009-3419.2021.102.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Detterbeck, F. C. & Zeeshan, A. Thymoma: Current diagnosis and treatment. Chin. Med. J. (Engl)126, 2186–2191 (2013). 10.3760/cma.j.issn.0366-6999.20130177 [DOI] [PubMed] [Google Scholar]
- 4.Wang, J. & Zhang, S. Advances on diagnosis and treatment of malignant thymic tumors. Zhongguo Fei Ai Za Zhi13, 985–991. 10.3779/j.issn.1009-3419.2010.10.10 (2010). 10.3779/j.issn.1009-3419.2010.10.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engels, E. A. & Pfeiffer, R. M. Malignant thymoma in the United States: Demographic patterns in incidence and associations with subsequent malignancies. Int. J. Cancer105, 546–551. 10.1002/ijc.11099 (2003). 10.1002/ijc.11099 [DOI] [PubMed] [Google Scholar]
- 6.Yuan, D. et al. Clinical study on the prognosis of patients with thymoma with myasthenia gravis. Zhongguo Fei Ai Za Zhi21, 1–7. 10.3779/j.issn.1009-3419.2018.01.01 (2018). 10.3779/j.issn.1009-3419.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travis, W. D. et al. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J. Thorac. Oncol.10, 1240–1242. 10.1097/JTO.0000000000000663 (2015). 10.1097/JTO.0000000000000663 [DOI] [PubMed] [Google Scholar]
- 8.Multidisciplinary Committee of Oncology, Chinese Physicians Association. Chinese guideline for clinical diagnosis and treatment of thymic epithelial tumors (2021 edition). Zhonghua Zhong Liu Za Zhi43, 395–404. 10.3760/cma.j.cn112152-20210313-00226 (2021). 10.3760/cma.j.cn112152-20210313-00226 [DOI] [PubMed] [Google Scholar]
- 9.Fang, W., Chen, W., Chen, G. & Jiang, Y. Surgical management of thymic epithelial tumors: A retrospective review of 204 cases. Ann. Thorac. Surg.80, 2002–2007. 10.1016/j.athoracsur.2005.05.058 (2005). 10.1016/j.athoracsur.2005.05.058 [DOI] [PubMed] [Google Scholar]
- 10.Liu, X., Li, X. & Li, J. Treatment of recurrent thymoma. Zhongguo Fei Ai Za Zhi23, 204–210. 10.3779/j.issn.1009-3419.2020.03.11 (2020). 10.3779/j.issn.1009-3419.2020.03.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, W. et al. Management of thymic tumors—Consensus based on the Chinese alliance for research in thymomas multi-institutional retrospective studies. Zhongguo Fei Ai Za Zhi19, 414–417. 10.3779/j.issn.1009-3419.2016.07.02 (2016). 10.3779/j.issn.1009-3419.2016.07.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomiyama, N. et al. Anterior mediastinal tumors: Diagnostic accuracy of CT and MRI. Eur. J. Radiol.69, 280–288. 10.1016/j.ejrad.2007.10.002 (2009). 10.1016/j.ejrad.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 13.Jiao, Y., Ren, Y. & Zheng, X. Quantitative imaging assessment of tumor response to chemoradiation in lung cancer. Zhongguo Fei Ai Za Zhi20, 407–414. 10.3779/j.issn.1009-3419.2017.06.07 (2017). 10.3779/j.issn.1009-3419.2017.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayoral, M. et al. Conventional and radiomic features to predict pathology in the preoperative assessment of anterior mediastinal masses. Lung Cancer178, 206–212. 10.1016/j.lungcan.2023.02.014 (2023). 10.1016/j.lungcan.2023.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian, D. et al. Machine learning-based radiomic computed tomography phenotyping of thymic epithelial tumors: Predicting pathological and survival outcomes. J. Thorac. Cardiovasc. Surg.165, 502-516.e9. 10.1016/j.jtcvs.2022.05.046 (2023). 10.1016/j.jtcvs.2022.05.046 [DOI] [PubMed] [Google Scholar]
- 16.Xiao, G. et al. MR imaging of thymomas: A combined radiomics nomogram to predict histologic subtypes. Eur. Radiol.31, 447–457. 10.1007/s00330-020-07074-3 (2021). 10.1007/s00330-020-07074-3 [DOI] [PubMed] [Google Scholar]
- 17.Feng, X.-L. et al. Optimizing the radiomics-machine-learning model based on non-contrast enhanced CT for the simplified risk categorization of thymic epithelial tumors: A large cohort retrospective study. Lung Cancer166, 150–160. 10.1016/j.lungcan.2022.03.007 (2022). 10.1016/j.lungcan.2022.03.007 [DOI] [PubMed] [Google Scholar]
- 18.Rao, A., Pang, M., Kim, J. et al. Assessing the Utility of ChatGPT Throughout the Entire Clinical Workflow. medRxiv 2023.02.21.23285886. 10.1101/2023.02.21.23285886 (2023). [DOI] [PMC free article] [PubMed]
- 19.Lu, C.-F. et al. Machine learning-based radiomics for molecular subtyping of gliomas. Clin. Cancer Res.24, 4429–4436. 10.1158/1078-0432.CCR-17-3445 (2018). 10.1158/1078-0432.CCR-17-3445 [DOI] [PubMed] [Google Scholar]
- 20.Hu, Y. et al. Assessment of intratumoral and peritumoral computed tomography radiomics for predicting pathological complete response to neoadjuvant chemoradiation in patients with esophageal squamous cell carcinoma. JAMA Netw. Open3, e2015927. 10.1001/jamanetworkopen.2020.15927 (2020). 10.1001/jamanetworkopen.2020.15927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, C. et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature483, 474–478. 10.1038/nature10860 (2012). 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambin, P. et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol.14, 749–762. 10.1038/nrclinonc.2017.141 (2017). 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 23.Hinkley, D. Bootstrap methods: Another look at the jackknife. In The Science of Bradley Efron. Springer Series in Statistics (Morris, C.N., Tibshirani, R. eds.). 10.1007/978-0-387-75692-9_9 (Springer, 2008).
- 24.Sipper, M. & Moore, J. H. Conservation machine learning: A case study of random forests. Sci. Rep.11, 3629. 10.1038/s41598-021-83247-4 (2021). 10.1038/s41598-021-83247-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pham, T. X., Siarry, P. & Oulhadj, H. Segmentation of MR brain images through hidden Markov random field and hybrid metaheuristic algorithm. IEEE Trans. Image Process.10.1109/TIP.2020.2990346 (2020). 10.1109/TIP.2020.2990346 [DOI] [PubMed] [Google Scholar]
- 26.Aerts, H. J. W. L. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun.5, 4006. 10.1038/ncomms5006 (2014). 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, C.-B. et al. Application of machine learning model to predict osteoporosis based on abdominal computed tomography images of the psoas muscle: A retrospective study. BMC Geriatr.22, 796. 10.1186/s12877-022-03502-9 (2022). 10.1186/s12877-022-03502-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang, Z. et al. A novel multi-stage residual feature fusion network for detection of COVID-19 in chest X-ray images. IEEE Trans. Mol. Biol. Multiscale Commun.8, 17–27. 10.1109/TMBMC.2021.3099367 (2022). 10.1109/TMBMC.2021.3099367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gafita, A. et al. Nomograms to predict outcomes after 177Lu-PSMA therapy in men with metastatic castration-resistant prostate cancer: An international, multicentre, retrospective study. Lancet Oncol.22, 1115–1125. 10.1016/S1470-2045(21)00274-6 (2021). 10.1016/S1470-2045(21)00274-6 [DOI] [PubMed] [Google Scholar]
- 30.Feng, S. T. et al. An individually optimized protocol of contrast medium injection in enhanced CT scan for liver imaging. Contrast Media Mol. Imaging2017, 7350429. 10.1155/2017/7350429 (2017). 10.1155/2017/7350429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang, B. et al. Comparison of computed tomography features of gastric and small bowel gastrointestinal stromal tumors with different risk grades. J. Comput. Assist. Tomogr.46(2), 175–182. 10.1097/RCT.0000000000001262 (2022). 10.1097/RCT.0000000000001262 [DOI] [PubMed] [Google Scholar]
- 32.Liu, W. et al. Development and validation of multi-omics thymoma risk classification model based on transfer learning. J. Digit. Imaging36, 2015–2024. 10.1007/s10278-023-00855-4 (2023). 10.1007/s10278-023-00855-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin, X. et al. Small cell lung cancer transformation: From pathogenesis to treatment. Semin. Cancer Biol.86, 595–606. 10.1016/j.semcancer.2022.03.006 (2022). 10.1016/j.semcancer.2022.03.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The other original contributions presented in the study were included in the article/Supplementary Material. For more inquiries can contact the corresponding authors.