Abstract
Cancer is one of the most concerning public health issues and breast cancer is one of the most common cancers in the world. The immune cells within the tumor microenvironment regulate cancer development. In this study, single immune cell data sets were used to identify marker gene sets for exhausted CD8 + T cells (CD8Tex) in breast cancer. Machine learning methods were used to cluster subtypes and establish the prognostic models with breast cancer bulk data using the gene sets to evaluate the impacts of CD8Tex. We analyzed breast cancer overexpressing and survival-associated marker genes and identified CD8Tex hub genes in the protein–protein-interaction network. The relevance of the hub genes for CD8 + T-cells in breast cancer was evaluated. The clinical associations of the hub genes were analyzed using bulk sequencing data and spatial sequencing data. The pan-cancer expression, survival, and immune association of the hub genes were analyzed. We identified biomarker gene sets for CD8Tex in breast cancer. CD8Tex-based subtyping systems and prognostic models performed well in the separation of patients with different immune relevance and survival. CRTAM, CLEC2D, and KLRB1 were identified as CD8Tex hub genes and were demonstrated to have potential clinical relevance and immune therapy impact. This study provides a unique view of the critical CD8Tex hub genes for cancer immune therapy.
Keywords: Immune cell, Breast cancer, Machine learning, Single-cell sequencing, Spatial sequencing
Subject terms: Computational biology and bioinformatics, Cancer, Breast cancer, Oncogenes, Tumour biomarkers, Tumour immunology
Introduction
Cancer is one of the most concerning public health issues in the world1. It is estimated that in 2024, there will be approximately 2,001,140 new cancer cases and 611,720 cancer-related deaths in the United States2. In China, it was estimated that approximately 4,800,000 new cancer cases occurred, causing about 3,200,000 cancer-related deaths3. Breast cancer is one of the most common cancers in the world 3. Much as the prevention and tumorigenesis of breast cancer have been studied intensively in the past decades4, the incidence of breast cancer increased by 0.5% each year from 2014 to 20185. The development of breast cancer was impacted by both genetic risk factors and environmental risk factors. Clinical breast cancer was subtyped by the expression level of certain breast cancer critical receptors: the estrogen receptor (ER), the progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Breast cancer was divided into the following 4 molecular subtypes: Luminal A, Luminal B, Triple negative (all called basal-like), and HER2-enriched.
It has been widely accepted that the immune cells within the tumor microenvironment regulate cancer development6. Tumor-infiltrating immune cells have emerged as clinically relevant and highly associated with prognosis and response to treatment for breast cancer as well as other cancer types 7. Checkpoint blockade therapies have demonstrated notable advancements in treating various human cancer types8–10. Breast cancer, previously considered poorly immunogenic, has been an exception11. Even though breast cancer isn't typically considered highly immunogenic due to its relatively low tumor mutational burden, the abundance of tumor-infiltrating lymphocytes in breast cancer correlates with markedly improved prognoses, both with and without PD-1 targeted immunotherapy12,13. Two checkpoint inhibitors targeting the PD-1/PD-L1 pathway, atezolizumab and pembrolizumab, have gained approval for treating triple-negative breast cancer patients14,15. Our comprehension of the mechanisms underlying resistance or response to immunotherapy remains incomplete, as does our understanding of the intricate cellular interactions within the tumor immune microenvironment. To develop new immunotherapies and utilize existing ones effectively for breast cancer patients, it is imperative to grasp the tumor immune microenvironment comprehensively. Although breast cancer tumor-infiltrating lymphocytes are mainly composed of CD3 + T cells16,17, recent research has identified a subset of CD8 + T cells that play a crucial role in breast cancer18. This finding is supported by a single-cell RNA sequencing study of CD3 + T cells isolated from human primary breast cancer18. This subset of CD8 + T cells exhibited elevated expression levels of immune checkpoint molecules like PDCD1 (PD-1), CTLA4, HAVCR2 (TIM-3), and LAG3. The transcriptional signature originating from these cells was linked to improved prognoses, irrespective of the total quantity of CD8 + T cells present and the administered treatment18,19. For immune cells and other cells in cancer, the unique biomarkers of cells can be used to evaluate the abundance of the cells in tumors. The discovery and investigation of these cell biomarkers in cancer facilitate the understanding of the role and function of the corresponding cells in tumors.
The inherent heterogeneity of breast cancer poses significant challenges for conventional diagnostic and therapeutic methods20. Typically, these approaches rely on analyzing bulk tumor tissue samples, which may obscure underlying heterogeneity due to their focus on average expression levels20. However, emerging technologies like single-cell analysis offer promising alternatives, already widely used in oncology research21,22. By examining gene expression, phenotypes, protein levels, and other cellular properties at an individual cell level, these techniques are well-suited to tackle tumor heterogeneity, particularly in highly diverse cancers like breast cancer23–25. Single-cell analysis can aid in predicting cellular evolution during tumor progression and enhance the precision of predicting treatment outcomes and patient prognosis23–25. Moreover, these techniques play a vital role in devising novel therapeutic strategies by enabling the detailed examination of genetic variations and phenotypic characteristics of tumor cells, leading to the identification of new therapeutic targets20. This, in turn, facilitates the development of highly targeted treatment strategies, improving our ability to predict treatment efficacy and potential drug resistance. Single-cell gene sequencing, utilizing next-generation sequencing (NGS), has become indispensable for studying breast cancer heterogeneity26. Unlike traditional Sanger sequencing, NGS systems employ massive parallel sequencing to generate billions of DNA reads, allowing for the detection of various genetic variations, including single-nucleotide mutations, small insertions/deletions, and copy number variations27. This comprehensive view aids in streamlining the development of targeted treatment strategies. For example, in HER2-positive breast cancer, single-cell sequencing identifies diversity in HER2 gene amplification across different cells, facilitating personalized treatment plans28. NGS versatility extends to RNA sequencing (RNA-seq), enabling quantitative and sequence analyses of diverse RNA types and their expression levels, enriching our understanding of breast cancer molecular mechanisms29. RNA sequencing has been wildly used in cancer research 30–39.
For transcriptomic data, generally, bulk sequencing data provides averaged expression levels of all cells in a tissue sample, while single-cell sequencing data has been used to decipher the cellular and molecular landscape at a single-cell resolution 40. The advantage of bulk sequencing data is that it often comes with the clinical information of the patients. Such patient levels data facilitate the analysis of diagnosis and prognosis as well as other clinical factors associated with cancers. In addition, spatial sequencing provides gene expression profiles of a sample with positional information, which is useful for studying heterogeneity within a tumor sample 41. Single-cell RNA sequencing (scRNA-seq) presents significant new prospects for systematically delineating the cellular landscape of tumors and uncovering fresh insights into cell biology, disease etiology, and drug response42,43. Numerous studies have effectively employed scRNA-seq to examine selected populations within human breast tumors, unveiling a spectrum of differentiation states within tumor-infiltrating lymphocytes44, highlighting the role of tissue-resident CD8 cells in breast cancer18, and shedding light on chemoresistance mechanisms in breast cancer neoplastic cells45. Recent endeavors have utilized mass cytometry with antibody marker panels to scrutinize breast cancer cell types and ecosystems across hundreds of patients46,47. Consequently, there is a pressing need for a more comprehensive transcriptional atlas of breast tumors at high molecular resolution, encompassing all subtypes and cell types. Such an atlas would aid in refining the disease's taxonomy, delineating heterotypic cellular interactions, and elucidating cellular differentiation processes. Equally crucial are data systematically mapping the spatial transcriptomic architecture of breast tumors, as this can unveil how cells in the tumor microenvironment (TME) are organized into functional units. A recent paper integrated single-nucleus RNA sequencing with microarray-based spatial transcriptomics to delineate cell populations and their spatial distribution within breast cancer tissues 48.
Our study focused on exhaust CD8 + T cells (CD8 + Tex). The recent surge in cancer immunotherapy, primarily based on checkpoint blockade, has been a breakthrough in treating various cancer types. However, certain factors are hindering the progress of these treatments, such as varying genetic make-up of individuals, resistant tumor sub-types, and immune-related adverse events. While the focus of immunotherapies has been on improving CD8 + T cells, the relationship between CD4 + T cells and CD8 + T cells is also gaining attention. The tumor-infiltrating T regulatory (Treg) cells are a major obstacle in the cross-talk between CD4 + T cells and CD8 + T cells since they are capable of inhibiting anti-tumour immunity49. CD8 + Tex, which is often seen in chronic infections and cancer, is a progressive process characterized by decreased effector function and upregulation of inhibitory receptors such as PD-1 and Tim-3. Although immunological checkpoint inhibitors have allowed for the eradication of tumors, a better understanding of the mechanisms by which T cell–exhaustion pathways work in tumors and the factors that drive them is needed. In this regard, the role of CD8 + Tex in immunosuppression is key to the resistance of cancer in immune therapy.
The study hypothesized that certain genes can be biomarkers of CD8 + T cells in breast cancer as well as other cancer types. In this study, we aimed to identify these genes and demonstrate their association with cancers. We believe this study provides a unique view of the critical T cell hub genes for cancer immune therapy.
Methods
Overview of the study
Single immune cell data sets were used to identify marker gene sets for CD8 + Tex cells in breast cancer. A machine learning method, consensus clustering, was used to cluster TCGA BRCA patients using the identified marker genes, hence, we constructed CD8 + Tex-based genetic subtypes based on the abound of CD8 + Tex in breast cancer samples. We compared the immune cell infiltration levels and predicted immune checkpoint blockade response rate among subtypes to demonstrate the potential clinical value of the subtype systems for immune therapy of breast cancer. The identified marker gene sets were also used to construct the prognostic models for breast cancer patients using a machine learning algorithm lasso (least absolute shrinkage and selection operator) with TCGA BRCA cohort, which further identified the critical genes for the subsequent study. We further analyzed the protein–protein interaction of these molecules and identified hub genes in the protein–protein-interaction network. The correlation between the hub genes and CD8 + T-cell infiltration levels of breast cancer was evaluated using different immune cell calculation algorithms. The clinical associations of these hub genes were analyzed using the clinical information of breast cancer patients and their expression differences in invasive ductal cancer and ductal cancer in situ were analyzed using spatial sequencing data. The pan-cancer cancer-non-cancer expression and survival association of these hub genes were analyzed. The correlation between the hub genes and CD8 + infiltration levels and the immune therapy predictive values of these hub genes were analyzed using immune checkpoint blockade sub-cohorts.
Data collection
Single-cell cohorts were accessed and analyzed from the TISCH2 platform50(http://tisch.comp-genomics.org/). In this study, 4 single-cell sequence data sets were included as shown in Table 1. This dataset comprises expression data from immune cells obtained through fluorescence-activated cell sorting (FACS), focusing on an enriched fraction of immune cells. The MAESTRO v1.1.0 workflow51 (https://github.com/liulab-dfci/MAESTRO/blob/master/README.md) employed PCA for dimension reduction and KNN and Louvain algorithms52,53 for clustering to identify 2000 variable features for each dataset; the number of principal components and the resolution for graph-based clustering were adjusted according to the cell number. Previous studies revealed that MAESTRO demonstrated superior consistency across nearly all cell types, regardless of whether the correlation was computed using all genes or solely the top 2000 variable genes51, hence in this study, only the top 2000 variable genes were used. UMAP was used to reduce the dimension and visualize the clustering results54, and the Wilcoxon test was used to identify differentially expressed (DE) genes of each cluster of cell type (|logFC|> = 0.25, FDR < 1e-05). Data from The Cancer Genome Atlas (TCGA, https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and GTEx (https://gtexportal.org/home/) were obtained, which included gene expression profiling data and clinical information on cancer tissues. This data was obtained in accordance with the guidelines and policies.
Table 1.
Information for single-cell data sets.
| Dataset Name | Species | Treatment | Patients number | Cells | CD8Tex cells | CD8Tex cell (%) | Platform | Primary or metastasis | PublicationS |
|---|---|---|---|---|---|---|---|---|---|
| GSE110686 | Human | None | 2 | 6,035 | 622 | 10.3 | 10 × Genomics | Primary and metastasis | 18 |
| GSE114727_10X | Human | None | 3 | 28,678 | 1389 | 4.8 | 10 × Genomics | Primary | 44 |
| GSE176078 | Human | None | 26 | 89,471 | 13,500 | 15.1 | 10 × Genomics | Primary | 55 |
| EMTAB8107 | Human | None | 14 | 33,043 | 5193 | 15.7 | 10 × Genomics | Primary | 56 |
Single-cell data quality control
A standardized analysis pipeline utilizing MAESTRO v1.1.051 was employed to process all gathered datasets. This workflow encompassed quality control, batch effect mitigation, cell clustering, differential expression analysis, cell-type annotation, and malignant cell classification. The raw count, TPM, or FPKM table served as input for this standardized workflow. Cell quality was assessed using two metrics: total counts (UMI) per cell (library size) and the number of detected genes per cell. Cells with low quality were excluded if the library size was < 1000 or the number of detected genes was < 500.
Single-cell data batch effect correction
To systematically assess batch effects across each dataset, an entropy-based metric 44,57 was utilized to quantify data mixing among batches. Typically, samples from different patients in most datasets are susceptible to batch effects. A k-NN graph (k = 30) was constructed based on the Euclidean distance between cells in UMAP coordinates for datasets with more than one patient. For each cell j, the distribution of patients in its nearest neighbors was computed. The measure of mixing between patients Hj is defined as:
here, represents the proportion of cells from patient t among the 30 nearest neighbors of cell j, while T denotes the total number of patients. High entropy, indicating that the most similar cells in a cell's neighborhood come from different patients, suggests potential batch effects. Conversely, low entropy suggests that the most similar cells originate from the same patient, indicating a potential batch effect. It is noteworthy that datasets primarily composed of malignant cells (malignant % > 75%) may exhibit low entropy due to the heterogeneity of malignant cell expression among different tumors 42. Consequently, the collected datasets were classified into three groups. Firstly, for datasets primarily containing malignant cells, there was no need to eliminate batch effects between different patients, as they reflect differences between distinct tumors. Secondly, datasets with a median entropy lower than 0.7 underwent batch effect correction using Seurat v3.1.258. Median entropies shifted towards higher values post-batch effect removal, indicating significant correction of potential batch effects. Thirdly, datasets with a median entropy higher than 0.7 were considered less affected by batch effects.
Single-cell clustering and differential gene analysis
For each dataset, the MAESTRO workflow identified the top 2000 variable features and conducted PCA for dimension reduction, followed by employing the KNN and Louvain algorithm for cluster identification52,53. To better capture cellular differences and variabilities across datasets with varying cell numbers, adjustments were made to the number of principal components and the resolution for graph-based clustering. Both parameters were increased with increasing cell numbers. The uniform manifold approximation and projection (UMAP) were utilized for further dimension reduction and visualization of clustering results59. The Wilcoxon test was applied to identify differentially expressed (DE) genes for each cluster compared to all other cells, based on criteria such as log-transformed fold change (|logFC|≥ 0.25) and false discovery rate (FDR < 1e−05). Clusters of cells were identified using a combination of three approaches. Firstly, cell-type annotations provided by the original studies were utilized. Secondly, the expression distribution of malignant cell markers from the initial research was assessed, including epithelial markers and EMT genes where available. Thirdly, we applied InferCNV (https://github.com/broadinstitute/infercnv) to predict cell malignancy based on predicted copy number variations was employed, segregating cells into malignant and non-malignant clusters. For the remaining normal clusters, an automated marker-based annotation method within MAESTRO was applied using the differentially expressed genes between clusters. Subsequently, the cell-type annotations based on the annotations provided by the original studies were manually verified and corrected.
Bulk data differential expression analysis
Differentially expressed genes (DEGs) between subtypes were identified using the Limma package with a cut-off fold change of 1.3 and a P-value of 0.05. DEGs are genes whose expression levels vary significantly between different groups. In this study, the goal is to identify genes that are differentially expressed between different subtypes. Limma (Linear Models for Microarray Data, https://bioconductor.org/packages/release/bioc/html/limma.html) is a statistical package in R used for analyzing gene expression data. It employs linear models and empirical Bayes methods to identify DEGs with high sensitivity and specificity.
Bulk data enrichment analysis
For the GO biological and KEGG pathway enrichment analyses, the ClusterProfiler package (version: 3.18.0, https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) in R was employed. The False discovery rate (FDR) and p.adjust were set at 0.25 and 0.05, respectively. Gene Ontology (GO, https://geneontology.org/) is a standardized system for annotating genes and their products with terms representing biological processes, molecular functions, and cellular components. GO biological pathway enrichment analysis involves taking a list of genes, often derived from experimental data such as gene expression studies or genome-wide association studies, and determining whether any particular biological processes represented by GO terms are significantly enriched in that gene list compared to what would be expected by chance. The Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) is a collection of databases that contain information about biological pathways, diseases, drugs, and other biological entities. KEGG pathway enrichment analysis involves mapping a list of genes to known biological pathways in the KEGG database and determining whether any pathways are significantly enriched in the gene list.
Analysis of the survival
The univariate Cox regression analysis, Kaplan–Meier (KM) plot, and log-rank analysis were used to assess the survival association and display the survival curves of genes. Univariate Cox regression analysis examines the association between a single predictor variable (such as a gene expression level or a clinical characteristic) and survival time. It calculates the hazard ratio, which represents the relative risk of experiencing the event of interest (such as death) between two groups defined by the predictor variable. The Cox proportional hazards model is commonly used for this analysis, allowing for the estimation of hazard ratios while accounting for censoring and other covariates. The Kaplan–Meier plot is a graphical method used to estimate the survival function (probability of survival) over time. It is particularly useful for visualizing survival differences between groups defined by categorical variables (such as sub-groups or biomarker expression levels). The plot displays the proportion of individuals surviving at each time point, along with confidence intervals. The log-rank test is a statistical test used to compare the survival curves of two or more groups. It assesses whether there are significant differences in survival times between the groups, taking into account censoring. The test evaluates whether the observed differences in survival are greater than would be expected by chance. If the p-value from the log-rank test is below a predetermined significance level (0.05), it indicates that there is a statistically significant difference in survival between the groups. All analyses were carried out using R (foundation for statistical computing 2020) version 4.0.3 (https://cran.r-project.org/bin/windows/base/old/4.0.3/). Statistical significance was defined as p < 0.05.
Consensus clustering analysis
Subtyping of the samples was carried out using the ConsensusClusterPlus package(https://bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html). The number of clusters was set at 1–6 for consistency analysis to optimize the best clustering number. Unsupervised class discovery involves identifying potential groups within a dataset based solely on inherent features without external guidance. Researchers using this technique typically aim to address two key questions: the number of groups present in the data and the confidence level associated with both the group quantity and their memberships. Consensus clustering60 serves as a valuable method for tackling these inquiries, particularly prominent in fields such as cancer research36,61. Consensus clustering offers both quantitative and visual indicators of “stability”, obtained through iterative subsampling and clustering. By synthesizing outcomes from multiple repetitions, consensus clustering generates a consensus that demonstrates resilience against sampling variations. While initially available within the GenePattern software62, the consensus clustering technique has been further developed into ConsensusClusterPlus, a package in the R language offering enhanced functionalities and visualizations. In this study, this method is suitable for distinguishing subsets of samples of breast cancer patients based on certain genes and comparing the overall effect of these genes on clinical phenotypes.
Immune analysis
The immune cell infiltration level was calculated using different algorithms, including TIMER63, XCELL64, CIBERSORT65, MCPCOUNTER66, QUANTISEQ67, and EPIC68. TIMER is a web server for the comprehensive analysis of tumor-infiltrating immune cells. It provides immune cell infiltration levels in various cancer types and their associations with clinical outcomes. TIMER (http://timer.cistrome.org/) utilizes gene expression data to estimate the abundance of immune cell subtypes within tumor samples. XCELL (https://github.com/dviraran/xCell) is another computational tool used for cell type enrichment analysis in gene expression data. It estimates the relative abundance of different cell types within a heterogeneous sample, including immune cells. XCELL utilizes gene expression signatures specific to various cell types to infer their proportions in the sample. CIBERSORT (Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts, https://cibersortx.stanford.edu/) is a computational method used to characterize cell composition in complex tissues based on gene expression data. It deconvolutes bulk tissue gene expression profiles to estimate the relative proportions of different cell types present. CIBERSORT is particularly useful for studying immune cell populations in tumor microenvironments. MCPCOUNTER (Microenvironment Cell Populations-counter, https://github.com/ebecht/MCPcounter) is a gene expression-based method for quantifying the abundance of specific immune and stromal cell populations in tumor samples. It uses predefined gene signatures associated with different cell types to estimate their relative proportions in the sample. QUANTISEQ (https://icbi.i-med.ac.at/software/quantiseq/doc/) is a computational tool for the deconvolution of gene expression profiles to estimate the proportions of immune cell subsets within a sample. It utilizes gene expression signatures specific to different immune cell types to infer their relative abundances. EPIC (Evaluating the Presence of Immune Cells, https://github.com/GfellerLab/EPIC) is a computational tool for quantifying immune cell infiltration in tumor samples based on DNA methylation data. It uses DNA methylation profiles to estimate the proportions of immune cell subsets present in the tumor microenvironment. Immune checkpoint blockade (ICB) responses of subtypes were predicted using the Tumor Immune Dysfunction and Exclusion (TIDE, http://tide.dfci.harvard.edu/) algorithm69 using the TIDE online analysis platform. TIDE represents a computational framework designed to assess the likelihood of immune evasion by tumors. It achieves this by analyzing the gene expression patterns present in cancer samples. The immune therapy sub-cohorts were accessed and analyzed with TIDE tool69 (http://tide.dfci.harvard.edu/setquery/). The TIDE score was designed to predict response to immune checkpoint blockade, including anti-PD1 and anti-CTLA4, for melanoma and NSCLC. The use of TIDE in this study is based on the assumption that breast cancer has a similar immune system as melanoma and NSCLC.
Prognostic model for identification of critical genes
The model was constructed using the glmnet (https://glmnet.stanford.edu/articles/glmnet.html) R package, which implemented the least absolute shrinkage and selection operator (LASSO) regression algorithm70 with tenfold cross-validation for gene signature selection. LASSO is a regression analysis method used for variable selection and regularization. It aims to find the subset of predictor variables that are most relevant for predicting the response variable while simultaneously performing variable selection and regularization to prevent overfitting. In LASSO regression, the ordinary least squares (OLS) objective function is augmented with a penalty term that is the sum of the absolute values of the coefficients multiplied by a regularization parameter (lambda). This penalty term encourages the coefficients of less important variables to be exactly zero, effectively performing variable selection by shrinking some coefficients to zero. In this study, LASSO was used to construct the prognostic model. A validation cohort from Xena-hubs Breast Cancer (Caldas)71 was used for validation of the model.
Protein–protein interaction network and hub gene
The protein–protein interaction network was constructed with STRING (https://string-db.org/), where interactions with a score greater than 0.4 were considered. The top 10 hub nodes in the network were identified using the Hubba72 (https://apps.cytoscape.org/apps/cytohubba) in Cytoscape73 (https://cytoscape.org/). The algorithm used included Maximum Clique Cardinality (MCC), Density of Maximum Neighborhood Component (DMNC), Maximum Neighborhood Component (MNC), and Degree Centrality (Degree). The combination of these algorithms offers a comprehensive approach to unsupervised class discovery. Each algorithm provides a unique perspective on the dataset. MCC focuses on identifying densely connected subgraphs (cliques), DMNC evaluates the density of the neighborhood around each node, MNC finds the largest connected component, and Degree Centrality measures the importance of nodes based on their connections. By incorporating these diverse perspectives, the combined approach can capture different aspects of the underlying structure of the data. The algorithms complement each other's strengths and weaknesses. For example, while MCC is effective at identifying densely connected subgroups, Degree Centrality can highlight nodes that serve as central hubs within the network. This complementary nature ensures that a wider range of structural features within the data are considered, leading to a more comprehensive analysis.
Spatial sequencing analysis
The expression differences in invasive ductal cancer and ductal cancer in situ were analyzed using spatial sequencing data with SpatialDB (http://www.spatialomics.org/SpatialDB/index.php). Spatial sequencing, also known as spatial transcriptomics or spatially resolved transcriptomics, is a technology that allows researchers to study gene expression patterns within tissues while preserving spatial information. Spatial sequencing methods integrate high-throughput sequencing techniques with spatially resolved imaging approaches to generate spatially resolved gene expression profiles. These methods enable us to analyze gene expression patterns within intact breast cancer tissue sections, providing insights into the spatial organization of cells and tissues and their functional implications. The spatial sequencing data used in this study were published in a previous paper74. The invasive ductal cancer and ductal cancer in situ were labeled by a licensed clinical pathologist Dr Jielin Weng in the Department of Pathology, The Second Affiliated Hospital of Guangzhou Medical University. The data were visualized using the SpatialDB tools75.
Results
CD8Tex marker gene identification based on single immune cell sequencing
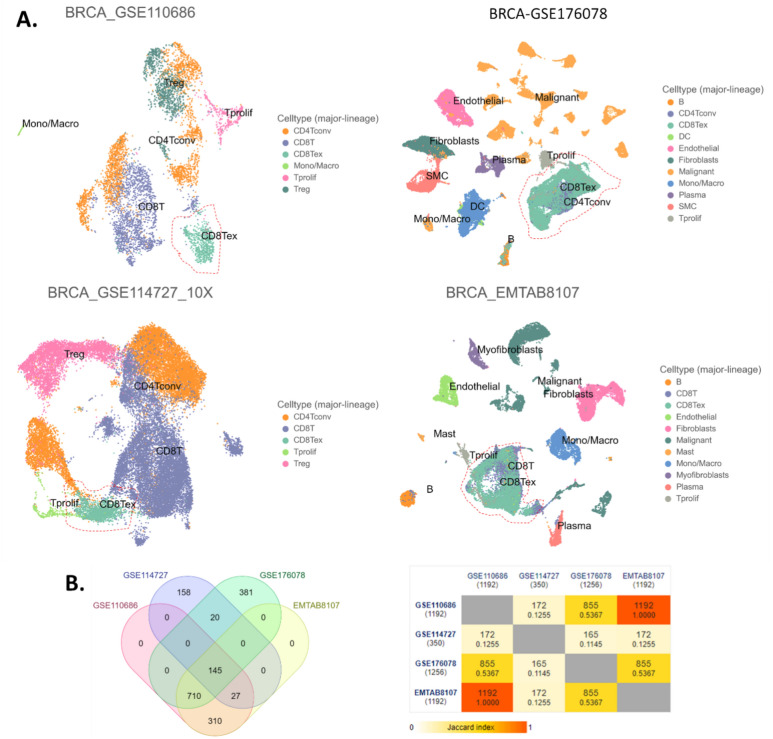
The UMAP plots were conducted for each single-cell sequence data set respectively (Fig. 1A). These cells were annotated and sorted into 17 cell types as shown in Table 2. Marker genes of CD8Tex cells were identified for each data set respectively, and the marker genes were cross-validated by intersection analysis of the marker gene sets obtained from different data sets (Fig. 1B left panel). The Jaccard index was calculated as shown in different colors in Fig. 1B right panel. Eventually, we obtained 145 marker genes for CD8Tex (Fig. 1B).
Figure 1.
CD8Tex marker genes identification based on single immune cell sequencing. (A) UMAP plot of breast cancer single immune cell sequencing data sets. (B) Intersection of marker genes identified by single immune cell sequencing data sets. Left panel: the Venn diagram. Right panel: pairwise intersections analysis.
Table 2.
Cell type abbreviation in single-cell data.
| Cell type | Abbreviation | Full name |
|---|---|---|
| Immune cells | B | B Cells |
| CD4T | CD4 T Cells | |
| CD4Tconv | Conventional CD4 T Cells | |
| CD8T | CD8 T Cells | |
| CD8Tex | Exhausted CD8 T Cells | |
| DC | Dendritic Cells | |
| Mono/Macro | Monocytes or Macrophages | |
| Mast | Mast Cells | |
| Neutrophils | Neutrophils | |
| NK | Natural Killer Cells | |
| Tprolif | Proliferating T Cells | |
| Treg | Regulatory T Cells | |
| Stromal cells | Endothelial | Endothelial CELLS |
| Fibroblasts | Fibroblasts | |
| Myofibroblasts | Myofibroblasts | |
| Cancer cells | Malignant | Malignant cells |
| Other cells | Oligodendrocyte | Oligodendrocytes |
Clustering of CD8Tex-based subtypes
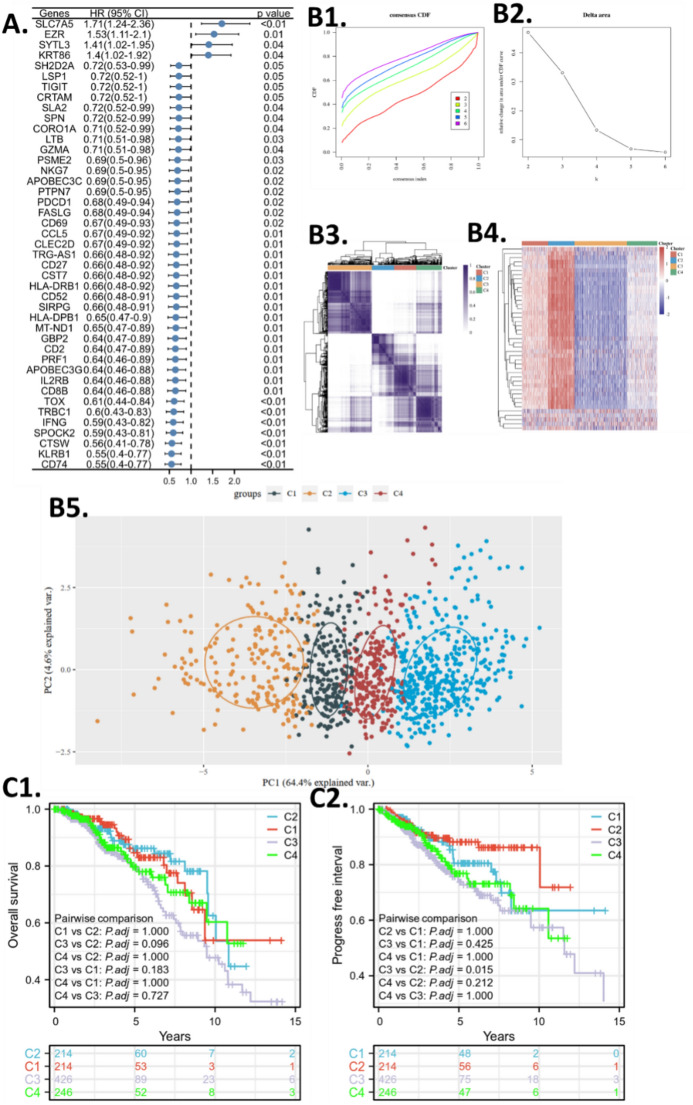
To further study the CD8Tex cells in breast cancer, we collected TCGA cohort to join-analyze the CD8Tex marker genes. The basic clinical information was provided in the Supplementary Table. All of these marker genes indeed have potential value for clinical application, however, the survival association of these genes revealed their practical value as a molecule that remarkably affects the progress of the tumor, hence, we believe that those genes that significantly impact patient survival are more likely to make a clinical difference when used as biomarkers or therapeutic targets. First, we excluded genes that do not affect the survival of breast cancer by conducting an overall survival comparison between high and low-gene expression groups (divided by expression medium). The univariate analysis was performed and the hazard ratio of significant genes was shown in Fig. 2A. Only four genes were risk factors for breast cancer patients, while the others were protective factors.
Figure 2.
CD8Tex-based subtype clustering. (A) Survival screening of the CD8Tex marker genes. The forest plot shows the hazard ratio (HR). (B1) Consensus Cumulative Distribution Function (CDF) plot of subtype numbers (k = 2–6). (B2) Delta area plot of the consensus CDF plot. (B3) Consensus matrix and cluster trees of subtypes. (B4) Gene expression heatmap of the subtypes. (B5) Principal Component Analysis (PCA) plot of the consensus clustering. (C1) Overall survival Kaplan–Meier plot (KM plot) of the subtypes. (C2) Progression-free interval KM plot of the subtypes.
We admitted that not all markers are highly specific for exhausted T-cells, the criteria to collect them is because their expression is significantly different from other cell types, thus, the expression of these marker genes can help define the distinctive expression patterns of cell infiltration features in bulk sequencing data. We aimed to investigate breast cancer with different CD8Tex infiltration levels, thus, consensus clustering was used to cluster TCGA BRCA patients into CD8Tex-based subtypes using the identified marker genes. Based on the consensus cumulative distribution function (CDF) plotting, the number of clusters (K) = 4 was the optimum cluster number for all of these clustering (Fig. 2B1, 2). The subtyping approach has been applied to help understand the role of a gene set76,77. By the NMF method, which is an effective dimension reduction method for cancer subtype identification, patients were clustered into four distinct subtypes (Fig. 2B3). The heatmap in Fig. 2B4 illustrates the differential expression patterns of marker genes across the breast cancer subtypes (C1, C2, C3, and C4), while the PCA plot in Fig. 2B5 demonstrates the clustering of patients based on their gene expression profiles within each subtype. (C1, C2, C3, and C4) (Fig. 2B4, 5). The heatmap and PCA plots in Fig. 2B4, B5 visually depict the distinct expression patterns among the identified breast cancer subtypes. The heatmap color scale ranges from low expression (blue) to high expression (red), facilitating the interpretation of differential expression patterns across subtypes. In Fig. 2B4, the heatmap visually represents the relative expression levels of marker genes across the identified breast cancer subtypes (C1, C2, C3, and C4). Each row in the heatmap corresponds to a gene, and each column represents a patient sample, with color intensity indicating the expression level of each gene. This visualization allows us to observe patterns of differential expression among the subtypes, highlighting potential molecular distinctions. In contrast, Fig. 2B5 utilizes PCA (Principal Component Analysis) to explore the overall variation in gene expression profiles among patients within each subtype. PCA is a dimensionality reduction technique that identifies patterns in data and visualizes these patterns by projecting patients onto a reduced-dimensional space defined by principal components. The PCA plot helps to visualize how patients cluster based on their gene expression profiles, providing insights into the similarities and differences among subtypes beyond individual gene expression levels. We analyzed the overall survival and progress-free interval of the subtypes and found that the subtyping failed to distinguish different survival patients except for the progress-free interval of C3 versus C2 (Fig. 2C1,2).
Immune difference of CD8Tex-based subtypes
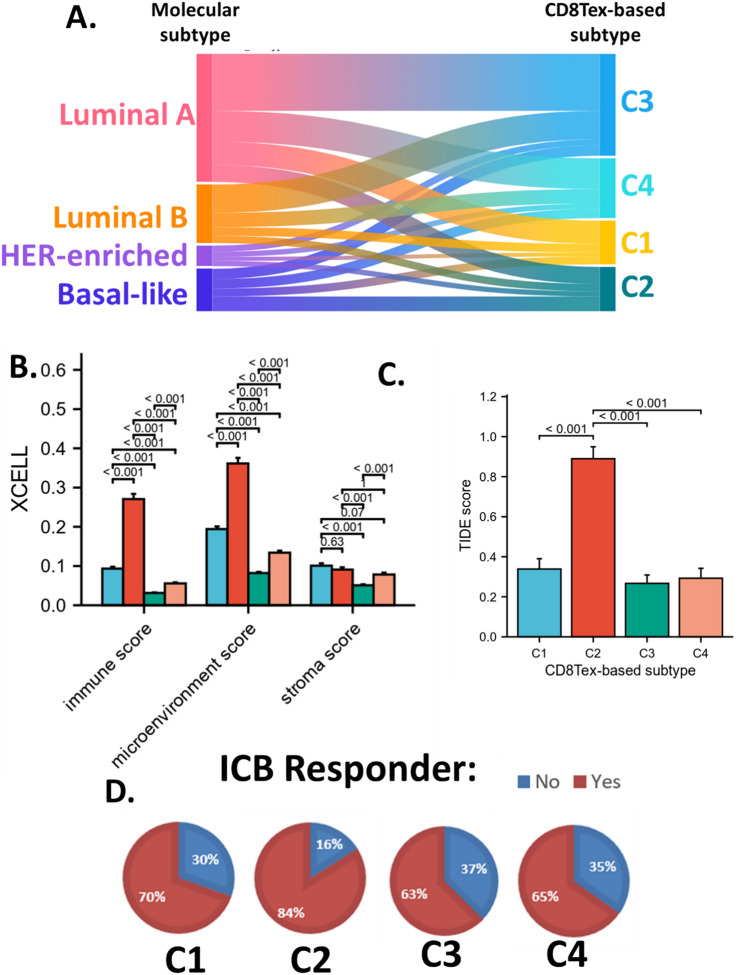
In addition, we also display the association between molecular subtypes and CD8Tex-based subtypes with a Sankey diagram (Fig. 3A). Generally, CD8Tex-based subtypes were not associated with the molecular subtypes. In addition, we also calculated the immune cell infiltration levels of the CD8Tex-based subtypes using multiple algorithms as provided in the supplementary materials. The XCELL algorithms suggested that the CD8Tex-based subtypes separated samples with different immune profiles. The immune score and microenvironment score were remarkably different among CD8Tex-based subtypes. The stroma scores were also significantly different among CD8Tex-based subtypes. The C2 subtypes had a very high immune score and microenvironment score. (Fig. 3B).
Figure 3.
Immune association of the CD8Tex-based subtypes. (A) Sankey diagram showing the association between breast cancer molecular subtypes and CD8Tex-based subtypes. (B) The XCELL scores in CD8Tex-based subtypes. (C) Tumor Immune Dysfunction and Exclusion (TIDE) scores in CD8Tex-based subtypes. (D) Predicted immune checkpoint blockade (ICB) response of the subtypes based on the TIDE score.
To demonstrate the potential clinical value of these subtyping systems, we calculated the TIDE score to predict the response of the samples for ICB therapy. Results revealed that the C2 subtype had a significantly higher TIDE score compared to the other subtypes (Fig. 3C). This indicates that C2 subtypes had low T cell response. Based on the TIDE score, we predicted the response of each sample for ICB and calculated the response ratio for each subtype. Results showed that C1 had a 70% response rate, C3 had a 63% response rate, and C4 had a 65% response rate, yet, C2 had an 84% response rate (Fig. 3D). It is not surprising that C2 with the highest immune score and microenvironment score also has highest response rate. We believe that this is because, although the results are derived from two distinct algorithms, there are common parameter genes utilized in their calculation. These results suggested that the subtyping systems performed well in the separation of patients with different immune relevance, especially identified C2 subtypes as the responder subtype for ICB, indicating that these marker-gene sets potentially provided clinical value for breast cancer immune therapies.
Expression difference of CD8Tex-based subtypes
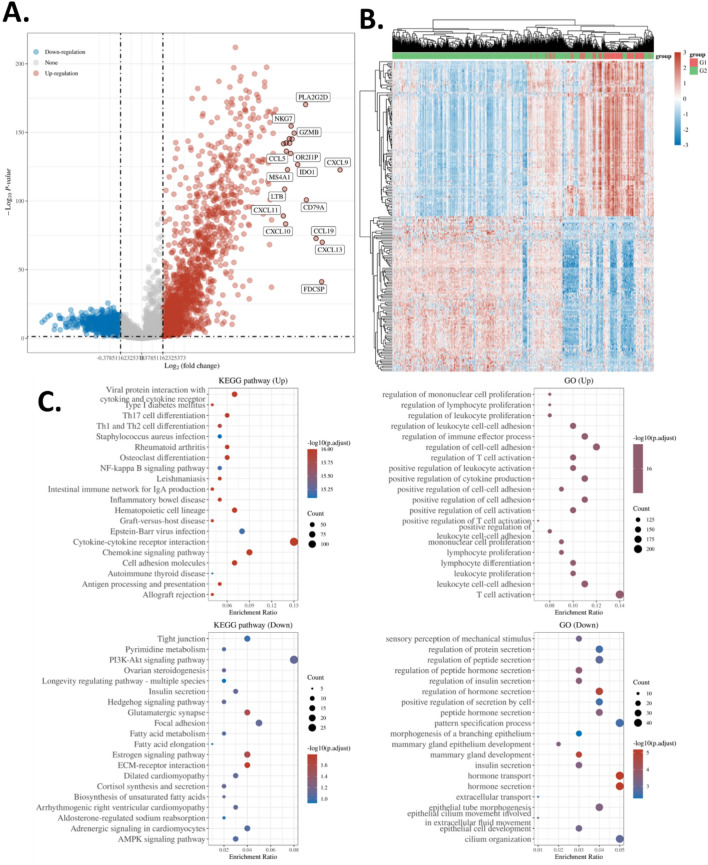
To further investigate the features of patients in C2 subtypes compared to the other patients, we conducted a differential expression gene analysis comparing C2 and the other subtypes. The analysis revealed 1552 up-regulated genes and 980 down-regulated genes in the C2 subtype as shown in the volcano plot (Fig. 4A) and heatmap with clustering (Fig. 4B). Figure 4B depicts a heatmap with hierarchical clustering illustrating the differential expression patterns of genes between the C2 subtype and other subtypes. The rows represent genes, while the columns represent patient samples. The color scale in the legend indicates gene expression levels, ranging from low (blue) to high (red), facilitating the interpretation of the heatmap. This visualization highlights distinct gene expression clusters associated with the C2 subtype, suggesting potential biomarkers or pathways specific to this subgroup.
Figure 4.
Differential expression gene enrichment. (A) Volcano plot of the differential genes in CD8Tex-based subtypes C2. TCGA BRCA cohorts were analyzed. C2 samples were compared with the other samples to identify C2 differential genes. (B) Heat map and clustering of the differential genes in CD8Tex-based subtypes C2. (C) GO and KEGG enrichment analysis78–80 of the differential genes in CD8Tex-based subtypes C2.
Subsequently, we enriched these genes in the GO database and KEGG pathways database. Results showed that the up-regulated genes were enriched in multiple terms that are related to the T cell activity. The KEGG pathway enrichment analysis revealed that the up-regulated genes were associated with cytokine interaction and Th1/Th2 differentiation. On the other hand, the down-regulated genes were associated with protein secretion and hormone secretion as well as the PI3K-Akt signaling pathway. (Fig. 4C) Although these DEGs and enrichment might related to the previously identified 145 biomarkers, the objective of this enrichment analysis is to broadly investigate the disparities within the C2 cluster and discern which pathways might underlie the diverse clinical phenotypes observed. Rather than pinpointing specific markers, this analysis aims to offer general insights into the biological mechanisms driving these clinical differences.
Construction of a CD8Tex-based survival model
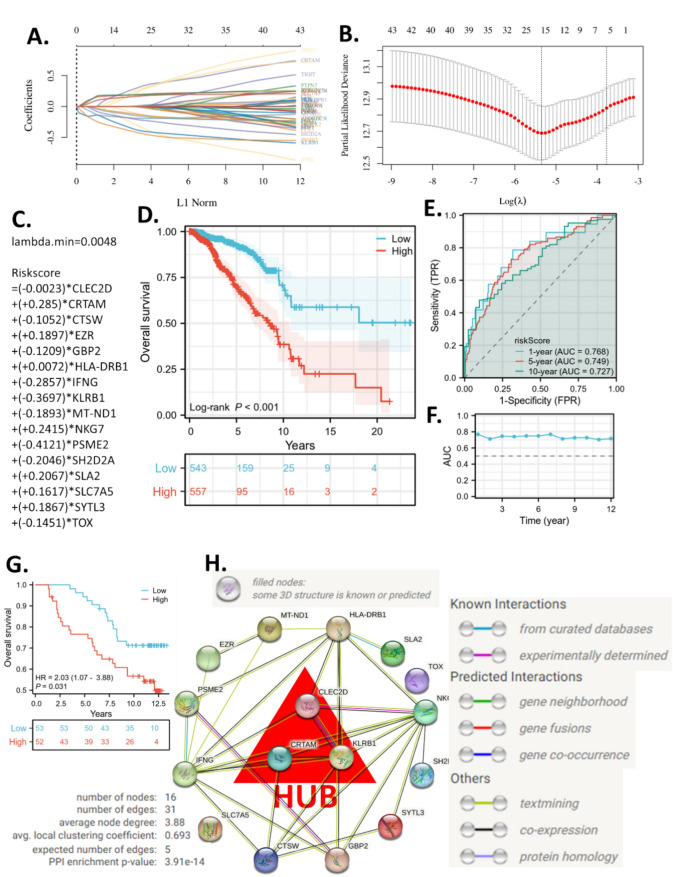
To further explore the prognostic value of the CD8Tex marker gene set and obtain critical genes for breast cancer patients, we trained a machine-learning prognostic model using the LASSO algorithm and TCGA BRCA cohort. The model suggested that the minimum lambda was 0.0048. When lambda was 0.0048, the model achieved the best performance. The risk score formula and the included genes are presented in Fig. 5A–C. The model includes risky genes (CLEC2D, CRTAM, EZR, HLA-DRB1, NKG7, SLA2, SLC7A5, and SYTL3) and protective genes (CTSW, GBP2, IFNG, KLRB1, MT-ND1, PSME2, SH2D2A, and TOX) which are discussed later. The KM plot suggested that the high-risk group had a significantly worse survival than the low-risk group (Fig. 5D). This model helped us narrow down the critical genes for CD8Tex in breast cancer. The time-dependent ROC analysis also revealed that the AUC of the ROC was over 7 for different years of prediction, indicating a good accuracy of the model for survival prediction (Fig. 5E, F). The model is validated with another independent cohort (Fig. 5G). The validation cohort results are consistent with those of the training cohort, demonstrating that the model can distinguish between patients with long survival and those with short survival.
Figure 5.
Machine learning prognostic models based on CD8Tex marker genes and hub genes identification. (A-C) The λ and coefficients of the model. (D) Overall survival KM plots of high- and low-risk groups in TCGA-BRAC. (E) Representative time-dependent receiver operating characteristic (ROC) curve of the risk model. (F) Area Under Curve (AUC) of the time-dependent ROC curve of the risk model. (G) Overall survival KM plots of high- and low-risk groups in a validation cohort from Xena-hubs Breast Cancer (Caldas 2007). (H) The protein–protein interaction network with hub genes identified.
Identification of the hub genes for the CD8Tex regulation network
The genes in the LASSO model were critical CD8Tex marker genes for breast cancer patients. We constructed a protein–protein interaction network using these genes and calculated the top three hub genes using MCC, DMNC, MNC, and Degree algorithms. The protein–protein interaction network presented 31 edges and the average node degree was 3.88 (Fig. 5H). The MCC, DMNC, MNC, and Degree of each gene were calculated as presented in Supplementary Table. We then ranked the scores and obtained the average rank of each gene. Eventually, CRTAM, CLEC2D, and KLRB1 stood out as the top three hub genes in critical CD8Tex marker genes for breast cancer patients.
CD8 T cell infiltration relevant to CD8Tex hub genes
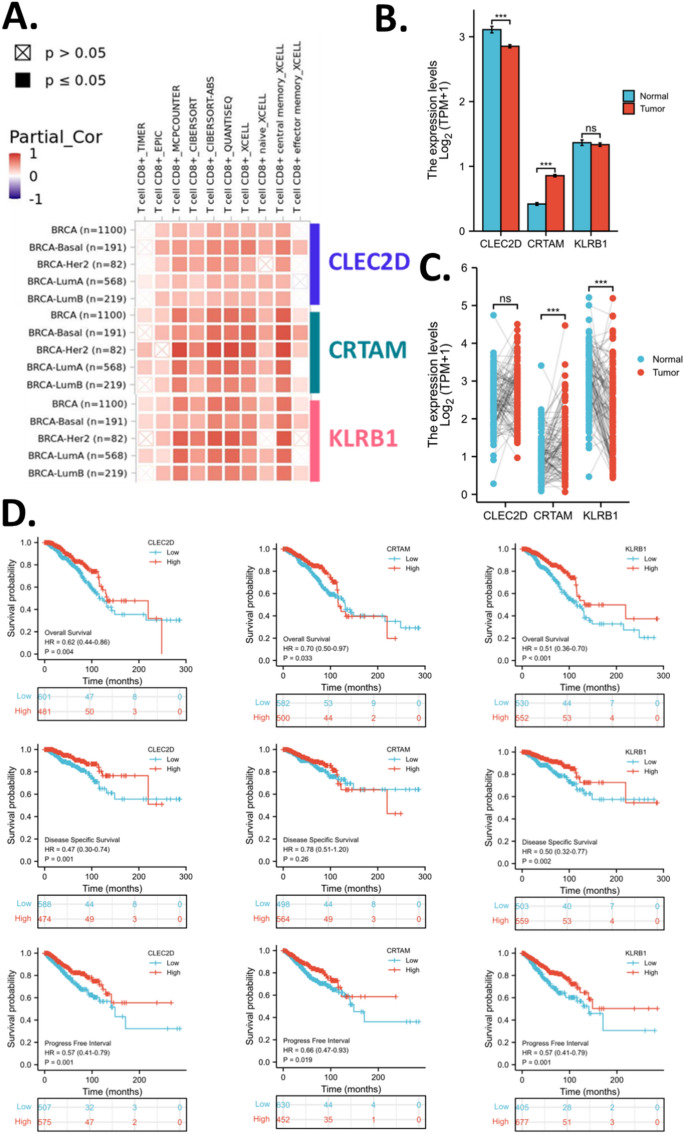
To evaluate the clinical value of the hub genes identified, CRTAM, CLEC2D, and KLRB1, for clinical evaluation of CD8 + T-cell infiltration levels in breast cancer, we calculated the different CD8 + T-cell infiltration levels and analyzed their correlation with CRTAM, CLEC2D, and KLRB1 in breast cancer and molecular subtypes. To avoid bias in some immune cell scores, we have employed multiple algorithms. Results showed that CRTAM, CLEC2D, and KLRB1 were positively correlated with T cell CD8 + in MCPCOUNTER, CIBERSORT, CIBERSORT-ABS, EPIC, and QUANTISEQ, but not in TIMER. (Fig. 6A).
Figure 6.
CD8Tex hub marker genes for breast cancer. (A) Correlation analysis between CD8Tex hub marker genes and CD8 + T cell infiltration scores. (B) Expression of CD8Tex hub marker genes in breast cancer and normal breast tissues. TCGA and GTEx data were analyzed. (C) Expression of CD8Tex hub marker genes in breast cancer and normal breast tissues. TCGA-paired samples were analyzed. (D) KM plot of the CD8Tex hub marker genes in breast cancer. Rolls 1–3 display overall survival, disease-free survival, and progress-free interval respectively. Columns 1–3 display CLEC2D, CRTAM, and KLRB1 respectively.
CD8Tex hub genes expression and survival analysis
Data also suggested that CLEC2D expression slightly decreased in tumors compared to normal breast tissue, while CRTAM expression increased in tumor tissue and KLRB1 expression had no difference in tumors compared to normal tissues (Fig. 6B). However, in the cancer-noncancer paired samples, the comparison suggested that CRTAM expression increased in tumor tissue while KLRB1 expression decreased in the tumor (Fig. 6C). Given the expression analysis results for CRTAM and KLRB1 are not consistent, we cannot determine the cancer-non-cancer expression pattern of these two genes definitively. However, based on consistent findings, we can conclude that tumors exhibit elevated expression of CRTAM. This suggests that tumors may possess a distinct CD8 + T-cell infiltration feature compared to normal tissues. Moreover, the expression levels of CRTAM and KLRB1 in normal breast tissue are comparable to those in breast cancer tissue. This suggests a potentially similar infiltration pattern of exhausted CD8 + T-cells in both cancer and non-cancer tissues. This similarity in pattern could hinder the development of cancer-specific targets for treatment.
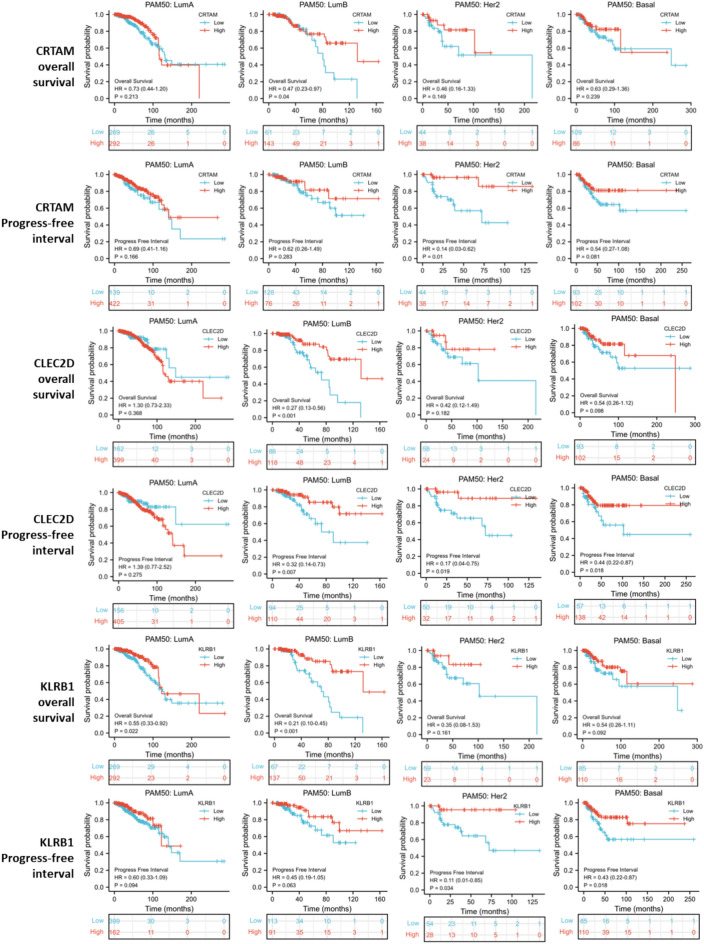
KM survival analysis revealed that CRTAM, CLEC2D, and KLRB1 were all significantly associated with better overall survival of breast cancer patients. For disease-free survival, CRTAM was not associated, while CLEC2D and KLRB1 were associated with better disease-free survival. As for the progress-free interval, all three genes were associated. (Fig. 6D) Survival association of CD8Tex hub marker genes for breast cancer subtypes was also analyzed. Results showed that CRTAM was a good prognostic biomarker for Luminal B, HER-enriched, and basal-like breast cancer, but not for Luminal A breast cancer. CLEC2D was associated with overall survival and progress-free interval in all subtypes of breast cancer. Similar to CRTAM, KLRB1 was a good prognostic biomarker for Luminal B, HER-enriched, and basal-like breast cancer, but not for Luminal A breast cancer. However, KLRB1 was not significant in the overall survival analysis of basal-like breast cancer. (Fig. 7).
Figure 7.
Survival association of CD8Tex hub marker genes for breast cancer subtypes.
Clinical association of the CD8Tex hub genes in breast cancers
The expression of CD8Tex hub genes has been demonstrated to be critical markers for CD8 + T cells in breast cancer. While CD8 + T cells are critical for breast cancer development, we proposed that CLEC2D, CRTAM, and KLRB1 are associated with clinical characteristics. The comparison of clinical information between high and low-expression groups suggested that CLEC2D was associated with T stage, age, and radiation therapy of breast cancer patients. CRTAM was associated with the N stage, race, age, PAM50 (molecular subtype), and radiation therapy of breast cancer patients. KLRB1 was associated with the M stage, race, age, PAM50, menopause status, and radiation therapy of breast cancer patients (Supplementary Table). In the high-expression group of CLEC2D, CRTAM, and KLRB1, more patients underwent radiation therapy. In general, radiation therapy is often recommended for patients who are in relatively good condition and able to tolerate treatment. However, it can also be administered to patients with more advanced disease or compromised health status, particularly if the potential benefits of treatment outweigh the risks. Therefore, the association inferred from our data suggests that high expression of CLEC2D, CRTAM, and KLRB1 may be associated with less severe cases where radiation therapy is recommended, possibly because patients are resistant to other types of treatment such as chemotherapy. CLEC2D was associated with the T stage indicating that it results in smaller tumors, which might reflect the impact of T cells on tumor growth. Tumor-infiltrating T cells have been associated with smaller tumor size81. CRTAM was associated with a slightly higher lymph node metastasis (N stage), which might reflect the impact of T cells on lymph node metastasis. Tumor-infiltrating lymphocytes, including cytotoxic CD8 + T cells, can recognize and eliminate cancer cells within lymph nodes, thereby inhibiting the spread of metastatic disease82. The association of KLRB1 with decreased tumor metastasis (M stage) suggests a potential role for CD8Tex cells in limiting breast cancer metastasis. Taken together, these results suggest that patients with CD8Tex may have smaller tumors with locoregional metastases, a clinical scenario commonly addressed with post-surgical radiotherapy. The association of CD8Tex hub genes with clinical characteristics, particularly the link with radiotherapy, raises intriguing questions about the immune activity status in breast cancer patients. The observed associations between CLEC2D, CRTAM, and KLRB1 expression levels and various clinical parameters, including tumor stage, lymph node involvement, molecular subtype, menopause status, and receipt of radiotherapy, suggest potential implications for patient management and treatment outcomes. However, it is important to note that the observed differences were slight and may not have significant clinical implications.
These findings underscore the importance of considering the immune context of tumors in clinical decision-making and the potential implications for personalized treatment approaches. However, further investigation, including validation in independent cohorts and functional studies, is needed to fully elucidate the role of exhausted T-cells and their associated biomarkers in breast cancer progression and response to therapy.
CD8Tex hub genes and breast cancer heterogeneity
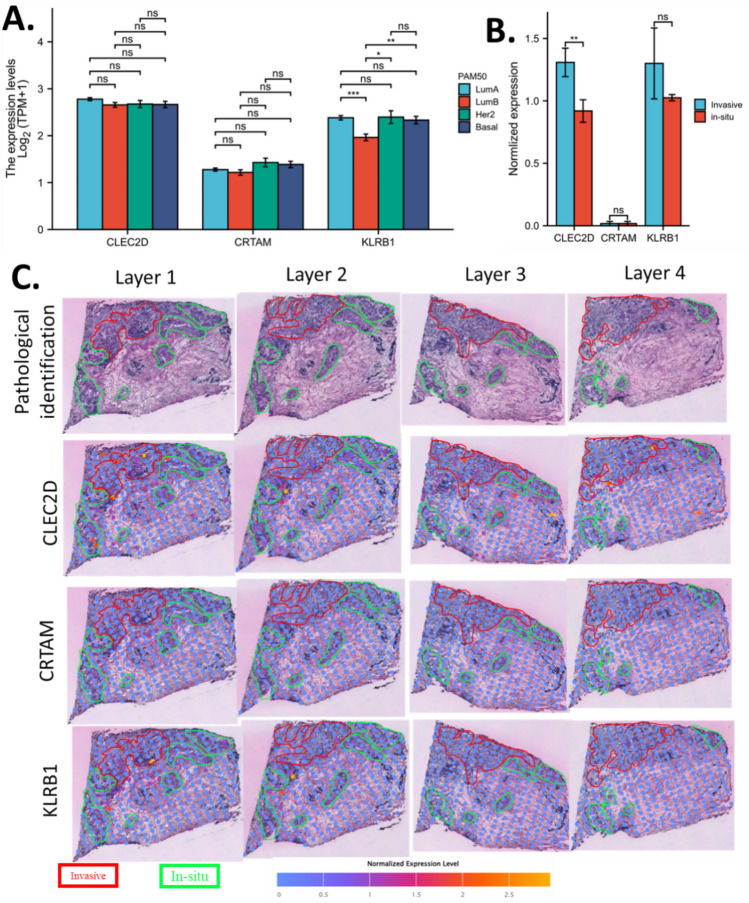
To investigate the CD8Tex hub genes and breast cancer extra-tumour heterogeneity, we plotted the expression level of CLEC2D, CRTAM, and KLRB1 in PAM50 breast cancer subtypes. Results revealed that CLEC2D and CRTAM do not have differences among PAM50 breast cancer subtypes, KLRB1 expression in Luminal B was significantly lower than the other subtypes. (Fig. 8A) To explore the CD8Tex hub genes and breast cancer intro-tumour heterogeneity, we also compared the levels of CLEC2D, CRTAM, and KLRB1 in invasive ductal cancer and ductal cancer in situ using spatial sequencing data. The invasive ductal cancer and ductal cancer in situ of breast cancer tissue were delineated by an experienced clinical pathologist and the expression of CLEC2D, CRTAM, and KLRB1 in invasive ductal cancer and ductal cancer in situ of breast cancer tissue were measured at 4 separate layers. We summed the results of 4 layers for analysis and results showed that CRTAM and KLRB1 expression was not significantly different between invasive ductal cancer and ductal cancer in situ, while CLEC2D expression was significantly higher in invasive ductal cancer over ductal cancer in situ(Fig. 8B). Images of the spatial sequencing slide were shown in Fig. 8C. These data suggested that KLRB1 could mark the difference between cancer types while CLEC2D could mark the difference between breast cancer tissue types within a sample. As CLEC2D and KLRB1 are two critical T cell genes, these data also reflect the potential role of T cell infiltration in breast cancer.
Figure 8.
Tumour heterogeneity association of CD8Tex hub marker genes. (A) Expression of CD8Tex hub marker genes in breast cancer subtypes. (B) Expression comparison of CD8Tex hub marker genes in invasive ductal cancer and ductal cancer in situ by spatial sequencing. C. Immages of the spatial expression of CD8Tex hub marker genes in invasive ductal cancer and ductal cancer in situ.
Pan-cancer expression and survival analysis of CD8Tex hub genes
To further expand the application of the CD8Tex hub genes for pan-cancer use, we systematically explored the expression of CLEC2D, CRTAM, and KLRB1 across cancer types, which is provided in the Supplementary results.
Discussion
Tumor-associated immune cells may promote or inhibit cancer cells depending on their function and role in immunity. Many previous studies have suggested the impact of the immune environment on cancers30,32,34,36–38,61,83–86. The novelty of this study lies in our approach to identifying marker genes for CD8Tex using multiple immune single-cell sequencing datasets. By conducting intersection analysis across multiple independent single-cell datasets, we aimed to ensure the stability and reproducibility of the identified biomarker gene sets. Furthermore, we identified hub genes for these biomarkers, namely CLEC2D, CRTAM, and KLRB1, which have been less explored in the context of breast cancer. Of significant importance is our discovery of the association between these hub genes and various aspects of breast cancer immunity, clinical features, and tumor heterogeneity. This comprehensive analysis sheds light on previously overlooked molecular players in breast cancer and highlights their potential relevance for understanding disease progression and treatment response. Overall, our study offers valuable insights into the immune landscape of breast cancer and uncovers potential targets for further investigation and therapeutic intervention.
In the CD8Tex-based survival model we constructed, the risky genes include CLEC2D, CRTAM, EZR, HLA-DRB1, NKG7, SLA2, SLC7A5, and SYTL3, while the protective genes include CTSW, GBP2, IFNG, KLRB1, MT-ND1, PSME2, SH2D2A, and TOX. Most of these genes are reported for the first time as prognostic biomarkers in breast cancer, although some have been previously associated with breast cancer treatment or the breast cancer microenvironment. CRTAM enhances the immune response against tumors in triple-negative breast cancer by increasing the infiltration of CD8 + T cells87. An integrative multi-omics analysis identifies a gene signature related to metastasis, highlighting the potential oncogenic role of EZR in breast cancer88. Breast cancer is associated with increased allele frequencies of HLA-DRB1*11:01 and HLA-DRB1*10:01 in a population of patients from Central Italy89; however, this pertains to genetic mutation rather than gene expression. Targeting the glutamine metabolic reprogramming mediated by SLC7A5 enhances the effectiveness of anti-PD-1 therapy in triple-negative breast cancer90. GBP2 serves as a prognostic biomarker and is linked to immunotherapeutic responses in gastric cancer91; however, its association with breast cancer has not been reported. As a single-cell signature, low expression of KLRB1 predicts poor survival outcomes and is associated with immune infiltration in breast cancer92. It is also reported that inhibiting KLRB1 expression is associated with impaired cancer immunity, leading to cancer progression and poor prognosis in patients with breast invasive carcinoma93. Data suggested that PSME2 was associated with immune-hot tumors in breast cancer and with a favorable therapeutic response to immunotherapy94. Overall, our results align with previous studies regarding some of these genes. However, we emphasize their prognostic value and have included additional novel biomarker genes to enhance the overall prognostic accuracy of our model.
The cell types identified with biomarker gene sets all play important roles in the immunity of cancers. Tumor-infiltrating B cells are an essential part of the antitumor action of T cells95. CD8 + T cells (CD8T) and CD4 + T cells have a similar differentiation process but once differentiated, the CD4 + cells or the CD8 + cells are fixed and function differently96. Conventional CD4 + T cells (CD4Tconv) lack phenotypic markers that distinguish these cells from FoxP3 + T regulatory cells (Treg)95. CD8T includes cytotoxic T cells, which directly target cancer cells and induce apoptosis of cancer cells97. During cancer progression, cancer-associated fibroblasts, M2 macrophage, and Treg might negatively regulate anti-cancer immune responses mediated by CD8 + T cells98. Macrophages are monocytes that have migrated from the bloodstream into tumor tissues, which serve as scavengers, maintaining homeostasis in tumors 99. The effect of myofibroblast on cancer immunity has been less reported. A study has shown that myofibroblast regulated type I collagen thereby impacting cancer immunity100. It was reported that the tumor-infiltrating T cells directly in contact with the cancer cells proliferate more frequently compared with T cells in the stroma101, hence, proliferating T cells (Tprolif) were thought to be a subline of T cells. Exhausted CD8 T Cells (CD8Tex) is a set of sub-cell lines of CD8 T Cells that persist in cancer but are unsuccessful in killing cancer cells102. The presence and the type of CD8Tex have been thought to be critical for the response of cancer to some immune checkpoint blockades103. Therefore, the abundance of these immune-related cells in tumors is a critical factor for tumorigenesis as well as cancer immune therapy.
The analysis of the immune association of subtypes based on marker genes for different cell types suggested that the subtyping systems performed well in the separation of patients with different immune relevance. The subtyping identified C2 as standing out from the other patients. Interestingly, the subtypes also performed well in the separation between ICB responders and ICB non-responders. These ICB predictions were based on TIDE scores. However, the TIDE score was designed to predict response to immune checkpoint blockade, including anti-PD1 and anti-CTLA4, for melanoma and NSCLC. The use of TIDE in this study is based on the assumption that breast cancer has a similar immune system as melanoma and NSCLC. Hence, one should be cautious in interpreting these results. Further study to define a prediction model specific to breast cancer is required when more breast cancer immune therapy cohort data are available for model training. In the comparison of survival models, we concluded that the Tprolif model was the best overall. However, the other model also provided a rather comparative prognostic power. Nevertheless, the main purpose of this analysis is to narrow down the critical genes for the subsequent study. Another critical limitation is the observations in special sequencing data might be subjected to bias. Firstly, it's essential to note that the resolution of the spatial sequencing results is relatively low, and some identified blocks may encompass both invasive ductal carcinoma and ductal carcinoma in situ within breast cancer tissue. Secondly, the classification of invasive ductal carcinoma and ductal carcinoma in situ is determined by a licensed clinical pathologist using her clinical expertise and knowledge, rather than relying on a standard computational pipeline for detection. Additionally, drawing conclusions solely based on these biomarkers for T cell infiltration levels may carry significant risk. Hence, our data offer only initial insights into the biomarkers, and further investigation into the association of CD8Tex heterogeneity is warranted.
In this study, three critical hub genes were identified, including CLEC2D, CRTAM, and KLRB1. NK cells play a central role in the immune system, particularly in the recognition and elimination of malignant and infected cells. 2B4 (CD244, SLAMF4) and CS1 (CD319, SLAMF7) are NK cell receptors that control their cytotoxic function. Lectin-like transcript 1 (LLT1), a member of the C-type lectin-like domain family 2 (CLEC2D), induces IFN-g production but does not directly regulate cytolysis. LLT1, which is expressed in other cells, acts as a ligand for the NK cell inhibitory receptor NKRP1A (CD161) and suppresses NK cytolytic activity. Research has been conducted on novel therapies that target these receptors to enhance NK cell effector functions104. Hence, CLEC2D is associated with the NK-CD8 + cell regulation. On the other hand, CD4( +)T cells possessing CRTAM can differentiate into CD4( +)CTLs. A study found that after activation, CRTAM( +) CD4( +) T cells secrete IFN-γ and express CTL-related genes such as eomesodermin, Granzyme B, and perforin, which suggest that CRTAM( +) T cells are the precursor of CD4( +)CTLs105. Additionally, ectopic expression of CRTAM in T cells induces IFN-γ production, expression of CTL-related genes, and cytotoxic activity. This is further supported by the fact that CRTAM-mediated intracellular signaling is required for the induction of both CD4( +)CTLs and IFN-γ production105. Furthermore, these CRTAM( +) T cells traffic to mucosal tissues and inflammatory sites and develop into CD4( +)CTLs, which can either mediate protection against infection or induce inflammatory response depending on the situation. These results demonstrate that CRTAM is a key factor in the differentiation of CD4( +)CTLs through the induction of Eomes and CTL-related genes105. As for KLRB1, this is a gene that encodes CD161 protein. CD161 has been proposed as a pan-cancer immune checkpoint106. Therefore, it is not surprising that, in this study, we demonstrated that these two genes were closely associated with the cancer immune environment and can be used to predict the immune therapy response.
The mining and analysis of multi-omic profiling data enable bioinformatic study with a rather comprehensive understanding of genes in cancers. In this study, combining single-cell RNA sequencing (scRNA-seq), bulk RNA sequencing (bulk RNA-seq), and spatial transcriptomics data would indeed constitute a multi-omic approach, provided that all of these datasets are derived from RNA sequencing (RNA-seq) technologies. While each type of RNA-seq data captures gene expression information at different scales and resolutions, integrating them allows for a more comprehensive understanding of biological systems. Integrating scRNA-seq, bulk RNA-seq, and spatial transcriptomics data allows researchers to analyze gene expression dynamics at multiple scales, from individual cells to tissue-level spatial organization. This multi-omic approach enables the identification of cell types, subpopulations, and spatially defined gene expression patterns, facilitating a deeper understanding of complex biological processes and disease mechanisms.
The perspective of this work is to deepen our understanding of the role of CD8Tex in breast cancer. While immune therapy is not yet widely utilized in breast cancer treatment, there is promise in using T cell-based immune therapy for some refractory breast cancer cases. CD8 + Tex cells might also be developed as a drug target, as numerous studies have reported interactions involving genes, drugs107, diseases, cells, and even the microbiome108–112. Whether CD8 + Tex cells play a role in these interactions can be explored in future research. A recent study found an association between breast cancer and menopause113. Further investigation is needed to determine whether menopause affects CD8 + T cells in breast cancer. One of the biggest issues in breast cancer is drug resistance114. Addressing the challenges in this field necessitates exploration. Our identification of hub genes for these biomarkers, such as CLEC2D, CRTAM, and KLRB1, and their potential association with known biomarkers of breast cancer cells, such as CHEK2115 and TP53116, lays the groundwork for potential clinical applications in breast cancer management. Further research is needed to explore how these biomarkers can be effectively incorporated into clinical practice to improve patient outcomes. Additionally, non-invasive early detection of breast cancer has recently shown promising advancements117. The biomarkers discovered may have the potential to contribute to the non-invasive detection of this cancer type.
Supplementary Information
Acknowledgements
The authors of the paper acknowledge the support and help of Yaqi Yang, Bryan Liu, Weifen Chen, Zongxiong Liu, and Kashvi Agarwal.
Author contributions
The analyses were done by H.L. J.W., P.W., and H.L. wrote the paper. A.D. and A.M.R. edited and polished the paper and contributed to the reanalysis of the results. J.W. and P.W. supervised the project.
Funding
Panpan Wang received funding from the K. C. Wong Education Foundation, the Natural Science Foundation of China (81603342), the Guangdong Basic and Applied Basic Research Foundation(2022A151501264, 2022A1515012641, 2024A1515012948), the Guangzhou Science and Technology Project (SL2023A03J00309, 2024A03J0154, 2023B01J1004), and the Guangdong Provincial Bureau of Traditional Chinese Medicine Research Project (20221107).
Data availability
Data were available from the open-access databases, and the links for the data sources were provided: TCGA (https://www.cancer.gov/ccg/research/genome-sequencing/tcga), GTEx (https://gtexportal.org/home/), STRING (https://string-db.org/), TISCH2(http://tisch.comp-genomics.org/home/), and SpatialDB (http://www.spatialomics.org/SpatialDB/index.php).
Code availability
All codes used in this study are open-source codes and are available from the author with a reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Panpan Wang, Email: wangpp@jnu.edu.cn.
Jieling Weng, Email: jieling_weng@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-70184-1.
References
- 1.Sonkin, D., Thomas, A. & Teicher, B. A. Cancer treatments: Past, present, and future. Cancer Genet.286–287, 18–24. 10.1016/j.cancergen.2024.06.002 (2024). 10.1016/j.cancergen.2024.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49. 10.3322/caac.21820 (2024). 10.3322/caac.21820 [DOI] [PubMed] [Google Scholar]
- 3.Xia, C. et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin. Med. J.135, 584–590. 10.1097/cm9.0000000000002108 (2022). 10.1097/cm9.0000000000002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britt, K. L., Cuzick, J. & Phillips, K. A. Key steps for effective breast cancer prevention. Nat. Rev. Cancer20, 417–436. 10.1038/s41568-020-0266-x (2020). 10.1038/s41568-020-0266-x [DOI] [PubMed] [Google Scholar]
- 5.Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72, 7–33. 10.3322/caac.21708 (2022). 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 6.Lei, X. et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett.470, 126–133. 10.1016/j.canlet.2019.11.009 (2020). 10.1016/j.canlet.2019.11.009 [DOI] [PubMed] [Google Scholar]
- 7.Dieci, M. V., Miglietta, F. & Guarneri, V. Immune infiltrates in breast cancer: Recent updates and clinical implications. Cells10.3390/cells10020223 (2021). 10.3390/cells10020223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med.363, 711–723 (2010). 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robert, C. et al. Durable complete response after discontinuation of pembrolizumab in patients with metastatic melanoma. J. Clin. Oncol.36, 1668–1674 (2018). 10.1200/JCO.2017.75.6270 [DOI] [PubMed] [Google Scholar]
- 10.Wang, J. et al. Role of immune checkpoint inhibitor-based therapies for metastatic renal cell carcinoma in the first-line setting: A Bayesian network analysis. EBioMedicine47, 78–88 (2019). 10.1016/j.ebiom.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vonderheide, R. H., Domchek, S. M. & Clark, A. S. Immunotherapy for breast cancer: What are we missing?. Clin. Cancer Res.23, 2640–2646 (2017). 10.1158/1078-0432.CCR-16-2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emens, L. A. et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer: biomarker evaluation of the IMpassion130 study. J. Natl. Cancer Inst.113, 1005–1016. 10.1093/jnci/djab004 (2021). 10.1093/jnci/djab004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loi, S. et al. Tumor-infiltrating lymphocytes and prognosis: A pooled individual patient analysis of early-stage triple-negative breast cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol.37, 559–569. 10.1200/jco.18.01010 (2019). 10.1200/jco.18.01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwapisz, D. Pembrolizumab and atezolizumab in triple-negative breast cancer. Cancer Immunol Immunother70, 607–617. 10.1007/s00262-020-02736-z (2021). 10.1007/s00262-020-02736-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latif, F. et al. Atezolizumab and pembrolizumab in triple-negative breast cancer: A meta-analysis. Exp. Rev. Anticancer Therapy22, 229–235. 10.1080/14737140.2022.2023011 (2022). 10.1080/14737140.2022.2023011 [DOI] [PubMed] [Google Scholar]
- 16.Ruffell, B. et al. Leukocyte composition of human breast cancer. Proc. Natl. Acad. Sci. USA109, 2796–2801. 10.1073/pnas.1104303108 (2012). 10.1073/pnas.1104303108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.König, L. et al. Dissimilar patterns of tumor-infiltrating immune cells at the invasive tumor front and tumor center are associated with response to neoadjuvant chemotherapy in primary breast cancer. BMC Cancer19, 120. 10.1186/s12885-019-5320-2 (2019). 10.1186/s12885-019-5320-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savas, P. et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med.24, 986–993. 10.1038/s41591-018-0078-7 (2018). 10.1038/s41591-018-0078-7 [DOI] [PubMed] [Google Scholar]
- 19.Byrne, A. et al. Tissue-resident memory T cells in breast cancer control and immunotherapy responses. Nat. Rev. Clin. Oncol.17, 341–348. 10.1038/s41571-020-0333-y (2020). 10.1038/s41571-020-0333-y [DOI] [PubMed] [Google Scholar]
- 20.Zhu, Z., Jiang, L. & Ding, X. Advancing breast cancer heterogeneity analysis: insights from genomics, transcriptomics and proteomics at bulk and single-cell levels. Cancers10.3390/cancers15164164 (2023). 10.3390/cancers15164164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, J., Guo, Z. & Wang, L. Progress and challenges of immunotherapy predictive biomarkers for triple negative breast cancer in the era of single-cell multi-omics. Life (Basel)10.3390/life13051189 (2023). 10.3390/life13051189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nolan, E., Lindeman, G. J. & Visvader, J. E. Deciphering breast cancer: from biology to the clinic. Cell186, 1708–1728. 10.1016/j.cell.2023.01.040 (2023). 10.1016/j.cell.2023.01.040 [DOI] [PubMed] [Google Scholar]
- 23.Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature512, 155–160 (2014). 10.1038/nature13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung, W. et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun.8, 15081 (2017). 10.1038/ncomms15081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. J., Liang, W., Kang, C.-C., Pegram, M. D. & Herr, A. E. Single-cell immunoblotting resolves estrogen receptor-α isoforms in breast cancer. Plos one16, e0254783 (2021). 10.1371/journal.pone.0254783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlinger, M. & Swanton, C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br. J. Cancer103, 1139–1143 (2010). 10.1038/sj.bjc.6605912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satam, H. et al. Next-generation sequencing technology: Current trends and advancements. Biology (Basel)10.3390/biology12070997 (2023). 10.3390/biology12070997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiovitz, S. & Korde, L. A. Genetics of breast cancer: A topic in evolution. Ann. Oncol.26, 1291–1299 (2015). 10.1093/annonc/mdv022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griseri, P. & Pagès, G. Regulation of the mRNA half-life in breast cancer. World J. Clin. Oncol.5, 323 (2014). 10.5306/wjco.v5.i3.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, H. & Weng, J. A comprehensive bioinformatic analysis of cyclin-dependent kinase 2 (CDK2) in glioma. Gene10.1016/j.gene.2022.146325 (2022). 10.1016/j.gene.2022.146325 [DOI] [PubMed] [Google Scholar]
- 31.Liu, H. & Tao, T. Pan-cancer genetic analysis of cuproptosis and copper metabolism-related gene set. Front. Oncol.10.3389/fonc.2022.952290 (2022). 10.3389/fonc.2022.952290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, H. Pan-cancer profiles of the cuproptosis gene set. Am. J. Cancer Res.12, 4074–4081 (2022). [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, H. Pan-cancer profiles of the cuproptosis gene set. Res. Square10.21203/rs.3.rs-1716214/v1 (2022). 10.21203/rs.3.rs-1716214/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Y. & Liu, H. Clinical powers of aminoacyl tRNA synthetase complex interacting multifunctional protein 1 (AIMP1) for head-neck squamous cell carcinoma. Cancer Biomark. Sect. A Dis. Mark.10.3233/cbm-210340 (2022). 10.3233/cbm-210340 [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., Liu, H. & Han, Y. Potential Roles of Cornichon Family AMPA Receptor Auxiliary Protein 4 (CNIH4) in Head and Neck Squamous Cell Carcinoma. Research Square (2021). [DOI] [PubMed]
- 36.Liu, H. & Tang, T. MAPK signaling pathway-based glioma subtypes, machine-learning risk model, and key hub proteins identification. Sci. Rep.13, 19055. 10.1038/s41598-023-45774-0 (2023). 10.1038/s41598-023-45774-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, H. & Tang, T. Pan-cancer genetic analysis of disulfidptosis-related gene set. Cancer Genet.278–279, 91–103. 10.1016/j.cancergen.2023.10.001 (2023). 10.1016/j.cancergen.2023.10.001 [DOI] [PubMed] [Google Scholar]
- 38.Liu, H. & Tang, T. A bioinformatic study of IGFBPs in glioma regarding their diagnostic, prognostic, and therapeutic prediction value. Am. J. Transl. Res.15, 2140–2155 (2023). [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, H. & Tang, T. Pan-cancer genetic analysis of disulfidptosis-related gene set. bioRxiv, 2023.2002. 2025.529997 (2023). [DOI] [PubMed]
- 40.Hong, M. et al. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol.13, 166. 10.1186/s13045-020-01005-x (2020). 10.1186/s13045-020-01005-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X. & Wang, C. Y. From bulk, single-cell to spatial RNA sequencing. Int. J. Oral Sci.13, 36. 10.1038/s41368-021-00146-0 (2021). 10.1038/s41368-021-00146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell171, 1611-1624.e1624. 10.1016/j.cell.2017.10.044 (2017). 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med.24, 1277–1289. 10.1038/s41591-018-0096-5 (2018). 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- 44.Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell174, 1293-1308.e1236. 10.1016/j.cell.2018.05.060 (2018). 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim, C. et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell173, 879-893.e813. 10.1016/j.cell.2018.03.041 (2018). 10.1016/j.cell.2018.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali, H. R. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer1, 163–175. 10.1038/s43018-020-0026-6 (2020). 10.1038/s43018-020-0026-6 [DOI] [PubMed] [Google Scholar]
- 47.Wagner, J. et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell177, 1330–1345. 10.1016/j.cell.2019.03.005 (2019). 10.1016/j.cell.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, S. Q. et al. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J. Hematol. Oncol.15, 19. 10.1186/s13045-022-01236-0 (2022). 10.1186/s13045-022-01236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McRitchie, B. R. & Akkaya, B. Exhaust the exhausters: Targeting regulatory T cells in the tumor microenvironment. Front. Immunol.13, 940052. 10.3389/fimmu.2022.940052 (2022). 10.3389/fimmu.2022.940052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han, Y. et al. TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucl. Acids Res.51, D1425-d1431. 10.1093/nar/gkac959 (2023). 10.1093/nar/gkac959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, C. et al. Integrative analyses of single-cell transcriptome and regulome using MAESTRO. Genome Biol.21, 198. 10.1186/s13059-020-02116-x (2020). 10.1186/s13059-020-02116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888-1902.e1821. 10.1016/j.cell.2019.05.031 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, C. & Su, Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics31, 1974–1980. 10.1093/bioinformatics/btv088 (2015). 10.1093/bioinformatics/btv088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.10.1038/nbt.4314 (2018). 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- 55.Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet.53, 1334–1347. 10.1038/s41588-021-00911-1 (2021). 10.1038/s41588-021-00911-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian, J. et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res.30, 745–762. 10.1038/s41422-020-0355-0 (2020). 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell181, 442–459. 10.1016/j.cell.2020.03.048 (2020). 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 58.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36, 411–420. 10.1038/nbt.4096 (2018). 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.37, 38–44 (2019). 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- 60.Monti, S., Tamayo, P., Mesirov, J. & Golub, T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn.52, 91–118 (2003). 10.1023/A:1023949509487 [DOI] [Google Scholar]
- 61.Liu, H. Expression and potential immune involvement of cuproptosis in kidney renal clear cell carcinoma. Cancer Genet.274–275, 21–25. 10.1016/j.cancergen.2023.03.002 (2023). 10.1016/j.cancergen.2023.03.002 [DOI] [PubMed] [Google Scholar]
- 62.Reich, M. et al. GenePattern 2.0. Nat. Genet.38, 500–501. 10.1038/ng0506-500 (2006). 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- 63.Li, T. et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res.77, e108–e110. 10.1158/0008-5472.Can-17-0307 (2017). 10.1158/0008-5472.Can-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aran, D., Hu, Z. & Butte, A. J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol.18, 220. 10.1186/s13059-017-1349-1 (2017). 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M. & Alizadeh, A. A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. (Clifton, N.J.)1711, 243–259. 10.1007/978-1-4939-7493-1_12 (2018). 10.1007/978-1-4939-7493-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol.17, 218. 10.1186/s13059-016-1070-5 (2016). 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finotello, F. et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med.11, 34. 10.1186/s13073-019-0638-6 (2019). 10.1186/s13073-019-0638-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Racle, J. & Gfeller, D. EPIC: A tool to estimate the proportions of different cell types from bulk gene expression data. Methods Mol. Biol. (Clifton, N.J.)2120, 233–248. 10.1007/978-1-0716-0327-7_17 (2020). 10.1007/978-1-0716-0327-7_17 [DOI] [PubMed] [Google Scholar]
- 69.Fu, J. et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med.12, 21. 10.1186/s13073-020-0721-z (2020). 10.1186/s13073-020-0721-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med.16, 385–395. 10.1002/(sici)1097-0258(19970228)16:4%3c385::aid-sim380%3e3.0.co;2-3 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Chin, S. F. et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol.8, R215. 10.1186/gb-2007-8-10-r215 (2007). 10.1186/gb-2007-8-10-r215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin, C. Y. et al. Hubba: Hub objects analyzer–a framework of interactome hubs identification for network biology. Nucl. Acids Res.36, W438-443. 10.1093/nar/gkn257 (2008). 10.1093/nar/gkn257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504. 10.1101/gr.1239303 (2003). 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science353, 78–82. 10.1126/science.aaf2403 (2016). 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 75.Fan, Z., Chen, R. & Chen, X. SpatialDB: A database for spatially resolved transcriptomes. Nucl. Acids Res.48, D233-d237. 10.1093/nar/gkz934 (2020). 10.1093/nar/gkz934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng, X. et al. Molecular subtyping and immune score system by a novel pyroptosis-based gene signature precisely predict immune infiltrating, survival and response to immune-checkpoint blockade in breast cancer. Cancer Genet.276–277, 60–69. 10.1016/j.cancergen.2023.07.007 (2023). 10.1016/j.cancergen.2023.07.007 [DOI] [PubMed] [Google Scholar]
- 77.Li, W., Wu, H. & Xu, J. Construction of a genomic instability-derived predictive prognostic signature for non-small cell lung cancer patients. Cancer Genet.278–279, 24–37. 10.1016/j.cancergen.2023.07.008 (2023). 10.1016/j.cancergen.2023.07.008 [DOI] [PubMed] [Google Scholar]
- 78.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucl. Acids Res.28, 27–30. 10.1093/nar/28.1.27 (2000). 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. Publ. Soc.28, 1947–1951. 10.1002/pro.3715 (2019). 10.1002/pro.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucl. Acids Res.51, D587-d592. 10.1093/nar/gkac963 (2023). 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eerola, A.-K., Soini, Y. & Pääkkö, P. A high number of tumor-infiltrating lymphocytes are associated with a small tumor size, low tumor stage, and a favorable prognosis in operated small cell lung carcinoma. Clin. Cancer Res.6, 1875–1881 (2000). [PubMed] [Google Scholar]
- 82.Rye, I. H. et al. Breast cancer metastasis: immune profiling of lymph nodes reveals exhaustion of effector T cells and immunosuppression. Mol. Oncol.16, 88–103 (2022). 10.1002/1878-0261.13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu, H. & Weng, J. A pan-cancer bioinformatic analysis of RAD51 regarding the values for diagnosis, prognosis, and therapeutic prediction. Front. Oncol.10.3389/fonc.2022.858756 (2022). 10.3389/fonc.2022.858756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu, H. & Tang, T. Pan-cancer genetic analysis of cuproptosis and copper metabolism-related gene set. Front. Oncol.12, 952290. 10.3389/fonc.2022.952290 (2022). 10.3389/fonc.2022.952290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu, H. & Li, Y. Potential roles of cornichon family AMPA receptor auxiliary protein 4 (CNIH4) in head and neck squamous cell carcinoma. Cancer Biomark. Sect. A Dis. Mark.10.3233/cbm-220143 (2022). 10.3233/cbm-220143 [DOI] [PubMed] [Google Scholar]
- 86.Liu, H., Dilger, J. P. & Lin, J. A pan-cancer-bioinformatic-based literature review of TRPM7 in cancers. Pharmacol. Ther.10.1016/j.pharmthera.2022.108302 (2022). 10.1016/j.pharmthera.2022.108302 [DOI] [PubMed] [Google Scholar]
- 87.Zheng, S. et al. CRTAM promotes antitumor immune response in triple negative breast cancer by enhancing CD8+ T cell infiltration. Int. Immunopharmacol.129, 111625. 10.1016/j.intimp.2024.111625 (2024). 10.1016/j.intimp.2024.111625 [DOI] [PubMed] [Google Scholar]
- 88.Xiao, G. et al. Integrative multiomics analysis identifies a metastasis-related gene signature and the potential oncogenic role of EZR in breast cancer. Oncol. Res.30, 35–51 (2022). 10.32604/or.2022.026616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aureli, A. et al. Breast cancer is associated with increased HLA-DRB1*11:01 and HLA-DRB1*10:01 allele frequency in a population of patients from central Italy. Immunol. Invest.49, 489–497. 10.1080/08820139.2020.1737539 (2020). 10.1080/08820139.2020.1737539 [DOI] [PubMed] [Google Scholar]
- 90.Huang, R. et al. Targeting glutamine metabolic reprogramming of SLC7A5 enhances the efficacy of anti-PD-1 in triple-negative breast cancer. Front. Immunol.14, 1251643. 10.3389/fimmu.2023.1251643 (2023). 10.3389/fimmu.2023.1251643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang, Y. et al. GBP2 is a prognostic biomarker and associated with immunotherapeutic responses in gastric cancer. BMC Cancer23, 925. 10.1186/s12885-023-11308-0 (2023). 10.1186/s12885-023-11308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu, X., Cui, Q. & Qin, N. Low expression of KLRB1 predicts poor survival outcomes and is associated with immune infiltration in breast cancer. Transl. Cancer Res.13, 1225–1240 (2024). 10.21037/tcr-23-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He, J. R. et al. Inhibiting KLRB1 expression is associated with impairing cancer immunity and leading to cancer progression and poor prognosis in breast invasive carcinoma patients. Aging15, 13265–13286 (2023). 10.18632/aging.205239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu, C., Zhong, R., Sun, X. & Shi, J. PSME2 identifies immune-hot tumors in breast cancer and associates with well therapeutic response to immunotherapy. Front. Genet.13, 1071270. 10.3389/fgene.2022.1071270 (2022). 10.3389/fgene.2022.1071270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Engelhard, V. et al. B cells and cancer. Cancer Cell39, 1293–1296. 10.1016/j.ccell.2021.09.007 (2021). 10.1016/j.ccell.2021.09.007 [DOI] [PubMed] [Google Scholar]
- 96.Overgaard, N. H., Jung, J. W., Steptoe, R. J. & Wells, J. W. CD4+/CD8+ double-positive T cells: More than just a developmental stage?. J. Leukocyte Biol.97, 31–38. 10.1189/jlb.1RU0814-382 (2015). 10.1189/jlb.1RU0814-382 [DOI] [PubMed] [Google Scholar]
- 97.van der Leun, A. M., Thommen, D. S. & Schumacher, T. N. CD8(+) T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer20, 218–232. 10.1038/s41568-019-0235-4 (2020). 10.1038/s41568-019-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farhood, B., Najafi, M. & Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell Physiol.234, 8509–8521. 10.1002/jcp.27782 (2019). 10.1002/jcp.27782 [DOI] [PubMed] [Google Scholar]
- 99.Mehla, K. & Singh, P. K. Metabolic regulation of macrophage polarization in cancer. Trends Cancer5, 822–834. 10.1016/j.trecan.2019.10.007 (2019). 10.1016/j.trecan.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen, Y. et al. Type I collagen deletion in αSMA(+) myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell39, 548-565.e546. 10.1016/j.ccell.2021.02.007 (2021). 10.1016/j.ccell.2021.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Golby, S. J., Chinyama, C. & Spencer, J. Proliferation of T-cell subsets that contact tumour cells in colorectal cancer. Clin. Exp. Immunol.127, 85–91. 10.1046/j.1365-2249.2002.01730.x (2002). 10.1046/j.1365-2249.2002.01730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dolina, J. S., Van Braeckel-Budimir, N., Thomas, G. D. & Salek-Ardakani, S. CD8(+) T cell exhaustion in cancer. Front. Immunol.12, 715234. 10.3389/fimmu.2021.715234 (2021). 10.3389/fimmu.2021.715234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thommen, D. S. & Schumacher, T. N. T cell dysfunction in cancer. Cancer Cell33, 547–562. 10.1016/j.ccell.2018.03.012 (2018). 10.1016/j.ccell.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buller, C. W., Mathew, P. A. & Mathew, S. O. Roles of NK Cell Receptors 2B4 (CD244), CS1 (CD319), and LLT1 (CLEC2D) in Cancer. Cancers10.3390/cancers12071755 (2020). 10.3390/cancers12071755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takeuchi, A. et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med.213, 123–138. 10.1084/jem.20150519 (2016). 10.1084/jem.20150519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou, X. et al. A pan-cancer analysis of CD161, a potential new immune checkpoint. Front. Immunol.12, 688215. 10.3389/fimmu.2021.688215 (2021). 10.3389/fimmu.2021.688215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li, R. et al. Effects of local anesthetics on breast cancer cell viability and migration. BMC Cancer18, 666 (2018). 10.1186/s12885-018-4576-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ou, L. et al. 1,3,6-Trigalloylglucose: A novel potent anti-helicobacter pylori adhesion agent derived from aqueous extracts of Terminalia chebula Retz. Molecules (Basel, Switzerland)29, 1161 (2024). 10.3390/molecules29051161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ou, L. et al.Terminalia chebula Retz. aqueous extract inhibits the Helicobacter pylori-induced inflammatory response by regulating the inflammasome signaling and ER-stress pathway. J. Ethnopharmacol.320, 117428. 10.1016/j.jep.2023.117428 (2024). 10.1016/j.jep.2023.117428 [DOI] [PubMed] [Google Scholar]
- 110.Peng, C. et al. Syzygium aromaticum enhances innate immunity by triggering macrophage M1 polarization and alleviates Helicobacter pylori-induced inflammation. J. Funct. Foods107, 105626 (2023). 10.1016/j.jff.2023.105626 [DOI] [Google Scholar]
- 111.Hengrui, L. An example of toxic medicine used in Traditional Chinese Medicine for cancer treatment. J. Tradit. Chin. Med.43, 209–210 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu, H. et al. Exploring the mechanism underlying hyperuricemia using comprehensive research on multi-omics. Sci. Rep.13, 7161. 10.1038/s41598-023-34426-y (2023). 10.1038/s41598-023-34426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Berkel, C. & Cacan, E. Half of most frequently mutated genes in breast cancer are expressed differentially between premenopausal and postmenopausal breast cancer patients. Cancer Genet.286–287, 11–17. 10.1016/j.cancergen.2024.06.001 (2024). 10.1016/j.cancergen.2024.06.001 [DOI] [PubMed] [Google Scholar]
- 114.Glaviano, A. et al. Mechanisms of sensitivity and resistance to CDK4/CDK6 inhibitors in hormone receptor-positive breast cancer treatment. Drug Resist. Updat.76, 101103. 10.1016/j.drup.2024.101103 (2024). 10.1016/j.drup.2024.101103 [DOI] [PubMed] [Google Scholar]
- 115.Mundt, E. et al. Breast and colorectal cancer risks among over 6,000 CHEK2 pathogenic variant carriers: A comparison of missense versus truncating variants. Cancer Genet.278–279, 84–90. 10.1016/j.cancergen.2023.10.002 (2023). 10.1016/j.cancergen.2023.10.002 [DOI] [PubMed] [Google Scholar]
- 116.Ward, A. et al. Clinical management of TP53 mosaic variants found on germline genetic testing. Cancer Genet.284–285, 43–47. 10.1016/j.cancergen.2024.04.002 (2024). 10.1016/j.cancergen.2024.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gonzalez, T., Nie, Q., Chaudhary, L. N., Basel, D. & Reddi, H. V. Methylation signatures as biomarkers for non-invasive early detection of breast cancer: A systematic review of the literature. Cancer Genet.282–283, 1–8. 10.1016/j.cancergen.2023.12.003 (2024). 10.1016/j.cancergen.2023.12.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were available from the open-access databases, and the links for the data sources were provided: TCGA (https://www.cancer.gov/ccg/research/genome-sequencing/tcga), GTEx (https://gtexportal.org/home/), STRING (https://string-db.org/), TISCH2(http://tisch.comp-genomics.org/home/), and SpatialDB (http://www.spatialomics.org/SpatialDB/index.php).
All codes used in this study are open-source codes and are available from the author with a reasonable request.