Abstract
Background and aim
As the presence of single nucleotide polymorphisms (SNPs) in the interleukin (IL)-10 gene continues to be a major challenge in the development of effective therapies for digestive cancers, this case-control study was conducted to assess the possible influence of genotype, haplotype and diplotype for two SNPs (−1082A/G (rs1800896) and −592A/C (rs1800872)) located in the promoter region of IL-10 gene on the incidence, severity and prognosis of colorectal cancer (CRC) in Tunisians.
Methods
IL-10 gene SNPs were analyzed in 130 CRC cases and 165 healthy subjects (HS) using PCR-SSP.
Results
For the IL-10 -1082A/G SNP, the comparison of genotype frequencies between cases and HS groups showed that the G allele significantly reduced CRC risk under the recessive model (GG vs. AA + AG: OR [95%CI] = 0.44 [0.21–0.93], p = 0.03). Conversely, a positive association was observed between the codominant model (AG vs. AA + GG) and high susceptibility (OR [95%CI] = 1.65 [1.02–2.63], p = 0.04). After stratification by disease site, the recessive model was also found to reduce susceptibility to colon cancer (OR [95%CI] = 0.18 [0.04–0.72], p = 0 0.01), while the homozygote model (AA vs. GG) was suggested as a risk factor (OR [95%CI] = 5.16 [1.31–23.26], p = 0.02). Furthermore, the codominant model (AG vs. AA + GG) doubled the risk of rectum cancer (OR [95%CI] = 1.98 [1.07–3.70], p = 0.03). For the IL-10 -592A/C SNP, the codominant model (AC vs. AA + CC) has a protective effect against the development of CRC (OR [95%CI] = 0.59 [0.36–0.94], p = 0.03). The IL-10 gene haplotype was not associated with CRC risk. A stratified analysis by disease site demonstrated that the presence of Hap3 (−1082G and −592C alleles) specifically reduced the risk of developing colon cancer (OR [95%CI] = 0.51 [0.32–0.80], p = 0.003). Moreover, homozygous Hap3/Hap3 diplotype significantly reduced susceptibility to CRC (OR [95%CI] = 0.35 [0.14–0.85], p = 0.02). Interestingly, this diplotype has not been identified in colon cancer patients. Kaplan-Meier analysis showed that the homozygous Hap2/Hap2 diplotype was significantly associated with decreased overall survival (Log-rank: p = 0.01). This association was also observed in the colon cancer subgroup (Log-rank: p = 0.001).
Conclusion
Our findings provide preliminary indications that the −1082A/G and −592/AC SNPs within the IL-10 gene may exhibit significant associations with the pathogenesis and prognostic outcomes of CRC. However, further investigations are still warranted to validate and establish the veracity of our findings.
Keywords: Colorectal cancer, IL-10, SNP, Genotype, Haplotype, Diplotype, Risk, Survival
1. Introduction
As of 2020, colorectal cancer (CRC) ranked as the third most prevalent condition worldwide in terms of morbidity, and it held the position of the second leading cause of mortality [1]. The 5-year relative survival rate of CRC stands at approximately 90 % when diagnosed at earlier stages, but this rate significantly declines to 32 % once the cancer metastasizes to distant tissues [2]. In Tunisia, CRC is recognized as a critical public health challenge. The incidence rate of CRC is expected to increase from 12.4 registered cases in 2009 to 39.3 new cases per 100,000 population by the year 2024 [3].
Cytokines play an important role in coordinating and regulating inflammation and immune responses. Their deregulation is associated with the development of various diseases including CRC [4]. Interleukin (IL)-10 is a potent anti-inflammatory cytokine produced by myeloid lymphoid, keratinocyte, and epithelial cells [5]. In addition to normal and immune cells, it can also be secreted by various tumor cell lines [5]. Intensive studies have shown that this interleukin may have bimodal functions. Indeed, the balance can be switched from an inflammatory response promoting tumor progression to an immunostimulatory microenvironment that promotes tumor regression [6].
IL-10 is a pleiotropic cytokine that mediates important tumor-inhibiting effects. It suppresses the local release of pro-inflammatory molecules that promote tumor growth, survival, and metastasis. Moreover, IL-10 can create an inhibitory microenvironment that enhances tumor escape and progression by recruiting and activation of cytotoxic CD8+ T cells and NK cells in the tumor site, by disrupting the inflammatory M1 macrophage-Th17 T cells axis, and by reducing the synthesis of pro-angiogenic factors [7].
On the other hand, IL-10 can act as a promoter of tumor growth. Elevated levels of IL-10 stimulate cancer cell proliferation via STAT3 activation and inhibit apoptosis [8,9]. Additionally, IL-10 can reduce or inhibit tumor antigen presentation via downregulation of the expression of MHC class II in antigen-presenting cells (APCs). Furthermore, IL-10 stimulates the expression of immunosuppressive molecules such as the unconventional human leukocyte antigen (HLA) molecules such as HLA-G and HLA-E [10]. These molecules inhibit the functions of various immune cells, including natural killer (NK) cell-mediated lysis, cytotoxicity of CD8+ T cells, and the maturation and proliferation of APCs [11].
IL-10 expression is tightly controlled by genetic regulatory mechanisms. The IL-10 gene contains five exons separated by four introns, all located on the long arm of chromosome 1 [12]. Several single nucleotide polymorphisms (SNPs), including-1082A/G (rs1800896) and −592A/C (rs1800872), are localized within the promoter region of the IL-10 gene [12]. These SNPs are considered as prominent regulators of IL-10 transcription [13,14]. To gain deeper insights into the genetic framework influencing the pathogenesis and progression of CRC, substantial attention has been directed toward investigating the oncogenic implications of IL-10 -1082A/G and −592A/C SNPs. Numerous studies have provided evidence linking mutations in the IL-10 gene at loci −1082 and −592 with CRC susceptibility [[15], [16], [17], [18]]. However, contradicting results have been observed in Scottish, Italian or Swedish populations, where no such associations were observed [[19], [20], [21]]. Consequently, these divergent findings have led to conflicting viewpoints. To address this issue and gain further insights, the present study investigates the distribution of genotypes, haplotypes, and diplotypes of the −1082 A/G (rs1800896) and −592 A/C (rs1800872) SNPs within the IL-10 gene promoter region. By comparing CRC cases and healthy controls, our objective is to assess the potential associations of these genetic variations with disease susceptibility, severity, and outcome in the Tunisian population. Through this investigation, we aim to contribute valuable knowledge and shed light on the role of IL-10 gene variations in CRC within the Tunisian population.
2. Research design and methods
Population enrollment: The study included a cohort of 130 Tunisian patients diagnosed with CRC collected between October 2016 and August 2019. The diagnosis of CRC was substantiated through radiological and histopathological assessments carried out at Salah Azaiez Institute (ISA) of Tunis, Tunisia. Extensive admission record reviews and face-to-face interviews were conducted to obtain comprehensive information on the participants’ personal demographics, medical history, family background, and tumor-specific characteristics (Table 1). The control group consisted of 165 healthy subjects (HS) with no history of pathological disease. This study was reviewed and approved by the Ethics Committee at ISA, Tunisia (ISA/03/2016) and informed consent was obtained from all participants.
Table 1.
Characteristics of the study population.
| Variable | CRC cases (N = 130) | Healthy subjects (N = 165) | |
|---|---|---|---|
| Age | Mean, years (SD) | 59.51 (12.19) | 55.47 (13.32) |
| Gender, N (%) | Male | 67 (52) | 79 (48) |
| Female | 63 (48) | 86 (52) | |
| Disease site, N (%) | Colon | 69 (53) | |
| Rectum | 61 (47) | ||
| Stage, N (%) | Early | 39 (30) | |
| Advanced | 91 (70) | ||
| Grade, N (%) | Low | 75 (58) | |
| High | 51 (39) | ||
| Unknown | 4 (3) | ||
N: Number, SD: standard deviation, %: percentage.
SNP detection: Genomic DNA was extracted from blood cells using a standard protocol [22,23]. IL-10 genotyping of the −1082A/G (rs1800896) and −592A/C (rs1800872) SNPs was performed using a slightly modified sequence-specific primer-polymorphism-polymerase chain reaction (SSP-PCR) assay. The human growth hormone gene was used as an internal control. The primer sets and their corresponding sizes used in this study have been previously described [24,25]. For IL-10 -1082A/G, we used the following primers: FA (sense) 5′-ACTACTAAGGCTTCTTTGGGAA-3, FG (sense) 5′-CTACTAAGGCTTCTTTGGGAG3, and a common antisense 5′-CAGTGCCAACTGAGAATTTGG-3′. For IL-10 -592A/C, we used the following primers: FA (sense) 5′-GACTGGCTTCCTACAGT-3′, FC (sense) 5′-CTGGCTTCCTACAGG-3′ and a common antisense 5′-GCTCACTATAAAAATAGAG ACGG-3′. Briefly, the reaction begins with a preheating step (94°C/2min) followed by 5 cycles (94°C/25s and 63°C/45s), followed by 26 cycles (94°C/25s, 55°C/55s for −1082AG or 54°C/50s for −592AC and 72°C/50s), and a final extension at 72°C for 3 min. Amplified fragments were separated by electrophoresis on a 2 % agarose gel.
Data interpretation: All statistical analyses were performed using two packages: Graphpad Prism 8.0 and SPSS 25.0. To test for deviations from Hardy-Weinberg equilibrium (HWE), we compared observed and expected genotype frequencies for each SNP in the control group. The 2 × 2 contingency chi-square test (or Fisher's exact test where appropriate) was used to estimate the association between two categorical variables. The degree of association between each allele, genotype, haplotype or diplotype and disease risk and/or severity was determined by calculating odds ratios with 95 % confidence intervals (OR [95% CI]). Overall survival (OS: time from disease onset until death) was estimated and compared using the Kaplan-Meier curve and log-rank test. Distributions of linkage disequilibrium (LD) and haplotype frequencies were analyzed using Haploview 4.2 and PHASE 2.1. A p-value <0.05 was considered statistically significant.
3. Results
3.1. Association between genotypes/alleles of IL-10 gene SNPs with CRC risk and its severity
For the IL-10 gene SNPs examined in this study, genotype frequencies, in the control group, were found to be consistent with the assumption of HWE (p-values: 0.40 and 0.91 for −1082A/G and −592A/C, respectively). Regarding IL-10 -1082A/G, when comparing the cases and healthy subjects (HS) groups, the G allele in the recessive model (GG vs. AA + AG) showed a significant reduction in the overall risk of CRC (OR [95%CI] = 0.44 [0.21–0.93], p = 0.03, Table 2). Additionally, the AG genotype was found to be more prevalent among CRC cases (63 %) compared to HS (51 %), and appeared to increase the susceptibility to CRC by 1.65-fold (p = 0.04, Table 2). In the stratified analyses based on the disease site, it was observed that the recessive model (GG vs. AA + AG) significantly decreased the susceptibility to colon cancer (OR [95%CI] = 0.18 [0.04–0.72], p = 0 0.01, Table 2), whereas the homozygote model (AA vs. GG) was associated with a five-fold increase in the risk of this disease (OR [95%CI] = 5.16 [1.31–23.26], p = 0.02, Table 2). Regarding rectum cancer, the codominant model (AG vs. AA + GG) contributed to approximately a 2-fold increased risk of this cancer (OR [95%CI] = 1.98 [1.07–3.70], p = 0.03, Table 2). Similarly, we found a significant association between the heterozygote model and a higher rectal cancer risk (OR [95%CI] = 2.02 [1.01–4.10], p = 0.04).
Table 2.
Distribution of alleles and genotypes of IL-10 gene SNPs among CRC, colon cancer, rectum cancer cases, and healthy subjects.
| Genetic Model | CRC cases N(%) | Colon cancer cases N(%) | Rectum cancer cases N(%) | HS N (%) | P1 | OR1 (95%CI) | P2 | OR2 (95%CI) | P3 | OR3 (95%CI) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| −1082 A/G (rs1800896) |
Allele model | 0.67 | 1.07(0.77–1.51) | 0.17 | 1.34(0.88–2.06) | 0.44 | 0.85(0.56–1.29) | ||||
| A | 162(62) | 93(67) | 69(57) | 200 (60) | |||||||
| G | 98(38) | 45(33) | 53(43) | 130 (40) | |||||||
| Dominant model | 0.43 | 0.82(0.51–1.35) | 0.71 | 1.11(0.61–1.96) | 0.08 | 0.55(0.28–1.06) | |||||
| AA | 40(31) | 26(38) | 14(23) | 58 (35) | |||||||
| GG + AG | 90(69) | 43(62) | 47(77) | 107 (65) | |||||||
| Recessive model | 0.03 | 0.44(0.21–0.93) | 0.01 | 0.18(0.04–0.72) | 0.41 | 0.67(0.27–1.64) | |||||
| GG | 8(6) | 2(3) | 6(10) | 23 (14) | |||||||
| AA + AG | 122(94) | 67(97) | 55(90) | 142 (86) | |||||||
| Codominant Model | 0.04 | 1.65(1.02–2.63) | 0.23 | 1.41(0.79–2.49) | 0.03 | 1.98(1.07–3.70) | |||||
| AG | 82(63) | 41(59) | 41(67) | 84 (51) | |||||||
| AA + GG | 48(37) | 28(41) | 20(33) | 81 (49) | |||||||
| Homozygote model | 0.13 | 1.98(0.84–4.73) | 0.02 | 5.16(1.31–23.26) | 0.89 | 0.93(0.31–2.77) | |||||
| AA | 40(83) | 26(93) | 14(70) | 58 (72) | |||||||
| GG | 8(17) | 2(7) | 6(30) | 23 (28) | |||||||
| Heterozygote model | 0.18 |
0.71(0.42–1.16) |
0.78 |
0.92(0.52–1.65) |
0.04 |
0.49(0.24–0.99) |
|||||
| AA | 40 (33) | 26(39) | 14(25) | 58 (41) | |||||||
| AG |
82 (67) |
41(61) |
41(75) |
84 (59) |
|||||||
| 592 A C (rs1800872) −592 A/C (rs1800872) |
Allele model | 0.90 | 1.02(0.73–1.43) | 0.76 | 1.07(0.70–1.63) | 0.91 | 0.98(0.62–1.52) | ||||
| A | 88(34) | 48(35) | 40(33) | 110 (33) | |||||||
| C | 172(66) | 90(65) | 82(67) | 220 (67) | |||||||
| Recessive model | 0.09 | 1.76(0.88–3.43) | 0.10 | 1.90(0.88–4.11) | 0.27 | 1.60(0.70–3.79) | |||||
| AA | 23(18) | 13(19) | 10(16) | 18 (11) | |||||||
| AC + CC | 107(82) | 56(81) | 51(84) | 147 (90) | |||||||
| Dominant model | 0.33 | 1.26(0.79–2.02) | 0.48 | 1.22(0.70–2.12) | 0.38 | 1.30(0.72–2.36) | |||||
| CC | 65(50) | 34(49) | 31(51) | 73 (44) | |||||||
| AA + AC | 65 (50) | 35(51) | 30(49) | 92 (56) | |||||||
| Co-dominant Model | 0.03 | 0.59(0.36–0.94) | 0.07 | 0.58(0.32–1.06) | 0.10 | 0.60(0.32–1.11) | |||||
| AC | 42(32) | 22(32) | 20(33) | 74 (45) | |||||||
| AA + CC | 88(68) | 47(68) | 41(67) | 91 (55) | |||||||
| Homozygote model | 0.31 | 1.44(0.71–2.79) | 0.29 | 1.55(0.67–3.38) | 0.55 | 1.31(0.53–3.12) | |||||
| AA | 23(26) | 13(28) | 10(25) | 18 (20) | |||||||
| CC | 65(74) | 34(72) | 31(75) | 73 (80) | |||||||
| Heterozygote model | 0.08 | 0.64 (0.38–1.04) | 0.16 | 0.64 (0.34–1.22) | 0.17 | 0.64 (0.34–1.21) | |||||
| AC | 42(39) | 22(39) | 20(39) | 74 (50.3) | |||||||
| CC | 65(61) | 34(61) | 31(61) | 73 (49.7) | |||||||
Regarding IL-10 -592A/C, a noteworthy reduction in CRC risk was observed under the codominant model (AC vs. AA + CC: OR [95%CI] = 0.59[0.36–0.94], p = 0.03, Table 2). Conversely, the other genetic models did not demonstrate any significant associations with the development of CRC in this study.
Furthermore, stratification analyses did not reveal any significant associations between the studied IL-10 gene SNPs and CRC clinicopathological characteristics including tumor infiltration depth, lymph node involvement, distant metastasis, TNM staging and grade (data not shown).
3.2. Association between haplotypes of IL-10 gene SNPs with CRC risk and its severity
In this study, IL-10 -1082A/G exhibited partial linkage disequilibrium with IL-10 -592A/C (D’ = 0.65, r2 = 0.13, Fig. 1a–b). Based on these two SNPs, four possible haplotypes (Hap1, Hap2, Hap3, and Hap4) were identified. Among these haplotypes, only the first three (Hap1, Hap2, and Hap3; Table 3) displayed frequencies above 1 % among both cases and controls, thus warranting further evaluation. No significant disparity in the overall distribution of IL-10 gene haplotypes was observed between the CRC and HS groups (Table 3). It is essential to note that a trend towards decreased CRC risk was evident in the presence of Hap3 which consisted of IL-10 -1082G and −592C alleles (OR [95%CI] = 0.74 [0.52–1.04], p = 0.08, Table 3). Subgroup analysis by disease site showed that Hap1, consisting of IL-10 -1082A and −592 C alleles, was more common among colon cancer cases (43.0 %) than in HS (27.7 %), resulting in increased susceptibility (OR [95%CI] = 1.72 [1.15–2.60], p = 0.01, Table 3). Furthermore, we found that Hap3 was significantly associated with a reduced risk of this type of cancer compared to other combinations (OR [95%CI] = 0.51[0.32–0.80], p = 0.003; Table 3). In rectal cancer, the distribution of haplotypes between cases and HS groups was similar (p > 0.05, Table 3).
Fig. 1.
HaploView linkage disequilibrium (LD) plots of -1082A/G (rs1800896) and -592A/C (rs1800872) SNPs in the promoter region of IL-10 gene. The LD pattern was derived from the combined study population (both cases and healthy subjects). Values in the LD blocks indicated the D’ (a.) and the r2 (b.) in percentages.
Table 3.
Distribution of haplotypes of IL-10 gene SNPs between CRC, colon cancer, rectum cancer cases, and HS.
| Haplotype IL-10 1082–592 | CRC cases |
Colon cancer cases |
Rectum cancer cases |
HS |
P1 | OR1 (95%CI) | P2 | OR2(95%CI) | P3 | OR3(95%CI) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N = 260 | Frequency | N = 138 | Frequency | N = 122 | Frequency | N = 330 | Frequency | |||||||
| Hap1 (A1082C592) vs. others | 95 | 0.364 | 59 | 0.430 | 34 | 0.277 | 100 | 0.303 | 0.11 | 1.32 (0.94–1.86) | 0.01 | 1.72 (1.15–2.60) | 0.61 | 0.89 (0.56–1.39) |
| Hap2 (A1082A592) vs. others | 67 | 0.259 | 34 | 0.244 | 35 | 0.290 | 100 | 0.303 | 0.22 | 0.80 (0.55–1.14) | 0.22 | 0.75 (0.48–1.19) | 0.74 | 0.93 (0.58–1.44) |
| Hap3 (G1082C592) vs. others | 77 | 0.298 | 31 | 0.222 | 48 | 0.396 | 120 | 0.364 | 0.08 | 0.74 (0.52–1.04) | 0.003 | 0.51 (0.32–0.80) | 0.56 | 1.14 (0.74–1.73) |
HS: healthy subjects. N: Number. OR: odds ratio. 95%CI: confidence interval.
P-values are estimated by the chi-square test or the Fisher's exact test when appropriate.
P1 and OR1= CRC cases vs. HS, P2 and OR2= Colon cancer cases vs. HS, P3 and OR3= Rectum cancer cases vs. HS.
Significant P-values are represented in Bold.
Associations between IL-10 SNPs and the clinicopathological features of CRC at the haplotype level were also evaluated. However, no significant results were found (data not shown).
3.3. Association between diplotypes of IL-10 gene SNPs with CRC risk and its severity
Based on the promising results in the preceding analysis, we proceeded to conduct further evaluations concerning the combined effect of haplotype pairs (diplotypes) within the IL-10 promoter region on the development and severity of CRC. The homozygous Hap3/Hap3 diplotype demonstrated a significant reduction in susceptibility to CRC (OR [95%CI] = 0.35[0.14–0.85], p = 0.02, Table 4). Nevertheless, consistent with our expectations, there were no distinct associations observed between all diplotypes tested and the clinicopathological characteristics of CRC (data not shown).
Table 4.
Distribution of diplotypes of IL-10 gene SNPs between CRC, colon cancer, rectum cancer cases, and HS.
| Diplotypes | CRC cases N (%) |
Colon cancer cases N (%) | Rectum cancer cases N (%) | HS N (%) |
P1 | OR1 (95%CI) | P2 | OR2 (95%CI) | P3 | OR3 (95%CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| Hap1/Hap1 vs. | 16 (12) | 12 (17) | 4 (7) | 15 (9) | 0.37 | 1.40 (0.67–3.00) | 0.07 | 2.10 (0.90–4.60) | 0.54* | 0.70 (0.25–2.10) |
| Hap1/others + Others/others | 114 (88) | 57 (83) | 57 (93) | 150 (91) | ||||||
| Hap2/Hap2 vs. | 14 (11) | 7 (10) | 7 (11) | 15 (9) | 0.63 | 1.21 (0.59–2.67) | 0.80 | 1.13 (0.44–2.93) | 0.59 | 1.30 (0.51–3.04) |
| Hap2/others + Others/others | 116 (89) | 62 (90) | 54 (89) | 150 (91) | ||||||
| Hap3/Hap3 vs. | 6 (5) | 0 (0) | 6 (10) | 20 (12) | 0.02 | 0.35 (0.14–0.85) | – | – | 0.63 | 0.79 (0.31–1.99) |
| Hap3/others + Others/others | 124 (95) | 69 (100) | 55 (90) | 145 (88) |
HS: healthy subjects. N: Number. OR: odds ratio. 95%CI: confidence interval.
P-values are estimated by the chi-square test or the Fisher's exact test when appropriate.
P1 and OR1: CRC cases vs. HS, P2, and OR2: Colon cancer cases vs. HS, P3 and OR3: Rectum cancer cases vs. HS.
Significant P-values are represented in Bold.
3.4. Effect of IL-10 gene SNPs genotype, haplotype, and diplotype on overall survival
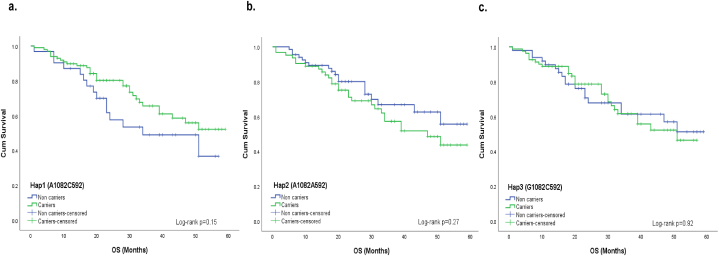
Upon analyzing all patients, we found a significant association between IL-10 -592A/C SNP and worse OS (AA genotype versus other genotypes; Log-rank p = 0.021, Fig. 2c; However, no association at genotype levels Fig. 2b). However, we revealed no significant effect of IL-10 -1082A/G on OS at genotype level (Log-rank p > 0.05, Fig. 2a). Similarly, no association was observed between the IL-10 haplotype and OS (Log-rank p > 0.05, Fig. 3a–c).
Fig. 2.
Effect of IL-10 gene SNPs on the overall survival in CRC cases Kaplan-Meier estimates showed no association between the genotypes of SNPs −1082A/G (Log-rank p = 0.53) (a.) and −592AC (Log-rank p = 0.066) (b.) in the promoter region of the IL-10 gene and the overall survival of CRC cases. Kaplan-Meier estimates showed an association between the IL-10 -592 AA genotype versus other genotypes (CC + AC) (Log-rank p = 0.021) (c.).
Fig. 3.
Effect of IL-10 gene haplotypes on the overall survival in CRC cases Kaplan-Meier estimates showed no association between all the combinations of haplotypes derived from the −1082A/G and −592A/C SNPs in the promoter region of the IL-10 gene and the overall survival in CRC cases (Log-rank p value > 0.05) (a. b. and c.).
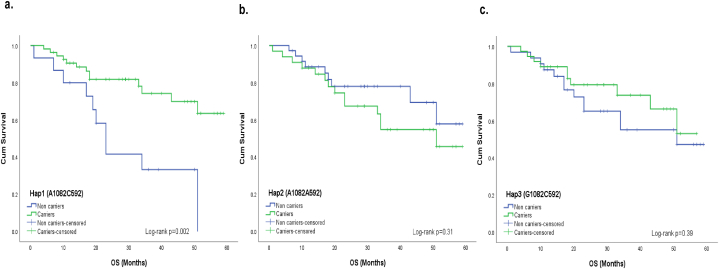
However, examining CRC cases based on disease sites revealed interesting results. Notably, Hap1 exhibited a significant association with improved OS in the colon cancer subgroup (Log-rank p = 0.002, Fig. 4a). No association was found between other analyzed haplotypes and patients’ survival (Fig. 4b-c). Furthermore, diplotype-based analysis revealed that patients with homozygous Hap2/Hap2 diplotype had shorter OS (Log-rank p = 0.01, Fig. 5b), and this finding was consistent in the colon cancer subgroup (Log-rank p = 0.001, Fig. 6b). Although the relationship was not statistically significant (Fig. 5a and c), there was a tendency for improved OS among CRC cases with the homozygous Hap3/Hap3 diplotype (Log-rank p = 0.21, Fig. 5c). In addition, it was noteworthy that none of the colon cancer cases exhibited the homozygous Hap3/Hap3 diplotype. No association was found between homozygous Hap1/Hap1 diplotype and OS among colon cancer patients (Fig. 6a).
Fig. 4.
Effect of IL-10 gene haplotypes on the overall survival in cases with colon cancer. In cases with colon cancer subgroup, Kaplan-Meier estimates showed that Hap1 carriers (consisting of the 1082A and 592C alleles) had a better overall survival (OS) compared with non-carriers (Log-rank p = 0.002) (a.). No significant results were observed for other haplotype combinations (Log-rank p values > 0.05) (b. and c.).
Fig. 5.
Effect of IL-10 gene diplotypes on the overall survival in CRC cases Kaplan-Meier estimates showed that carriers of the homozygous Hap2/Hap2 diplotype had a lower overall survival (OS) compared with Hap2/Others + Others/Others carriers (Log-rank p = 0.01) (b.). No significant association was observed between all the other combinations of diplotype and OS in CRC cases (Log-rank p values > 0.05) (a. and c.).
Fig. 6.
Effect of IL-10 gene diplotypes on the overall survival in cases with colon cancer. In cases with colon cancer subgroup, Kaplan-Meier estimates showed that carriers of homozygous Hap2/Hap2 diplotype had a significantly shorter OS compared to Hap2/Others + Others/Others carriers (Log-rank p = 0.001) (b.). No association was observed between homozygous Hap1/Hap1 diplotype and patients' outcome (Log-rank p = 0.21) (a.). None of cases with colon cancer had the homozygous Hap3/Hap3 diplotype.
4. Discussion
In this study, the frequencies of the IL-10 -1082G and −592C alleles in the healthy controls were 0.38 and 0.66, respectively. These frequencies were very similar to those previously reported in various studies involving healthy North African subjects [[26], [27], [28]]. Based on the NCBI database of short genetic variation (dbSNP), this consistency was also observed in healthy Caucasian subjects (0.47 and 0.76, respectively). The −1082G allele is very rare (0.06) in Asian populations. However, 592C appears to be less common in Asians than in Caucasians and Africans (0.27, 0.76, and 0.59, respectively).
The genetic SNPs −1082A/G and −592A/C are situated in the proximal promoter region of the IL-10 gene and are recognized as critical determinants in regulating the levels of IL-10. Studies have demonstrated that the −1082G allele, along with the GCC haplotypes (comprising IL-10-1082G, −819C, and −592C), as well as the GCC/GCC diplotype, are strongly associated with higher expression levels of IL-10. Conversely, the ACC haplotype (comprising IL-10-1082A, −819C, and −592C) and ATA haplotype (comprising IL-10-1082A, −819T, and −592A) are considered as intermediate- and low-producers of IL-10, respectively [13]. To the best of our knowledge, this study represents the first investigation into the influence of genotypes, haplotypes, and diplotypes of-1082A/G (rs1800896) and −592A/C (rs1800872) SNPs within the IL-10 gene promoter on CRC susceptibility, progression and outcome within the Tunisian population. Our findings revealed that the presence of the genotype AG at position −1082 serves as a risk factor for CRC (OR = 1.65, p = 0.04). Conversely, we observed that −1082G allele in the recessive model is associated with a reduced risk of CRC (OR = 0.44, p = 0.03). After stratification according to the disease site, this association persisted when comparing colon cancer patients with the HS group (OR = 0.18, p = 0.01). These results are consistent with previous studies from the US and Romania supporting a protective role of −1082G allele for the development of CRC [17,29]. However, Miteva et al.(2014) reported no genetic association between the IL-10 -1082A/G SNP and the occurrence of CRC in the Bulgarian population [30]. They suggested that the G allele may be involved in leading the disease to a more aggressive phenotype. In contrast, no discernible association between IL-10 -1082A/G and CCR risk was observed in various other ethnic groups [19,20].
In this study, a positive association was observed between IL-10 -592A/C SNP and CRC. Indeed, the codominant model (AC vs. AA + CC) was associated with a low CRC risk (OR = 0.59, p = 0.03). Although these results contradict a previous study [21], they agree with what was reported in a recent Tunisian study [31]. Moreover, it has been shown that Chinese carrying AC or AC/CC genotypes are less prone to CRC compared with the AA genotype [32,33]. There is no consensus on the role of the IL-10 -592AA variant in CRC. On the contrary, in a study conducted on 142 CRC patients from Kashmir, the authors reported that the −592A variant is a protective factor against the development of CRC at both allelic and genotypic levels [34]. In an attempt to address the inconsistency observed in various investigating the role of IL-10 -1082A/G and −592A/C SNPs in the development of CRC, Zhang et al. (2012) conducted a comprehensive meta-analysis that encompassed a total of 1469 CRC patients and 2566 controls [35]. They reported that none of the aforementioned SNPs had a significant effect on CRC risk. However, through stratified analysis of controls, they found that the −592A allele may potentially increase susceptibility to CRC. More recently, a meta-analysis conducted by Mirjalili et al. (2018) involving a larger dataset of 5647 CRC patients and 6908 controls, did not reveal any relevant association between IL-10 gene SNPs and CRC [36].
Interestingly, haplotype and diplotype-based studies consider the interactions and cumulative effects of different genetic variants and aim to advance our understanding of the multigenic basis underlying disease susceptibility, progression, and response to treatment. These approaches go beyond the analysis of individual genotypes and allow for more comprehensive assessments. In our study, we examined the involvement of haplotypes and diplotypes derived from the two SNPs of the IL-10 gene in relation to the development of CRC and its severity. Interestingly, we observed that carrying Hap3, formed by the combination of −1082G and −592C alleles, provided significant protection against the onset of CRC compared to other haplotypes. Additionally, individuals who are homozygous for the Hap3/Hap3 diplotype showed a reduced CRC risk (OR = 0.35, p = 0.02). In the subset of patients with colon cancer, we found that Hap1, formed by the combination of IL-10-1082A and −592C alleles, increased the susceptibility for this type of cancer compared to other combinations (OR = 1.72, p = 0.01). It was noteworthy that our findings differ from those reported in studies examining gastric and nasopharyngeal cancers, wherein no association was observed between any of the IL-10 haplotypes derived from these two SNPs and cancer susceptibility [27,37]. However, El-Omar et al. (2003) showed that the haplotype ATA (consisting of IL-10-1082A, −819T and −592A) may increase the risk of noncardia gastric cancer 2.5-fold compared to the GCC haplotype (composed of IL-10-1082G, −819C and −592C alleles) [38].
In the overall population with CRC, carriers of the Hap2 haplotype (composed of IL-10-1082A and −592A alleles) exhibited a worse OS. Furthermore, when considering only patients with colon cancer, we observed a significant association between the homozygous Hap2/Hap2 diplotype and worse OS. It is important to note that there is limited research investigating the relationship between CRC outcome and these two SNPs at the haplotype level, and as a result, available data in this regard are scarce [19,20,39]. Supporting our findings, a study on Australian CRC patients also reported a shorter OS in patients who were homozygous for the A allele at position −1082, and those who carried at least one copy of the A allele at position −592 in the promoter region of the IL-10 gene compared to individuals with −1082G and −592C alleles, respectively [40]. Additionally, this study found that the −592A variant is associated with a higher production of IL-10. In another study conducted in Spain, a borderline association was observed between the −1082GG genotype and a favorable OS among patients with lymphoid cancers (p = 0.05) [41].
Martinez-Escribano et al. revealed that low-producer genotypes of the IL-10 gene, particularly the ACC/ATA genotype (representing genotypic variations at positions −1082, −819, and −592 in the IL-10 gene promoter), were linked to poorer survival in Spanish patients with melanoma patients [42]. Similarly, in Hodgkin's lymphoma, the −592AA genotype and IL-10 haplotype (−3575T/-2849G/-2763C/-1082A/-592A) were associated with a worse prognosis [43]. IL-10 plays a critical role as a regulatory cytokine in the immune system and its differential expression patterns may influence the occurrence or protection against diseases, although this role remains controversial. Some studies have reported a positive association between low IL-10 expression and the development of CRC [44]. However, adenomas and serrated adenomas have been found to exhibit increased concentrations of IL-10 compared to normal cells [45]. and metastatic colon cancer has shown higher IL-10 expression than primary tumors [46]. Furthermore, IL-10 deficiency has been linked to intestinal mucosal lesions, leading to chronic inflammation and potentially paving the way for CRC development [6]. In intestinal epithelial cells, loss of IL-10 has been associated with decreased expression of the immunosuppressive gene Bcl3, and increased expression of IL-17, IFN-γ, and TNF-α expression, which can cause damage to the intestinal mucosal surface [47]. Notably, IL-10 high expression or treatment with pegylated IL-10 has been found to induce cancer regression and confer a persistent and effective immune response in murine models [6]. However, contrasting relationships have been observed in other studies between IL-10 expression and CRC onset and outcome. It has been shown a significant association between low IL-10 levels and disease susceptibility, while elevated levels were correlated with an unfavorable prognosis [44]. These findings further emphasize the duality of IL-10 function in the pathogenesis of CRC, demanding extensive attention to fully understand its complex role in cancer development and progression.
Taken together, our results indicated that the −1082A/G and −592A/C SNPs in the promoter region of the IL-10 gene are associated with CRC and may be valuable factors in disease susceptibility and prognosis. Further studies in a larger population are needed to advance our understanding of the pathogenesis of CRC, which may lead to improved diagnosis and treatment strategies.
CRediT authorship contribution statement
Sabrine Dhouioui: Writing – original draft, Visualization, Methodology, Investigation, Formal analysis. Sana Baroudi: Investigation, Formal analysis. Ines Zemni: Writing – review & editing, Validation, Resources, Formal analysis. Fadia Mahdhi: Investigation, Formal analysis. Afef Najjari: Writing – review & editing. Hanen Chelbi: Writing – review & editing, Methodology. Houyem Khiari: Validation, Methodology. Nadia Boujelbene: Writing – review & editing, Validation, Resources, Methodology, Conceptualization. Ines Zidi: Writing – review & editing, Validation, Supervision, Resources, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Ines Zidi is currently serving as an Associate Editor for Heliyon Immunology. Although she was not involved in the review of this specific manuscript, she is disclosing this position to ensure transparency and uphold the integrity of the review process for this submission.
Acknowledgements
This study was supported by the Ministry of Higher Education and Scientific Research of Tunisia. We are grateful to all participants for their valuable contributions to this research.
References
- 1.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso R., et al. Overall and stage-specific survival of patients with screen-detected colorectal cancer in European countries: a population-based study in 9 countries. The Lancet Regional Health - Europe. 2022;21 doi: 10.1016/j.lanepe.2022.100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khiari H., et al. Colorectal cancer incidence trend and projections in Tunisia (1994 - 2024) Asian Pac. J. Cancer Prev. APJCP : Asian Pac. J. Cancer Prev. APJCP. 2017;18(10):2733–2739. doi: 10.22034/APJCP.2017.18.10.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West N.R., et al. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 2015;15(10):615–629. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 5.Weiss E., et al. The role of interleukin 10 in the pathogenesis and potential treatment of skin diseases. J. Am. Acad. Dermatol. 2004;50(5):657–675. doi: 10.1016/j.jaad.2003.11.075. [DOI] [PubMed] [Google Scholar]
- 6.Oft M. IL-10: master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol. Res. 2014;2(3):194–199. doi: 10.1158/2326-6066.CIR-13-0214. [DOI] [PubMed] [Google Scholar]
- 7.Li Q., Anderson C.D., Egilmez N.K. Inhaled IL-10 suppresses lung tumorigenesis via abrogation of inflammatory macrophage–Th17 cell Axis. J. Immunol. 2018;201(9):2842–2850. doi: 10.4049/jimmunol.1800141. [DOI] [PubMed] [Google Scholar]
- 8.Ibrahim M.L., et al. Myeloid-derived suppressor cells produce IL-10 to elicit DNMT3b-dependent IRF8 silencing to promote colitis-associated colon tumorigenesis. Cell Rep. 2018;25(11):3036–3046.e6. doi: 10.1016/j.celrep.2018.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi L., et al. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget. 2016;7(44) doi: 10.18632/oncotarget.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonçalves A.S., et al. Relevance of HLA-G, HLA-E and IL-10 expression in lip carcinogenesis. Hum. Immunol. 2016;77(9):785–790. doi: 10.1016/j.humimm.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Contini P., et al. HLA-G expressing immune cells in immune mediated diseases. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trifunović J., et al. Pathologic patterns of interleukin 10 expression--a review. Biochem. Med. 2015;25(1):36–48. doi: 10.11613/BM.2015.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrey C., et al. Genotyping for polymorphisms in interferon-γ, interleukin-10, transforming growth factor-β1 and tumour necrosis factor-α genes: a technical report. Transpl. Immunol. 1998;6(3):193–197. doi: 10.1016/s0966-3274(98)80045-2. [DOI] [PubMed] [Google Scholar]
- 14.Suárez A., et al. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms1. Transplantation. 2003;75(5):711–717. doi: 10.1097/01.TP.0000055216.19866.9A. [DOI] [PubMed] [Google Scholar]
- 15.Althubyani S.A., et al. A preliminary study of cytokine gene polymorphism effects on Saudi patients with colorectal cancer. Saudi Med. J. 2020;41(12):1292–1300. doi: 10.15537/smj.2020.12.25543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen V., et al. Interaction between interleukin-10 (IL-10) polymorphisms and dietary fibre in relation to risk of colorectal cancer in a Danish case-cohort study. BMC Cancer. 2012;12(1):183. doi: 10.1186/1471-2407-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burada F., et al. Cytokine promoter polymorphisms and risk of colorectal cancer. Clin. Lab. 2013;59(7–8):773–779. doi: 10.7754/clin.lab.2012.120713. [DOI] [PubMed] [Google Scholar]
- 18.Čačev T., et al. Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis. 2008;29(8):1572–1580. doi: 10.1093/carcin/bgn164. [DOI] [PubMed] [Google Scholar]
- 19.Basavaraju U., et al. Cytokine gene polymorphisms, cytokine levels and the risk of colorectal neoplasia in a screened population of Northeast Scotland. Eur. J. Cancer Prev. 2015;24(4):296–304. doi: 10.1097/CEJ.0000000000000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crivello A., et al. Regulatory cytokine gene polymorphisms and risk of colorectal carcinoma. Ann. N. Y. Acad. Sci. 2006;1089(1):98–103. doi: 10.1196/annals.1386.002. [DOI] [PubMed] [Google Scholar]
- 21.Wilkening S., et al. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis. 2008;29(6):1202–1206. doi: 10.1093/carcin/bgn101. [DOI] [PubMed] [Google Scholar]
- 22.Dhouioui S., et al. Association of HLA-G 3′UTR polymorphisms and haplotypes with colorectal cancer susceptibility and prognosis. Hum. Immunol. 2022;83(1):39–46. doi: 10.1016/j.humimm.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3) doi: 10.1093/nar/16.3.1215. 1215-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talaat R.M., et al. Interleukin 10 (- 1082 G/A) and (- 819 C/T) gene polymorphisms in Egyptian women with polycystic ovary syndrome (PCOS) Meta Gene. 2016;9:254–258. doi: 10.1016/j.mgene.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zammiti W., et al. Association of -592C/A, -819C/T and -1082A/G interleukin-10 promoter polymorphisms with idiopathic recurrent spontaneous abortion. Mol. Hum. Reprod. 2006;12(12):771–776. doi: 10.1093/molehr/gal084. [DOI] [PubMed] [Google Scholar]
- 26.Bahgat N.A., et al. Interferon-γ and interleukin-10 gene polymorphisms are not predictors of chronic hepatitis C (Genotype-4) disease progression. Asian Pac. J. Cancer Prev. APJCP. 2015;16(12):5025–5030. doi: 10.7314/apjcp.2015.16.12.5025. [DOI] [PubMed] [Google Scholar]
- 27.Moumad K., et al. The involvement of interleukin-10 promoter genetic polymorphism in epstein-barr virus-associated nasopharyngeal carcinoma from North africa. EJMO. 2022;6(3):232–240. [Google Scholar]
- 28.Zidi S., et al. IL-10 gene promoter and intron polymorphisms as genetic biomarkers of cervical cancer susceptibility among Tunisians. Cytokine. 2015;76(2):343–347. doi: 10.1016/j.cyto.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Tsilidis K.K., et al. Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control. 2009;20(9):1739–1751. doi: 10.1007/s10552-009-9427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miteva L.D., et al. Significance of −1082A/G polymorphism of IL10 gene for progression of colorectal cancer and IL-10 expression. Tumor Biol. 2014;35(12):12655–12664. doi: 10.1007/s13277-014-2589-2. [DOI] [PubMed] [Google Scholar]
- 31.Hazgui M., et al. Interleukin-1β, interleukin-6 and interleukin-10 polymorphisms in Tunisian patients with colorectal cancer and liver metastasis. Arch. Biol. Sci. 2022;74(4):333–345. [Google Scholar]
- 32.Cai J., Zhang Z. An analysis of IL-10/IL-10r genetic factors related to risk of colon cancer and inflammatory bowel disease in a han Chinese population. Clin. Lab. 2016;62(6):1147–1154. doi: 10.7754/clin.lab.2015.151120. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y., et al. Polymorphisms of inflammation-related genes and colorectal cancer risk: a population-based case–control study in China. Int. J. Immunogenet. 2014;41(4):289–297. doi: 10.1111/iji.12119. [DOI] [PubMed] [Google Scholar]
- 34.Banday M.Z., et al. Interleukin-10 −592C/A, but not −1082A/G promoter single nucleotide polymorphism, is associated with a decreased risk of colorectal cancer in an ethnic Kashmiri population: a case–control study. Eur. J. Cancer Prev. 2017;26(6):476–490. doi: 10.1097/CEJ.0000000000000370. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y.M., et al. Meta-analysis of epidemiological studies of association of two polymorphisms in the interleukin-10 gene promoter and colorectal cancer risk. Genet. Mol. Res. 2012;11(3):3389–3397. doi: 10.4238/2012.September.25.7. [DOI] [PubMed] [Google Scholar]
- 36.Mirjalili S.A., et al. Association of promoter region polymorphisms of interleukin-10 gene with susceptiblity to colorectal cancer: a systematic review and meta-analysis. Arq. Gastroenterol. 2018;55(3):306–313. doi: 10.1590/S0004-2803.201800000-66. [DOI] [PubMed] [Google Scholar]
- 37.Pan X.F., et al. Interleukin-10 gene promoter polymorphisms and risk of gastric cancer in a Chinese population: single nucleotide and haplotype analyses. Asian Pac. J. Cancer Prev. APJCP. 2013;14(4):2577–2582. doi: 10.7314/apjcp.2013.14.4.2577. [DOI] [PubMed] [Google Scholar]
- 38.El-Omar E.M., et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124(5):1193–1201. doi: 10.1016/s0016-5085(03)00157-4. [DOI] [PubMed] [Google Scholar]
- 39.Macarthur M., et al. The role of cytokine gene polymorphisms in colorectal cancer and their interaction with aspirin use in the northeast of Scotland. Cancer Epidemiol. Biomarkers Prev. 2005;14(7):1613–1618. doi: 10.1158/1055-9965.EPI-04-0878. [DOI] [PubMed] [Google Scholar]
- 40.Sharma R., et al. Systemic inflammatory response predicts prognosis in patients with advanced-stage colorectal cancer. Clin. Colorectal Cancer. 2008;7(5):331–337. doi: 10.3816/CCC.2008.n.044. [DOI] [PubMed] [Google Scholar]
- 41.Eva D.-D., et al. Impact of interleukin-10 polymorphisms (−1082 and −3575) on the survival of patients with lymphoid neoplasms. Haematologica. 2007;92(11):1475–1481. doi: 10.3324/haematol.11350. [DOI] [PubMed] [Google Scholar]
- 42.Martínez-Escribano J.A., et al. Interleukin-10, interleukin-6 and interferon-γ gene polymorphisms in melanoma patients. Melanoma Res. 2002;12(5) doi: 10.1097/00008390-200209000-00008. [DOI] [PubMed] [Google Scholar]
- 43.Hohaus S., et al. Polymorphism in cytokine genes as prognostic markers in Hodgkin's lymphoma. Ann. Oncol. 2007;18(8):1376–1381. doi: 10.1093/annonc/mdm132. [DOI] [PubMed] [Google Scholar]
- 44.Abtahi S., et al. Dual association of serum interleukin-10 levels with colorectal cancer. J. Cancer Res. Therapeut. 2017;13(2):252–256. doi: 10.4103/0973-1482.199448. [DOI] [PubMed] [Google Scholar]
- 45.Marszałek A., et al. Impact of COX-2, IL-1β, TNF-α, IL-4 and IL-10 on the process of carcinogenesisin the large bowel. Pol. J. Pathol. 2012;63(4):221–227. doi: 10.5114/pjp.2012.32768. [DOI] [PubMed] [Google Scholar]
- 46.Townsend M.H., et al. Metastatic colon adenocarcinoma has a significantly elevated expression of IL-10 compared with primary colon adenocarcinoma tumors. Cancer Biol. Ther. 2018;19(10):913–920. doi: 10.1080/15384047.2017.1360453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarry A., et al. Mucosal IL-10 and TGF-β play crucial roles in preventing LPS-driven, IFN-γ–mediated epithelial damage in human colon explants. J. Clin. Investig. 2008;118(3):1132–1142. doi: 10.1172/JCI32140. [DOI] [PMC free article] [PubMed] [Google Scholar]