Abstract
The deletion of geranylgeranyl diphosphate synthase 1 (GGPS1) has been reported to inhibit the proliferation of multiple cells. Although emerging evidence has demonstrated a correlation between GGPS1 and cancer, no pan-cancer analysis has been conducted to date. This study explored the potential tumorigenesis of GGPS1 using data from the cancer genome atlas human clinical database. GGPS1 expression was considerably upregulated at both the RNA and protein levels in several cancer types, especially in breast carcinoma (BRCA), (liver) hepatocellular carcinoma (LIHC/HCC) and lung adenocarcinoma (LUAD). Amplification is the most common form of genetic alteration observed in invasive BRCA, ovarian epithelial tumor and HCC. Additionally, elevated GGPS1 expression was markedly related to poor patient prognosis and overall survival in several cancer types including LIHC. GGPS1 expression was also linked to cancer-associated fibroblasts (CAFs) infiltration in several cancer types, such as BRCA and LUAD. Moreover, GGPPS-interacting proteins and GGPS1-correlated genes in cancers were functionally enriched in terpenoid backbone biosynthesis, steroid biosynthesis, and metabolic pathways. These results indicate that GGPS1 may play a role in promoting the tumorigenesis and tumor development, particularly in BRCA and LUAD, and may play a role in steroid biosynthesis and metabolic pathways.
Keywords: Geranylgeranyl diphosphate synthase 1, Pan-cancer analysis, Tumorigenesis
1. Background
Tumors are multifactorial diseases associated with high mortality rates. Understanding the mechanisms of tumorigenesis and tumor development is essential for identifying potential targets for tumor prevention and treatment. Online public oncology databases, such as the cancer genome atlas (TCGA), contain a wealth of information suitable for pan-cancer analysis of the association between genes and tumors [[1], [2], [3]].
Geranylgeranyl diphosphate synthase (GGPPS) is a key enzyme in the mevalonate (MVA) pathway and is responsible for the condensation of 15-carbon farnesyl pyrophosphate into 20-carbon geranylgeranyl pyrophosphate (GGPP) [4]. GGPP belongs to the isoprene hydrophobic groups that can covalently bind to conserved cysteine residues (such as CAAX and CCXX) at the C-terminus of proteins through transferase catalysis. This modification increases the lipophilicity of the substrate, enabling the protein to interact and bind to the membrane of other proteins [5]. However, it generally does not affect the stability and activity of the proteins [6].
We previously found that Ggps1 knockout inhibits the proliferation of young vascular smooth muscle cells and causes apoptosis in mice [7]. Additionally, other studies have found that GGPPS/GGPS1 expression is involved in the occurrence of hepatocellular carcinoma (HCC) [8] and tumor metastasis of in lung adenocarcinoma (LUAD) [9]. These findings suggested that GGPPS may be involved in cancer progression and is a potential target of cancer treatment.
However, more evidences is required to determine whether GGPS1 is worth further studying for its role in tumors. Therefore, we conducted a pan-cancer analysis of GGPS1 using clinical data from sources such as TCGA database. We evaluated RNA expression, protein expression, gene alteration, and immune infiltration. We enriched pathways from GGPS1-correlated genes in cancers and GGPPS-interacting proteins from experiments to investigate the correlation and potential molecular mechanism of GGPS1 in the pathogenesis and clinical prognosis of various cancers.
2. Materials and methods
2.1. Gene expression analysis
The “Gene_DE” module in tumor immune estimation resource, version 2 (TIMER2) [[10], [11], [12]] [cited 2022 Jun 29] was used to evaluate the RNA expression of GGPS1 in tumor and adjacent normal tissues among diverse tumor subtypes from the TCGA database. The University of Alabama at Birmingham cancer data analysis portal (UALCAN) [cited 2022 Jun 29] provided a protein expression level analysis option using data from the clinical proteomic tumor analysis consortium (CPTAC) [13,14]. Protein expression of human GGPPS (NP_001032354) in diverse tumor and normal tissues was observed. Immunohistochemistry staining images of GGPS1 in breast carcinoma (BRCA), LUAD, liver cancer, colon adenocarcinoma (COAD), prostate cancer, and normal tissues were obtained from the human protein atlas (HPA) database [cited 2024 Jun 29]. The default conditions of the respective websites were used to generate the results.
2.2. Survival prognosis analysis
The “Survival Map” module under “Survival Analysis” in gene expression profiling interactive analysis 2 (GEPIA2) [15], was used to analyze the correlation of GGPS1 expression and patient prognosis, including overall survival (OS) and disease-free survival (DFS) across all TCGA tumors. The relevant thresholds were set as follows: significance level at “0.05”, “median” for group cutoff, and “50 %” for both cutoff-high and cutoff-low. Cancers with a significant correlation (p < 0.05) in OS and DFS were marked with a black dark frame. Kaplan-Meier curves were plotted for cancers with these significant correlations. All other conditions used to output the results were set to the default parameters of the GEPIA2 sites.
2.3. Genetic alteration analysis
In cBioPortal website [16,17] [cited 2022 Jun 29], the “TCGA Pan Cancer Atlas Studies” under “Quick select” section was used to obtain alteration frequency of GGPS1. The results of the mutation, amplification, deep deletion, and multiple alterations were observed in the “Cancer Types Summary” module across all TCGA tumors. In “Mutations” module, the number of GGPS1 alterations was observed along with the exon pattern. Additionally, “Survival” module under “Comparison/Survival” section revealed the survivorship curves for TCGA cancer cases with or without GGPS1 genetic alteration in OS, progression free survival (PFS), disease-specific survival (DSS), and DFS. All other conditions used to output the results were set to the default conditions of the cBioPortal sites.
2.4. Immune infiltration analysis
The “Immune” module under “Gene” section of the TIMER2 website was used to analyze the correlation between GGPS1 expression and immune infiltration. Cancer-associated fibroblasts (CAFs) were selected to assess infiltration levels and obtain a related heatmap using EPIC, MCPCOUNTER, XCELL, and TIDE for immune infiltration estimations. The GGPS1 expression and infiltration levels in esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), and HNSC-HPV (human papillomavirus)- were visualized as scatter plots. The p-values for purity and infiltration levels were obtained using the purity-adjusted Spearman's rank correlation test.
2.5. GGPS1-related gene enrichment analysis
To identify GGPPS-interacting proteins, we searched for “GGPPS” as the protein name and “Homo sapiens” as the organism on the search tool for the retrieval of interacting genes/proteins (STRING) website [18] [cited 2022 Jun 29]. The following parameters were configured under “Settings” mudule: “full STRING network” for network type; “evidence” for meaning of network edges; “Experiments” for active interaction sources; “low confidence (0.150)” for minimum required interaction score; “no more than 50 interactors” for max number of interactors to show; and “interactive svg” for network display options. We obtained a networkof GGPPS and the top 50 GGPPS-interacting proteins.
The “Similar Gene Detection” module under “Similar Genes” section in GEPIA2 website provided GGPS1-correlated genes with similar expression patterns across cancer types and tissues. Moreover, the “Gene_Corr” module of TIMER2 was used to create a heatmap depicting the relationship between GGPS1 and several genes, such as component of oligomeric golgi complex 2 (COG2), SCY1-like pseudokinase 3 (SCYL3), signal recognition particle 9 (SRP9), translin associated factor X (TSNAX), and zinc finger and BTB domain containing 41 (ZBTB41) in various cancers.
We used Venny2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) to generate an interactive Venn diagram composing of GGPPS-interacting proteins and GGPS1-correlated genes in various cancers from the TCGA database. We entered the combined proteins into the OmicShare website (https://www.omicshare.com/tools/Home/Task/index) for Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment. The first 20 signaling pathways were depicted.
3. Results
3.1. Gene expression analysis data
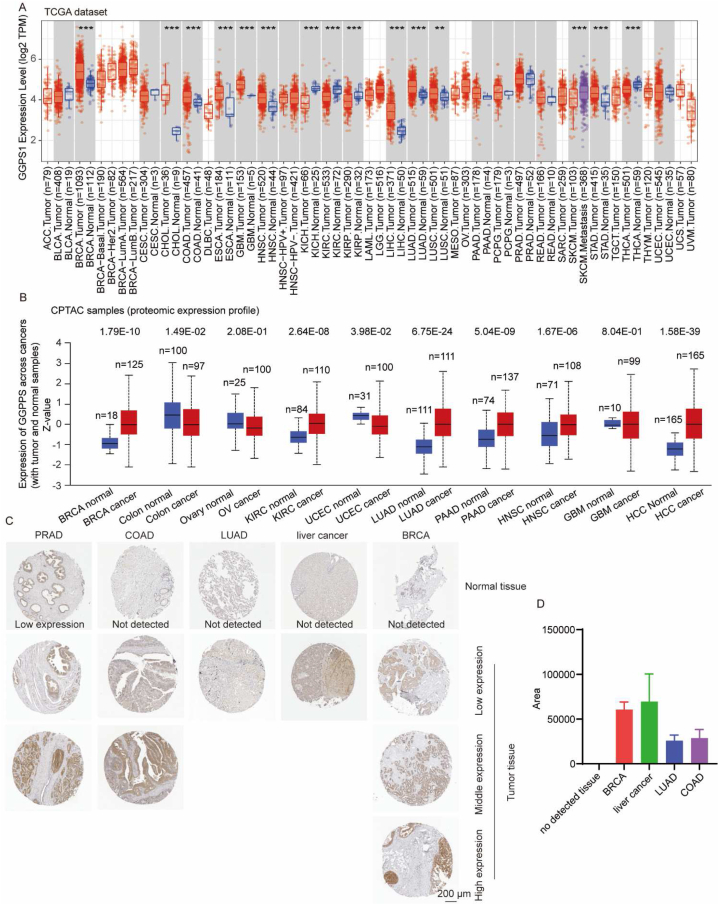
We compared the RNA expression differences of GGPS1 between cancer tissues and adjacent normal tissues in various cancers using the TCGA data in the TIMER2 website. The results revealed that the RNA expression levels of GGPS1 in the tumor tissues of BRCA, cholangiocarcinoma, COAD, ESCA, glioblastoma multiforme, HNSC, kidney chromophobe, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), LIHC, LUAD, lung squamous cell carcinoma (LUSC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), and thyroid carcinoma were significantly (p < 0.001) higher than that of corresponding normal tissues (Fig. 1A).
Fig. 1.
Expression level of GGPS1 gene in diverse tumors A) Expression status of the geranylgeranyl diphosphate synthase 1 (GGPS1) gene in diverse cancer types or specific subtypes analyzed usingTIMER2. The statistical significance computed using the Wilcoxon test is annotated by the number of stars. *: p < 0.05; **: p < 0.01; ***: p < 0.001. B) Analysis of the expression level of GGPPS total protein between normal and cancer tissues based on the CPTAC dataset. The p values were calculated using one-sided Fisher's exact test. C) Immunohistochemistry staining image of GGPS1 in LUAD, BRCA, COAD, liver cancer, and prostate cancerwere obtained from the HPA database. Scale bar: 200 μm. D) Quantitative histogram of C. n = 5∼11.
We used the UALCAN website to further evaluate the total GGPPS protein expression in cancer samples compared to normal tissues based on the CPTAC database. The results indicated significant upregulation of GGPPS protein expression in BRCA, KIRC, LUAD, pancreatic adenocarcinoma, HNSC, and HCC (p < 0.0001) compared to that in normal tissues (Fig. 1B). These results were consistent with the RNA expression results. However, GGPPS protein expression decreased in colon cancer (p < 0.05), which was inconsistent with the RNA results (Fig. 1B). The RNA expression compensates for the increase when protein expression decreases. Therefore, we speculated that this inconsistency may be due to negative feedback regulation. We further evaluated GGPPS protein expression in BRCA, LUAD, liver cancer, COAD, prostate cancer, and normal tissues using immunohistochemical results from the HPA database. GGPPS protein expression was detected in normal prostate tissue but not liver, breast, colon, and lung tissues. The results indicated that the GGPPS protein expression levels in the BRCA, LUAD, liver cancer, COAD and prostate cancer were increased compared to adjacent normal tissue (Fig. 1C). This suggests that high GGPS1 expression may be involved in the tumorigenesis of various cancers, especially BRCA, LUAD and liver cancer.
3.2. Survival analysis data
To determine whether GGPS1 expression level was related to patient prognosis, cancer cases from the TCGA database were divided into high- and low-expression groups based on a 50 % cutoff of GGPS1 expression. As depicted in Fig. 2A and B, high GGPS1 expression was significantly linked to poor OS in several cancers: adrenocortical carcinoma (ACC)(hazard ratio [HR] > 1; p = 0.0052); LIHC (HR > 1; p = 0.0021); and uveal melanoma(HR > 1; p = 0.026). Similarly, it was associated with poor DFS in ACC (HR > 1; p = 0.0002); bladder urothelial carcinoma (HR > 1; p = 0.03), cervical squamous cell carcinoma (HR > 1; p = 0.0064); and KIRP (HR > 1; p = 0.026). Additionally, low GGPS1 expression was associated with the poor prognosis of OS (HR < 1; p = 0.0033) and DFS (HR < 1; p = 0.0041) for KIRC (Fig. 2A and B). These findings suggest a significant association between GGPS1 expression and cancer prognosis, with high GGPS1 expression predicting poor outcomes across various cancer types.
Fig. 2.
Correlation between GGPS1 gene expression and survival prognosis of cancers in TCGA A) Overall survival (OS) and B) disease-free survival (DFS) analyses of diverse tumors in the cancer genome atlas (TCGA) based on GGPS1 expression using GEPIA2 tool. Red label indicates positively correlation of GGPS1 expression to hazard ratio (HR). Blue label indicates negatively correlation. The cancers with significant correlation (p < 0.05) between GGPS1 expression and patient prognosis of OS and DFS in survival map are marked with black dark frame. Kaplan-Meier curves depict survival outcomes of these cancers. p values estimated using Mantel–Cox test. HR calculated based on Cox proportional hazards Model. HR > 1 represents high GGPS1 expression with reduced survival rate. HR < 1 represents high GGPS1 expression with increased survival rate.
3.3. Genetic alteration analysis data
To determine how GGPS1 changes at the genetic level in diverse types of cancer, we used the cBioPortal website to evaluate the genetic alteration status of GGPS1 based on data from the TCGA database. The results revealed that amplification was the most common alteration observed in most cancers. The frequency of amplification was up to 10 % in BRCA (Fig. 3A), indicating an increased number of GGPS1 DNA copies in the genome. Additionally, this likely explains the upregulation of GGPS1 protein and RNA expression in BRCA.
Fig. 3.
Mutation features of GGPS1 in different tumors based on TCGA A) The alteration frequency with mutation types and B) mutation sites of GGPS1 gene in diverse cancers analyzed based on TCGA in the cBioPortal website. C) The correlation between GGPS1 genetic alteration and OS, DSS, DFS, and PFS analyzed in cBioPortal website. CNA: copy number alterations. VUS: variants of uncertain significance. p values were estimated using Mantel–Cox test.
Mutations were as the primary alteration type in endometrial carcinoma (up to 3 % alteration frequency), mature B-cell neoplasms, and COAD (Fig. 3A). We further evaluated the mutation types, mutation sites and case numbers based on 10 953 patients/10 967 samples from 32 studies. We identified only 47 variants (Fig. 3B). These results suggest that mutations are a relatively uncommon type of genetic alteration in cancer. Additionally, we explored the correlation between GGPS1 genetic alterations and clinical survival prognosis based on TCGA database. However, no statistically significant differences between GGPS1 genetic alterations and the survival across various cancers were observed (Fig. 3C).
3.4. Immune infiltration analysis data
Immune invasion into the tumor microenvironment (TME) is an critical feature associated with tumor development. CAFs are involved in regulating the function of various tumor infiltrating immune cells [19]. TIMER2.0 was used to generate a heatmap table of the Spearman's correlations between the GGPS1 expression and the abundance of CAFs in the 59-cell hierarchy across all cancer types. The EPIC, MCPCOUNTER, XCELL, and TIDE algorithms were used to assess immune infiltration. The cor less than 0.2 indicated that no correlation between tumor purity and GGPS1 expression. In other words, the tumor purity didn't influence the correlation between GGPS1 expression and immune infiltration of CAFs. The results showed that GGPS1 expression was slightly positively correlated with CAF infiltration in BRCA, ESCA, HNSC, HNSC-HPV-, LUSC, and STAD (Cor >0.2; p < 0.05) (Fig. 4A and B).
Fig. 4.
Correlation analysis between GGPS1 expression and immune infiltration of CAFs A) Correlation analysis between GGPS1 expression and immune infiltration of CAFs using EPIC, MCPCOUNTER, XCELL, and TIDE for immune infiltration estimations across all types of cancer based on TCGA database. Red label indicates positive correlation (p < 0.05, Cor >0). Blue label indicates negative correlation (p < 0.05, Cor >0). Gray label indicates not significant correlation (p > 0.05). B) Correlation of GGPS1 expression with tumor purity (the proportion of cancer cells in a sample) (left) and with the infiltration level of CAFs in ESCA, HNSC, HNSC-HPV estimated by TIMER (right) were shown as scatter plot. Cor: The Spearman's rank correlation coefficients.
3.5. Enrichment analysis of GGPS1-related functional pathways
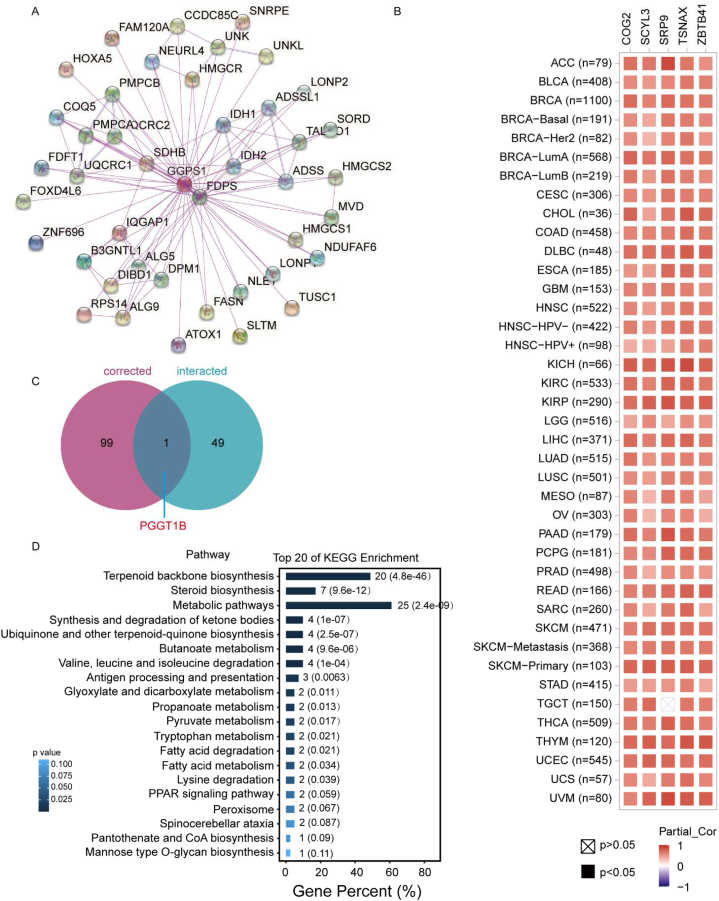
To elucidate the mechanism of action of GGPS1 in cancer development, we utilized the GGPPS-interacting proteins obtained from experiments and GGPS1-correlated genes in cancers to conduct functional enrichment analysis. Fifty GGPPS-interacting proteins were screened using STRING (Fig. 5A). The top 100 proteins sharing similarly expression patterns with GGPS1 in cancers were screened using the GEPIA2 tool (Fig. 5B). One common gene, the protein geranylgeranyltransferase type I beta subunit (PGGT1B), was found among the GGPPS-interacting proteins and GGPS1-correlated genes (Fig. 5C). PGGT1B, which encodes encodes a prenylation-catalyzing enzyme (geranylgeranyltransferase type I; GGTase I), is responsible for transferring GGPP to substrate proteins and is involved in inflammation and metabolic processes [20,21]. We used the 149 genes to conduct functional enrichment analysis on the OmicShare website. The results highlighted that the top Kyoto encyclopedia of genes and genomes (KEGG) pathways were related to metabolic pathways, ranking second only to terpenoid backbone and steroid biosynthesis.
Fig. 5.
GGPS1-related gene enrichment analysis A) Co-expression network of 50 genes interacting with GGPPS obtained from the STRING tool. B) Top 100 GGPS1- correlated genes in TCGA projects obtained using GEPIA2. The top genes whose expression are associated with GGPS1, including COG2, SCYL3, SRP9, TSNAX, and ZBTB41 are displayed as heatmap. The Spearman's rank correlation coefficients were calculated. C) Correlation of GGPS1-interacting proteins and correlated genes are presented as a Venn-diagram. D) Top 20 signaling KEGG pathways analyzed using OmicShare based on the GGPPS-interacting proteins and GGPS1-correlated genes.
4. Discussion
Recent reports have highlighted that the MVA pathway is involved in several aspects of tumorigenesis including the control of cell proliferation, survival, invasion and metastasis [[22], [23], [24]]. Our findings also demonstrated that the RNA and protein expression of GGPS1 was significantly upregulated in most tumors, including BRCA, LUAD, PAAD and GBM. Amplification was also the primary GGPS1 genetic alteration pattern in BRCA and HCC samples. Additionally, high GGPS1 expression was significantly correlated with the poor OS and DFS outcomes in cancers, such as HCC. The GGPS1 expression was significantly and positively correlated with CAFs in several tumors including BRCA. However, GGPPS protein expression was decreased in a few cancers, such as colon cancer and UCEC. The inconsistent of GGPPS expression revealed the complexity of tumor formation. The specific reasons need to be studied in the future.
GGPPS is a key enzyme in the MVA pathway, suggesting that the GGPPS expression/MVA pathway is highly associated with cancers progression and patient prognosis in several tumors types, such as BRCA, HCC and LUAD. Statins, which are mevalonate synthesis inhibitors, are potential drugs for cancer treatment [25]. However, the relationship between GGPS1 expression and cancer requires further investigation. GGPPS deletion caused cell death through eicosanoids metabolism disorders and inflammation accumulation [7]. Inflammation within the TME is regarded as a cancer marker and has been studied as a key target for cancer treatment in recent years. We applied multiple algorithms (EPIC, XCELL, TIDE and MCPCOUNTER) and observed a statistically significant positive correlation between GGPS1 expression and CAFs in BRCA, ESCA, HNSC, HNSC-HPV-, LUSC, and STAD tumors.
Cancer is associated with substantial changes in the cellular metabolism. Recent studies reported that arachidonic acid (AA) was enhanced in PIK3CA-induced BRCA. PIK3CA-induced tumorigenesis can be rescued by inhibiting cPLA2 synergy and limiting fatty acid intake [26]. AA, synthesized from dietary polyunsaturated fatty acids (PUFA), are the key substrates for eicosanoids metabolism. Bioactive metabolites exert both proinflammatory and anti-inflammatory effects [27]. Multiple studies have reported that eicosanoids metabolism is associated with tumor growth, progression and metastasis [28,29]. Despite the multiplex network of eicosanoid metabolites, there are no well-defined targets for eicosanoids metabolism in cancer treatment. Our previous findings suggested that GGPPS and/or CYB5R3 [7], which are eicosanoid metabolism upregulators, are potentially effective targets for cancer treatment concerning inflammation. Our analysis identified GGTase I as a major downstream target of GGPPS that not only binds to GGPPS but also correlates with GGPS1 in cancers. GGTase I is responsible for the protein prenylation during the post-translational modification of several target proteins containing CAAX at the C-terminus, especially the geranylgeranylation of small G proteins, such as Rac1(20). This may be the key mechanism through which GGPS1 and PGGT1B are involved in cancer development.
We found that the top KEGG pathways enriched from GGPS1 interacting proteins and correlated genes were also related to metabolic pathways. These results suggest that eicosanoid metabolism may be an emerging perspective on the effect of GGPPS on cancers. In summary, we revealed a significantly correlation of GGPS1 and cancer development, clinical prognosis, and immune infiltration across diverse cancer types using a pan-cancer analysis based on a lot of public clinical data. We hypothesize that metabolism, especially eicosanoids metabolism, plays a key role in the involvement of GGPPS in cancer. However, the underlying mechanism requires further studies.
Abbreviations: GGPS1: geranylgeranyl diphosphate synthase 1; TCGA: The Cancer Genome Atlas; MVA pathway: mevalonate pathway; FPP: farnesyl pyrophosphate; GGPP: geranylgeranyl pyrophosphate; CPTAC: Clinical Proteomic Tumor Analysis Consortium; HPA: Human Protein Atlas; OS: overall survival; DFS: disease-free survival; DSS: disease-specific survival; PFS: progress-free survival; ACC: adrenocortical carcinoma; BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma; CHOL: cholangio carcinoma; COAD: colon adenocarcinoma; DLBC: diffuse large B cell lymphoma; ESCA: esophageal carcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LAML: acute myeloid leukemia; LGG: low-grade glioma; LIHC (HCC): liver hepatocellular carcinoma (hepatocellular carcinoma); LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MESO: malignant mesothelioma; OV: ovarian cancer; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectal cancer; SARC: sarcoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; UCEC: uterine corpus endometrial cancer; UCS: uterine carcinosarcoma; UVM: uveal melanoma; EC: endometrial carcinoma; EOC: epithelial ovarian cancer; HNSC-HPV(human papillomavirus)-; COG2: component of oligomeric golgi complex 2; SCYL3: SCY1-like pseudokinase 3; SRP9: signal recognition particle 9; TSNAX: translin associated factor X; ZBTB41: Zinc finger and BTB domain containing 41; KEGG: Kyoto encyclopedia of genes and genomes; TME: tumor microenvironment; CAFs: tumor-associated fibroblasts; PGGT1B: protein geranylgeranyl transferase type I beta subunit; GGTase I: geranylgeranyltransferase type I; AA: arachidonic acid; PUFA: polyunsaturated fatty acids.
Funding
This study was supported by National Natural Science Foundation of China Grants 82200976 (to Lisha Wei).
Data sharing statement
Publicly available datasets were analyzed in this study. These data can be found as follows. The gene expression analysis was available from TIMER2 (http://timer.cistrome.org/), UALCAN (http://ualcan.path.uab.edu/analysis-prot.html) and GEPIA2 (http://gepia2.cancer-pku.cn/#index). Survival prognosis analysis was available from GEPIA2 (http://gepia2.cancer-pku.cn/#index). Genetic alteration analysis was available from cBioPortal (https://www.cbioportal.org/). Immune infiltration analysis was available from TIMER2 (http://timer.cistrome.org/). GGPS1-related gene enrichment analysis was available from STRING (https://string-db.org/), GEPIA2 (http://gepia2.cancer-pku.cn/#index), Venny2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) and OmicShare website (https://www.omicshare.com/tools/Home/Task/index).
Ethics statement
No experiments were performed in this study. Therefore, ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
CRediT authorship contribution statement
Lisha Wei: Writing – review & editing, Writing – original draft, Visualization, Software, Resources, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
All authors disclosed no relevant relationships.
References
- 1.Blum A., Wang P., Zenklusen J.C. SnapShot: TCGA-analyzed tumors. Cell. 2018;173:530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 2.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clough E., Barrett T. The gene expression omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 5.McTaggart S.J. Isoprenylated proteins. Cell. Mol. Life Sci. 2006;63:255–267. doi: 10.1007/s00018-005-5298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F.L., Casey P.J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 7.Wei L., Zheng Y.Y., Sun J., Wang P., Tao T., Li Y., et al. GGPP depletion initiates metaflammation through disequilibrating CYB5R3-dependent eicosanoid metabolism. J. Biol. Chem. 2020;295:15988–16001. doi: 10.1074/jbc.RA120.015020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu D.C., Liu J., Chen J., Shao J.J., Shen X., Xia H.G., et al. GGPPS1 predicts the biological character of hepatocellular carcinoma in patients with cirrhosis. BMC Cancer. 2014;14:248. doi: 10.1186/1471-2407-14-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Xu W., Zhan P., Xu T., Jin J., Miu Y., et al. Overexpression of geranylgeranyl diphosphate synthase contributes to tumour metastasis and correlates with poor prognosis of lung adenocarcinoma. J. Cell Mol. Med. 2018;22:2177–2189. doi: 10.1111/jcmm.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B., Severson E., Pignon J.C., Zhao H., Li T., Novak J., et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.Can-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–w514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat. Commun. 2022;13:2669. doi: 10.1038/s41467-022-30342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–w560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.Cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei X., Lei Y., Li J.K., Du W.X., Li R.G., Yang J., et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Chen G.P., Yang J., Qian G.F., Xu W.W., Zhang X.Q. Geranylgeranyl transferase-I knockout inhibits oxidative injury of vascular smooth muscle cells and attenuates diabetes-accelerated atherosclerosis. J. Diabetes Res. 2020;2020 doi: 10.1155/2020/7574245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Posadas R., Fastancz P., Martinez-Sanchez L.D.C., Panteleev-Ivlev J., Thonn V., Kisseleva T., et al. Inhibiting PGGT1B disrupts function of RHOA, resulting in T-cell expression of integrin alpha4beta7 and development of colitis in mice. Gastroenterology. 2019;157:1293–1309. doi: 10.1053/j.gastro.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Clendening J.W., Pandyra A., Boutros P.C., El Ghamrasni S., Khosravi F., Trentin G.A., et al. Dysregulation of the mevalonate pathway promotes transformation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15051–15056. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fatehi Hassanabad A. Current perspectives on statins as potential anti-cancer therapeutics: clinical outcomes and underlying molecular mechanisms. Transl. Lung Cancer Res. 2019;8:692–699. doi: 10.21037/tlcr.2019.09.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen P.J., Yu R., Longo J., Archer M.C., Penn L.Z. The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer. 2016;16:718–731. doi: 10.1038/nrc.2016.76. [DOI] [PubMed] [Google Scholar]
- 25.Juarez D., Fruman D.A. Targeting the mevalonate pathway in cancer. Trends Cancer. 2021;7:525–540. doi: 10.1016/j.trecan.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koundouros N., Karali E., Tripp A., Valle A., Inglese P., Perry N.J.S., et al. Metabolic fingerprinting links oncogenic PIK3CA with enhanced arachidonic acid-derived eicosanoids. Cell. 2020;181:1596–1611.e1527. doi: 10.1016/j.cell.2020.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B., Wu L., Chen J., Dong L., Chen C., Wen Z., et al. Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets. Signal Transduct. Targeted Ther. 2021;6:94. doi: 10.1038/s41392-020-00443-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panagiotopoulos A.A., Kalyvianaki K., Castanas E., Kampa M. Eicosanoids in prostate cancer. Cancer Metastasis Rev. 2018;37:237–243. doi: 10.1007/s10555-018-9750-0. [DOI] [PubMed] [Google Scholar]
- 29.Cathcart M.C., Lysaght J., Pidgeon G.P. Eicosanoid signalling pathways in the development and progression of colorectal cancer: novel approaches for prevention/intervention. Cancer Metastasis Rev. 2011;30:363–385. doi: 10.1007/s10555-011-9324-x. [DOI] [PubMed] [Google Scholar]