Abstract
The occurrence of harmful algal blooms (HABs) in freshwater environments has been expanded worldwide with growing frequency and severity. HABs can pose a threat to public water supplies, raising concerns about safety of treated water. Many studies have provided valuable information about the impacts of HABs and management strategies on the early-stage treatment processes (e.g., pre-oxidation and coagulation/flocculation) in conventional drinking water treatment plants (DWTPs). However, the potential effect of HAB-impacted water in the granular media filtration has not been well studied. Biologically-active filters (BAFs), which are used in drinking water treatment and rely largely on bacterial community interactions, have not been examined during HABs in full-scale DWTPs. In this study, we assessed the bacterial community structure of BAFs, functional profiles, assembly processes, and bio-interactions in the community during both severe and mild HABs. Our findings indicate that bacterial diversity in BAFs significantly decreases during severe HABs due to the predominance of bloom-associated bacteria (e.g., Spingopyxis, Porphyrobacter, and Sphingomonas). The excitation-emission matrix combined with parallel factor analysis (EEM-PARAFAC) confirmed that filter influent affected by the severe HAB contained a higher portion of protein-like substances than filter influent samples during a mild bloom. In addition, BAF community functions showed increases in metabolisms associated with intracellular algal organic matter (AOM), such as lipids and amino acids, during severe HABs. Further ecological process and network analyses revealed that severe HAB, accompanied by the abundance of bloom-associated taxa and increased nutrient availability, led to not only strong stochastic processes in the assembly process, but also a bacterial community with lower complexity in BAFs. Overall, this study provides deeper insights into BAF bacterial community structure, function, and assembly in response to HABs.
Keywords: Harmful algal bloom, Biologically-active filters, Bacterial community structure, Network analysis, Drinking water treatment process
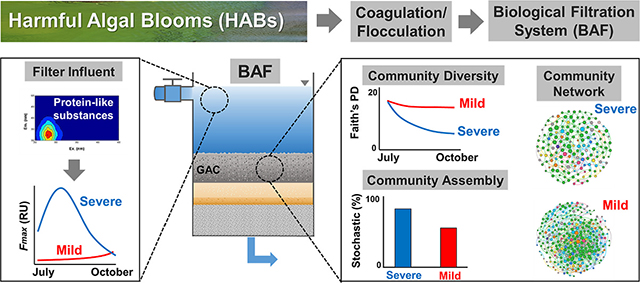
GRAPHICAL ABSTRACT
1. Introduction
Cyanobacteria play a critical role in recycling carbon and nitrogen as the primary producers in most aquatic ecosystems (Sánchez-Baracaldo et al., 2022). Harmful algal blooms (HABs) are massive, uncontrolled growth of cyanobacteria, which can have adverse effects on aquatic organisms and humans. The main concerns of water utilities facing HABs include deteriorated water quality by algal organic matter (AOM) containing cyanotoxins, taste and odor causing compounds, and the formation of disinfectant byproducts (DBPs) (Liu et al., 2020). Especially, in 1998, the World Health Organization proposed a drinking-water guideline for microcystins-LR (MC-LR), one of the most toxic cyanotoxins. Since then, many countries have implemented appropriate regulatory guidelines to manage the risk of cyanotoxins in drinking water (Ibelings et al., 2014). Meanwhile, various physical, chemical, and biological treatment methods have been evaluated to mitigate the global increase of HABs (Gallardo-Rodríguez et al., 2019; Paerl et al., 2018), but no practical solution has emerged yet. For that reason, the annual recurrence of HABs still poses significant challenges for water utilities (Brooks et al., 2016).
Most drinking water treatment plants (DWTPs) rely on a conventional treatment process, consisting of coagulation/flocculation (C/F), sedimentation, filtration, and disinfection (Shannon et al., 2009). To manage the influx of algal cells during HABs, DWTPs have implemented C/F processes coupled with high doses of coagulants and pre-oxidation (e.g., permanganate and chlorine) (Piezer et al., 2021; Qi et al., 2021). The pre-oxidation step has been shown to be effective for algal cell removal by enhancing coagulation (Lin et al., 2021). Despite its effectiveness, the treatment process requires a relatively greater amount of the oxidant or a longer contact time to attain satisfactory removal efficiency during heavy loads of algal blooms (Jia et al., 2018). This can lead to the release of additional intracellular AOM, including cyanotoxins, from damaged algal cells. In addition, AOM includes large portions of hydrophilic and low molecular weight compounds, which are difficult to be removed by the C/F treatment (Joh et al., 2011; Naceradska et al., 2019). Consequently, AOM possibly affects the downstream granular-media filtration system, which is the last physical unit process of a DWTP. However, much remains unknown about how filtration is affected during HABs.
Biologically-active filters (BAFs) with granular activated carbon (GAC) have been adopted for the removal of natural organic matter (NOM) and various contaminants (e.g., DBPs, pharmaceutically active compounds) in DWTPs (Golea et al., 2020; Kennedy et al., 2015; Piai et al., 2020). In BAFs, microorganisms colonized on the large surface area of GAC play an essential role in degrading contaminants (Yuan et al., 2022). With the development of next-generation sequencing technologies, recent studies have improved our understanding of microorganisms in terms of diversity and community structure in BAFs (Abkar et al., 2024). These have provided us with a deeper insight into the biofilm-associated processes involved in the transformation of target contaminants in BAFs (Gibert et al., 2013). However, the underlying mechanisms are still not well understood and are considered a black box. Identifying ecological mechanisms and forces governing microbial assemblages can facilitate to unveil key parameters in managing BAF performance in DWTPs. Based on ecological principles, the structure of microbial communities is determined by deterministic and stochastic processes, which are both complex and dynamic (Chase and Myers, 2011; Vellend et al., 2014; Zhou and Ning, 2017). Deterministic processes are the result of non-random, niche-based mechanisms imposed by both environmental filtering (e.g., eutrophication and drought) and species interactions (e.g., mutualism, predation, and competition), whereas stochastic processes involve random ecological processes that generate chance colonization and random extinction. Previously, studies have focused on understanding microbial succession in natural ecosystems (e.g., soil and groundwater) (Ferrenberg et al., 2013; Zhou et al., 2014), demonstrating that the initial succession of microbial communities is governed by stochastic processes, and further determined by environmental factors. Hence, applying the ecological approach to engineered water systems can help better explain complicated microbial community assembly. To the best of our knowledge, however, no studies have assessed the dynamics of BAF bacterial communities, functional profiles, and ecological assembly processes subjected to the influence of HABs.
The primary goal of the study was to uncover the impact of the severity of HABs on the dynamics of bacterial communities within BAFs, focusing on bacterial community assembly and function. Specifically, the specific objectives of this study were: (1) to investigate the bacterial community diversity and composition of BAFs during mild and severe HABs, (2) to examine the correlation between BAF community functions and the severity level of HABs, and (3) to unravel governing assembly processes shaping BAF communities and species interaction patterns during HABs.
2. Materials and methods
2.1. Study site, sampling, and sample analysis
The studied DWTP is located in the City of Toledo (Ohio, USA) and takes source water from Lake Erie, which experiences chronic HABs dominated by Microcystis. The DWTP utilizes conventional treatment processes, including potassium permanganate pretreatment, coagulation, flocculation, sedimentation, GAC/sand filtration, and chlorine disinfection. Field samplings were performed once a month for two consecutive years during the typical HAB season (July through October 2017 and 2018). National Oceanic and Atmospheric Administration (NOAA) annually provides information for seasonal HABs in Lake Erie based on the Severity Index (SI). SI is calculated by the amount of bloom biomass captured over the peak (for 30 days) of a bloom (Leadbetter et al., 2018). This is further correlated with the cyanobacteria index (CI) (Wynne and Stumpf, 2015). In the case of Lake Erie, the CI corresponds to Microcystis biomass, with CI of 0.001 sr−1 corresponding to 105 cells mL−1. The calculations made in this are based on the nadir pixel view of about 1 km by both Moderate Resolution Imaging Spectroradiometer and reduced-resolution Medium Resolution Imaging Spectrometer data. If the surface concentration is assumed to be one meter deep (the maximum depth of detection), then an accumulated CI of 1.0 units corresponds to 1020 cells in the area (Stumpf et al., 2012). When the SI is in the range of 2–4, blooms were regarded as mild, with severe blooms above 4. Thus, the mild bloom indicates that there are <4 × 1020 cells in the area. The largest bloom observed by NOAA was 10.5 in 2015. According to the SI, the 2017 (SI: 8) and 2018 (SI: 3.6) bloom events were categorized as severe and mild HABs, respectively (Wynne et al., 2013). It should be pointed out that either a severe or a mild bloom caused by toxic cyanobacteria has occurred in Lake Erie every year since 2007 (Wynne et al., 2021).
The sampling was performed once a month from BAFs, which consisted of a layer of GAC (effective size: 1.0–1.2 mm). Before filter media sampling, BAFs were drained and GAC filter media were collected at depths of 2 and 6 in. The collected samples were stored in sterile bags and then transported in a cooler to the laboratory within 2 h. On the same day, 1 L of filter influent was also collected in a sterile container and subsamples were used to analyze water quality parameters. Raw water quality information of Lake Erie (e.g., temperature, MCs, chlorophyll, and dissolved phosphorus) was obtained from the NOAA’s Archived Lake Erie Water Quality and Microcystin Data (https://www.glerl.noaa.gov/res/HABs_and_Hypoxia/wle-weekly/; accessed on Dec 1, 2019) (Table S1). The station name was WE12 (latitude: 44.7°, longitude: −83.26°), which was the closest station from the Toledo WTP intake crib (Fig. S1). Turbidity and alkalinity data were obtained from the Toledo WTP. The total organic carbon (TOC) and total nitrogen (TN) of the filter influent samples were obtained using a TOC analyzer (TOC-VCSH, Shimadzu, Japan). The remaining media samples were stored at −80 °C for DNA extraction and sequencing.
2.2. Fluorescence emission-excitation matrix (EEM) spectra and parallel factor analysis (PARAFAC)
Fluorescence EEM spectra of collected filter influents were gathered using a spectrofluorometer (RF-6000, Shimadzu, Japan). Excitation wavelengths of 250–400 nm in 5 nm intervals and emission wavelengths of 300–600 nm in 2 nm intervals were scanned. Each sample was analyzed in triplicate. Deionized water was scanned for collecting blank EEM spectra prior to sample analyses. Inner filter effects were corrected by a matrix of correction factors, which were generated from the absorbance spectra (200–600 nm) of each sample, measured by a UV–visible spectrophotometer (UV-1800, Shimadzu, Japan). Fluorescence index (FIX) was obtained by the ratio of intensities between Em 450 and 500 nm at Ex 370 nm (Jiang et al., 2022). Biological index (BIX) was the ratio of Em intensities at 380 nm and the intensity maximum in 420–435 nm, obtained at Ex 310 nm (DeVilbiss et al., 2016). The FIX provides information about the source of samples (e.g., microbial or terrestrial). When FIX is above 1.9, it implies that the dissolved organic matter (DOM) sources are dominated by microorganisms and algae. High values of BIX (>1.0) indicate that the DOM of samples is predominantly originated from an autochthonous source (Huguet et al., 2009). Data processing of 140 EEM spectra and PARAFAC modeling were performed and sample-specific maximum fluorescence (Fmax) was obtained using Matlab (MATLAB R2018a, MathWorks) and DOMFluor Toolbox (Stedmon and Bro, 2008). More details are available at Text S1.
2.3. DNA extraction, Illumina 16S rRNA sequencing, and sequences data processing
DNA was extracted from the GAC media samples (0.5 g) using the PowerSoil DNA isolation kit (Mo-Bio Laboratories, USA). DNA extracts were used to generate bacterial 16S rRNA gene metabarcoded libraries using primers (515F-806R) targeting the V4 region (Caporaso et al., 2012), as described elsewhere (Kapoor et al., 2016). The libraries were sequenced using Illumina Miseq coupled with PE250 kits at a DNA Core facility. Raw sequences were processed using QIIME2 (v2019.1) (Bolyen et al., 2019). Briefly, DADA2 was used for processing the sequences (e.g., quality filtering, denoising, merging, and chimera filtering) to generate unique amplicon sequence variants (ASVs). Representative sequences were taxonomically assigned using q2-feature-classifier based on Silva SSU database release 132 (Bokulich et al., 2018; Quast et al., 2013). Sequence alignment and construction of phylogenetic trees were conducted by q2-phylogeny. Alpha and beta diversity analyses were performed by q2-diversity. All sequence data have been deposited in the NCBI under PRJNA1040411.
To estimate the relative abundance of functional metabolic pathways, a phylogenetic investigation of communities was used via the reconstruction of unobserved states (PICRUSt2) that yielded pathway abundance output based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Clusters of Orthologous Groups (COG) databases (Douglas et al., 2020). For pathway profile visualization and comparison, the software STAMP was used (Parks et al., 2014).
Microbial ecological networks at genus level were constructed to reveal the bacteria associations in response to HABs within BAF communities using SPIEC-EASI package (1.0.2) (Kurtz et al., 2015). For the network construction, the sparse graphical lasso-based setting was used, and optimal sparsity parameter was selected based on the Stability Approach to Regularization Selection (StARS) with a variability threshold of 0.05 for all networks(Liu et al., 2010). The interactive platform Gephi (0.9.2) Bastian et al. (2009) was used to visualize the network maps and obtain network-based topological properties, such as modularity and clustering coefficient (Mendes et al., 2018). A modularity value represents how well the constructed network is separated into modules, where the value is proportionally correlated with an extent of niche differentiation. Betweenness indicates how many times a node acted as a bridge between two other nodes on their geodesic or shortest path. Average clustering coefficient is used to indicate the complexity of the network (Deng et al., 2012). Generally, the more complex a network is, the stronger the microbial interaction is.
In general, community assembly is governed by a combination of stochastic and deterministic processes (Adler et al., 2007). To examine the relative contribution of community assembly processes in BAFs under HABs, normalized stochasticity ratio (NST) was estimated with the NST package (Ning et al., 2019). This approach employs Bray-Curtis metrics for NST calculation. The principle of NST is assessing stochasticity through a comparison of the observed dissimilarity in community structures with the random patterns generated by a null model. The permutational multivariate analysis of variance (PERMANOVA) test was conducted on the model analysis with 999 permutations (N = 999). NST values, which range from 0 to 1, serve as indicators of the processes governing community assembly. A value above 0.5 indicates that the assembly is largely influenced by stochastic forces. Conversely, a value below 0.5 indicates a system in which deterministic processes are more dominant than stochastic ones.
2.4. Statistical analysis
Principal coordinate analysis (PCoA) was performed after constructing weighted UniFrac distance matrices based on the phylogenetic tree. Non-metric multidimensional scaling (NMDS) analysis based on Bray-Curtis distances was used to ordinate the BAF samples using the R package Vegan (2.5) and relationships between environmental factors and ASVs were examined after permutation (n = 999).
3. Results
3.1. Water characteristic parameters of original lake water and filter influent
The filter influent samples were obtained before collecting filter media, and their water quality parameters were analyzed (Table 1). During the severe HAB period, temperature decreased from 23.8±1.2 °C (July) to 18.2±1.9 °C (October). Turbidity ranged from 0.3±0.1 NTU to 0.6±0.1NTU, and no MC was found. TOC remained at 1.2±0.3 mg/L, but TN decreased from 1.0±0.3 mg/L to 0.1±0.05 mg/L. In the case of alkalinity, it increased from 43.5±4.3 mg/L in July to 52.2±7.3 mg/L in September, before dropping back down to 46±3.9 mg/L in October. For pH, it remained between 9.7±0.2 and 9.61±0.2. Compared to the severe HAB, no significant differences were observed during the mild HAB.
Table 1.
Water quality parameters of filter influent water in the study. Data are shown as the mean±SD.
| Severe HAB (2017) |
Mild HAB (2018) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| July | August | September | October | July | August | September | October | ||
|
| |||||||||
| WTP filter influent | Temperature (°C) | 23.8 ± 1.2 | 23.4 ± 0.8 | 20.3 ± 1.7 | 18.2 ± 1.9 | 24.5 ± 0.8 | 24.7 ± 0.5 | 23.0 ± 1.5 | 16.4 ± 3.5 |
| Turbidity (NTU) | 0.3 ± 0.1 | 0.5 ± 0.3 | 0.6 ± 0.1 | 0.5 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.04 | 0.4 ± 0.2 | 0.4 ± 0.3 | |
| Total MCs (μg/L)a | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | |
| TOC (mg/L) | 1.2 ± 0.3 | 1.1 ± 0.2 | 1.3 ± 0.1 | 1.2 ± 0.2 | 1.3 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.4 ± 0.3 | |
| TN (mg/L) | 1.0 ± 0.3 | 0.7 ± 0.5 | 0.1 ± 0.1 | 0.1 ± 0.05 | 0.7 ± 0.02 | 0.44 ± 0.1 | 0.3 ± 0.01 | 0.29 ± 0.04 | |
| Alkalinity mg/L as CaCO3 | 43.5 ± 4.3 | 48.7 ± 5.2 | 52.2 ± 7.3 | 46 ± 3.9 | 41.3 ± 2.2 | 42.1 ± 2.4 | 43.3 ± 3.1 | 46.3 ± 3.5 | |
| PH | 9.7 ± 0.2 | 9.64 ± 0.2 | 9.65 ± 0.2 | 9.61 ± 0.2 | 9.61 ± 0.1 | 9.67 ± 0.1 | 9.67 ± 0.1 | 9.65 ± 0.1 | |
Total MCs were analyzed by ELISA and its detection limit is 0.1 μg/L.
During the same time period, data from the buoy located closest to the intake crib of the DWTP and the laboratory at the DWTP monitored various parameters of Lake Erie (Table S1). Among these parameters, the concentration of particulate MC-LR showed a gradual increase starting in July, reaching its peak at 3.09±3.11 μg/L in September during the severe HAB. A similar pattern was observed during the mild HAB, with the concentration increasing to 1.47±0.16 μg/L in August before decreasing in September. A more significant contrast between the two HABs was evident in the case of phycocyanin and chlorophyll a. During the severe HAB, phycocyanin increased substantially from 1.18 ±0.85 μg/L to 141±263 μg/L, whereas in the mild HAB, it showed an increase from 2.23±2.68 μg/L to 11.4±3.57 μg/L. Likewise, chlorophyll a increased to 62.2±79.8 μg/L in September during the HAB, while the highest recorded in mild HABs was 19.0±3.13 μg/L. Overall, a notable distinction was observed between the two HABs in the source water, with the severe HAB reaching their peak in September.
3.2. Changes in fluorescence components of filter influent during HABs
Based on the EEM spectra data, FIX and BIX were obtained to characterize the source of DOM in filter influent (Fig. S2). The FIX values for the samples were >1.9, indicating that DOM of filter influent samples during both HABs was predominantly originated by microorganisms and algae. The BIX represent the contribution of autochthonous DOM. In the study, the samples from August were >1.0, indicating that they were mostly associated with an autochthonous DOM. Except for the FIX in July, the greater values were observed for severe HAB.
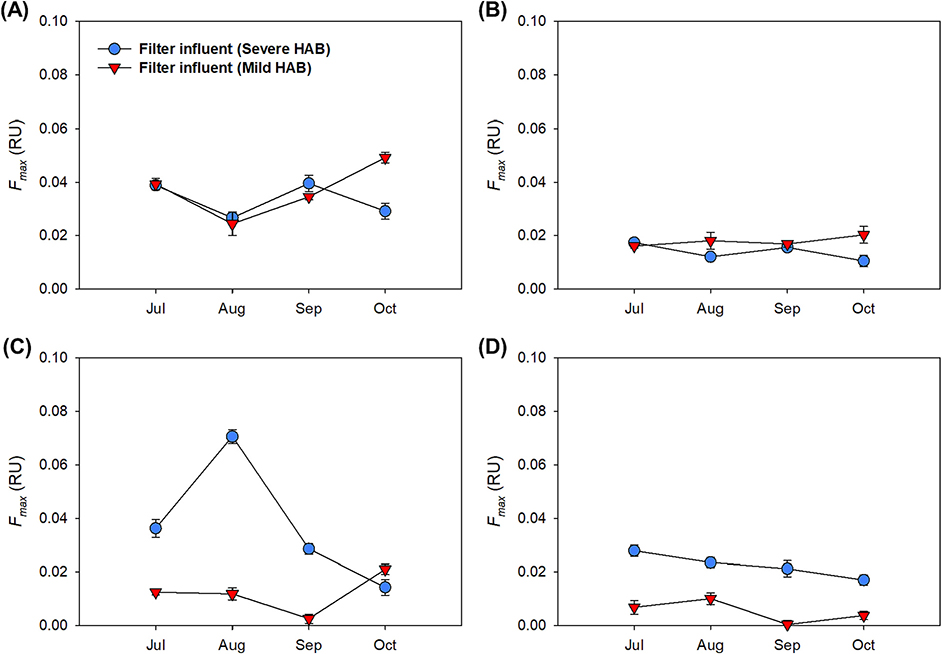
Using PARAFAC, 140 EEM spectra collected from filter influent samples of the DWTP during HABs were modeled. As illustrated in Fig. S3, four components were identified by the PARAFAC analysis. Component 1 (C1) was made up of a single peak at Ex/Em (300 nm/420 nm) (Table 2), representing a low molecular weight marine humic-like substances (Jaffé et al., 2014). Component 2 (C2) was characterized by two excitation maxima at 270 nm/370 nm with the emission maxima at 466 nm, which is related to high molecular weight terrestrial humic-like substances (Wünsch and Murphy, 2021). Component 3 (C3) and Component 4 (C4) had a fluorescent peak of Ex/Em at 280 nm/322 nm and 250 nm/308 nm, respectively. These components are related to protein-like substances. C3 is associated with tryptophan-like substances while C4 is related to tyrosine-like substances (Chen et al., 2003; Villacorte et al., 2015). As shown in Fig. 1, the Fmax values for C1 and C2 (humic-like substances) showed similar trends at the two different HAB conditions. C1 (marine humic-like substances) was more abundant than C2 (terrestrial humic-like substances) in filter influent. Meanwhile, we found that C3 and C4 (protein-like substances) showed relatively higher Fmax values in the severe HAB than in the mild HAB, with the exception of the October sample. In addition, the sharp spike of Fmax value for C3 (tryptophan-like substances) was observed in August. It’s important to note that samplings were conducted during the last week of the month. As severe bloom lasted through the end of August to mid-September, higher AOM components were observed in the August sample.
Table 2.
Spectral characteristics of the four components obtained by PARAFAC in the study and their comparison with previously identified components.
| Component | Coordinate intervals (ex/em) (nm) | Description and source assignment |
|---|---|---|
|
| ||
| C1 | 320/394 | Marine humic-like substances (Yamashita et al., 2008) |
| C2 | 270 (370)/466 | Terrestrial humic substances (Murphy et al., 2008; Stedmon et al., 2003) |
| C3 | 280/322 | Proteins, tryptophan-like substances including autochthonous protein-like substances (Chen et al., 2003; Stedmon and Markager, 2005a,b) |
| C4 | 250/308 | Protein, Tyrosine-like substances (Murphy et al., 2011) |
Fig. 1.
The Fmax values of C1 (marine humic-like substances) (A), C2 (terrestrial humic-like substances) (B), C3 (tryptophan-like substances) (C), and C4 (tyrosine-like substances) (D) in filter influent under the severe (blue) and the mild (red) HABs.
3.3. Bacterial compositions and phylogenetic dynamics of BAF microbiota during HABs
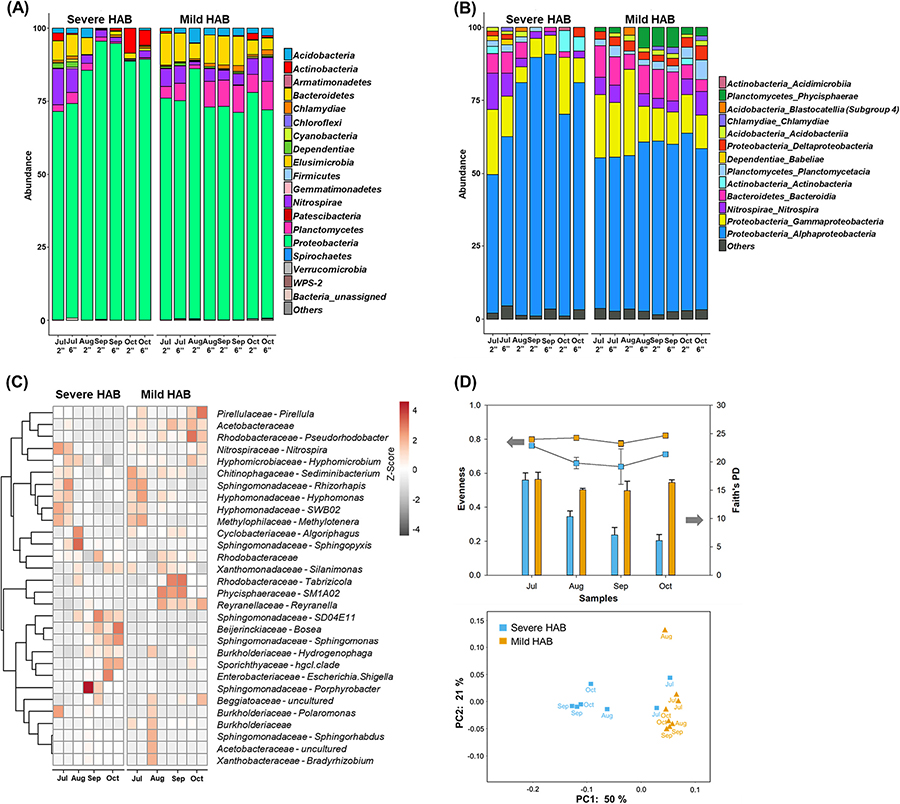
A total of 279,816 high-quality filtered sequences for GAC filter media samples were retrieved by Illumina high-throughput sequencing. Proteobacteria predominated during both HAB seasons (85.5±8.8% for severe HAB and 75.2±4.4% for mild HAB; Fig. 2A). The highest relative abundance of Proteobacteria was observed during the severe HAB (i.e., 95.1% in September and 85.6% in August). Acidobacteria (2.32±0.1%), Bacteroidetes (8.0±2.6%), Nitrospirae (4.7±2.0%), and Plantomycetes (6.7±2.8%) were prevalently enriched during the mild HAB. At the class level (Fig. 2B), Alphaproteobacteria was the most abundant group in all samples, showing a similar abundance pattern as that of Proteobacteria. Alphaproteobacteria in the severe HAB reached its maximum in September, accounting for 88.0±0.5%, and its minimum in July with 52.7±5.3%. For the mild HAB, its abundance ranged from 52.2% to 58% over the sampling period. Gammaproteobacteria was the second most abundant class, with 11.7±6.3% (severe HAB) and 16.0±6% (mild HAB).
Fig. 2.
Taxonomic and diversity analysis of bacterial communities at the WTP filtration system during the severe and the mild HABs. Relative abundance of bacterial community composition at Phylum (A) and Class level (B). The heatmap of genera with relative abundances >2% (C) (Both family and genus levels are shown. If the genus level is not available, only the family level is displayed). Differences in alpha and beta diversity (D); evenness (dot plots, left vertical axis) and Faith’s PD (bar graphs, right vertical axis) (top) and plot of principal coordinate analysis (PCoA) of the microbial communities based on the weighted Unifrac distance (bottom).
At the genus level (Fig. 2C), Sediminibacterium (2.81% and 3.91%), Rhizorhapis (3.77% and 4.81%), Hyphomonas (2.85% and 3.22%), SWB02 (3.5% and 3.0%), and Methylotenera (8.67% and 9.72%) were enriched in July during the severe and the mild HABs, respectively. In August, Sphingopyxis was dominant with a relative abundance of 33.8%, but later decreased to 3.9±2.2% in September and October. In contrast, its average relative abundance during the mild HAB was consistently low at 3.49±1.7%. In September, bacterial communities were dominated by Porphyrobacter during the severe HAB, with a relative abundance of 59.9% and 20.0% at the 2 and 6-in. GAC layers, respectively. In October 2017, Bosea, Sphingomonas, and Hydrogenophaga were dominant with averages of 5.2±1.8%, 25.8±0.8%, and 2.3±0.3%, respectively. On the other hand, Reyranella was enriched (6.0±1.2%) from August to October 2018 in the mild HAB.
To further compare the phylogenetic relationship of bacterial communities developed in BAFs during HABs, alpha and beta diversity analysis were performed (Fig. 2D). The overall alpha diversity analysis revealed that indices such as phylogenetic diversity, chao, and Shannon significantly decreased during the severe HAB but were relatively stable for the mild HAB (Fig. 2D and Table S2). PCoA based analysis on weighted Unifrac dissimilarity (beta diversity) showed two distinct groups (Fig. 2D). Samples from the mild HAB were clustered together on the right, while those from the severe HAB were gathered in the middle, with the exception of July samples. This result indicates that the community structure is initially similar, but shifts in significantly different directions due to the impact of the severe HAB. Additionally, given the differences in the community structure, NMDS analysis was performed to examine which environmental variables drove the shape of BAF bacterial community during HABs (Fig. S4).
3.4. Functional profiles of HAB-impacted microbiomes in BAFs
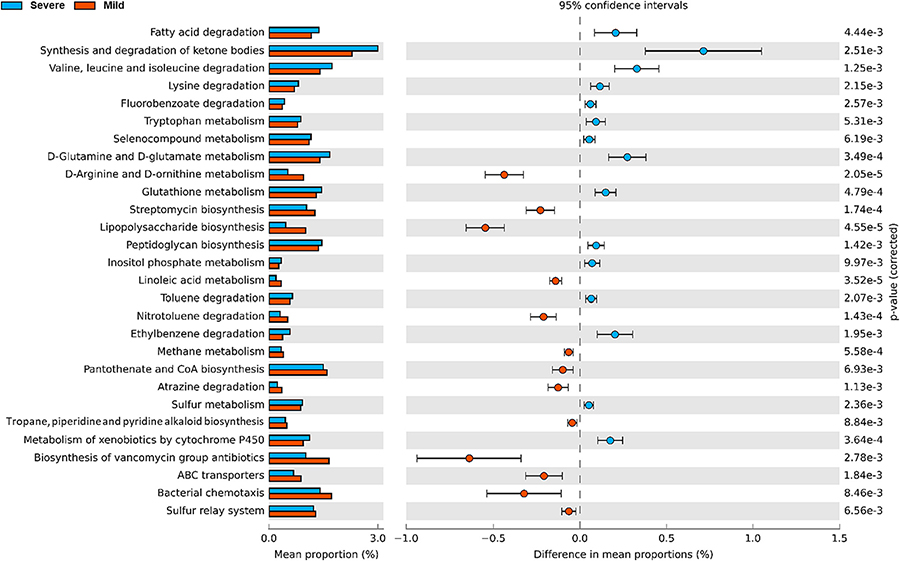
Functional pathways of filter media samples were predicted by PICRUST2 to compare functional characteristics of bacteria communities in BAFs under the two distinct HABs. Based on differences in functional abundance average (p < 0.01), we obtained 28 differentially-abundant KEGG pathways (Fig. 3). Higher gene abundance involved in lipid metabolism (fatty acid degradation, synthesis and degradation of ketone bodies) was predicted under the severe HAB. Moreover, gene functions related to the amino acid degradation/metabolism categories such as valine/leucine/isoleucine/lysine degradation and tryptophan/glutamine/glutamate metabolism were also high compared to the mild HAB. Meanwhile, abilities for D-arginine/D-ornithine metabolism, biosynthesis of vancomycin group antibiotics, lipopolysaccharide biosynthesis, and bacterial chemotaxis were higher in the mild HAB.
Fig. 3.
Comparison of functional pathway abundances in BAFs during the severe (blue) and the mild (red) HABs. The left panel indicates the relative abundance of the predicted functional pathways, and the right panel shows the mean differences at the 95% confidence interval with p-values (<0.01).
3.5. Network topological features and modular structure of bacterial communities in BAFs
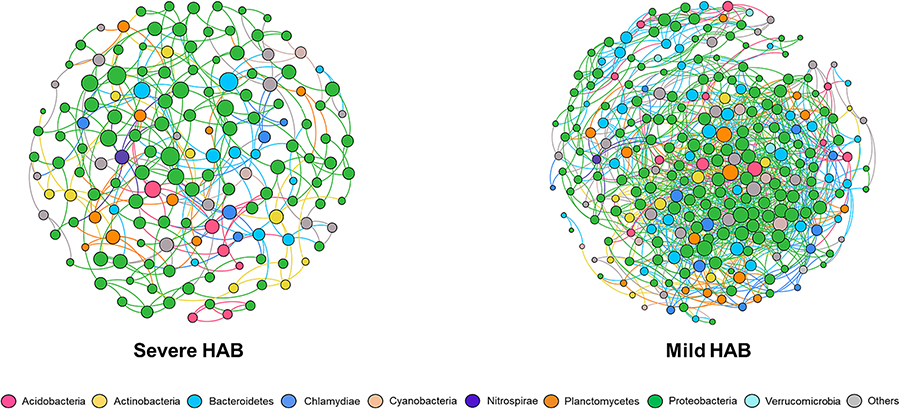
Two SPIEC-EASI-based networks were constructed to explore the ecological interaction of co-occurring species in BAF microbiomes under the two different HAB conditions (Fig. 4). The topological features of the obtained networks are summarized in Table 3. Briefly, 157 nodes with 286 edges and 266 nodes with 1058 edges were obtained for the severe and the mild HABs, respectively. The network of the mild HAB showed higher connections per node (average degree: 4.01) but longer network distances (average path length: 3.03). Both networks presented high modularity (> 0.4) and were composed of modules (12 for severe HAB and 8 for mild HAB), indicating that the connections between communities were strong (Newman and Girvan, 2004). However, the mild HAB-associated network was more complex with higher average degree, average clustering coefficient, and average betweenness.
Fig. 4.
Networks of BAF bacterial communities during the severe (left) and the mild HABs (right). Each node represents taxa affiliated at phylum level (based on 16S rRNA), and edges between nodes represent a predicted interaction. The size of node is proportional to the number of connections (i.e. degree).
Table 3.
Topological properties of WTP filter microbiome networks.
| Severe HABs | Mild HABs | |
|---|---|---|
|
| ||
| Number of nodes | 157 | 266 |
| Number of edges | 286 | 1058 |
| Network diameter | 5 | 12 |
| Average path length | 1.89 | 3.03 |
| Average degree | 1.81 | 4.01 |
| Modularity | 0.68 | 0.51 |
| Module number | 12 | 8 |
| Average clustering coefficient | 0.20 | 0.23 |
| Average betweenness | 282.32 | 350.69 |
| Eigenvector centrality distribution | 0.033 | 0.046 |
3.6. Predicting bacterial community assembly patterns associated with HABs in BAFs
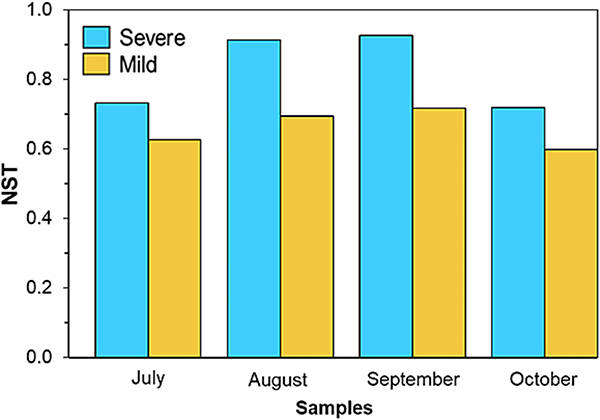
Since the severe HAB had significant impacts on bacterial diversity and composition in BAFs, the major forces shaping bacterial community composition and structure were examined (Fig. 5). Analysis results revealed that all samples had an NST value of 0.5 or higher, indicating that stochastic processes were the major process across the period of HABs in bacterial community assembly. Moreover, the average NST for the severe HAB was 0.82±0.09, whereas it was 0.65±0.05 for the mild HAB. This suggests that community assembly in BAFs is more influenced by the stochastic processes within the severe HAB compared to the mild HAB. While the contribution of stochastic processes exhibited dynamic changes under both HAB conditions, the prevalence of stochastic processes tended to rise consistently from July to September. In particular, the highest NST values were observed in September, reaching 0.92 for the severe HAB and 0.71 for the mild HAB, respectively.
Fig. 5.
The assembly progresses governing the BAF over the period of severe and mild HABs, represented by the estimated NST over examined time points; severe HAB (blue) and mild HAB (yellow). A threshold of 0.5 distinguishes between more deterministic (<0.5) and more stochastic (>0.5) processes.
4. Discussion
4.1. Bacterial community dynamics of BAF media during HABs
In this study, we observed a gradual decrease of bacterial community diversity in BAFs during the severe HAB, while no significant reduction was found in the mild HAB (Fig. 2D). Specifically, we found that the severe HAB significantly influenced the bacterial community composition in BAFs through the enrichment of bloom-associated taxa. For example, Sphingopyxis accounted for 33% of the community composition in the August sample under the severe HAB (Fig. 2C). Several species within this genus become dominant by degrading MC-LR during a bloom event (Zhang et al., 2020b). In this respect, they proliferated rapidly due to a high concentration of MCs in August, leading to the accumulation of the genus in BAFs. Later, however, the relative abundance of Sphingopyxis significantly decreased to 3.3% in September. This may be because the nutrients for the genus were limited in both source water and BAFs. It should be noted that MCs were not detected in filter influent during the mild and severe HAB (Table 1).
Meanwhile, Porphyrobacter was dominant (59.9% and 20% at 2 in. and 6 in., respectively) in September 2017 when the bloom was severe and fully developed. Porphyrobacter has been suggested to stimulate Microcystis aeruginosa to release extracellular polymeric substances, which make cyanobacteria aggregated with heterotrophic bacterial cells by forming colonies (Yan et al., 2021). Thus, the predominance of Porphyrobacter indicates that a more condensed bloom was formed by cyanobacterial colonies in September compared to August. These results were validated by the data obtained from NOAA which indicated that in 2017, the severe bloom continued from late August to mid-September. In addition, the relative abundance of Sphingomonas also significantly increased from September to October (7.9±4.2% to 25.8±0.8%) during the severe HAB. Compared to Sphingopyxis and Porphyrobacter, the genus was dominant in BAFs even after the end of blooms. Since the genus Sphingomonas has abilities like assimilating various complex organic carbons in low-nutrient conditions and forming biofilms (Jeong et al., 2016; Sun et al., 2013), its predominance indicates that the genus survives in conditions where biotic (e.g., competition for nutrient acquisition) and abiotic stresses (e.g., regular backwashing with chlorinated water for BAFs) coexist. While the dominance of these genera, including Porphyrobacter, was temporary, they are known to possess the ability to degrade MCs (McCartney et al., 2020). Consequently, future BAF studies should investigate their potential as biomarkers for HABs and their role in degrading secondary metabolites, including MCs.
4.2. Functional pathways of BAF bacterial community during HABs
Environmental factors such as source water quality, process configurations, and filter media type are known to influence the bacterial community structure and function in BAFs (Hu et al., 2020; Vignola et al., 2018). In this study, we demonstrated that the severe HAB event creates circumstances where the portion of AOM-related compounds increases in filter influent. Further analyses by PICRUST2 revealed that the severe HAB affected the bacterial community function, which possibly corresponded to the utilization of AOM in BAFs (Fig. 3). For example, gene proportions for lipid metabolism (the fatty acids degradation and the synthesis and degradation of ketone bodies) in the severe HAB were higher than in the mild HAB. Generally, fatty acids are converted to acetyl-coenzyme A (CoA) by β-oxidation and then the produced acetyl-CoA is used to synthesize ketone bodies, which are one of the vital energy sources for all domains (i.e. Archaea, Eukarya, and Bacteria) (Puchalska and Crawford, 2017). It should be noted that fatty acids and nitrogen-containing compounds are abundant in DOM from lysed Microcystis sp. (Gonsior et al., 2019). Therefore, it can be inferred that the increased lipid metabolism in the severe HAB may be linked to the production of fatty acids from Microcystis sp.
In addition, a noticeable increase in functional abundance during the severe HAB was observed in amino acid degradation and tryptophan metabolism. Within intracellular AOM of Microcystis sp., tryptophan-like fluorophores constitute >70% of total component fraction, followed by approximately 20% of terrestrial humic-like material (Lee et al., 2018). In this study, we observed the high intensity of C3 (tryptophan-like substances) in the severe HAB (especially in August), but no distinctive increase of C2 (humic-like substances) was not detected (Fig. 1). The major reason for the low C2 may be because large carbonaceous organic compounds such as humic-like and fulvic-like substances are efficiently removed by C/F process, while low molecular weight substances (<1 kDa) remain during the process (Lin et al., 2018; Yang et al., 2019). Furthermore, considering that lysine, arginine, valine, and glutamine are major amino acids in the cellular organic matter of Microcystis sp., we conclude that the enhanced metabolisms at the severe HAB are closely correlated with intracellular AOM uptake by BAF bacterial community (Wang et al., 2023).
4.3. BAF community assembly and bacterial interactions during HABs
Our results revealed that community assembly in BAFs was mostly driven by stochastic processes in the period of HABs (Fig. 5). These are in line with a recent study demonstrating that stochastic processes were more closely associated with BAF bacterial community dynamics and assembly than deterministic processes throughout the annual monitoring period (Li et al., 2021). In addition, compared to the mild HAB, stochastic processes significantly increased from August to September at the severe HAB. There are several possible reasons for the high contribution of stochastic processes during the severe HAB. Firstly, nutrient input is believed to generate stochastic variations by increasing ecological drift while weakening niche-selection (deterministic) (Chase and Myers, 2011). Given that the fraction of AOM-associated substances in source water was relatively high during the severe HAB, it is presumed that the presence of AOM, an easily biodegraded substrate, contributed to the increase of stochasticity by enhancing ecological drift (via stimulating the growth of various bacteria) and undermining niche selection (via reducing competitions for resources). Another reason is the influence of taxa arriving randomly via dispersal on the BAF bacterial community. Previous studies showed that sudden changes in environmental conditions, such as river water intrusion into an aquifer, can be a factor in opening new niches by hindering or erasing resident taxa, and thus randomly arrived taxa can fill the niches, promoting the relative contribution of stochastic processes (Fillinger et al., 2021; Shade et al., 2012). Accordingly, the massive appearance of various bloom-associated bacteria (e.g., Spingopyxis, Porphyrobacter, and Sphingomonas) accompanying with the decrease of alpha diversity is undoubtedly deemed as the sudden changes in bacterial community composition. This may lead to increasing the influence of stochastic processes during the severe HAB. Lastly, the network developed during the mild HAB exhibited a more clustered distribution than that formed during the severe HAB (Fig. 4). This indicates that deterministic processes contributed more to the network established in the mild HAB, compared to the severe HAB (Xu et al., 2019). Moreover, the analysis results (higher modularity with lower average degree and clustering coefficient) in the severe HAB implies the creation of high niche differentiation, which occurs in weaker interactions among microorganisms (Lin et al., 2017). It is noteworthy that direct (e.g., competition and mutualism) and indirect (e.g., shared niches) interactions between species represent deterministic processes, rather than stochastic processes (Barberan et al., 2012´ ). As a result, deterministic processes are expected to contribute more to community assembly than stochastic processes during the mild HAB in BAFs.
The network structure from communities with the severe HAB had smaller, but more numerous modules, while communities with the mild HAB had more complex, but fewer modules (Table 3). When the environment is harsh and nutrient availability is limited, microorganisms tend to have complex interactions because they are likely to be dependent on nutrient acquisition (Zhang et al., 2020a, 2020b). Nutrient availability especially becomes a more important driving force affecting the community network structure than other physicochemical parameters (e.g., temperature) of filter influent in summer (Li et al., 2021). Therefore, our result could be interpreted as that more nutrient availability during the severe HAB resulted in the formation of less complex networks.
5. Engineering implications
With an annual reoccurrence of HABs in aquatic ecosystems worldwide, there are still no standard guidelines for DWTPs utilizing the conventional treatment train to control cyanobacterial cells and cyanotoxins. Our findings can offer insight into understanding the ecological effects of severe HABs on BAF community structure and function, and can help optimize BAF-based treatment processes for DWTPs impacted by HABs. While the severe HAB significantly decreased bacterial community diversity of BAFs, we identified various bloom-associated bacteria, including potential MC-degraders that colonized the GAC filter media. A previous study on cyanotoxin removal in BAFs showed that initial breakthrough of MC-LR occurred right after the injection of 5 μg/L of MC-LR at a slow filtration condition (Jeon et al., 2020). Similarly, it took approximately 7 days (including 3 days of lag time) to completely remove 20 μg/L of MC-LR in a sand filter (Ho et al., 2006). These results indicate that indigenous bacterial communities in BAFs may require adaptation time for MC removal. In this study, we showed that potential MC-degrading bacteria, such as Sphingopyxis sp. and Sphingomonas sp., can be enriched in BAFs. Thus, a bioaugmentation approach can be considered to prevent the initial outbreak of MCs in the early stage of HABs. However, more studies focused on elucidating the relationship between the abundance of immigrated MC-degraders from source water and MC-LR removal, and the effect of operational conditions (e.g., backwashing and flow rate) on their adaptability in BAFs are required in the future.
In general, taxonomically diverse communities comprising unique members are functionally redundant but they are critical to maintain system function and stability against environmental stress (e.g., pH, substrate, and temperature) (Xue et al., 2018). In this work, bacterial diversity in BAFs significantly decreased during the severe bloom by stochastic events such as immigration. This may negatively affect treatability of AOM by decreasing the system function and stability (Zhang et al., 2020a, 2020b). The PICRUST result revealed that that microorganisms in BAFs can utilize AOM-associated compounds, but further studies focusing on how the diversity and functional stability in BAF bacterial communities are correlated with the performance of BAFs for AOM removal during HABs are required.
The major shift in physiochemical and community structures of biofilms may occur when the bacteria in the BAFs are introduced to a new nutrient source, AOM (Li et al., 2019, 2021). This alteration is anticipated to intensify biological activity, resulting in increased biofilm formation in BAF (Jeon et al., 2020; Li et al., 2019). Extracellular polymeric substances (EPS), comprising approximately 90% of the biofilm, primarily consist of proteins, polysaccharides, and nucleic acids, which can provide a protective barrier against disinfection Previously, BAFs have been linked to the release of EPS and other biofilm compounds, such as detached biofilm clusters, into the effluent, impacting both biological and chemical stability in drinking water distribution systems (DWDSs) (Pinto et al., 2012; Li et al., 2020; Wang et al., 2021; Xue et al., 2014). During disinfection, released EPS and biofilm components may consume disinfectants and act as precursors for the formation of carbonaceous DBP (C-DBP) and N-DBP (Wang et al., 2012, 2013). Studies have indicated that a higher organic nitrogen content within biofilms tends to promote the formation of N-DBPs, which are significantly more toxic than C-DBPs (Pérez and Susa, 2017; Wang et al., 2021). Additionally, more recent studies reported that unremoved AOM could contribute to the formation of DBPs and microbial biofilms in DWDSs (Li et al., 2019, 2020). Therefore, characterizing BAF communities and functions during the HABs would assist the DWTP operator in predicting the performance of BAF and effluent water quality. Furthermore, careful monitoring of the water quality of BAF effluent, along with the optimization of BAF operation, will be crucial for the DWTP to minimize the release of biofilms and potential DBP formation while complying with current regulatory requirements for BAF systems (Korotta-Gamage and Sathasivan, 2017; Liu et al., 2017).
6. Conclusions
The main conclusions of this study are as follows:
Bacterial community structure in BAFs was closely linked to the bloom severity level and bloom progression. Accordingly, bacterial diversity in BAFs decreased with selective enrichment of bloom-associated bacteria (e.g., Spingopyxis, Porphyrobacter, and Sphingomonas) during the severe HAB, while it was stable under the mild HAB.
The severe HAB increased the portion of protein-like substances in filter influent. In addition, metabolism associated with the utilization of lipids and amino acids in BAF community function was more enriched at the severe HAB compared to the mild HAB.
Stochastic processes played more important roles in driving the community assembly in BAFs and the relative contribution of stochastic processes significantly increased during the severe HAB.
The severe HAB lead to the development of less complex networks from the BAF communities by providing more nutrients associated with AOM, compared to the mild HAB.
Supplementary Material
HIGHLIGHTS.
Effects of HAB severity on bacterial community structure in BAFs were examined.
Dominance of bloom-associated bacteria was observed in BAFs during the severe HAB.
Severe HAB led to increased metabolism of bacteria related to AOM utilization.
Community assembly processes were mainly governed by stochastic processes in BAFs.
The relative contribution of stochastic processes increased during the severe HAB.
Acknowledgements
This study was supported by the National Science Foundation of United States (CBET-1605185), the Ohio Water Development Authority (7174 and 8675), and the Ohio Department of Higher Education (R/SDW-2-BOR). The study was also funded in part by the U.S. Environmental Protection Agency and the U.S. Army Corps of Engineers (W912HZ2120015). Any opinions expressed do not reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Youchul Jeon: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Lei Li: Data curation, Formal analysis, Validation, Writing – review & editing. Mudit Bhatia: Data curation, Validation, Writing – review & editing. Hodon Ryu: Methodology, Resources, Validation, Writing – review & editing. Jorge W. Santo Domingo: Methodology, Resources, Validation, Writing – review & editing. Jess Brown: Funding acquisition, Project administration, Writing – review & editing. Jake Goetz: Data curation, Resources, Validation. Youngwoo Seo: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2024.171301.
Data availability
Data will be made available on request.
References
- Abkar L, Moghaddam HS, Fowler SJ, 2024. Microbial ecology of drinking water from source to tap. Sci. Total Environ. 908, 168077 10.1016/j.scitotenv.2023.168077 [DOI] [PubMed] [Google Scholar]
- Adler PB, HilleRisLambers J, Levine JM, 2007. A niche for neutrality. Ecol. Lett. 10, 95–104. 10.1111/j.1461-0248.2006.00996.x [DOI] [PubMed] [Google Scholar]
- Bastian M, Heyman S, Jacomy M, 2009. Gephi: An Open Source Software for Exploring and Manipulating Networks. Third International AAAI Conference on Weblogs and Social Media. https://ojs.aaai.org/index.php/ICWSM/article/view/13937 [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N, 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. 10.1038/ismej.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J, 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG, 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BW, Lazorchak JM, Howard MD, Johnson MVV, Morton SL, Perkins DA, Reavie ED, Scott GI, Smith SA, Steevens JA, 2016. Are harmful algal blooms becoming the greatest inland water quality threat to public health and aquatic ecosystems? Environ. Toxicol. Chem. 35 (1), 6–13. https://setac.onlinelibrary.wiley.com/doi/10.1002/etc.3220 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 (8), 1621–1624. https://www.nature.com/articles/ismej20128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JM, Myers JA, 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B 366, 2351–2363. 10.1098/rstb.2011.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Westerhoff P, Leenheer JA, Booksh K, 2003. Fluorescence excitation-emission matrix regional integration to quantify spectra for dissolved organic matter. Environ. Sci. Technol. 37, 5701–5710. 10.1021/es034354c [DOI] [PubMed] [Google Scholar]
- Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J, 2012. Molecular ecological network analyses. BMC Bioinform. 13, 113. 10.1186/1471-2105-13-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVilbiss SE, Zhou Z, Klump JV, Guo L, 2016. Spatiotemporal variations in the abundance and composition of bulk and chromophoric dissolved organic matter in seasonally hypoxia-influenced Green Bay, Lake Michigan, USA. Sci. Total Environ. 565, 742–757. 10.1016/j.scitotenv.2016.05.015 [DOI] [PubMed] [Google Scholar]
- Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI, 2020. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. 10.1038/s41587-020-0548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrenberg S, O’Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR, 2013. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 7, 1102–1111. 10.1038/ismej.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger L, Hug K, Griebler C, 2021. Aquifer recharge viewed through the lens of microbial community ecology: initial disturbance response, and impacts of species sorting versus mass effects on microbial community assembly in groundwater during riverbank filtration. Water Res. 189, 116631 10.1016/j.watres.2020.116631 [DOI] [PubMed] [Google Scholar]
- Gallardo-Rodríguez JJ, Astuya-Villalón A, Llanos-Rivera A, Avello-Fontalba V, Ulloa-Jofré V, 2019. A critical review on control methods for harmful algal blooms. Rev. Aquac. 11, 661–684. 10.1111/raq.12251 [DOI] [Google Scholar]
- Gibert O, Lefèvre B, Fernández M, Bernat X, Paraira M, Calderer M, Martínez-Lladó X, 2013. Characterising biofilm development on granular activated carbon used for drinking water production. Water Res. 47, 1101–1110. 10.1016/j.watres.2012.11.026 [DOI] [PubMed] [Google Scholar]
- Golea DM, Jarvis P, Jefferson B, Moore G, Sutherland S, Parsons SA, Judd SJ, 2020. Influence of granular activated carbon media properties on natural organic matter and disinfection by-product precursor removal from drinking water. Water Res. 174, 115613 10.1016/j.watres.2020.115613 [DOI] [PubMed] [Google Scholar]
- Gonsior M, Powers LC, Williams E, Place A, Chen F, Ruf A, Hertkorn N, Schmitt-Kopplin P, 2019. The chemodiversity of algal dissolved organic matter from lysed Microcystis aeruginosa cells and its ability to form disinfection by-products during chlorination. Water Res. 155, 300–309. 10.1016/j.watres.2019.02.030 [DOI] [PubMed] [Google Scholar]
- Ho L, Meyn T, Keegan A, Hoefel D, Brookes J, Saint CP, Newcombe G, 2006. Bacterial degradation of microcystin toxins within a biologically active sand filter. Water Res. 40, 768–774. 10.1016/j.watres.2005.12.009 [DOI] [PubMed] [Google Scholar]
- Hu W, Liang J, Ju F, Wang Q, Liu R, Bai Y, Liu H, Qu J, 2020. Metagenomics unravels differential microbiome composition and metabolic potential in rapid sand filters purifying surface water versus groundwater. Environ. Sci. Technol. 54, 5197–5206. 10.1021/acs.est.9b07143 [DOI] [PubMed] [Google Scholar]
- Huguet A, Vacher L, Relexans S, Saubusse S, Froidefond JM, Parlanti E, 2009. Properties of fluorescent dissolved organic matter in the Gironde estuary. Org. Geochem. 40, 706–719. 10.1016/j.orggeochem.2009.03.002 [DOI] [Google Scholar]
- Ibelings BW, Backer LC, Kardinaal WEA, Chorus I, 2014. Current approaches to cyanotoxin risk assessment and risk management around the globe. Harmful Algae 40, 63–74. https://www.sciencedirect.com/science/article/abs/pii/S1568988314001735?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé R, Cawley KM, Yamashita Y, 2014. Applications of Excitation Emission Matrix Fluorescence with Parallel Factor Analysis (EEM-PARAFAC) in assessing environmental dynamics of natural Dissolved Organic Matter (DOM) in aquatic environments: a review. In: Rosario-Ortiz F (Ed.), ACS Symposium Series. American Chemical Society, Washington, DC, pp. 27–73. 10.1021/bk-2014-1160.ch003 [DOI] [Google Scholar]
- Jeon Y, Li L, Calvillo J, Ryu H, Santo Domingo JW, Choi O, Brown J, Seo Y, 2020. Impact of algal organic matter on the performance, cyanotoxin removal, and biofilms of biologically-active filtration systems. Water Res. 184, 116120 10.1016/j.watres.2020.116120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Cho K, Bae H, Keshvardoust P, Rice SA, Vigneswaran S, Lee S, Leiknes T, 2016. Effect of microbial community structure on organic removal and biofouling in membrane adsorption bioreactor used in seawater pretreatment. Chem. Eng. J. 294, 30–39. 10.1016/j.cej.2016.02.108 [DOI] [Google Scholar]
- Jia P, Zhou Y, Zhang X, Zhang Y, Dai R, 2018. Cyanobacterium removal and control of algal organic matter (AOM) release by UV/H2O2 pre-oxidation enhanced Fe(II) coagulation. Water Res. 131, 122–130. 10.1016/j.watres.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Jiang H, Tang J, Li J, Zhao S, Mo Y, Tian C, Zhang X, Jiang B, Liao Y, Chen Y, Zhang G, 2022. Molecular signatures and sources of fluorescent components in atmospheric organic matter in South China. Environ. Sci. Technol. Lett. 9, 913–920. 10.1021/acs.estlett.2c00629 [DOI] [Google Scholar]
- Joh G, Choi YS, Shin J-K, Lee J, 2011. Problematic algae in the sedimentation and filtration process of water treatment plants. J. Water Supply Res. Technol. AQUA 60, 219–230. 10.2166/aqua.2011.035 [DOI] [Google Scholar]
- Kapoor V, Elk M, Li X, Impellitteri CA, Santo Domingo JW, 2016. Effects of Cr(III) and Cr(VI) on nitrification inhibition as determined by SOUR, function-specific gene expression and 16S rRNA sequence analysis of wastewater nitrifying enrichments. Chemosphere 147, 361–367. 10.1016/j.chemosphere.2015.12.119 [DOI] [PubMed] [Google Scholar]
- Kennedy AM, Reinert AM, Knappe DRU, Ferrer I, Summers RS, 2015. Full- and pilot-scale GAC adsorption of organic micropollutants. Water Res. 68, 238–248. 10.1016/j.watres.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Korotta-Gamage SM, Sathasivan A, 2017. A review: potential and challenges of biologically activated carbon to remove natural organic matter in drinking water purification process. Chemosphere 167, 120–138. 10.1016/j.chemosphere.2016.09.097 [DOI] [PubMed] [Google Scholar]
- Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA, 2015. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11, e1004226 10.1371/journal.pcbi.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter A, Silke J, Cusack C, 2018. Creating a Weekly Harmful Algal Bloom Bulletin (Technical Report). Marine Institute. In: https://oar.marine.ie/bitstream/handle/10793/1344/HAB-Bulletin-Process_11April2018.pdf [Google Scholar]
- Lee D, Kwon M, Ahn Y, Jung Y, Nam S-N, Choi I, Kang J-W, 2018. Characteristics of intracellular algogenic organic matter and its reactivity with hydroxyl radicals. Water Res. 144, 13–25. 10.1016/j.watres.2018.06.069 [DOI] [PubMed] [Google Scholar]
- Li L, Jeon Y, Lee S-H, Ryu H, Santo Domingo JW, Seo Y, 2019. Dynamics of the physiochemical and community structures of biofilms under the influence of algal organic matter and humic substances. Water Res. 158, 136–145. 10.1016/j.watres.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jeon Y, Ryu H, Santo Domingo JW, Seo Y, 2020. Assessing the chemical compositions and disinfection byproduct formation of biofilms: application of fluorescence excitation-emission spectroscopy coupled with parallel factor analysis. Chemosphere 246, 125745. 10.1016/j.chemosphere.2019.125745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ning D, Jeon Y, Ryu H, Santo Domingo JW, Kang D-W, Kadudula A, Seo Y, 2021. Ecological insights into assembly processes and network structures of bacterial biofilms in full-scale biologically active carbon filters under ozone implementation. Sci. Total Environ. 751, 141409 10.1016/j.scitotenv.2020.141409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, De Vrieze J, Li C, Li Jiaying, Li Jiabao, Yao M, Hedenec P, Li H, Li T, Rui J, Frouz J, Li X, 2017. Temperature regulates deterministic processes and the succession of microbial interactions in anaerobic digestion process. Water Res. 123, 134–143. 10.1016/j.watres.2017.06.051 [DOI] [PubMed] [Google Scholar]
- Lin J-L, Hua L-C, Hung SK, Huang C, 2018. Algal removal from cyanobacteria-rich waters by preoxidation-assisted coagulation–flotation: effect of algogenic organic matter release on algal removal and trihalomethane formation. J. Environ. Sci. 63, 147–155. 10.1016/j.jes.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Lin J-L, Nugrayanti MS, Ika AR, Karangan A, 2021. Removal of Microcystis Aeruginosa by oxidation-assisted coagulation: effect of algogenic organic matter fraction changes on algae destabilization with Al hydrates. J. Water Process Eng. 42, 102142 10.1016/j.jwpe.2021.102142 [DOI] [Google Scholar]
- Liu H, Roeder K, Wasserman L, 2010. Stability Approach to Regularization Selection (StARS) for High Dimensional Graphical Models. 10.48550/arXiv.1006.3316 [DOI] [PMC free article] [PubMed]
- Liu C, Ersan MS, Wagner E, Plewa MJ, Amy G, Karanfil T, 2020. Toxicity of chlorinated algal-impacted waters: Formation of disinfection byproducts vs. reduction of cyanotoxins. Water Res. 184, 116145. https://www.sciencedirect.com/science/article/abs/pii/S0043135420306825 [DOI] [PubMed] [Google Scholar]
- Liu C, Olivares CI, Pinto AJ, Lauderdale CV, Brown J, Selbes M, Karanfil T, 2017. The control of disinfection byproducts and their precursors in biologically active filtration processes. Water Res. 124, 630–653. 10.1016/j.watres.2017.07.080 [DOI] [PubMed] [Google Scholar]
- McCartney AT, Yeo J-Y, Blomquist TM, Huntley JF, 2020. Whole-genome sequencing of bacterial isolates that degrade the cyanobacterial toxin microcystin-LR. Microbiol. Resour. Announc. 9, e00959–20 10.1128/MRA.00959-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM, 2018. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224. 10.1038/ismej.2017.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KR, Hambly A, Singh S, Henderson RK, Baker A, Stuetz R, Khan SJ, 2011. Organic matter fluorescence in municipal water recycling schemes: toward a unified PARAFAC model. Environ. Sci. Technol. 45 (7), 2909–2916. https://pubs.acs.org/doi/10.1021/es103015e [DOI] [PubMed] [Google Scholar]
- Murphy KR, Stedmon CA, Waite TD, Ruiz GM, 2008. Distinguishing between terrestrial and autochthonous organic matter sources in marine environments using fluorescence spectroscopy. Mar. Chem. 108 (1–2), 40–58. https://www.sciencedirect.com/science/article/abs/pii/S0304420307002356 [Google Scholar]
- Naceradska J, Novotna K, Cermakova L, Cajthaml T, Pivokonsky M, 2019. Investigating the coagulation of non-proteinaceous algal organic matter: optimizing coagulation performance and identification of removal mechanisms. J. Environ. Sci. (China) 79, 25–34. 10.1016/j.jes.2018.09.024 [DOI] [PubMed] [Google Scholar]
- Newman MEJ, Girvan M, 2004. Finding and evaluating community structure in networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 69, 026113 10.1103/PhysRevE.69.026113 [DOI] [PubMed] [Google Scholar]
- Ning D, Deng Y, Tiedje JM, Zhou J, 2019. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. U. S. A. 116, 16892–16898. 10.1073/pnas.1904623116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl HW, Otten TG, Kudela R, 2018. Mitigating the expansion of harmful algal blooms across the freshwater-to-marine continuum. Environ. Sci. Technol. 52, 5519–5529. 10.1021/acs.est.7b05950 [DOI] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG, 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez ML, Susa MR, 2017. Exopolymeric substances from drinking water biofilms: dynamics of production and relation with disinfection by products. Water Res. 116, 304–315. [DOI] [PubMed] [Google Scholar]
- Piai L, Blokland M, van der Wal A, Langenhoff A, 2020. Biodegradation and adsorption of micropollutants by biological activated carbon from a drinking water production plant. J. Hazard. Mater. 388, 122028 10.1016/j.jhazmat.2020.122028 [DOI] [PubMed] [Google Scholar]
- Piezer K, Li L, Jeon Y, Kadudula A, Seo Y, 2021. The application of potassium permanganate to treat Cyanobacteria-laden water: a review. Process. Saf. Environ. Prot. 148, 400–414. 10.1016/j.psep.2020.09.058 [DOI] [Google Scholar]
- Pinto AJ, Xi C, Raskin L, 2012. Bacterial community structure in the drinking water microbiome is governed by filtration processes. Environ. Sci. Technol. 46, 8851–8859. 10.1021/es302042t [DOI] [PubMed] [Google Scholar]
- Puchalska P, Crawford PA, 2017. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 25, 262–284. 10.1016/j.cmet.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Ma B, Miao S, Liu R, Hu C, Qu J, 2021. Pre-oxidation enhanced cyanobacteria removal in drinking water treatment: a review. J. Environ. Sci. (China) 110, 160–168. 10.1016/j.jes.2021.03.040 [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO, 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–596 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Baracaldo P, Bianchini G, Wilson JD, Knoll AH, 2022. Cyanobacteria and biogeochemical cycles through Earth history. Trends Microbiol. 30, 143–157. 10.1016/j.tim.2021.05.008 [DOI] [PubMed] [Google Scholar]
- Shade A, Peter H, Allison S, Baho D, Berga M, Buergmann H, Huber D, Langenheder S, Lennon J, Martiny J, Matulich K, Schmidt T, Handelsman J, 2012. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3. https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2012.00417/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon MA, Bohn PW, Elimelech M, Georgiadis JG, Marinas BJ, Mayes AM, ˜ 2009. Science and technology for water purification in the coming decades. In: Nanoscience and Technology. Co-Published with Macmillan Publishers Ltd, UK, pp. 337–346. 10.1142/9789814287005_0035 [DOI] [Google Scholar]
- Stedmon CA, Bro R, 2008. Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial. Limnology & Ocean Methods 6, 572–579. 10.4319/lom.2008.6.572b [DOI] [Google Scholar]
- Stedmon CA, Markager S, 2005a. Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnol. Oceanogr. 50 (2), 686–697. https://aslopubs.onlinelibrary.wiley.com/doi/abs/10.4319/lo.2005.50.2.0686 [Google Scholar]
- Stedmon CA, Markager S, 2005b. Tracing the production and degradation of autochthonous fractions of dissolved organic matter by fluorescence analysis. Limnol. Oceanogr. 50 (5), 1415–1426. https://aslopubs.onlinelibrary.wiley.com/doi/abs/10.4319/lo.2005.50.5.1415 [Google Scholar]
- Stedmon CA, Markager S, Bro R, 2003. Tracing dissolved organic matter in aquatic environments using a new approach to fluorescence spectroscopy. Mar. Chem. 82 (3–4), 239–254. https://www.sciencedirect.com/science/article/abs/pii/S0304420303000720 [Google Scholar]
- Stumpf RP, Wynne TT, Baker DB, Fahnenstiel GL, 2012. Interannual variability of cyanobacterial blooms in Lake Erie. PLoS One 7, e42444. 10.1371/journal.pone.0042444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Liu W, Cui L, Zhang M, Wang B, 2013. Characterization and identification of a chlorine-resistant bacterium, Sphingomonas TS001, from a model drinking water distribution system. Sci. Total Environ. 458–460, 169–175. 10.1016/j.scitotenv.2013.04.030 [DOI] [PubMed] [Google Scholar]
- Vellend M, Srivastava DS, Anderson KM, Brown CD, Jankowski JE, Kleynhans EJ, Kraft NJB, Letaw AD, Macdonald AAM, Maclean JE, Myers-Smith IH, Norris AR, Xue X, 2014. Assessing the relative importance of neutral stochasticity in ecological communities. Oikos 123, 1420–1430. 10.1111/oik.01493 [DOI] [Google Scholar]
- Vignola M, Werner D, Wade MJ, Meynet P, Davenport RJ, 2018. Medium shapes the microbial community of water filters with implications for effluent quality. Water Res. 129, 499–508. 10.1016/j.watres.2017.09.042 [DOI] [PubMed] [Google Scholar]
- Villacorte LO, Ekowati Y, Neu TR, Kleijn JM, Winters H, Amy G, Schippers JC, Kennedy MD, 2015. Characterisation of algal organic matter produced by bloom-forming marine and freshwater algae. Water Res. 73, 216–230. 10.1016/j.watres.2015.01.028 [DOI] [PubMed] [Google Scholar]
- Wang Z, Kim J, Seo Y, 2012. Influence of bacterial extracellular polymeric substances on the formation of carbonaceous and nitrogenous disinfection byproducts. Environ. Sci. Technol. 46, 11361–11369. 10.1021/es301905n [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi O, Seo Y, 2013. Relative contribution of biomolecules in bacterial extracellular polymeric substances to disinfection byproduct formation. Environ. Sci. Technol. 47, 9764–9773. 10.1021/es402067g [DOI] [PubMed] [Google Scholar]
- Wang Z, Li L, Ariss RW, Coburn KM, Behbahani M, Xue Z, Seo Y, 2021. The role of biofilms on the formation and decay of disinfection by-products in chlor (am) inated water distribution systems. Sci. Total Environ. 753, 141606. https://www.sciencedirect.com/science/article/abs/pii/S0048969720351354?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- Wang X, Qian Y, Chen Y, Liu F, An D, Yang G, Dai R, 2023. Application of fluorescence spectra and molecular weight analysis in the identification of algal organic matter-based disinfection by-product precursors. Sci. Total Environ. 882, 163589 10.1016/j.scitotenv.2023.163589 [DOI] [PubMed] [Google Scholar]
- Wünsch UJ, Murphy K, 2021. A simple method to isolate fluorescence spectra from small dissolved organic matter datasets. Water Res. 190, 116730 10.1016/j.watres.2020.116730 [DOI] [PubMed] [Google Scholar]
- Wynne TT, Stumpf RP, 2015. Spatial and temporal patterns in the seasonal distribution of toxic Cyanobacteria in Western Lake Erie from 2002–2014. Toxins 7, 1649–1663. 10.3390/toxins7051649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne TT, Stumpf RP, Tomlinson MC, Fahnenstiel GL, Dyble J, Schwab DJ, Joshi SJ, 2013. Evolution of a cyanobacterial bloom forecast system in western Lake Erie: development and initial evaluation. J. Great Lakes Res. Remote Sens. Great Lakes Other Inl. Waters 39, 90–99. 10.1016/j.jglr.2012.10.003 [DOI] [Google Scholar]
- Wynne TT, Stumpf RP, Litaker RW, Hood RR, 2021. Cyanobacterial bloom phenology in Saginaw Bay from MODIS and a comparative look with western Lake Erie. Harmful Algae 103, 101999. 10.1016/j.hal.2021.101999 [DOI] [PubMed] [Google Scholar]
- Xu R, Yu Z, Zhang S, Meng F, 2019. Bacterial assembly in the bio-cake of membrane bioreactors: stochastic vs. deterministic processes. Water Res. 157, 535–545. 10.1016/j.watres.2019.03.093 [DOI] [PubMed] [Google Scholar]
- Xue Y, Chen H, Yang JR, Liu M, Huang B, Yang J, 2018. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 12 (9), 2263–2277. https://www.nature.com/articles/s41396-018-0159-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Lee WH, Coburn KM, Seo Y, 2014. Selective reactivity of monochloramine with extracellular matrix components affects the disinfection of biofilm and detached clusters. Environ. Sci. Technol. 48, 3832–3839. 10.1021/es405353h [DOI] [PubMed] [Google Scholar]
- Yamashita Y, Jaffe R, Maie N, Tanoue E, 2008. Assessing the dynamics of dissolved ´ organic matter (DOM) in coastal environments by excitation emission matrix fluorescence and parallel factor analysis (EEM-PARAFAC). Limnol. Oceanogr. 53 (5), 1900–1908. https://aslopubs.onlinelibrary.wiley.com/doi/abs/10.4319/lo.2008.53.5.1900 [Google Scholar]
- Yan P, Guo J, Zhang P, Xiao Y, Li Z, Zhang S, Zhang Y, He S, 2021. The role of morphological changes in algae adaptation to nutrient stress at the single-cell level. Sci. Total Environ. 754, 142076 10.1016/j.scitotenv.2020.142076 [DOI] [PubMed] [Google Scholar]
- Yang Z, Hou J, Miao L, Yang Y, You G, Jia D, Gao M, 2019. Removing specific extracellular organic matter from algal bloom water by Tanfloc flocculation: performance and mechanisms. Sep. Purif. Technol. 212, 65–74. 10.1016/j.seppur.2018.11.008 [DOI] [Google Scholar]
- Yuan J, Passeport E, Hofmann R, 2022. Understanding adsorption and biodegradation in granular activated carbon for drinking water treatment: a critical review. Water Res. 210, 118026 10.1016/j.watres.2021.118026 [DOI] [PubMed] [Google Scholar]
- Zhang B, Ning D, Yang Y, Van Nostrand JD, Zhou J, Wen X, 2020a. Biodegradability of wastewater determines microbial assembly mechanisms in full-scale wastewater treatment plants. Water Res. 169, 115276. https://www.sciencedirect.com/science/article/abs/pii/S0043135419310504 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang F, Chen L, Feng H, Yin S, Chen M, 2020b. Insights into ecological roles and potential evolution of Mlr-dependent microcystin-degrading bacteria. Sci. Total Environ. 710, 136401 10.1016/j.scitotenv.2019.136401 [DOI] [PubMed] [Google Scholar]
- Zhou J, Ning D, 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002–17 10.1128/MMBR.00002-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP, 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl. Acad. Sci. U. S. A. 111 10.1073/pnas.1324044111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.