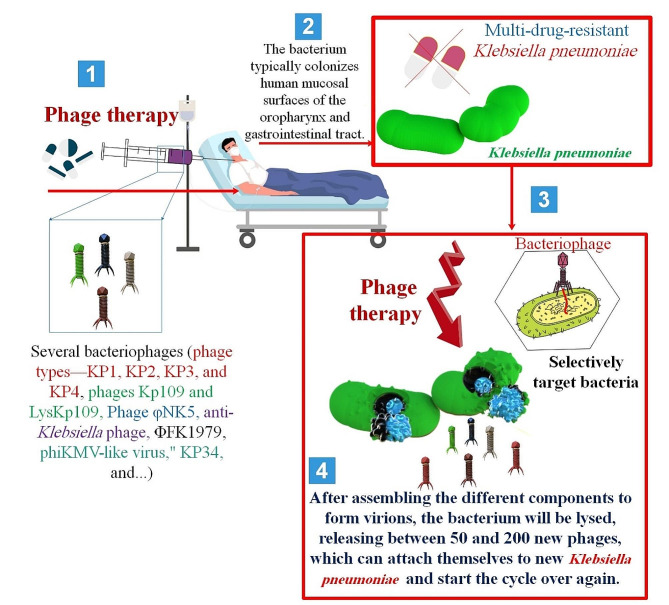
Abstract
One of the most common bacteria that cause nosocomial infections is Klebsiella pneumonia (K. pneumoniae), especially in patients who are very sick and admitted to the intensive care unit (ICU). The frequency of multi-drug-resistant Klebsiella pneumoniae (MDRKP) has dramatically increased worldwide in recent decades, posing an urgent threat to public health. The Western world’s bacteriophage (phage) studies have been revitalized due to the increasing reports of antimicrobial resistance and the restricted development and discovery of new antibiotics. These factors have also spurred innovation in other scientific domains. The primary agent in phage treatment is an obligately lytic organism (called bacteriophage) that kills the corresponding bacterial host while sparing human cells and lessening the broader effects of antibiotic usage on commensal bacteria. Phage treatment is developing quickly, leading to many clinical studies and instances of life-saving medicinal use. In addition, phage treatment has a few immunological adverse effects and consequences in addition to its usefulness. Since K. pneumoniae antibiotic resistance has made treating multidrug-resistant (MDR) infections challenging, phage therapy (PT) has emerged as a novel therapeutic strategy. The effectiveness of phages has also been investigated in K. pneumoniae biofilms and animal infection models. Compared with antibiotics, PT exhibits numerous advantages, including a particular lysis spectrum, co-evolution with bacteria to avoid the emergence of phage resistance, and a higher abundance and diversity of phage resources than found in antibiotics. Moreover, phages are eliminated in the absence of a host bacterium, which makes them the only therapeutic agent that self-regulates at the sites of infection. Therefore, it is essential to pay attention to the role of PT in treating these infections. This study summarizes the state of knowledge on Klebsiella spp. phages and provides an outlook on the development of phage-based treatments that target K. pneumoniae in clinical trials.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-024-02450-7.
Keywords: Klebsiella pneumoniae, Antibacterial, Bacteriophages, Antibiotic resident
Introduction
Multidrug-resistant (MDR) bacteria are one of the most important current threats to public health. The rod-shaped gram-negative bacteria K. pneumoniae is one of the most frequent causes of infections linked to healthcare that affect people, including sepsis, pneumonia, meningitis, and urinary tract infections (UTIs). K. pneumoniae is an opportunistic pathogen most likely to infect immunocompromised individuals in a hospital environment. Acquisition of antibiotic-resistance factors is a severe threat to infected patients [1, 2]. In the last several decades, polymyxins have been the most widely used antibiotic choice for treating K. pneumoniae that is resistant to carbapenem. Polymyxin E (colistin) is often the only antibiotic to produce sufficient serum levels and minimum inhibitory concentrations (MIC), making it a “last resort” treatment for multidrug-resistant K. pneumoniae (MDRKP) infections [3, 4]. One of the biggest challenges facing physicians in MDRKP is managing antibiotic resistance. The best course of action for treating MDRKP infections is still debated. High-dose meropenem, colistin, fosfomycin, tigecycline, and aminoglycosides are common combination therapies, but they don’t always work as well as they should. Over the last several decades, new antimicrobials that target MDRKP have been created and are undergoing different clinical testing phases [5].
An important risk to managing infectious illnesses is antibiotic resistance in bacterial pathogens [6]. There is a great need for novel antimicrobial medicines, but the development of new classes of antibiotics has stalled recently owing to demands of cost and market viability. There have been several anecdotal stories of success supporting the long-standing consideration of bacteriophages (phages) for medicinal application. Recent clinical achievements with customized phage mixtures have rekindled interest in phage treatment, and many clinical trials are now underway [7, 8]. The use of phage therapy (PT) is one such option. Phages are a kind of virus that targets explicitly and infects, or kills, in the case of lytic phages, the bacterial host. A few also exhibit anti-biofilm properties, which might be beneficial in managing device infections. The synergy between phage-antibiotic combinations may also have the benefit of directly lysing bacterial host cells, applying selection pressure to reduce virulence (such as biofilm development), and/or re-sensitizing bacteria to certain antibiotics [9].
The emergence of bacterial resistance to the phage poses a challenge to effectively implementing phage treatment. Several methods, such as changing surface phage receptors, breaking down phage nucleic acids, and initiating abortive infection systems, may lead to resistance to a specific phage. [10] Therefore, a phage cocktail comprising several distinct phages that not only jointly infect a significant percentage of circulating strains but also target various receptor binding proteins (RBPs) on host strains to minimize viable routes to evolved resistance would constitute an ideal broad-spectrum phage medicine [10, 11]. Phage compounds can expand the host range and reduce the emergence of phage-resistant strains compared to single phages despite the possibility of encountering phage-resistant bacterial mutants in a single-phage treatment. The most effective phage cocktails are typically composed of phages with a complementary host range and similar antigenicity [12–14]. For instance, three Klebsiella lytic phages combined to form a phage cocktail exhibited bactericidal solid action. In a mouse mammary gland infected with K. pneumoniae, the bacterial load was decreased by phage cocktail. Furthermore, phage cocktails reduced the expression of inflammatory factors in the mouse mammary gland infected with K. pneumoniae [15]. A phage cocktail will probably be a helpful treatment for infections in humans caused by this bacterial clone that is becoming more and more common and often quite resistant. Researchers [16] aim to develop a phage-based solution to the rising incidence of extensively drug-resistant clinical K. pneumoniae sequence type (ST16) infections starting from a set of phages recently characterized against this lineage. A phage cocktail (Katrice-16) composed of eight lytic phages was characterized for potential use in humans. Katrice-16 was highly active in vitro against researchers’ K. pneumoniae ST16 collection. Katrice-16 additionally exhibited good anti-biofilm activity, especially at 12-h post-treatment. In addition, Katrice-16 maintained high activity in human body fluids [16].
Seongjun Yoo et al. [17] goal was to create a phage cocktail that is both new and potent enough to take out phage-resistant mutants as well as wild-type bacteria. To do this, researchers used phage-resistant bacteria to isolate four phages (U2874, phi_KPN_H2, phi_KPN_S3, and phi_KPN_HS3) that could detect various bacterial surface chemicals. They created three phage cocktails and evaluated each one’s capacity to reduce phage resistance against MDRKP. Investigators contend that the phage cocktail that causes phage susceptibility to resensitize shows better phage resistance-suppressive capacity. Furthermore, they saw trade-off effects in phage-resistant bacteria that gradually became apparent. Also, they speculate that these trade-off effects may increase the effectiveness of treatment. To increase the effectiveness of phage treatment, the researcher also advises collecting phage host range data against phage-resistant mutants and wild-type bacteria when creating phage banks. The significance of phage host range information in creating potent phage mixtures for therapeutic use is highlighted by their research [17].
Jokar et al. [18] showed that four distinct phage types—KP1, KP2, KP3, and KP4—were identified and isolated. Due to the emergence of mutant strains, bacterial fast regrowth was seen after every individual phage-host contact but not after phage cocktails. Without any bacterial regrowth, the phage cocktail showed much better antibacterial action than any single phage. Phage cocktail treatment showed promise in eliminating MDRKP isolates. Phage treatment, and phage cocktail, in particular, is a potential strategy for eliminating MDRKP identified from diabetic foot ulcer (DFU). Researchers demonstrated that it is possible to use a particular phage cocktail to eradicate different illnesses [18].
Phages may be found in bodily fluids and many other sites where bacteria are present [19]. Isolating phages from urine samples of individuals suffering from UTIs was the researcher’s goal of the investigation [20]. Three phages (33%) were identified from ten urine samples that had positive cultures; two of these phages underwent further investigation. K. pneumoniae and Escherichia coli (E. coli) were infected by the Klebsiella phage GADU21 and Escherichia phage GADU22, respectively. The Klebsiella phage GADU21 was able to infect two species, namely K. pneumoniae and Proteus mirabilis. In contrast, the Escherichia phage GADU22 was able to infect four species, namely Shigella flexneri, Shigella sonnei, and E. coli, out of the 14 species that were examined for host range study. Researchers showed that it is possible to extract new phages by using infected bodily fluids as phage sources. The first Klebsiella phage to be identified from human bodily fluid is GADU21. The phages have the potential to be used as therapeutic tools against illnesses since their genomes lack genes associated with virulence and drug resistance [20].
In this article, we have investigated the essential features of antimicrobial resistance (AMR) in multidrug-resistant Klebsiella pneumoniae (MDRKP). In addition, we have investigated the characteristics of bacteriophages and PT used against Klebsiella pneumoniae (K. pneumoniae). Finally, we have comprehensively reviewed the future of PT and its advantages and disadvantages for controlling K. pneumoniae.
Characterization of Klebsiella pneumoniae
It is known that the Gram-negative bacteria K. pneumoniae is an opportunistic pathogen that may infect people with various illnesses [21]. In the past, K. pneumoniae has been connected to UTIs, pneumonia, and bacteremia in immunocompromised or often hospitalized people. Furthermore, the resistance of K. pneumoniae isolates to carbapenems, a widely used broad-spectrum antibiotics, characterizes them as members of the carbapenem-resistant Enterobacteriaceae (CRE) family [22]. The World Health Organization (WHO) has classified these isolates as a “critical concern,” indicating the urgent need for new antibiotic research and development to combat this significant danger to human health. One of the few Gram-negative rods that may cause primary pneumonia is K. pneumoniae, which is the leading cause of nosocomial pneumonia. Nosocomial infections are the hallmark of classical K. pneumonia (cKP), and they are most often seen in elderly or immunocompromised individuals [23, 24].
K. pneumoniae is often found in mucosal tissues and the natural flora of the gut. The most frequent location of a Klebsiella infection is the lung, characterized by bronchopneumonia and bronchiectasis brought on by the germs’ aspiration into the lower respiratory tract from the oropharyngeal mucosa. Even when treated with antibiotics, Klebsiella infections result in opportunistic infectious episodes that have a high fatality risk because they primarily affect individuals with compromised immune systems. K. pneumoniae is also one of the most prevalent co-infecting organisms of COVID-19 during the previous pandemic. In addition to causing lung infections, K. pneumoniae may also cause infections in the lower biliary system, the urethra, and surgical incisions. Notably, during the last 10 years, the genus Klebsiella has rapidly evolved, giving rise to MDR strains. These strains are linked to severe infections that are acquired in hospitals and communities and sometimes need sophisticated antibiotics or maybe no medication at all for treatment [25].
Due to an increase in severe infections and a growing lack of effective therapies, K. pneumoniae has lately garnered popularity as an infectious agent. The advent of K. pneumoniae strains that have gained extra genetic features and become either hypervirulent (HV) or antibiotic-resistant is the cause of these alarming situations. Initially known as Friedlander’s bacteria, K. pneumoniae was originally isolated in the late 1800s. This bacterium is encapsulated, nonmotile, and Gram-negative. It may be found in soil, surface waters, and medical equipment. Crucially, K. pneumoniae easily colonizes human mucosal surfaces, such as those in the oropharynx and gastrointestinal (GI) tract, where its colonization seems to have benign side effects [26].
Based on genomic analyses, the K. pneumoniae pan-genome spans 5 to 6 Mbp and contains 5 to 6 kilogenes that need encoded. About 17,000 genes are identified as core genes out of this total number of encodable genes. Among the K. pneumoniae bacterial species, the core genome is conserved. Usually, ≥ 95% of the individuals belonging to a particular species have the core genes. Accessory genes are present in the remaining genetic pool, nevertheless. Put another way, the term “auxiliary genome,” which differs across Klebsiella spp., refers to the dispensable, flexible, adaptable, or additional genome. For a particular species, the accessory genes are usually found in less than 95% of the individuals. Based on genome-wide phylogenetic structure, the K. pneumoniae spp., the complex is divided into seven major phylogenetic groups: Kp1 (K. pneumoniae subsp. pneumoniae or K. pneumoniae sensu stricto); Kp2 (K. quasipneumoniae subsp. quasipneumoniae); Kp3 (K. variicola subsp. variicola); Kp4 (K. quasipneumoniae subsp. similipneumoniae); Kp5 (K. variicola subsp. tropica); Kp6 (K. quasivariicola); and Kp7 (K. africana). Seven housekeeping genes—gapA, infB, mdh, pgi, phoE, rpoB, and tonB—have been sequenced in this respect. Furthermore, wzi gene sequencing or serotyping techniques may be used to accomplish K-typing or capsule typing [27].
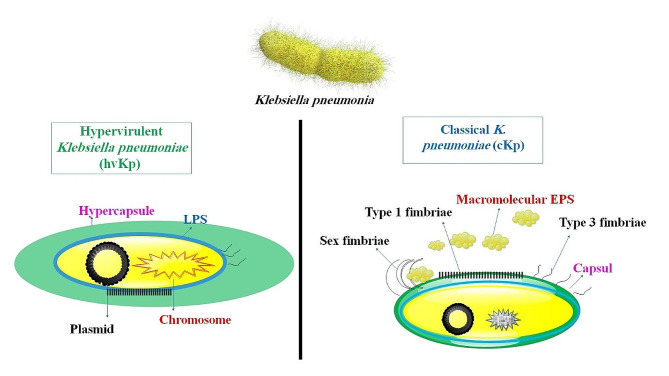
To successfully infect a host, K. pneumoniae must first overcome chemical and mechanical barriers and avoid being recognized by the host’s cellular and humoral defenses. Currently, lipopolysaccharide (LPS), iron carriers, pili, and capsules have been identified as the four virulent factors. Adhesins, namely type 1 and type 3 pili, assemble K. pneumoniae and facilitate bacterial adherence to abiotic surfaces, immune cells, and epithelium (Fig. 1). K. pneumoniae’s virulence factor RmpA is plasmid-located and controls the production of capsular polysaccharides (CPS). Strains expressing RmpA were substantially linked to purulent tissue infections, including liver abscesses and the high-mucus phenotype of hypervirulent K. pneumoniae (hvKP). Although the capsule is essential in shielding K. pneumoniae from the host’s immune responses, other elements may also contribute to the pathogen’s pathogenicity. Some of the primary strains of K. pneumoniae may have partially altered LPS, which prevents the bacteria from being identified by the host cell. On the other hand, some strains may employ the capsule to conceal LPS to evade being recognized as toll-like receptor 4 (TLR4). These alterations decrease the clearance of microorganisms and limit the inflammatory response. Simultaneously, bacterial growth and replication depend on the availability of iron. K. pneumoniae has four iron-absorbing molecules, or iron carriers: aerobactin, salmochelin, yersiniabactin, and enterobactin. Enteromycin, present in both common and highly pathogenic strains, is thought to be the primary mechanism for absorbing iron because of its strong affinity for the metal. Gastrin and yersinide are more common in hvKp than in cKp, in contrast to enteromycin [26, 28].
Fig. 1.
A schematic representation of K. pneumoniae illustrating the distinction between classical K. pneumoniae (cKp) and HvKP. An evolving pathotype, hypervirulent K. pneumoniae (hvKp), is considerably more virulent than cKp. Typically, healthy members of the community are infected with hvKp
Whereas ST23 hvKp is more common in community-acquired infections in Asia, the ST258 cKp strain is more common in hospital-acquired illnesses in North America and Europe. To better understand K. pneumoniae mucosal pathophysiology, researchers created symptomatic mouse mucosal infection models that mimic human natural infections. It was shown that the nasal cavity serves as the site of cKp replication, rather than the lungs. This early infection event is essential for the development of chronic colonization in the colon and cecum. On the other hand, hvKp multiplies directly in the lungs to produce a fatal bacterial burden, and early esophageal infection promoted subsequent transitory colonization in the ileum and cecum. Here, researchers have created an in vivo model demonstrating how variations in K. pneumoniae tropism cause virulence and disease phenotype in cKp and hvKp, laying the groundwork for more mechanistic research [29] (Fig. 1).
Molecular characterization of multidrug-resistant Klebsiella pneumoniae
Developing extensive, self-conjugating plasmids has been primarily linked to K. pneumoniae’s AMR. These extrachromosomal DNA molecules maintain a consistent copy number per bacterial cell and control their replication rate. They encode components that promote the commencement of replication independently of bacterial chromosome replication. The term “basic replicon” refers to the smallest segment of a plasmid that regulates the replication start while preserving the copy number of the parent plasmid. Plasmid replicon type was not identified in early bacterial typing investigations of the emerging K. pneumoniae ST258 clone because most K. pneumoniae plasmids were not typable using the conventional polymerase chain reaction (PCR)-based replicon typing technique [30, 31]. The whole sequencing of Klebsiella plasmids was recently acquired, thanks to genomics, which showed that K. pneumoniae was endowed with a novel set of plasmids containing hitherto unidentified replicons. The majority of these new plasmid varieties, nonetheless, were connected to the plasmid family known as IncF. All Enterobacteriaceae spp., have been shown to have IncF plasmids, often linked to virulence genes. FII-related replicons are carried by IncF-related plasmids, which have a low copy number and vary in size from 60 to 200 kb. The repA gene, whose transcription is regulated by antisense RNAs that operate in trans by sequence complementarity on target sense mRNAs, makes up the normal FII replicon [31].
Prolonged outbreaks, a higher illness burden, and high death rates are linked to members of the troublesome ESKAPE (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii (A. baumannii), P. aeruginosa (P. aeruginosa), Enterobacter) group of pathogens, particularly cKp. Since 2005, there has been a consistent rise in the incidence of cKp infections, mainly due to strains gaining enzyme-synthesis-like bacteria (ESBLs) and carbapenemases that confer resistance to drugs such as carbapenems and third-generation cephalosporins [32].
The typical K. pneumoniae pathogen is an opportunistic one that often affects people with compromised immune systems and frequently results in nosocomial infections. Pastly healthy individuals may be afflicted by a subgroup of hvKp serotypes that produce more capsule polysaccharide, which may lead to potentially fatal community-acquired illnesses such as pyogenic liver abscess, meningitis, necrotizing fasciitis, endophthalmitis, and severe pneumonia. K. pneumoniae uses a range of virulence factors, including iron acquisition and nitrogen source utilization determinants, fimbriae, LPS, capsule polysaccharide, and outer membrane proteins, to survive and evade the immune system during infection [33]. Antimicrobial-resistant opportunistic infections in hospitalized patients are often caused by K. pneumoniae. The population usually carries acquired resistance to many antimicrobials, and the species is innately resistant to penicillins. However, nothing is known about the ecology, population dynamics, or pathogenicity of K. pneumoniae. Because MDR strains that produce extended-spectrum β-lactamases and/or carbapenemases are increasingly responsible for healthcare-associated infections, K. pneumoniae has become a serious clinical and public health problem during the last 10 years. Severe community-acquired infections due to hvKp have also been reported, and these illnesses are linked to strains that express acquired virulence factors [34]. It may be inferred that rigorous monitoring is necessary to control the obvious and present threat of antibiotic resistance in K. pneumoniae in Asia. Public healthcare institutions should monitor and report any changes in isolates resistant to antibiotics. About one-third of all Gram-negative infections, including endocarditis, septicemia, cystitis, pneumonia, surgical site infections, and UTIs, are caused by these bacteria, claim Shiri and colleagues. Necrotizing pneumonia, pyogenic liver abscesses, and endogenous endophthalmitis are further effects. Infections produced by this bacterium are often linked to high costs, prolonged hospital stays, and high death rates. As MDR and extremely drug-resistant (XDR) Enterobacteriaceae infections are common natural inhabitants of the human and animal microbiome, their sharp increase in frequency is a severe economic issue. Despite its clinical significance, little is known about K. pneumoniae [31]. K. pneumoniae constantly acquires antibiotic resistance genes (ARGs) due to antibiotic selection. This occurs via de novo mutations, plasmid acquisition, and transferable genetic elements and results in XDR strains that have a “super resistome.” Over the last twenty years, several high-risk (HiR) MDR and XDR K. pneumoniae sequence variants have surfaced, demonstrating an enhanced capacity to initiate multicontinental epidemics and persistent worldwide transmission. The findings demonstrate the intricate development of ARG transfer and dissemination, as well as the presence of epidemic plasmids in highly spreading effective clones in MDR and XDR K. pneumoniae. In light of the global crisis of antibiotic resistance and the pressing need to identify the primary pathogens that will continue to threaten future infectious disease outbreaks, further research is necessary to ascertain the epidemic characteristics and plasmid acquisition in K. pneumoniae [31]. Based on current knowledge, the mechanisms underlying antibiotic resistance may include the production of extended-spectrum beta-lactamases, the formation of biofilms, the lack of parenchyma porin antimicrobial agents, and mutations in the genes gryA and parC. These mechanisms render conventional drugs ineffective, necessitating the urgent development of novel treatments [35].
Global public health is under a significant threat from the rise of carbapenem-resistant K. pneumoniae (CRKP), which may produce incurable infections since there are few effective treatment options. One of the antibiotics used as a last option to treat CRKP infections is tigecycline. Nonetheless, reports of tigecycline resistance in CRKP have increased recently. The most common mechanisms of tigecycline resistance have been linked to mutations in the ribosomal S10 protein encoded by the rpsJ gene, which has no horizontal transfer ability and only mildly endangers the public’s health, or to overexpression of non-specific efflux pumps encoded by chromosomes. However, there is a chance that tigecycline resistance would spread horizontally as a result of the identification of plasmid-mediated tigecycline resistance determinants such tet(A) and tet(X) variations. Low-level tigecycline resistance is said to be mediated by tet(A) mutations, which are primarily seen in K. pneumoniae [36].
The AcrAB and OqxAB efflux pumps have been linked to MDR in several bacteria during the last several decades, most notably K. pneumoniae. The acrAB and oqxAB efflux pumps’ higher expression is correlated with a rise in antibiotic resistance. Since ΔΔCT = 0 and 2 to the power of 0 = 1, reference sample relative gene expression is often set to 1. Findings: Cefotaxime (100%), cefuroxime (100%), cefepime (100%), levofloxacin (98%), trimethoprim-sulfamethoxazole (80%), and gentamicin (72%), were shown to have the most incredible rates of resistance, while imipenem (34%) had the lowest. Ciprofloxacin-resistant isolates overexpressed the genes acrA and acrB, oqxA and oqxB, regulators marA, soxS, and rarA compared to the reference strain (strain A111). Additionally, there was a moderate relationship found between the MIC of ciprofloxacin and the expression of the acrAB and oqxAB genes. Through this study, the roles of transcriptional regulators marA, soxS, and rarA, and efflux pump genes, namely acrAB and oqxAB, in bacterial resistance to ciprofloxacin are better understood [37].
The host bacterium’s CPS type primarily determines the K. pneumoniae phage’s effectiveness. One of the main components of K. pneumoniae’s pathogenicity that shields the bacteria from external influences, such as human immunity, is its polysaccharide capsule. Currently, the conventional serological method and the method of sequencing individual genes of the cps gene cluster are used to distinguish between over 100 different polysaccharide capsule types; some of these (КL1, ^L2, \L8, ^L20, ^L39, ^L41, ^L47, ^L53, ^L57, \L64, ^L102, и КL107) are linked to increased virulence and antibiotic resistance [38].
An emerging pathogen that infects both immunocompromised and healthy individuals is hypervirulent K. pneumoniae (hvKp), which causes endophthalmitis, liver abscess, osteomyelitis, meningitis, and necrotizing soft tissue infections. The acquisition of antibiotic-resistance genes by hvKp has emerged as a global concern in recent times. In the Behera et al. investigation [39], 74 K. pneumoniae isolates were gathered for this investigation and identified via blaSHV gene amplification and VITEK2. 18.91% (14/74) of these isolates were determined to be hvKp by utilizing both the phenotypic string test and genotypic iucA PCR amplification. Antibiotic susceptibility testing identified 57.14% (8 of 14) MDR isolates and 35.71% (5 of 14) XDR isolates. Each isolate exhibited resistance to the β-lactam and β-lactamase + inhibitor classes of antibiotics, with colistin demonstrating the lowest resistance. IroB is present in every single one of the fourteen hvKp isolates (100%), followed by the genes iutA (92.85%), peg344 (85.71%), rmpA (57.14%), and magA (21.42%). K1 was the most prevalent serotype among those examined, comprising 21.4% (3/14), with K5 following at 14.3% (2/14). blaOXA-48 was the most prevalent carbapenemase gene (78.57%), followed by blaNDM (14.28%) and blaKPC (14.28%), both of which were found to co-carry multiple resistance genes, including blaSHV (100%), blaCTX-M (92.55%), and blaTEM (78.57%). Strong biofilm production was observed in approximately 92.85% (13/14) of the hvKp isolates, whereas only one isolate (hvKp 10) exhibited moderate biofilm production. Molecular typing with (GTG)5-PCR identified a significant variety of hvKp isolates at the tertiary care facility. MDR-hvKp appears to be an emerging pathogen and a challenge to clinical practice, according to researchers’ findings. To prevent epidemics of the hvKp strain in hospital settings, it is crucial to implement stringent infection control measures and efficient surveillance systems [39].
4. Isolation and characterization of phages specific toKlebsiella pneumoniae Before a novel medication can be licensed for sale in the UK, it must first be produced in compliance with good manufacturing practice (GMP) guidelines and subjected to randomized controlled trials to evaluate its effectiveness and safety. In the UK, PT medication manufacturers must spend heavily and take time to reach GMP standards. When patients with MDR infections have tried every other treatment and need PT, there is a compelling justification for an alternate, quick-turnaround, reasonably inexpensive route [40–42]. When it comes to being an antibiotic substitute, phage treatment has benefits. Combining it with different agents is an emerging strategy. This application approach has shown efficacy in managing intricate microbial biofilms. Antibiotics will be a clear combination. Phages and antibiotics work well together to treat infections brought on by planktonic cells and microbial biofilms. however, their use in treating other illnesses has not been as successful. This increase in effectiveness for biofilms may be explained by the structural modifications that one or both of the agents produced to the cell. The P. aeruginosa biofilm study conducted in 2017 by Chaudhry et al. provided evidence of this phenomenon. They discovered that the bacteria’s peripheral cells were eliminated by the phage’s activity, leaving it vulnerable to the antibiotic that was administered in addition to it. This might also occur in reverse, with the antibiotic upsetting the microbial film’s structure and making it easier for the phages to infiltrate [43].
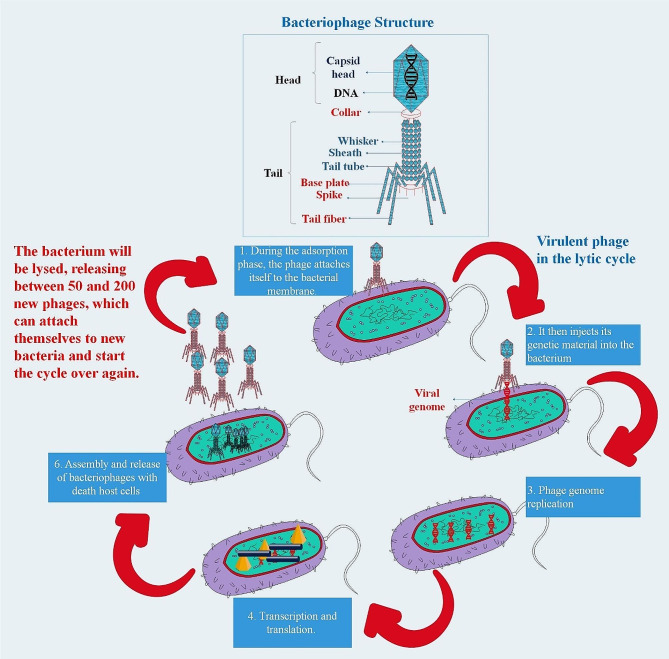
The two primary life cycles or dynamic processes by which phages multiply are crucial to their therapeutic applications. While temperate or lysogenic phages may undergo a lytic life cycle or integrate permanently into their host genome, virulent or obligatory lytic phages infect and rapidly kill their bacterial hosts. To lyse bacterial infections, PT is defined as the direct injection of lytic phages into patients. As an alternate antibacterial approach to treat bacteria that develop biofilms and are resistant to antibiotics, PT has garnered a lot of attention recently [44, 45]. The use of phages in the management of bacterial illnesses has several advantages. One example of a bactericidal agent that may help determine its effective dosages is Phages (auto-dosing). Furthermore, phages are easily separated and have minimal intrinsic toxicities. Generally speaking, wastewater may be used to isolate enterococcal Phages [46] (Fig. 2).
Fig. 2.
Adsorption, replication, and discharge of phage progeny by bacteriophages. Adsorption is the mechanism by which bacteriophages become adhered to the cell membranes of bacteria. This is achieved when phage receptor binding proteins (RBPs) situated on the phage tail engage in interaction with specific receptors located on the bacterial cell surface. Lipopolysaccharide (the most prevalent receptor found in Gram-negative bacteria), polysaccharides, proteins, flagella, and pili are all examples of receptors. In the case of lytic bacteriophages, genetic material is transferred from the phage head into the bacterial cell upon binding, and the molecular apparatus of the host is utilized to generate new phage progeny. Phage release is facilitated by the interaction between holin and endolysin, which induces bacterial cell lysis, culminating in the rupture of the cell membrane and subsequent cell demise [47]
The host range of a phage may dictate how useful the phage is for PT. A few phages have a polyvalent host range, which allows them to infect many species, whereas others have a more extensive host range and can only infect a few strains. It seems that there is no host range selection in standard isolation techniques. Instead, the host range is typically determined after the phage is isolated, which is usually done on a single strain of bacteria. however, some people prefer to use a combination of many host strains for phage isolation [48]. Since Felix d’Herelle invented phage isolation, the fundamental procedure has not altered. This process is often referred to as enrichment. An ambient sample and a sample of bacteria are combined, and the combination is then incubated for a while—usually overnight. After removing the remaining bacteria from the culture using centrifugation, filtering, or both, the filtrate is tested for the presence of phages. After identification, any phages are characterized to see whether they possess the necessary characteristics for PT [48, 49]. The same procedures should be followed for PT, including figuring out how to mix phages into multi-phage combinations called phage cocktails [50]. Phage-based antibacterial treatments play a crucial role in combating diseases for which there are few antibiotic therapy alternatives. As shown by Mattila et al. [51], enriching samples from environmental reservoirs may facilitate the on-demand isolation of matching phages. It’s noteworthy to note that there are wide variations in the effectiveness of enrichment-based phage isolation from municipal sewage; P. aeruginosa, Salmonella, and ESBL-producing E. coli and K. pneumoniae show the greatest results [50–52].
Furthermore, phage virulence, host range, obligatorily lytic vs. alternative life cycles, and other factors may influence their use. While there are few exceptions, screening for many of these qualities during isolation is often challenging, thus testing for them subsequently is done. For instance, lipid-containing and filamentous phages are usually inactivated by chloroform treatment, which may be used to remove filamentous phages from isolation even if they tend to infect animals chronically [48]. In a different study, water samples were treated with phages obtained using a double agar overlay technique. After dilution in SM buffer and filtration via a 0.22 μm filter, isolated phages were purified. The spot technique was used to analyze the host range of the purified lysate. They tested its host range against 20 different bacterial strains, including MDR. Eight distinct host species were targeted by 67 distinct phages that were identified. 47 phages were chosen from among them to examine their host range. Among these, Serratia phages (ΦSER) could infect 17 distinct bacterial strains, including MDR, that had the ESBL and MBL genotypes. In contrast to other phages, Klebsiella phages (ΦKP) have a limited host range. Isolated phages exhibited a broader host range and the capacity to affect clinical strains of bacteria. Above all, the study’s encouraging results against MDR pathogens have increased the likelihood that these phages may be helpful for the biological control of bacterial infections, particularly those caused by MBL and ESBL strains [53].
Two novel fast assays based on NanoSight Limited (NS) technology and quantitative real-time PCR (QPCR) have been suggested recently to determine phage quantities. The QPCR approach measures the total fluorescence produced during each PCR cycle as the phage-containing samples are amplified; that is, time-dependent measurement of the buildup of phage DNA molecules. The fundamental theory of the test is that one may use the former to calculate phage concentrations in aqueous solutions by establishing the connection between DNA counts (ascertained by the QPCR) and the number of viable phage particles (ascertained by plaque assays). The NTA-based method shows nanoparticles (NPs) in a transparent liquid directly and in real-time using laser-illuminated optical microscopy. In seconds or minutes, the NPs are counted when light-scattering centers moving under Brownian motion are recognized. Nanoparticle tracking analysis (NTA) with NanoSight Limited (NS) technology has been used for the analysis of several NPs in suspension and, more recently, for the study of at least one phage preparation [54].
Intranasal and topical use of phages against lung and wound infections caused by K. pneumoniae was beneficial in reducing bacterial loads and death rates in mice. Two lytic phages that can decolonize K. pneumoniae’s intestinal tract in a mouse model were also discovered in recent research [55].
Because of their wide spectrum host-range effectiveness against various bacteria, phages with a large host range are often selected for therapeutic usage. The best phages for treatment are those from the order Caudovirales (Family-Myoviridae, Siphoviridae, and Podoviridae) that have a proteinaceous tail and solely follow the lytic route [56].
Tail fiber proteins, tail spikes, and tail tips are examples of RBPs that are produced by phages and are involved in the first stage of phage infection. These proteins provide several benefits over complete phage particles, including excellent sensitivity and specificity, a reduced probe size, great stability at high pH and temperature levels, strong resistance to proteases, and simplicity in recombinant overexpression. RBPs are being used as recognition elements by several researchers to identify microorganisms. For example, scientists have discovered and described a novel RBP from the K. pneumoniae phage [57]. Recently, metagenomics or viromics has been used by researchers to examine phages in poultry and animals. The rumen of ruminants contains 397,180 viral operational taxonomic units (vOTUs), of which 10.29% are predicted to be phages. Of these, 3.8% are capable of infecting multiple bacterial species, which is a significantly higher percentage than the 0.13% of phages capable of doing so in the human gut [58].
The terminal α-galactosyl moieties of glycolipids and glycoproteins are hydrolyzed by the exoglycosidase enzyme α-Galactosidase (α-D-galactoside galactohydrolase; EC 3.2.1.22). It is widely used in the food, pharmaceutical, and biotechnology sectors and everywhere in nature. The investigators’ goal was to purify α-galactosidase from K. pneumoniae, a bacterium isolated from the human oral cavity. A 70% ammonium sulfate precipitation, dialysis, ion exchange chromatography on a DEAE-cellulose column, and affinity monolith chromatography were the purification procedures. To ascertain the molecular weight of the isolated enzyme, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was used. Using p-nitrophenyl-α-D-galactopyranoside as a substrate, the kinetic constants for this enzyme were determined, including the maximum velocity (Vmax) and Michaelis constant (Km). The purification fold, specific activity, and yield were found to be 126.52, 138.58 units/mg, and 21.5%, respectively, according to the data. The isolated enzyme’s molecular weight was determined by SDS-PAGE to be 75 kDa. The pure α-galactosidase function best at pH 6.0 and 50 °C, respectively. For this enzyme, the corresponding kinetic constants were 769.23 U/ml and 4.6 mM, or the Michaelis constant (Km) and maximum velocity (Vmax). K. pneumoniae’s α-galactosidase was isolated and described. The isolated enzyme manifested as a single band with a molecular weight of 75 kDa, according to (SDS-PAGE) analysis. This investigation aims to extract, purify, and characterize α-galactosidase from K. pneumoniae in the oral cavity of humans. Using chromatographic techniques, such as affinity chromatography and anion-exchanger DEAE-cellulose, is one of the purification processes. The activity of the refined enzyme was assessed over various pH values and temperatures. Analyses of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and kinetic parameter (Km and Vmax) determination were also performed [59].
A single lytic phage, KPO1K2, that is specific to K. pneumoniae B5055 and has a wide host range was chosen by Verma et al. KPO1K2 has an icosahedral head with a pentagonal nature and an apex-to-apex head diameter of about 39 nm, as shown by TEM analysis. The lytic phage known as T7-like was proposed to be classified within the family Podoviridae due to the presence of a short, noncontractile tail (10 nm). Further evidence of this phage’s T7-like traits came from its phage development cycle, which included a latent period of 15 min, a burst size of around 140 PFU per infected cell, a genome of 42 kbps, and a structural protein pattern. Phage showed its highest activity at 37 °C and was stable throughout a pH range of 4–11. Six hours after the mice were injected, a high phage titer was seen in the blood, kidney, and bladder; however, the titers in the kidney and bladder were more significantthan those in the blood. Phage was entirely removed from the blood after 36 h, although it took longer to clear from the kidneys and bladder. It is suggested that this phage, KPO1K2, be employed as a preventive or therapeutic agent, particularly for the treatment of UTIs linked with catheter usage that are caused by K. pneumoniae [60].
Phages can potentially be an effective tool for treating patients, decolonizing colonized persons, and limiting the spread of isolates resistant to antibiotics. Ciacci et al. [61] have described vB_Kpn_F48, a lytic phage unique to K. pneumoniae isolates from clonal group 101. Transmission electron microscopy examination led to the classification of Phage vB_Kpn_F48 as a member of the Myoviridae, order Caudovirales. vB_Kpn_F48’s physiological characterization revealed that it has a small host range, a brief latent time, and a small burst size, and it is very stable to changes in pH and temperature. The phage is characterized by a 171 Kb dsDNA genome that is devoid of genes that are undesirable from a therapeutic standpoint, including integrases, antibiotic resistance genes, and toxin-encoding genes, according to high throughput sequencing and bioinformatics research. vB_Kpn_F48, which we have provisionally dubbed “F48virus,” is a T4-like phage that seems to belong to a new genus within the Tevenvirinae subfamily, according to phylogenetic research. Given its limited host range, phage vB_Kpn_F48’s genomic characteristics and general physiological factors make it a viable option for use alone or in combinations for PT purposes [61].
Morozova et al. [62] provided the genome, partial structural proteome, and biological characteristics and suggested the taxonomy of KP8, a new lytic N4-like phage unique to K. pneumoniae. According to electron microscopy, KP8 is a member of the Podoviridae family. The genome of KP8 was 73,679 base pairs in size and included 97 putative open reading frames. According to a comparative genome study, the N4-like podoviruses Enquatrovirus and Gamaleyavirus phages have genomes most comparable to the KP8 genome. Furthermore, the KP8 genome included the gene encoding a large virion-encapsulated RNA polymerase and had gene synteny characteristic of the N4-like podoviruses. The KP8 genome underwent phylogenetic analysis, which showed that it belonged to a separate branch within the clade that also included members of the Enquatrovirus and Gamaleyavirus genera. Similar to the evolutionary divergences between the Enquatrovirus and Gamaleyavirus genera (0.468 substitutes/site), the average evolutionary divergences between KP8/Enquatrovirus and KP8/Gamaleyavirus were 0.466 and 0.447 substitutions per site (substitutes/site), respectively. According to the information gathered, Klebsiella phage KP8 is distinct from other phages that are related to it and could perhaps be a new genus of N4-like phages [62].
In a study [63], by enrichment, the phage KP34, which is unique to K. pneumoniae, was recovered from a sewage sample. A sewage sample was centrifuged at 15,000×g for 15 min, and the supernatant was filtered using a Millipore 0.22-µm filter. The propagation of phages adhered to the modified Ślopek et al. approach. This research focused on the new “phiKMV-like virus,” KP34, which was propagated on an ESBL-producing MDRKP strain. Regarding host specificity, activity spectrum, morphological, biological, molecular, and genomic characteristics, phage KP34 was thoroughly characterized. This novel member of the Podoviridae family of phages is being investigated for possible use in PT. The new virus known as phage KP34 is lytic for extended-spectrum β-lactamase-producing K. pneumoniae strains and is a member of the Autographivirinae subfamily. Its shape, activity spectrum, burst size, host specificity, vulnerability to chemical and physical agents, and biological characteristics were identified. KP34 molecular characteristics, such as its protein composition and genome sequencing, were investigated as possible antibacterial agents for therapeutic application. The KP34 phage genome sequences’ clustering and phylogenetic analysis demonstrated their strong connections to “phiKMV-like viruses.” Concurrently, whole-genome analysis allowed all phages belonging to the Podoviridae family with fully sequenced genomes to be grouped and classified [63].
Tehran’s hospital waste-water treatment facility provided water samples for the study. After filtering and combining the samples with an overnight-grown K. pneumoniae (ATCC 10031) culture, the samples were incubated for the whole night at 37 °C. The K. pneumoniae ATCC strain was used to perform phage titration, assess the latent period, and determine burst size using the double-layer agar technique [64]. Transmission electron microscopy (TEM), temperature, pH, and chloroform stability measurements were used to characterize the isolated phage. The susceptibility of 51 MDRKP isolates, A. baumannii, P. aeruginosa, ESBL-producing K. pneumoniae, and E. coli was determined by incubating 20 µl of the phage suspension (108 PFU) at 37 °C for a whole night on bacterial lawns. Clear zones that formed suggested vulnerability. With a latent period of 40 min and a burst time of 52 min, the isolated lytic phage produced tiny, transparent plaques infected with 35–40 phage particles per infected cell. The phage was given the designation vB_KpnS-Teh.1, resembled the tailed Siphoviridae family according to TEM data. It was found that the phage vB_KpnS-Teh.1 was resistant to chloroform and most stable at 37 °C and pH 7. In conclusion, the lytic phage that was isolated demonstrated specificity for K. pneumoniae. Its potential for treating K. pneumoniae infections will be further investigated [64].
Six lytic Klebsiella phages were identified as potential candidate bacteria for further testing after being isolated from Georgian river water tainted by sewage. More research was done on two of the phages [65]. For each of the six phages, biological characteristics such as shape, host range, growth phenotype, nucleic acid content, stability in temperature and pH, and growth phenotype were examined. To determine the phylogeny of the phages vB_Klp_5 and vB_Klox_2, which are unique to K. pneumoniae and K. oxytoca, respectively, limited sample sequencing was carried out. The last two phages were stable in unfavorable environments and had huge burst sizes and effective adsorption rates. Double-stranded DNA bacterial viruses from the families Podoviridae and Siphoviridae are the phages described in this research. Approximately 63% of the Klebsiella bacteria in 123 clinical isolates from Georgia and the UK could be effectively lysed by one or more of the six phages. Several characteristics of these phages point to their possible use in PT cocktails [65].
Investigators identified and described a phage (NTUH-K2044-K1-1) that infects K. pneumoniae NTUH-K2044 (capsular type K1) [66]. None of the strains with different capsular types were infected by the phage; only the K1 strains were. This phage did not lyse mutants with deleted capsules, indicating that the capsule was necessary for phage infection. A new phiKMV-like virus was identified as the phage by whole genome sequencing. It was determined which gene encodes capsule depolymerase. The K1 capsule showed selective lysis by the recombinant enzyme. Mice infected with the NTUH-K2044 strain, including one treated after imaging revealed a neck abscess, showed markedly improved survival rates when treated with the phage or recombinant enzyme. After this phage was given to mice, no overt illness was seen. Two days after injection, mice’s liver, spleen, brain, and blood still contained measurable amounts of phage. These findings show that this phage and its capsule depolymerase are specific for capsular type K1, indicating that K1 K. pneumoniae infections may be diagnosed and treated with them [66].
Ciacci et al. reported the isolation, physical, biological, and molecular characterization of a new lytic phage, as well as its effectiveness in controlling MDRKP. Using K. pneumoniae CCCD-K001 as the host, the phage vB_KpnS_Uniso31, also known as phage Kpn31, was identified from hospital wastewater [61]. Based on the whole genome sequence, Phage Kpn31 was categorized as a Sugarlandvirus (Demerecviridae) due to its siphovirus-like morphotype. There are no known toxins, AMR genes, or sequences linked to depolymerizes encoded in the 113,444 bp Kpn31 genome. The data from Phage Kpn31’s eclipse time of 15 min and burst size of 9.12 PFU/host cell allowed us to infer that, given a 30-minute latency period, it replicates successfully in K. pneumoniae CCCD-K001. In vitro antibacterial activity studies demonstrated the efficacy of phage Kpn31 against a minimum of six clinical isolates of MDRKP patients. Phage Kpn31 has characteristics that suggest the possibility of managing infections by MDRKP [61].
Therapeutic potential of Klebsiella phages
K. pneumoniae-related UTIs have the potential to produce clinical severe problems, including death. For patients who are infected, phages provide an additional therapeutic option. In a past investigation, researchers used K. pneumoniae as a host to isolate the phage KP-1 from sewage water [67]. A whole genome study revealed that the genome lacked virulence or antibiotic resistance genes and was a double-stranded linear DNA molecule measuring 176,096 base pairs with a 41.8% GC content. Phage KP-1’s inactivation capacity was evaluated in broth at a multiplicity of infection (MOI) of 1 and 10, with maximal inactivation of 4.9 and 5.4 log CFU/mL after 9 h, respectively. To examine the phage’s performance in an acidic environment, the MOI of 10 effectiveness was also measured in urine. After nine hours, a maximum inactivation of 3.8 log CFU/mL was observed. The findings suggest that phage KP-1 may be helpful in managing UTIs caused by K. pneumoniae; however, additional research is required to expand the phage’s spectrum of activity through phage cocktails and to prevent phage viability from being negatively impacted by urine’s low pH by enclosing the phages in micro- or nanocarriers [67].
Researchers set out to determine how a phage specific to Klebsiella that was isolated from sewage affected the growth of clinical isolates of MDRKP using the time-kill assay, as well as how phages and phage-antibiotic (imipenem) combinations affected the formation of biofilms [68]. There were 40 MDRK isolates in all. It was using K. pneumoniae subspp. K. pneumoniae ATCC 33,495 as the host, a phage unique to Klebsiella was identified from sewage. When phage was given alone or in conjunction with imipenem, the in vitro time-kill test revealed a 0.4 log reduction and a 0.5 log decline in K. pneumoniae colony counts at 4 h, respectively. Combinations of phage and phage-imipenem decreased K. pneumoniae’s capacity to form biofilm by 38% and 53%, respectively. The results of this investigation indicated that PT may have inhibitory effects on MDRK. Although it does not have a completely cidal effect, it is reported to lower the number of bacteria and the production of biofilms. However, a phage cocktail, or phage combined with another antibiotic(s), is recommended to get a significant response [68].
Here, scientists [69] have identified and described phages based on their genomes that may infect all 77 reference serotypes of Klebsiella spp., including capsular types often seen in HiR K. pneumoniae clones that cause nosocomial infections. A few phages showed the ability to infect a wide range of targets, but most taxonomically related phages showed strong capsular specificity. The researchers isolated phages for every kind of capsular in the collection. Investigators concentrated on the receptor-binding proteins of these phages, paying special attention to their depolymerase domains to unravel the factors that determine their specificity. They also looked at the potential for creating a broad-spectrum phage cocktail using phages found in strains of reference capsular type and assessing its capacity to kill pertinent clinical isolates. It’s interesting to note that a panel of clinical isolates of K. pneumoniae resistant to carbapenem was used to evaluate a combination of 12 phages that could infect 60% of the reference serotypes of Klebsiella spp. According to their findings, phage hunting in a very changeable encapsulated bacterial host has to be focused on the particular Klebsiella isolates. This study advances knowledge of the intricacy of phage-host interactions and emphasizes the significance of applying accurate and phage-specific treatment approaches to K. pneumoniae infections globally [69].
CRKP is one of the primary pathogens responsible for challenging-to-treat wound infections linked to healthcare. Phages are a different strategy to combat CRKP. The CRKP clinical strain of sequence type 11 and capsular type KL64, which is the predominant type in China and has genes encoding KPC-2 and NDM-1 carbapenemases, were used by investigators to develop murine wound infection models. On day 7 post-treatment, the phage treatment group’s wound bacterial load was 4.95 × 102 CFU/g lower than the infection control groups, and wounds healed on day 10 instead of day 14 in the latter group. There were no negative phage-related occurrences noted. The phage cocktail accelerated wound healing while safely lowering the bacterial burden in the wound [70].
In another study [71], from hospital sewage, phage vB_KpP_HS106 against MDR-hv K. pneumoniae strains were identified. It may remain constant in activity between 4 and 12 pH and between 4 and 50 °C in temperature. It was discovered that the phage HS106 had a maximum adsorption rate of around 84.2% at 6 min. According to a one-step growth curve study, HS106 had a latent period of 10 min and a burst size of around 183 PFU/cell. Moreover, complete genome analysis revealed that the phage HS106 genome was a linear, double-stranded DNA molecule 76,430 bp in length and 44% GC. The HS106 genome included a total of 95 annotated open reading frames; no virulence or antibiotic resistance genes were found in these frames. At 25 °C, phage HS106 decreased MDRKP by around 1.6 log10 CFU/mL in milk and by almost 2 log10 CFU/cm3 in chicken. For this reason, vB_KpP_HS106 is a viable biocontrol agent against MDRKP in food as an alternative to antibiotics [71].
Using KP 1513, a clinical MDRKP isolate, as the host, Phage 1513 was isolated and described [72]. It was assigned to the Siphoviridae family after producing a transparent plaque with a halo. Its burst size was 264, its latent period was 30 min, and its ability to suppress K. pneumoniae 1513 development in vitro followed a dose-dependent pattern. Mice were protected against fatal pneumonia by intranasal delivery of a single dose of 2 × 109 PFU/mouse two hours after K. pneumoniae 1513 inoculation. Phage-treated mice showed reduced lung K. pneumoniae load in a sublethal pneumonia paradigm compared to the untreated control group. These mice showed decreased levels of inflammatory cytokines in their lungs and lost less weight overall. PT improved the lung lesion conditions. As a result, phage 1513 has strong effects both in vitro and in vivo, suggesting that it may be used as a substitute for antibiotics in treating pneumonia brought on by the MDRKP [72].
A mouse model of K. pneumoniae-mediated lobar pneumonia was used to assess the therapeutic potential of phage SS specific for K. pneumoniae B5055, which was identified and described. Intranasal (i.n.) injection of mice with bacteria (108 PFU. ml− 1) posed a problem. Following an intraperitoneal injection of 1010 PFU/ml− 1 phage, 100% of the animals were successfully rescued from respiratory infections caused by K. pneumoniae. Infected mice showed considerable protection when the phage preparation was given three hours before the intracellular bacterial challenge; however, the PT was useless when the phage preparation was administered even six hours after the infection was induced. According to findings, the timing of the PT after the illness first appears has a major impact on how well the therapy works [73].
One significant component of K. pneumoniae’s pathogenicity is its CPS. There is a correlation between the pathogenicity of K. pneumoniae and the 78 capsular types identified so far. Using transmission electron microscopy, a recently reported lytic phage specific for KN2 K. pneumoniae, called 0507-KN2-1, was isolated and studied. A 159 991 bp double-stranded DNA genome with a G + C content of 46.7% and at least 154 open reading frames was discovered by whole-genome sequencing of 0507-KN2-1. 0507-KN2-1’s morphological and genetic features led to its classification as a phage belonging to the Myoviridae family. After this phage was further examined, a 3738-bp gene encoding a potential polysaccharide depolymerase was found. To verify this protein’s enzymatic activity and specificity to KN2 CPS, a recombinant version was created and subjected to assays. Spot analysis revealed that KN2 K. pneumoniae strains were more sensitive to this depolymerase than the homologous phage. Here, investigators report the discovery of a new Klebsiella capsular type in a collection of clinical strains. Researchers discovered a polysaccharide depolymerase-producing KN2-specific phage that may be used for effective capsular typing. In treating K. pneumoniae infections, the lytic phage and depolymerase also hold promise as substitute therapeutic agents to antibiotics, particularly when treating strains of the illness that are resistant to medicines [74].
The need for novel treatments that are effective against MDRKP sequence type 258 (ST258) is highlighted by severe infections produced by this pathogen. In this work [75], researchers evaluated the microbiologic, histopathologic, and survival results in ST258-infected mice after systemic phage injection. Investigators discovered mice with K. pneumoniae ST258 bacteremia were saved when they received timely therapy with two phages, either alone or together. The mice in the combined PT therapy group showed the highest increase in survival and the lowest prevalence of phage resistance among the germs retrieved from the blood and tissue of the mice among the three treatment groups. Their results confirm the effectiveness of PT as a therapy for refractory ST258 infections and highlight how this modality may be improved by carefully choosing which phages to use [75].
The four virulent phages from the Siphoviridae, Myoviridae, and Podoviridae families are described in this article in terms of their microbiological, physicochemical, and genomic makeup [76]. In the Zurabov et al. [76] study, the genomes of all four phages were studied by restriction digestion and whole genome sequencing. Phages were analyzed by electron microscopy, which allowed researchers to identify their host range, lytic activity, adsorption rate, burst size, latent period, frequency of phage-resistant forms generation, lysis dynamics, and sensitivity of phage particles to temperature and pH. The phages under study demonstrated a broad host range and excellent stability at various pH and temperature levels. A combination of phages destroyed every strain of bacteria investigated, as opposed to individual phages. Moreover, no instances of the formation of phage-resistant bacterial colonies were found. The absence of virulence, lysogenic genes, and antibiotic resistance in isolated viruses was shown by genomic data. Polysaccharide depolymerases, which break down biofilms and capsules, are encoded by three of the four phages. The phages examined in this paper show promise for more in vivo investigations and may be used in PT as a component of sophisticated preventive and therapeutic phage preparation. The results of the experiments indicate that the complex preparation works better than the individual phages. By using a complex phage cocktail, the lytic spectrum may be expanded and the likelihood of phage-resistant forms arising is significantly decreased [76].
In the Szeloch et al. research [77], eight of the 32 isolated phages were members of the Myoviridae family, eight of the Siphoviridae family, and sixteen of the Podoviridae family. These phages were tested against 254 clinical Enterobacteriaceae strains, including isolates of MDRKP that produced ESBLs. Six phages were further characterized for burst size, physicochemical features, and susceptibility to restriction endonuclease digestion based on their lytic potential. Furthermore, five had complete sequencing. Numerous host resistance mechanisms encoded by phages were discovered. Low numbers of host restriction sites were present in the Siphoviridae phage genomes (KP16 and KP36), which is comparable to the T7-like phages’ (KP32) tactic. Furthermore, phage KP36 encoded an Adenine Methyltransferase for itself. This study’s φKMV-like KP34 phage was susceptible to every endonuclease used. Dam methylation of KP34 DNA was found, although this occurred without the presence of a methyltransferase that could be identified in the phage. The Dam and Dcm methyltransferase genes, along with other anti-restriction mechanisms clarified in earlier research, were carried out by the Myoviridae phages KP15 and KP27. Other research is necessary to fully understand the unknown anti-restriction mechanism that the Myoviridae phage KP27 encodes. No other anti-restriction mechanisms, such as unusual nucleotides (hmC or glucosyl hmC), were discovered [77].
In vitro tests were conducted to assess the lytic activity of phage ZCKP1, which was isolated from freshwater in Giza, Egypt, against an MDRKP KP/01 that was isolated from a diabetic patient’s foot wound in Egypt. The ZCKP1 phage’s characterization revealed that it has a 150.9 kb ds-DNA genome and belongs to the Myoviridae family. Numerous osteomyelitis pathogenic organisms, such as isolates of E. coli, Proteus spp., and Klebsiella spp., were lysed by the phage ZCKP1. When treated at high multiplicity of infection (50 PFU/CFU), the phage decreased the bacterial counts of the host bacteria by ≥ 2 log10 CFU/ml at 25 °C. It also showed the capacity to reduce bacterial counts and biofilm biomass (> 50%). Because of these qualities, the ZCKP1 phage may be used therapeutically to cure K. pneumoniae and related bacteria that are prevalent in patients with diabetic foot [78].
Up to 80 °C, phage VTCCBPA43 was discovered to have a high-temperature tolerance. It showed a limited host range, peaked at pH 5, and had a burst size of 172 PFU/mL. The phage was recognized as KP36-like by the use of shotgun proteomics. After a single dosage (2 × 109 PFU/mouse) of virulent K. pneumoniae was administered intranasally, there was a significant decrease in the lung bacterial burden at all time points and the presence of physiologically active phage in vivo. In the pneumonic mouse model, VTCCBPA43 PT seemed to have generally positive effects based on decreased lesion severity [79]. Phage-resistant genotypes of K. pneumoniae emerged after the second phage treatment. A comparative genomic analysis revealed that phage-resistant strains possess fabF deletion. The LPS structure of the host receptor for therapeutic phages was observed to be altered by the fabF knockout strain (Kp7450ΔfabF). Virulence evaluations in zebrafish and mice revealed that LPS was the principal determinant of virulence in Kp7450 and that modifications in the structure of LPS in Kp7450ΔfabF inhibited virulence. However, phage-resistant strains exhibited a reduction in virulence at a cost [80] (Table 1).
Table 1.
The role of several bacteriophages as an alternative method against K. pneumoniae antibiotic resistance
| Bacteriophage | Function against K. pneumoniae | REF |
|---|---|---|
| Kp7450ΔfabF | Virulence assays conducted on zebrafish and rodent models revealed that LPS was the predominant determinant impacting the virulence of Kp7450 and modifications in the structure of LPS in Kp7450ΔfabF. Concurrently, the virulence of the phage-resistant strains diminished at a cost. | [80] |
| VB_KPM_KP1LMA (KP1LMA) | Researchers hypothesize that phage KP1LMA may be able to treat a UTI brought on by this strain of K. pneumoniae, suggesting that if further strain-specific phages are discovered, the same process may be used to treat UTIs brought on by other strains. Even though phage KP1LMA has a limited host range, attempts may be undertaken to broaden its spectrum of action and mix it with other phages to possibly make it effective against additional strains of K. pneumoniae that are linked to UTIs. | [85] |
| Phage M1 | Phage M1, isolated from sewage water, has shown potential in treating infections brought on by K. pneumoniae that is resistant to drugs when used in conjunction with medicines. This strategy has shown promise in clinical settings by dramatically lowering the occurrence of bacterial phage resistance. Furthermore, studies on patients with primary sclerosing cholangitis (PSC) demonstrated the therapeutic potential of bacteriophage combinations in inhibiting the development of certain strains of K. pneumoniae. | [86] |
| phage vB_KshKPC-M | One major clinical issue is carbapenem-resistant K. pneumoniae (CRKP). Furthermore, research looking at CRKP strains from COVID-19 patients who had pneumonia related to using a ventilator emphasizes how crucial it is to create specific bacteriophage treatments for complicated illnesses. Phage vB_KshKPC-M is a good option for phage treatment applications in the future due to its antibacterial activity and broad host range. Most of the clinical isolates resistant to antibiotics could be lysed by this isolated phage. To control and suppress respiratory infections produced by these bacteria, particularly when treating respiratory diseases caused by resistant strains in ill patients, this phage may thus be utilized alone or in combination with other phages in future research. | [87] |
| KPW-1, KPW-2, KPW-3 | The isolated phages (KPW-1, KPW-2, and KPW-3) were shown by researchers to have a highly selective host range and to be stable in various pH and temperature settings. The phages successfully inhibited the development of biofilms and bacterial growth. Researchers showed how to isolate and characterize phages that may be used against strains of K. pneumoniae that were resistant to drugs. To treat bacterial illnesses resistant to many drugs, these phages show encouraging potential as substitute therapeutic agents. | [88] |
| EKq1 | Phage EKq1 exhibits lytic activity towards certain clinical isolates of MDRKP. | [89] |
| PG14 | It has been shown that antibiofilm phages are beneficial against MDRKP infections. PG14, a phage of Klebsiella, has significant antibiofilm efficacy by lysing carbapenem-resistant K. pneumoniae G14 and exhibiting 80% biofilm suppression and 71% biofilm disruption. | [90] |
| vB_Kpn_ZCKp20p | Isolated from medical and industrial effluent, phage vB_Kpn_ZCKp20p exhibited the capacity to lyse biofilm-forming MDRKP isolates while preventing cytotoxicity to human skin fibroblasts. Additionally, it disrupted mature biofilms and impeded biofilm formation. | [91] |
| vB_KpnS_FZ10, vB_KpnP_FZ12 and vB_KpnM_FZ14 | Zurabov et al. documented the successful suppression of MDRKP Kl 315 biofilms through the utilization of a depolymerase-active cocktail comprising three bacteriophages (vB_KpnS_FZ10, vB_KpnP_FZ12, and vB_KpnM_FZ14). | [92] |
Singh et al. [81] have shown the isolation, microbiological characterization, and genetic analysis of phage Kp109, which can infect K. pneumoniae. Phage Kp109 demonstrated strong resistance to a wide range of pH values (3–9) and temperatures (4–60 °C). According to genomic study and transmission electron microscopy, phage Kp109 is a member of the Drexlerviridae family and genus Webervirus. The Kp109 has a 51,630 bp long double-stranded DNA genome with a 51.64% GC content, according to genomic research. Phage Kp109 is a safer biocontrol agent for phage treatment and other uses since its genome does not include any known virulence, lysogenic, or antibiotic-resistant genes (ARGs). Based on computational study, the putative endolysin gene has the potential to be exploited as an enzymatic, as shown by the binding energy of − 6.23 kcal/mol between LysKp109 and ligand NAM-NAG. In the Singh et al. study, phenotypic, genomic, and computational characterizations indicate that phages Kp109 and LysKp109 are viable options for in vivo investigations in the future and may have applications in the management of K. pneumoniae infection [81].
Using an intragastric model, a recently identified phage (φNK5) with lytic activity for K. pneumoniae was employed to cure an illness [82]. A single dose of φNK5 administered intraperitoneally or intragastrically at a dose less than 2 × 108 PFU at 30 min post-infection with K. pneumoniae was found to protect mice against death in a dose-dependent manner; however, the more significant protection was obtained with a low dose of φNK5 administered intragastrically. Because the germs were still spreading into the bloodstream 24 h after the K. pneumoniae inoculation, intraperitoneal PT was more effective than intragastric treatment. Phage φNK5 remained protective even when given as late as 24 h after the K. pneumoniae inoculation. Bacterial counts were surveyed in mice administered φNK5 intraperitoneally, and the results showed that the germs were successfully removed from the liver and blood tissues. Treatment with φNK5 dramatically reduced the generation of inflammatory cytokines, as well as aspartate and alanine aminotransferases in the blood and liver harm caused by K. pneumoniae, such as liver necrosis. These findings imply that a modest dosage of φNK5 may be a helpful treatment for liver infections brought on by K. pneumoniae [82].
Little progress has been achieved in the last several decades in using virulent phages in place of or in addition to antibiotics for treating K. pneumoniae infections. This paper tackles the problem of anti-Klebsiella phages’ limited host range and shows the potential benefits of an anti-Klebsiella phage separation technique. Anti-Kd phages may function at infection sites where capsule expression is sporadic or suppressed, or they can work in tandem with anti-K phages, which often cause escape mutants to lose their capsules [83].
Here, scientists showed that a K2-specific lysis phage, ΠFK1979, to contain polysaccharide depolymerase, quickly evolved resistance to it [84]. This phage is one of the primary pandemic lineages of hvKP with thick capsules. The expression of the capsule was significantly reduced in the phage-resistant mutants. In the mutants, single nucleotide polymorphisms were found in the genes encoding sugar glycosyltransferase, RfaH, galU, and polysaccharide deacetylase family protein, according to WGS. It was also shown that phage sensitivity and capsule formation depend on RfaH and galU. In the mutants, there was a substantial reduction in the expression of genes related to the manufacture or control of LPS and/or capsules. Phage-resistant mutants of hvKP were more vulnerable to the immune system, demonstrated by the diminution of virulence and fitness in vitro and in vivo, despite the fast and frequent development of phage resistance being a disadvantage. It’s interesting to note that the shape of the viral particles and plaques of the recently identified phages that target mutants altered dramatically. Compared to ΦFK1979, their genomes were both substantially more giant and distinct. Compared to FK1979, they have far broader host spectrums and far more functional proteins. According to researchers, K2-specific phage may be used to combat hvKP-relevant illnesses as an anti-virulence agent or as a component of phage cocktails targeting phage-resistant bacteria [84].
Antibiotic with bacteriophage againstKlebsiella pneumoniae
The majority of phage preparations being studied and created by Western businesses are called “phage cocktails,” which appear to ignore several unique characteristics of phages, including hostile coevolution and target selectivity. These static phage mixtures performed poorly in recent randomized controlled trials, in contrast to a growing number of case studies that used preadapted (or even modified) phages that are more efficient against the infecting bacteria or phages used as supplementary treatment. A fresh perspective on PT may be gained from the current case study, which advocates using phage-antibiotic combinations customized for each patient. It’s still unclear whether using phages and antibiotics in that order or concurrently is best. Strategies akin to “Trojan horse” tactics may be explored, in which phages are used first to demolish biofilms and stimulate dormant cells, increasing their susceptibility to antibiotics administered in a subsequent stage. Furthermore, several phages possess the capacity to reverse antibiotic resistance [93].
Investigators evaluated the effectiveness of phage, either alone or in conjunction with amoxicillin, in eliminating the biofilm of K. pneumoniae B5055 [94]. Planktonic cells and the biofilm of K. pneumoniae B5055, which were cultivated in 96-well microtiter plates, were subjected to three different antibiotic doses (512, 256, and 128 µg/ml) as well as phage at varying multiplicities of infections (MoIs). Following exposure to 256 µg/ml (MIC) of amoxicillin, the bacterial load of the 1-day-old biofilm and planktonic culture was reduced by a log factor of 1.24 ± 0.27 and 4.1 ± 0.31, respectively; however, the bacterial load of the mature (8-day-old) biofilm showed no significant reduction. Eight-day-old biofilm showed a log decrease of 2.97 ± 0.11 and 3.51 ± 0.19 when exposed to a higher dosage of antibiotics (512 µg/ml) or phage alone (MoI = 0.01). Following exposure to a combination of amoxicillin and phage, a noteworthy decrease in the biofilm’s bacterial load was observed. Therefore, a more vital degradation of the biofilm structure when an antibiotic was used in conjunction with a particular phage revealed that the phages might be utilized in conjunction with antibiotic treatment with success. Combination treatment also has the additional benefit of preventing the development of resistant mutants, which would otherwise be simple to do when using phage or antibiotics alone [94].
It was investigated if lytic phage [95], alone or in conjunction with ciprofloxacin, was an effective therapeutic agent against K. pneumoniae biofilm. The resistant variations that developed throughout therapy were assessed for their pathogenic potential. The K. pneumoniae B5055 biofilm was treated with a lytic phage and ciprofloxacin. Following the corresponding treatments, estimates were made of the effectiveness and frequency of resistant variant generation. Their potential for pathogenicity was assessed for the resistant types. The biofilm was successfully removed by phage on its own, and there was no discernible change in its capacity to remove biofilm when used in conjunction with ciprofloxacin. Nevertheless, the growth of resistant genotypes was dramatically halted by combined therapy using phage and ciprofloxacin. The few variations that emerged generated little levels of cell-associated CPS were more susceptible to mice peritoneal macrophages, and could not build biofilms. Additionally, it was noted that the outer membrane proteins of the bacterial isolates had altered shape and pattern [95].
It has been shown that ciprofloxacin therapy alone may substantially destroy the K. pneumoniae juvenile biofilm. In the current investigation [96], the age of the biofilm plays a critical role in deciding how well antibiotic therapy works. Therefore, the biofilm of K. pneumoniae that had formed for lengthy periods was treated with ciprofloxacin and/or lytic phage that produces depolymerase (KPO1K2). When the two treatments were applied together, the number of bacteria in the older biofilm decreased more since ciprofloxacin by itself was unable to reduce the bacterial biomass in older biofilms substantially. Following KPO1K2 treatment, confocal imaging revealed that the biofilm matrix underwent structural alterations, and the size of the micro-colonies shrank. The little biofilm removal by a virus that did not produce depolymerase but did remove the biofilm when combined with pure depolymerase highlighted the function of phage-associated depolymerase. These results show that older biofilms may be considerably eliminated by a lytic phage alone and that this lytic phage’s primary mode of action is depolymerase-mediated. However, applying the phage and antibiotic together led to a marginal improvement in biofilm removal, supporting the theory that phage might boost the effectiveness of antibiotics [96].
Investigators assessed the effectiveness of phage and antibiotic combinations against MDRKP. Two MDR strains of K. pneumoniae (GenBank nos. MF953600 & MF953599) were used. Using a twofold agar overlay technique, phages were separated from hospital sewage samples and identified using transmission electron microscopy. The host range test and death assay were used to characterize phages better. Phages (7.5 × 104 PFU/mL) were used in conjunction with different antibiotics to evaluate the effectiveness of the treatment. Recording OD650nm values demonstrated that the phage-cefepime and tetracycline combinations showed potential therapeutic effects, limiting the development of K. pneumoniae isolates [97].
Clinical trial of Klebsiella phages
In another study, a total of 223 patients with lung and pleural infections were treated with phages; the outcomes were assessed in comparison to 117 cases treated with antibiotics. Of the patients in the phage-treated group, 82% achieved complete recovery, whereas only 64% did so in the antibiotic-treated group [98]. Because they can differentiate between strains of the same bacterial species, phages are also therapeutically important despite being mostly replaced by newer technology. Similar to how people may get numerous viruses, most examined bacterial species have multiple phage pathogens. Compared to the number of in vivo studies and case reports, there are very few clinical trials using PT against the four identified microorganisms. The majority are phase I/II clinical studies focusing on safety rather than effectiveness. Some have been carried out mainly to combat a single germ, while others concentrate on a concrete infectious illness brought on by several bacteria [99].
Any scientific activity must pass through a crucial shift from the observable and anecdotal to the reproducible and quantified. Numerous new clinical investigations suggest that PT is still at a turning point in its development from qualitative to quantitative. Clinicaltrials.gov lists seven PT clinical studies carried out between 2000 and 2015. On the other hand, in 2022 alone, 18 clinical studies were started. Clinicaltrials.gov now lists 45 clinical studies as of March 2023. Additionally, a phase I/II clinical trial completed in 2015 and looked at PT for P. aeruginosa burn wound infections was posted on the European equivalent website, clinical trials register.eu (accessed on 18 April 2023). Of these, twenty-five are clinical studies in either phase I or phase II, and fourteen of them are in combination. There are five phase III studies and none in phase IV. Preclinical, observational, and extended-use studies comprise the remaining research [100].
The phagebiotic composition’s therapeutic and preventive efficacy was shown in the treatment and prevention of experimental acute K. pneumoniae infection in mice. According to the research, the preparation has an excellent therapeutic efficiency and is quite competitive with ciprofloxacin, which is particularly efficient in treating the K. pneumoniae infection. The purpose of investigators small-scale clinical experiment was to assess the therapeutic efficacy of the phagebiotic composition in an epidemiological emergency caused by multi-resistant strains of P. aeruginosa, K. pneumoniae, and A. baumannii in an intensive care unit (ICU). Blood, urine, and endotracheal aspirates from 14 patients comprised 79% of the original samples that were contaminated. The contamination level decreased to 21% 24 h after the three-day PT (20 ml of cocktail at a titer for each phage 108 pfu/ml were administered intragastrically via a tube once a day). Because of the findings, researchers were able to develop a novel phagebiotic composition that might be utilized to treat various illnesses linked to healthcare instead of using antibiotics [101].
Bochkareva et al. described various clinical examples of 42 patients receiving preventive long-term lung ventilation in a Russian neurological ICU. Two phages of each species of bacterium were utilized in a cocktail of phages against A. baumannii, K. pneumoniae, P. aeruginosa, and S. aureus. 54–62.5% of the contaminated loci in the patients receiving phage treatment were resolved. Crucially, phages were administered orally before anti-phage IgGs were produced, and phage treatments after that did not eliminate bacteria. The scientists concluded that the immune response may prevent the same phage from working after many treatments. Evaluating PT as a therapy against E. Coli and P. aeruginosa wound infections in burnt patients was the goal of the PhagoBurn (NCT02116010) clinical study, which included 27 patients. Although the bacterial burden of P. aeruginosa in the majority of infected wounds was successfully reduced at the end of physical therapy (PT), the clinical trial was canceled before it could be completed because the median time to achieve this endpoint was significantly longer for those in the PT group (143 h) than for those in the standard care group (47 h). The titer of the phage cocktail dropped after manufacture, according to the scientists, and the patients got lower-than-expected quantities (from 106 to 102 PFU/mL) [99, 102, 103].
NCT04803708, a clinical study with 26 individuals, is now accepting new volunteers. Its objective is to assess the safety of PT (TP-102) in treating DFUs against various infections, such as P. aeruginosa, A. baumannii, and S. aureus. Topically applied phage containing 109 PFU/mL of phages will be used. Examining the safety, effectiveness, and tolerability of PT (BACTELIDE) in conjunction with routine therapy for pressure ulcers colonized by P. aeruginosa, K. pneumoniae, or S. aureus is the goal of another study (NCT04815798) with 69 patients. A biodegradable polymer encasing 14 phages makes up the cocktail known as BACTELIDE. The safety and effectiveness of the phage cocktail-SPK in conjunction with the standard of care (xeroform and kenacomb topical antibiotic cream) for the treatment and prevention of burns susceptible to infection by P. aeruginosa, S. aureus, and K. pneumoniae spp. are the main focus of the clinical trial NCT04323475 (n = 12 participants). With an effective dose of 2.5 × 105 PFU/cm2 of burnt area, PT uses a dosage-metered airless spray that contains a cocktail of 14 phages at a concentration of 1.4 × 108 PFU/mL. The COVID-19 pandemic has led to the development of a clinical study (NCT04636554) aimed at providing tailored PT for COVID-19 patients who have bacterial co-infections resulting in pneumonia or bacteremia/septicemia. This clinical trial’s objectives are to assess the safety of phage administration in conjunction with traditional antimicrobial treatments and the viability of creating a customized phage treatment for COVID-19 patients who experience pneumonia, bacteremia, or septicemia brought on by P. aeruginosa, Acinetobacter, or S. aureus [99].
Phage screening using clinically isolated strains is also necessary for PT to determine which phage is best for therapy. Because phages have a high selectivity, bacterial variability within a population may potentially contribute to PT’s failure. As a result, phages with a wide spectrum of activity should be chosen for therapy. In this work, researchers describe a case of a customized phage cocktail-treated polyclonal heterogenous bacterial UTI. A phage cocktail combined with a mix of antibiotics and PCN ultimately healed the patient of a long-term MDRKP UTI after four rounds of phage screening against clinical isolates. In this instance, the bladder and renal pelvis were colonized by heterogeneous K. pneumoniae before PT. Nevertheless, researchers could only isolate the K. pneumoniae strain from the urine for their first PT, and researchers ran phage screening against these isolates. During PT, the urine culture of the patient still contained both phage-sensitive and phage-resistant bacteria. Later, it was shown that K. pneumoniae colonized in the renal pelvis was not responsive to the therapeutic phage ФJD902 and that isolates from the bladder and the renal pelvis were distinct clones. Phage-resistant bacteria from the patient’s urine were quickly identified, even after the phage cocktail therapy. These resistant strains may arise from mutations or small, unsuspected resistant populations. In this instance, pathogenic bacteria had also colonized the patient’s renal pelvis, making it challenging for phages to get there by bladder irrigation. Researchers hypothesize that these are the two primary reasons treatments don’t work. As the patient in this instance had a UTI, researchers decided to address the patient’s bladder effusion as our first course of treatment because it was less intrusive and more accessible to administer. However, because of bacterial heterogeneity and renal pelvis colonization, the K. pneumoniae infection did not go away. To accomplish simultaneous perfusion of the bladder and renal pelvis, investigators used phage cocktails that are active against various pathogens. For the patient, researchers created a customized strategy that included the preparation and delivery of a phage cocktail. The outcome of this instance offers insightful suggestions and fixes for customized PT for complicated illnesses [104].
One potentially fatal side effect of complete knee replacement is prosthetic joint infection (PJI). Targeting microbial biofilms and managing diseases that include microorganisms resistant to antibiotics are two potential uses for PT. There is little experience with PT for infections brought on by hardware that has been kept. A 62-year-old man with diabetes who had undergone a right total knee arthroplasty 11 years earlier was offered limb amputation due to a persistent right prosthetic knee infection caused by K. pneumoniae complex. Despite multiple surgeries and prolonged courses of antibiotics, the man experienced episodes of prosthetic knee infection, which worsened over time, and developed severe allergies to antibiotics. PT administered intravenously was started as a limb-salvaging treatment. In addition to getting continuous minocycline (which he had been taking when he suffered growing pain, edema, and erythema before PT commencement), the patient got 40 intravenous doses of a single phage (KpJH46Φ2) targeting his bacterial isolate. Before, during, and after therapy, serial cytokine and biomarker tests were carried out. Safranin staining was used to assess the in vitro anti-biofilm efficacy of KpJH46Φ2, minocycline, and their combination against a constructed biofilm of the patient’s isolate. With physical therapy, local discomfort and infection indicators subsided, and function returned. 34 weeks after finishing therapy and continuing to take minocycline, the patient did not have any side effects from the medication. 22 h after exposure to KpJH46Φ2, a trend in the decrease of biofilm biomass was observed. The use of phage led to a good result in this instance of an uncontrollably severe biofilm-related prosthetic knee infection. PT has potential in treating device-associated infections, pending more research to determine its safety and effectiveness [105].
Researchers describe a 63-year-old female patient who had recurrent UTIs caused by extensively drug-resistant K. pneumoniae (ERKp), a bacterium that is highly resistant to drugs. Within days, phage-resistant mutants emerged in the first two rounds of PT. Even while ERKp strains were resistant to sulfamethoxazole-trimethoprim, the patient’s UTI was effectively treated when sulfamethoxazole-trimethoprim and phage cocktail were combined. This combination suppressed the development of phage-resistant mutants in vitro. Therefore, non-active antibiotic and bacteriophage synergism (NABS) is suggested by researchers as a potential substitute tactic for customized physical therapy. From the patient’s urine, researchers identified an ERKP strain (CX7224). This isolate’s antibiotic susceptibility pattern was the same as that of the patient’s earlier isolates, and its sequence type (ST) was identified as ST11. From our collection of more than 200 lytic phages, researchers screened prospective phages against K. pneumoniae using a phage spot test utilizing the double-layer agar technique. Cocktail I for the first round of PT consisted of five lytic phages: SZ-1, SZ-2, SZ-3, SZ-6, and SZ-8. These phages exhibit effective lytic activity against the CX7224 strain. Researchers implemented a 5-day PT and antibiotic withdrawal protocol from May 9–13, 2019. Every day, researchers used bladder irrigation to deliver 50 milliliters (mL) of cocktail I containing evenly mixed phages (5 × 108 PFU/mL of each phage) [106] (Table 2).
Table 2.
The effectiveness of antibiotics with bacteriophage against K. pneumoniae
| Antibiotics | Bacteriophage | The effects of combined therapy | Ref |
|---|---|---|---|
| Ceftazidime/avibactam was administered at the recommended dosage (2 g/0.5 g, q8h) | Kp040741 and Kp040762 | Objective improvements are seen in the patient’s general state and wounds on a clinical, microbiological, and radiological level as a consequence of this salvage treatment. In vitro, in 7-day mature biofilms and suspensions, the bacteriophage and antibiotic combination is highly efficient against the patient’s strain of K. pneumoniae. | [93] |
| Sulfamethoxazole-trimethoprim | Phage cocktail (lytic phages, namely, SZ-1, SZ-2, SZ-3, SZ-6, and SZ-8) (5 × 108 plaque forming units per mL of each phage) | The patient’s UTI was successfully treated with sulfamethoxazole-trimethoprim and phage cocktail, although ERKp strains were resistant to this combination. This combination also prevented the emergence of phage-resistant mutants in vitro. Therefore, researchers suggested a different approach to tailored phage treatment might be non-active antibiotic and bacteriophage synergism (NABS). | [106] |
| Gentamicin | Phage vB_KpnM_P-KP2 | Mice were given P-KP2 after concentration (1.0 × 107 PFU/mouse, 1.0 × 108 PFU/mouse, or 1.0 × 109 PFU/mouse) or P-KP2 (1.0 × 109 PFU/mouse) in combination with gentamicin (gentamicin was given intranasally 30 min after P-KP2 administration) intraperitoneally at 1-hour post-infection. When treating fatal pneumonia brought on by K. pneumoniae W-KP2 (K47 serotype), P-KP2 was similar to gentamicin. Moreover, the mice with the infection were fully recovered by the combination therapy of gentamicin and P-KP2. To battle MDR infections, researchers’ work not only adds a new entry to the phage therapeutic library but also acts as a reference for future phage-antibiotic combinations. | [111] |
| Ciprofloxacin | KPO1K2 (Podoviridae, T7-like virus) | The biofilm was successfully removed by bacteriophage, and there was no discernible change in its capacity to remove biofilm when used in conjunction with ciprofloxacin. Nevertheless, the growth of resistant genotypes was dramatically halted by combined therapy using bacteriophage and ciprofloxacin. The few variations that did arise generated less cell-associated capsular polysaccharide, were more susceptible to mice peritoneal macrophages, and had a decreased potential to form biofilms. The outer membrane proteins of the bacterial isolates were also found to have altered shape and pattern. When compared to separate treatments, the combined therapy considerably inhibited the production of resistant variations and killed the bacteria. | [95] |
| Mitomycin C and imipenem | Lytic phage vB_KpnM-VAC13 | In vitro and in vivo, the lytic phage vB_KpnM-VAC13 and mitomycin C exhibited synergistic effects on imipenem-resistant and persister isolates. The persisters were eradicated by the phage-imipenem combination, but the imipenem-resistant isolate that contained OXA-245 β-lactamase was not affected. Intriguingly, the combinations reduced the emergence of in vitro-resistant mutants of both isolates. Mitomycin C and imipenem were efficacious against the persister K. pneumoniae isolate when combined with the lytic phage vB_KpnM-VAC13. The imipenem-resistant K. pneumoniae strains that contain OXA-245 β-lactamase were also susceptible to the lytic phage-mitomycin C combination. | [112] |
| Amoxicillin | K. pneumoniae B5055 specific phages | In another study, a log reduction of 2.97 ± 0.11 and 3.51 ± 0.19 was observed when 8-day-old biofilm was exposed to a higher antibiotic concentration (512 µg/ml) or phage alone (Multiplicity of Infections (MoIs) = 0.01). The biofilm’s bacterial burden was significantly reduced when exposed to a combination of amoxicillin and phage. Therefore, the fact that the biofilm structure was more effectively destroyed when antibiotics were used with specific phages indicated that the phages could be effectively used in conjunction with antibiotic therapy. One additional benefit of combination therapy is its capacity to prevent the development of resistant mutations, which would otherwise be readily induced by phage or antibiotics alone. | [94] |
Researchers have isolated, read, and assembled the genome sequences of four phages that infect the clinical disease K. pneumoniae: vB_KpnP_FBKp16, vB_KpnP_FBKp27, vB_KpnM_FBKp34, and Jumbo phage vB_KpnM_FBKp24. Their goal is to increase the collection of phage genomes that are now accessible. The identity of the four phages to genomic phage sequences stored in the GenBank database is relatively low (0–13%). Three of the four phages are tRNA-encoding and differ significant from the host regarding GC content. Significantly, separate clades of tubulin spindle and nucleus shell proteins that some phages use to compartmentalize viral replication are revealed by the genome sequences of the phages, along with perhaps new DNA packing processes. Overall, this work offers new options for PT applications against antibiotic-resistant K. pneumoniae infections and sheds light on various previously unidentified viruses [107].
In this instance, the extracellular matrix capsule of K. pneumoniae is very diverse, displaying several variations. Here, researchers describe the isolation of four new phages that infect K. pneumoniae capsular type K22 from environmental samples in Valencia, Spain. These phages are members of the Podoviridae family, as shown by genome sequencing, and they encode putative depolymerases that break down certain K22 K. pneumoniae capsules. Based on their restricted infectivity in a panel of clinical isolates of K. pneumoniae, the researchers’ findings support the capsular type-specificity of the phages. However, this study is a step toward characterizing various phages, and it might lead to the use of vast panels of phages for typing and/or fighting drug-resistant K. pneumoniae in the future [108].
Researchers have published the whole genomic sequences, insertion sites, and phylogenetic analysis of 150 parasites discovered in 40 clinical isolates of K. pneumoniae that were recovered during an epidemic in a Portuguese hospital. Every strain carried a minimum of one prophage, and 104 (69.3%) strains were found to be intact. The prophage codes for between 32 and 78 putative genes, with sizes ranging from 29.7 to 50.6 kbp. Compared to the typical GC level of 57.1% in K. pneumoniae, the prophage GC content is 51.2%. In the order Caudolovirales, complete prophages were divided into three families: Podoviridae (1.9%), Siphoviridae (38.5%), and Myoviridae (59.6%). Nine different groups were also found via phylogenetic analysis and alignment. Some prophages showed evidence of recombination in their genomes. Still, most of the time, the proteins essential for the structure, transcription, replication, and control (lysogenic/lysis) of the virus were preserved. These findings confirm that prophages are varied and extensively distributed across K. pneumoniae genomes, aiding the species’ development and bestowing extra characteristics. Additionally, K. pneumoniae was discovered to be present in a group of endolysin genes that coded for lysozyme-active proteins that cleaved the β-1,4 linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in the peptidoglycan network, indicating genes that may be involved in lysin PT [109].
Surgeons have a significant problem when dealing with nonhealing ulcers since they might sometimes result in the amputation of the afflicted region. The main cause of wounds that do not heal is infection by bacteria resistant to antibiotics, which leads to the production of biofilms. Personalized physical therapy, however, could overcome both of the obstacles above. Researchers designed a study that included 48 participants, ages 12 to 70, who had at least one full-thickness wound that did not heal after six weeks of standard care. Individuals suffering from systemic illnesses, such as burns, cancer, skin conditions, and ulcers caused by Tuberculosis or leprosy, were not included in the study. Nonetheless, the research included participants with hypertension and diabetes. On different days, the wound surface was covered with a cocktail of phages specific to two or more invading bacteria and a personalized monophage for a single bacterial infection. Until all harmful germs were removed from the wound, five to seven treatments were done. August 2018 to May 2019 was the extended research period. After starting treatment, the research participants were monitored for three months by researchers. It was possible to achieve an 81.2% cure rate, of which 90.5% (19/21) were non-diabetic, and 74.1% (20/27) were. Researchers were seen that the K. pneumoniae-infected wounds healed somewhat more slowly. Also, investigators demonstrated that the proportion of lymphocytes and the mean hemoglobin level rose sharply after PT. For ulcers that don’t heal, tailored local physical therapy is very promising. [110].
Future perspective
The number of studies showing how to use phages to treat MDR bacterial infections has continuously risen since 2018. The worldwide phage market, which is anticipated to reach a value of USD 1,441.3 million by 2028, is expected to equal this. The testing of various phages to treat drug-resistant infections has advanced significantly due to the growing urgency of MDR bacterial infections and the decrease in the discovery of new antibiotics, making this a crucial advancement in the treatment of complex cases of drug-resistant bacterial infections. A rising number of papers also discuss giving phages and antibiotics at the same time to individuals who have severe MDR bacteria [113].
For instance, a lytic phage has been isolated in a few recent PT reports against the widely drug-resistant A. baumannii. This common opportunistic nosocomial pathogen causes MDR infections, such as blood, urinary tract, pneumonia, and wound infections in other body parts. A second paper has shown how phage-antibiotic synergy may be used in a novel way to treat fracture-related infections caused by pan-drug-resistant K. pneumoniae. This involves using antibiotics in conjunction with pre-adapted physical therapy. K. pneumoniae belongs to the Enterobacteriaceae family and is often linked to endogenous infections, including meningitis, pneumonia, bloodstream infections, and urinary tract infections. These infections may also occur outside of clinical settings. During the first five years of the Israeli PT Center, the compassionate use of phages for severe persistent illnesses revealed that the number of registered requests increased yearly, with MDR bacteria making up 38% of all phage requests [113, 114].
Specifications and standards for phage use in industry have been continually improved over the last 20 years, with the establishment of phage research and development centers in China, the US, and the EU. The China Phage Research Alliance has released a standard document on PT for the first time, which further refines the operational processes and technological criteria to close the gap in rules and standards for phage clinical application. Several months ago, the European Medications Agency officially approved the world’s first scientific guideline on phage veterinary medications in applications related to cattle and poultry production. The guideline was titled Quality, safety, and effectiveness of phages as veterinary medicines—Scientific guideline. The guideline establishes certain standards for animal clinical trials and describes phage processing as a new treatment. It also emphasizes the safety and residue testing of phage therapies. PT has proven to be of utmost strategic significance to the cattle and poultry sector [115].
At the University of California, San Diego, phage preparation was used in March 2016 to treat a severe multi-antibiotic-resistant A. baumannii, marking the first documented practical clinical application of intravenous PT in the United States. This instance catalyzed the national resurgence of interest in PT, leading to official public lectures, a book, and other initiatives to raise awareness of the potential benefits of phage treatments. Transcripts from a conference on the “Science and Regulation of PT” were made public by the Food and Drug Administration (FDA) in August 2021. During the meeting, prominent scientists and healthcare professionals engaged in a thorough discussion about the practicalities of phage use in clinical settings. The U.S. FDA has awarded PT an emergency use authorization (EUA) because of the widespread effects of antibiotic-resistant bacterial infections. On a case-by-case basis, this EUA permits the compassionate use of this treatment. Expanded access, sometimes called compassionate use of experimental medicines, refers to the provision of goods that regulatory authorities have not completely authorized to patients recognized by a clinician as having no other therapeutic choices. These instances of compassionate care take place in a monitored clinical setting. However, since they don’t occur in clinical trial settings, case series of specific individuals are used to assess them rather than larger cohorts of targeted groups [116].
Phages have several unique benefits over conventional antibiotic substitutes. First off, because phages interact with host genes to promote co-evolution, carefully crafted phage cocktails may effectively prevent bacterial resistance; secondly, because phages are more precise in their target point of action, they can quickly adsorb and kill the host bacteria without upsetting the normal bacterial balance in the body. Third, because they cannot multiply in eukaryotic cells, they do not directly cause pharmacological effects in animals (unlike cytokines, hormones, and autoantibodies), so there are no mechanism-based concerns regarding toxicity in animals. To prevent bacteria from becoming resistant to a single phage variety, phages are typically applied in the form of " phage cocktails,” which are mixtures of multiple phages or the addition of various types of phages at different times. To attain optimal efficiency, phage cocktails combine various phage invasions and lysis mechanisms directed at the same or related bacterial hosts [115, 117, 118].
It is crucial to emphasize that phage treatment experiments carried out in animal models are not exact replicas of human illnesses [119]. For example, animals are frequently given fatal amounts of bacteria to show phage effectiveness, which causes them to die quickly. The PT is usually started almost away after the animals are challenged with bacteria. Furthermore, there are no mouse strains that exhibit elevated mucus blockages in the lungs and airways, which might significantly affect the ability of phages to manage infections linked to cystic fibrosis illnesses. Furthermore, the physiologies of animals and humans vary in terms of microbiomes, secretions (such as gut pH), and structure. Animal studies are often safe and effective, even if they do not precisely mimic the human state. Furthermore, consistent with mouse data, no adverse effects related to the use of phages have been identified in human clinical studies yet. Phage treatment has gone a long way, and there are plenty of compelling instances of its effectiveness, but before it is widely used in clinical settings, there are still a lot of obstacles to be addressed. It is necessary to establish precise regulations to approve phage-based goods, both naturally occurring and manufactured. This might encourage businesses to increase their meager investments in phage research [120].
Information on patient opinions on phages and public knowledge of them is scarce. This aligns with the idea that phage therapies represent a relatively new area of clinical infection control. 2020 saw the completion of the first study on phage receptivity and public awareness in Scotland. The research assessed patient knowledge of AMR and PT after therapy for DFUs, and it gave important information about patient responsiveness to phage treatments despite having a small sample size and poor statistical power. While most patients understood the idea of antibiotic resistance and voiced worries about its deteriorating effects, many were unaware of PT (less than 10% of participants) or phages (less than 25% of participants). When presented as a possible means of addressing AMR, patients were typically enthusiastic about PT and showed no more worry for clinical phage applications than antibiotic treatment. Interestingly, the survey also identified recurring patterns in patient queries and worries. To effectively reintroduce and progress phages in a therapeutic context, further research must examine patient viewpoints and concerns about this innovative treatment. The issues and experiences that patients have will undoubtedly influence the creation and use of these therapies. Therefore, further research has to be done to assess the opinions and worries of those taking part in clinical trials connected to phages [100, 121].
The issue of patenting phage preparations could be resolved by significant technical advancements. This may help to improve PT’s availability and specificity. Moreover, phages can be altered through genome editing methods (such as sequencing, homologous recombination, CRISPR/Cas-based phage engineering, and phage genomic DNA assembly) to create preparations that can exclusively eradicate bacteria resistant to antibiotics (without compromising the treated patient’s commensal microflora). It is easier to patent and sell such unusual phages or phage mixtures. Phage libraries created by different research teams and PT centers worldwide may satisfy the need for easily accessible phage products. Validated quality control procedures are necessary for the manufacturing of safe phage therapeutics, whether they are produced on a larger scale for individualized treatment or on a smaller one. In any event, the medicinal preparation’s sterility, stability, and lack of endo- and exotoxins or other dangerous contaminants must be considered. Aspects including the lifespan of individual phages or combinations of phages, the antagonistic and synergistic effects of phages and/or antibiotics, pharmacokinetics, pharmacodynamics, pharmacogenomics, and host reactions to phage preparations make this treatment very difficult to use at the moment. Creating a sizable, publicly accessible phage bank and efficient screening methods for target bacterial isolation—that is, germs resistant to antibiotic treatment—is the first and most crucial stage in this strategy. Another is getting rid of the patient’s bacterial infection (via cell lysis), ideally without interfering with the microbiota’s natural balance or producing uncomfortable side effects. Because phages cannot proliferate in the treated organism’s cells and are eliminated by the patient’s kidneys after the antibiotic-resistant pathogen is destroyed, phages are usually regarded as safe for human use. Phage lysates, however, have the potential to include a wide range of toxic substances that pose a threat to human health and survival. These include protein toxins generated by pathogenic bacterial strains and endotoxins (LPS) from Gram-negative bacteria. Therefore, techniques for isolating, enriching, and purifying phage preparations into safe and effective forms that may be used in human and animal treatments must constantly be improved. It should be able to acquire phage products in specialized GMP facilities as a result of technical advancements in fields like as genetic engineering, synthetic biology, throughput sequencing, and metagenomics. Even though it is easy to separate naturally occurring phages from many diseases, there are still a lot of bacterial species for which phages are few or nonexistent. This implies that finding new sources to acquire and isolate phages with a broad host range is required [122, 123].
Conclusion
K. pneumoniae is a significant new pathogen that affects both people and animals and may have detrimental clinical effects. K. pneumoniae strains that produce ESBLs and are resistant to carbapenem have become more prevalent due to increased antibiotic usage. In the future, PT could signify a novel, widely used strategy for battling diseases with resistance. Numerous publications, both in vitro and in vivo, have shown the potential use of lytic phages in the fight against K. pneumoniae superbug stains. To prevent or delay the development of phage resistance and broaden the host range, phage compounds, which consist of multiple phages, are frequently used. By broadening the host bacterial range, reducing the severity of infections, preventing phage elimination by the immune system, and minimizing the development of phage-resistant mutations, phage compounds enhance the long-term efficacy of the drug. Before phages can be employed to treat pneumonia caused by K. pneumoniae, additional research is required on phage cocktails and the evolution of phage resistance [124]. Inhibiting action of the PT is shown against MDRKP. It is discovered to have a partial cidal action, lowering the number of bacteria and the production of biofilms. Therefore, it may be necessary to use a phage cocktail, or a mixture of phage types, to eradicate bacterial cells and biofilms. One possible tool against MDR bacterial infections is PT. A physical or virtual “biobank” of identified phages against prevalent AMR bacteria is essential for PT readiness. Phage characterization is necessary to minimize adverse effects and provide early, efficient therapy. Therefore, for PT to become a widely accessible option for patients who have not responded to antibiotic therapy, logistical and regulatory challenges must be addressed. In addition, more clinical research is also required to supplement clinical data about phage biology, phage preparation dosage, and PT delivery techniques.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank all researchers who contributed to the advancement of science.
Author contributions
O.G., H.E.G., M.T., R.R., R.D., writing–original draft. All authors participated in the manuscript in the critical review process of the manuscript and approved the final version.
Funding
There is no Funding.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Credit authorship contribution statement
O.G., H.E.G., M.T., R.R., R.D., writing–original draft. All authors participated in the manuscript in the critical review process of the manuscript and approved the final version.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu M, Li X. Klebsiella pneumoniae and Pseudomonas aeruginosa, in molecular medical microbiology. Elsevier; 2015. pp. 1547–64.
- 2.Cruz-Córdova A, et al. Pathogenic determinants of clinical Klebsiella pneumoniae strains associated with their persistence in the hospital environment. Boletín médico Del Hosp Infantil De México. 2014;71(1):15–24. [Google Scholar]
- 3.Sugden R, Kelly R, Davies S. Combatting antimicrobial resistance globally. Nat Microbiol. 2016;1(10):1–2. 10.1038/nmicrobiol.2016.187 [DOI] [PubMed] [Google Scholar]
- 4.Petrosillo N, Taglietti F, Granata G. Treatment options for colistin resistant Klebsiella pneumoniae: present and future. J Clin Med. 2019;8(7):934. 10.3390/jcm8070934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassetti M, et al. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Expert Rev anti-infective Therapy. 2018;16(10):749–61. 10.1080/14787210.2018.1522249 [DOI] [PubMed] [Google Scholar]
- 6.Okeke IN, et al. The scope of the antimicrobial resistance challenge. Lancet. 2024;403(10442):2426–38. 10.1016/S0140-6736(24)00876-6 [DOI] [PubMed] [Google Scholar]
- 7.Hatfull GF, Dedrick RM, Schooley RT. Phage therapy for antibiotic-resistant bacterial infections. Annu Rev Med. 2022;73:197–211. 10.1146/annurev-med-080219-122208 [DOI] [PubMed] [Google Scholar]
- 8.Petrovic Fabijan A, et al. Translating phage therapy into the clinic: recent accomplishments but continuing challenges. PLoS Biol. 2023;21(5):e3002119. 10.1371/journal.pbio.3002119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu A, et al. Assessment of the microbiome during bacteriophage therapy in combination with systemic antibiotics to treat a case of staphylococcal device infection. Microbiome. 2021;9:1–8. 10.1186/s40168-021-01026-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu J, et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE. 2012;7(3):e31698. 10.1371/journal.pone.0031698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina F, et al. Systematic analysis of putative phage-phage interactions on minimum-sized phage cocktails. Sci Rep. 2022;12(1):2458. 10.1038/s41598-022-06422-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malik DJ, et al. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv Colloid Interface Sci. 2017;249:100–33. 10.1016/j.cis.2017.05.014 [DOI] [PubMed] [Google Scholar]
- 13.Zurabov F, Zhilenkov E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol J. 2021;18:1–20. 10.1186/s12985-020-01485-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dąbrowska K, Abedon ST. Pharmacologically aware phage therapy: pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol Mol Biol Rev. 2019;83(4):00012–19. 10.1128/mmbr. 10.1128/mmbr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang B, et al. Effective of phage cocktail against Klebsiella pneumoniae infection of murine mammary glands. Microb Pathog. 2023;182:106218. 10.1016/j.micpath.2023.106218 [DOI] [PubMed] [Google Scholar]
- 16.Martins WM, et al. Effective phage cocktail to combat the rising incidence of extensively drug-resistant Klebsiella pneumoniae sequence type 16. Volume 11. Emerging Microbes & Infections; 2022. pp. 1015–23. 1. [DOI] [PMC free article] [PubMed]
- 17.Yoo S et al. Designing phage cocktails to combat the emergence of bacteriophage-resistant mutants in multidrug-resistant Klebsiella pneumoniae. Microbiol Spectr, 2023: p. e01258–23. [DOI] [PMC free article] [PubMed]
- 18.Jokar J et al. Antibacterial effects of single phage and phage cocktail against multidrug-resistant Klebsiella pneumoniae isolated from diabetic foot ulcer. Virus Genes, 2023: p. 1–8. [DOI] [PubMed]
- 19.Łusiak-Szelachowska M, Weber-Dąbrowska B, Żaczek M. The Presence of Bacteriophages in the Human Body: Good, Bad or Neutral? 2020. 8(12). [DOI] [PMC free article] [PubMed]
- 20.Salih Doğan H et al. Two novel phages, Klebsiella phage GADU21 and Escherichia phage GADU22, from the urine samples of patients with urinary tract infection Virus Genes, 2024: pp. 1–14. [DOI] [PubMed]
- 21.Effah CY, et al. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19:1–9. 10.1186/s12941-019-0343-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzouvelekis L, et al. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magill SS, et al. Changes in prevalence of health care–associated infections in US hospitals. N Engl J Med. 2018;379(18):1732–44. 10.1056/NEJMoa1801550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abbas R, et al. General Overview of Klebsiella pneumonia: epidemiology and the role of Siderophores in its pathogenicity. Biology. 2024;13(2):78. 10.3390/biology13020078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang L, et al. Single-dose of hybrid eukaryotic-prokaryotic nanovesicles to drive a rapid and controllable immune response against Klebsiella pneumoniae induced nosocomial pneumonia. Nano Today. 2024;54:102135. 10.1016/j.nantod.2023.102135 [DOI] [Google Scholar]
- 26.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–61. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karampatakis T, Tsergouli K, Behzadi P. Carbapenem-resistant Klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics. 2023;12(2):234. 10.3390/antibiotics12020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, et al. The characteristic of virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17(17):6278. 10.3390/ijerph17176278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teo T-H et al. Differential mucosal tropism and dissemination of classical and hypervirulent Klebsiella pneumoniae infection. iScience, 2024. [DOI] [PMC free article] [PubMed]
- 30.Carattoli A, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–28. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 31.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–75. 10.1093/femsre/fux013 [DOI] [PubMed] [Google Scholar]
- 32.Martin MJ et al. A panel of diverse Klebsiella pneumoniae clinical isolates for research and development. Microb Genomics, 2023. 9(5). [DOI] [PMC free article] [PubMed]
- 33.Li B, et al. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–81. 10.2217/fmb.14.48 [DOI] [PubMed] [Google Scholar]
- 34.Wyres KL, Lam MM, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–59. 10.1038/s41579-019-0315-1 [DOI] [PubMed] [Google Scholar]
- 35.Pu Q, et al. Know your enemy: Klebsiella pneumoniae, in Molecular Medical Microbiology. Elsevier; 2024. pp. 1491–501.
- 36.Yao H, et al. Conjugative plasmids facilitate the transmission of tmexCD2-toprJ2 among carbapenem-resistant Klebsiella pneumoniae. Sci Total Environ. 2024;906:167373. 10.1016/j.scitotenv.2023.167373 [DOI] [PubMed] [Google Scholar]
- 37.Albarri O, et al. Antimicrobial Resistance of Clinical Klebsiella pneumoniae isolates: involvement of AcrAB and OqxAB Efflux Pumps. Curr Mol Pharmacol. 2024;17(1):e310323215266. [DOI] [PubMed] [Google Scholar]
- 38.Gorodnichev R et al. ISOLATION AND CHARACTERIZATION OF VIRULENT BACTERIOPHAGES AGAINST KLEBSIELLA PNEUMONIAE OF SIGNIFICANT CAPSULAR TYPES
- 39.Behera B et al. Genotypic characterization of hypervirulent Klebsiella pneumoniae (hvKp) in a tertiary care Indian hospital. Int Microbiol, 2024: p. 1–10. [DOI] [PubMed]
- 40.Fletcher J, et al. The Citizen Phage Library: Rapid isolation of phages for the Treatment of Antibiotic Resistant Infections in the UK. Microorganisms. 2024;12(2):253. 10.3390/microorganisms12020253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verbeken G, Pirnay J-P. European regulatory aspects of phage therapy: magistral phage preparations. Curr Opin Virol. 2022;52:24–9. 10.1016/j.coviro.2021.11.005 [DOI] [PubMed] [Google Scholar]
- 42.Abedon ST, Danis-Wlodarczyk KM, Wozniak DJ. Phage cocktail development for bacteriophage therapy: toward improving spectrum of activity breadth and depth. Pharmaceuticals. 2021;14(10):1019. 10.3390/ph14101019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogungbe BA, et al. Progress of phage therapy research as an alternative to antibiotics: current status, challenges, and the future of phage therapeutics. J Med Surg Public Health. 2024;2:100042. 10.1016/j.glmedi.2023.100042 [DOI] [Google Scholar]
- 44.Rose T, et al. Experimental phage therapy of burn wound infection: difficult first steps. Int J Burns Trauma. 2014;4(2):66. [PMC free article] [PubMed] [Google Scholar]
- 45.Viertel TM, Ritter K, Horz H-P. Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother. 2014;69(9):2326–36. 10.1093/jac/dku173 [DOI] [PubMed] [Google Scholar]
- 46.Elahi Y, Nowroozi J, Fard RMN. Isolation and characterization of bacteriophages from wastewater sources on Enterococcus spp. isolated from clinical samples. Iran J Microbiol. 2021;13(5):671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hitchcock NM. and D. Devequi Gomes Nunes. Curr Clin Landsc Global Potential Bacteriophage Therapy 2023. 15(4). [DOI] [PMC free article] [PubMed]
- 48.Hyman P. Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals (Basel), 2019. 12(1). [DOI] [PMC free article] [PubMed]
- 49.Van Twest R, Kropinski AM. Bacteriophage enrichment from water and soil Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions, 2009: pp. 15–21. [DOI] [PubMed]
- 50.Abedon ST et al. Phage therapy: past, present and future. 2017, Front Media SA. p. 981. [DOI] [PMC free article] [PubMed]
- 51.Mattila S, Ruotsalainen P, Jalasvuori M. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol. 2015;6:1271. 10.3389/fmicb.2015.01271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, et al. SLPW: a virulent bacteriophage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo. Front Microbiol. 2016;7:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhetwal A et al. Isolation of potential phages against multidrug-resistant bacterial isolates: promising agents in the rivers of Kathmandu, Nepal BioMed Research International, 2017. 2017. [DOI] [PMC free article] [PubMed]
- 54.Anderson B, et al. Enumeration of bacteriophage particles: comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays. Bacteriophage. 2011;1(2):86–93. 10.4161/bact.1.2.15456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirza KA, et al. Bacteriophage-mediated decolonization of Klebsiella pneumoniae in a novel Galleria mellonella gut colonization model with Enterobacteriaceae. Sci Rep. 2024;14(1):318. 10.1038/s41598-023-50823-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manohar P, et al. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol. 2019;10:574. 10.3389/fmicb.2019.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu R, et al. Bacteriophage long tail fibre proteins as a biorecognition element in electrochemical biosensors for the detection of Salmonella in lake water. Sens Actuators B. 2024;403:135148. 10.1016/j.snb.2023.135148 [DOI] [Google Scholar]
- 58.Yan M, et al. Interrogating the viral dark matter of the rumen ecosystem with a global virome database. Nat Commun. 2023;14(1):5254. 10.1038/s41467-023-41075-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kareem ZG, Al-Zamily, Al-Khafaji NS. Purification and characterization of α-galactosidase isolated from Klebsiella pneumoniae in the human oral cavity. Int J Biol Macromol, 2024: p. 129550. [DOI] [PubMed]
- 60.Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol. 2009;59:274–81. 10.1007/s00284-009-9430-y [DOI] [PubMed] [Google Scholar]
- 61.Ciacci N, et al. Characterization of vB_Kpn_F48, a newly discovered lytic bacteriophage for Klebsiella pneumoniae of sequence type 101. Viruses. 2018;10(9):482. 10.3390/v10090482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morozova V, et al. Isolation and characterization of a novel Klebsiella pneumoniae N4-like bacteriophage KP8. Viruses. 2019;11(12):1115. 10.3390/v11121115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drulis-Kawa Z, et al. Isolation and characterisation of KP34—a novel φKMV-like bacteriophage for Klebsiella pneumoniae. Appl Microbiol Biotechnol. 2011;90:1333–45. 10.1007/s00253-011-3149-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sasani MS, Eftekhar F, Hosseini SM. Isolation and characterization of Klebsiella pneumoniae specific lytic bacteriophage from. J Med Microbiol. 2019;7(1–2):6–11. [Google Scholar]
- 65.Karumidze N, et al. Isolation and characterisation of lytic bacteriophages of Klebsiella pneumoniae and Klebsiella oxytoca. Curr Microbiol. 2013;66:251–8. 10.1007/s00284-012-0264-7 [DOI] [PubMed] [Google Scholar]
- 66.Lin T-L, et al. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J Infect Dis. 2014;210(11):1734–44. 10.1093/infdis/jiu332 [DOI] [PubMed] [Google Scholar]
- 67.Duarte J et al. Potential of a New Bacteriophage to Control Klebsiella pneumoniae: Preliminary Studies to Control Urinary Tract Infections 2024. [DOI] [PMC free article] [PubMed]
- 68.Shree S, et al. Effect of Klebsiella-specific phage on multidrug-resistant Klebsiella pneumoniae-an experimental study. Ind J Med Microbiol. 2024;47:100515. 10.1016/j.ijmmb.2023.100515 [DOI] [PubMed] [Google Scholar]
- 69.Ferriol-Gonzalez C et al. Targeted phage hunting to specific Klebsiella pneumoniae clinical isolates is an efficient antibiotic resistance and infection control strategy bioRxiv, 2024: p. 2024.01. 07.574526.
- 70.Feng Y et al. Safety and efficacy of a phage cocktail on murine wound infections caused by carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents, 2024: p. 107088. [DOI] [PubMed]
- 71.Chen C, et al. Isolation and characterization of novel bacteriophage vB_KpP_HS106 for Klebsiella pneumonia K2 and applications in foods. Front Microbiol. 2023;14:1227147. 10.3389/fmicb.2023.1227147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao F et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice BioMed Research International, 2015. 2015. [DOI] [PMC free article] [PubMed]
- 73.Chhibber S, Kaur S, Kumari S. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. J Med Microbiol. 2008;57(12):1508–13. 10.1099/jmm.0.2008/002873-0 [DOI] [PubMed] [Google Scholar]
- 74.Hsu C-R, et al. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS ONE. 2013;8(8):e70092. 10.1371/journal.pone.0070092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hesse S et al. Bacteriophage treatment rescues mice infected with multidrug-resistant Klebsiella pneumoniae ST258 MBio, 2021. 12(1): p. 10.1128/mbio. 00034 – 21. [DOI] [PMC free article] [PubMed]
- 76.Zurabov F, Zhilenkov E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol J. 2021;18(1):1–20. 10.1186/s12985-020-01485-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kęsik-Szeloch A, et al. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol J. 2013;10:1–12. 10.1186/1743-422X-10-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taha OA, et al. Bacteriophage ZCKP1: a potential treatment for Klebsiella pneumoniae isolated from diabetic foot patients. Front Microbiol. 2018;9:2127. 10.3389/fmicb.2018.02127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anand T, et al. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J Global Antimicrob Resist. 2020;21:34–41. 10.1016/j.jgar.2019.09.018 [DOI] [PubMed] [Google Scholar]
- 80.Li J, et al. Development of phage resistance in multidrug-resistant Klebsiella pneumoniae is associated with reduced virulence: a case report of a personalised phage therapy. Clin Microbiol Infect. 2023;29(12):e16011–7. 10.1016/j.cmi.2023.08.022 [DOI] [PubMed] [Google Scholar]
- 81.Singh D et al. Characterization and complete genome analysis of Klebsiella phage Kp109 with lytic activity against Klebsiella pneumoniae. Virus Genes, 2024: p. 1–13. [DOI] [PubMed]
- 82.Hung C-H, et al. Experimental phage therapy in treating Klebsiella pneumoniae-mediated liver abscesses and bacteremia in mice. Antimicrob Agents Chemother. 2011;55(4):1358–65. 10.1128/AAC.01123-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lourenço M et al. Phages against Noncapsulated Klebsiella pneumoniae: Broader Host range, Slower Resistance Microbiology Spectrum, 2023: pp. e04812-22. [DOI] [PMC free article] [PubMed]
- 84.Tang M, et al. Phage resistance formation and fitness costs of hypervirulent Klebsiella pneumoniae mediated by K2 capsule-specific phage and the corresponding mechanisms. Front Microbiol. 2023;14:1156292. 10.3389/fmicb.2023.1156292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duarte J, et al. Potential of an isolated bacteriophage to inactivate Klebsiella pneumoniae: preliminary studies to control urinary tract infections. Antibiotics. 2024;13(2):195. 10.3390/antibiotics13020195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bai R, Guo J. Interactions and implications of Klebsiella pneumoniae with Human Immune responses and metabolic pathways: a Comprehensive Review. Infect Drug Resist. 2024;17(null):449–62. 10.2147/IDR.S451013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohammadi M, et al. Isolation, characterization, therapeutic potency, and genomic analysis of a novel bacteriophage vB_KshKPC-M against carbapenemase-producing Klebsiella pneumoniae strains (CRKP) isolated from Ventilator-associated pneumoniae (VAP) infection of COVID-19 patients. Ann Clin Microbiol Antimicrob. 2023;22(1):18. 10.1186/s12941-023-00567-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei H et al. Phage: Tackling Drug-Resistant Klebsiella Pneumoniae Available at SSRN 4713374.
- 89.Bird Jordan T, et al. Genome sequence of the Klebsiella quasipneumoniae bacteriophage EKq1 with activity against Klebsiella pneumoniae. Microbiol Resource Announcements. 2023;13(1):e00954–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mulani Mansura S, Shital NK, Pardesi Karishma R. Characterization of Novel Klebsiella Phage PG14 and its Antibiofilm Efficacy. Microbiol Spectr. 2022;10(6):e01994–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zaki BM, et al. Characterization and comprehensive genome analysis of novel bacteriophage, vB_Kpn_ZCKp20p, with lytic and anti-biofilm potential against clinical multidrug-resistant Klebsiella pneumoniae. Front Cell Infect Microbiol. 2023;13:1077995. 10.3389/fcimb.2023.1077995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zurabov F, et al. Bacteriophages with depolymerase activity in the control of antibiotic resistant Klebsiella pneumoniae biofilms. Sci Rep. 2023;13(1):15188. 10.1038/s41598-023-42505-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eskenazi A, et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat Commun. 2022;13(1):302. 10.1038/s41467-021-27656-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bedi MS, Verma V, Chhibber S. Amoxicillin and specific bacteriophage can be used together for eradication of biofilm of Klebsiella pneumoniae B5055. World J Microbiol Biotechnol. 2009;25:1145–51. 10.1007/s11274-009-9991-8 [DOI] [Google Scholar]
- 95.Verma V, Harjai K, Chhibber S. Restricting ciprofloxacin-induced resistant variant formation in biofilm of Klebsiella pneumoniae B5055 by complementary bacteriophage treatment. J Antimicrob Chemother. 2009;64(6):1212–8. 10.1093/jac/dkp360 [DOI] [PubMed] [Google Scholar]
- 96.Verma V, Harjai K, Chhibber S. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling. 2010;26(6):729–37. 10.1080/08927014.2010.511196 [DOI] [PubMed] [Google Scholar]
- 97.Qurat-ul-Ain H et al. Efficacy of phage-antibiotic combinations against multidrug-resistant Klebsiella pneumoniae clinical isolates. Jundishapur J Microbiol, 2021. 14(1).
- 98.Sulakvelidze A, Alavidze Z, Morris JG Jr. Bacteriophage therapy. Antimicrob Agents Chemother. 2001;45(3):649–59. 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Broncano-Lavado A, et al. Advances in bacteriophage therapy against relevant multidrug-resistant pathogens. Antibiotics. 2021;10(6):672. 10.3390/antibiotics10060672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hitchcock NM, et al. Current Clinical Landscape and global potential of bacteriophage therapy. Viruses. 2023;15(4):1020. 10.3390/v15041020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aleshkin A, et al. Phagebiotics in treatment and prophylaxis of healthcare-associated infections. Bacteriophage. 2016;6(4):40–6. 10.1080/21597081.2016.1251379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bochkareva S, et al. Anti-phage аntibody response in phage therapy against healthcare-associated infections (HAIs). Infekc Bolezni. 2017;15:35–40. 10.20953/1729-9225-2017-1-35-40 [DOI] [Google Scholar]
- 103.Jault P, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis. 2019;19(1):35–45. 10.1016/S1473-3099(18)30482-1 [DOI] [PubMed] [Google Scholar]
- 104.Qin J, et al. Heterogeneous Klebsiella pneumoniae co-infections complicate personalized bacteriophage therapy. Front Cell Infect Microbiol. 2021;10:608402. 10.3389/fcimb.2020.608402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cano EJ, et al. Phage therapy for limb-threatening prosthetic knee Klebsiella pneumoniae infection: case report and in vitro characterization of anti-biofilm activity. Clin Infect Dis. 2021;73(1):e144–51. 10.1093/cid/ciaa705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bao J, et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Volume 9. Emerging microbes & infections; 2020. pp. 771–4. 1. [DOI] [PMC free article] [PubMed]
- 107.Estrada Bonilla B, et al. Genomic characterization of four novel bacteriophages infecting the clinical pathogen Klebsiella pneumoniae. DNA Res. 2021;28(4):dsab013. 10.1093/dnares/dsab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Domingo-Calap P, et al. Isolation of four lytic phages infecting Klebsiella pneumoniae K22 clinical isolates from Spain. Int J Mol Sci. 2020;21(2):425. 10.3390/ijms21020425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marques AT, et al. Genomic analysis of prophages from Klebsiella pneumoniae clinical isolates. Microorganisms. 2021;9(11):2252. 10.3390/microorganisms9112252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Patel DR, et al. Use of customized bacteriophages in the treatment of chronic nonhealing wounds: a prospective study. Int J Low Extrem Wounds. 2021;20(1):37–46. 10.1177/1534734619881076 [DOI] [PubMed] [Google Scholar]
- 111.Wang Z, et al. Combination therapy of phage vB_KpnM_P-KP2 and gentamicin combats acute pneumonia caused by K47 serotype Klebsiella pneumoniae. Front Microbiol. 2021;12:674068. 10.3389/fmicb.2021.674068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pacios O, et al. Enhanced antibacterial activity of repurposed mitomycin C and imipenem in combination with the lytic phage vB_KpnM-VAC13 against clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2021;65(9). 10.1128/aac. 00900 – 21. [DOI] [PMC free article] [PubMed]
- 113.Mboowa G. Reviewing the journey to the clinical application of bacteriophages to treat multi-drug-resistant bacteria. BMC Infect Dis. 2023;23(1):654. 10.1186/s12879-023-08621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Onallah H, Hazan R, Nir-Paz R. Compassionate use of bacteriophages for failed persistent infections during the first 5 years of the Israeli phage Therapy Center. Open Forum Infectious diseases. Oxford University Press US; 2023. [DOI] [PMC free article] [PubMed]
- 115.Jiang A, et al. Prospects and challenges of Bacteriophage Substitution for antibiotics in Livestock and Poultry Production. Biology. 2024;13(1):28. 10.3390/biology13010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McCallin S, Brüssow H. Clinical trials of bacteriophage therapeutics. Bacteriophages: Biology, Technology, Therapy,; 2021. pp. 1099–127. [Google Scholar]
- 117.Hampton HG, Watson BN, Fineran PC. The arms race between bacteria and their phage foes. Nature. 2020;577(7790):327–36. 10.1038/s41586-019-1894-8 [DOI] [PubMed] [Google Scholar]
- 118.Federici S, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 2022;185(16):2879–98. e24. 10.1016/j.cell.2022.07.003 [DOI] [PubMed] [Google Scholar]
- 119.Penziner S, Schooley RT, Pride DT. Animal models of phage therapy. Front Microbiol. 2021;12:631794. 10.3389/fmicb.2021.631794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Melo LD, et al. Phage therapy efficacy: a review of the last 10 years of preclinical studies. Crit Rev Microbiol. 2020;46(1):78–99. 10.1080/1040841X.2020.1729695 [DOI] [PubMed] [Google Scholar]
- 121.Macdonald KE, et al. Patient perceptions of phage therapy for diabetic foot infection. PLoS ONE. 2020;15(12):e0243947. 10.1371/journal.pone.0243947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ali Y et al. The current status of phage therapy and its advancement towards establishing standard antimicrobials for combating multi drug-resistant bacterial pathogens Microbial Pathogenesis, 2023: p. 106199. [DOI] [PubMed]
- 123.Zalewska-Piątek B. Phage Therapy-Challenges, opportunities and Future prospects. Pharmaceuticals (Basel), 2023. 16(12). [DOI] [PMC free article] [PubMed]
- 124.Gan L et al. Bacteriophage effectively rescues Pneumonia caused by prevalent multidrug-resistant Klebsiella pneumoniae in the early stage. 2022. 10(5): p. e0235822. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.