Abstract
Background
ACSS3 (acyl-CoA synthetase short-chain family member 3) is found in numerous tissues and is linked to tumor cell type development and metastasis.
Methods
We conducted a comprehensive pan-cancer analysis of ACSS3. The TCGA (Cancer Genome Atlas), CPTAC (Clinical Proteomic Tumor Analysis Consortium), and HPA databases were used to ascertain the connection between ACSS3 and various types of tumors. Genes in the TCGA database would be identified using cBioPortal queries, and their transcriptome expression would then be verified using GEO data. ACSS3 expression and cellular localization in various tumor tissues of most cancer types were analyzed using single-cell sequencing data from the TISCH database. According to HPA and CPTAC databases, we analyzed and evaluated protein expression levels. Predictive analysis based on precise survival data of ACSS3 expression levels for 26 cancer types predicted using the TCGA database. Furthermore, we investigated the relationship between ACSS3 and immune microenvironments in different tumor tissues using the TIMER and TISCH databases. CellMiner, GDSC, and CTRP data would clarify the relationship between ACSS3 and drug resistance and explore the chemicals that affect ACSS3 expression. The final part of our study explored and validated the role ACSS3 played in glioma proliferation, migration, and invasion.
Results
ACSS3 is differentially expressed in various tumors and exhibits early diagnostic value. ACSS3 expression is associated with clinical features, and high ACSS expression anticipates a worse prognosis in multiple tumors and may impact drug sensitivity. The changes in the immunosuppressive microenvironment of gliomas are closely related to the upregulation of ACSS3.
Conclusions
ACSS3 is a novel biomarker for forecasting different human cancer prognoses, as it can influence the biological process by modulating the immune microenvironment. ACSS3 is a critical prognostic factor for glioma and is related to its proliferation, migration, and invasion.
Keywords: ACSS3, Pan-cancer, Glioma, Biomarker, Cell proliferation, Prognosis, Immunotherapy
1. Introduction
Cancer is a global public health issue that presents a significant health threat to patients and reduces their life quality [1,2]. The continuous increase in the number of new cancer cases is the leading cause of global mortality [3]. The biological processes of cancer are complex, related to multiple signaling pathways, and closely related to the tumor microenvironment (TME). The new immunotherapy has changed the treatment pattern of various tumor types, and tumor immunotherapy possesses an involvement in the tumor occurrence, development, diagnosis, and prognosis [4,5]. Therefore, identifying novel biomarkers related to tumor immunity is crucial. It is significant to execute a pan-cancer investigation on a specific gene and study its connection with cancer-related immunity, prognosis, and chemotherapy.
The massive metabolism of cancer cells leads to the continuous collection of many metabolic enzymes and metabolic intermediates in the TME [6]. AcCoA (Acetyl-Coenzyme A) is a critical metabolic intermediate that is essential for the anabolic and catabolic pathways [7,8]. ACSS1, ACSS2, and ACSS3 are family members of ACSS (the acetyl-CoA synthetase short-chain family). Among them, the research on ACSS2 is the most extensive. Cancer cells use Acetate as a nutrient source in an ACSS2-dependent manner [9]. Studies have shown that ACSS2 is associated with high biosynthesis and bioenergetics, which may be necessary for the growth of intracranial malignancies [10]. However, there are relatively few studies on ACSS3, and little is known about its function. Recent studies have shown that ACSS3 is an acetyl-CoA synthase that can affect acetate utilization and histone acetylation, affecting prostate, gastric, and bladder cancer cell proliferation [[11], [12], [13]]. ACSS3 possesses biological function in some human cancers. However, due to significant differences between different tumors, the role of ACSS3 is still being determined and has yet to be systematically analyzed. Immunotherapy and targeted therapy have received increasing attention in the past decade. There has not yet been a systematic study of ACSS3 as a component of the tumor immune microenvironment nor its prognostic significance in cancer prognosis and response to immunotherapy. Therefore, we will conduct a pan-cancer analysis of ACSS3. Glioma is a fatal malignancy of the primary central nervous system (CNS) with high intracranial incidence [14]. Very few biomarkers can reflect immune cell infiltration in the TME at the moment. Identifying specific immune-related genes as therapeutic targets for gliomas will be of great interest, especially in the absence of reliable and practical gliomas for biomarkers [15,16].
We executed a pan-cancer analysis depending on web tools and R language to study various public databases and clinical data from the Second Hospital of Hebei Medical University. In addition to exploring the potential mechanisms and clinical prognosis of ACSS3 in different cancers, the possible functional expression of ACSS3 and its impact on immune cell infiltration in pan-cancer cells were also explored. We manifested a significant correlation between ACSS3 and many cancer attributes, including immunological microenvironment, immune cell infiltration, treatment resistance, and patient survival. In addition, we conducted in vitro cell experiments using glioma specimens and cell lines to investigate its role in glioma's occurrence, development, and immune microenvironment of ACSS3. These outcomes emphasize the crucial contribution of ACSS3 in cancer and offer strategies and directions for future anti-tumor treatment research based on ACSS3(Fig. 17, workflow of our study).
Fig. 17.
Workflow of our study.
2. Method
2.1. Genomic profiling and clinical information analysis of tumor databases
Firstly, we gathered expression data for 33 types of tumors and normal tissues (ACC, BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, and UCEC) from the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) and GTEx databases (http://www.gtexportal.org) [17,18]. The ACSS3 mRNA expression was analyzed in both normal and malignant tissues (N = 17244). The ACSS3 expression in tumors and associated normal tissues (n = 11,060) was determined using data from UCSC XENA (https://xenabrowser.net/datapages/). We used the Wilcoxon rank sum test to assess disparities in gene mRNA expression between the tumor and surrounding normal tissues in TCGA. Furthermore, we conducted the Wilcoxon rank sum test on paired specimens categorized by TCGA cancer, and a significance level of P < 0.05 was used. The expression data of TCGA comes from the corrected TCGA dataset RNA seq data. From the (EBPlusPlusAdjustPANCAN-IlluminaHiSeq-RNASeqV2.geneExp.tsv) file provided by PanCanAtlas. TCGA-GTEx: To expand the normal sample size, we paired the TPM (transcripts per million)expression levels of GTEx normal samples with TCGA tumor TPM expression levels (from the tcga-RSEM_gene_tpm and gtex-RSEM_gene_tpm datasets in the USCS Xena database). The expression distribution of genes in various organs was visualized using the gganatogram package. We validated the gene protein expression at the transcriptional level based on the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and the protein level based on the mass spectrometry data of CPTAC (https://cptac-data-portal.georgetown.edu). The GEO and CPTAC mass spectrometry data are in the supplementary documents. The mRNA logistic analysis of pan-cancer ACSS3 was based on TCGA, TCGA-GTEx, GEO, and CPTAC databases further to validate the accuracy of the Wilcoxon rank sum test.
The analysis steps of R language. Using the R4.2.2 version for analysis, convert the data into unincorporated Z-Score scores by tumor using (x - μ)/σ. Normalize gene expression levels using the scale function for zscore, remove missing or empty values using the na.omit function, and calculate the significance of Wilcoxon Rank Sum Tests using the Wilcox.test function,
Visualize the ggplot function. Visualize the expression distribution of genes in various organs using the gganatogram software package. Based on the pROC package, evaluate the receiver operating characteristic (ROC) curve to ascertain the gene's significance in pan-cancer diagnosis, with an area under the curve (AUC) value of 0.5–1.
2.2. Analysis of somatic copy-number alteration (SCNA), mutation, and DNA methylation
For the analysis and visualization of cancer genomics data derived from several sources, the cBioPortal site (http://www.cbioportal.org) was used. The cBioPortal website offers a range of investigations, incorporating mutation-related analysis and visualization [19]. Upon accessing the cBioPortal website, go to the “Quick Selection” section and choose the “TCGA Pan-Cancer Atlas Study.” Proceed by entering “ACSS3” to start a search for genetic alterations in genes. The frequency of changes, kinds of mutations, and copy number alterations (CNA) statistics for all TCGA tumors were acquired with the “Cancer Type Summary” feature on the website. The precise position of gene mutations may be illustrated in a two-dimensional (2D) schematic representation of protein structure using the “mutation” tool available on the website. SCNA and mutation analysis enhance the amplification and deletion of gene copy numbers, affecting both heterozygous and homozygous states. SCNA with a frequency over 5 % is called high-frequency. Compute the Spearman correlation coefficient between each gene expression value and the copy number fragment value in order to ascertain the relationship between SCNA and gene expression. Utilize the Bioconductor platform to import the R package “IlluminaHumanMmethylation-450kanno.ilmn12.hg19” in order to provide annotation for the methylation probes associated with each gene promoter. A P-value threshold of 0.05 was applied to find genes that were significantly hypomethylated or hypermethylated using the Wilcoxon rank test. Determine the Spearman correlation coefficient between the level of gene transcription expression and the Beta value of promoter DNA methylation. In addition, we calculated the Spearman correlation of genes with 10 genomic features.
The variations in gene methylation patterns indicate the intricate nature of gene regulation and the unique characteristics of cancer types. The beta value represents DNA methylation. Regulatory potential (RP) ratings are calculated using the BETA algorithm [20]. The BETA method calculates regulatory potential (RP) scores to predict the likelihood of a factor regulating genes. BETA employs a distance-weighted metric to assess the regulatory capacity of all binding sites of the factor within a specified proximity to a target gene in ChIP-seq, DNase-seq, or ATAC-seq samples. Genes are likely to be regulated by factors that have high RP scores.
2.3. The connection between elevated ACSS3 expression and immune infiltration in cancer
Hence, we assessed the association between the ACSS3 expression and genes relevant to the immune system, comprising immune activation, immunological suppression, chemokine, chemokine receptor, and major histocompatibility complex (MHC) genes [21]. We utilized the identical GSVA algorithm and parameters as previously employed to assess the anticancer impacts in the seven stages of the cancer immune cycle: the cancer cell antigens release (step 1), the cancer antigens presentation (step 2), the initiation and activation (step 3), the immune cells transportation to the tumor (step 4), the immune cells infiltration into the tumor (step 5), the cancer cells recognition by T cells (step 6), and the killing of cancer cells (step 7). The immune status was evaluated by assigning scores to each step, and the differences between high and low gene expression groups were calculated [22]. TIMER2.0 useed seven advanced algorithms to precisely evaluate immune infiltration levels in the TCGA tumor spectrum, exploring the association between ACSS3 expression and classic disease/psychological phenotypes [23,24].
The TISCH database was used to obtain a sizeable single-cell dataset of ACSS3, thereby validating the TME results from single-cell resolution. The immunotherapy data of glioma utilized in this study were downloaded from the GEO (Gene Expression Omnibus) dataset. They were the GSE35640 and GSE78220 datasets, respectively.
The relationship between ICIs (immune checkpoint inhibitors) and ACSS3 expression levels was predicted by the Cancer Immunomics Atlas (TCIA, https://tcia.at). Patients with a high immune phenol score (IPS) were related to better clinical outcomes after ICI treatment. TIDE [25] (Tumor Immune Dysfunction and Exclusion) and ImmuCellAI [26] (Immune Cell Abundance Identifier) examinations were also utilized to assess the association of ACSS3 expression levels with treatment with ICIs.
2.4. Pathways and functional mechanisms analysis
The cluster profile package will be applied to conduct KEGG enrichment analysis on ACSS3 to identify its function or involvement in related biological pathways in pan-cancer [27]. The Spearman correlation analysis system was implemented to ascertain the correlation between ACSS3 mRNA expression and protein content determined by the RPPA method from the TCPA database [28]. Using the Sankey plot, visualize the results with a correlation coefficient greater than 0.4 in all tumors. CancerSEA has redefined a grand total of 14 distinct functional states. The Z-score method was devised by Lee et al. [29]. It incorporates gene expression features to accurately represent the level of activity in a certain pathway. We applied the z-score parameter in the R package GSVA to compute 14 gene sets representing functional states.
2.5. Identifying chemical substances that interact with ACSS3
The GSCA database (http://bioinfo.life.hust.edu.cn/web/GSCALite/) has a collection of 750 small molecule pharmaceuticals obtained from GDSC and CTRP. These medications were used for the purpose of analyzing gene expression and drug sensitivity [30]. We extract significant small-molecule compounds linked to gene expression data. In order to investigate the connection between mRNA expression and drug sensitivity zscore, the NCI-60 expression data was obtained from CellMiner, and to compute the Spearman correlation coefficient [31].
Determine the differentially expressed gene levels between high and low gene specimens in various forms of cancer. Collect 150 genes with the highest upregulation or downregulation as gene-related markers. The CMAP_gene_signatures The RData file encompasses 1288 compound-linked characteristics obtained from the database website (https://www.pmgenomics.ca/bhklab/sites/default/files/downloads). Download and use for computing matching scores. Herein, the investigation adhered to the methodologies described in earlier papers [32,33]. The findings for the top 5 rankings of 32 different forms of cancer were condensed and visually shown using the R programming language.
2.6. Survival and clinical outcome analysis
We utilized the “survival” and “surveyor” R packages to analyze gene expression and prognostic indicators. These powerful and widely used packages offer a range of statistical tools and functions essential for relevant analyses, including survival analysis, Cox proportional hazards regression, and graphical representation of survival curves. By incorporating these packages into our analysis pipeline, we were able to estimate the relationship between gene expression levels and various prognostic factors across different cancer types. By using these specific R packages, we were able to assure the precision and resilience of our outcomes, allowing us to make significant conclusions about the prognostic importance of the gene under investigation.
2.7. Tissue samples
This study involved the collection of 78 glioma and normal tissues from consenting patients previously treated at the Second Hospital of Hebei Medical University. All tissue samples were pathology-verified, excluding those who had undergone radiation or chemotherapy prior to surgery. Consent for participation was waived by the ethics committee.
2.8. Cell cultures and reagent
Normal human astrocytes (NHA), U251, T98G, LN229, and U87 were purchased from Procell Life Science. RPMI-1640 medium (GibcoTM, 11875093) was utilized to cultivate U251 cells. DMEM (GibcoTM, 10,564,011) was utilized to cultivate LN229 cells. MEM (GibcoTM, 11090081) was suitable for cultivating U87 and T98G cell lines, while NHA cells need to be cultured in a specific Astrocyte Medium (AM, ScienCellTM, #1801). 10 % FBS (GibcoTM, 10099141) and Pen-Strep Solution (BI, 2114091) were added to all the above media. Culture conditions at 37 °C with 5 % CO2 were essential for the survival of glioma cell lines.
2.9. Cell transfection
The U251 and U87 cells were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences in Shanghai, China. They were cultivated in RPMI 1640 media enriched with 10 % fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) and 1 % penicillin-streptomycin. The cells were kept at 37 °C and a CO2 concentration of 5 %. We synthesized the ACSS3 gene overexpressing lentivirus(pcACSS3) and lentiviral vectors for negative control (NC-ACSS3) through Shanghai Gene Chemical Co., Ltd. and took MOI = 5 according to the lentivirus instructions. The ACSS3 siRNA(siACSS3) had the following sequence: 5′- GCAACUAAUGAGAUGUGGCGA-3′. The cells, with a density of 5 × 103 per well, were subcultured in 96-well culture plates and then infected with pcACSS3/siACSS3. The infected cells were chosen by incubating them to a concentration of 2 μg/mL puromycin for a period of 48 h. The efficacy of the ACSS3 knockdown and overexpression was assessed by western blotting.
2.9.1. RT-PCR
TRIzol (Invitrogen) was utilized for obtaining total RNA from glioma tissues and cell lines. The process of reverse transcription was used to convert total RNA into cDNA with PrimeScript RT Reagent (Takara). The SYBR Premix ExTaq (Takara) was used for RT-PCR. The primer sequences used were as follows (5′-3′): forward GCCGTTGATCGTCATATTGAAA and reverse TGTAGATAACCACAGTGTCACC for the target gene, and forward GGACGGAGATCCCCTCCAAAAT and reverse GGCTGTTGTCATACTTCTCATGG for the GAPDH gene. The 2-△△Cq technique was applied to ascertain the relative expression level.
2.10. Western blot
A cocktail of protease and phosphatase inhibitors (K1015, APExBIO) was added to the RIPA Lysis Buffer (Epizyme Biomedical Technology), which was then utilized to lyse human tissues and cells. The protein samples were separated using SDS-polyacrylamide gel electrophoresis and then placed on PVDF membranes. Following a 20-min blocking at ambient temperature with Protein Free Rapid Blocking Buffer (Epizyme Biomedical Technology), PVDF membranes were subsequently incubated with primary antibodies versus GAPDH (ab8245, 1:8000) and ACSS3 (1:1000; 16204-1) at 4 °C overnight. The following day, following washes with TBST, the secondary antibody (1:10,000; Abcam; ab216773) was incubated for approximately 2 h. The images were finally detected utilizing the Odyssey system (LI-COR, USA).
2.11. CCK-8 assay
Cell specimens were first cultivated in each well of a 96-well plate at a standard of 5 × 103 cells/well. Cell proliferation rates were estimated on days 0, 1, 2, 3, and 4 of seeding cells. Supplement each well in a 96-well plate with 10 μl of CCK-8 (reporter) and hatch for an additional 2 h at 37 °C. Finally, absorbance at 450 nm was estimated utilizing a strip reader.
2.12. Transwell migration and invasion assays
The U251 and U87 cell's invasion and migration were evaluated with Transwell (Corning Incorporated). The top chamber was either pre-coated or left untreated with Matrigel. Serum-free medium containing glioma cell line cells was cultivated in the upper chamber. Introduce a medium containing 10 % FBS into the bottom chamber of the Transwell in order to evaluate the cells' capacity for invasion and migration. Examine the outcomes during a 24-h incubation period at 37 °C and a CO2 concentration of 5 %. The cells are first removed from the top chamber by scraping. Fix the specimen with paraformaldehyde for a duration of 10 min prior to applying 0.1 % crystal violet stain.
2.13. Wound healing
Glioma cells were seeded when the growth of the cells reached 90 % confluence; the following experiment was carried out. We used a pipette tip to create scratches on these cell monolayers. Scratch pictures at 0 h and 24 h were analyzed using ImageJ.
2.14. Xenograft Experiment
Male athymic BALB/c nude mice (4 weeks) were utilized for in vivo experiments. Each nude mouse received a subcutaneous injection of 5 × 106 U251 cells in the axilla. The volume was estimated using Vernie's equation: V = (W2 × L)/2 (length (L) and width (W)). Twenty-eight days after inoculation, BALB/c nude mice were sacrificed. Photographs were taken after the subcutaneous xenografts were removed. Each xenograft was weighed.
2.15. Statistical analyses
The experiment was replicated thrice. The statistical data are presented as the mean ± the standard deviation (SD). The statistical data analysis was conducted using Prism 9 (GraphPad Inc, USA) and R (version 3.6.3). The statistical procedures used were the T-test and one-way variance. The predictive significance was evaluated with the Kaplan-Meier curve. Overall survival factors were analyzed with the log-rank test. A p-value below 0.05 was deemed to have statistical significance.
3. Results
3.1. Aberrant Expression of ACSS3 among cancers
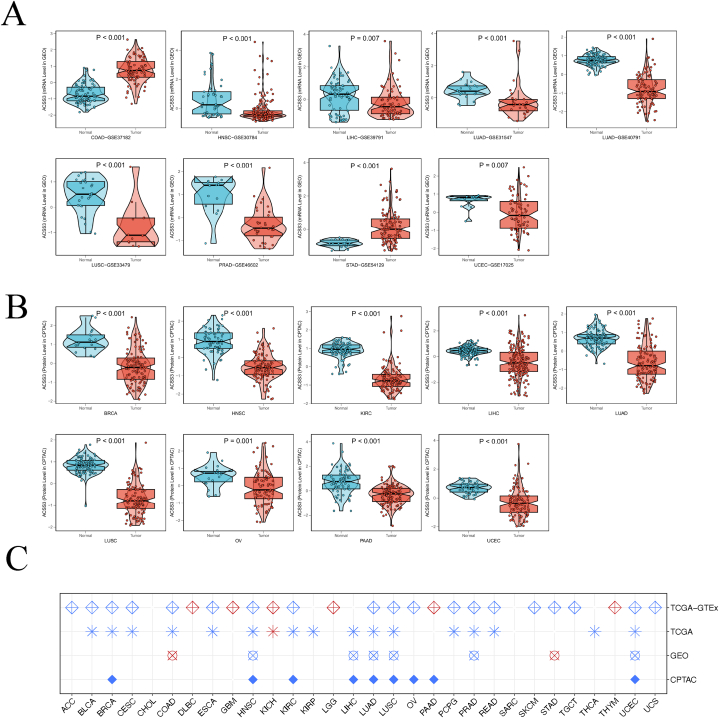
We performed a pan-cancer investigation to determine the ACSS3 expression patterns in different cancers. By integrating and mining data from the TCGA and GTEx databases, we can first estimate the ACSS3 differential expression level across a pan-cancer perspective (P < 0.05, Fig. 1A). As a result, we manifested that the ACSS3 gene is differentially expressed in various cancer types, and it is significantly upregulated in DLBC, GBM, KICH, LGG, PAAD, and THYM. Afterward, we performed a pan-cancer differential analysis using samples from the TCGA database. This analysis manifested an elevation in expression in GBM and KICH (P < 0.05, Fig. 1B). Based on the study of the TCGA dataset, ACSS3 demonstrated differential expression in tumor tissues and paired with surrounding normal tissues in several types of cancer, encompassing BLCA, HNSC, UCEC, KIRC, PRAD, KIRP, LUSC, COAD, READ, CHOL, LIHC, BRCA, KICH, THCA, LUAD, KICH, and STAD (P < 0.05, Fig. 1C). Present the differential expression distribution pattern of ACSS3 in an organ diagram (Fig. 1D). ACSS3 has satisfactory sensitivity and specificity in diagnosing tumor patients, such as CHOL (AUC = 1.000), CESC (AUC = 0.983), PCPG (AUC = 0.973), and LUSC (AUC = 0.972), as determined by ROC curves calculated from TCGA data (Fig. 1E). The ROC results remained stable after combining the typical sample size in the GTEx database, including CHOL (AUC = 0.988), CESC (AUC = 0.975), PCPG (AUC = 0.919), and LUSC (AUC = 0.946) (Fig. 1F). We applied data from the GEO database to confirm the ACSS3 mRNA expression levels in tumors of COAD, HNSC, LIHC, LUAD, LUSC, PRAD, STAD, and UCEC (Fig. 2A). Also, ACSS3 protein levels were validated using the CPTAC database for different tumors (Fig. 2B), BRCA, HNSC, KIRC, LIHC, LUAD, LUSC, OV, PAAD, and UCEC. We further conducted logistic regression analysis using four datasets: TCGA, TCGA-GTEx, GEO, and CPTAC, and found that ACSS3 was negatively correlated with HNSC, LUAD, LUSC, and UCEC in all datasets. In TCGA-GTEx, high mRNA expression of ACSS3 was associated with DLBC, GBM, KICH, LGG, PAAD, and THYM. (Fig. 2C). Through multi-omics and multi-database data analysis, it was found that the ACSS3 expression trend in different tumors is consistent. In summary, we have found that dysregulated ACSS3 expression is involved in various tumors using multiple omics, databases, tumors, and methods.
Fig. 1.
Pan-cancer analysis of ACSS3. (A) Pan-cancer mRNA data of ACSS3 from TCGA and GTEx datasets; (B) Pan-cancer mRNA data of ACSS3 from TCGA; (C)Paired samples grouped by cancer from the TCGA. (D) Expression and distribution of ACSS3s in various organs. (E) Diagnostic value of ACSS3 to distinguish tumor tissues from normal tissues in TCGA; (F) Diagnostic value of ACSS3 to distinguish tumor tissues from normal tissues in TCGA-GTEx‥ (TCGA: The Cancer Genome Atlas, CPTAC: Clinical Proteomic Tumor Analysis Consortium, GTEx: Genotype-Tissue Expression Project, AUC values of 0.5–0.7, 0.7–0.9, and >0.9 represent low, deterministic, and high accuracy, respectively. The closer the AUC value is to 1, the better the diagnostic performance.)
Fig. 2.
Aberrant Expression of ACSS3 among Cancer. (A) External validation of mRNA levels was performed using the GEO database; (B) External validation of protein levels were performed using a CPTAC database. (C) Logistic regression analysis of TCGA, TCGA-GTEx, GEO, CPTAC.
3.2. Genetic alterations of ACSS3 in cancers
These alterations can encompass various molecular events, including mutations, copy number variations, and DNA methylation changes. The frequency and types of genetic alterations can vary across different cancer types. For instance, in some cancers, such as breast and lung cancer, ACSS3 mutations have been reported, while in others, like colorectal cancer, copy number variations are more prevalent. Additionally, alterations in the DNA methylation patterns of the ACSS3 gene have been observed in specific cancer types. These genetic alterations can potentially impact the expression and function of ACSS3, leading to dysregulated cellular processes and contributing to tumorigenesis. Understanding these genetic alterations in ACSS3 provides crucial insights into the molecular mechanisms underlying cancer development and may offer opportunities for targeted therapeutic approaches.
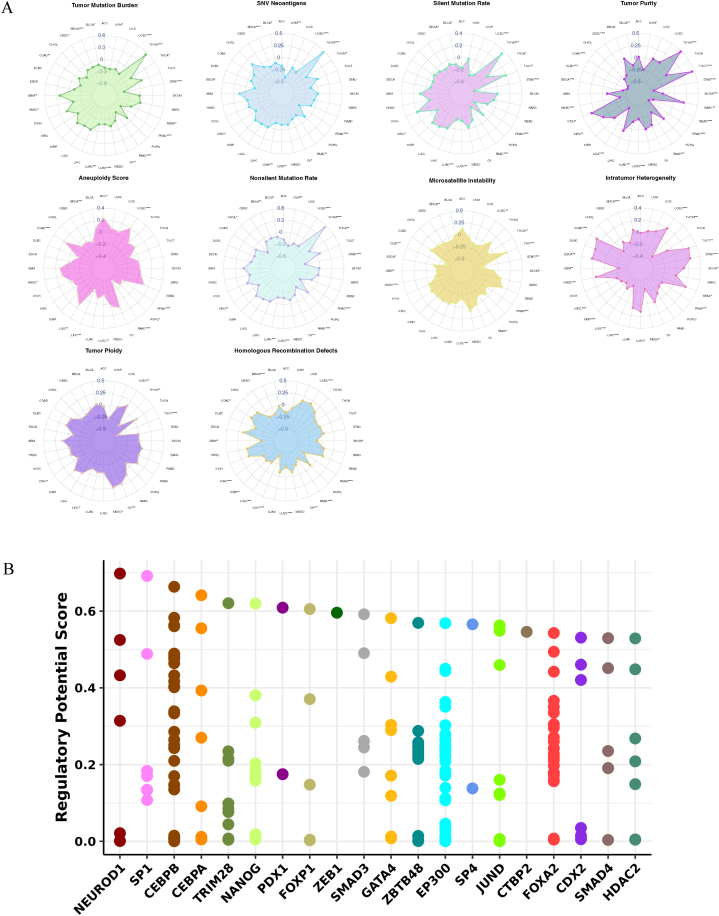
We performed a comprehensive analysis utilizing genomic data from TCGA tumors and normal tissues to investigate genetic variations, SCNAs, mRNA expression, and DNA methylation patterns of the ACSS3 gene. Through the cBioPortal database, we examined the 2D structure of ACSS3 gene mutation sites and identified amplifications and mutations occurring in various cancers (Fig. 3A). The mutation rate of ACSS3 was observed in 25 cancer types, with the highest occurrence (>10 %) observed in melanoma cases (Fig. 3B). Analysis of somatic copy number alterations revealed that the ACSS3 gene regulates gene expression (Fig. 3C). Furthermore, we examined the percentage of SCNAs and found that they occurred frequently (over 5 % of all specimens) in several cancer types (Fig. 3E). ACSS3 was significantly elevated in 19 cancer types while mitigated in 7 (Fig. 3D). Additionally, we investigated the DNA methylation patterns of ACSS3, as changes in promoter methylation are often linked to tumor development. We observed a consistent methylation pattern of ACSS3 in the pan-cancer cohort, with tumor tissues exhibiting high or low methylation compared to normal tissues (Fig. 3F). Additionally, a detrimental association was identified between the ACSS3 mRNA expression and DNA methylation (Fig. 3G), revealing that the methylation status of ACSS3 is complex and cancer-specific. Furthermore, we analyzed the Regulatory Potential (RP) score, which estimates the likelihood of a factor regulating a gene. We selected the top 20 transcription factors with high RP scores for peak annotation. Our outcomes demonstrated that these elements are often situated in the gene promoter region, suggesting their possible involvement in regulating gene expression (Fig. 4B). According to scoring results, ACSS3 correlated with 10 genomic features (Fig. 4A). The outcomes offer significant insight into the genetic alterations and regulatory components linked to ACSS3 in cancer.
Fig. 3.
Genetic Alterations of ACSS3 in Cancers. (A) Sites and number of cases with ACSS3 genetic alterations in pan-cancer from cBioPortal; (B) Frequency of ACSS3 mutations in different tumor types; (C) Relationship between ACSS3 mRNA expression and genetic alterations; (D) The Spearman's correlation between somatic copy number alterations and the expression of ACSS3; (E) Histogram shows the frequency of somatic copy number alterations for ACSS3 in each cancer type; (F) Heatmap shows the differential methylation of ACSS3 in cancers; (G) Spearman's correlation of ACSS3 between transcriptional expression and promoter methylation.
Fig. 4.
Gauge the regulatory potential of all the factor's binding sites. (A) Radar map visualization of Spearman correlation coefficients of ACSS3 and 10 genomic features. (B) The top 20 factors are shown in this plot.
3.3. High ACSS3 expression correlates with immune infiltration in cancer
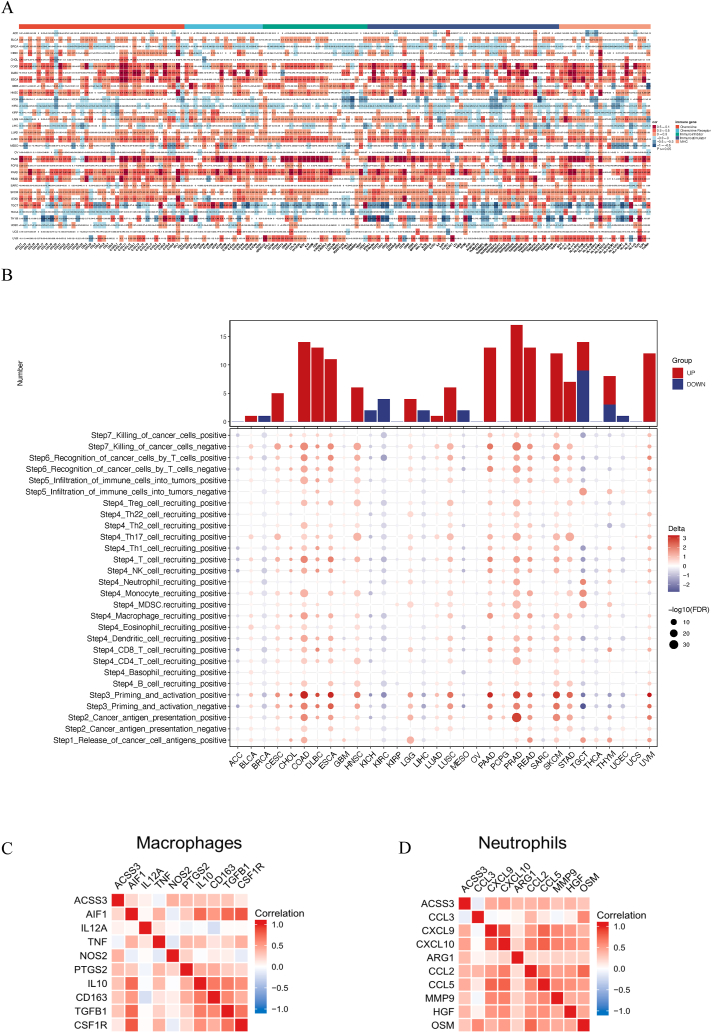
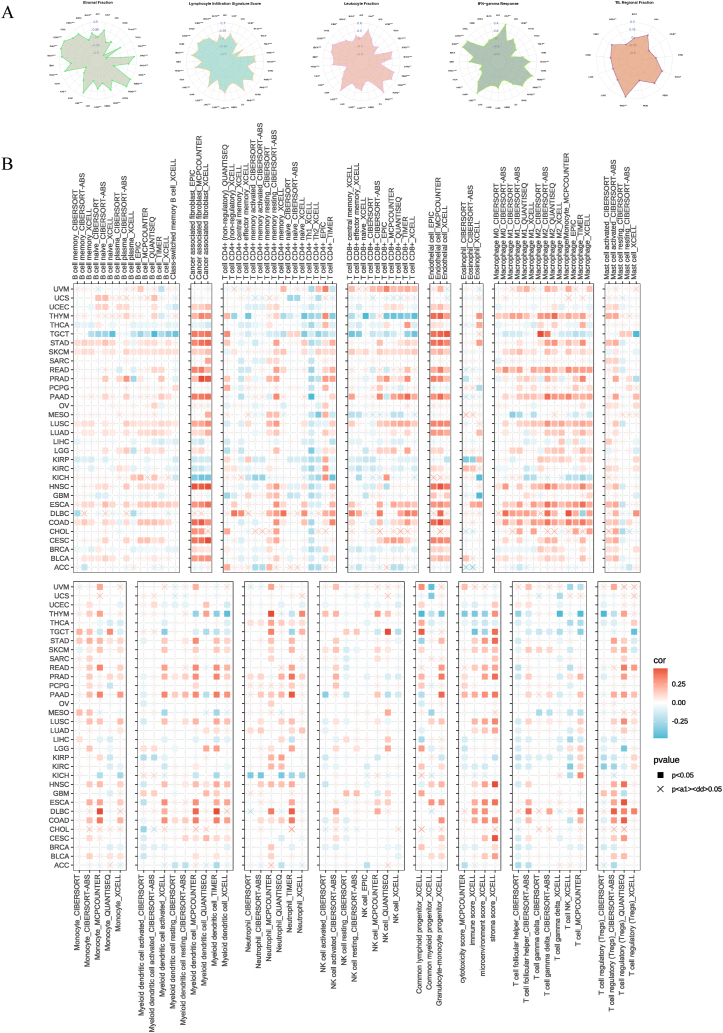
To estimate the connection between ACSS3 and immune infiltration in cancer types, we first assessed the association between ACSS3 and genes relevant to the immune system (Fig. 5A). We used the GSVA algorithm and parameters to ascertain the ACSS3 score in different types of cancer and the seven-step immune cycle of cancer. We measured the distinctions between the high and low-expression groups of ACSS3 (Fig. 5B). Cancer cells' survival, metastasis, and immune escape depend on the dysregulation of the tumor's immune microenvironment. Analyzing the connection between ACSS3 expression and the classical macrophage and neutrophil phenotype in the TCGA database, we manifested that ACSS3 was linked to the M0 marker AIF1 and the M2 marker IL10, CD163, TGFB1, and CSF1R of TAM (M2 type macrophages promote tumor progression) (Fig. 5C). Likewise, ACSS3 was also positively correlated with the N2 phenotypic markers CCL2, CCL5, MMP9, HFG, and OSM of TAN (Fig. 5D). We investigated five immune-related scores in the TME. Spearman correlation analysis was applied to investigate the immune status of ACSS3 in pan-cancer patients (Fig. 6A). Using 7 state-of-the-art algorithms from the TIMER 2.0 database, calculate which immune cell types may be affected by the ACSS3 expression in whole cancer. It was found that ACSS3 showed different correlation trends in different immune cells of different cancers (Fig. 6B). It is worth noting that this may be attributed to various percentages of immune infiltration and unique TME in different cancers.
Fig. 5.
ACSS3 is associated with immune infiltration in various cancers. (A) The correlation between ACSS3 and immune-related genes; (B) Fact sheet on the cancer immune cycle. (C) Correlation of ACSS3 with M0, M1, and M2. (D) Correlation of ACSS3 with N1 and N2.
Fig. 6.
The relationship between ACSS3 and tumor microenvironment (TME). (A) Spearman correlation analysis of the immune status of ACSS3 in pan-cancer. (B) Seven software were used to evaluate the correlation between ACSS3 expression and cancer immune infiltration.
3.4. Prediction of response to immunotherapy in patients with high and Low-ACSS3 expression
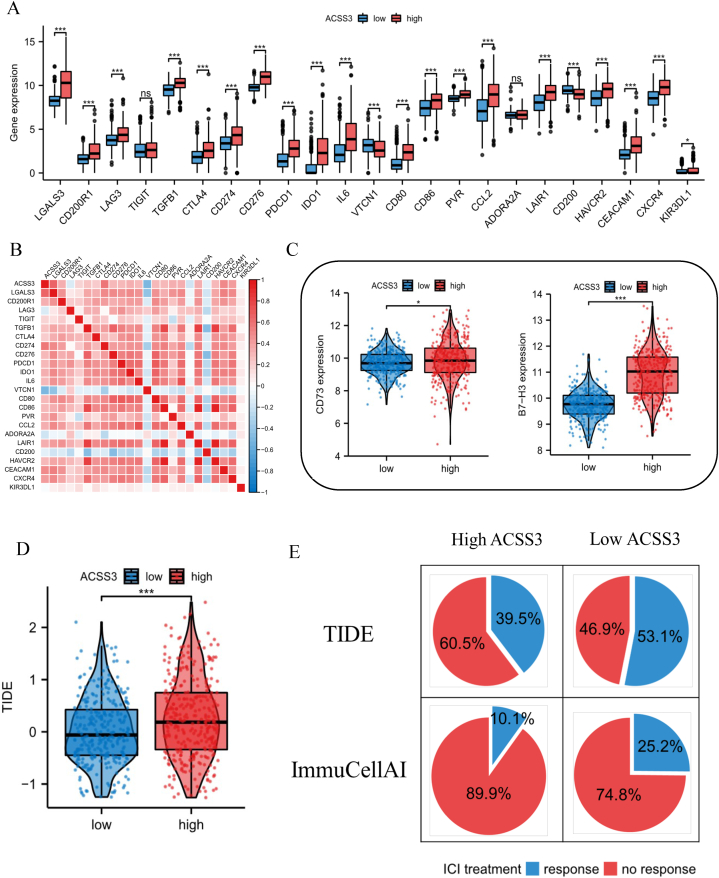
Current research shows that the continued development of immunotherapy and novel targeted therapies is necessary to prolong the 5-year survival rate of cancer patients. Currently, ICIs are a potential new immunotherapy type mainly characterized by relatively low toxicity and no apparent side effects. We detected the expression levels of low- and high-expressing ACSS3 groups and 23 ICI-linked genes in the TCGA database. Boxplots demonstrate that the ICI expression in the high-ACSS3 group was greater than in the low-ACSS3 group (Fig. 7A and B). Recent studies have shown that B7–H3 and CD73 are essential markers predicting glioma ICI response [34,35]. Our results showed that patients in the high-ACSS3 group had higher B7–H3 and CD73 expression in glioma patients (Fig. 7C). This suggested that ICIs would have a role in patients with lower ACSS3 expression. These results were consistent with the TIDE score (Fig. 7D). We utilized the TIDE and ImmuCellAI platforms to predict responses to ICIs in high and low ACSS3 groups. The outcomes demonstrated that ICI treatment might exert a more significant effect on patients in the low-ACSS3 group than those in the high-ACSS3 group (Fig. 7E).
Fig. 7.
Prediction of immunotherapy with ICIs. (A–B) Association of ACSS3 with immune checkpoints. (C) Relationship of B7–H3 and CD73 to ACSS3. (D) TIDE score. (E) Prediction results utilizing TIDE and ImmuCellAI.
3.5. Single-cell expression levels of ACSS3 in multiple cancer tissues
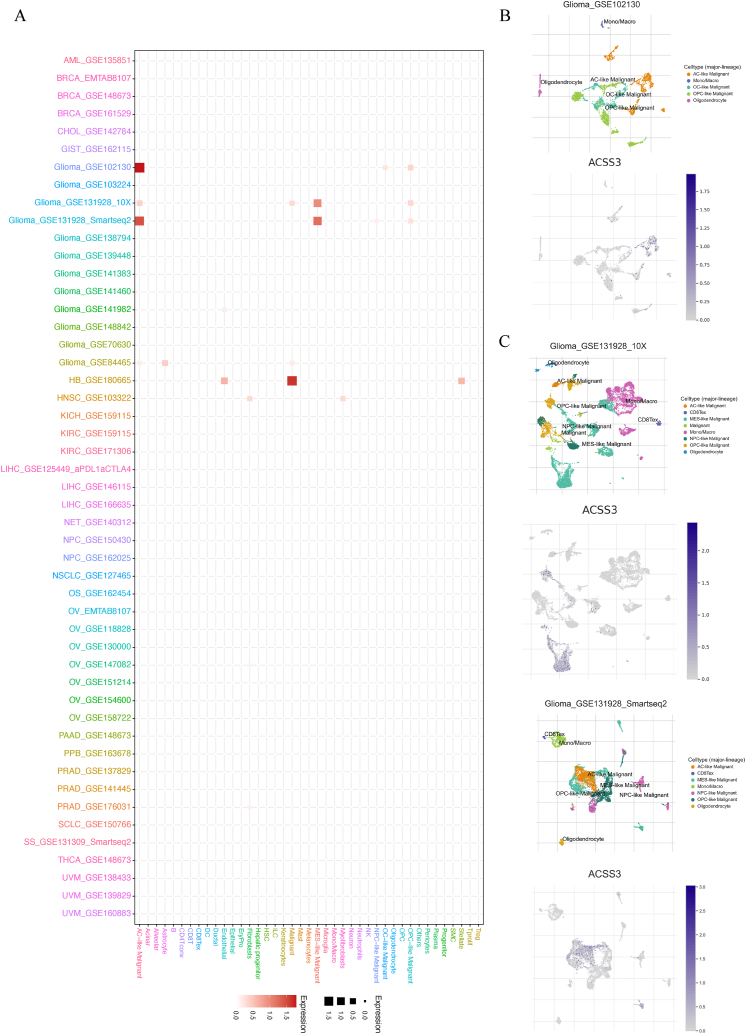
We used the TISCH database to obtain the expression of ACSS3 on many single-cell datasets. Then, the heatmap presented the relative expression level and visualization of ACSS3 in the TME and immune infiltration in pan-cancer (Fig. 8A). This indicates that ACSS3 is widely expressed in various immune and malignant cells. For example, in glioma (glioma_GSE1102130), the UMAP map shows the expression of ACSS3 in glioma cells such as AC-like Malignant and OC-like Malignant (Fig. 8B). And use 10 × Genomics and Smartseq2 platforms were implemented to ascertain the ACSS3 expression in gliomas (glioma_GSE131928) (Fig. 8C).
Fig. 8.
Single-cell expression levels of ACSS3 in multiple cancer tissues. (A) Relative expression levels of ACSS3 in tumor microenvironment and immune infiltration in pan-cancer; (B) UMAP map shows the expression of ACSS3 in gliomas; (C) 10 × Genomics and Smartseq2 platforms display the expression of ACSS3 in gliomas.
3.6. Association between ACSS3 and Pathways in Cancer
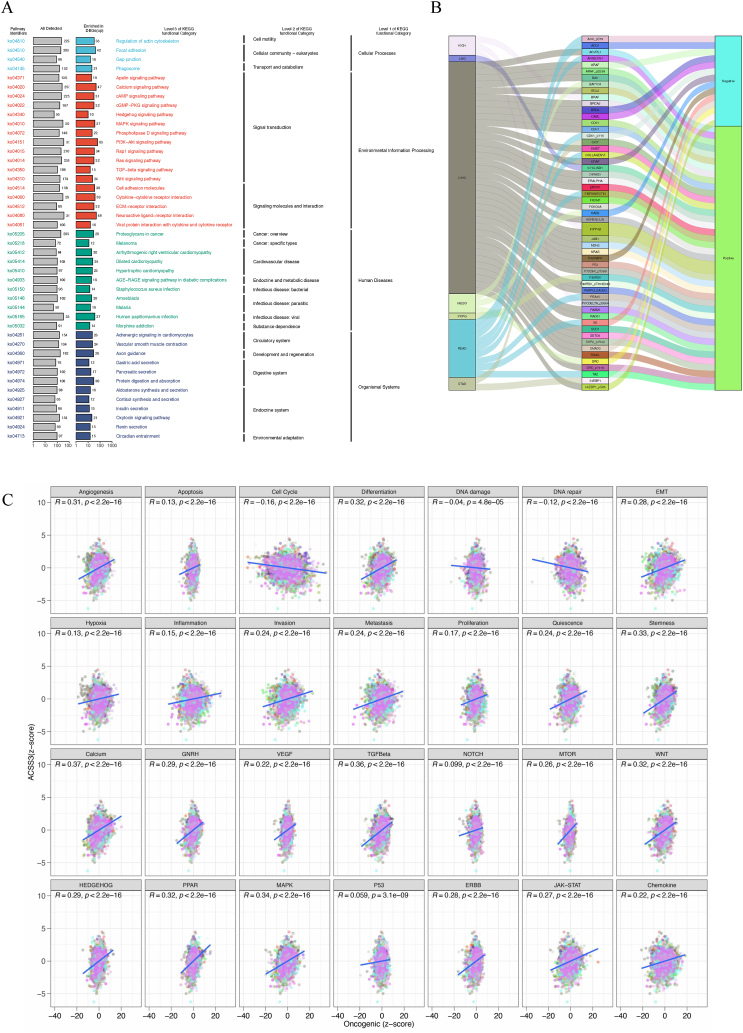
Differential genes were selected for KEGG analysis according to the high and low ACSS3 expression levels to identify conserved functions or related biological pathways involved in pan-cancer (Fig. 9A). Immune, metabolic, and signaling pathways are significantly activated. Functional proteomics, such as expression and modification, are crucial in studying complex diseases such as cancer. Studying functional proteomics will contribute substantially to understanding the causes of cancer and other cancers and determining effective treatment methods. According to the data from the TCPA database, we adopted the method of using RPPA(reverse phase protein array) [36]. It was found that ACSS3 mRNA is correlated with many functional proteins. Visualize the results with a correlation coefficient greater than 0.4 in all tumors using the Sankey plot (Fig. 9B). The Z-score method was proposed by Lee [29]. We applied Pearson correlation analysis to analyze the link between 14 cancer markers and 14 tumor-related pathway scores and ACSS3 (Fig. 9C). Among them, the scores for Angiogenesis, Apoptosis, Invasion, and Metastasis are higher and positively correlated with other scores. Based on the 30 % differential expression of ACSS3 at the top and bottom, specimens of each tumor type were allocated into two groups and subjected to Gene Set Enrichment Analysis (GSEA) to emphasize the activation or suppression of 85 metabolic gene sets of 50 hallmark gene sets relative to the low expression group in different tumors (Fig. 10). Indicating that ACSS3 may be involved in metabolic disorders in tumors.
Fig. 9.
Association between ACSS3 and Pathways in Cancer. (A) Perform KEGG analysis to verify the functionality of ACSS3; (B) The correlation between ACSS3 mRNA expression and functional proteins; (C) The relationship between 14 cancer markers and 14 tumor-related pathway scores and ACSS3.
Fig. 10.
Enrichment differences of ACSS3 in 50 HALLMARK and 85 metabolism gene sets.
3.7. Association between ACSS3 and chemotherapy
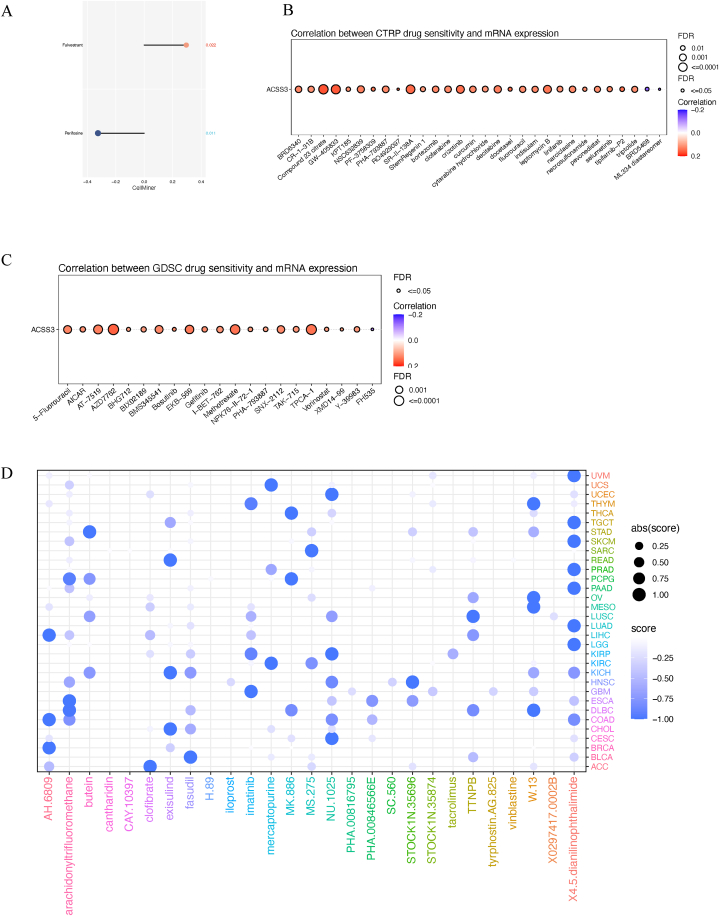
We conducted an analysis to determine the possible relationship between the sensitivity of chemotherapeutic drugs and the ACSS3 expression using three databases (CTRP, GDSC, and Cellminer). We conducted an analysis with the Cellminer database to investigate the link between ACSS3mRNA expression and drug sensitivity z-score. We then estimated the Spearman correlation coefficient. ACSS3 had a positive connection with Fulvestrant and an adverse relationship with Periposine (Fig. 11A). Using the CTRP (Fig. 11B) and GDSC (Fig. 11C) databases, we discovered a significant and positive connection between ACSS3 and numerous medicines. ACSS3 has the potential to be a gene that is sensitive to chemotherapy. By creating gene signatures that consist of the 150 genes that are most significantly elevated and the 150 genes that are most significantly diminished [37], the optimal feature matching method XSum (eXtreme Sum) was used to compare ACSS3-related features with CMap gene features. X4.5. dianilinophthalimide was found to exhibit significantly lower scores in most cancer types, manifesting that they may hinder gene-mediated cancer-promoting consequences (Fig. 11D)
Fig. 11.
ACSS3 and chemotherapy. Based on the Cellminer database (A), CTRP (B), and GDSC (C), we analyzed the relationship between ACSS3mRNA expression and drug sensitivity; (D) the optimal feature matching method XSum (eXtreme Sum) was used to compare ACSS3-related features with CMap gene features.
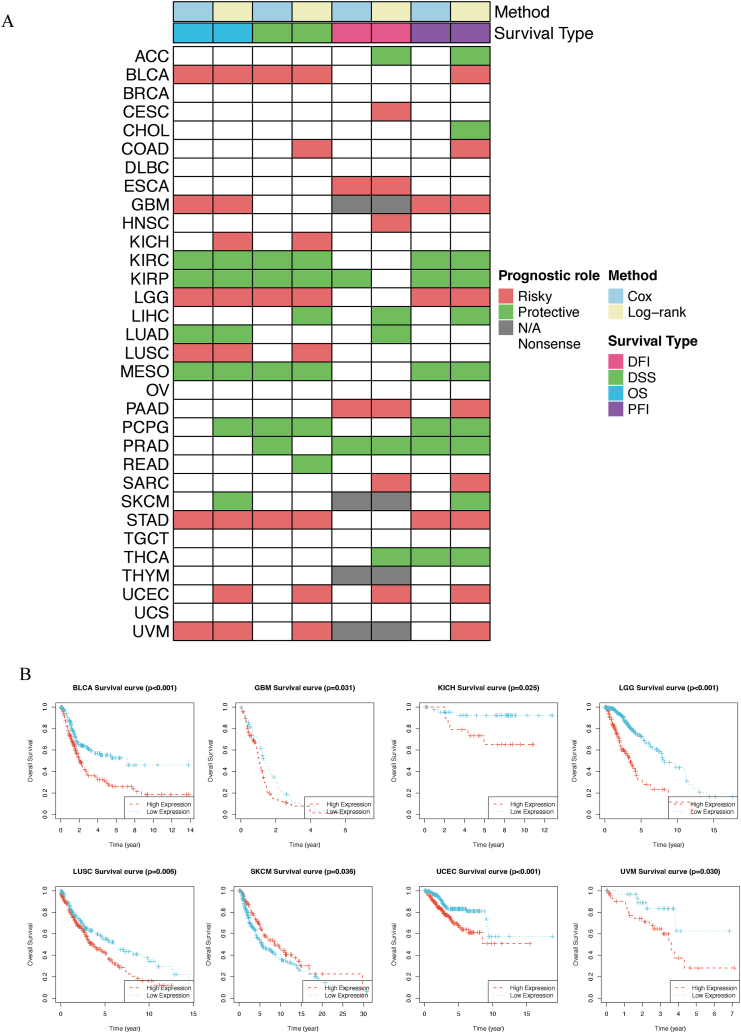
3.8. Prognostic role of ACSS3 in human cancers
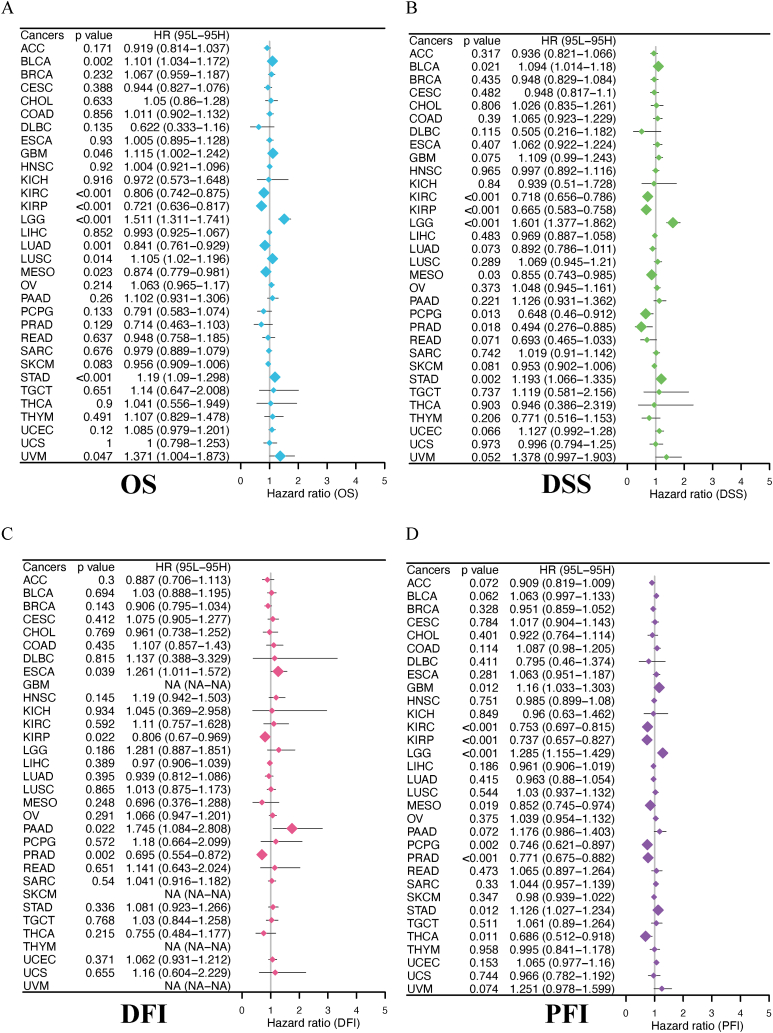
The role of genes in cancer survival was analyzed to additionally analyze the clinical relevance of ACSS3 in different types of cancer in humans. The heatmap clustering analysis revealed that the ACSS3 expression level is a prognostic indicator for 26 cancer kinds (Fig. 12A). And in most cases, ACSS3 can serve as a risk factor for different types of cancer. Especially in UCEC, ACSS3 is a risk factor in all four survival periods. Some tumors also present as protective factors; for example, patients with elevated gene expression have improved survival rates in KIRP and other conditions. This indicates that the gene ACSS3 plays different roles in different cancers, and its functional role in cancer survival should be further explored. We used Kaplan Meier curves to display the results of BLCA, GBM, and KICH (Fig. 12B). To supplement the pan-cancer map further, we used forest plots to display the Cox survival analysis results for four different survival periods (Fig. 13).
Fig. 12.
Clinical Relevance of ACSS3. (A) Summary of the correlation between expression of ACSS3 with overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI); (B) Kaplan-Meier survival analysis; The forest plot exhibited the prognostic role of ACSS3 in cancers by univariate Cox regression method.
Fig. 13.
The forest plot exhibited the prognostic role of ACSS3 in cancers by univariate Cox regression method.
3.9. High expression of ACSS3 predicted poor survival in glioma patients
The ACSS3 expression levels in normal brain and glioma tissue were quantified with Western Blot, RT-qPCR, and IHC, respectively. The outcomes showed that ACSS3 was significantly higher in four different WHO-grade glioma patients than in normal brain tissue (Fig. 14A–D). The expression levels gradually increased with increasing grades. Similarly, ACSS3 expression was significantly higher than NHA in 4 different glioma cell lines (Fig. 14E–G). This further recommended that ACSS3 could be an oncogene in glioma with potential as a biomarker.
Fig. 14.
Expression of ACSS3 in tissues and cells. (A) Presentation of ACSS3 mRNA in normal brain samples and glioma samples (NB = 16; LGG = 16; GBM = 16). (B and C) Determination of ACSS3 protein expression in normal brain samples and glioma samples (D) Immunohistochemical profile of ACSS3 in glioma tissues. (E–G) Determination of ACSS3 expression levels in HA and four glioma cell lines. (H) Kaplan-Meier curve of overall survival of ACSS3 glioma patients based on 78 glioma clinical data, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In addition, we conducted a Kaplan-Meier survival analysis using clinical information from 78 patients. Typically, 78 cases were diagnosed by highly skilled pathologists. The patients were categorized into two groups depending on the median cutoff value for ACSS3 expression. Herein, we demonstrated that patients with glioma in the high-ACSS3 group experienced a significantly worse overall survival rate contrasted with those in the low-ACSS3 group (Fig. 14H).
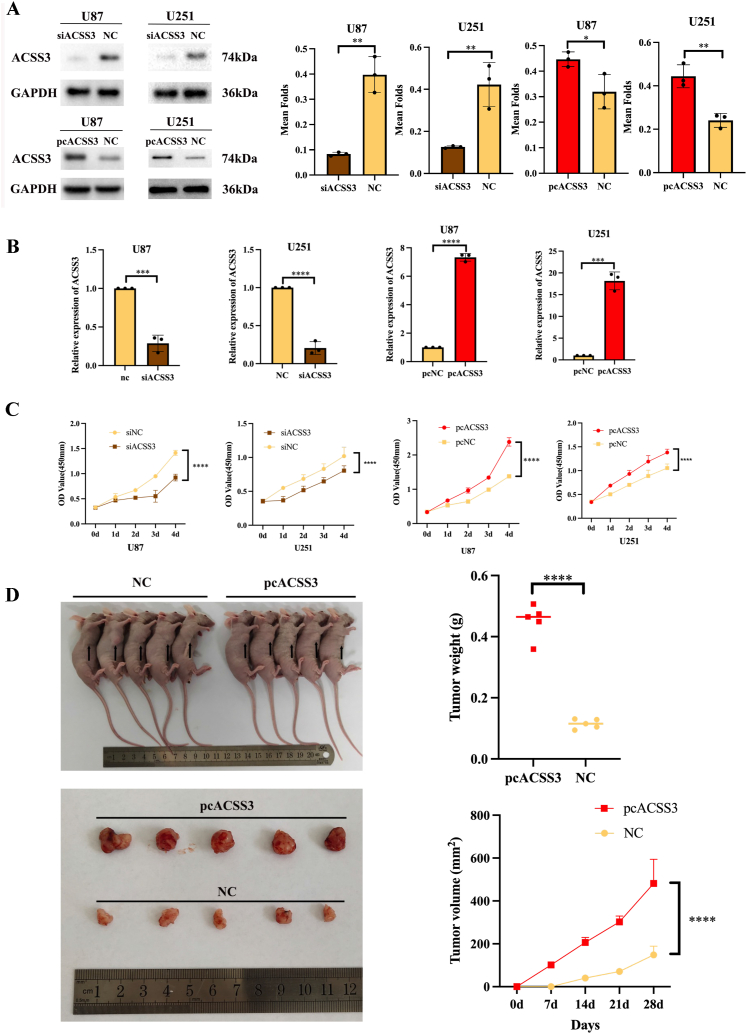
3.10. ACSS3 promotes glioma cell proliferation in vitro
U251 and U87 cells were transfected with virus or siRNA, and the ACSS3 expression level in the cells was identified utilizing RT-qPCR and Western Blotting. The ACSS3 expression level was elevated in the overexpression virus-transfected group contrasted with pcNC and siNC. Conversely, siACSS3-transfected glioma cells were significantly downregulated (Fig. 15A and B). Overexpression of ACSS3 significantly promoted cell proliferation as determined by CCK-8; on the other hand, siACSS3 led to the opposite result (Fig. 15C).
Fig. 15.
ACSS3 enhanced glioma cell proliferation. (A, B) ACSS3 was amplified and knocked out in glioma cells. (C) The CCK8 assay measured the effect of ACSS3 on glioma cell growth. (D) Xenograft Experiment of ACSS3. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
3.11. Overexpression of ACSS3 promotes the growth of glioma cells in vivo
We further performed in vivo experiments by inoculating untreated and transfected ACSS3-overexpressing U251 cells into the axilla of immunodeficient mice. Our findings indicate that the tumor volume and weight of the ACSS3 overexpression group indicated a significant elevation contrasted with the control group. After 28 days of inoculation, the tumor from the mice was removed, weighed, and photographed after being euthanized. The tumor volume of the ACSS3 overexpression group was 481.6 ± 112.60 mm3, and that of the control group was 148.00 ± 40.76 mm3. The weight of the ACSS3 overexpression group was 45.10 ± 5.53 mg, and that of the control group was 11.47 ± 1.58 mg. This indicated that the tumor weight and volume of the ACSS3 overexpression group exhibited a significant rise compared to the control group (Fig. 15D). This represented that ACSS3 promoted the U251 glioma cells' proliferative capacity.
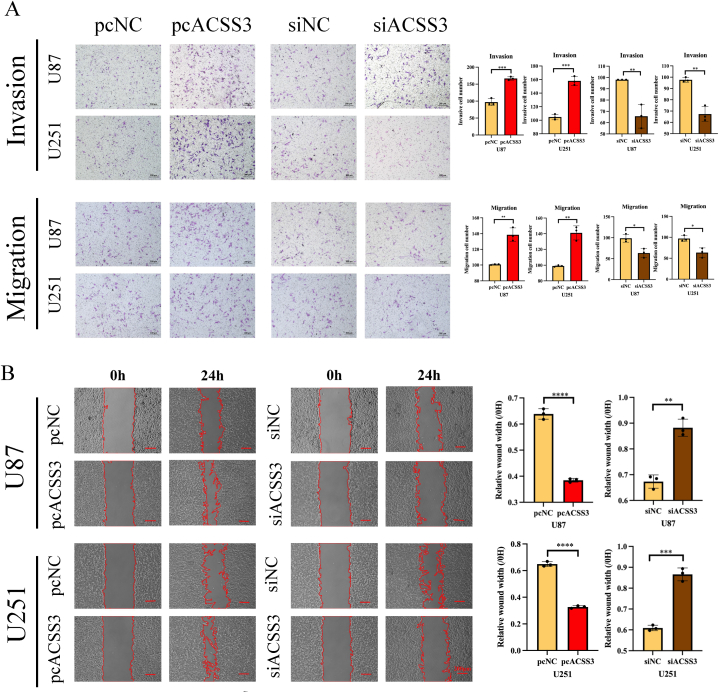
3.12. ACSS3 promotes invasion and metastasis of glioma cells in vitro
To determine ACSS3's role in cell invasion and migration, we utilized scratch wound healing, transwell invasion, and migration assays. The results showed that siACSS3 attenuated glioma cells' invasive and migratory abilities. Meanwhile, ACSS3 overexpression enhanced cell migration and invasion contrasted cells in the pcNC group (Fig. 16A and B). This suggested ACSS3 was indispensable for glioma cells' proliferative, invasive, and migratory abilities.
Fig. 16.
Effects of ACSS3 on glioma cell invasion and migration. (A) Transwell assay to evaluate invasion and migration. (B) Wound healing assay. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
4. Discussion
The metabolic needs and conditions vary among different cancer cells. Cancer often represents high metabolism, which leads to the release of many metabolic enzymes and intermediates [38]. Among them, acetyl CoA (AcCoA) is an essential metabolic intermediate in both the creation and catabolic pathways, and it is involved in generating biomass [39,40].
The balance of AcCoA is crucial for histone acetylation levels, especially in high metabolic cancers where it can participate in epigenetics, post-translational modifications, and regulation of cancer cell biomass [41]. The ACSS family primarily comprises three members: ACSS1/2/3. Nevertheless, the precise biological function of ACSS3 in cancer cells is yet to be properly validated.
ACSS3 is distributed in mitochondria and is an important prognostic biomarker for cancer [25]. We can first obtain the differential expression level of ACSS3 from a pan-cancer perspective by integrating and mining resources from TCGA and GTEx databases. We found that the ACSS3 gene is differentially expressed in different types of cancer, typically downregulated in cancer cross-analysis but significantly upregulated in gliomas. This may be because different tumor cells activate other genes, and the activation of genes varies at various stages of tumor development.
Through multi-omics and multi-database data analysis, it was found that the ACSS3 expression trend in various tumors is consistent. Based on TCGA data, we calculated the ROC curves of ACSS3 in different tumors and found that ACSS3 has satisfactory sensitivity and specificity in diagnosing various tumor patients. The 2D structure of ACSS3 gene mutation sites and the amplification and mutation of ACSS3 gene in various cancers were studied using the cBioPortal database. We observed ACSS3 mutation rates in 25 types of cancer. Melanoma has the highest mutation rate (>10 %). Through SCNA, it was found that the ACSS3 gene possesses an indispensable contribution in regulating gene expression. Subsequently, for ChIP seq samples, factors with high RP scores may regulate a given gene. We have chosen 20 transcription factors that have the greatest peak annotation scores. Our findings indicate that these elements are often situated in the promoter region of genes, suggesting their potential role in regulating gene expression.
Immunotherapy can overcome the limitations of standard treatment in various tumors. Immunotherapy is expected to become an alternative therapy when tumor patients experience recurrence or develop intolerance to traditional therapies [26,42]. Immune cells infiltrating tumors regulate tumor invasion and immune evasion, thereby influencing cancer treatment outcomes [43,44]. The Tumor Microenvironment (TME) is a complex biological system composed of tumor cells, fibroblasts, immune cells, infiltrating lymphocytes, Th2 cells, and endothelial cells. TIIC is crucial in biological processes such as tumor occurrence, malignant progression, and metastasis [43,45]. The TME contains both tumor-associated macrophages (TAMs) and neutrophils (TANs). TAMs polarize into anti-tumor (M1) or pro-tumor (M2) phenotypes based on their response to various stimuli within the microenvironment [46,47].
Similarly, the anti-tumor (N1) or primary tumor (N2) phenotype is a result of neutrophil polarization [48]. Multiple studies have shown that glioma patients' TAM and TAN grading significantly correlate with clinical prognosis [[49], [50], [51]]. ACSS3 helps to form an immunosuppressive microenvironment by regulating drug resistance phenotype, immune cell infiltration, and processes related to the cancer immune cycle. Current research indicates that Th2 cells inhibit anti-cancer immunity by secreting various cytokines [52]. Our further analysis manifests that the high ACSS3 expression is closely connected with the role of immunosuppressive cells (macrophages, Th2 cells, and neutrophils). At the same time, it was found that an increase in ACSS3 can promote tumor development, leading to enhanced tumor immune suppression, which is related to ACSS3 blocking specific steps of the cancer immune cycle. This indicates that ACSS3 can promote tumor growth by regulating the cancer immune cycle and immune cells.
ICIs as immunotherapy are a hot research topic [53,54]. CD73 and B7–H3 are presently identified as predictors of response to ICIs in GBM [34,35].
Recent studies have shown that B7–H3 and CD73 are essential markers predicting glioma ICI response [34,35]. Lower CD73 and B7–H3 are effective for prolonging survival. In our study, ACSS3 was positively correlated with CD73 and B7–H3. This suggested that ICIs would have a role in patients with lower ACSS3 expression. Utilizing TIDE to examine data from public datasets (GSE35640 and GSE78220), it was calculated that patients with lower ACSS3 responded better to immunotherapy. This was the same result in our validation utilizing ImmuCellAI and TIDE databases.
We obtained the expression of ACSS3 on many single-cell datasets using the TISCH database. Then, the heatmap showed the relative expression level and visualization of ACSS3 in the TME and immune infiltration in pan-cancer cells. This indicates that ACSS3 is widely expressed in various immune and malignant cells. For example, in gliomas (glioma _GSE1102130), the UMAP plot shows the expression of ACSS3 in glioma cells, such as AC-like and OC-like malignancy. And use 10 × Genomics and Smartseq2 platform were implemented to determine the ACSS3 expression in gliomas (glioma _GSE131928).
Based on the high and low ACSS3 expression levels, differential genes were selected for KEGG analysis to determine the conserved functions or related biological pathways involved in pan-cancer. Immune, metabolic, and signaling pathways are significantly activated. Functional proteomics, such as expression and modification, is crucial in studying complex diseases like cancer. We used Pearson correlation analysis to analyze the relationship between 14 cancer biomarkers and 14 tumor-related pathway scores, as well as ACSS3. Among them, the scores for angiogenesis, cell apoptosis, invasion, and metastasis were higher and positively correlated with other scores. Based on the 30 % differential expression of ACSS3 at the top and bottom, each tumor type specimens were allocated into two groups, and GSEA was conducted, which identified the activation or suppression of 85 metabolic gene sets out of 50 marker gene sets. All of these indicate that ACSS3 may be related to metabolic disorders in tumors.
We conducted an analysis to determine the possible relationship between the sensitivity of chemotherapeutic drugs and the ACSS3 expression using three databases (CTRP, GDSC, and Cellminer). Using the CTRP and GDSC datasets, we discovered a significant and favorable association between ACSS3 and several medications. ACSS3 may be a potential chemotherapy-sensitive gene. X4.5. dianilinophthalimide exhibited significantly mitigated scores in most cancer types, indicating that they may suppress gene-mediated cancer-promoting implications. This discovery will help guide clinical drug selection and patient prognosis.
The ACS family possessed an involvement in lipid metabolism [55]. The abnormal accumulation of lipids in cells usually occurs in lipid droplets (LDs), which contain triglycerides and cholesterol esters in the core. Literature has shown that ACSS3 affects tumor progression by reducing lipid accumulation within the tumor [11]. Abnormal metabolism contributes to glioma pathogenesis [56]. The latest literature shows that acss3 may involved in the glioblastoma (GBM) progression and recurrence [57]. However, no further in vitro and in vivo experiments were conducted based on the outcomes of differential analysis, combined with our own experience in neurosurgery and pathological tissues from 78 cases of glioma at the Second Hospital of Hebei Medical University. We chose glioma specimens as the basis for further research. Gliomas are the most well-known and lethal primary brain tumors, are highly heterogeneous and aggressive, and, as a rule, have a poor prognosis. Although traditional treatments and new immunotherapeutic technologies are developing continuously, molecular diagnosis plays an indispensable role in treating glioma. Be that as it may, more reliable biomarkers are still needed for early diagnosis and prediction of treatment effects.
Consequently, finding new biomarkers is the ongoing concentration and difficulty of glioma treatment. Studies have shown that ACSS3 is essential to tumor development and metastasis in various cancers. ACSS3 is thought to be distributed in mitochondria and is an important prognostic biomarker for cancer [24]. This study is the first to systematically evaluate the role of ACSS3 in glioma and explore its function in the glioma-associated immune microenvironment.
Using the outcomes of bioinformatics analysis, we confirmed the ACSS3 expression in glioma tissues and cell lines. The findings demonstrated that ACSS3 had elevated expression levels in several glioma cell lines as contrasted with Normal human astrocytes (NHA). Furthermore, the ACSS3 expression in glioma tissue is significantly higher as compared to normal brain tissue next to the tumor. The significant rise in this expression indicates that ACSS3 may have a function not only in the glioma growth but also in the recurrence phase of glioma. Meanwhile, we experimentally verified that knocking down ACSS3 with siRNA can inhibit glioma cell migration and invasion, while ACSS3 overexpression produces the opposite consequences.
Further analysis of 78 cases of GBM revealed that patients with glioma in the high-ACSS3 group showed a significantly superior overall survival rate contrasted with those in the low-ACSS3 group. This aligns with the previously documented role of ACSS3 in some types of cancer. ACSS3 has high expression levels in gastric cancer tumors, and the ACSS3 downregulation hampers the advancement of gastric cancer [12]. ACSS3 expression in mitochondria is capable of particularly identifying a subtype of hepatocellular carcinoma known as iHCC2, which is associated with poor clinical survival. This suggests that increased ACSS3 expression may contribute to the advancement of hepatocellular carcinoma [58]. Nevertheless, investigations have shown that ACSS3 overexpression might impede the PC3 cells proliferation. ACSS3 can hinder the progression of CRPC and reverse the resistance of Enzalutamide by reducing the deposition of intertumoral lipid droplets (LD) [11]. This illustrates the diverse roles that ACSS3 plays in various kinds of malignancies. The variability in the activity of ACSS3 in different malignancies is likely due to its dependence on distinct activation states and substrate binding. This variability ultimately impacts the functions and signaling pathways associated with the substrate. This may be related to gene interactions, epigenetic changes, environmental factors, and changes in penetrance caused by randomness. The abnormal mRNA expression of ACSS3 in various cancers may cause metabolic abnormalities in tumors. Therefore, further research is needed on the other ACSS3 functions in various tumor forms.
The precise molecular mechanism behind the ACSS3 function in normal tissues and its role in the development and advancement of cancer remain incompletely comprehended and necessitate more investigation. Nevertheless, the correlation between ACSS3 and certain tumor progression and tumor-related pathways might provide vital insights into comprehending its distinct functioning processes in cancer. The functional outcomes of GSEA suggest that ACSS3 has the capacity to impact the development and advancement of cancer via diverse methods, including inflammatory responses, such as L6 JAK STAT3 Signaling and L2 STAT5 Signaling. It is related to propionic acid metabolism, tryptophan metabolism, and tyrosine metabolism in metabolic pathways. At the same time, ACSS3 positively correlates with KRAS Signaling regulation in some tumors, indicating that ACSS3 can regulate cancer cells' migration and invasion ability. Moreover, the raised ACSS3 expression is positively linked to EPITHELIAL MESENCHYMAR TRANSATION (EMT) in BLCA, CESC, COAD, ESCA, GBM, HNSC, LAML, LGG, LUSC, PAAD, PRAD, READ, SKCM, STAD, TGCT, THYM, and UCEC patients. The link between high levels of ACSS3 and cancer recurrence may be attributed to the increased susceptibility of individuals with elevated ACSS3 levels in certain forms of cancer. In addition, our Gene Set Variation Analysis (GSVA) revealed that ACSS3 is widely involved in multiple pathways. It is worth noting that ACSS3 plays a role in stress-induced tumor responses, such as Apoptosis, Angiogenesis, Hypoxia, Inflammation, Metastasis, EMT, and Stemness signals. These cumulative findings indicate the multifaceted involvement of ACSS3 in cancer development.
ACSS3, as a member of the short-chain family of acetyl CoA synthase, acetyl CoA (AcCoA) is a crucial metabolic intermediate in synthetic and catabolic pathways. During the development of cancer, abnormal metabolic conditions often accompany it, which may lead to stress caused by hypoxia and nutrient deficiency. Cancer cells must adapt to the nutrient-deficient TME [59,60]. Therefore, metabolism-related acetyl CoA has received particular attention in cancer research. Under energy-deficient conditions in cancer, hypoxia-inducible factor 1 (HIF-1) can reduce acetyl CoA [61]. Mitochondria are the “metabolic checkpoint” organelles that perceive cellular energy fluctuations [62]. Among them, ACSS3 from mitochondria may assist in re-supplying acetyl CoA to cancer cells. We found in GSVA that ACSS3 is negatively correlated with oxidative pathways in various tumors and positively correlated with Hypoxia. On the other hand, ACSS3 is essential for utilizing the environment and intracellular acetate [13]. We will further investigate its specific mechanisms of action in normal tissues and various cancers in the future.
Due to the primary localization of ACSS3 in mitochondria, its high expression in gliomas may differ from other tumors in its mechanism of utilizing acetate to promote histone acetylation. The possible reason may be that the recycling mechanism of endogenous acetate is different, which requires further exploration.
Although we have described in detail the ACSS3 function in pan-cancer, some limitations should be considered. Firstly, most of our analysis is based on bioinformatics analysis, and ACSS3 expression has only been experimentally validated in gliomas. Secondly, our study did not investigate the molecular mechanism of ACSS3 in cancer. Additional investigation is needed to ascertain the expression mechanism of ACSS3 in tumors in the future. At the same time, we need extensive multicenter prospective studies to improve the clinical application of our research results.
5. Conclusion
Our study comprehensively elucidates the role of ACSS3 expression as a novel biomarker in forecasting the prognosis of different human cancers, as it can influence the biological process by modulating the immune microenvironment. ACSS3 was also a critical prognostic factor for glioma and related to its proliferation, migration, and invasion.
Ethics statement
The Ethics Committee of the Second Hospital of Hebei Medical University reviewed and approved studies involving human participants (2022-R704, 2022/09/01). The ethics committee waived the requirement of written informed consent for participation. The Ethics Committee of the Second Hospital of Hebei Medical University reviewed and approved the animal study (2022-AE276, 2022/09/01) and strictly followed National Institutes of Health guidelines for animal care and use. This study is reported following the ARRIVE guidelines (https://arriveguidelines.org).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Funding
This research was supported by the Youth Fund of the Hebei Natural Science Foundation of China (H2021206027) and the Hebei Medical Science Research Project of China (20230033).
Abbreviations
The information is shown in Table 1.
CRediT authorship contribution statement
Zhanzhan Zhang: Writing – original draft, Visualization, Supervision, Investigation, Data curation. Hongshan Yan: Software, Resources, Methodology, Formal analysis, Data curation. Hao Tong: Validation, Software, Investigation, Data curation. Kai Guo: Validation, Software. Zihan Song: Supervision, Software, Data curation. Qianxu Jin: Validation, Software, Resources. Zijun Zhao: Software, Resources. Zongmao Zhao: Writing – review & editing, Validation, Supervision. Yunpeng Shi: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Methodology, Data curation.
Declaration of competing interest
We declare that we have no financial or personal relationships with others or organizations that can inappropriately influence our work. There is no professional or other personal interest of any nature or kind in any product, service, and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled, “ACSS3 is a potential biomarker in glioma and is related to the tumor immune microenvironment”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35231.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ferlay J., et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019 Apr 15;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.A Chukasemrat N., Charakorn C., Lertkhachonsuk A.A. The use of complementary and alternative medicine in Thai gynecologic oncology patients: influencing factors. Evid Based Complement Alternat Med. 2021 Nov 10;2021 doi: 10.1155/2021/1322390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farolfi A.etal. Immune system and DNA repair defects in ovarian cancer: implications for locoregional approaches. Int. J. Mol. Sci. 2019 May 25;20(10):2569. doi: 10.3390/ijms20102569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P., Hu-Lieskovan S., Wargo J.A., Primary A. Ribas. Adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raemaekers T., Ribbeck K., Beaudouin J., Annaert W., Van Camp M., Stockmans I., et al. NuSAP is a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 2003;162:1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbet C., Feron O. Tumour acidosis: from the passenger to the driver's seat. Nat. Rev. Cancer. 2017 Oct;17(10):577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 7.Shi L., Tu B.P. Acetyl-CoA, and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metabol. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Schug Z.T., Peck B., Jones D.T., Zhang Q., et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015 Jan 12;27(1):57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mashimo T., Pichumani K., Vemireddy V., Hatanpaa K.J., Singh D.K., Sirasanagandla S., Nannepaga S., Piccirillo S.G., Kovacs Z., Foong C., Huang Z., Barnett S., Mickey B.E., DeBerardinis R.J., Tu B.P., Maher E.A., Bachoo R.M. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014 Dec 18;159(7):1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L., Song Z., Hu J., Liu L., Hou Y., Zhang X., Yang X., Chen K. ACSS3 represses prostate cancer progression through downregulating lipid droplet-associated protein PLIN3. Theranostics. 2021 Jan 1;11(2):841–860. doi: 10.7150/thno.49384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang W., Cheng W., Cheng B., et al. Mitochondrial acetyl- CoA synthetase 3 is a biosignature of gastric cancer progression. Cancer Med. 2018;7(4):1240–1252. doi: 10.1002/cam4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Duan H., Feng Z., Han X., Gu C. Acetyl-CoA synthetase 3 promotes bladder cancer cell growth under metabolic stress. Oncogenesis. 2020;9(5):1–10. doi: 10.1038/s41389-020-0230-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Louis D.N., Perry A., Reifenberger G., et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016 Jun;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 15.Zachariah M.A., Oliveira-Costa J.P., Carter B.S., Stott S.L., Nahed B.V. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol. 2018;20(9):1155–1161. doi: 10.1093/neuonc/noy074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T.C., da Fonseca C.O., Schönthal A.H. Intranasal perillyl alcohol for glioma therapy: molecular mechanisms and clinical development. Int. J. Mol. Sci. 2018;19(12):3905. doi: 10.3390/ijms19123905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng S., Xu Z., Liang Q., Thakur A., Liu Y., Zhou S., Yan Y. The prognostic gene CRABP2 affects drug sensitivity by regulating docetaxel-induced apoptosis in breast invasive carcinoma: a pan-cancer analysis. Chem. Biol. Interact. 2023 Mar 1;373 doi: 10.1016/j.cbi.2023.110372. [DOI] [PubMed] [Google Scholar]
- 18.Dastjerdi S., Haghparast A., Amroabadi J.M., Dolatabadi N.F., Mirzaei S., Zamani A., Hashemi M., Mahdevar M., Ghaedi K. Elevated CDK5R1 expression associated with poor prognosis, proliferation, and drug resistance in colorectal and breast malignancies: CDK5R1 as an oncogene in cancers. Chem. Biol. Interact. 2022 Dec 1;368 doi: 10.1016/j.cbi.2022.110190. [DOI] [PubMed] [Google Scholar]
- 19.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 May;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013 Dec;8(12):2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ru B., Wong C.N., Tong Y., Zhong J.Y., Zhong S.S.W., Wu W.C., Chu K.C., Wong C.Y., Lau C.Y., Chen I., Chan N.W., Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019 Oct 15;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 22.Xu L., Deng C., Pang B., Zhang X., Liu W., Liao G., Yuan H., Cheng P., Li F., Long Z., Yan M., Zhao T., Xiao Y., Li X. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res. 2018 Dec 1;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M., Li X., Chen Y., Wang S. MD2 is a potential biomarker associated with immune cell infiltration in gliomas. Front. Oncol. 2022 Mar 17;12 doi: 10.3389/fonc.2022.854598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020 Jul 2;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimura Y., Araki A., Maruta H., Takahashi Y., Yamashita H. Molecular cloning of rat ACSS3 and characterization of mammalian propionyl-CoA synthetase in the liver mitochondrial matrix. J. Biochem. 2017 Mar 1;161(3):279–289. doi: 10.1093/jb/mvw067. [DOI] [PubMed] [Google Scholar]
- 26.Preusser M., Lim M., Hafler D.A., Reardon D.A., Sampson J.H. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat. Rev. Neurol. 2015;11:504–514. doi: 10.1038/nrneurol.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021 Jul 1;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Akbani R., Zhao W., Lu Y., Weinstein J.N., Mills G.B., Liang H. Explore, visualize, and analyze functional cancer proteomic data using the cancer proteome Atlas. Cancer Res. 2017 Nov 1;77(21):e51–e54. doi: 10.1158/0008-5472.CAN-17-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E., Chuang H.Y., Kim J.W., Ideker T., Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput. Biol. 2008 Nov;4(11) doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C.J., Hu F.F., Xie G.Y., Miao Y.R., Li X.W., Zeng Y., Guo A.Y. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic, and immunogenomic levels. Briefings Bioinf. 2023 Jan 19;24(1) doi: 10.1093/bib/bbac558. [DOI] [PubMed] [Google Scholar]
- 31.Reinhold W.C., Wilson K., Elloumi F., Bradwell K.R., Ceribelli M., Varma S., Wang Y., Duveau D., Menon N., Trepel J., Zhang X., Klumpp-Thomas C., Micheal S., Shinn P., Luna A., Thomas C., Pommier Y. CellMinerCDB: NCATS is a web-based portal integrating public cancer cell line databases for pharmacogenomic explorations. Cancer Res. 2023 Jun 15;83(12):1941–1952. doi: 10.1158/0008-5472.CAN-22-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N., Kamińska B., Huelsken J., Omberg L., Gevaert O., Colaprico A., Czerwińska P., Mazurek S., Mishra L., Heyn H., Krasnitz A., Godwin A.K., Lazar A.J., Cancer Genome Atlas Research Network, Stuart J.M., Hoadley K.A., Laird P.W., Noushmehr H., Wiznerowicz M. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018 Apr 5;173(2):338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C., Zhang H., Chen M., Wang S., Qian R., Zhang L., Huang X., Wang J., Liu Z., Qin W., Wang C., Hang H., Wang H. A survey of optimal strategy for signature-based drug repositioning and an application to liver cancer. Elife. 2022 Feb 22;11 doi: 10.7554/eLife.71880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goswami S., Walle T., Cornish A.E., et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat. Med. 2020;26:39–46. doi: 10.1038/s41591-019-0694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duerinck J., Schwarze J.K., Awada G., et al. Intracerebral administration of CTLA-4 and PD-1 immune checkpoint blocking monoclonal antibodies in patients with recurrent glioblastoma: a phase I clinical trial. J Immunother Cancer. 2021 Jun;9(6) doi: 10.1136/jitc-2020-002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Akbani R., Zhao W., Lu Y., Weinstein J.N., Mills G.B., Liang H. Explore, visualize, and analyze functional cancer proteomic data using the cancer proteome Atlas. Cancer Res. 2017 Nov 1;77(21):e51–e54. doi: 10.1158/0008-5472.CAN-17-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malta T.M., Sokolov A., Gentles A.J., et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018 Apr 5;173(2):338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corbet C., Feron O. Tumour acidosis: from the passenger to the driver's seat. Nat. Rev. Cancer. 2017 Oct;17(10):577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 39.Shi L., Tu B.P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol. 2015 Apr;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metabol. 2015 Jun 2;21(6):805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Falkenberg K.J., Johnstone R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases, and immune disorders. Nat. Rev. Drug Discov. 2014 Sep;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 42.Xu S., Tang L., Li X., Fan F., Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12. doi: 10.1016/j.canlet.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Anderson N.M., Simon M.C. vol. 30. 2020. pp. R921–R925. (The Tumor Microenvironment. CurrBiol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinshaw D.C., Shevde L.A. The tumor MicroenvironmentInnately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017 Mar 13;31(3):326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J., Bae J.S. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediat. Inflamm. 2016;2016 doi: 10.1155/2016/6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guadagno E., Presta I., Maisano D., Donato A., Pirrone C.K., Cardillo G., et al. Role of macrophages in brain tumor growth and progression. Int. J. Mol. Sci. 2018;19:1005. doi: 10.3390/ijms19041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fridlender Z.G., Albelda S.M. Tumor-associated neutrophils: friend or foe? Carcinogenesis. 2012;33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- 49.Rahbar A., Cederarv M., Wolmer-Solberg N., Tammik C., Stragliotto G., Peredo I., et al. Enhanced neutrophil activity is associated with shorter time to tumor progression in glioblastoma patients. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2015.1075693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fossati G., Ricevuti G., Edwards S.W., Walker C., Dalton A., Rossi M.L. Neutrophil infiltration into human gliomas. Acta Neuropathol. 1999;98:349–354. doi: 10.1007/s004010051093. [DOI] [PubMed] [Google Scholar]
- 51.Zhai H., Heppner F.L., Tsirka S.E. Microglia/macrophages promote glioma progression. Glia. 2011;59:472–485. doi: 10.1002/glia.21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen T., Guo J., Cai Z., et al. Th9 cell differentiation and its dual effects in tumor development. Front. Immunol. 2020;11:1026. doi: 10.3389/fimmu.2020.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong Y., Ma Z., Wang F., et al. In vivo molecular imaging for immunotherapy using ultra-bright near-infrared-IIb rare-earth nanoparticles. Nat. Biotechnol. 2019;37:1322–1331. doi: 10.1038/s41587-019-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agrawal L., Bacal A., Jain S., et al. Immune checkpoint inhibitors and endocrine side effects, a narrative review. PGM (Postgrad. Med.) 2020;132:206–214. doi: 10.1080/00325481.2019.1709344. [DOI] [PubMed] [Google Scholar]
- 55.Jia Z., Chen X., Chen J., Zhang L., Oprescu S.N., Luo N., Xiong Y., Yue F., Kuang S. ACSS3 in brown fat drives propionate catabolism and its deficiency leads to autophagy and systemic metabolic dysfunction. Clin. Transl. Med. 2022 Feb;12(2) doi: 10.1002/ctm2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W., Jia H., Li Q., Cui J., Li R., Zou Z., Hong X., Glycerophosphatidylcholine P.C. (36:1) absence and 3'-phosphoadenylate (pAp) accumulation are hallmarks of the human glioma metabolome. Sci. Rep. 2018 Oct 3;8(1) doi: 10.1038/s41598-018-32847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui X., Zhao J., Li G., Yang C., Yang S., Zhan Q., Zhou J., Wang Y., Xiao M., Hong B., Yi K., Tong F., Tan Y., Wang H., Wang Q., Jiang T., Fang C., Kang C. Blockage of EGFR/AKT and mevalonate pathways synergize the antitumor effect of temozolomide by reprogramming energy metabolism in glioblastoma. Cancer Commun. 2023 Dec;43(12):1326–1353. doi: 10.1002/cac2.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bidkhori G., Benfeitas R., Klevstig M., Zhang C., Nielsen J., Uhlen M., Boren J., Mardinoglu A. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc. Natl. Acad. Sci. U.S.A. 2018 Dec 11;115(50):E11874–E11883. doi: 10.1073/pnas.1807305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamphorst J.J., Chung M.K., Fan J., Rabinowitz J.D. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals a substantial contribution from acetate. Cancer Metabol. 2014 Dec 11;2:23. doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., Kelleher J.K., Vander Heiden M.G., Iliopoulos O., Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011 Nov 20;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang D., Li T., Li X., Zhang L., Sun L., He X., Zhong X., Jia D., Song L., Semenza G.L., Gao P., Zhang H. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014 Sep 25;8(6):1930–1942. doi: 10.1016/j.celrep.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 62.Green D.R., Galluzzi L., Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014 Sep 19;345(6203) doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.