Abstract
The Illuminating the Druggable Genome (IDG) consortium generated reagents, biological model systems, data, informatic databases, and computational tools. The Resource Dissemination and Outreach Center (RDOC) played a central administrative role, organized internal meetings, fostered collaboration, and coordinated consortium-wide efforts. The RDOC developed and deployed a Resource Management System (RMS) to enable efficient workflows for collecting, accessing, validating, registering, and publishing resource metadata. IDG policies for repositories and standardized representations of resources were established, adopting the FAIR (findable, accessible, interoperable, reusable) principles. The RDOC also developed metrics of IDG impact. Outreach initiatives included digital content, the Protein Illumination Timeline (representing milestones in generating data and reagents), the Target Watch publication series, the e-IDG Symposium series, and leveraging social media platforms.
Keywords: druggable genome, understudied proteins, FAIR data, metadata management, resource dissemination, outreach
Teaser
The RDOC was responsible for administering the IDG consortium and disseminating its resources. Best practices for resource management, outreach, and evaluating impacts are reported.
Introduction
The Illuminating the Druggable Genome (IDG) program was initiated by the National Institutes of Health (NIH) Common Fund with the goal of characterizing and illuminating the functions and properties of proteins and genes that have been historically understudied but have the potential to become targets for drug development.1 The Resource Dissemination and Outreach Center (RDOC) of the IDG consortium served as the internal coordinator and facilitated the external dissemination of IDG resources generated by the Data and Resource Generation Centers (DRGCs). As the administrative core for IDG, the RDOC organized and coordinated internal IDG meetings, such as steering committee and working group teleconferences, as well as the annual IDG consortium meetings, with the aim of building a cohesive, collaborative, and well-managed program. This was the basis from which the RDOC was then able to display the outcomes for the IDG consortium to the larger scientific community in practical, useful, and reproducible workflows and mechanisms. To achieve this, the RDOC developed and deployed informatics infrastructure and systems to streamline the end-to-end management of IDG-developed products and tools. This included policies and reporting guidelines for standardized representations of resources and preferred repositories for data and reagents. The RDOC also monitors the impact of IDG’s work and resources. The RDOC tracks the number of references to IDG publications and their impact factor. Additionally, the RDOC keeps records of orders from commercial vendors, attendance at webinars and symposiums, and the initiation of additional collaborations and research opportunities, such as the IDG-DREAM competition and international foundation funding calls. The RDOC also tracks the usage and access of IDG’s web-based tools.
Resource submission and management infrastructure
The IDG consortium and RDOC have strived to adhere to the best practices for the management and stewardship of data, including the findable, accessible, interoperable, and reusable (FAIR) principles2 and numerous policies to govern data annotations, submissions, sharing, and quality control (QC) metrics. To facilitate the FAIRness of the IDG resources, the RDOC has led the development of metadata standards specifications for a variety of resources generated by the consortium. These specifications were stored and managed within a PostgreSQL Metadata Specifications Database, and they describe the resources in terms of: (i) the consortium-wide metadata standards specifications, (ii) resource descriptors for the Pharos representation of the IDG resources,3,4 and (iii) information specific to the Protein Illumination Timeline (PIT)5 milestones to report the IDG progress to communities. The metadata standard specifications were further formalized into human- and machine-readable JSON schemas to encode required data fields and their allowed values, including terminology and ontologies, along with other information.
The RDOC has developed and deployed the Resource Submission System (RSS) and Resource Management System (RMS) to collect, validate, process, and manage the metadata for generated resources, and to enable their dissemination via Pharos and the IDG Website (https://druggablegenome.net/).6 The RSS enables submission of metadata and standardized descriptions of IDG resources in two ways: (i) the information can be captured via Center for Expanded Data Annotation and Retrieval (CEDAR)7 forms (this option is suitable for single-record submissions); and (ii) as batch submissions, where the RSS offers a set of CSV templates that can be stored locally, populated, and uploaded to the RSS database. For batch submission, the RSS enables mapping of the headers in the submission file to the established IDG metadata specifications to ensure data standardization and harmonization. The JSON schemas were used to generate the submission forms (implemented via the CEDAR platform) and CSV templates, and to enforce the metadata validation based on the encoded rules (e.g. required vs optional metadata specification, controlled vocabulary term, ontology term, etc.). Validation is enabled via integration with BioPortal for the ontologies [e.g. Phenotypic Quality Ontology (PATO), Experimental Factor Ontology (EFO), BioAssay Ontology (BAO)] and internally in the RSS and CEDAR for controlled vocabularies (e.g. WB, IHC, IF, IP, or Flow term for antibody usage; Monoclonal or Polyclonal for antibody clonality, etc.).
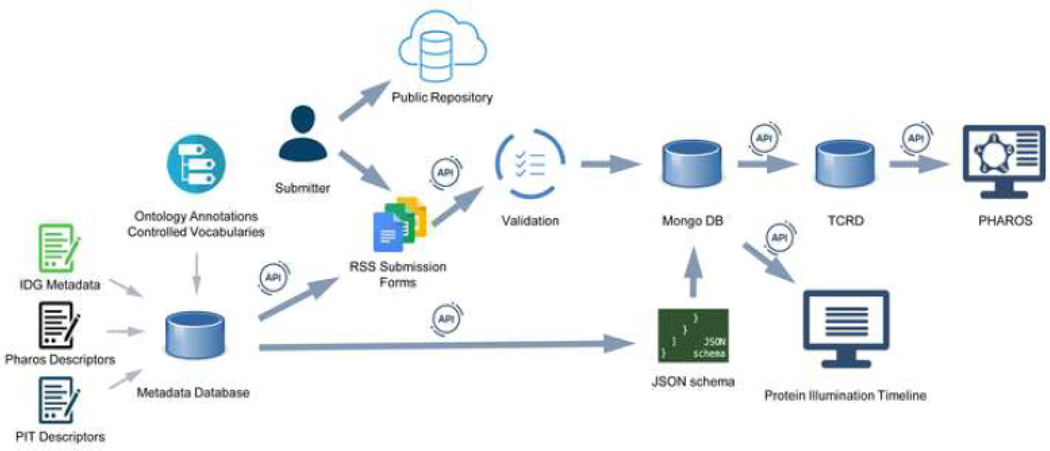
The submitted and validated information is further stored in Mongo DB, where each submitted record is saved as a formalized JSON for Linking Data (JSON-LD) instance that is based on the corresponding JSON schema. The information is disseminated via RSS application programming interface (API) endpoints that enable database queries in terms of genes, resource types, resource names, and completeness of the resource descriptions required for Pharos representation (Pharos-ready predefined annotations required during submission). The database is exported to the Target Central Resource Database (TCRD),8 and the records are subsequently displayed in Pharos for end-user access. The RSS also provides an API to PIT to visualize available data by utilizing metadata specifications related to specific illumination timelines and their corresponding milestones and statuses. Figure 1 illustrates the data flow through the RSS and RMS infrastructure.
FIGURE 1.
Illustration of metadata management and submission workflow, supported by the IDG RSS and RMS infrastructure and disseminated via Pharos and PIT.
In addition, and as a part of the implementation of the FAIR principles, the IDG metadata standard specifications have been released and shared via the IDG Website and registered at FAIRsharing.org.9,10 The IDG project is registered under FAIRsharing record ID 3523.11 This record is associated with the other IDG FAIRsharing-registered records, such as the IDG Consortium policies12 and corresponding metadata standards for IDG resources, such as small-molecules standards,13 mouse strains standards,14 antibody standards,15 etc.
Resource repositories and tracking
The choice of repository can have a significant impact on the availability of material and digital resources. For example, a data repository should provide reliable and consistent access to datasets and their descriptions, implement stable and well-formed identifiers, and facilitate the FAIR principles. The RDOC, together with the IDG consortium, has considered various repositories for reagents and datasets. The agreed-upon repositories are well established across industry and research communities and align with best practices in terms of resource handling, storage, accessibility, and security. Specifically, for the reagents, commonly used vendors that register and distribute the resources were chosen as the primary reagent repositories in the IDG consortium, e.g. for mouse strains, the repository of choice was the Mutant Mouse Resource and Research Center (MMRRC),16,17 while for the genetic constructs (i.e. plasmids) the repository of choice was AddGene,18 commonly used across communities. In addition to reagent distribution, most of the selected repositories also generate reagent identifiers (e.g. AddGene or MMRRC ID) that are integrated via the SciCrunch19 platform as Research Resource Identifiers (RRIDs).20 The RRIDs are unique identifiers assigned to research resources, including antibodies, cell lines, model organisms, and software tools. The RRIDs are composed of an abbreviation of the repository name and the corresponding resource ID. By including RRIDs in publications, authors can provide unambiguous references to the research materials and tools used, thereby making it easier for others to replicate their experiments and build upon the research findings. For the IDG reagent resources, the RRIDs have been inherently implemented by registering mouse strains, antibodies, genetic constructs, and cell reagents in the corresponding repositories. The RDOC worked directly with these repositories and SciCrunch to annotate the IDG resources for unique identification and tracking. In addition to unique identification via SciCrunch RRIDs, novel small molecules were distributed via well-known and established commercial vendors, and registered in public chemical databases such as PubChem, Chemical Entities of Biological Interest (ChEBI),21,22 and ZINC.23–25 Small-molecule chemical probes, defined by the criteria established by the Structural Genomics Consortium,26 were registered in the Chemical Probes Portal27,28 along with the supporting data and information. IDG-generated peptides were distributed via the commercial vendor New England Peptide29 and tracked by the vendor catalog identifiers, while NeuroMab30 was used for the antibodies in the same manner. Cells were distributed via the University of California San Francisco (UCSF) Darkmatter website31 and registered under the Cell Line Ontology (CLO)32 and Cellosaurus.33
The selection of data repositories received the same scrutiny by the RDOC and IDG consortium members and, similar to reagent repositories, well-established repositories such as PRoteomics IDEntifications (PRIDE),34 Gene Expression Omnibus (GEO),35 International Mouse Phenotyping Consortium (IMPC),36 and protocols.io37 were chosen to submit IDG-generated datasets. The complete list of repositories by IDG resource types (reagents and data) is shown in Table 1.
TABLE 1.
Selected repositories of IDG resources
| IDG resource category | IDG resource repository | Resource repository link |
|---|---|---|
| Antibodies | NeuroMab | neuromab.ucdavis.edu/ |
| Cells | UCSF Darkmatter; CLO, Cellosaurus | darkmatter.ucsf.edu/cells; ontobee.org/ontology/CLO; cellosaurus.org |
| Small molecules | Commercial vendors; ChEBI/ZINC | www.ebi.ac.uk/chembl/; zinc15.docking.org/ |
| Genetic construct | AddGene | www.addgene.org/ |
| Mouse | MMRRC | www.mmrrc.org/ |
| Peptides | New England Peptide | www.newenglandpeptide.com/ |
| Probes | Chemical Probes Portal | www.chemicalprobes.org/ |
| Affinity purification mass spectrometry | PRIDE | www.ebi.ac.uk/pride/ |
| Chemical tool | ChEBI | www.ebi.ac.uk/chebi/ |
| Expression | GEO | www.ncbi.nlm.nih.gov/gds |
| Immunohistochemistry | UCSF Darkmatter | darkmatter.ucsf.edu/ |
| Ion channel activity | UCSF Darkmatter | darkmatter.ucsf.edu/ |
| Mouse image-based expression | AMIS; UCSF Darkmatter | amis.docking.org/;_darkmatter.ucsf.edu/ |
| Mouse phenotype | IMPC | www.mousephenotype.org/ |
| NanoBRET | Synapse | www.synapse.org/ |
| Proteomics | PRIDE | www.ebi.ac.uk/pride/ |
| Protocol | Protocols.io | www.protocols.io/ |
AMIS, A Mouse Imaging Server.
The IDG consortium required DRGCs to submit and register their generated resources to the selected above-listed repositories and to subsequently submit the resource descriptions and corresponding metadata, including the repositories and vendor identifiers, to the RDOC via the RSS. The repositories, submission processes, and timelines were specified in the IDG consortium policies to govern data sharing including resource tracking and dissemination.
Protein illumination timeline
As the IDG initiative commenced and the different DRGCs started producing various outcomes, it was requested that the RDOC aid in sharing these results in a manner that provided a general overview and details of the progress toward specific milestones related to illuminating understudied targets that were studied by the DRGCs. The visualization of progress was required to be amenable to the evolving methodologies and technologies utilized by the various DRGCs. The PIT was conceived to be a solution allowing the scientific community to observe the progress toward illumination of the different IDG targets based on the protein family. PIT tables were developed in collaboration with the various DRGC awardees to outline the specifications for achieving illumination milestones. The RDOC coordinated this process, which included modifications of metadata to accurately reflect the progress toward a protein’s illumination in accordance with feedback from DRGC awardees. The milestone specifications defined during this process were further organized and formally described, conforming to the IDG metadata specifications. The RDOC integrated these requirements into the RMS for the collection of PIT annotation elements via RSS templates for each stage of the target illumination journey. Specifically, each milestone could be described as ‘In progress’ or ‘Complete’. Validation functionality was integrated into the RSS and included terminology, milestone specifications, and requirement-related rules. Together, these components and functionalities of the RSS enabled submission of verified relevant PIT information. Once the information had been submitted and verified as valid, the JSON-LD records were generated and shared via the RSS API. The RSS JSON-LD conforms to the W3C Simple Knowledge Organization System (SKOS) vocabulary,38 as well as the Dublin Core39 and the W3C Resource Description Framework (RDF) schema.40 For example, the RSS tissue JSON-LD code is in the following form:
{
…
“skos”: “http://www.w3.org/2004/02/skos/core#”,
“rdfs”: “http://www.w3.org/2000/01/rdf-schema#”,
“Tissue_ID”: “http://purl.org/dc/elements/1.1/identifier”,
“Name”: “http://purl.org/dc/elements/1.1/title”,
…
}
The user interface for the PIT takes advantage of the RSS API for dynamic updating and incorporation of the various IDG-generated resources in the display of PIT on the IDG website. Users can search, filter, sort, and download PIT tables as CSV files, allowing seamless integration of IDG data across multiple platforms in compliance with the FAIR criteria. In addition, the information is also available directly via the API as JSON files.
Outreach venues
To represent IDG in the wider community of scientists and beyond, the RDOC branded the IDG consortium with a lightbulb logo having a DNA helix and acquired the domain DruggableGenome.net for the main IDG consortium website. During the early phase of the IDG program, it was also pertinent to create a social media presence by establishing X (formerly Twitter; @DruggableGenome) and LinkedIn (DruggableGenome IDG) accounts. These platforms allowed for further dissemination of IDG announcements concerning resource development and outreach events. Additionally, a YouTube channel was created (@DruggableGenomeIDG)41 as a mechanism to showcase IDG presentations and tutorials. Currently the channel hosts 67 videos, with a total watch time greater than 220 hours. Some of these videos are grouped into playlists based on IDG-organized events, such as the e-IDG Symposium series and IDG Digital Tool Fest. In association with editors at Nature Reviews Drug Discovery, the RDOC was able to initiate a series of short reviews or Biobusiness Briefs focusing on highlighting the research on understudied proteins. This collection is referred to as ‘Target Watch’, and contributions to this series came from across the IDG consortium, highlighting probe development for different kinases, diseases associated with ion channels, and G protein-coupled receptor (GPCR) involvement in metabolism and the immune response. Since its inception in 2019, there have been 21 articles. One of the recurring themes for Target Watch, led by Dr. Tudor Oprea, was an annual assessment of novel drug targets. Five articles have reported the trends of small-molecule, peptide, and antibody drugs, among other types of drugs, and their mechanisms of action and targets, ranging from human to viral proteins.
Beginning in July 2019, the RDOC began direct marketing of IDG by writing and distributing monthly IDG newsletters. Via Intuit MailChimp, an established web marketing service, the RDOC was able to: (i) communicate updates regarding target illumination as new resources or manuscripts from IDG members were published; (ii) announce specific IDG-sponsored outreach events; and (iii) share funding opportunities from the NIH. The recipients of the IDG newsletters are IDG interested parties, or ‘IDG customers’, who expressed a desire to be informed of IDG developments and gave permission to receive these newsletters. Compliance with the collection of contacts and the Terms of Use, formulated by MailChimp in adherence with the General Data Protection Regulation (GDPR), were implemented and included the audience’s right to be removed from the contact list. In all, 50 monthly newsletters have been shared.
Another aim for the RDOC was to expand IDG outreach by facilitating and coordinating meetings with other consortia and research groups. Due to the diversity of the IDG consortium constituents, there were a wide variety of experimental and informatics groups with which the RDOC forged alliances. These meetings evolved into an assortment of multiday in-person symposia with oral and poster presentations, as well as virtual events with multiple short demonstrations. The main objectives for these meetings were to allow attendees to share their illumination work and facilitate the exchange of ideas to enrich the potential of future collaborations. The success of these meetings was in part due to the active participation by IDG principal investigators (PIs), who served as leads or cohosts. The various categories of outreach that the RDOC organized42 are listed below.
Showcase/community building
Public meetings with presentations from all consortium members allowed for a complete display of all elements of the program. These types of meetings serve as great introductions to both internal and external group members. An example of this type of outreach meeting was the IDG Outreach meeting at the University of North Carolina (UNC) in September 2020.
Conference exposure
To raise awareness of a consortium of specific scientific societies or communities, meetings can be organized adjacent to conferences and organizations with connections to consortium members. Examples include two IDG-associated Federation of American Societies for Experimental Biology (FASEB) conferences: ‘The Understudied Druggable Proteome Conference (#UDP21)’ in August 2021 and ‘The Illuminating the Understudied Druggable Proteome Conference (#UDPSRC)’ in June 2023; a social event at the Society for Neuroscience (SfN) Global Connectome in January 2021; and a webinar in the Target2035 series entitled ‘Drugging the dead – selective targeting of pseudokinases’ in June 2021.
Relationship/collaborations
Identifying and connecting to other research consortia or organizations with related interests can leverage synergies and develop fruitful collaborations. Such consortia collaborations can be initiated at symposia with presentations from the different groups to assess and prioritize new opportunities for collaboration. Consideration should be given to facilitating interactions across the different groups, such as breakout sessions, set-aside time for discussions, and/or poster sessions for sharing individual research outcomes. Some examples of IDG outreach meetings of this sort include the ‘Finding Targets for Drug Discovery’ IDG/OpenTarget Meeting in November 2019 and the ATOM – IDG Symposium Series (ATOM: Accelerating Therapeutics for Opportunities in Medicine) in October 2020.
Training/workshop
By providing opportunities to educate, demonstrate, and offer hands-on training to the scientific community, the utility and reach of the informatic tools and data analysis products developed by the consortium can be increased. The IDG Digital Tool Fest 2021 presented 14 informatic tools to the public. In addition, there were four IDG Hackathons (2018,43 2020,44−46 2021,47−50 and 202351−54) arranged with the RDOC’s assistance. These were attended largely by IDG members but also included invited collaborators with the aim to develop and improve informatics workflows across the IDG consortium and beyond. Participation in these Hackathons benefitted from shared GitHub repositories.
Scientific webinars
With disease-agnostic consortia such as NIH Common Fund-sponsored programs, the consortium members span many different scientific areas, as was the case for IDG. Facilitating scientific presentations on specific topics highlights these areas of interest and the expertise of members and thereby brings in other interested parties from those specific fields of study. In addition, utilizing online webinars for such scientific presentation series lowers costs and facilitates engagement for all. The e-IDG Symposium Series was born in the spring of 2021. Since then, five series of these virtual webinars have been hosted by the RDOC on the Zoom platform (spring 2021, fall 2021, spring 2022, fall 2022, spring 2023). Each series consisted of five or six 1-hour sessions, with speakers presenting their research on understudied GPCRs, ion channels, kinases, and/or informatics. In total, there have been 27 sessions with a broad reach; each session had an average of 50 attendees from five different countries, resulting in an average of 14 new ‘IDG customers’ per session. With speakers’ permission, certain presentations were shared via the DruggableGenome IDG YouTube channel, allowing for additional audiences in locations where the timing of the live e-IDG symposium session was unfavorable.
Another platform for outreach was the IDG-DREAM Drug Kinase Binding Prediction Challenge, announced on October 2, 2018 as part of the Sage Bionetworks DREAM challenges. This challenge solicited predictions by machine learning algorithms of kinase binding affinities, which were experimentally tested by the Kinase DRGC. The outcomes from this IDG DREAM challenge were published as ‘Crowdsourced mapping of unexplored target space of kinase inhibitors’,55 and the winners were recognized at the RECOMB/ISCB RSG conference held in New York, NY in November 2019.56
Concluding remarks
Impact
Overall accomplishments of the consortium include the generation of highly diverse resources such as small-molecule chemicals, genetic constructs, mouse models, and computational/digital resources. The metrics of success for the RDOC are continual growth of engagement via monthly newsletters, social media outreach, and expanding attendance of webinars and presentations. Several recommendations and best practices (Box 1) guided the initiation, coordination, and administration of the newly formed consortium and were incorporated into its metrics of success.
BOX 1. RDOC recommendations and best practices.
Establishing consortium-wide policies in the early phase of the project
Building personal relationships among RDOC, KMC, DRGCs, CEITs, and R03s
Establishing channels of communication between the groups
Assigning specific tasks to individuals (e.g. submission task)
Exhibiting flexibility during COVID-19 regarding travel and hybrid meeting formats
Implementing regular outreach and communications (i.e. monthly newsletters), social media, and establishing reoccurring opportunities for scientific publication such as the Nature Reviews Drug Discovery series for Target Watch
Cross-tracking users and customers, e.g. e-IDG attendees and IDG mailing lists
Holding various outreach events to engage the scientific community, industry, and other consortia
Planning for an evolving consortium (e.g. new CEITs) to accommodate new members, data, resources, project needs, outreach, etc.
Building modular processes and software tools to enable rapid implementation of new products and functionalities (e.g. PIT)
Defining quality control metrics for generated resources to maintain highest quality of consortium- generated resources
Developing metadata standards and resource representations compatible with the FAIR guiding principles for scientific data management and stewardship
Selecting well-established repositories to enable seamless maintenance and longevity of generated resources
When applicable, tracking resources via SciCrunch and the corresponding RRIDs
Implementing robust tools and infrastructure to support resource information submission, validation, processing, and dissemination
Accounting and planning for flexibility of software and processes to accommodate new resource types over the project lifecycle (e.g. newly joined IDG groups generating new types of resources that needed to be captured through the same submission system) CEITs, Cutting-Edge Informatic Tools; KMC, Knowledge Management Center.
Resources
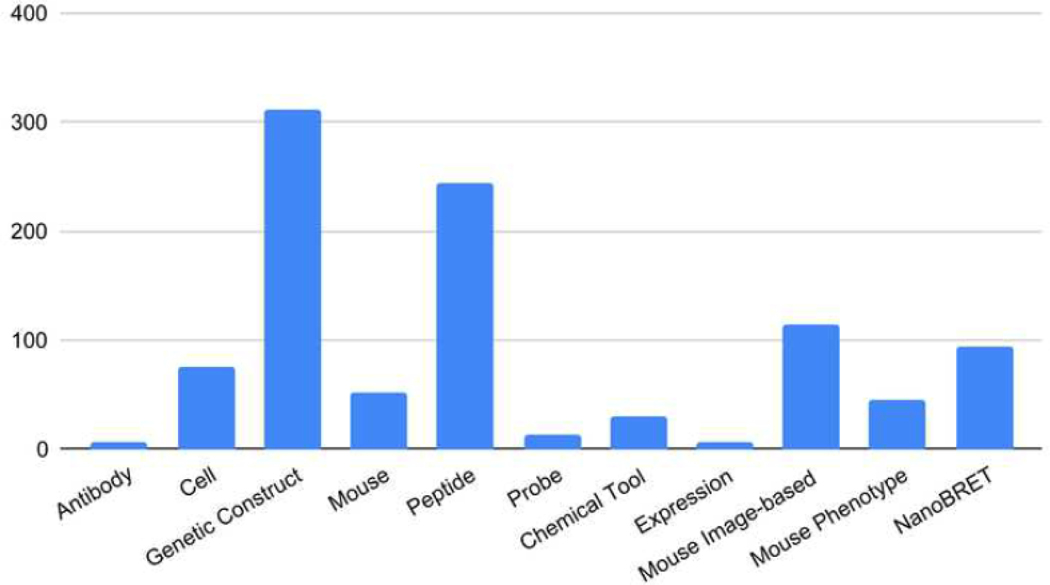
The major resources generated by the DRGCs were divided into two main categories: reagents and datasets (for the full list of reagents and data types, see Table 1). The corresponding information about these resources was submitted by DRGCs to the RSS and processed via the RMS for further dissemination through Pharos and PIT. The RDOC-developed tools enabled easy submission and, as of now, the DRGCs have submitted 994 records to the RSS. Among these records, 373 fulfill completeness and annotation criteria (Pharos-ready) and thus can be shown on Pharos. Most of the IDG-generated resources are reagents, with genetic constructs developed for the GPCRs, kinases, and ion channels being the majority. Besides the genetic constructs, the DRGCs also devoted effort to generating other resources: cells with overexpressed genes, mouse strains, and analysis of mouse phenotypes for ion channels; peptides and chemical tools, and analysis of their effectiveness for kinases as measured by the NanoBRET assay; and chemical probes, mouse strains, and mouse image-based expression for GPCRs, as additional major resource categories.
The numbers of IDG records reported to the RDOC are shown in Figure 2.
FIGURE 2.
Numbers of IDG resources submitted to RDOC by resource type.
In addition to tracking the publications published by the IDG members,57 the RDOC was also able to track publications utilizing the IDG resources when referenced by their RRIDs. These publications were identified via SciCrunch integration. As of now, SciCrunch has identified 259 publications mentioning 31 genetic constructs, 2 publications mentioning two cells, and at least 31 publications referring to the IDG project.
Additionally, IDG resource utilization by external institutions was tracked via direct reporting from the corresponding vendors to the RDOC, and consequently to the NIH. These reports did not reveal the specific institutions, but they included details such as the ordering interval (typically quarterly) and the quantities of the corresponding resources.
Sustainability
To facilitate long-term IDG data accessibility and tracking, the IDG consortium led by the RDOC implemented (i) data policies to govern the resource repositories and vendors, (ii) registration of resources across established repositories and databases (e.g. ZINC and/or ChEBI for small molecules), (iii) metadata standards to describe and identify IDG-generated resources, (iv) integration of the RRIDs between repositories/vendors and the SciCrunch platform, and (v) infrastructure to host the metadata and information sufficient for resource identification and tracking. The RDOC thus enabled handshakes and information exchange between various entities and institutions to ensure that IDG resources are FAIR and sustainable.
As the IDG project comes to an end, the RDOC continues to maintain informatics infrastructure and support the implemented processes and best practices to enable consortium members to submit and register their resources. Long-term data and resource repositories, along with annotations based on established and documented data standards and unique resource identifiers, support the sustainability of IDG resources. To assure their findability and accessibility, the RDOC will generate catalogs of IDG resources with annotations, persistent identifiers (generated at identifiers.org), and links that can be maintained with minimal effort for an extended period. These catalogs can be hosted at the IDG-supported infrastructure (e.g. DruggableGenome.net) or easily transferred to cloud platforms (e.g. AWS cloud) or other hosting solutions (e.g. GitHub). Due to the RDOC’s proximity to the IDG-associated NIH Common Fund Data Ecosystem (CFDE) project at the University of New Mexico (UNM), we are poised to continue collaborating and coordinating to enhance IDG resources that will be highlighted and integrated with the NIH CFDE portal.
Highlights.
The IDG consortium has generated numerous reagent and digital resources
RDOC implemented infrastructure and processes for FAIR resource management
IDG resources are accessible via a number of established repositories
The Protein Illumination Timeline serves as a live catalog of IDG resources
Several outreach mechanisms facilitated awareness of IDG resources
Acknowledgments
We are very grateful to all the awardees of the IDG Consortium and our NIH working group for their engagement with outreach events, contributions to developing policies and metadata standards, and participation in discussions for enhancing our abilities to disseminate IDG resources for target illumination. We would like to recognize Dr. Tudor Oprea for his service as a co-lead of the RDOC at UNM for 5 years. We would also like to acknowledge and express our appreciation to Amar Koleti (University of Miami) for his work in developing the RSS and Nicholas Stanford (UNM) for implementing the user interface of Protein Illuminating Timelines.
Funding
This work was supported by NIH grant U24TR002278 (Illuminating the Druggable Genome Resource Dissemination and Outreach Center, IDG RDOC) awarded by the National Center for Advancing Translational Sciences (NCATS). IDG (https://druggablegenome.net/) is an NIH Common Fund program.
In addition, L.A.S. was supported by NIH grant P20GM121176 (AIM), and S.C.S. and D.V. were supported by grants U01LM012630 and R01LM013391.
Footnotes
Declarations of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, Yu FH. Too many roads not taken. Nature. 2011;470(7333):163–165. 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen DT, et al. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017;45(D1):D995–D1002. 10.1093/nar/gkw1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelleher KJ, et al. Pharos 2023: an integrated resource for the understudied human proteome. Nucleic Acids Res. 2023;51(D1):D1405–D1416. 10.1093/nar/gkac1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.IDG Protein Illumination Timeline (PIT). https://druggablegenome.net/ProteinTimeline. [Google Scholar]
- 6.Illuminating the Druggable Genome (IDG) Project Website. https://druggablegenome.net/. [Google Scholar]
- 7.Musen MA, et al. The center for expanded data annotation and retrieval. J Am Med Inform Assoc. 2015;22(6):1148–1152. 10.1093/jamia/ocv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheils TK, et al. TCRD and Pharos 2021: mining the human proteome for disease biology. Nucleic Acids Res. 2021;49(D1):D1334–D1346. 10.1093/nar/gkaa993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sansone SA, et al. FAIRsharing as a community approach to standards, repositories and policies. Nat Biotechnol. 2019;37(4):358–367. 10.1038/s41587-019-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.FAIRsharing.org. A curated, informative and educational resource on data and metadata standards, inter-related to databases and data policies. https://fairsharing.org/. [Google Scholar]
- 11.Illuminating the Druggable Genome Project - FAIRsharing record. https://fairsharing.org/3523. [Google Scholar]
- 12.Illuminating the Druggable Genome Consortium Policies - FAIRsharing record. 10.25504/FAIRsharing.ZXDCn7. [DOI] [Google Scholar]
- 13.IDG Small Molecule Resource Reporting Standards. 10.25504/FAIRsharing.LaaxUg. [DOI] [Google Scholar]
- 14.IDG Mouse Resource Reporting Standards. 10.25504/FAIRsharing.iUR9zW. [DOI] [Google Scholar]
- 15.IDG Antibody Resource Reporting Standards. 10.25504/FAIRsharing.stYji6. [DOI] [Google Scholar]
- 16.Mutant Mouse Resource and Research Centers (MMRRC). https://www.mmrrc.org/. [Google Scholar]
- 17.Amos-Landgraf J, et al. The Mutant Mouse Resource and Research Center (MMRRC): the NIH-supported National public repository and distribution archive of mutant mouse models in the USA. Mamm Genome. 2022;33(1):203–212. 10.1007/s00335-021-09894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Addgene. https://www.addgene.org/. [Google Scholar]
- 19.SciCrunch. https://scicrunch.org/. [Google Scholar]
- 20.RRID Portal. https://scicrunch.org/resources. [Google Scholar]
- 21.Chemical Entities of Biological Interest (ChEBI). https://www.ebi.ac.uk/chebi/. [Google Scholar]
- 22.Hastings J, et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44(D1):D1214–D1219. 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ZINC. Irwin and Shoichet Laboratories in the Department of Pharmaceutical Chemistry at the University of California, San Francisco (UCSF). https://zinc15.docking.org/. [Google Scholar]
- 24.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52(7):1757–1768. 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sterling T, Irwin JJ. ZINC 15--ligand discovery for everyone. J Chem Inf Model. 2015;55(11):2324–2337. 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrowsmith CH, et al. The promise and peril of chemical probes. Nat Chem Biol. 2015;11(8):536–541. 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.. Increasing the quality and robustness of biomedical research using chemical probes. https://www.chemicalprobes.org/. [Google Scholar]
- 28.Antolin AA, et al. The Chemical Probes Portal: an expert review-based public resource to empower chemical probe assessment, selection and use. Nucleic Acids Res. 2023;51(D1):D1492–D1502. 10.1093/nar/gkac909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.New England Peptide. http://www.newenglandpeptide.com/. [Google Scholar]
- 30.Gong B, Murray KD, Trimmer JS. Developing high-quality mouse monoclonal antibodies for neuroscience research - approaches, perspectives and opportunities. N Biotechnol. 2016;33(5 Pt A):551–564. 10.1016/j.nbt.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.IDG. Dark ion channel overexpression cell lines. https://darkmatter.ucsf.edu/cells. [Google Scholar]
- 32.Sarntivijai S, et al. CLO: The cell line ontology. J Biomed Semantics. 2014;5:37. 10.1186/2041-1480-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29(2):25–38. 10.7171/jbt.18-2902-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Riverol Y, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50(D1):D543–D552. 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–514. 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teytelman L, Stoliartchouk A, Kindler L, Hurwitz BL. Protocols.io: virtual communities for protocol development and discussion. PLoS Biol. 2016;14(8):e1002538. 10.1371/journal.pbio.1002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.W3C. SKOS simple knowledge organization system reference, W3C recommendation. World Wide Web Consortium. https://www.w3.org/TR/skos-reference/. [Google Scholar]
- 39.Dublin Core. Dublin core metadata initiative. https://www.dublincore.org/resources/glossary/dublin_core/. [Google Scholar]
- 40.W3C. RDF schema, W3C recommendation. World Wide Web Consortium. https://www.w3.org/TR/rdf-schema/. [Google Scholar]
- 41.Druggable Genome IDG, Youtube Channel. IDG RDOC. https://www.youtube.com/c/DruggableGenomeIDG. [Google Scholar]
- 42.IDG Events. https://druggablegenome.net/IDG-Events. [Google Scholar]
- 43.IDG 2018. Hackathon - Druggable Genome github repository. https://github.com/druggablegenome/hackathon2018. [Google Scholar]
- 44.IDG 2020. Hackathon - Monarch Initiative SLDBGen github repository. https://github.com/monarch-initiative/SLDBGen. [Google Scholar]
- 45.IDG 2020. Hackathon - ProteinGraphML github repository. https://github.com/unmtransinfo/ProteinGraphML. [Google Scholar]
- 46.IDG 2020. Hackathon - ProKino KinView github repository. https://prokino.github.io/kinview/. [Google Scholar]
- 47.IDG 2021. Hackathon - AnacletoLab github repository. https://github.com/AnacletoLAB/ensmallen. [Google Scholar]
- 48.IDG 2021. Hackathon - Monarch Initiative Embiggen github repository. https://github.com/monarch-initiative/embiggen. [Google Scholar]
- 49.IDG 2021. Hackathon - Grape Appyter github repository. https://github.com/caufieldjh/grape_appyter. [Google Scholar]
- 50.IDG 2021. Hackathon - Maayan Lab Appyter github repository. https://github.com/MaayanLab/appyter. [Google Scholar]
- 51.IDG 2023. Hackathon - Druggable Genome github repository. https://github.com/druggablegenome/hackathon2023. [Google Scholar]
- 52.IDG 2023. Hackathon - Pharos Community Data API github repository. https://github.com/ncats/pharos-community-data-api. [Google Scholar]
- 53.IDG 2023. Hackathon - Pharos Graphql Server github repository. https://github.com/ncats/pharos-graphql-server. [Google Scholar]
- 54.IDG 2023. Hackathon - Pharos Frontend github repository. https://github.com/ncats/pharos_frontend. [Google Scholar]
- 55.Cichonska A, et al. Crowdsourced mapping of unexplored target space of kinase inhibitors. Nat Commun. 2021;12(1):3307. 10.1038/s41467-021-23165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.RECOMB/ISCB RSG conference. https://www.iscb.org/recomb-regsysgen2019-program/recomb-regsysgen2019-oral-schedule. [Google Scholar]
- 57.Sharma KR, Colvis CM, Rodgers GP, Sheeley DM. Illuminating the druggable genome: pathways to progress. Drug Discov Today. 2023;29(3):103805. 10.1016/j.drudis.2023.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]