Abstract
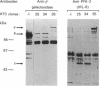
In liver, the 470-residue bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFK-2/FBPase-2) catalyses the synthesis and degradation of fructose 2,6-bisphosphate, a potent stimulator of glycolysis. In rat hepatoma (HTC) cells, this enzyme has kinetic, antigenic, and regulatory properties, such as insensitivity to cyclic AMP-dependent protein kinase and lack of associated FBPase-2 activity, that differ from those in liver. To compare the sequence of the HTC enzyme with that of the liver enzyme, we have cloned the corresponding fully-coding cDNA from HTC cells. This cDNA predicts a protein of 448 residues in which the first 32 residues of liver PFK-2/FBPase-2 including the cyclic AMP target sequence have been replaced by a unique N-terminal decapeptide. The rest of the protein is identical with the liver enzyme. An N-terminally truncated recombinant peptide of 380 residues containing the PFK-2 and FBPase-2 domains was expressed in Escherichia coli as a beta-galactosidase fusion protein. It was recognized by anti-PFK-2 antibodies but its enzymic activities were barely detectable. In contrast, a cDNA fully-coding for the HTC enzyme could be expressed in E. coli as a beta-galactosidase-free peptide that exhibited both PFK-2 and FBPase-2 activities. This peptide had those PFK-2 kinetic properties of the HTC enzyme that differ from the liver enzyme. These data, together with immunoblot experiments, suggest that the lack of associated FBPase-2 activity in HTC cells results from a post-translational modification of the enzyme rather than from the difference in amino acid sequence. As well as this peculiar type of PFK-2/FBPase-2 mRNA, HTC cells also contained low concentrations of the liver-type mRNA. Unlike in liver, neither mRNA was induced by dexamethasone in these cells.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrameas S., Ternynck T. The cross-linking of proteins with glutaraldehyde and its use for the preparation of immunoadsorbents. Immunochemistry. 1969 Jan;6(1):53–66. doi: 10.1016/0019-2791(69)90178-5. [DOI] [PubMed] [Google Scholar]
- Bartrons R., Hue L., Van Schaftingen E., Hers H. G. Hormonal control of fructose 2,6-bisphosphate concentration in isolated rat hepatocytes. Biochem J. 1983 Sep 15;214(3):829–837. doi: 10.1042/bj2140829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca L., Rousseau G. G., Hue L. Phorbol 12-myristate 13-acetate and insulin increase the concentration of fructose 2,6-bisphosphate and stimulate glycolysis in chicken embryo fibroblasts. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6440–6444. doi: 10.1073/pnas.82.19.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Colosia A. D., Marker A. J., Lange A. J., el-Maghrabi M. R., Granner D. K., Tauler A., Pilkis J., Pilkis S. J. Induction of rat liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase mRNA by refeeding and insulin. J Biol Chem. 1988 Dec 15;263(35):18669–18677. [PubMed] [Google Scholar]
- Coutelier J. P., Van Roost E., Lambotte P., Van Snick J. The murine antibody response to lactate dehydrogenase-elevating virus. J Gen Virol. 1986 Jun;67(Pt 6):1099–1108. doi: 10.1099/0022-1317-67-6-1099. [DOI] [PubMed] [Google Scholar]
- Crepin K. M., Darville M. I., Hue L., Rousseau G. G. Characterization of distinct mRNAs coding for putative isozymes of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Eur J Biochem. 1989 Aug 1;183(2):433–440. doi: 10.1111/j.1432-1033.1989.tb14946.x. [DOI] [PubMed] [Google Scholar]
- Crepin K. M., Darville M. I., Hue L., Rousseau G. G. Starvation or diabetes decreases the content but not the mRNA of 6-phosphofructo-2-kinase in rat liver. FEBS Lett. 1988 Jan 25;227(2):136–140. doi: 10.1016/0014-5793(88)80884-6. [DOI] [PubMed] [Google Scholar]
- Darville M. I., Crepin K. M., Vandekerckhove J., Van Damme J., Octave J. N., Rider M. H., Marchand M. J., Hue L., Rousseau G. G. Complete nucleotide sequence coding for rat liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase derived from a cDNA clone. FEBS Lett. 1987 Nov 30;224(2):317–321. doi: 10.1016/0014-5793(87)80476-3. [DOI] [PubMed] [Google Scholar]
- El-Maghrabi M. R., Pate T. M., Murray K. J., Pilkis S. J. Differential effects of proteolysis and protein modification on the activities of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. J Biol Chem. 1984 Nov 10;259(21):13096–13103. [PubMed] [Google Scholar]
- Hue L., Rider M. H. Role of fructose 2,6-bisphosphate in the control of glycolysis in mammalian tissues. Biochem J. 1987 Jul 15;245(2):313–324. doi: 10.1042/bj2450313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lively M. O., el-Maghrabi M. R., Pilkis J., D'Angelo G., Colosia A. D., Ciavola J. A., Fraser B. A., Pilkis S. J. Complete amino acid sequence of rat liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. J Biol Chem. 1988 Jan 15;263(2):839–849. [PubMed] [Google Scholar]
- Loiseau A. M., Rider M. H., Foret D., Rousseau G. G., Hue L. Rat hepatoma (HTC) cell 6-phosphofructo-2-kinase differs from that in liver and can be separated from fructose-2,6-bisphosphatase. Eur J Biochem. 1988 Jul 15;175(1):27–32. doi: 10.1111/j.1432-1033.1988.tb14161.x. [DOI] [PubMed] [Google Scholar]
- Loiseau A. M., Rousseau G. G., Hue L. Fructose 2,6-bisphosphate and the control of glycolysis by glucocorticoids and by other agents in rat hepatoma cells. Cancer Res. 1985 Sep;45(9):4263–4269. [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkis S. J., Regen D. M., Stewart H. B., Pilkis J., Pate T. M., El-Maghrabi M. R. Evidence for two catalytic sites on 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase. Dynamics of substrate exchange and phosphoryl enzyme formation. J Biol Chem. 1984 Jan 25;259(2):949–958. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schamhart D. H., van de Poll K. W., van Wijk R. Comparative studies of glucose metabolism in HTC, RLC, MH1C1, and Reuber H35 rat hepatoma cells. Cancer Res. 1979 Mar;39(3):1051–1055. [PubMed] [Google Scholar]
- Tauler A., Rosenberg A. H., Colosia A., Studier F. W., Pilkis S. J. Expression of the bisphosphatase domain of rat liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6642–6646. doi: 10.1073/pnas.85.18.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse S. The Warburg hypothesis fifty years later. Z Krebsforsch Klin Onkol Cancer Res Clin Oncol. 1976;87(2):115–126. doi: 10.1007/BF00284370. [DOI] [PubMed] [Google Scholar]
- Woodgett J. R., Gould K. L., Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986 Nov 17;161(1):177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- van Schaftingen E., Davies D. R., Hers H. G. Fructose-2,6-bisphosphatase from rat liver. Eur J Biochem. 1982 May;124(1):143–149. doi: 10.1111/j.1432-1033.1982.tb05917.x. [DOI] [PubMed] [Google Scholar]